Abstract
Long non-coding RNAs (lncRNAs) have been recently emerged as critical modulators of oxidative stress pathway. Likewise, rising evidence currently highlights dysfunction of oxidative stress pathways in bipolar disorder (BD) patients.
In the current study, we evaluated the expression levels of H19, SCAL1 (LUCAT1), RMST, MEG3 and MT1DP lncRNAs in the PBMC from 50 patients with BD and 50 control subjects (male/female ratio in each group: 70%/30%). Expression levels of SCAL1, RMST and MEG3 but not H19 and MT1DP were considerably decreased in BD patients compared with healthy individuals. Such significant decrease in the expression of MEG3, RMST and SCAL1 was only reported in male BD patients compared with male controls. Substantial pairwise correlations were observed between expression levels of these lncRNAs in BD subjects. The area under curve values for RMST, MEG3 and SCAL1 were 0.70, 0.63 and 0.61 respectively. On the basis of this finding, RMST had the best efficiency in the discrimination of disease status between BD patients and controls. Taken together, the current results suggest a role for MEG3, RMST and SCAL1 lncRNAs in the pathogenesis of BD. In addition, peripheral expression levels of these lncRNAs might serve as potential peripheral markers for BD.
Keywords: RMST lncRNA, MEG3, Stress oxidative, Bipolar, Biomarker, Brain
Introduction
Bipolar disorder (BP) is a prevalent psychiatric disorder [1]. Genetics and environmental parameters have roles in the etiology of this disorder. Although researchers have not yet understood how genetics and environment interact with each other to cause this disorder, there are several studies that have reported dysregulated expression of genes in association with the pathogenesis of BD [2–4]. Oxidative stress that triggers cell death is one of the pathways implicated in this disorder. The brain is susceptive to oxidative stress, and it is essential to maintain a redox state for normal function of brain [5]. Long noncoding RNAs (lncRNA) are a kind of regulatory RNAs that modulate the expression of several genes in the oxidative stress pathway [6–8]. These RNAs contain more than 200 nucleotides and critically partake in transcriptional and post-transcriptional modulation of gene expression. They can regulate various biological pathways, including cell differentiation, apoptosis, cell growth, oxidative stress, and many more [9]. Several lines of evidence have indicated dysregulation of lncRNAs in brain tissues and the peripheral blood of patients with BD [2, 10, 11]. Yet, the function of many other lncRNAs in the etiology of this mental illness has not been investigated. In the present research project, we analyzed the expression of five lncRNAs namely H19, SCAL1 (LUCAT1), RMST, MEG3 and MT1DP in peripheral blood of cases with BD and healthy controls. H19 is one of the lncRNAs that regulate cell stress and harmonize cell fate and differentiation [12]. Research has shown that H19 is expressed in hippocampal neurons and promotes apoptosis in these neurons [12]. Moreover, H19 has been found to decrease defects of dopaminergic neurons through modulation of Wnt/β-catenin signaling [13], a pathway which is dysregulated in mood disorders and has a critical role in the development of these conditions [14]. SCAL1 (LUCAT1) is an lncRNA whose knockdown has decreased cell proliferation while inducing apoptosis and cellular stress [15, 16]. This lncRNA has been suggested to be an essential intermediate molecule in the process of regulation of antioxidant molecules by Nrf2 [17]. Nrf2 partakes in the pathogenesis of mood disorders. Knock-out this gene has resulted in induction of depression-like symptoms in the mice in association with increased concentrations of pro-inflammatory cytokines in the serum while decreased levels of BDNF in the prefrontal cortex and hippocampus of animals [18]. RMST as an lncRNA that has a role in neuronal differentiation through interaction with SOX2 [19]. Moreover, RMST expression has been found to be enhanced during differentiation of neural crest cells. This lncRNA has been suggested as a regulator of GnRH neurons [20]. Overexpression of MEG3 lncRNA has decreased neuronal activity and increased cell apoptosis [21]. MEG3 regulates hypoxia-mediated neuron apoptosis through affecting lipoxygenase signaling [22], a pathway which is found to be disturbed in the brain tissues of BD patients [23]. RMST and MEG3 lncRNAs contribute in controlling oxidative stress [24]. LncRNA MT1DP protects cells against oxidative stress, particularly the toxicity of cadmium to hepatocytes [25]. Consequently, aberrant expression of these lncRNAs may partake in the pathogenesis of BD or might be used as biomarker of this disease.
