ABSTRACT
Marine bacteria usually contain polysaccharide utilization loci (PUL) for metabolizing red algae polysaccharides. They are of great significance in the carbon cycle of the marine ecosystem, as well as in supporting marine heterotrophic bacterial growth. Here, we described the whole κ-carrageenan (KC), ι-carrageenan (IC), and partial λ-carrageenan (LC) catabolic pathways in a marine Gram-negative bacterium, Flavobacterium algicola, which is involved carrageenan polysaccharide hydrolases, oligosaccharide sulfatases, oligosaccharide glycosidases, and the 3,6-anhydro-d-galactose (d-AHG) utilization-related enzymes harbored in the carrageenan-specific PUL. In the pathways, the KC and IC were hydrolyzed into 4-sugar-unit oligomers by specific glycoside hydrolases. Then, the multifunctional G4S sulfatases would remove their nonreducing ends’ G4S sulfate groups, while the ι-neocarratetrose (Nι4) product would further lose the nonreducing end of its DA2S group. Furthermore, the neocarrageenan oligosaccharides (NCOSs) with no G4S and DA2S groups in their nonreducing ends would completely be decomposed into d-Gal and d-AHG. Finally, the released d-AHG would enter the cytoplasmic four-step enzymatic process, and an l-rhamnose-H+ transporter (RhaT) was preliminarily verified for the function for transportation of d-AHG. Moreover, comparative analysis with the reported carrageenan metabolism pathways further implied the diversity of microbial systems for utilizing the red algae carrageenan.
IMPORTANCE Carrageenan is the main polysaccharide of red macroalgae and is composed of d-AHG and d-Gal. The carrageenan PUL (CarPUL)-encoded enzymes exist in many marine bacteria for decomposing carrageenan to provide self-growth. Here, the related enzymes in Flavobacterium algicola for metabolizing carrageenan were characterized for describing the catabolic pathways, notably, although the specific polysaccharide hydrolases existed that were like previous studies. A multifunctional G4S sulfatase also existed, which was devoted to the removal of G4S or G2S sulfate groups from three kinds of NCOSs. Additionally, the transformation of three types of carrageenans into two monomers, d-Gal and d-AHG, occurred outside the cell with no periplasmic reactions that existed in previously reported pathways. These results help to clarify the diversity of marine bacteria using macroalgae polysaccharides.
KEYWORDS: red algae, polysaccharides, polysaccharide utilization loci, Flavobacterium algicola
INTRODUCTION
Photosynthetic carbon sequestration by macroalgae, as one of the foundations of marine primary productivity, plays a critical role carbon cycle of the marine ecosystem (1). Algae polysaccharides would be synthesized by taking carbon dioxide from the atmosphere or the ocean. Additionally, with biological evolution, some marine heterotrophic bacteria attached to macroalgae have evolved a series of enzymes, including hydrolases, transporters, modifying enzymes, etc., to degrade algae polysaccharides for self-growth requirements, as well as releasing carbon dioxide. These enzymes are usually present in polysaccharide utilization loci (PUL), which are adjacent and mutually adjustable (2). Thus, the polysaccharide synthesis by macroalgae and polysaccharide metabolism by marine heterotrophic bacteria constitute the carbon cycle of the marine ecosystem (3).
Macroalgae are classified into green, red, and brown macroalgae (4). Therein, the red macroalgae have much higher carbohydrate content than those of brown and green macroalgae. Moreover, the monomeric sugar yields of red macroalgae were the highest among the three types of macroalgae (5–7). The main polysaccharides in red algae are classified into agar and carrageenans based on their monomer composition. Therein, carrageenans are linear sulfated polysaccharides with repeating disaccharide subunits composed of d-galactose (d-Gal) and 3,6-anhydro-d-galactose (d-AHG), which are alternately linked by β-1,4- and α-1,3-glycosidic linkages, respectively (8). According to the number and position of sulfate groups, natural carrageenans are divided into kappa (κ)-, iota (ι)-, and lambda (λ)-carrageenan. κ-Carrageenan (KC) only contains a C-4 hydroxyl sulfated d-Gal residue (G4S), and ι-carrageenan (IC) also contains a C-2 hydroxyl sulfated d-AHG residue (DA2S), while λ-carrageenan (LC) contains three types of sulfate groups, including C-2 hydroxyl sulfated d-Gal residue (G2S), DA2S, and C-6 hydroxyl sulfated d-AHG residue (DA6S), in its disaccharide unit (9–11).
The first carrageenan PUL (CarPUL) have been described in the marine bacterium Zobellia galactanivorans DsijT (12). In this carrageenan utilization pathway, KC and IC were transformed into β-neocarrageenan oligosaccharides (NβCOSs) with no sulfated residues by two specific pathways, respectively. Herein, KC was first decomposed into κ-neocarrageenan oligosaccharides (NκCOSs) by a GH16 family (http://www.cazy.org/) κ-carrageenase CgkA (13), and then the sulfate group from G4S of NκCOS was removed by an S1_7 (http://abims.sb-roscoff.fr/sulfatlas/) κCOS G4S sulfatase ZGAL_3146 to produce NBCOSs (14), while IC was first hydrolyzed into ι-neocarrageenan oligosaccharides (NιCOSs) under the coaction of three GH82 ι-carrageenases (CgiA1, CgiA2, and CgiA3) (15, 16). Furthermore, the transformation of NιCOSs products into NβCOSs was achieved by two sulfatases. S1_19 NιCOS, G4S sulfatase ZGAL_3145 removed the G4S sulfate group from NιCOSs, and then the DA2S sulfate group was removed by S1_17 DA2S-sulfatase ZGAL_3151. The further bioconversion of NβCOS into two monomers, d-AHG and d-Gal, relied on the coaction of exo-α-3,6-anhydro-d-galactosidase (ADAG) (GH127 ZGAL_3150, GH129-like ZGAL_3152) and exo-β-galactosidase (BG) (GH2 ZGAL_3633 and ZGAL_4655). Finally, d-Gal and d-AHG were transported into the cytoplasm for metabolism.
Also, other nonspecific KC and IC catabolic pathways have been revealed in five marine bacteria Pseudoalteromonas species (17). Different from the CarPUL in Z. galactanivorans DsijT, the CarPUL-encoded enzymes deployed in Pseudoalteromonas include three GH16 endo-acting carrageenases (GH16A, -B, and -C), an endo-acting KC and IC G4S sulfatase S1_19A, an exo-acting DA2S sulfatase S1_NC, an exo-acting G4S sulfatase S1_19B, and a GH167 β-neocarrabiose (Nβ2) releasing exo-β-galactosidases. In this carrageenan metabolism pathway, on the one hand, KC can be hydrolyzed into NκCOSs by GH16A/C; on the other hand, they can also be transformed into β-carrageenan (BC) and α-carrageenan (AC) by G4S sulfatase S1_19A. BC and AC were further degraded into NβCOSs and α-neocarrageenan oligosaccharides (NαCOSs) under the action of GH16B, respectively. In the periplasm, four types of neocarrageenan oligosaccharides (NCOSs) were completely decomposed into d-Gal and d-AHG. Therein, GH167 can cut the first β-1,4-glycosidic linkage from the nonreducing end of NCOSs to release the Nβ2 motif. Thus, NβCOSs can be directly hydrolyzed by GH167, while the sulfated NCOSs, including NκCOSs, NιCOSs, and NαCOSs, need extra steps under the action of sulfates to generate an Nβ2 nonreducing terminal; continued GH167 action would release the Nβ2 unit. However, the hydrolase activity toward Nβ2 to produce monosaccharides was not detected. This carrageenan catabolic pathway displays a completely different degradation model from the Z. galactanivorans DsijT, which suggested the diversity of carrageenan utilization pathways in these microbes.
For better illustrating this carrageenan metabolic versatility in the carrageenan-degrading microbes, here, the marine Gram-negative bacterium Flavobacterium algicola (18), which can grow with KC, IC, and LC as the sole carbon source, was selected for further studying. We first acquire the whole genome of F. algicola. Then, the bioinformatics approach was used for obtaining the CarPUL-encoding genes that may involve carrageenan metabolism. Furthermore, related encoded enzymes were expressed. Combined with activity verification and biochemical analysis, the KC and IC catabolic pathway was successfully proposed, as well as the partial LC pathway. These carrageenan metabolism pathways are not exactly the same as previously reported pathways, which would likely further reveal the diversity of microbial utilization systems of carrageenan.
RESULTS AND DISCUSSION
The conserved genes encode κ/ι/λ-carrageenan-specific enzymes.
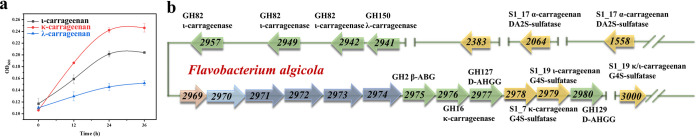
The gradually increased optical density at 600 nm (OD600) suggested the marine bacterium F. algicola could grow with KC, IC, and LC as the sole carbon source (Fig. 1a), which indicated the κ/ι/λ-CarPUL may exist. Consequently, the genome of F. algicola has been sequenced to further reveal the carrageenan utilization pathway. By using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and BioEdit tools (19), the KC-, IC-, and LC-related gene cluster was identified (Fig. 1b). This gene segment contains various carrageenan polysaccharide hydrolases (one GH16 κ-carrageenase is encoded by 2976, three GH82 ι-carrageenases are encoded by 2942, 2949, and 2957, and one GH150 λ-carrageenase is encoded by 2941), oligosaccharide or polysaccharide sulfatases (two S1_17 α-carrageenan DA2S-sulfatases are encoded by 1558 and 2064, two S1_19 IC G4S sulfatases are encoded by 2979 and 3000, and one S1_7 KC, G4S sulfatase is encoded by 2978), oligosaccharide glycosidases (one GH127 ADAG is encoded by 2977, one GH129 ADAG is encoded by 2980, and one GH2 BG is encoded by 2975), and the d-AHG utilization-related enzymes (d-AHG dehydrogenase is encoded by 2972, 3,6-anhydro-d-galactonate cycloisomerase is encoded by 2971, 2-keto-3-deoxy-d-galactonate kinase is encoded by 2973, and 2-keto-3-deoxy-d-galactonate aldolase is encoded by 2974). In addition, an AraC family transcriptional regulator is encoded by 2969 located in this segment. Additionally, this CarPUL also contains an l-rhamnose-H+ transporter (RhaT), which is encoded by gene 2970, which was speculated as the transporter for uptake of 3,6-anhydro-l-galactose (l-AHG) in previous research (20, 21). Thus, it is inferred that the KC, IC, and LC utilization pathways are located in the gene cluster from genes 1558 to 3000 in F. algicola.
