ABSTRACT
Pseudomonas putida KT2440 has long been studied for its diverse and robust metabolisms, yet many genes and proteins imparting these growth capacities remain uncharacterized. Using pooled mutant fitness assays, we identified genes and proteins involved in the assimilation of 52 different nitrogen containing compounds. To assay amino acid biosynthesis, 19 amino acid drop-out conditions were also tested. From these 71 conditions, significant fitness phenotypes were elicited in 672 different genes including 100 transcriptional regulators and 112 transport-related proteins. We divide these conditions into 6 classes, and propose assimilatory pathways for the compounds based on this wealth of genetic data. To complement these data, we characterize the substrate range of three promiscuous aminotransferases relevant to metabolic engineering efforts in vitro. Furthermore, we examine the specificity of five transcriptional regulators, explaining some fitness data results and exploring their potential to be developed into useful synthetic biology tools. In addition, we use manifold learning to create an interactive visualization tool for interpreting our BarSeq data, which will improve the accessibility and utility of this work to other researchers.
IMPORTANCE Understanding the genetic basis of P. putida’s diverse metabolism is imperative for us to reach its full potential as a host for metabolic engineering. Many target molecules of the bioeconomy and their precursors contain nitrogen. This study provides functional evidence linking hundreds of genes to their roles in the metabolism of nitrogenous compounds, and provides an interactive tool for visualizing these data. We further characterize several aminotransferases, lactamases, and regulators, which are of particular interest for metabolic engineering.
KEYWORDS: nitrogen, Pseudomonas putida, RB-TnSeq, transposon, metabolism, BarSeq, t-SNE, aminotransferase, lactam, biosensor, amino acid, nucleotide, nitrate, nitrite, polyamine, aminotransferases, biosensors, nitrogen metabolism
INTRODUCTION
As a free-living soil bacterium, Pseudomonas putida encounters many different organic and inorganic nitrogen sources, and its responses to these conditions have been the target of recent study. During rhizosphere colonization, P. putida displays chemotaxis toward plant root exudates, which contain nitrogenous compounds such as benzylamines, polyamines, pyrrole derivatives, nucleotide derivatives, amino acids, and phenylpropanoids (1, 2). It also demonstrates chemotaxis toward and degradation of the phytotoxic and insecticidal benzoxazinoids exuded from the roots of maize seedlings (3, 4). Not only is P. putida able to withstand and metabolize these varied compounds in a nitrogen-rich rhizosphere, it is also capable of adapting to nitrogen-scarce conditions by increasing polyhydroxyalkanoate (PHA) production, repressing carbon catabolism, and increasing expression of transporters for nitrogen uptake (5).
Given its ability to adapt to the varied environment of the rhizosphere and its burgeoning role as a host for sustainable chemical bioproduction, it is not surprising that P. putida’s nitrogen metabolism has also been examined in the context of metabolic engineering. Due to the utility of PHAs as a next-generation bioplastic, the transcriptomic, proteomic, and metabolomic response of P. putida strains to nitrogen-limited growth conditions has been analyzed in order to better understand how PHA synthesis is triggered by nitrogen scarcity (6, 7). P. putida’s nitrogen metabolism and its regulation have also been a source of metabolic engineering parts. Some of its 39 predicted aminotransferases have been heterologously expressed as part of benzylamine derivative and glutaric acid production pathways (8, 9). The transcription factor regulating capro- and valerolactam degradation has been developed into a highly sensitive biosensor with the potential to be applied to increasing lactam production titers and used as an inducible system in pathway engineering (10). Moreover, many nitrogenous compounds are relevant building blocks for commodity chemicals, and understanding P. putida’s metabolism of them can enable more flux to be directed toward the desired product (11).
Despite its relevance to both metabolic engineering and basic scientific research, our understanding of P. putida’s nitrogen metabolism is far from complete. Many gene functions have been assigned through homology predictions with limited functional evidence, which can hamper the accuracy of metabolic modeling (12). Furthermore, P. putida has multiple paralogs of many of its enzymes, each potentially with different substrate preferences. Functional genomics can provide evidence for assigning gene functions and illuminate the specific roles of genes that have multiple paralogs (13).
In previous studies, we have employed barcoded transposon sequencing (BarSeq) to interrogate lysine, fatty acid, alcohol, and aromatic degradation in P. putida (13–15). In this work, we use BarSeq to study P. putida KT2440’s metabolism of 52 different nitrogen-containing compounds, nearly doubling the amount of publicly available BarSeq data for this bacterium. We provide evidence for many known nitrogen assimilation pathways and their regulatory systems, and also assign function to genes whose exact roles in nitrogen metabolism were not previously known. Due to their relevance in recent metabolic engineering efforts, we further examine the substrate specificity of P. putida’s 5-oxoprolinases and aminotransferases (8, 9, 16). To understand the regulation of these enzymes in vivo, we further characterized their cognate regulators. This work will bolster P. putida’s utility both as a host microorganism and as a source of metabolic engineering parts for sustainable chemical production.
RESULTS AND DISCUSSION
BarSeq reveals the genetic bases of diverse nitrogen metabolisms.
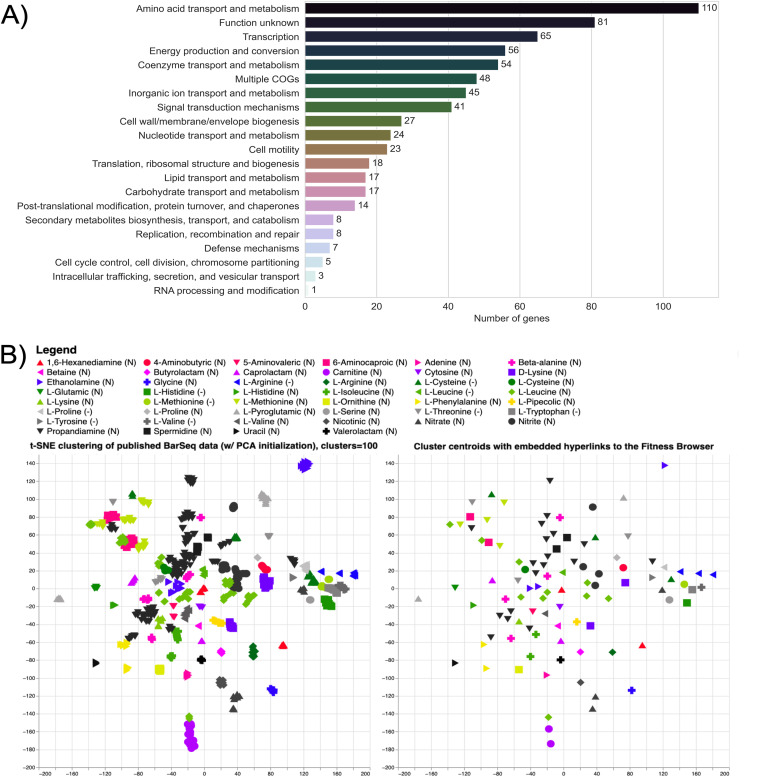
Genes involved in nitrogen utilization from natural and unnatural compounds were identified using BarSeq. In these assays, a library of barcoded transposon insertion mutants was cultured in minimal media with glucose and a variety of sole nitrogen sources (Fig. S1). A list of these nitrogen sources and the section in which each is discussed can be found in Table 1. Wild-type growth on these substrates over 96 h is shown in Fig. S2. These assays revealed 672 genes with strong (|fitness| > 1) and significant (|t | > 5) fitness phenotypes. These include 100 transcription factors, 112 transport proteins, and numerous other enzymes with applications to engineered systems, such as aminotransferases (17, 18) (Fig. 1A, Fig. S3). Additionally, 529 of these genes encode proteins that are currently unreviewed in the Uniprot database, and 256 have not shown significant phenotypes in previous BarSeq studies. To visualize the fitness data, the manifold learning method, t-distributed stochastic neighbor embedding (t-SNE), was employed to cluster genes based on their fitness values in the tested conditions (Fig. 1B, Fig. S3) (19). The clusters were named based on the condition that elicited the largest and most frequent changes in fitness scores for genes within the cluster. In this visualization, we can identify genes that may take part in similar metabolisms. An interactive version that contains embedded hyperlinks to the Fitness Browser (20) can be accessed via this link: https://ppnitrogentsne.lbl.gov. Although genes with specific phenotypes to one condition are easy to identify, genes essential across many conditions are more challenging to assign a specific function. This is particularly problematic for genes that are essential in the majority of the tested conditions, like those involved in amino acid biosynthesis. To our surprise, clusters of genes involved in tryptophan, arginine, methionine, and branched chain amino acid biosynthesis were resolved by t-SNE (Fig. I1-2 available at https://ppnitrogentsne.lbl.gov). While not as exhaustive as pairwise comparisons of conditions, t-SNE provides a useful visualization of this diverse data set.
