Abstract
From genomic DNA of Ralstonia pickettii isolate PIC-1, a β-lactamase gene was cloned that encodes the oxacillinase OXA-22. It differs from known oxacillinases, being most closely related to OXA-9 (38% amino acid identity). The hydrolytic spectrum of OXA-22 is limited mostly to benzylpenicillin, cloxacillin, and restricted-spectrum cephalosporins. OXA-22-like genes were identified as single chromosomal copies in five other R. pickettii clinical isolates. The expression of OXA-22-like β-lactamases was inducible in R. pickettii.
Ralstonia pickettii is a nonfermenting gram-negative rod that is an occasional pathogen of nosocomial septicemia and tissue infections (10, 17, 20). In 1992, this organism was transferred from the genus Pseudomonas RNA homology group II to the genus Burkholderia (8) and recently to the novel genus Ralstonia (22).
The β-lactam resistance mechanism(s) of R. pickettii isolates is not known in detail. In this report, we describe the genetic and biochemical characterization of inducible oxacillinases that occur naturally in R. pickettii and that may explain part of its β-lactam resistance profile.
Bacterial strains, PFGE, plasmids, and conjugation assays.
R. pickettii clinical isolates PIC-1, PIC-2, and PIC-3 were from the hospitals Bicêtre and Antoine Béclère (Paris area, France), R. pickettii reference strains CIP 103413 and CIP 74.22 were from the strain collection of the Pasteur Institute (Paris, France), and strain ATCC 27511 was from the American Type Culture Collection. These strains were identified by standard biochemical techniques (8).
Comparison of R. pickettii whole-cell DNAs was performed by a pulsed-field gel electrophoresis (PFGE) technique as previously reported (5, 14). PFGE of either XbaI- or SpeI-restricted DNAs of R. pickettii strains showed that they are not clonally related, except for R. pickettii strains PIC-1 and PIC-3 (data not shown).
Plasmid DNA extractions (3, 13, 14) failed to detect any plasmid in R. pickettii strains. Direct transfer (13, 14) of an amoxicillin resistance marker from R. pickettii strains to rifampin-resistant E. coli JM109 also failed.
Susceptibility testing.
MICs of selected β-lactams were determined as described previously (14). R. pickettii isolates were resistant or had decreased susceptibility to aminopenicillins, ureidopenicillins, restricted-spectrum cephalosporins, ceftazidime, and aztreonam (Table 1), as previously reported (7). Addition of clavulanic acid and tazobactam did not significantly modify this resistance profile. Only R. pickettii CIP 103413 was more resistant to piperacillin, cephalothin, and cefepime (Table 1); tazobactam decreased the piperacillin MIC 16-fold.
TABLE 1.
MICs of β-lactams for R. pickettii isolates, E. coli DH10B harboring recombinant plasmid pSC13, and reference strain E. coli DH10B
| β-Lactama | MIC (μg/ml)
|
|||
|---|---|---|---|---|
| R. pickettii PIC-1, PIC-2, PIC-3, ATCC 2751, CIP 74.22 | R. pickettii CIP 103413 | E. coli DH10B (pSC13)b | E. coli DH10B | |
| Amoxicillin | 8–64 | 128 | 64 | 4 |
| Amoxicillin + CLA | 8–64 | 128 | 8 | 4 |
| Ticarcillin | 8–64 | 256 | 64 | 4 |
| Ticarcillin + CLA | 8–64 | 256 | 16 | 4 |
| Piperacillin | 2–4 | 32 | 64 | 1 |
| Piperacillin + TZB | 0.5–1 | 2 | 32 | 1 |
| Cephalothin | 4–8 | 64 | 16 | 2 |
| Cephalothin + CLA | 4–8 | 64 | 8 | 2 |
| Cefuroxime | 8 | 8 | 16 | 0.5 |
| Cefoxitin | 1–4 | 4 | 4 | 1 |
| Ceftazidime | 4–16 | 16 | 0.5 | 0.5 |
| Ceftazidime + CLA | 4–16 | 16 | 0.5 | 0.5 |
| Cefotaxime | 0.5–2 | 2 | 0.12 | 0.12 |
| Cefotaxime + CLA | 0.5–2 | 2 | 0.12 | 0.12 |
| Cefepime | 0.5–1 | 8 | 0.25 | 0.03 |
| Cefepime + CLA | 0.5–1 | 4 | 0.12 | 0.03 |
| Moxalactam | 8–16 | 32 | 0.5 | 0.12 |
| Aztreonam | 128–256 | 256 | 1 | 0.25 |
| Aztreonam + CLA | 128–256 | 256 | 0.5 | 0.25 |
| Imipenem | 0.5–1 | 1 | 0.25 | 0.12 |
CLA, clavulanic acid at a fixed concentration of 2 μg/ml; TZB, tazobactam at a fixed concentration of 4 μg/ml.