FIG 1.
(a) Growth curves of F. algicola in M9 minimal medium with additives containing 3 g/L KC, IC, or LC as the sole carbon source. (b) Genes in the κ/ι/λ-CarPUL were annotated by using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and BioEdit tools. Carrageenan polysaccharide hydrolases, oligosaccharide or polysaccharide sulfatases, oligosaccharide glycosidases, d-AHG metabolism-related enzymes, and transporter are shown. KC, κ-carrageenan; IC, ι-carrageenan; LC, λ-carrageenan; κ/ι/λ-CarPUL, κ/ι/λ-carrageenan polysaccharide utilization loci; d-AHG, 3,6-anhydro-d-galactose.
The production of neocarrageenan oligosaccharides from hydrolyzing carrageenan.
There are a total of five GHs involved in the process of oligosaccharides production in F. algicola, including one κ-carrageenase, OUC-FaKC16A (encoded by gene 2976); one λ-carrageenase, OUC-FaLC150A (encoded by gene 2941); and three ι-carrageenases, OUC-FaLC82A (encoded by gene 2942), OUC-FaLC82B (encoded by gene 2949), and OUC-FaLC82C (encoded by gene 2957). These GHs have been successfully cloned and expressed in Escherichia coli BL21(DE3) for exploring their main products and to further purify them with an Ni2+-nitrilotriacetic acid (NTA) column to obtain pure enzymes (Fig. S1 in the supplemental material).
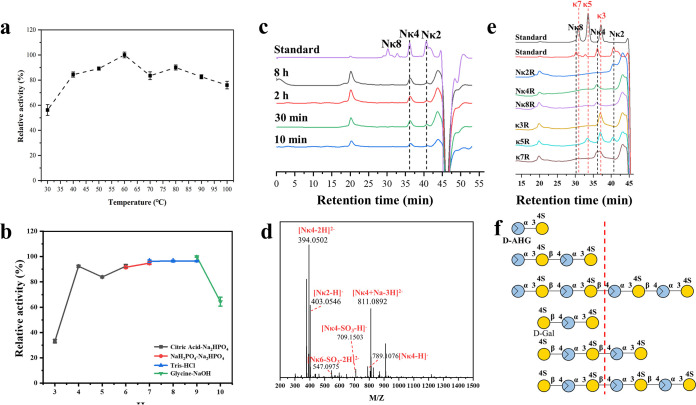
The OUC-FaKC16A was identified as κ-carrageenase and shows an ∼60% amino acid (aa) sequence similarity with the characterized κ-carrageenase ZgCgKA (GenBank accession no. CAZ94309.1) from Z. galactanivorans DsijT (22) (Table S1), but the sequence coverage is only 68%, which indicates its novelty. Phylogenetic analysis suggested that it belongs to the GH16 family and showed obvious affinity with the characterized κ-carrageenases (Fig. S2). Its enzymatic properties were further studied. Our data indicate the OUC-FaKC16A showed its highest activity at 60°C and pH 9.0 (Fig. 2a and b). Moreover, its products from decomposing KC were detected by high-performance liquid chromatography (HPLC); the results suggest that the products consisted of Nκ4 and Nκ2, of which Nκ4 is the main product (Fig. 2c). Mass spectrometry (MS) results further confirm this conclusion (Fig. 2d). Furthermore, the degradation pattern of OUC-FaKC16A toward NκCOSs and κCOSs has been analyzed. As shown in Fig. 2e, it can be concluded that the minimum actionable substrate of OUC-FaKC16A is κ-carrapentaose (κ5), and its minimum identifiable units are Nκ4 and κ-carratriose (κ3) from the nonreducing ends of NκCOSs and κCOSs, respectively. Compared with the reported κ-carrageenases, the OUC-FaKC16A possessed a relatively high optimum temperature, while most of the characterized κ-carrageenases were below to 55°C (22–26). Meanwhile, it exhibited wide temperature and pH reactivity. What’s more, the 24-h hydrolysate of OUC-FaKC16A contained ∼84% of Nκ4. These results indicated that the potential of OUC-FaKC16A is applied in functional NCOSs industry production.
FIG 2.
Functional characterization of GH16 κ-carrageenase OUC-FaKC16A from F. algicola. (a) Optimum temperatures of OUC-FaKC16A for hydrolyzing KC. (b) Optimum reaction pH levels of OUC-FaKC16A for hydrolyzing KC. (c) HPLC for analyzing the main products of OUC-FaKC16A from hydrolyzing KC. (d) MS for analyzing the end products of OUC-FaKC16A from hydrolyzing KC for 8 h. (e) HPLC for analyzing the products of OUC-FaKC16A from hydrolyzing Nκ2, Nκ4, Nκ8, κ3, κ5, and κ7. (f) Scheme of the oligosaccharide degradation model of OUC-FaKC16A. KC, κ-carrageenan; Nκ2, κ-neocarrabiose; Nκ4, κ-neocarratetrose; Nκ6, κ-neocarrahexaose; Nκ8, κ-neocarraoctaose; κ3, κ-carratriose; κ5, κ-carrapentaose; κ7, κ-carranonaose; d-AHG, 3,6-anhydro-d-galactose; d-Gal, d-galactose.
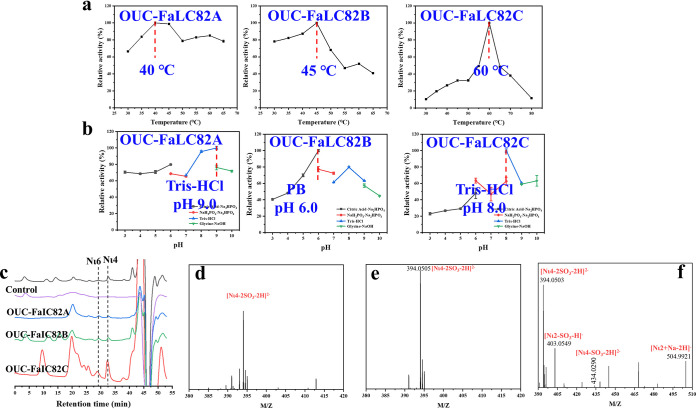
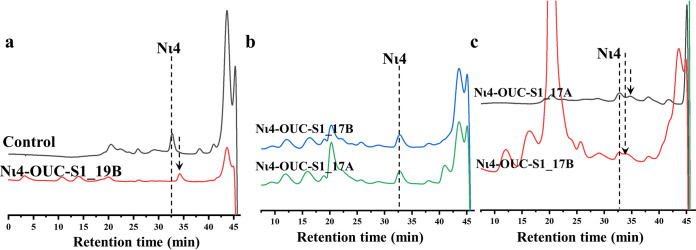
In addition, the three ι-carrageenases OUC-FaLC82A, OUC-FaLC82B, and OUC-FaLC82C all belong to the GH82 family from the phylogenetic analysis with characterized GH81, GH82, and GH83 hydrolyses (Fig. S3). Therein, OUC-FaLC82A shows an ∼56% aa sequence identity with the characterized ι-carrageenase CgiB_Ce (GenPept accession no. AGN70890.1) from the Cellulophaga sp. strain QY3 (27), while OUC-FaLC82B and OUC-FaLC82C show ∼28% and ∼37% aa sequence identity with the characterized ι-carrageenase CgiF (GenPept accession no. APX55175.1) from Flavobacterium sp. strain YS-80-122, respectively (Table S1) (28). The optimal temperatures of OUC-FaLC82A, OUC-FaLC82B, and OUC-FaLC82C were determined at 40, 45, and 60°C, while their optimal pH levels were 6.0, 9.0, and 8.0, respectively. Furthermore, their final products from hydrolyzing IC at the optimal reaction conditions were analyzed by HPLC and MS, which indicate that OUC-FaLC82A, OUC-FaLC82B, and OUC-FaLC82C would produce Nι4 and ι-neocarrahexaose (Nι6) during the reaction process, in which Nι4 is the main product (Fig. 3c to f).
FIG 3.
Functional characterization of GH82 ι-carrageenases OUC-FaIC82A, OUC-FaIC82B, and OUC-FaIC82C from F. algicola. (a) Optimum temperatures of OUC-FaIC82A, OUC-FaIC82B, and OUC-FaIC82C for hydrolyzing IC. (b) Optimum reaction pH levels of OUC-FaIC82A, OUC-FaIC82B, and OUC-FaIC82C for hydrolyzing IC. (c) HPLC for analyzing the main products of OUC-FaIC82A, OUC-FaIC82B, and OUC-FaIC82C from hydrolyzing IC. (d) MS for analyzing the end products of OUC-FaIC82A from hydrolyzing IC for 24 h. (e) MS for analyzing the end products of OUC-FaIC82B from hydrolyzing IC for 24 h. (f) MS for analyzing the end products of OUC-FaIC82C from hydrolyzing IC for 24 h. IC, ι-carrageenan; Nι2, ι-neocarrabiose; Nι4, ι-neocarratetrose; Nι6, ι-neocarrahexaose.
Additionally, the λ-carrageenase OUC-FaLC150A exhibits an ∼45% aa sequence identity with the two of reported λ-carrageenases, which are from the bacteria Pseudoalteromonas carrageenovora (GenPept accession no. CAL37005.1) and Pseudoalteromonas sp. strain CL19 (GenPept accession no. BAF35571.1), respectively (Table S1) (29, 30). Phylogenetic tree results further proved that it belongs to the GH150 family (Fig. S4). Its hydrolysis products were further detected; the HPLC and MS results suggest that it take the λ-neocarratetrose (Nλ4) as the main product (Fig. S5a and b). In general, the first step of metabolizing KC, IC, and LC in F. algicola is carried out, and KC, IC, and LC are transformed into Nκ4, Nι4, and Nλ4 by three types of specific carrageenases, respectively.
The processes of removing sulfate groups in neocarrageenan oligosaccharides.