Materials and methods
Subjects
The Ethical Committee of Shahid Beheshti University of Sciences has approved this project. All participants in this study signed the written informed consent and the study protocol was performed in accordance with the ethical guidelines. Blood specimens of 50 patients and 50 healthy people were collected from Farshchian and Imam Hussein hospitals during 2016–2019. Patients were diagnosed assessed based on the presence of manic and depressive episodes according to the Diagnostic and Statistical Manual of Mental Disorders-5 [26]. All subjects were assessed using a semi-structured interview by experienced psychiatrists. Disease duration, disease onset and medication history were obtained. All BD patients were under treatment with standard dose of Carbamazepine. With the intention of decreasing the heterogeneity of patients’ cohort, patients who were under treatment with other drugs were omitted from the study. None of the patients and controls had a background of head trauma, encephalitis or other mental illnesses, neurodegenerative diseases, cancer, Epstein–Barr virus infection, or other infections.
Sample collection and RNA extraction
Four ml of peripheral blood was gathered from all people in EDTA tubes. The blood samples were centrifuged at 3000 rpm for 10 min and the Buffy coat was separated. Total RNA was drawn out from the PBMC using RNX kit (EX6101, Cinnagen, Tehran, Iran) according to the manufacturer’s guidelines. Qualitative and quantitative analyses of extracted RNA were performed by gel electrophoresis and the spectrophotometer.
cDNA production and real-time PCR assay
cDNA was produced using 3 μg of purified total RNA and High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems, PN: 4375575), based on the manufacturer’s rules. The expression levels of lncRNAs were measured in comparison with GAPDH as an internal control using appropriate primers (Table 1). Quantitative Real-time PCR was executed in the ABI 7500 sequence detection system (Applied Biosystem, Foster City, CA, USA) using 10 μl of BIOFACT™ 2X Real-Time PCR Master Mix, 10 ng cDNA and 200 nM of each primer. All experiments were performed at least in duplicate. Means of ΔCT for cases and controls were calculated and, lastly, the fold changes of expressions of genes were measured by ratio = 2-ΔΔCt as explained by Livak [27].
Table 1.
Primers used in RT-qPCR
| Gene | Forward Primer | Reverse Primer | Product Size (bp) |
|---|---|---|---|
| H19 | 5′-TGCTGCACTTTACAACCACTG-3′ | 5′-ATGGTGTCTTTGATGTTGGGC-3′ | 105 |
| MEG3 | 5′-TGGCATAGAGGAGGTGAT-3′ | 5′-GGAGTGCTGTTGGAGAATA-3′ | 111 |
| RMST | 5′-CAGGATGGCAGTGGGTGA-3′ | 5′-GTCCCTTGTGATCTCTGTGAC-3′ | 137 |
| SCAL1 | 5′-CCCAATGAAAAGGAACAAAACC-3’ | 5′-ATTTGTGAGGGGATGAGAATAC-3’ | 217 |
| MT1DP | 5′-CAAGAAGAACTGCTGCTCCT-3’ | 5′-TTGTAGGGGTTGCGTTATTTAC-3’ | 147 |
| GAPDH | 5′-CCATGAGAAGTATGACAAC-3’ | 5′-GAGTCCTTCCACGATACC-3’ | 105 |
Statistical method
Statistical analysis was accomplished in the GraphPad Prism 8 (GraphPad Software, Inc., San Diego, CA, USA). Kolmogorov–Smirnov test was used for assessment of normality of distribution of data. The t-test was employed to compare the differences in expression amounts of lncRNAs between patients and controls. Pearson’s coefficient of correlation was measured for appraisal of correlation between lncRNAs expression and the clinical features. P-value < 0.05 was considered as significant. Receiver Operating Characteristic (ROC) curve was used to evaluate the specificity and sensitivity of the genes selected as potential biomarkers.
Results
Cases and controls
In this study, 50 BD type I patients and 50 healthy individuals were enrolled. General data of patients and healthy controls are presented in Table 2.
Table 2.