E. coli DH10B harboring recombinant plasmid pSC13 produced the β-lactamase OXA-22.
Cloning and sequencing of the β-lactamase OXA-22 gene.
Genomic DNA of R. pickettii PIC-1 was partially digested with Sau3AI and ligated into BamHI-digested phagemid pBK-CMV as previously described (13). Only one Escherichia coli DH10B strain that contained recombinant plasmid pSC13 was obtained. Sequence analysis (3) of the 1.2-kb insert of pSC13 revealed an open reading frame (ORF) of 828 bp (data not shown). The G+C content of this ORF was 65%, which is within the range of G+C contents of Ralstonia genes (60.1 to 69.5%; GenBank database).
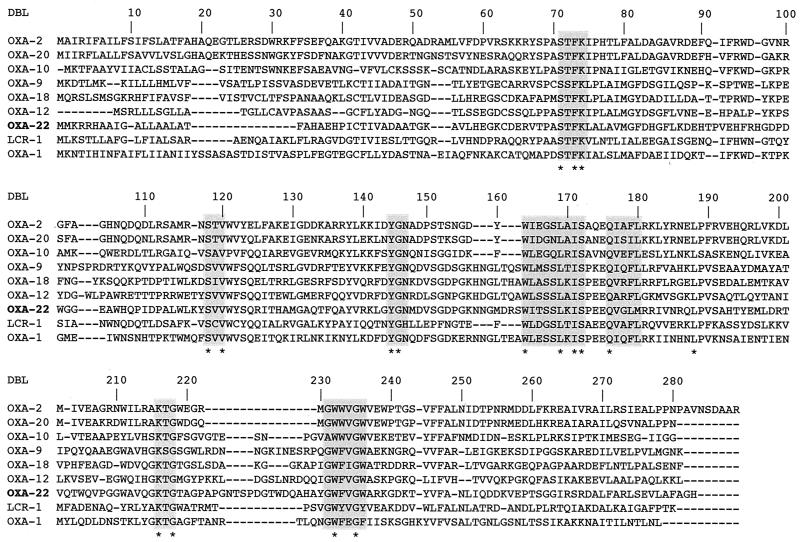
Within the deduced protein of this ORF (275 amino acids), an S-T-F-K tetrad was found at class D β-lactamase (DBL) (6) positions 71 to 75 (Fig. 1). Four structural elements characteristic of DBLs (2) were found in this novel enzyme; they were named OXA-22 (Y-G-N at DBL positions 144 to 146, W-X-E-X-X-L-X-I-S at DBL positions 164 to 172, Q-X-X-X-L at positions 176 to 180, and K-T-G at positions 216 to 218 (Fig. 1) (11). In addition, another stretch of amino acids, at DBL position 231 to 236, which is conserved in class D enzymes was also present (Fig. 1). OXA-22 had low amino acid identity with other Ambler DBLs, ranging from 19 to 38% for OXA-20 to OXA-9, respectively (12, 19). The highest percentages of identity were with OXA-9, OXA-18, and OXA-12, at 38, 37, and 34%, respectively (1, 6, 13, 19).
FIG. 1.
Alignment of the OXA-22 amino acid sequence with those of the most closely related DBLs and some oxacillinases taken as representatives of each phylogenetic subgroup of DBLs (6, 10). The shaded boxes indicate regions conserved among DBLs, and stars indicate highly conserved residues. Dashes indicate gaps in the alignment.
Biochemical properties of the OXA-22 β-lactamase.