After NCOS production, the next step for catabolizing carrageenan is to remove the sulfate groups for producing NβCOSs. Taking the reported sulfatases involved in the carrageenan metabolism, from which the Z. galactanivorans DsijT and Pseudoalteromonas species (12, 17) are used as the templates for sequence alignment in the proteome of F. algicola, results show that the OUC-S1_7A (encoded by gene 2978) exhibits an ∼55% aa sequence identity with the S1_7 KC G4S sulfatase ZGAL_3146 (GenPept accession no. CAZ97285.1) from Z. galactanivorans DsijT. OUC-S1_19A (encoded by gene 1510) exhibits a 45% aa sequence identity with the exo-acting G4S NκCOS sulfatase S1_19B (GenPept accession no. KAA1157118.1) from Pseudoalteromonas fuliginea. OUC-S1_19B (encoded by gene 2979) and OUC-S1_19C (encoded by gene 3000) exhibit ∼65% and ∼59% aa sequence identity with the S1_19 NιCOS G4S sulfatase ZGAL_3145 (GenPept accession no. CAZ97284.1) from Z. galactanivorans DsijT, as well as ∼53% and ∼56% identity with the KC/IC G4S sulfatase S1_19A (GenPept accession no. KAA1157105.1) from P. fuliginea, respectively. Also, OUC-S1_17A (encoded by gene 1558) and OUC-S1_17B (encoded by gene 2064) exhibit ∼37% and ∼36% aa sequence identity with the S1_17 DA2S sulfatase ZGAL_3151 (GenPept accession no. CAZ97290.1) from Z. galactanivorans DsijT, respectively (Table S2). To test these sulfatases’ activity, the recombinant proteins were obtained and further purified by affinity chromatography.
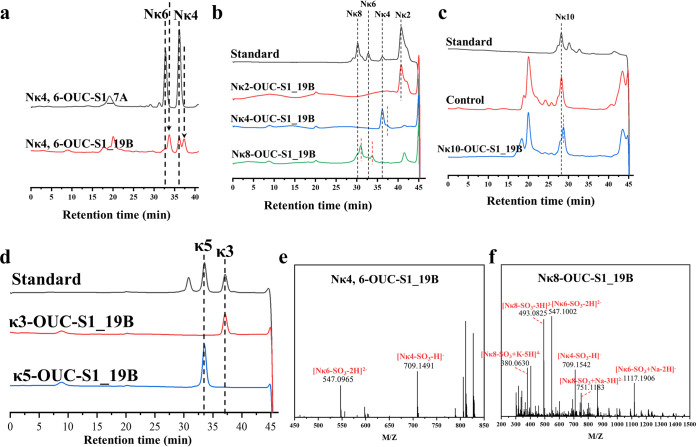
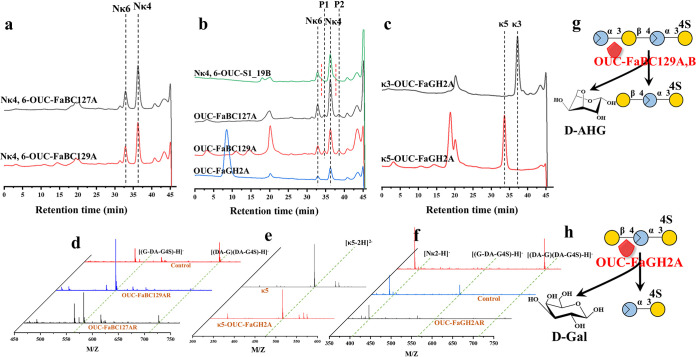
The predicted KC or NκCOS G4S sulfatases, including OUC-S1_7A, OUC-S1_19A, OUC-S1_19B, and OUC-S1_19C, have had their effects on KC or NκCOS with different degree of polymerization (DP) verified. The results indicated that OUC-S1_19B (encoded by gene 2979) showed activity for removing G4S sulfate groups from the mixture of Nκ4 and Nκ6 (Fig. 4a), while OUC-S1_19C (encoded by gene 3000) also showed activity to desulfate the G4S group of Nκ6 (Fig. S6a), but its activity was obviously less than OUC-S1_19B. Moreover, pure Nκ2, κ3, Nκ4, κ-neocarrapentaose (κ5), κ-neocarraoctaose (Nκ8), and κ-neocarradecaose (Nκ10) were used as the substrates for further activity tests. OUC-S1_19B exhibited obvious activity toward Nκ4, Nκ8, and Nκ10 (Fig. 4b to d), while it had no effects on Nκ2, κ3, and κ5 (Fig. 4b and d). Thus, these results allowed us to understand that the substrates of G4S sulfatase OUC-S1_19B should take the d-AHG as the nonreducing end. In order to further illustrate its action mode, the products acting on the mixture Nκ4, Nκ6, and Nκ8 were analyzed by MS that further proved the activity of OUC-S1_19B for removing the G4S sulfate groups from NκCOSs (Fig. 4e and f). Surprisingly, the MS result suggested that Nκ2 also would be desulfated under the action of excess OUC-S1_19B (Fig. S7). Moreover, it suggested that OUC-S1_19B could only remove one G4S sulfate group to produce the products with one less sulfate group than Nκ2, Nκ4, Nκ6, and Nκ8.
FIG 4.
Functional characterization of G4S sulfatase from F. algicola. (a) HPLC analysis of the activity of OUC-S1_7A and OUC-S1_19B for removing the G4S sulfate group from the mixture of Nκ4 and Nκ6. (b) HPLC analysis of the activity of OUC-S1_19B for removing the G4S group from Nκ2, Nκ4, and Nκ8. (c) HPLC analysis of the activity of OUC-S1_19B for removing the G4S group from Nκ10. (d) HPLC analysis of the activity of OUC-S1_19B for removing the G4S sulfate group from κ3 and κ5. (e) MS for analyzing the desulfated products from OUC-S1_19B acting on the mixture of κ4 and Nκ6. (f) MS for analyzing the desulfated products from OUC-S1_19B acting on the Nκ8. G4S, C-4 hydroxyl sulfated d-Gal residue; Nκ2, κ-neocarrabiose; Nκ4, κ-neocarratetrose; Nκ6, κ-neocarrahexaose; Nκ8, κ-neocarraoctaose.
Considering the high sequence similarities of OUC-S1_19B and OUC-S1_19C with S1_19 NιCOS G4S sulfatase ZGAL_3145 (GenBank accession no. CAZ97284.1) (Table S2), we also tested them with Nι4 as the substrate to check their activity for removing G4S sulfate groups from NιCOS. HPLC results showed that the new product with a smaller molecular weight (MW) was obtained after the reactions that illustrated the sulfatase activity of OUC-S1_19B and OUC-S1_19C (Fig. 5a and Fig. S6b). OUC-S1_17A (encoded by gene 1558) and OUC-S1_17B (encoded by gene 2064) were further tested the activity to remove the DA2S sulfate groups; only OUC-S1_17A successfully catalyzed the formation of a smaller new product from OUC-S1_19B-pretreated Nι4 (Fig. 5b). HPLC detected that the retention time of the OUC-S1_17A-catalyzed product was between OUC-S1_19B-pretreated Nι4 and Nκ4, which indicated the OUC-S1_17A could only remove one DA2S group from OUC-S1_19B-pretreated Nι4. What’s more, we also verified their catalytic ability on Nι4, but it exhibited no effects. The results above suggest that G4S sulfatase OUC-S1_19B and DA2S sulfatase OUC-S1_17A were involved in the process of removing sulfate groups in NιCOSs in which the removal of G4S sulfate group relies on G4S sulfatase OUC-S1_19B as the first step. We also tried to remove the G2S sulfate group in NλCOSs; HPLC indicated there had produced a new product with smaller MW from Nλ4 (Fig. S5c). However, the remaining two steps for further removing the DA2S and DA6S groups have not been resolved successfully in our research. Thus far, the processes of removing sulfate groups in NκCOS and NιCOS have been described.
FIG 5.
Functional characterization of G4S sulfatase OUC-S1_19B and DA2S sulfatase OUC-S1_17A/B from F. algicola. (a) HPLC analysis of the activity of OUC-S1_19B for removing the G4S sulfate group from the Nι4. (b) HPLC analysis of the activity of OUC-S1_17A and OUC-S1_17B for removing the DA2S sulfate group from Nι4. (c) HPLC analysis of the activity of OUC-S1_17A and OUC-S1_17B for removing the DA2S sulfate group from OUC-S1_19B-treated Nι4. G4S, C-4 hydroxyl sulfated d-Gal residue; DA2S, C-2 hydroxyl sulfated d-AHG residue; Nι4, ι-neocarratetrose.
The d-AHG and d-Gal production from neocarrageenan oligosaccharides.
Compared with the monosaccharide production pathway from Z. galactanivorans DsijT, the GH127 ADAG OUC-FaBC127A (encoded by gene 2977) and GH129 OUC-FaBC129A (encoded by gene 2980) in F. algicola were found, which exhibit ∼67% and ∼64% aa sequence identity with GH127 ADAG ZGAL_3150 (GenPept accession no. CAZ97289.1) and GH129-like ADAG ZGAL_3152 (GenPept accession no. CAZ97291.1), respectively (Table S2). GH2 BG OUC-FaGH2A (encoded by gene 2975) was selected for further in vitro assay because it is adjacent to the predicted CarPUL. In the beginning, the mixture of Nκ4 and Nκ6 was used as the substrate for OUC-FaBC127A and OUC-FaBC129A hydrolysis; HPLC showed no product formation (Fig. 6a). However, there were new, smaller products when taking the pretreated mixture of Nκ4 and Nκ6 by using OUC-S1_19B (Fig. 6b), which indicated that OUC-S1_19B tends to remove the G4S sulfate group of nonreducing d-Gal residue. The OUC-S1_19B-treated Nκ4 was further used as their substrate; MS analysis indicated oligosaccharides with DP3 (G-DA-G4S) and d-AHG were produced after the hydrolysis process (Fig. 6d; Fig. S8a). Additionally, the product from Nι4 that was treated successively with OUC-S1_19B and OUC-S1_17A also can be hydrolyzed by OUC-FaBC129A (Fig. S9), indicating that the action site of DA2S sulfate OUC-S1_17A was the first DA2S group of NιCOSs from its nonreducing end. The above results revealed that OUC-FaBC127A and OUC-FaBC129A tended to act on the first α-1,3-glycosidic linkage at the nonreducing end of NCOSs with no G4S group to release d-AHG and the remaining odd-numbered oligocarrageenans (Fig. 6g). This type of hydrolysis pattern is very similar to that of α-neoagarobiose hydrolase (NABH) involved in the agar metabolic pathway, which is devoted to releasing 3,6-anhydro-l-galactose (l-AHG) from the nonreducing end of neoagarooligosaccharides (NAOSs) (31).
FIG 6.