TABLE 1.
Compounds used as nitrogen sources in BarSeq experiments and the section of the paper they are discussed in nitrogen sources indicated by (N) and amino acid dropout conditions indicated by (-)
| Section | Experiments included | |
|---|---|---|
| Inorganic nitrogen sources and urea | ammonium (N) | nitrite (N) |
| nitrate (N) | urea (N) | |
| Proteinogenic amino acids | l-isoleucine (N) | glycine (N) |
| l-leucine (N) | l-histidine (N) | |
| d-lysine (N) | l-alanine (-) | |
| l-lysine (N) | l-arginine (-) | |
| l-methionine (N) | l-asparagine (-) | |
| l-ornithine (N) | l-aspartate (-) | |
| l-pipecolic (N) | l-cysteine (-) | |
| l-proline (N) | l-glutamate (-) | |
| l-threonine (N) | l-glutamine (-) | |
| l-phenylalanine (N) | glycine (-) | |
| l-serine (N) | l-serine (-) | |
| l-valine (N) | l-histidine (-) | |
| 4-guanidinobutyrate (N) | l-isoleucine (-) | |
| d-alanine (N) | l-leucine (-) | |
| l-alanine (N) | l-lysine (-) | |
| l-arginine (N) | l-methionine (-) | |
| l-asparagine (N) | l-proline (-) | |
| l-aspartate (N) | l-threonine (-) | |
| l-cysteine (N) | l-tyrosine (-) | |
| l-glutamate (N) | l-tryptophan (-) | |
| l-glutamine (N) | l-valine (-) | |
| Quaternary amines and ethanolamine | betaine (N) | choline (N) |
| carnitine (N) | ethanolamine (N) | |
| Purines and pyrimidines | adenine (N) | hydantoin (N) |
| cytosine (N) | uracil (N) | |
| Lactams | butyrolactam (N) | 5-oxoproline (N) |
| caprolactam (N) | valerolactam (N) | |
| γ-aminobutyric (N) | ||
| Polyamines, ω-amino acids, and GABA isomers | 1,3 diaminopropane (N) | 6-aminocaproic acid (N) |
| 1,6-diaminohexane (N) | β-alanine (N) | |
| dl-2-aminobutyrate (N) | cadaverine (N) | |
| dl-3-aminoisobutyrate (N) | putrescine (N) | |
| 3-aminobutyrate (N) | spermidine (N) | |
| 5-aminovalerate (N) | ||
| Not applicable | nicotinic acid (N) | |
FIG 1.
Global analysis of the P. putida KT2440 BarSeq data. (A) Significant genes (|fitness| > 1 and |t | > 5) from all 71 tested conditions sorted by their cluster of orthologous groups (COGs) based on the eggNOG database (149, 153, 154). “Multiple COGs” indicates that there was more than one COG assigned. (B) Image of the interactive t-SNE visualization (Fig. I1 available at https://ppnitrogentsne.lbl.gov) showing the legend, t-SNE clustering (left) and cluster centroids (right). By clicking on a substrate in the legend, the corresponding cluster (left) and centroid (right) is highlighted, opening a list of cluster members and additional information. By clicking the highlighted centroid (right), the user is redirected to the Fitness Browser (https://fit.genomics.lbl.gov) (20), where the fitness data for all significant genes in the condition cluster is shown. An additional t-SNE visualization including COG identifiers is presented in Fig. I3 (available at https://ppnitrogentsne.lbl.gov). More information about the interactive figures can be found in the description of Fig. S3.
Global effectors of nitrogen metabolism.
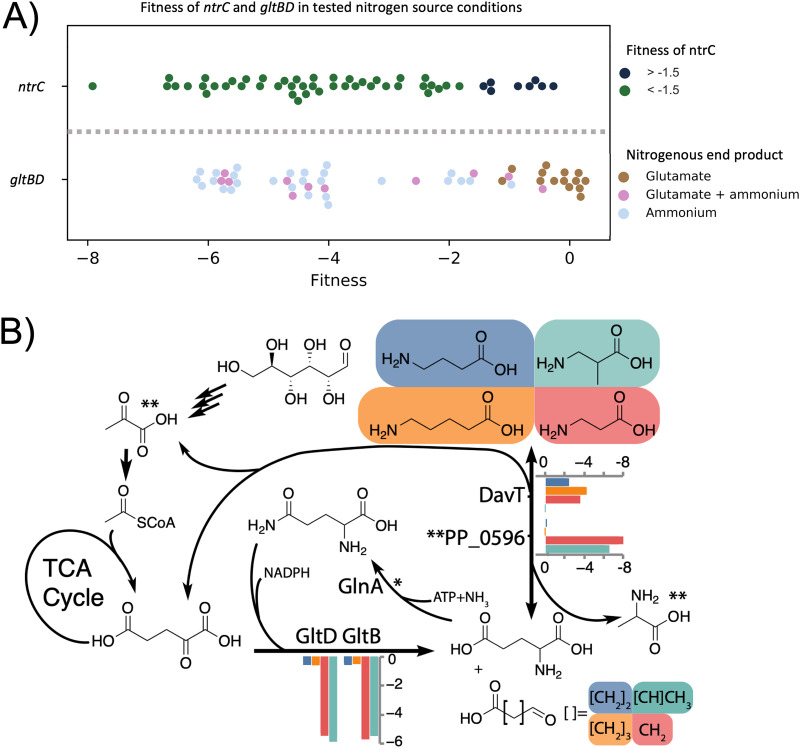
Mutants affected in the global regulators NtrB (PP_5047) and NtrC (PP_5048) have diverse phenotypes in the conditions tested. NtrBC is a two-component system that regulates the expression of numerous nitrogen assimilatory genes in P. putida (5). When the cell is nitrogen starved, the uridylyltransferase protein GlnD (PP_1589) modulates the activity of GlnK (PP_5234, protein PII). GlnK then activates NtrB, which affects the phosphorylation state of NtrC. Subsequently, NtrC modulates expression of its target regulon (5). While the library used for these assays has no insertions in glnK, the three other members of the signaling cascade are represented in the data set and occupy the same cluster determined by t-SNE (Fig. I1 available at https://ppnitrogentsne.lbl.gov). It is unclear why certain nitrogen sources trigger a strong NtrC response (Fig. 2A). The general role of NtrC is to counteract nitrogen starvation by activating the majority of nitrogen assimilatory genes, such as transporters (5). By setting an arbitrary fitness score cutoff at >−1.5, we identified at least 8 out of 52 nitrogen sources that might not rely on a functional copy of ntrC (Table 2). The utilization pathways for those compounds seem to be less dependent on NtrC activation, suggesting specific regulation systems or the presence of constitutively expressed transporter and degradation pathways for these nitrogen sources. The fitness profile of mutants in the extracytoplasmic function sigma factor SigX (PP_2088) also seems to be correlated with that of ntrBC mutants. Previously, we found sigX to be partially essential for growth on d-lysine as a carbon source (15). While no clear pattern in the fitness data could be determined for NtrC, another global factor involved in nitrogen metabolism, GltBD, illustrates a clear relationship between the conditions tested and their downstream metabolites.