Cultures of E. coli DH10B(pSC13) were grown overnight at 37°C in 6 liters of Trypticase soy broth with amoxicillin at 30 μg/ml and kanamycin at 30 μg/ml. Unpurified extract of OXA-22 was obtained in 30 ml of sodium phosphate buffer as previously described (3). This extract was dialyzed in 20 mM H2SO4-Tris (pH 8) overnight at 4°C and then loaded onto a preequilibrated Q-Sepharose column (Amersham Pharmacia Biotech) in Tris buffer-H2SO4. The β-lactamase was eluted with a linear K2SO4 gradient (0 to 500 mM). The elution peak containing the highest β-lactamase activity was subsequently dialyzed overnight against 50 mM phosphate buffer, pH 7.0, prior to 10-fold concentration with a Centrisart-C30 microcentrifuge filter (Sartorius, Goettingen, Germany). Kinetic parameters were obtained as described previously (3) using a UV spectrophotometer with 100 μM cephalothin as the substrate for inhibition studies, and one unit of enzyme activity was defined (15) as the activity which hydrolyzed 1.0 μmol of cephalothin per min.
The specific activity of OXA-22 was 9.1 mU/mg, and its purification coefficient was 20-fold. OXA-22 showed its strongest hydrolytic activity against benzylpenicillin, cephalothin, cephaloridine, and cloxacillin (Table 2). The lack of oxacillin hydrolysis by OXA-22 was peculiar for an oxacillinase (4, 11). Further OXA-22 purification may help to detect oxacillin hydrolysis as described for LCR-1 (Y. Yang and K. Bush, Letter, Antimicrob. Agents Chemother. 39:1209, 1995). AmpS, an oxacillinase related to OXA-12 (90% amino acid identity) from Aeromonas jandaei, and LCR-1 strongly hydrolyze oxacillin but not cloxacillin (9, 21; Yang and Bush; letter). Inhibition studies that measured 50% inhibitory concentrations (IC50) (14) showed that OXA-22 activity was inhibited partially by clavulanic acid (IC50, 1.2 μM) and less by tazobactam (IC50, 6.5 μM). These results contrasted with those found for the other oxacillinases, for which the tazobactam inhibitory property is equal to or higher than that of clavulanic acid (4, 11). OXA-22 activity was inhibited by NaCl, as were other oxacillinases (IC50, 80 mM) (4).
TABLE 2.
Steady-state kinetic parameters of the β-lactamase OXA-22
| β-Lactam | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) |
|---|---|---|---|
| Amoxicillin | 0.1 | 6 | 0.016 |
| Aztreonam | —a | — | — |
| Benzylpenicillin | 0.1 | <2 | >0.04 |
| Cephaloridine | 5.5 | 10 | 0.55 |
| Cephalothin | 0.9 | 3 | 0.33 |
| Cefotaxime | — | — | — |
| Cefoxitin | 0.05 | 10 | 0.004 |
| Ceftazidime | — | — | — |
| Cefuroxime | 0.05 | 5 | 0.01 |
| Cloxacillin | 0.7 | 5 | 0.14 |
| Oxacillin | — | — | — |
| Piperacillin | — | — | — |
| Ticarcillin | 0.1 | 9 | 0.01 |
—, not determinable (the initial hydrolytic rate [kcat] was lower than 0.001 s−1).
Analytical isoelectric focusing (IEF), performed as previously reported (14), revealed that a culture of R. pickettii PIC-1 gave two β-lactamase activities with pI values of 7.0 and 7.1, like E. coli DH10B(pSC13) cultures (data not shown). These pI values may correspond to proteolytic cleavage, partial unfolding, and/or monomer-dimer conversion of OXA-22. The relative molecular mass of OXA-22, determined with an E. coli DH10B(pSC13) culture as previously described (3), was 28 kDa.
OXA-22-like β-lactamases in R. pickettii isolates.
IEF analysis revealed that the unpurified β-lactamase extract of each R. pickettii culture gave different pI values: for PIC-3 (as for R. pickettii PIC-1), 7.0 and 7.1; for R. pickettii PIC-2 and ATCC 27511, 7.4; for R. pickettii CIP 74.22, 7.5. An R. pickettii CIP 103413 culture gave two very different pI values of 6.8 and 7.5, likely corresponding to two β-lactamases.