Demographic and clinical features of BD patients and controls
| Cases | Controls | |
|---|---|---|
| Number | 50 | 50 |
| Sex | ||
| Male (%) | 70 | 70 |
| Female (%) | 30 | 30 |
| Mean age (range) | 36 (17–56) | 34 (14–52) |
| Disease duration (range) | 4 (1–14) | – |
| Onset age (range) | 32 (15–48) | – |
Gene expression levels in participants
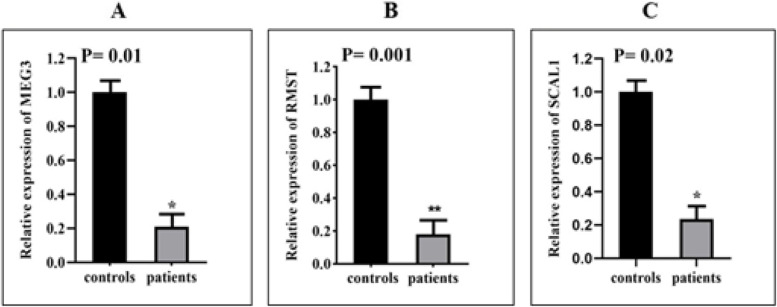
The expression levels of lncRNAs MEG3 (P = 0.0115), RMST (P = 0.0013), and SCAL1 (P = 0.0221) were significantly different between BD patients and healthy individuals (Fig. 1 A-C), although, there was no significant difference in the expression levels of lncRNAs H19 and MT1DP between controls and patients (Table 3). In fact, results demonstrated that the expression level of MEG3, RMST, and SCAL1 in BD patients decreased 4.76, 5.50, and 4.34 times compared to controls respectively. Furthermore, expressions of lncRNAs MEG3 (P = 0.009), RMST (P = 0.009), and SCAL1 (P = 0.048) were different between male BD patients and male controls. Instead, there was no important difference in expression of these lncRNAs in female patients compared to control females except for the lncRNA RMST (P = 0.04). After correction for multiple comparisons, the obtained P values remained significant for RMST expression in total patients compared with total controls (P = 0.005) and in male patients compared with male controls (P = 0.005). Moreover, expression of MEG3 in male patients compared with male controls remained significant (P = 0.005). Table 3 displays the outlines of relative expression (fold change) analysis of lncRNAs in BD patients and healthy controls.
Fig. 1.
Expression analysis of lncRNAs in the PBMCs. Relative expression (Fold change) of MEG3 (A), RMST (B), and SCAL1 (C). The expression of lncRNAs was down-regulated in BD patients compared with controls. Expression levels of lncRNAs in each sample were normalized to GAPDH expression. The relative expression of lncRNAs was obtained using the formula 2 -ΔΔCt and t-test
Table 3.
Relative expression of lncRNAs in BD patients and healthy controls
| lncRNAs | Parameters | Total patients (n = 50)/ total controls (n = 50) | Male patients (n = 35)/ male controls (n = 35) | Female patients (n = 15)/ female controls (n = 15) |
|---|---|---|---|---|
| MEG3 | 1/Fold change | 4.76 | 7.69 | 3.33 |
| P-value | 0.011* | 0.009** | 0.35 | |
| RMST | 1/Fold change | 5.50 | 6.25 | 12.50 |
| P-value | 0.001** | 0.009** | 0.04* | |
| SCAL1 | 1/Fold change | 4.34 | 5.00 | 4 |
| P-value | 0.022* | 0.048* | 0.36 | |
| H19 | 1/Fold change | 1.08 | 1.33 | 1.31 |
| P-value | 0.90 | 0.84 | 0.75 | |
| MT1DP | 1/Fold change | 1.12 | 1.16 | 1.31 |
| P-value | 0.83 | 0.95 | 0.94 |
Correlation analysis
Significant positive correlations were observed between expression amounts of all pairs of lncRNAs. Table 4 displays the results of pairwise correlation analysis between expression levels of genes. There was no noteworthy correlation between the level of expressions of genes in BD patients and age, disease duration, and onset age of disease (Table 5).
Table 4.
Pairwise correlation between expression levels of lncRNAs in cases group
| Correlation | MT1DP | H19 | RMST | MEG3 |
|---|---|---|---|---|
| SCAL1 | P < 0.0001 | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| r = 0.8517 | r = 0.6760 | r = 0.865 | r = 0.8811 | |
| MEG3 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |
| r = 0.8574 | r = 0.7316 | r = 0.8296 | ||
| RMST | P < 0.0001 | P < 0.0001 | ||
| r = 0.9107 | r = 0.7493 | |||
| H19 | P < 0.0001 | |||
| r = 0.7997 |
Table 5.