FIG 2.
(A) Scatterplot of the average fitness values (n = 2) for ntrC and gltBD in all the tested nitrogen conditions. For ntrC, nitrogen conditions are grouped based on whether the fitness phenotype of ntrC is < −1.5 (blue) or > −1.5 (green). Conditions where ntrC fitness is >−1.5 (green) may be less dependent on ntrC activation and are shown in Table 2. gltBD phenotypes are sorted based on putative glutamate (brown), glutamate+ammonium (pink), or ammonium (gray) release during nitrogen source utilization. Fitness values for conditions resulting in gltBD fitness >−1.5 are shown in Table 3. (B) The role of the GS/GOGAT cycle in the beta-alanine (red), 3-aminoisobutyrate (green), 4-aminobutyrate (blue), and 5-aminovalerate (orange) nitrogen source conditions. Average fitness values (n = 2) are shown for gltBD, davT, and the pyruvate dependent transaminase PP_0596. *GlnA lacks fitness data because the library has no insertions. **Pyruvate and alanine are the specific nitrogen acceptor and product of PP_0596.
TABLE 2.
Nitrogen source conditions in which regulation may be less dependent on NtrC activation, indicated by ntrC fitness > −1.5 and illustrated by the dark blue points in Fig. 2A
| Conditions in which NtrC fitness is >−1.5 | |
|---|---|
| Condition | NtrC fitness |
| l-glutamine | −0.27 |
| putrescine | −0.46 |
| l-asparagine | −0.57 |
| ammonium chloride | −0.66 |
| 4-aminobutyric acid | −0.86 |
| 4-guanidinobutyric acid | −1.30 |
| l-histidine | −1.31 |
| adenine | −1.43 |
GltB (PP_5076) and GltD (PP_5075) comprise the glutamate synthase (GOGAT) of P. putida, which plays an important role in the regulation of nitrogen assimilation. Interestingly, gltBD has diverse phenotypes across the conditions we tested (Fig. 2). However, the second component of the central GS/GOGAT cycle, glutamine synthetase (glnA, PP_5046), has no insertions in our BarSeq library, indicating that it might have been essential during library construction due to glutamine auxotrophy of the mutants. We were able to observe particularly strong fitness defects for gltBD in nitrogen conditions that either produce free ammonium or do not yield l-glutamate (Fig. 2A) (21). For example, gltBD exhibits a very strong fitness defect in the nitrate (−6.17) and l-serine (−5.95) conditions but not in the l-phenylalanine condition (Fig. 2A, Table 3). Another example is in the comparison of 5-AVA (5-aminovalerate), 4-ABA (4-aminobutyrate), β-alanine, and 3-AIBA (3-aminoisobutyrate). 3-AIBA utilization likely uses pyruvate as an amino-acceptor (via PP_0596), 4-ABA and 5-AVA require 2-oxoglutarate (via PP_0214/DavT), and β-alanine requires both aminotransferases (Fig. 2B, Fig. S4). An exemption to this rule is the nitrogen source ammonium chloride. The fitness defects for gltBD in this condition were much lower (|fitness| < 2), but still significant (|t | > 5). It has been shown that at high concentrations of ammonium (>10 mM), its assimilation is achieved by both the glutamate dehydrogenase (GdhA) PP_0675 and GltBD (5, 22, 23).
TABLE 3.
Nitrogen source conditions in which gltBD fitness > −1.5, illustrated by the scatterplot in Fig. 2A
| Conditions in which GltBD fitness is >−1.5 | |
|---|---|
| Condition | GltBD fitness |
| l-isoleucine | +0.20 |
| butyrolactam | +0.12 |
| l-pyroglutamic acid | +0.10 |
| l-valine | +0.08 |
| l-phenylalanine | +0.04 |
| l-aspartate | −0.07 |
| l-glutamic acid | −0.15 |
| l-proline | −0.16 |
| l-glutamine | −0.32 |
| 4-aminobutyric acid | −0.45 |
The regulators gacS (PP_1650) and gacA (PP_4099) show strong fitness phenotypes in our data set. This two-component system is homologous to the well-studied barA/uvrY system of Escherichia coli, and it has been identified as a global regulator of cellular physiology in diverse organisms (24, 25). The conditions that elicited the strongest negative fitness phenotypes for gacSA were l-alanine (−1.3), β-alanine (−1.8), spermidine (−2.0), and propanediamine (−7.4). The direct targets of GacSA regulation are the rsm noncoding RNAs which modulate the activity of translational repressors, resulting in global changes in gene expression (26–28). Through this mechanism, GacSA may be required for regulation of some portions of l-alanine, β-alanine, propanediamine, and spermidine metabolism.
Inorganic nitrogen sources and urea.