Functional characterization of ADAGs (OUC-FaBC127A and OUC-FaBC129A) and BG OUC-FaGH2A from F. algicola. (a) HPLC analysis of the activity of OUC-FaBC127A and OUC-FaBC129A for hydrolyzing the mixture of Nκ4 and Nκ6. (b) HPLC analysis of the activity of OUC-FaBC127A and OUC-FaBC129A for hydrolyzing the OUC-S1_19B-treated mixture of Nκ4 and Nκ6, and the activity of OUC-FaGH2A for acting on the products from OUC-FaBC129A hydrolysis of OUC-S1_19B-treated mixture of Nκ4 and Nκ6. (c) HPLC analysis of the activity of OUC-FaGH2A for hydrolyzing κ3 and κ5. (d) MS analysis of the products from OUC-FaBC127A and OUC-FaBC129A hydrolysis of the OUC-S1_19B-treated Nκ4. (e) MS analysis of the activity of OUC-FaGH2A for hydrolyzing κ5. (f) MS analysis of the products from OUC-FaGH2A hydrolysis of the COS with DP3 (G-DA-G4S). (g) The reaction scheme of ADAGs (OUC-FaBC127A and OUC-FaBC129A) toward NCOSs with no G4S group in the nonreducing end. (h) Reaction scheme of BG OUC-FaGH2A toward oligocarrageenans with no G4S group in the nonreducing end. ADAG, α-3,6-anhydro-d-galactosidase; BG, β-galactosidase; Nκ4, κ-neocarratetrose; Nκ6, κ-neocarrahexaose; κ3, κ-carratriose; κ5, κ-carrapentaose.
Furthermore, the ability of the gene 2975 encoding GH2 BG OUC-FaGH2A to release d-Gal had been explored. The data indicated that OUC-FaGH2A was inactive on κ3 and κ5 (Fig. 6c and e), but it could catalyze the degradation of products from OUC-FaBC127A or OUC-FaBC129A, which hydrolyzed the OUC-S1_19B-pretreated mixture of Nκ4 and Nκ6 (Fig. 6b). Also, these results further demonstrated that the G4S sulfatase OUC-S1_19B could obtain DA-G-DA-G4S from Nκ4. The oligosaccharide with DP3 (G-DA-G4S) was further used as a substrate for OUC-FaGH2A hydrolysis; the result suggested the DP3 was converted to Nκ2 (Fig. 6f) and d-Gal (Fig. S8b). All of these results allowed us to know that OUC-FaGH2A was an exo-lytic hydrolase that acts on the first β-1,4-glycosidic linkage at the nonreducing end of odd-numbered oligocarrageenans with no G4S group to release d-Gal and the remaining even-numbered NCOSs (Fig. 6h).
The metabolic pathway and transportation of d-AHG.
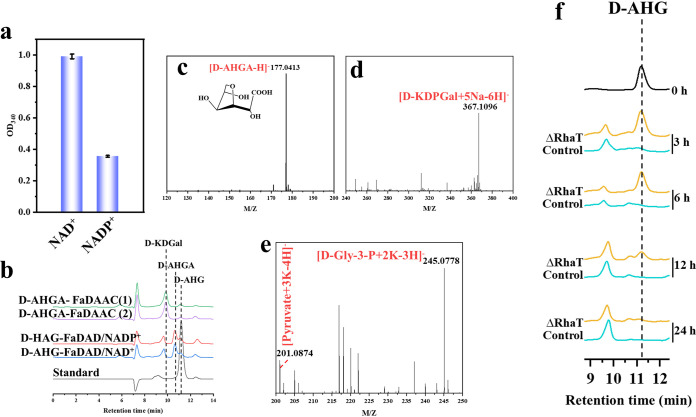
The above results demonstrated that KC and IC could be completely transformed into d-Gal and d-AHG under the coaction of GHs and sulfatases in F. algicola. Consequently, d-Gal and d-AHG would further enter the in vivo metabolic pathway and be decomposed and utilized for growth. The d-AHG metabolic pathway in carrageenan-degrading microorganisms was initially reported by Lee et al. (32). It revealed that the d-AHG pathway was composed of four enzymatic steps. d-AHG is first converted to 3,6-anhydro-d-galactonate (d-AHGA) by 3,6-anhydro-d-galactose dehydrogenase (d-AHGD), and then 3,6-anhydro-d-galactonate cycloisomerase (d-AHGAC) cyclizes d-AHGA into 2-keto-3-deoxy-d-galactonate (KDGal). The remaining two steps are consistent with the DeLey-Doudoroff pathway and d-galactonate pathway, including phosphorylation of d-KDGal and aldol cleavage of 2-keto-3-deoxy-6-phospho-d-galactonate (d-KDPGal) via KDGal kinase and KDPGal aldolase, respectively. Pyruvate and d-glyceraldehyde-3-phosphate (d-Gly-3-P) are eventually generated from d-AHG through the above-described pathway. Ficko-Blean et al. further proved that the same d-AHG pathway involved four enzymes (d-AHGD ZGAL_3155, d-AHGAC ZGAL_3156, KDGal kinase ZGAL_3154, and KDPGal aldolase ZGAL_3153) in Z. galactanivorans by in vitro experiments (12).
By bioinformatic analysis, the putative genes related to d-AHG catabolic pathway were found, including gene 2972 encoding d-AHGD (named FaDAD), 2971 encoding d-AHGAC (named FaDAAC), 2973 encoding KDGal kinase (named FaKDGK), and 2974 encoding KDPGal aldolase (named FaKDPGA). They exhibit ∼73%, ∼55%, ∼48%, and ∼64% aa sequence identities with the corresponding enzymes in Z. galactanivorans, respectively (Table S2). First, FaDAD’S oxidative activity toward d-AHG was tested, and the HPLC and MS results suggested d-AHGA was successfully obtained with NAD+ or NADP+ as cofactor (Fig. 7b and c), and it showed a higher activity when NAD+ existed (Fig. 7a). Then, the activity of FaDAAC to cyclize d-AHGA for producing d-KDGal was proven by HPLC (Fig. 7a). The d-KDPGal product of the third step was determined by MS (Fig. 7d), which indicated phosphorylation activity of FaKDGK in the presence of ATP. Finally, FaKDPGA could catalyze the formation of pyruvate and d-Gly-3-P from d-KDPGal; the end products pyruvate and d-Gly-3-P were also detected by our MS result (Fig. 7e).
FIG 7.
The metabolism and transportation analyses of d-AHG. (a) Comparison of the FaDAD’s (encoded by gene 2972) oxidative activity toward d-AHG with NAD+ or NADP+ as cofactor. (b) HPLC analysis of the activity of FaDAD for producing d-AHGA from d-AHG and FaDAAC (encoded by gene 2971) for converting d-AHGA into d-KDGal. (c) MS analysis of d-AHGA product by using FaDAD to oxidize d-AHG. (d) MS detection of d-KDPGal production from phosphorylation of d-KDGal using FaKDGK (encoded by gene 2973). (e) MS detection of final products pyruvate and d-Gly-3-P from a four-step enzymatic reaction. (f) HPLC for comparing the d-AHG’s consumption by the strains BL-DA and BL-DAΔRhaT over time. d-AHG, 3,6-anhydro-d-galactose; d-AHGA, 3,6-anhydro-d-galactonate; d-KDGal, 2-keto-3-deoxy-d-galactonate; d-KDPGal, 2-keto-3-deoxy-6-phospho-d-galactonate.
Before d-AHG enters the intracellular metabolic pathway, it is required to transport the d-AHG. Near the d-AHG metabolic gene cluster (2971 to 2974), gene 2970 was annotated as sugar transporter. In the Z. galactanivorans’s CarPUL, there is a gene, ZGAL_3157, with the same functional annotation, which exhibits a ∼52% aa sequence identity with gene 2970. Therefore, we speculate that this sugar symporter may be dedicated to the transport of d-AHG. To verify our hypothesis, the strains BL-DA and BL-DAΔRhaT, based on Gram-negative host Escherichia coli BL21(DE3), were constructed which carry the fragment from genes 2970 to 2974 and from genes 2971 to 2974, respectively. Then, their d-AHG utilization rates have been compared in the medium with 0.5 g/L d-AHG as the sole carbon source. The HPLC results suggested that the strain BL-DA could consume almost all d-AHG in 3 h, while BL-DAΔRhaT needed a 24-h consumption process. This phenomenon indicates the vital role of gene 2970 encoding RhaT in the d-AHG metabolism. In the agarose metabolic pathway of human bacterium Bacteroides uniformis NP1, a RhaT was speculated to be committed to the transmembrane transport of l-AHG (20). Likewise, a RhaT-encoding gene also existed in the l-AHG metabolic cluster that may be involved in metabolizing l-AHG in marine bacterium Z. galactanivorans (31). Combined with our experimental results, this further indicated the transport action of RhaT-like protein that is involved in d-AHG and l-AHG metabolism.
The KC and IC metabolic pathways in F. algicola.
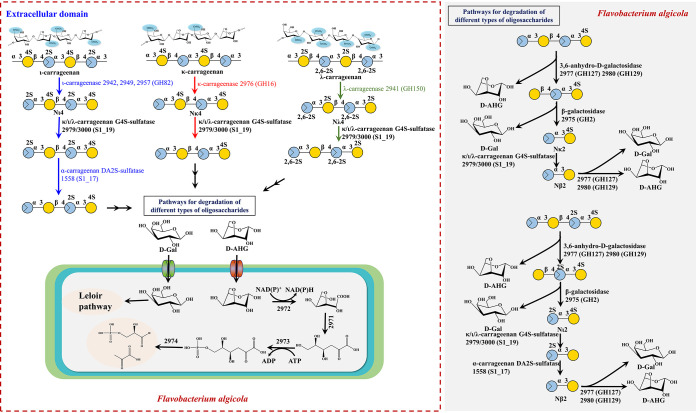
Based on the bioinformatics and biochemical data given here, the whole KC and IC and partial LC metabolic routes could be described (Fig. 8). In the first process, the three types of carrageenan were hydrolyzed into the corresponding NCOS of DP4 under the action of the specific polysaccharide hydrolases, including GH16 κ-carrageenase (OUC-FaKC16A), GH82 ι-carrageenases (OUC-FaIC82A, OUC-FaIC82B, and OUC-FaIC82C), and GH150 λ-carrageenase (OUC-FaLC150A). This process was consistent with the KC/IC catabolic pathways from marine bacterium Z. galactanivorans. The oligosaccharides further enter the next stage for removal of sulfate groups to produce NCOS with no sulfate groups in its nonreducing end. Therein, the Nκ4 only carries one type of sulfate group that would be desulfated by the G4S sulfatase OUC-S1_19B, while both Nι4 and Nλ4 were the substrates for OUC-S1_19B to remove their G4S and G2S sulfate groups from the nonreducing end, respectively, and the DA2S-sulfatase OUC-S1_17A acted on the OUC-S1_19B-treated Nι4 to remove its nonreducing end’s DA2S sulfate group. However, the sulfatases for removing the other two types of sulfate groups in Nλ4 have not been determined in this research. Then, the NCOSs of DP4 (DA-G-DA2S-G4S and DA-G-DA-G4S) would be sequentially hydrolyzed to generate d-AHG and d-Gal under the alternating action of ADAGs (OUC-FaBC127A and OUC-FaBC129A) and BG (OUC-FaGH2A), and the retained Nι2 and Nκ2 would transform into DP2 of DA2S-G and Nβ2, respectively, under the action again of OUC-S1_19B in which the DP2 of DA2S-G would further lose its DA2S group to produce Nβ2 by OUC-S1_17A. Then, the OUC-FaBC127A and OUC-FaBC129A completely decompose Nβ2 into d-Gal and d-AHG (Fig. 8). Finally, the released d-Gal was metabolized by the Leloir pathway; at the same time, the d-AHG would enter the cytoplasmic four-step enzymatic process to transform into d-Gly-3-P and pyruvate by a RhaT-like protein.