Appraisal of correlation between expression levels of lncRNAs and clinical data (R is the correlation coefficient or the Pearson’s correlation which measures the closeness of association of the points in the scatter plots to the linear regression line)
| MEG3 | RMST | SCAL1 | MT1DP | H19 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R | P value | R | P value | R | P value | R | P value | R | P value | |
| Patient Age | 0.087 | 0.551 | 0.148 | 0.309 | 0.215 | 0.136 | 0.097 | 0.504 | 0.041 | 0.776 |
| Age at onset | 0.200 | 0.166 | 0.111 | 0.447 | 0.191 | 0.188 | 0.156 | 0.284 | 0.142 | 0.330 |
| Disease duration | 0.160 | 0.270 | 0.204 | 0.158 | 0.277 | 0.053 | 0.190 | 0.189 | 0.180 | 0.215 |
ROC curve analysis
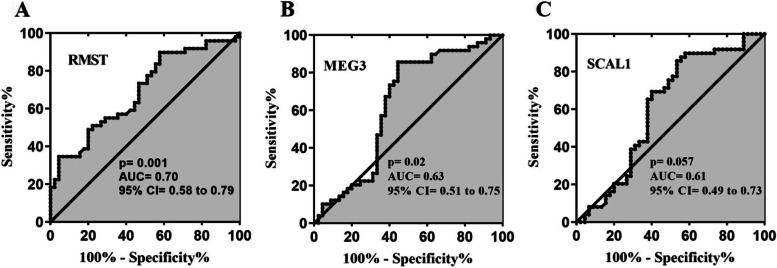
The sensitivity and specificity of expression levels of lncRNAs MEG3, SCAL1, and RMST as biomarkers were evaluated using the ROC curve analysis. The results showed that lncRNA RMST (AUC = 0.70, Sensitivity = 74%, Specificity = 63%, P = 0.001) and lncRNA MEG3 (AUC = 0.63, Sensitivity = 73%, Specificity = 60%, P = 0.028) could differentiate between patients and controls (Fig. 2 A, B). The AUC value for lncRNA SCAL1 was 0.61 (Sensitivity = 69%, Specificity = 60%, P = 0.0575) (Fig. 2 C). According to AUC values, RMST lncRNA had better performance in differentiating disease status in the study individuals compared with other genes.
Fig. 2.
ROC curve analysis of RMST (A), MEG3, (B) and SCAL1 (C). AUC: area under curve
Figure 3 shows the ROC curve for combination of expression levels of RMST, MEG3, and SCAL1 (AUC = 0.70, Sensitivity = 70%, Specificity = 61%, P = 0.001).
Fig. 3.
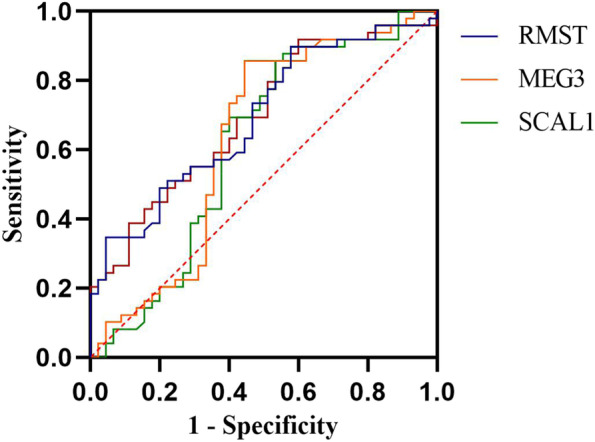
ROC curve for combination of expression levels of RMST, MEG3, and SCAL1
Discussion
The exact mechanism of BD is unclear. BD is a multifactorial disease, and various factors such as genetics and environmental parameters are involved in its etiology. There is strong evidence that oxidative stress partakes in the pathogenesis of the BD [28]. Also, some lncRNAs play an essential role in regulating oxidative stress [29]. As a result, in this research project, we examined the expression of five lncRNAs involved in the oxidative stress pathway in healthy and BD patients. Our study showed that expression levels of lncRNAs MEG3, RMST and SCAL1 were down-regulated in cases, but no significant difference was shown in expression levels of lncRNAs H19 and MT1DP between controls and patients. Similar results were observed in men. However, there was no significant difference in expression of lncRNAs between female BD cases and healthy women except for the RMST lncRNA.
MEG3 is a maternally imprinted gene being expressed in many normal tissues. This lncRNA is involved in the differentiation of GABAergic neurons [30]. Interestingly, up-regulation of MEG3 expression increases proinflammatory cytokines, decreases oxidative stress and apoptosis, and increases hippocampal neurons survival by activating the PI3K/AKT/mTOR pathway [31]. Meanwhile, this gene mediates ischemic nerve death by activating p53 and apoptosis [32]. Decreased expression of MEG3 lncRNA has been reported in schizophrenia, Huntington’s disease and epilepsy [33]. Consistent with these studies, decreased expression of MEG3 lncRNA in the present study may lead to increasing oxidative stress and reducing hippocampal neurons survival by inactivating the PI3K/AKT/mTOR pathway. Consequently, MEG3 might act as a regulatory factor in the pathogenesis of BD through this axis.
SCAL1 is located on chromosome 5 and is involved in tumorigenesis. Bhattacharjee et al. have shown that decreased SCAL1 gene expression was associated with inhibition of cell proliferation and increased apoptosis through decreasing expression levels of Bcl-2, while increasing Bax, Bad, and caspase-3 [34]. Studies have shown that SCAL1 is strictly related to Nrf2, and decreased expression of SCAL1 or Nrf2 significantly increases cigarette smoke toxicity [34]. In addition, downregulated expression of SCAL1 inhibits Nrf2 and cellular activity and induces oxidative stress via the Nrf2/Keap1 pathway [17]. Although the mechanism of SCAL1 partake in BD is not known, we suggest that decreased expression of lncRNA SCAL1 might affect the etiology of this disorder through enhancing oxidative stress and activating the Nrf2/Keap1 pathway.