PFGE of XbaI-restricted genomic DNAs of R. pickettii strains and reference strains Pseudomonas putida CIP 55.5, Brevundimonas diminuta CIP 63.27T, and Comamonas acidovorans 103685 (Pasteur Institute strain collection) produced a template used in a Southern transfer experiment (14, 18) with a PCR-obtained 622-bp internal fragment of blaOXA-22 as a labeled probe (3). blaOXA-22-like genes were identified in all of the R. pickettii isolates but in none of related gram-negative species (data not shown). The hybridizing XbaI DNA fragment differed for each R. pickettii isolate, except for R. pickettii isolates PIC-1 and PIC-3. OXA-22-like genes were found at a single copy on a large DNA fragment (>ca. 250 kb), further underlining the chromosomal origin of these genes (data not shown).
Most of the OXA-22-like β-lactamase genes were PCR amplified (14) from R. pickettii genomic DNAs (622 out of 828 bp) as a result of the choice of internal PCR amplification primers of blaOXA-22 (OXA-22A, 5′-TTGCATGAAGGCAAGTGCGACGAG-3′; OXA-22B, 5′-TCAACCTTGTCGTCCTGGATC-3′). The deduced proteins had 96 to 100% amino acid identity with OXA-22. We identified OXA-22 in R. pickettii PIC-1 and PIC-3, OXA-22a in R. pickettii PIC-2 and ATCC 27511, and OXA-22b in R. pickettii CIP 7422 (Table 3). Amplification of an OXA-22-like gene from R. pickettii CIP 103413 failed, despite the use of other primer combinations and different experimental setups.
TABLE 3.
Amino acid differences among OXA-22-like β-lactamases compared to OXA-22
| β-Lactamase | Amino acid at position:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 90 | 95 | 96 | 104 | 126 | 189 | 193 | 198 | |
| OXA-22 | H | R | H | E | I | P | H | L |
| OXA-22a | K | R | D | V | S | R | V | |
| OXA-22b | Y | K | R | D | S | R | V | |
DBL amino acid numbering is shown (6).
Induction studies.
Induction studies with cefoxitin at 0.5 μg/ml as a β-lactam inducer (15) and 100 μM cephaloridine as the substrate identified induced β-lactamase expression in cultures of all of the R. pickettii isolates tested. The induction rates ranged from 25- to 64-fold depending on the strain. In induced cultures, no other β-lactamase activity appeared on an IEF gel, compared to noninduced cultures, thus confirming that induced β-lactamase activities corresponded to those of OXA-22-like enzymes. Regulation of oxacillinase expression is known for OXA-12 and AmpS from A. jandaei, which are chromosomally located and occur naturally (1, 9, 16, 21). Further work will be directed toward the identification of the regulatory system of OXA-22-like β-lactamases and its comparison with the two-component regulon identified for coordinated expression of β-lactamases of different Ambler classes of A. jandaei (1, 16).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the GenBank nucleotide database under accession no. AF064820.
Acknowledgments
This work was funded by a grant from the Ministère de l'Education Nationale et de la Recherche (grant UPRES-JE 2227), Faculté de Médecine Paris-Sud, Université Paris XI, Paris, France.