FIG 8.
Schematic model of carrageenan metabolism in F. algicola. In the first process, the KC, IC, and LC were decomposed into Nκ4, Nι4, and Nλ4 under the action of κ-carrageenase (OUC-FaKC16A), ι-carrageenases (OUC-FaIC82A, OUC-FaIC82B, and OUC-FaIC82C), and λ-carrageenase (OUC-FaLC150A), respectively. Then, the sulfate groups’ removal steps in different types of NCOSs were performed with G4S sulfatase OUC-S1_19B and DA2S sulfatase OUC-S1_17A. Furthermore, the NCOSs with no G4S and DA2S groups of the nonreducing end would completely be decomposed into d-Gal and d-AHG under the combined action of ADAGs (OUC-FaBC127A and OUC-FaBC129A), BG (OUC-FaGH2A), and sulfatases. Finally, the released d-Gal was metabolized by the Leloir pathway, while d-AHG would enter the cytoplasmic four-step enzymatic process to transform into d-Gly-3-P and pyruvate by a RhaT-like protein. KC, κ-carrageenan; IC, ι-carrageenan; LC, λ-carrageenan; Nκ4, κ-neocarratetrose; Nι4, ι-neocarratetrose; Nλ4, λ-neocarratetrose; Nκ2, κ-neocarrabiose; Nι2, ι-neocarrabiose; ADAG, α-3,6-anhydro-d-galactosidase; BG, β-galactosidase; d-Gal, d-galactose; d-AHG, 3,6-anhydro-d-galactose.
Moreover, the fermented supernatant of F. algicola collected from M9 medium with KC as the sole carbon source was analyzed by HPLC and MS (Fig. S10). The results suggested that the supernatant was composed of d-AHG, d-Gal, and Nβ2. It was indicated that the process for transforming KC into d-AHG and d-Gal occurred outside the cells, and it was also demonstrated that the related enzymes existed in the extracellular domain of F. algicola. Therefore, it allowed us to know that the d-AHG and d-Gal production from IC and LC also occurred in the extracellular environment.
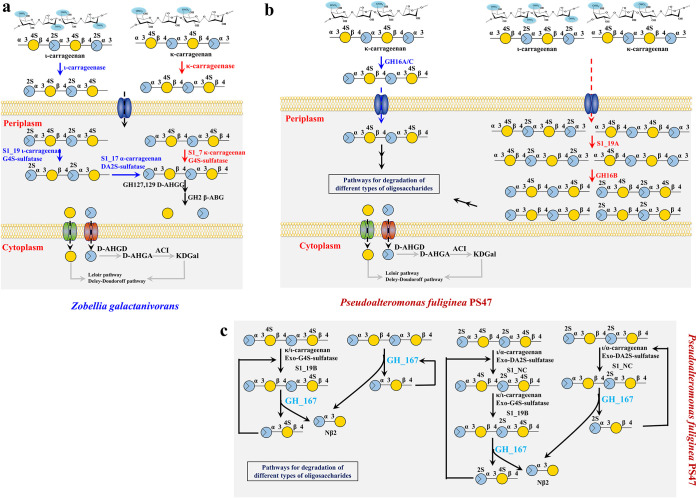
The carrageenan metabolic pathways have been illustrated in marine bacteria Z. galactanivorans DsijT and P. fuliginea (Fig. 9). In Z. galactanivorans DsijT, the specific KC and IC catabolic routes existed, including the specific polysaccharides’ degradation process and specific sulfate groups’ removal process, which relied on specific polysaccharide hydrolases and specific sulfatases. In addition, the transformation of KC or IC into NκCOSs or NιCOSs occurred outside the cell, while the removal of the sulfate groups’ and production of monomers (d-Gal and l-AHG) occurred in the periplasm (12). In another bacterium, P. fuliginea, the utilization of KC and IC was reliant on the nonspecific pathways based on the bifunctional hydrolases and sulfatases. Moreover, the κ-carrageenases (GH16A/C)-coding gene also existed in the CarPUL of P. fuliginea, which suggested the κ-carrageenan also can be utilized by specific pathways in P. fuliginea. Furthermore, it also involved the periplasm catabolic pathway for completely hydrolyzing different types of oligosaccharides into d-Gal and d-AHG (17). In earlier research about the red algal polysaccharide degradation pathway in the marine bacterium Paraglaciecola hydrolytica S66T, n GH16 κ-carrageenase and three GH16 β-carrageenases were proven to participate in the metabolic pathway of κ/β-carrageenan. Therein, two of the GH16 β-carrageenases, Ph1656 and Ph1663. hydrolyzed the BC or partially sulfated furcellaran to release oligosaccharides (33). Considering the high sequence identity (∼61%) of Ph1633 with the nonspecific carrageenase GH16B from P. fuliginea, it was speculated that Ph1663 can also act on the AC (or hybrid α/β-carrageenans). Additionally, the protein (GenBank accession no. WP_068376979.1) with high similarity (∼69%) to the multifunctional G4S sulfatase S1_19A of P. fuliginea was also found in P. hydrolytica S66T. Thus, the nonspecific catabolic pathways of KC and IC, consisting of polysaccharides’ desulfating process and sulfated polysaccharides’ degradation process, may also be present in P. hydrolytica S66T.
FIG 9.
Schematic model of KC and IC metabolism in Z. galactanivorans DsijT (a) and P. fuliginea PS47 (b). (a) In Z. galactanivorans DsijT, the specific KC and IC catabolic routes existed, including the specific polysaccharides’ degradation process and specific sulfate groups’ removal process, which relied on specific polysaccharide hydrolases and specific sulfatases. In addition, the transformation of KC or IC into NκCOSs or NιCOSs occurred outside the cell, while the removal of the sulfate groups and production of monomers (d-Gal and l-AHG) occurred in the periplasm. (b) In another bacterium P. fuliginea PS47, the KC and IC would be transformed into four types of NCOSs under the action of GH16A/C or the coaction of multifunctional G4S sulfatase S1_19A and GH16B. Then, the four types of NCOS were completely decomposed into d-Gal and d-AHG. (c) Therein, GH167 can cut the first β-1,4-glycosidic linkage from the nonreducing end of NCOSs to release the Nβ2 motif. Thus, BNCOSs can be directly hydrolyzed by GH167, while the sulfated NCOSs, including NκCOSs, NιCOSs, and NαCOSs, need extra steps under the action of sulfates to generate a Nβ2 nonreducing terminal; continued GH167 action would release the Nβ2 unit. KC, κ-carrageenan; IC, ι-carrageenan; Nβ2, β-neocarrabiose; NCOS, neocarrageenan oligosaccharides.
Compared with Z. galactanivorans DsijT, P. fuliginea, and P. hydrolytica S66T, the carrageenan-utilized pathways in F. algicola displayed different characteristics, although the specific polysaccharide hydrolases exist that are like Z. galactanivorans DsijT. A multifunctional G4S sulfatase, OUC-S1_19B, also existed that was devoted to the removal of G4S sulfate groups from different types of oligocarrageenans. The same multifunctional G4S sulfatase, S1_19A, had been discovered in P. fuliginea (34), but its role was to remove the G4S sulfate groups from KC and IC polysaccharides for further decomposing (17). Moreover, the OUC-S1_19B also proved the capacity to desulfate the sulfate group from NλCOS. The partial LC-utilized route described here may help the complete analysis of the LC degradation pathway in algal polysaccharide-degrading bacteria. What’s more, all the processes for completely converting KC into two monomers may happen in the extracellular environment of F. algicola, which is quite different from the pathways in Z. galactanivorans DsijT and P. fuliginea.
In this study, we found that F. algicola possesses the KC, IC, and LC metabolic pathways that were composed of a specific polysaccharide’s degradation process and a nonspecific sulfate group’s removal process. Furthermore, the RhaT protein involving the transmembrane transportation of d-AHG was preliminarily studied. Our results further reveal the diversity of microbial utilization systems of the red algae carrageenan, which will help to understand the significant role of algal polysaccharide-degrading bacteria in the carbon balance of the marine ecosystem.
MATERIALS AND METHODS
Materials.
F. algicola (catalog no. 1.12076) was purchased from China General Microbiological Culture Collection Center. The expression plasmids pET-28a(+) and pCold-SUMO have been kept in our lab. The clone host, E. coli DH5α, and expression host, E. coli BL21(DE3), were obtained from Tsingke Biotechnology Co., Ltd. (Beijing, China). Another expression host, RTS BL21(DE3) chaperone, was obtained from HaiGene (Qingdao, China). The KC and IC polysaccharides were obtained commercially from Sigma-Aldrich, and LC polysaccharide was ordered from TCI (Shanghai, China). κ3, κ5, κ-carranonaose (κ7), Nκ2, Nκ4, Nκ8, and Nκ10 used for HPLC analysis were purchased from Bz Oligo Biotech (Qingdao, China).
Growth of F. algicola on κ/ι/λ-carrageenan.
F. algicola was first grown in LB (1% tryptone, 0.5% yeast extract, and 1% NaCl) fluid medium at 25°C for 48 h. We washed the cells twice using sterilized water and then transferred them into M9 minimal medium (0.24 g/L MgSO4, 0.011 g/L CaCl2, 3.0 g/L Na2HPO4, 0.5 g/L NaCl, and 1.0 g/L NH4Cl) with additives (0.001 g/L vitamin, 0.001 g/L biotin, 0.05 g/L EDTA, 0.0083 g/L FeCl3·6 H2O, 0.00084 g/L ZnCl2, 0.00013 g/L CuCl2·6H2O, 0.0001 g/L CoCl2·2H2O, 0.0001 g/L CoCl2, 0.0001 g/L H3BO3, 0.000016 g/L MnCl2·6H2O, and 0.0003 g/L Na2MoO4·H2O) containing 3 g/L KC or IC or LC as the sole carbon source at 25°C. The OD600 was determined by using a spectrophotometer (Thermo Scientific, USA) for drawing of growth curves.
Sequence analyses.