The RMST gene is highly expressed in dopaminergic neurons in the midbrain [35]. Recent studies have shown that the RMST gene is controlled by REST and is involved in the differentiation of neurons in collaboration with the transcription factor SOX2 [19, 36]. RMST indirectly activates the p53/miR-107 signaling through interaction with the heterogeneous nuclear protein ribonucleus K (hnRNPK), which leads to an increase in BCL2 and eventually neuronal apoptosis [37]. Decreased expression of RMST lncRNA is also associated with increased oxidative stress. Silencing RMST expression reduces the size of the infarction and improves the results of the neural function test. Also, a decrease in markers of cerebral microgliosis and astrocytosis was also observed in the hippocampal region [38]. Considering the decreased lncRNA RMST expression in our study and previous information about this gene, such as its high expression in the brain, its role in neuronal differentiation, and oxidative stress regulation, this gene may be involved in the neurobiology of BD.
H19 is involved in neuronal differentiation [39], and epigenetic variation in ICR Igf2/H19 is associated with cerebellar development [40]. Research has shown that increasing the expression of H19 lncRNA increases Bax and caspase-3 expression and decreases levels of Bcl-2, and induces apoptosis of hippocampal neurons by suppressing the expression of let-7b microRNA [41]. Also, up-regulation of expression of H19 induces oxidative stress [12]. This gene is related with the initiation and prognosis of ischemic stroke [42]. In addition, expression of this lncRNA is increased in schizophrenia compared to healthy individuals. H19 might partake in the pathogenesis of psychiatric disorders by increasing neuronal apoptosis.
MT1DP is a pseudo-gene from the metallothionein (MT) family. Gao et al. have shown that MT1DP stimulates cadmium-induced oxidative stress by suppressing Nrf2 and increasing miR-365 expression. Increased expression of MT1DP lncRNA leads to decreased cell proliferation and increased apoptosis [41]. Also, this gene interacts with the RhoC protein. Following cadmium-associated stress, the MT1DP/RhoC complex rapidly activates the RhoC-CCN1/2-AKT signaling. It enhances the flow of Ca2 + into the cell, leading to increased cadmium uptake and toxicity, and eventually results in cell death. MT1H, meanwhile, binds to miR-214 as a competitive endogenous RNA (ceRNA) to prevent suppression of MT1DP [43, 44].
We demonstrated significant differences of the expression levels of lncRNAs MEG3 and SCAL1 only between male BD patients and male controls, which might show distinctive roles of these lncRNAs among males and females. There are a bulk of evidence suggesting a potential differential expression of lncRNAs in BD and other neuropsychiatric disorders based on the sex. Firstly, a study in post-mortem brain tissues has shown dysregulation of XIST in female BD cases [45]. Moreover, cortical transcriptome analyses have shown sex-biased pathways in BD [46]. Sexually dimorphic transcriptomic networks have also been identified in BD cases, particularly in the cholinergic system [47]. A former study has shown sex-specific dysregulation of HOXA-AS2, Linc-ROR, MEG3, SPRY4-IT1 and UCA1 lncRNAs in patients with schizophrenia [48].
In the period of study, we recruited all patients with the mentioned criteria. This resulted in the male/female ratio of 70%/30% in cases. Then, we recruited the controls with this male/female ratio. Thus, an alternative explanation for lack of significance among female subgroups might be the smaller sample size of these subjects.
In addition, we displayed several pairwise correlations between expressions of lncRNAs in patients with BD suggesting similar functional roles in same signaling pathways and processes and their regulation by similar epigenetic mechanisms.
We also reported that RMST and MEG3 may be useful for differentiating disease status in the study individuals. However, larger sample size is required for verifying these results and supporting the role of lncRNAs as diagnostic biomarkers in BD. In fact, we state small sample size as a limitation of our study.
Conclusion
As patients have been under treatment with Carbamazepine, one might speculate that differences in expression of genes might be due to the effects of medication. However, if all expression differences were due to medication effects, analysis of expression of these genes might help in the assessment of patients’ adherence to administered treatments or even risk of disease-related complications.
In brief, our study demonstrated that the expression levels of lncRNA MEG3 and lncRNA RMST were down-regulated in peripheral blood of BD patients.
Acknowledgements
Not applicable.