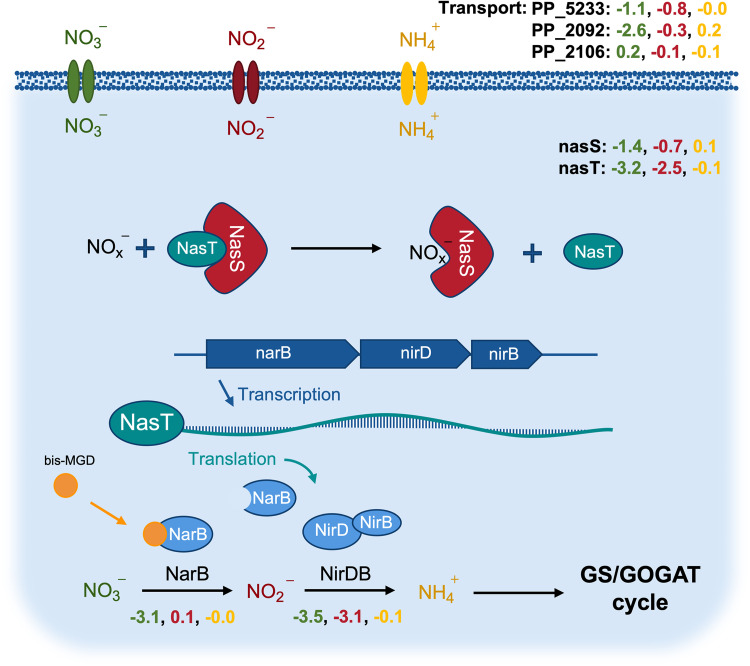
P. putida KT2440 is a known obligate aerobe, and cannot use alternative terminal electron acceptors during oxidative phosphorylation (29). Because of this, oxidized nitrogen species can only be used as nitrogen sources. Although the preferred inorganic nitrogen source for many bacteria is ammonium (30), other inorganic nitrogen species such as nitrate and nitrite can be utilized via the assimilatory nitrate reduction system, often organized in a single gene cluster (31, 32). The enzymes associated with the initial steps of the assimilatory nitrogen system in P. putida are the reductases NarB and NirDB, for which we see significant growth phenotypes (Fig. 3). In Pseudomonads, the two-component system NasS/T is a common regulator of this operon (32–34). In the absence of oxidized nitrogen sources, NasS and NasT form a complex that represses production of nitrate and nitrite reductases. When nitrate or nitrite are present, NasS dissociates from the NasS/T complex and the free RNA-binding antiterminator, NasT, enables full translation of the nitrate reduction operon (35) (Fig. 3).
FIG 3.
The assimilatory nitrate reduction system in P. putida KT2440. Average fitness values (n = 2) exhibited in the nitrate (green), nitrite (red), and ammonium (yellow) sole nitrogen source experiments. Shown are putative transporters, action of the NasST regulatory system, the assimilatory pathway, and the role of the bis-molybdopterin guanine dinucleotide (bis-MGD) cofactor. bis-MGD is the required cofactor for NarB (155). The fitness phenotypes for bis-MGD biosynthesis cluster together with the nitrate phenotypes and can be found in the interactive t-SNE visualization (Fig. I1 available at https://ppnitrogentsne.lbl.gov).
Our competitive growth assay using BarSeq indicated that the same system may also operate in P. putida KT2440. In the nitrate condition, the specific growth phenotypes for nasT (PP_2093) and nasS (PP_2094) are represented by the significant fitness values of −3.2 and −1.4, respectively. We were also able to identify a distinct nitrate transporter PP_2092 (−2.6). However, no significant phenotype could be detected for this gene in the nitrite condition, suggesting that it may be specific to nitrate. Because there were no mutants in predicted transporters with strong fitness phenotypes in the nitrite condition, it is also possible that PP_2092 or other unidentified transporters are responsible for nitrite transport. At pHs less than 7.2 and concentrations greater than 100 μM, nitrite is also known to be passively transported into the cell via diffusion of nitrous acid, likely explaining the lack of fitness data for this transport reaction (36).
Overall, the nitrite condition demonstrates a pattern of fitness phenotypes that resembles a stress response. In its free acid form, nitrite has been previously shown to have antimicrobial properties and was hypothesized to have wide-ranging mechanisms of action, including DNA damage, collapse of the proton motive force, and deleterious nitrosylation of cofactors and proteins (37). Our fitness data indicate that disruption of the recently identified RES-Xre toxin-antitoxin (TA) module (PP_2433-4) is detrimental to growth in the nitrite condition. In this system, the toxin (PP_2434) rapidly degrades NAD+ to halt bacterial growth and give the organism time to adapt its survival strategies, while the antitoxin (PP_2433) inhibits the toxin to allow restoration of NAD+ levels (38, 39). The significant fitness defect of mutants in the antitoxin PP_2433 (−6.4) might indicate that either the RES-Xre system is part of P. putida’s stress response to nitrite, or that nitrite disturbs the redox homeostasis of the cell and further disruption of redox balance by the PP_2434 toxin is lethal in strains lacking a functional PP_2433 antitoxin. We also observed significant phenotypes for several pathways that oxidize NAD(P)H, which supports the idea that NAD+ is depleted in the nitrite condition. One example is P. putida’s altered glucose utilization strategy. Fitness data indicates that glucose oxidation to 2-ketogluconate is preferred over the gluconate phosphorylation or direct glucose uptake pathways (40–42). Indicators for the accumulation of 2-ketogluconate in the nitrate condition include the specific fitness phenotypes for the transporter kguT (PP_3377; −1.5) and the NAD(P)H-dependent dehydrogenase kguD (PP_3376; −2.65). Compared with the other two pathways, glucose utilization via 2-ketogluconate produces more NAD(P)+. Other conditions that seemed to lead to similar phenotypes for kguT were 1,6-hexanediamine (−1.3), caprolactam (−1.55), 2-aminobutyric acid (−1.5), and uracil (−1.2).
Urea, the simplest organic nitrogen source we tested, only elicited significant fitness phenotypes in gltBD (−5.45) and ntrC (−1.8). Transcriptome analysis has previously revealed that the expression of a putative urease operon in P. putida (PP_2842-9) is controlled by NtrC (5). However, we observed no significant phenotypes in this operon. The presence of a second urea degradation pathway via urea carboxylase is unlikely, because it has been shown that organisms that possess the carboxylase pathway typically lack an urease (43–46). More research is necessary to further characterize urea metabolism in P. putida.
Proteinogenic amino acids.
The amino acid metabolism of microorganisms is of special interest for the food and bulk chemical industries, and has therefore been studied extensively for many decades (47). In contrast to the genus Corynebacterium, Pseudomonas spp. play a minor role in the industrial production of amino acids (48). However, their exceptional tolerance toward organic solvents makes them an excellent host for production of aromatic amino acid-derived compounds (11, 49–58).
Here, we have tested the 20 proteinogenic amino acids and two d-stereoisomers as the sole nitrogen source in a competitive growth assay using BarSeq. We also included BarSeq data from experiments where l-arginine, l-histidine, l-lysine, and d-lysine were used as sole carbon sources, and ammonium chloride was provided as a nitrogen source. No fitness data could be obtained for tyrosine and tryptophan due to insufficient biomass for DNA extraction and BarSeq analysis. Furthermore, although it has been reported that P. putida is unable to grow with the L-stereoisomers of methionine, threonine, or leucine as the sole source of carbon and nitrogen, we observed that P. putida could grow with these as sole nitrogen sources (Fig. S2) (59). The flux of their carbon backbones into the TCA cycle might be too inefficient for these three amino acids to simultaneously be the sole source of both carbon and nitrogen.
Microorganisms may possess multiple pathways for the degradation of the same amino acid, especially in the case of l-arginine, for which there are five known degradation pathways in Pseudomonas aeruginosa (60–62). Our nitrogen BarSeq experiments confirmed the presence of two of these pathways, and carbon source BarSeq data confirmed the presence of a third. Although another confirmed pathway in P. aeruginosa, the deiminase pathway, could not be identified in P. putida using BarSeq, the P. putida arginine deiminase AraA (PP_1001) has been successfully characterized in vitro (63).