Carbohydrate-active enzymes were identified by using DIAMOND (35) for alignment of amino acid sequences from F. algicola with the Carbohydrate-Active Enzymes database (CAZy). Predicted carbohydrate-active enzymes sequences were further compared with homologs at NCBI using BLASTp (https://blast.ncbi.nlm.nih.gov/) searches against the nonredundant (nr) databases; the closest characterized enzyme was selected to be described in Table S1 in the supplemental material. For discovering the related sulfatases, oligosaccharide glycosidases, and d-AHG utilization-related enzymes in F. algicola, the verified sulfatases involved in the carrageenan metabolism of the Z. galactanivorans DsijT and Pseudoalteromonas species were used as the template for sequence alignment using BioEdit tools to find the closest sequence in the proteome of F. algicola.
In order to further prove the GHs of five carrageenases, phylogenetic analyses were performed with MEGA version 6.0.
Gene cloning, expression, and purification of related enzymes.
Genomic DNA was extracted from F. algicola using a TIANamp bacteria DNA kit (Tiangen Biotech, Beijing, China). The related genes were further amplified with the genomic DNA of F. algicola as a template, using the primers listed in Table 1 and the 2× Phanta Max master mix (Vazyme, China) DNA polymerase. All the primers were synthesized by BGI (Beijing, China). The plasmids pET-28a(+) and pCold-SUMO were also linearized by PCR. The PCR products and linearized plasmids were linked by ClonExpress Ultra one-step cloning kit (Vazyme, China) and then transformed into E. coli DH5α for obtaining the recombinant vector. Therein, the genes 2941, 2957, and 2978 were linked to pET-28a(+), while other genes were linked to pCold-SUMO for the prevention of inclusion bodies. The inserted genes were further confirmed by sequencing (BGI, China). Afterward, the recombinant vectors were transformed into expression hosts; this procedure could be divided into four different situations. In the first case, the pET-28a(+)-carrying gene 2941 or 2957 was transformed into E. coli BL21(DE3) for protein expression. In the second case, the pET-28a(+)-carrying gene 2978 and pBAD/myc-his A Rv0712 (FGE) (Addgene; plasmid no. 16132), which encoded a formylglycine-generating enzyme to promote sulfatase maturation, were cotransformed into E. coli BL21(DE3) for sulfatase OUC-S1_7A expression. In the third case, each pCold-SUMO-carrying gene that was not encoding sulfatase was transformed into RTS BL21(DE3) chaperone for soluble expression. In the final case, each pCold-SUMO-carrying sulfatase-expressed gene and pBAD/myc-his K Rv0712 (FGE), which was modified from pBAD/myc-his A Rv0712 (FGE) by replacing its ampicillin-encoding gene with a kanamycin-encoding gene, were cotransformed into RTS BL21(DE3) chaperone for other sulfatases’ soluble expression.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′–3′) | Usage |
|---|---|---|
| 1558ColdF | gtaccctcgagggatccCAAGTTTCTCAATTAAAAAATAGTC | Cloning gene 1558 to ligate into linearized pCold-SUMO |
| 1558ColdR | gactgcaggtcgacaagcttGTAGGTATCAGGAGCCC | Cloning gene 1558 to ligate into linearized pCold-SUMO |
| 2064ColdF | cggtaccctcgagggatccCAAACTGCAAAAAAACCAAAC | Cloning gene 2064 to ligate into linearized pCold-SUMO |
| 2064ColdR | ctgcaggtcgacaagcttATTAAAAGCACTGATGCG | Cloning gene 2064 to ligate into linearized pCold-SUMO |
| 2941F | gtgcggccgcaagcttAAAAGCTTTCTCTAATTCCAC | Cloning gene 2941 to ligate into linearized pET28a(+) |
| 2941R | aatgggtcgcggatccGCAAATGGAACTCCACC | Cloning gene 2941 to ligate into linearized pET28a(+) |
| 2942ColdF | cggtaccctcgagggatccTTATGTTTTTCGCAAGACCG | Cloning gene 2942 to ligate into linearized pCold-SUMO |
| 2942ColdR | agtgcggccgcaagcttTGCTTTCTCTTTTTTAACTTTTG | Cloning gene 2942 to ligate into linearized pCold-SUMO |
| 2949ColdF | gtaccctcgagggatccGCTGATAATGTGTTTTATAATCC | Cloning gene 2949 to ligate into linearized pCold-SUMO |
| 2949ColdR | gcaggtcgacaagcttATTATCCTTTTTTTTTTTTCCTTT | Cloning gene 2949 to ligate into linearized pCold-SUMO |
| 2957F | aatgggtcgcggatccGCGCAAATTGATAAAGAGAG | Cloning gene 2957 to ligate into linearized pET28a(+) |
| 2957R | gcaggtcgacaagcttTTTTTTCATTTTCTTAGTTTCTTTTG | Cloning gene 2957 to ligate into linearized pET28a(+) |
| 2971ColdF | gtaccctcgagggatccATGAAAATTAAATCAATTGAATGTTTTATTC | Cloning gene 2972 to ligate into linearized pCold-SUMO |
| 2971ColdR | ctgcaggtcgacaagcttTTCTTTTAAAGCAAATTTATATAATGC | Cloning gene 2972 to ligate into linearized pCold-SUMO |
| 2972ColdF | ggtaccctcgagggatccATGGATTTTGGACATAAAAAATTATATATAG | Cloning gene 2949 to ligate into linearized pCold-SUMO |
| 2972ColdR | ctgcaggtcgacaagcttCTATAAAGCTTTTGTAACGCG | Cloning gene 2949 to ligate into linearized pCold-SUMO |
| 2973ColdF | ggtaccctcgagggatccATGAAACTTCCAAAATATTTTATTAG | Cloning gene 2973 to ligate into linearized pCold-SUMO |
| 2973ColdR | ctgcaggtcgacaagcttTTATTTTTCATTAAGTACAATTATTTTC | Cloning gene 2973 to ligate into linearized pCold-SUMO |
| 2974ColdF | cggtaccctcgagggatccATGAAAAATAATTCAGGATTTTCG | Cloning gene 2974 to ligate into linearized pCold-SUMO |
| 2974ColdR | gactgcaggtcgacaagcttTTATTTTATTTCGTTTTTAACACGTATG | Cloning gene 2974 to ligate into linearized pCold-SUMO |
| 2975ColdF | ggtaccctcgagggatccCAAAATGATTGGGAGAATGAG | Cloning gene 2975 to ligate into linearized pCold-SUMO |
| 2975ColdR | gactgcaggtcgacaagcttATTCATGAAATTCATTCCG | Cloning gene 2975 to ligate into linearized pCold-SUMO |
| 2976ColdF | cggtaccctcgagggatccGCGCAACATGCACC | Cloning gene 2976 to ligate into linearized pCold-SUMO |
| 2976ColdR | ctgcaggtcgacaagcttCTGTATTAAAAGTTTTTGGG | Cloning gene 2976 to ligate into linearized pCold-SUMO |
| 2977ColdF | ggtaccctcgagggatccCAGAATAAAGGGATTGTGG | Cloning gene 2977 to ligate into linearized pCold-SUMO |
| 2977ColdR | ctgcaggtcgacaagcttATCCCAAATAATTGGCATG | Cloning gene 2977 to ligate into linearized pCold-SUMO |
| 2978F | gagtgcggccgcaagcttaTTTGAAAAAACTACTTTCTGAC | Cloning gene 2978 to ligate into linearized pET28a(+) |
| 2978R | caaatgggtcgcggatcctGTAGCCAAGGACAAAAAAAAC | Cloning gene 2978 to ligate into linearized pET28a(+) |
| 2979ColdF | cggtaccctcgagggatccGCACAAACCTCAAAGCC | Cloning gene 2979 to ligate into linearized pCold-SUMO |
| 2979ColdR | gcaggtcgacaagcttTTTTTTCTGCTTTTCAGTCC | Cloning gene 2979 to ligate into linearized pCold-SUMO |
| 2980ColdF | ggtaccctcgagggatccGTAAAGGAAGGTGTAAAAC | Cloning gene 2980 to ligate into linearized pCold-SUMO |
| 2980ColdR | ctgcaggtcgacaagcttTTTATAACTAACCTGAAAGTTCC | Cloning gene 2980 to ligate into linearized pCold-SUMO |
| 3000ColdF | ggtaccctcgagggatccGCAAAACCAAATATTTTGG | Cloning gene 3000 to ligate into linearized pCold-SUMO |
| 3000ColdR | gactgcaggtcgacaagcttATCCAACAAGAATGCTTTG | Cloning gene 3000 to ligate into linearized pCold-SUMO |
| 28aF | ggatccgcgacccatttgc | To linearize the plasmid pET28a(+) |
| 28aR | aagcttgcggccgcactcg | To linearize the plasmid pET28a(+) |
| ColdF | aagcttgtcgacctgcagt | To linearize the plasmid pCold-SUMO |
| ColdR | ggatccctcgagggtaccg | To linearize the plasmid pCold-SUMO |
| 2970-2974F | gagctaacttacattaattAATATAGATATAAATTAGGATTCTGTTTTAG | Cloning gene from 2970 to 2974 to ligate into linearized pCold-SUMO |
| 2970-2974R | cgcgatcgattatttatttTTTTAAAGGATTGTAATCAATATTATAAC | Cloning gene from 2970 to 2974 to ligate into linearized pCold-SUMO |
| Cold-7074F | aattaatgtaagttagctcactc | To linearize the plasmid pCold-SUMO that only retained f1 ori, AmpR, and ori regions |
| Cold-7074R | aaataaataatcgatcgcg | To linearize the plasmid pCold-SUMO that only retained f1 ori, AmpR, and ori regions |
| Cold-7174F | ctaacttacattaattAAGAATAGTATTTATATAATATAGAAC | For defecting the 2970 from pCold-2970-2974 to construct pCold-2971-2974 |
| Cold-7174R | gatttaattttcatcttaaGTTCTATATTATATAAATACTATTCTT | For defecting the 2970 from pCold-2970-2974 to construct pCold-2971-2974 |
There are four corresponding types of expression procedures. First, for OUC-FaLC150A and OUC-FaIC82C recombinase production, the recombinant strains were grown in LB fluid medium containing 50 μg/mL kanamycin sulfate at 37°C with agitation at 220 rpm after the OD600 reached 0.6, a final concentration of 0.1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) was added into medium, and the culture temperature was changed to 16°C for 24 h for inducing protein production. Second, the expression of sulfatase OUC-S1_7A was performed according to the procedures from previous literature (17). Third, for OUC-FaKC16A, OUC-FaIC82A, OUC-FaIC82B, OUC-FaBC129A, OUC-FaBC129B, OUC-FaGH2A, FaDAD, FaDAAC, FaKDGK, and FaKDPGA recombinase production, the recombinant strains were grown in LB fluid medium containing 100 μg/mL ampicillin, 17 μg/mL chloramphenicol, and 0.5 mg/mL l-arabinose at 37°C with agitation at 220 rpm; 2 ng/mL tetracycline (induction of chaperone expression) was added when the OD600 reached 0.3, and then it continued to cultivate at 37°C after the OD600 reached 0.6. A final concentration of 0.1 mM IPTG was added into medium, and the culture temperature was changed to 16°C for 24 h for inducing protein production. Finally, for sulfatases OUC-S1_19A, OUC-S1_19B, OUC-S1_19C, OUC-S1_17A, and OUC-S1_17B expression, it was only necessary to modify the addition concentration of l-arabinose into 1 mg/mL for inducing formylglycine expression.