Abbreviations
- lncRNAs
Long non-coding RNAs
- BD
Bipolar disorder
- ROC
Receiver Operating Characteristic
- MT
Metallothionein
- ceRNA
Competitive endogenous RNA
Authors’ contributions
ZSF and MT wrote the manuscript and revised it. ZSF analyzed the data. ZM and SGF performed the experiment and bioinformatics analysis. All the authors contributed equally and are fully aware of submission. The author(s) read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participant
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent forms were obtained from all study participants. Informed consent forms were obtained from all study participants. The study protocol was approved by the ethical committee of Shahid Beheshti University of Medical Sciences (IR.SBMU.MSP.REC.1400.565). All methods were performed in accordance with the relevant guidelines and regulations.
Consent of publication
Not applicable.
Competing interests
The authors declare they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Soudeh Ghafouri-Fard, Email: s.ghafourifard@sbmu.ac.ir.
Zeinab Shirvani-Farsani, Email: z_shirvani@sbu.ac.ir.
References
- 1.Harrison PJ. Molecular neurobiological clues to the pathogenesis of bipolar disorder. Curr Opin Neurobiol. 2016;36:1–6. doi: 10.1016/j.conb.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naghavi-Gargari B, Zahirodin A, Ghaderian SMH, Shirvani-Farsani Z. Significant increasing of DISC2 long non-coding RNA expression as a potential biomarker in bipolar disorder. Neurosci Lett. 2019;696:206–211. doi: 10.1016/j.neulet.2018.12.044. [DOI] [PubMed] [Google Scholar]
- 3.Ghafouri-Fard S, Oskooei VK, Omrani MD, Taheri M. Dysregulation of cytokine coding genes in peripheral blood of bipolar patients. J Affect Disord. 2019;1(256):578–583. doi: 10.1016/j.jad.2019.06.028. [DOI] [PubMed] [Google Scholar]
- 4.Guan J, Cai JJ, Ji G, Sham PC. Commonality in dysregulated expression of gene sets in cortical brains of individuals with autism, schizophrenia, and bipolar disorder. Transl Psychiatry. 2019;9(1):152. doi: 10.1038/s41398-019-0488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim Y, Santos R, Gage FH, Marchetto MC. Molecular mechanisms of bipolar disorder: progress made and future challenges. Front Cell Neurosci. 2017;11:30. doi: 10.3389/fncel.2017.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghafouri-Fard S, Shoorei H, Taheri M. Non-coding RNAs are involved in the response to oxidative stress. Biomed Pharmacother. 2020;127:110228. doi: 10.1016/j.biopha.2020.110228. [DOI] [PubMed] [Google Scholar]
- 7.Zeng R, Zhang R, Song X, Ni L, Lai Z, Liu C, et al. The long non-coding RNA MALAT1 activates Nrf2 signaling to protect human umbilical vein endothelial cells from hydrogen peroxide. Biochem Biophys Res Commun. 2018;495(4):2532–2538. doi: 10.1016/j.bbrc.2017.12.105. [DOI] [PubMed] [Google Scholar]
- 8.Ghafouri-Fard S, Badrlou E, Taheri M, Dürsteler KM, Beatrix Brühl A, Sadeghi-Bahmani D, et al. A comprehensive review on the role of non-coding RNAs in the pathophysiology of bipolar disorder. Int J Mol Sci. 2021;22(10):5156. doi: 10.3390/ijms22105156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng W, Li L, Xu X, Jiao Y, Du W. Up-regulation of the long non-coding RNA RMRP contributes to glioma progression and promotes glioma cell proliferation and invasion. Arch Med Sci. 2017;13(6):1315. doi: 10.5114/aoms.2017.66747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hosseini E, Bagheri-Hosseinabadi Z, De Toma I, Jafarisani M, Sadeghi I. The importance of long non-coding RNAs in neuropsychiatric disorders. Mol Asp Med. 2019;70:127–140. doi: 10.1016/j.mam.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Shirvani Farsani Z, Zahirodin A, Ghaderian SMH, Shams J, Naghavi GB. The role of long non-coding RNA MALAT1 in patients with bipolar disorder. Metab Brain Dis. 2020;35(7):1077–1083. doi: 10.1007/s11011-020-00580-9. [DOI] [PubMed] [Google Scholar]