The use of l-arginine as the sole nitrogen source led to significant growth phenotypes in both the common arginine succinyltransferase (AST) pathway (< −2.7) and the arginine decarboxylase/agmatine deiminase pathway. Ornithine can also be incorporated into the AST pathway, however, the only significant fitness defect we observed in this condition was for the dehydrogenase AstD (PP_4478; −2.3) (64, 65). Interestingly, the required succinyltransferase (PP_4479-80) to channel ornithine into the AST pathway exhibits a positive fitness value (+1.5). Under these conditions, ornithine degradation through the AST pathway may not be the optimal pathway. An alternative route, such as direct deamination by the cyclodeaminase PP_3533 (−0.85) and conversion of proline to glutamate (PP_4947; −5.15), might be more favorable (61).
The presence of a functional arginine decarboxylase/agmatine deiminase (ADAD) pathway in P. putida is indicated by the fitness defect of the arginine decarboxylase PP_0567 (−0.75). The amidase family protein PP_2932 has a strong fitness defect (−1.85) in the arginine condition, suggesting that it acts on N-carbomylputrescine as part of the ADAD pathway and should be more specifically annotated as an N-carbomylputrescine amidase.
When l-arginine is used as the sole carbon source, significant phenotypes for the arginine:pyruvate transaminase (APT) pathway appear as well. This pathway consists of the transaminase AruH (PP_3721, −1.2), the decarboxylase AruL (PP_3723, −0.9), the dehydrogenase KauB (PP_5278, −1.6), and a guanidinobutyrase (PP_4523, −1.5). The putative guanidinobutyrase PP_4523 not only has a fitness defect with l-arginine as the carbon source, but also with 4-guanidinobutyric acid as the nitrogen source (−0.95).
Due to a slight fitness defect (−1.0) of the predicted alanine racemase (Alr) PP_3722 and its localization in the APT operon, it is unclear whether AruH acts on the D- or L-stereoisomer of arginine, or on both. In a previous study, the deletion of alr led to significantly decreased growth of P. putida with arginine as the sole carbon and nitrogen source (66). This might indicate the presence of another catabolic route for arginine via its d-stereoisomer.
Carbon and nitrogen availability could determine the preference for one pathway over another. Because the AST pathway requires succinate for the conversion of arginine to glutamate, it is less efficient under carbon-limiting conditions, whereas the products of the other pathways can feed directly into the TCA cycle via putrescine and gamma aminobutyric acid (GABA) degradation (60).
The predicted histidine-lysine-arginine-ornithine ABC transport system (PP_4483-PP_4486) shows significant phenotypes (> −2) only in the arginine condition, while the ornithine condition results in mild fitness defects (−0.44). Furthermore, a different transporter, PP_5031, seems to be the most important transporter for histidine (−0.55). Interestingly, PP_5031 has a much stronger phenotype in the histidine carbon source condition (−4.5). Because NtrC responds to nitrogen-scarce conditions by upregulating nitrogen transporters (5), it may be challenging to identify transporters that are distinctly expressed in the presence of a specific substrate. These mild fitness defects for transporters in the histidine and ornithine nitrogen source conditions suggest that multiple transporters capable of accepting histidine or ornithine are expressed as part of the NtrC response. This limitation of nitrogen source BarSeq assays might be circumvented by adding ammonium in excess or complementing nitrogen source BarSeq data with carbon source experiments.
Another interesting interaction between D- and L-stereoisomers can be observed in the catabolism of lysine and alanine. Lysine catabolism in P. putida has been extensively studied and was fully characterized by Thompson et al., who used d- and l-lysine as a carbon source for BarSeq experiments (15, 59, 66–71).
The key difference between the lysine carbon and nitrogen source data sets is in the genes PP_4493, PP_0213, and gltBD. PP_0213 and PP_4493 are involved in the flux of the carbon skeleton of d- and l-lysine into the tricarboxylic acid (TCA) cycle and are therefore critical under carbon-limiting conditions (Fig. S5). Because the substrates of both gene products are fully deaminated, their fitness values in nitrogen-limiting conditions are negligible. The strong phenotype for gltBD (−5.8) is likely caused by the release of free ammonium in the first deamination step.
Along with d-glutamate, d-alanine is an important compound for the synthesis of microbial peptidoglycan and therefore necessary for bacterial survival and growth (72). The main racemase in P. putida for alanine appears to be DadX (59, 73). Our BarSeq data for alanine suggest that the conversion reaction was essential for P. putida’s survival when the library was constructed because there are no dadX (PP_5269) mutants present in our mutant library. The dadX paralog alr may be able to convert alanine as well, but its insignificant growth phenotype implies that it only plays a minor role in alanine degradation (59, 66, 73). Furthermore, neither of the alanine transaminase homologs show a growth defect when either l- or d-alanine is used as the sole nitrogen source. The only significant growth defect under these conditions is that of the deaminating d-amino acid oxidoreductase PP_5270 (dadA) and interestingly, its fitness defect is only significant with l-alanine (−3.6). For d-alanine, it only has a slight fitness defect of −0.65. An indicator that d-alanine deamination is still dependent on PP_5270 is the significant growth defect caused by the transcriptional regulator PP_5271 (−2.05). The gene PP_5271 has high identity (93% with an E-value of < 9e-88) with the regulator lrp from P. aeruginosa PA14. In P. aeruginosa PA14, lrp is a transcriptional activator of the dad operon, which is required for l-alanine catabolism. The regulator is highly induced by l-alanine and also slightly less induced by d-alanine and l-valine (74). The strong negative fitness value of −2.05 also supports the theory that lrp is an activator rather than a repressor in P. putida (75).
Pseudomonads are well known for their ability to degrade and synthesize aromatic compounds (76). Therefore, it is not surprising that they have also developed a large repertoire of aminotransferases with activity on aromatic substrates (77, 78). The only aminotransferase we identified in an aromatic amino acid condition (l-phenylalanine) is PP_3590 (−4.0). Its gene product is annotated as the d-lysine aminotransferase AmaC, although it plays a minor role in d-lysine degradation (71). Our BarSeq experiments suggest that PP_3590 has a broad substrate range, with fitness defects appearing on other substrates as well. For example, it has a fitness defect of −1.25 on l-pipecolate, indicating potential l-2-aminoadipate transaminase activity. The homolog, PP_1972, seems to have little effect on phenylalanine (−0.35) or pipecolate (−0.15) growth. In a previous study, a double knockout mutant of PP_3590 and PP_1972 did not lead to phenylalanine auxotrophy, underlining the redundancy of aromatic aminotransferases in P. putida (78).
Overlaps between aromatic amino acid transferases and branched-chain amino acid (BCAA) metabolism are also common in bacteria (79). Our data indicate that the aromatic aminotransferase PP_3590 also seems to be involved in the transamination of l-isoleucine (−1.75). However, the main transaminase in BCAA degradation and biosynthesis is PP_3511 (ilvE). It shows a significant fitness defect (< −4) in almost all the tested conditions, including ammonium chloride (−5.55). This highlights the importance of PP_3511 in minimal media conditions and its unique role in the metabolism of BCAAs. In fact, IlvE-deficient strains require gene complementation or supplementation of all three BCAAs (valine, isoleucine, leucine) to restore growth in minimal media (80, 81). We observe a similar behavior for the histidinol-phosphate aminotransferase PP_0967, which is essential for all the tested conditions except histidine. Surprisingly, heterologous expression of PP_0967 has been used to facilitate the deamination of l-phenylalanine to phenylpyruvate (54).