REFERENCES
- 1.Alksne L E, Rasmussen B A. Expression of the AsbA1, OXA-12, and AsbM1 β-lactamases in Aeromonas jandaei AER14 is coordinated by a two-component regulon. J Bacteriol. 1997;179:2006–2013. doi: 10.1128/jb.179.6.2006-2013.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambler R P. The structure of β-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 3.Bellais S, Poirel L, Naas T, Girlich D, Nordmann P. Genetic-biochemical analysis and distribution of the Ambler class A β-lactamase CME-2, responsible for extended-spectrum cephalosporin resistance in Chryseobacterium (Flavobacterium) meningosepticum. Antimicrob Agents Chemother. 2000;44:1–9. doi: 10.1128/aac.44.1.1-9.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chetoui H, Melin P, Struelens M J, Delhalle E, Mutro Nigo M, De Ryck R, De Mol P. Comparison of biotyping, ribotyping, and pulsed-field gel electrophoresis for investigation of a common-source outbreak of Burkholderia pickettii bacteremia. J Clin Microbiol. 1997;35:1398–1403. doi: 10.1128/jcm.35.6.1398-1403.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Couture F, Lachapelle J, Levesque R C. Phylogeny of LCR-1 and OXA-5 with class A and class D β-lactamases. Mol Microbiol. 1992;6:1693–1705. doi: 10.1111/j.1365-2958.1992.tb00894.x. [DOI] [PubMed] [Google Scholar]
- 7.Fung-Tomc J, Bush K, Minassian B, Kolek B, Flamm R, Gradelski E, Bonner D. Antimicrobial activity of BMS-180680, a new catechol-containing monobactam. Antimicrob Agents Chemother. 1997;41:1010–1016. doi: 10.1128/aac.41.5.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilligan P H. Pseudomonas and Burkholderia. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of Clinical Microbiology. 6th ed. Washington, D.C.: American Society for Microbiology; 1995. pp. 509–519. [Google Scholar]
- 9.Iaconis J P, Sanders C C. Purification and characterization of inducible β-lactamases in Aeromonas spp. Antimicrob Agents Chemother. 1990;34:44–51. doi: 10.1128/aac.34.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahan A, Philippon A, Paul G, Weber S, Richard C, Hazebroucq G, Degeorges M. Nosocomial infection by chlorhexidine solution contaminated with Pseudomonas pickettii (biovar VA-I) J Infect. 1983;7:256–263. doi: 10.1016/s0163-4453(83)97196-7. [DOI] [PubMed] [Google Scholar]
- 11.Naas T, Nordmann P. OXA-type β-lactamases. Curr Pharmaceut Design. 1999;5:865–879. [PubMed] [Google Scholar]
- 12.Naas T, Sougakoff W, Casetta A, Nordmann P. Molecular characterization of OXA-20, a novel class D β-lactamase, and its integron from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:2074–2083. doi: 10.1128/aac.42.8.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Philippon L N, Naas T, Bouthors A-T, Barakett V, Nordmann P. OXA-18, a class D clavulanic-acid inhibited extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2188–2195. doi: 10.1128/aac.41.10.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poirel L, Guibert M, Bellais S, Naas T, Nordmann P. Integron- and carbenicillinase-mediated reduced susceptibility to amoxicillin-clavulanic acid in isolates of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 from French patients. Antimicrob Agents Chemother. 1999;43:1098–1104. doi: 10.1128/aac.43.5.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poirel L, Guibert M, Girlich D, Naas T, Nordmann P. Cloning, sequence analyses, expression, and distribution of ampC-ampR from Morganella morganii clinical isolates. Antimicrob Agents Chemother. 1999;43:769–776. doi: 10.1128/aac.43.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasmussen B A, Keeney D, Yang Y, Bush K. Cloning and expression of a cloxacillin-hydrolyzing enzyme and a cephalosporinase from Aeromonas sobria AER 14M in Escherichia coli: requirement for an E. coli chromosomal mutation for efficient expression of the class D enzyme. Antimicrob Agents Chemother. 1994;38:2078–2085. doi: 10.1128/aac.38.9.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raveh D, Simhon A, Gimmon Z, Sacks T, Shapiro M. Infections caused by Pseudomonas pickettii in association with permanent indwelling intravenous devices: four cases and review. Clin Infect Dis. 1993;17:877–880. doi: 10.1093/clinids/17.5.877. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Tolmasky M E, Crosa J H. Genetic organization of antibiotic resistance genes (aac(6′)-Ib, aadA and oxa9) in the multiresistance transposon Tn1331. Plasmid. 1993;29:31–40. doi: 10.1006/plas.1993.1004. [DOI] [PubMed] [Google Scholar]
- 20.Vershraegen G, Claeys G, Meeus G, Delanche M. Pseudomonas pickettii as a cause of pseudobacteremia. J Clin Microbiol. 1985;21:278–279. doi: 10.1128/jcm.21.2.278-279.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsh T R, Hall L, MacGowan A P, Bennett P. Sequence analysis of two chromosomally mediated inducible β-lactamases from Aeromonas sobria, strain 163a, one a class D penicillinase, the other an AmpC cephalosporinase. J Antimicrob Chemother. 1995;36:41–52. doi: 10.1093/jac/36.1.41. [DOI] [PubMed] [Google Scholar]
- 22.Yaabuchi E, Kosako Y, Yano I, Hotta H, Nishuichi Y. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni and Doudouroff 1973) comb. nov., Ralstonia solanoacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]