After induction, the cells were collected from centrifugation (4°C, 8,000 × g) for 15 min, resuspended in ultrapure water, and then disrupted by sonication. Subsequently, the crude enzyme supernatant was obtained from centrifugation (4°C, 9,000 × g) for 15 min. Then, these crude enzymes were purified with a Ni2+-NTA column in accordance with the instructions (TransGen Biotech, China). The target proteins were eluted with gradient concentrations of imidazole (10, 20, 40, 50, 80, 120, 200, and 500 mM imidazole). Finally, the pure enzymes were obtained by using the specific size (30 and 50 kDa) of ultrafiltration tubes for concentration and buffer replacement; meanwhile, these enzymes were analyzed by SDS-PAGE, and their concentrations were determined using a bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, USA) with bovine serum album (BSA) as the standard.
κ/ι/λ-carrageenase activity assays.
The κ-carrageenase OUC-FaKC16A and λ-carrageenase OUC-FaLC150A activity was determined by as previously described using the 3,5-dinitrosalicylic acid (DNS) method (22, 30). Briefly, a total of 200 μL reaction volume containing 0.3% (wt/vol) substrate (KC or LC), 20 mM Tris-HCl buffer, pH 7.0, and 20 μL enzyme (OUC-FaKC16A or OUC-FaLC150A) was incubated for 30 min at optimum temperature and then boiled for 10 min for stopping the reaction. Three hundred microliters of DNS reagent (Solarbio, China) were added into reaction volume, boiled immediately for 5 min, and then cooled in a cold-water bath. The absorbance was determined at 540 nm, and inactivated enzyme was used as a control. One unit of enzymatic activity was defined as the amount of enzyme required to obtain 1 μmol of reducing sugar per min.
The ι-carrageenases OUC-FaIC82A, OUC-FaIC82B, and OUC-FaIC82C were determined by the para-hydroxybenzoic acid hydrazide (pHBAH) method (36). The pHBAH chromogenic reagent was comprised of 1 volume of 20% (wt/vol) pHBAH (Sigma, USA) dissolved in 2 M HCl and 9 volumes of 2 M NaOH. The 200-μL reaction volume contained 0.3% (wt/vol) IC, 20 mM Tris-HCl buffer, pH 7.0, and 20 μL enzyme. It was incubated for 30 min at optimum temperature and then boiled for 10 min for stopping the reaction. Six hundred microliters of pHBAH chromogenic reagent were added into reaction volume, boiled immediately for 5 min, and then cooled in a cold-water bath. The absorbance was determined at 405 nm, and inactivated enzyme was used as a control.
Characterization of optimum reaction conditions for κ/ι-carrageenases.
The optimal temperature of κ-carrageenase OUC-FaKC16A was determined in the temperature range of 30 to 100°C, while the three ι-carrageenases (OUC-FaIC82A, OUC-FaIC82B, and OUC-FaIC82C) were determined in the temperature ranges of 30 to 65°C with 20 mM Tris-HCl buffer, pH 7.0, for incubation for 30 min. The optimal pH levels of OUC-FaKC16A, OUC-FaIC82A, OUC-FaIC82B, and OUC-FaIC82C were determined in a pH range of 3.0 to 10.0 by using different kinds of buffers at their optimal temperatures for 30 min. Each reaction was performed in triplicate.
HPLC and MS methods for detecting the κ/ι/λ-carrageenases’ products.
We treated 0.3% (wt/vol) KC with purified OUC-FaKC16A in glycine-NaOH buffer, pH 9.0, at 60°C for different times. Samples taken over time were detected by HPLC with a Superdex 30 10/300 gel filtration column (GE Health, Marlborough, MA, USA) with 0.2 M ammonia bicarbonate as the mobile phase at a flow rate of 0.4 mL/min; the detector was a refractive index detector (RID) (Shimadzu, Japan). Meanwhile, the 8-h product was analyzed by using the electrospray ionization mass spectrometry (ESI-MS) method on microTOF-Q II equipment (Agilent, USA) in a negative mode with ion spray voltage of 4 kV and source temperature of 350°C. The products from hydrolysis of κ3, κ5, κ7, Nκ2, Nκ4, and Nκ8 using purified OUC-FaKC16A were also detected by HPLC.
Also, the 24-h hydrolysis products of OUC-FaIC82A, OUC-FaIC82B, OUC-FaIC82C, and OUC-FaLC150A were also detected by HPLC and MS as described above.
Sulfatase activity assays.
Purified OUC-S1_7A, OUC-S1_19A, OUC-S1_19B, and OUC-S1_19C were incubated with the mixture of Nκ4 and Nκ6 over 24 h in Tris-HCl buffer, pH 7.0, at 35°C. Afterward, the products were analyzed by HPLC as described above. Furthermore, the reactions of purified OUC-S1_19B with κ3, κ5, κ7, Nκ2, Nκ4, Nκ8, and Nκ10 were analyzed by HPLC. Therein, the reaction of OUC-S1_19B with Nκ2, Nκ4, Nκ6, and Nκ8 was also detected by MS described above.
The detection of reactions about the removal of sulfate groups from Nι4, including OUC-S1_19B acting on Nι4, OUC-S1_17A acting on OUC-S1_17B with Nι4, and OUC-S1_17A and OUC-S1_17B acting on OUC-S1_19B-treated Nι4, were performed with above-mentioned HPLC method.
α-3,6-Anhydro-d-galactosidase and β-galactosidase activity assays.
Taking the mixture of Nκ4 and Nκ6 and OUC-S1_19B-treated mixture of Nκ4 and Nκ6 as the substrates for testing the hydrolysis activity of OUC-FaBC127A and OUC-FaBC129A, the products were analyzed by the above-mentioned HPLC and MS methods. Moreover, the reactions of OUC-FaGH2A with κ3 and κ5 were also detected by the same HPLC method.
Also, the above-mentioned MS method was applied to detect the reaction of OUC-FaBC127A and OUC-FaBC129A with OUC-S1_19B-treated pure Nκ4, as well as the reaction of OUC-FaGH2A with κ5 and odd-numbered oligocarrageenan with DP3 (G-DA-G4S).
3,6-Anhydro-d-galactose dehydrogenase activity assays.
A total of 200-μL reaction volume composed of 0.1% (wt/vol) d-AHG (Carbosynth, England), 10 μg purified FaDAD, 20 mM Tris-HCl, pH 7.0, and 1.5 mM NAD+ or NADP+ (Solarbio, China) was incubated at 35°C for 20 min. The inactivated enzyme was used as control, and the absorbance was determined at 340 nm. Moreover, the d-AHGA product was also analyzed by HPLC using an Aminex HPX-87H column (Bio-Rad, USA) and 0.5 mM H2SO4 as mobile phase. The column temperature was 50°C, the flow velocity was 0.5 mL/min, and the detector was a refractive index detector (Shimadzu, Japan). It was further analyzed by the above-mentioned MS method.
Detection of enzymatic products from FaDAAC, FaKDGK, and FaKDPGA.
The purified FaDAAC was incubated with FaDAD-treated d-AHG with NAD+ or NADP+ as cofactor at pH 7.0 and 35°C for 20 min. The reaction mixtures were analyzed by HPLC described in the last section. Then, we took the FaDAAC’s enzymatic product as the substrate for testing the phosphorylation activity of FaKDGK. The 200-μL reaction volume contained 100 μL FaDAAC enzymatic product, 10 μg purified FaKDGK, 20 mM Tris-HCl, pH 7.0, and 0.97 mM adenosine 5-triphosphate (ATP) at 35°C for 20 min. The product was derived by the above-mentioned MS method. Furthermore, the product of the final process was prepared in a 200-μL reaction volume that consisted of 100 μL FaKDGK’s enzymatic product, 10 μg purified FaKDPGA, and 20 mM Tris-HCl, pH 7.0, which was performed at 35°C for 30 min. The produced d-Gly-3-P and pyruvate were also derived by the same MS method.
Construction of BL-DA and BL-DAΔRhaT strains for illustrating the transportation function of RhaT.
The plasmid pCold-SUMO was used as the backbone to link the gene fragment from 2970 to 2974, which only retained its f1 ori, AmpR, and ori regions by PCR using designed primers (Table 1). The linearized backbone and PCR product (gene fragments from 2970 to 2974) were linked by ClonExpress Ultra one-step cloning kit. The details of construction process were described above. Then, based on the vector pCold-2970-2974, we designed primers for deleting gene 2970 to construct pCold-2971-2974. The successfully built plasmids pCold-2970-2974 and pCold-2971-2974 were further transformed into E. coli BL21(DE3) to construct BL-DA and BL-DAΔRhaT strains, respectively.
The BL-DA and BL-DAΔRhaT strains were first cultured in 5 mL LB liquid medium at 37°C overnight. Before transfer into M9 minimal medium with 0.5 g/L d-AHG as the sole carbon source, the cells were washed twice by M9 minimal medium and were made to reach the same initial OD600 of around 0.150 to further culture at 35°C with shaking at 200 rpm. The fermented samples taken over time were analyzed by the above-mentioned HPLC method using the Aminex HPX-87H column.
Data availability.
The draft genome sequences of F. algicola were available in the NCBI Reference Sequence database under the accession number JAJTUT010000001-JAJTUT010000006.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31922072), Natural Science Foundation of Shandong Province (ZR2020JQ15), Taishan Scholar Project of Shandong Province (tsqn201812020), and Fundamental Research Funds for the Central Universities (201941002).
Footnotes
Supplemental material is available online only.
Contributor Information
Xiangzhao Mao, Email: xzhmao@ouc.edu.cn.
Laura Villanueva, Royal Netherlands Institute for Sea Research.