Many steps of the amino acid catabolism in P. putida are already known or precisely predicted based on homology models. However, by using BarSeq, we were able to generate functional evidence for the pathways of at least eight additional amino acids beyond the ones discussed here. To further expand our understanding of the amino acid metabolism in P. putida, we also included drop-out growth experiments for all proteinogenic amino acids. By supplying all but one of the 20 proteinogenic amino acids, we created conditions where biosynthesis of amino acids is essential for growth. We refer to the Fitness Browser (https://fit.genomics.lbl.gov) and the interactive t-SNE visualization (Fig. I1-2 available at https://ppnitrogentsne.lbl.gov) for more details.
Quaternary amines and ethanolamine.
Choline is a trimethylated, positively charged amine and a common component of ionic liquids used in the depolymerization of lignin (82). In nature, choline is metabolized by most bacteria to serve as a precursor for betaine, which is an important osmoprotectant (83). While biosynthetic pathways for those compounds are rarely present in bacteria, transporters and catabolic routes for quaternary amines are ubiquitous in these organisms (84, 85).
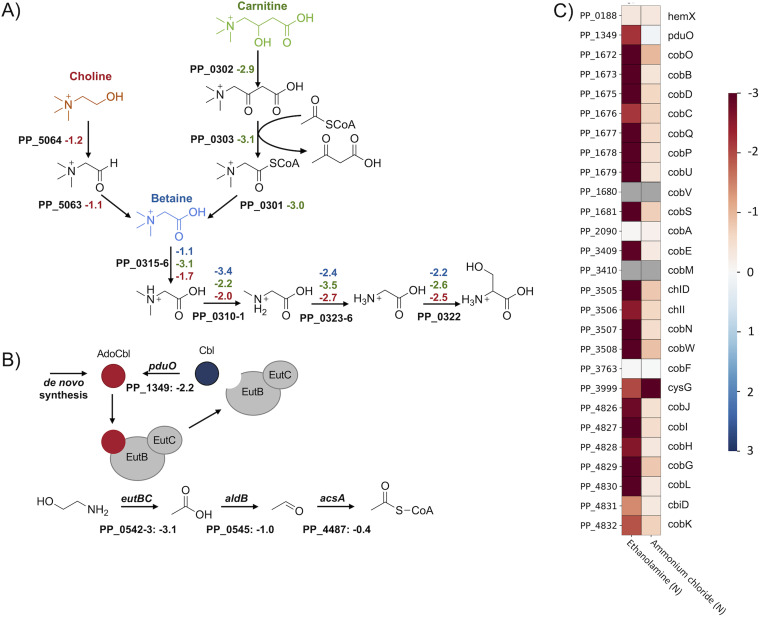
It is predicted that choline, betaine (trimethylglycine), and carnitine are metabolized through convergent pathways, with carnitine and choline entering betaine metabolism following thiolase cleavage and alcohol oxidation, respectively (86) (Fig. 4A). In the subsequent reactions, betaine is demethylated to produce glycine. Even though the glycine cleavage system (PP_0986, PP_0988, and PP_0989) has a significant phenotype when glycine is used as the sole source of nitrogen (< −4), it is not essential for growth in the quaternary amine conditions (> −0.3). Instead, conversion into serine is more efficient due to the generation of the required methyl-group donor 5,10-methylenetetrahydrofolate during demethylase activity of PP_0310-1 (87). As indicated by the strong fitness phenotypes elicited in the carnitine, choline, and betaine conditions, this reaction is likely catalyzed by PP_0322 (a predicted glycine/serine hydroxymethyltransferase) (< −2). Although PP_3144 (l-serine dehydratase) appears essential for growth on l-serine as a sole nitrogen source (−2.3), it is non-essential for growth on the tested quaternary amines. This suggests that other serine dehydratases (PP_0297 or PP_0987) or deaminating enzymes may be expressed in these conditions to release ammonium from serine.
FIG 4.
Quaternary amine and ethanolamine degradation in P. putida. (A) Putative routes for the quaternary amine catabolism in P. putida KT2440. The figure shows the degradation of choline (red), carnitine (green), and betaine (blue). The corresponding average fitness scores (n = 2) are shown next to each gene. (B) Ethanolamine degradation pathway, shown with fitness values (n = 2) and regeneration of the AdoCbl cofactor. (C) Heatmap with average fitness scores (n = 2) of genes that are putatively involved in P. putida’s adenosylcobalamin biosynthesis. No fitness scores (gray) could be obtained for the genes PP_1680 (cobV) and PP_3410 (cobM).
Based on the results of the BarSeq assay and sequence homology, the initial steps in carnitine metabolism were also identified. We propose that the beta-oxidation of carnitine to betainyl-CoA proceeds through the genes PP_0301 (a putative thioesterase), PP_0303 (a dehydrocarnitine cleavage enzyme), and PP_0302 (l-carnitine dehydratase) (Fig. 4A). The carnitine metabolism of P. aeruginosa was originally believed to proceed via CoA activation of oxidized carnitine (88). Enzymatic characterization of a PP_0303 homolog has since suggested that this protein acts upon oxidized carnitine and acetyl-CoA, releasing acetoacetate and betainyl-CoA (89).
We also observed fitness phenotypes for the transporters and regulators involved in quaternary amine metabolism. Although all three quaternary amines cause a strong NtrC response, which tends to mask distinct transporters, PP_0294-6 (choline/betaine/carnitine ABC transporter) exhibits fitness defects for choline (−1.13) and carnitine (−2.55). The metabolism of betaine and its precursors is regulated by the repressor BetI (PP_5719) and the activators GbdR (PP_0298) and CdhR (PP_0305) (84, 90). While gbdR is essential in all three conditions (< −1.0), cdhR is specific to carnitine (−2.5) and betI is specific to choline (+1.15). The negative and positive fitness defects correlate with the predicted mode of action for these regulators (90). The reason for the significant fitness phenotypes of PP_0308-9 in these conditions and their role in this operon are still unknown. Transcriptomic analysis suggested that they might be involved in the formation of filamentous biofilms (91).
Ethanolamine is a common molecule in nature and is involved in the choline and serine metabolism of plants (92, 93). Therefore, it is not surprising that several Pseudomonads are able to use it as a source for carbon and nitrogen (94). Bacterial ethanolamine degradation can be divided into two routes: (i) via acetyl-CoA or (ii) via ethanol (94, 95). However, the first step in both pathways is catalyzed by the adenosylcobalamin-dependent ethanolamine ammonia-lyase (EAL) EutBC (95–97). In P. putida, we observed a strong fitness phenotype for eutBC (PP_0542-3; −3.1) in the ethanolamine condition (Fig. 4B). Furthermore, we were also able to support the requirement for its cofactor adenosylcobalamin (AdoCbl). Because there is no exogenous AdoCbl in our minimal media and no specific phenotypes for the corrinoid-specific transport system PP_0524-5, P. putida is likely capable of the de novo synthesis of AdoCbl (98–101). This is also supported by the strong fitness phenotypes we observed for the putative cob genes in this condition (Fig. 4C). The additional requirement of the adenosyltransferase PduO (PP_1349; -2.2) is most likely caused by the release of Cbl during EAL activity (102–105). Re-adenylation of Cbl by PduO is likely less metabolically demanding than the de novo synthesis of AdoCbl.