- 12.Pang H-l, Zhao Q-q, Ma Y, Song Y-l, Min J, Lu J-r, et al. Long noncoding RNA H19 participates in the regulation of adipose-derived stem cells cartilage differentiation. Stem Cells Int. 2019;2019:2139814. doi: 10.1155/2019/2139814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang J, Piao X, Hu S, Gao J, Bao M. LncRNA H19 diminishes dopaminergic neuron loss by mediating microRNA-301b-3p in Parkinson's disease via the HPRT1-mediated Wnt/β-catenin signaling pathway. Aging (Albany NY) 2020;12(10):8820–8836. doi: 10.18632/aging.102877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muneer A. Wnt and GSK3 Signaling Pathways in Bipolar Disorder: Clinical and Therapeutic Implications. Clin Psychopharmacol Neurosci. 2017;15(2):100–114. doi: 10.9758/cpn.2017.15.2.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoon J-H, You B-H, Park CH, Kim YJ, Nam J-W, Lee SK. The long noncoding RNA LUCAT1 promotes tumorigenesis by controlling ubiquitination and stability of DNA methyltransferase 1 in esophageal squamous cell carcinoma. Cancer Lett. 2018;417:47–57. doi: 10.1016/j.canlet.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 16.Thai P, Statt S, Chen CH, Liang E, Campbell C, Wu R. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am J Respir Cell Mol Biol. 2013;49(2):204–211. doi: 10.1165/rcmb.2013-0159RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Shen C, Zhu J, Shen G, Li Z, Dong J. Long noncoding RNAs in the regulation of oxidative stress. Oxidative Med Cell Longev. 2019;2019:1318795. doi: 10.1155/2019/1318795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hashimoto K. Essential Role of Keap1-Nrf2 Signaling in Mood Disorders: Overview and Future Perspective. Front Pharmacol. 2018;9:1182. doi: 10.3389/fphar.2018.01182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng S-Y, Bogu GK, Soh BS, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell. 2013;51(3):349–359. doi: 10.1016/j.molcel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 20.Stamou M, Ng S-Y, Brand H, Wang H, Plummer L, Best L, et al. A balanced translocation in Kallmann syndrome implicates a long noncoding RNA, RMST, as a GnRH neuronal regulator. J Clin Endocrinol Metab. 2020;105(3):e231–ee44. doi: 10.1210/clinem/dgz011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, Wang H. Long non-coding RNA in CNS injuries: a new target for therapeutic intervention. Mol Ther Nucleic Acids. 2019;17:754. doi: 10.1016/j.omtn.2019.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Hou L, Huang W, Gao Y, Lv X, Tang J. The Mechanism of Long Non-coding RNA MEG3 for Neurons Apoptosis Caused by Hypoxia: Mediated by miR-181b-12/15-LOX Signaling Pathway. Front Cell Neurosci. 2016;10:201. doi: 10.3389/fncel.2016.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim H-W, Rapoport SI, Rao JS. Altered arachidonic acid cascade enzymes in postmortem brain from bipolar disorder patients. Mol Psychiatry. 2011;16(4):419–428. doi: 10.1038/mp.2009.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohamed DI, Khairy E, Khedr SA, Habib EK, Elayat WM, El-kharashi OA. N-acetylcysteine (NAC) alleviates the peripheral neuropathy associated with liver cirrhosis via modulation of neural MEG3/PAR2/NF-ҡB axis. Neurochem Int. 2020;132:104602. doi: 10.1016/j.neuint.2019.104602. [DOI] [PubMed] [Google Scholar]
- 25.Gao M, Chen M, Li C, Xu M, Liu Y, Cong M, et al. Long non-coding RNA MT1DP shunts the cellular defense to cytotoxicity through crosstalk with MT1H and RhoC in cadmium stress. Cell discovery. 2018;4(1):5. doi: 10.1038/s41421-017-0005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Severus E, Bauer M. Bipolar disorders in DSM-5. Nervenarzt. 2014;85(5):543–547. doi: 10.1007/s00115-013-3987-1. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001 Dec;25(4):402–8 PubMed PMID: 11846609. Epub 2002/02/16. eng. [DOI] [PubMed]
- 28.Brown NC, Andreazza AC, Young LT. An updated meta-analysis of oxidative stress markers in bipolar disorder. Psychiatry Res. 2014;218(1–2):61–68. doi: 10.1016/j.psychres.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Giannakakis A, Zhang J, Jenjaroenpun P, Nama S, Zainolabidin N, Aau MY, et al. Contrasting expression patterns of coding and noncoding parts of the human genome upon oxidative stress. Sci Rep. 2015;5:9737. doi: 10.1038/srep09737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, et al. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11(1):14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Tao J, Zhang S, Lv X. LncRNA MEG3 reduces hippocampal neuron apoptosis via the PI3K/AKT/mTOR pathway in a rat model of temporal lobe epilepsy. Neuropsychiatr Dis Treat. 2020;16:2519. doi: 10.2147/NDT.S270614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan H, Yuan J, Gao L, Rao J, Hu J. Long noncoding RNA MEG3 activation of p53 mediates ischemic neuronal death in stroke. Neuroscience. 2016;337:191–199. doi: 10.1016/j.neuroscience.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 33.Johnson R. Long non-coding RNAs in Huntington's disease neurodegeneration. Neurobiol Dis. 2012;46(2):245–254. doi: 10.1016/j.nbd.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 34.Bhattacharjee S, Li J, Dashwood RH. Emerging crosstalk between long non-coding RNAs and Nrf2 signaling. Cancer Lett. 2020;490:154–164. doi: 10.1016/j.canlet.2020.07.011. [DOI] [PubMed] [Google Scholar]