REFERENCES
- 1.Hedges JI, Baldock JA, Gélinas Y, Lee C, Peterson M, Wakeham SG. 2001. Evidence for non-selective preservation of organic matter in sinking marine particles. Nature 409:801–804. 10.1038/35057247. [DOI] [PubMed] [Google Scholar]
- 2.Hemsworth G, Déjean G, Davies G, Brumer H. 2016. Learning from microbial strategies for polysaccharide degradation. Biochem Soc Trans 44:94–108. 10.1042/BST20150180. [DOI] [PubMed] [Google Scholar]
- 3.Azam F, Malfatti F. 2007. Microbial structuring of marine ecosystems. Nat Rev Microbiol 5:782–791. 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 4.Yanagisawa M, Kawai S, Murata K. 2013. Strategies for the production of high concentrations of bioethanol from seaweeds: production of high concentrations of bioethanol from seaweeds. Bioengineered 4:224–235. 10.4161/bioe.23396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jang S-S, Shirai Y, Uchida M, Wakisaka M. 2012. Production of mono sugar from acid hydrolysis of seaweed. Afr J Biotechnol 11:1953. 10.5897/AJB10.1681. [DOI] [Google Scholar]
- 6.Kim NJ, Li H, Jung K, Chang HN, Lee PC. 2011. Ethanol production from marine algal hydrolysates using Escherichia coli KO11. Bioresour Technol 102:7466–7469. 10.1016/j.biortech.2011.04.071. [DOI] [PubMed] [Google Scholar]
- 7.Yoon JJ, Yong JK, Sang HK, Ryu HJ, Choi JY, Kim GS, Shin M. 2010. Production of polysaccharides and corresponding sugars from red seaweed. Amr 93-94:463–466. 10.4028/www.scientific.net/AMR.93-94.463. [DOI] [Google Scholar]
- 8.Anderson NS, Dolan TC, Rees DA. 1965. Evidence for a common structural pattern in the polysaccharide sulphates of the Rhodophyceae. Nature 205:1060–1062. 10.1038/2051060a0. [DOI] [PubMed] [Google Scholar]
- 9.Usov A. 2011. Polysaccharides of the red algae. Adv Carbohydr Chem Biochem 65:115–217. 10.1016/B978-0-12-385520-6.00004-2. [DOI] [PubMed] [Google Scholar]
- 10.Ai U. 1998. Structural analysis of red seaweed galactans of agar and carrageenan groups. Food Hydrocolloids 12:301. 10.1016/S0268-005X(98)00018-6. [DOI] [Google Scholar]
- 11.Zhu B, Ni F, Sun Y, Zhu X, Yin H, Yao Z, Du Y. 2018. Insight into carrageenases: major review of sources, category, property, purification method, structure, and applications. Crit Rev Biotechnol 38:1261–1276. 10.1080/07388551.2018.1472550. [DOI] [PubMed] [Google Scholar]
- 12.Ficko-Blean E, Prechoux A, Thomas F, Rochat T, Larocque R, Zhu Y, Stam M, Genicot S, Jam M, Calteau A, Viart B, Ropartz D, Perez-Pascual D, Correc G, Matard-Mann M, Stubbs KA, Rogniaux H, Jeudy A, Barbeyron T, Medigue C, Czjzek M, Vallenet D, McBride MJ, Duchaud E, Michel G. 2017. Carrageenan catabolism is encoded by a complex regulon in marine heterotrophic bacteria. Nat Commun 8:1685. 10.1038/s41467-017-01832-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbeyron T, Brillet-Gueguen L, Carre W, Carriere C, Caron C, Czjzek M, Hoebeke M, Michel G. 2016. Matching the diversity of sulfated biomolecules: creation of a classification database for sulfatases reflecting their substrate specificity. PLoS One 11:e0164846. 10.1371/journal.pone.0164846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbeyron T, Michel G, Potin P, Henrissat B, Kloareg B. 2000. iota-Carrageenases constitute a novel family of glycoside hydrolases, unrelated to that of kappa-carrageenases. J Biol Chem 275:35499–35505. 10.1074/jbc.M003404200. [DOI] [PubMed] [Google Scholar]
- 16.Rebuffet E, Barbeyron T, Jeudy A, Jam M, Czjzek M, Michel G. 2010. Identification of catalytic residues and mechanistic analysis of family GH82 iota-carrageenases. Biochemistry 49:7590–7599. 10.1021/bi1003475. [DOI] [PubMed] [Google Scholar]
- 17.Hettle AG, Hobbs JK, Pluvinage B, Vickers C, Abe KT, Salama-Alber O, McGuire BE, Hehemann JH, Hui JPM, Berrue F, Banskota A, Zhang J, Bottos EM, Van Hamme J, Boraston AB. 2019. Insights into the kappa/iota-carrageenan metabolism pathway of some marine Pseudoalteromonas species. Commun Biol 2:474. 10.1038/s42003-019-0721-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyashita M, Fujimura S, Nakagawa Y, Nishizawa M, Tomizuka N, Nakagawa T, Nakagawa J. 2010. Flavobacterium algicola sp. nov., isolated from marine algae. Int J Syst Evol Microbiol 60:344–348. 10.1099/ijs.0.009365-0. [DOI] [PubMed] [Google Scholar]
- 19.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser (Oxf) 41:95. [Google Scholar]
- 20.Pluvinage B, Grondin JM, Amundsen C, Klassen L, Moote PE, Xiao Y, Thomas D, Pudlo NA, Anele A, Martens EC, Inglis GD, Uwiera RER, Boraston AB, Abbott DW. 2018. Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont. Nat Commun 9:1043. 10.1038/s41467-018-03366-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yun EJ, Lee S, Kim HT, Pelton JG, Kim S, Ko HJ, Choi IG, Kim KH. 2015. The novel catabolic pathway of 3,6-anhydro-L-galactose, the main component of red macroalgae, in a marine bacterium. Environ Microbiol 17:1677–1688. 10.1111/1462-2920.12607. [DOI] [PubMed] [Google Scholar]
- 22.Matard-Mann M, Bernard T, Leroux C, Barbeyron T, Larocque R, Prechoux A, Jeudy A, Jam M, Nyvall Collen P, Michel G, Czjzek M. 2017. Structural insights into marine carbohydrate degradation by family GH16 kappa-carrageenases. J Biol Chem 292:19919–19934. 10.1074/jbc.M117.808279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu X, Li S, Yang X, Yu W, Han F. 2015. Cloning and characterization of a new κ-carrageenase gene from marine bacterium Pseudoalteromonas sp. QY203. J Ocean Univ China 14:1082–1086. 10.1007/s11802-015-2652-7. [DOI] [Google Scholar]
- 24.Wang L, Li S, Zhang S, Li J, Yu W, Gong Q. 2015. A new κ-carrageenase CgkS from marine bacterium Shewanella sp. Kz7. J Ocean Univ China 14:759–763. 10.1007/s11802-015-2713-y. [DOI] [Google Scholar]
- 25.Zhang Y, Lang B, Zeng D, Li Z, Yang J, Yan R, Xu X, Lin J. 2019. Truncation of kappacarrageenase for higher kappa-carrageenan oligosaccharides yield with improved enzymatic characteristics. Int J Biol Macromol 130:958–968. 10.1016/j.ijbiomac.2019.02.109. [DOI] [PubMed] [Google Scholar]
- 26.Zhao D, Jiang B, Pu Z, Sun W, Zhang Y, Bao Y. 2021. Module function analysis of a full-length kappa-carrageenase from Pseudoalteromonas sp. ZDY3. Int J Biol Macromol 182:1473–1483. 10.1016/j.ijbiomac.2021.05.110. [DOI] [PubMed] [Google Scholar]
- 27.Ma S, Tan YL, Yu WG, Han F. 2013. Cloning, expression and characterization of a new iota-carrageenase from marine bacterium, Cellulophaga sp. Biotechnol Lett 35:1617–1622. 10.1007/s10529-013-1244-0. [DOI] [PubMed] [Google Scholar]
- 28.Li S, Hao J, Sun M. 2017. Cloning and characterization of a new cold-adapted and thermo-tolerant iota-carrageenase from marine bacterium Flavobacterium sp. YS-80-122. Int J Biol Macromol 102:1059–1065. 10.1016/j.ijbiomac.2017.04.070. [DOI] [PubMed] [Google Scholar]
- 29.Guibet M, Colin S, Barbeyron T, Genicot S, Kloareg B, Michel G, Helbert W. 2007. Degradation of λ-carrageenan by Pseudoalteromonas carrageenovora λ-carrageenase: a new family of glycoside hydrolases unrelated to κ- and ι-carrageenases. Biochem J 404:105–114. 10.1042/BJ20061359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohta Y, Hatada Y. 2006. A novel enzyme, lambda-carrageenase, isolated from a deep-sea bacterium. J Biochem 140:475–481. 10.1093/jb/mvj180. [DOI] [PubMed] [Google Scholar]
- 31.Jiang C, Liu Z, Cheng D, Mao X. 2020. Agarose degradation for utilization: enzymes, pathways, metabolic engineering methods and products. Biotechnol Adv 45:107641. 10.1016/j.biotechadv.2020.107641. [DOI] [PubMed] [Google Scholar]
- 32.Lee SB, Kim JA, Lim HS. 2016. Metabolic pathway of 3,6-anhydro-D-galactose in carrageenan-degrading microorganisms. Appl Microbiol Biotechnol 100:4109–4121. 10.1007/s00253-016-7346-6. [DOI] [PubMed] [Google Scholar]
- 33.Schultz-Johansen M, Bech PK, Hennessy RC, Glaring MA, Barbeyron T, Czjzek M, Stougaard P. 2018. A novel enzyme portfolio for red algal polysaccharide degradation in the marine bacterium Paraglaciecola hydrolytica s66(t) encoded in a sizeable polysaccharide utilization locus. Front Microbiol 9:839. 10.3389/fmicb.2018.00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hettle AG, Vickers C, Robb CS, Liu F, Withers SG, Hehemann JH, Boraston AB. 2018. The molecular basis of polysaccharide sulfatase activity and a nomenclature for catalytic subsites in this class of enzyme. Structure 26:747–758.e4. 10.1016/j.str.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 35.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 36.Shen J, Chang Y, Dong S, Chen F. 2017. Cloning, expression and characterization of a iota-carrageenase from marine bacterium Wenyingzhuangia fucanilytica: a biocatalyst for producing iota-carrageenan oligosaccharides. J Biotechnol 259:103–109. 10.1016/j.jbiotec.2017.07.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S10 and Tables S1 and S2. Download aem.00256-22-s0001.pdf, PDF file, 0.9 MB (917.9KB, pdf)
Data Availability Statement
The draft genome sequences of F. algicola were available in the NCBI Reference Sequence database under the accession number JAJTUT010000001-JAJTUT010000006.