Purines and pyrimidines.
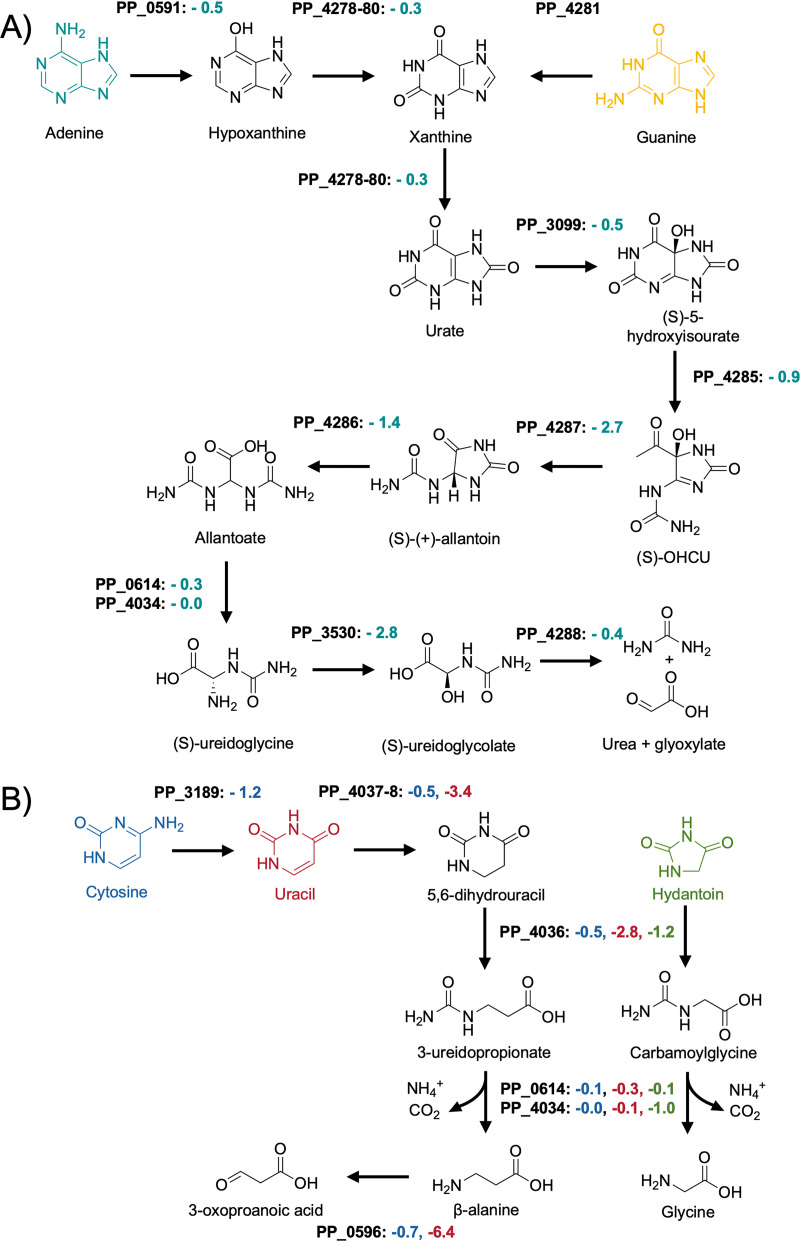
Purines and pyrimidines are widely distributed chemical structures in nature, and many organisms also salvage them as nitrogen sources. In this study, we have tested the purine base adenine and the pyrimidine bases cytosine and uracil as sole nitrogen sources using BarSeq. Similar to many other purines, the first steps in the degradation of adenine and guanine are the conversion into the shared intermediate xanthine (Fig. 5A). While adenine is converted to xanthine via the intermediate hypoxanthine, guanine is directly converted to xanthine by the guanine deaminase PP_4281 (106). The slight fitness defect of the adenosine deaminase PP_0591 (-0.45) might indicate that it can also act on adenine as a substrate.
FIG 5.
Putative routes for purine (A) and pyrimidine (B) catabolism in P. putida KT2440. Shown are the average fitness scores (n = 2) for genes involved in adenine (teal), guanine (yellow), cytosine (blue), uracil (red), and hydantoin (green) degradation.
The deamination products of the purine nucleobases, hypoxanthine and xanthine, are then both converted by the xanthine dehydrogenase complex xdhABC (PP_4178-80) to yield urate. For many years it was believed that the urate oxidation reaction yields (S)-allantoin in a single step (107). However, it was recently discovered that in addition to urate oxidase, this reaction actually involves two other enzymes and proceeds through the intermediate 5-hydroxyisourate, releasing hydrogen peroxide (108). To date, no urate oxidase has been experimentally verified in P. putida, and the only potential urate oxidase (PP_3099) exhibits no growth phenotype in any of the tested conditions. An indirect indicator for this reaction could be the significant fitness defect of the LysR-type transcriptional regulator PP_2250 (−2.35) and the conserved membrane protein of unknown function PP_2251 (−2.25). PP_2251 has high identity with the proteobacterial antimicrobial compound efflux (PACE) transporters A1S_1053 (57% with an E-value of 6e-45) from Acinetobacter baumannii and PFL_4585 (64% with an E-value of 5e-56) from Pseudomonas protegens Pf-5. It is possible that PP_2251 might also be a PACE transporter that functions as part of a defense mechanism against hydrogen peroxide or spontaneous peroxide radicals formed during urate oxidation (109, 110). The downstream reactions of 5-hydroxyisourate conversion to (S)-allantoin were strongly indicated by BarSeq. These reactions and further processing of (S)-allantoin to glyoxylate and urea are shown in Fig. 5A
The degradation of the pyrimidine base cytosine begins with the removal of the amine group by the cytosine deaminase CodB (PP_3189; −1.2) (Fig. 5B). Furthermore, we were able to confirm the significance of the designated cytosine transporter CodA (PP_3187; −1.2). Unexpectedly, the same transporter and deaminase cause fitness defects in the 3-aminobutyric acid (3ABA) condition (−1.15 and −1.15, respectively). The expression of the codBA operon and many other transport systems for nitrogenous compounds is controlled by the global regulator NtrC (5, 111). A general survival strategy for P. putida under nitrogen-limiting conditions seems to be the expression of numerous transporters for nitrogen-containing compounds that could potentially be present in the medium (5). Because 3ABA is an uncommon metabolite in nature, it might activate NtrC to such an extent that the cell starts scavenging nucleotides as a first response to nitrogen starvation (5, 111). Fitness defects for the hydantoinase PP_4036 (−1.8) and the dihydropyrimidine dehydrogenase PP_4037-8 (−2.78) are additional indicators for the degradation of nucleotides.
Given the deamination product of cytosine is uracil (Fig. 5A), similarly strong fitness values in both conditions could be expected. However, the fitness values associated with uracil degradation were at least 5-fold smaller in the cytosine condition. Our data suggest that in some cases subsequent deaminations of the same compound lead to less significant phenotypes; other examples include lysine and arginine nitrogen source experiments (Fig. S5).