- 35.Uhde CW, Vives J, Jaeger I, Li M. Rmst is a novel marker for the mouse ventral mesencephalic floor plate and the anterior dorsal midline cells. PLoS One. 2010;5(1):e8641. doi: 10.1371/journal.pone.0008641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Izuogu OG, Alhasan AA, Mellough C, Collin J, Gallon R, Hyslop J, et al. Analysis of human ES cell differentiation establishes that the dominant isoforms of the lncRNAs RMST and FIRRE are circular. BMC Genomics. 2018;19(1):1–18. doi: 10.1186/s12864-018-4660-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng H, Sun M, Wang Z-L, Wu Q, Yao J, Ren G, et al. LncRNA RMST-mediated miR-107 transcription promotes OGD-induced neuronal apoptosis via interacting with hnRNPK. Neurochem Int. 2020;133:104644. doi: 10.1016/j.neuint.2019.104644. [DOI] [PubMed] [Google Scholar]
- 38.Hou X-X, Cheng H. Long non-coding RNA RMST silencing protects against middle cerebral artery occlusion (MCAO)-induced ischemic stroke. Biochem Biophys Res Commun. 2018;495(4):2602–2608. doi: 10.1016/j.bbrc.2017.12.087. [DOI] [PubMed] [Google Scholar]
- 39.Farzi-Molan A, Babashah S, Bakhshinejad B, Atashi A, Fakhr TM. Down-regulation of the non-coding RNA H19 and its derived miR-675 is concomitant with up-regulation of insulin-like growth factor receptor type 1 during neural-like differentiation of human bone marrow mesenchymal stem cells. Cell Biol Int. 2018;42(8):940–948. doi: 10.1002/cbin.10960. [DOI] [PubMed] [Google Scholar]
- 40.Pidsley R, Fernandes C, Viana J, Paya-Cano JL, Liu L, Smith RG, et al. DNA methylation at the Igf2/H19 imprinting control region is associated with cerebellum mass in outbred mice. Mol Brain. 2012;5(1):1–9. doi: 10.1186/1756-6606-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao M, Li C, Xu M, Liu Y, Cong M, Liu S. LncRNA MT1DP aggravates cadmium-induced oxidative stress by repressing the function of Nrf2 and is dependent on interaction with miR-365. Adv Sci. 2018;5(7):1800087. doi: 10.1002/advs.201800087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Cao B, Gao Y, Han D, Zhao H, Chen Y, et al. Long non-coding RNA H19 positively associates with aspirin resistance in the patients of cerebral ischemic stroke. Front Pharmacol. 2020;11:1532. doi: 10.3389/fphar.2020.580783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao M, Chen M, Li C, Xu M, Liu Y, Cong M, et al. Long non-coding RNA MT1DP shunts the cellular defense to cytotoxicity through crosstalk with MT1H and RhoC in cadmium stress. Cell Discov. 2018;4(1):1–19. doi: 10.1038/s41421-017-0005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao M, Dong Z, Sun J, Liu W, Xu M, Li C, et al. Liver-derived exosome-laden lncRNA MT1DP aggravates cadmium-induced nephrotoxicity. Environ Pollut. 2020;258:113717. doi: 10.1016/j.envpol.2019.113717. [DOI] [PubMed] [Google Scholar]
- 45.Ji B, Higa KK, Kelsoe JR, Zhou X. Over-expression of XIST, the master gene for X chromosome inactivation, in females with major affective disorders. EBioMedicine. 2015;2(8):909–918. doi: 10.1016/j.ebiom.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lobentanzer S, Hanin G, Klein J, Soreq H. Integrative transcriptomics reveals sexually dimorphic control of the cholinergic/neurokine interface in schizophrenia and bipolar disorder. Cell Rep. 2019;29(3):764–77. e5. doi: 10.1016/j.celrep.2019.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simchovitz-Gesher A, Soreq H. Pharmaceutical implications of sex-related RNA divergence in psychiatric disorders. Trends Pharmacol Sci. 2020;41(11):840–850. doi: 10.1016/j.tips.2020.09.003. [DOI] [PubMed] [Google Scholar]
- 48.Fallah H, Azari I, Neishabouri SM, Oskooei VK, Taheri M, Ghafouri-Fard S. Sex-specific up-regulation of lncRNAs in peripheral blood of patients with schizophrenia. Sci Rep. 2019;9(1):1–8. doi: 10.1038/s41598-019-49265-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.