The ring-opening reaction for 5,6-dihydrouracil is catalyzed by PydB (PP_4036; −2.8), and the resulting amide (S)-ureidoglycolate is then hydrolyzed by hyuC (PP_4034) or its predicted paralog PP_0614. Both (S)-ureidoglycolate hydrolyzing genes have no significant fitness defect, likely due to functional redundancy. Surprisingly, the only significant growth phenotype we identified for hyuC was in the hydantoin condition.
Hydantoin was discovered as a reduction product of allantoin (112), and its derivatives are used as anticonvulsants and pesticides (113, 114). Because it is a xenobiotic compound, the ability of P. putida to utilize hydantoin is probably due to its structural similarity to 5,6-dihydrouracil. The pyrimidine permease PP_4035 (-1.05) appears to be involved in hydantoin transport, and the same dihydropyrimidinase (PydB) that acts on 5,6-hydrouracil is likely responsible for opening its 5-membered ring (−1.15), producing carbamoylglycine. The amidohydrolase hyuC has a fitness defect of −1.0 in hydantoin sole nitrogen source experiments, which is the strongest fitness defect we observed for this gene. This could be explained if hydantoin was not within the substrate range of its paralog PP_0614 (−0.15). It is known that R-substituted hydantoins are converted by bacteria to the corresponding d-amino acid (115). Although comparing the hydantoin condition with the glycine condition did not reveal any common phenotypes involved in these metabolisms, it is likely that the products of the carbamoylglycine hydrolysis are glycine, ammonia, and carbon dioxide.
Even though we have no data for thymine, we tested a racemic mixture of 3-aminoisobutyric acid (3-AIBA) as the sole source of nitrogen. Its d-stereoisomer is the final product of thymine degradation (116). To date, no aminotransferases have been identified in P. putida that show either d- or l-3-AIBA activity. However, the strong fitness phenotypes for the pyruvate transaminase PP_0596 (−6.3) and the dehydrogenase PP_0597 (−1.7) are most likely caused by the presence of the L-stereoisomer, which is a common intermediate in valine degradation (117). Another potential indicator that PP_0596 has 3-AIBA transaminase activity is the fitness defect for the transcriptional regulator lrp (−2.15) and the deaminating d-amino acid oxidoreductase PP_5270 (−1.4). As described earlier, these genes are part of the dad operon which is involved in alanine metabolism and therefore an indicator for pyruvate transaminase activity (74).
Lactams.
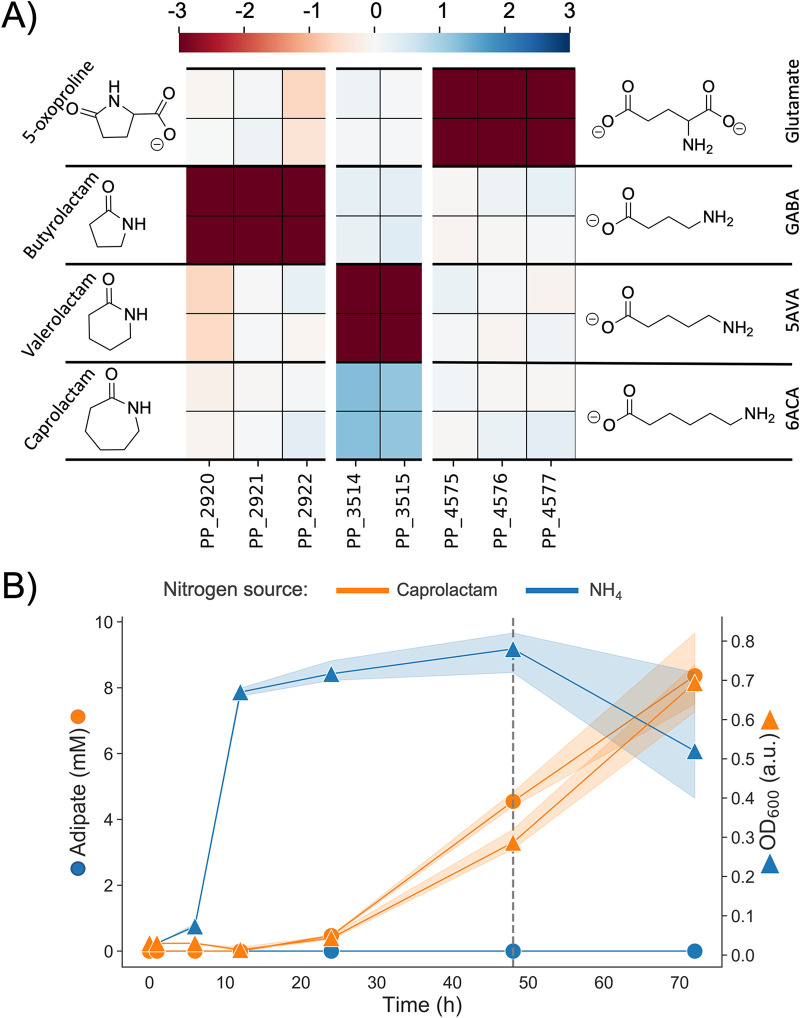
With applications ranging from manufacturing solvents to precursors for plastics and pharmaceuticals, lactams are an industrially relevant class of chemicals (118). Here, we tested four different lactams as nitrogen sources in our BarSeq experiments, gaining functional evidence for two previously uncharacterized lactam hydrolases, PP_2920-2 and PP_4575-7. Fitness data indicate that these lactam hydrolases are responsible for the hydrolysis of butyrolactam and 5-oxoproline (Fig. 6A). In-frame deletions of each of these hydrolases abolished growth when their respective lactam was provided as the sole nitrogen source. Growth could then be restored by complementation with a pBADT-based plasmid containing the lactam hydrolase (Fig. S6).
FIG 6.
(A) Heatmap with fitness scores (n = 2) of genes putatively involved in the hydrolysis of caprolactam, valerolactam, butyrolactam, and 5-oxoproline in P. putida KT2440. (B) LC-MS analysis of caprolactam degradation in P. putida KT2440. Wild-type cells were grown in MOPS minimal media with ammonium (blue) and caprolactam (orange) as the sole source of nitrogen. Shown is the OD (squares) and concentration of adipate in the supernatant (circles) over a time course of 72 h. The dashed line marks the time point at which caprolactam was no longer detected in the media.
The three subunits of PP_2920-2 and PP_4575-7 are annotated by UniProt as paralogs of pxpABC, the previously described widespread prokaryotic 5-oxoprolinase (119). Formerly, 5-oxoprolinases were only known to exist in eukaryotes and bacteria possessing a γ-glutamyl cycle. Niehaus et al. hypothesized that prokaryotes must have a way to combat the spontaneous cyclization of glutamate and glutamine to 5-oxoproline and identified pxpABC (119). Our fitness data suggests that only PP_4575-7 acts on 5-oxoproline, although it is also possible that spontaneous metabolite cyclization is the reason for the evolution of P. putida’s two other lactam hydrolases, PP_2920-2 and the previously identified valerolactam hydrolase OplBA (PP_3514-5) (16). The ω-amino acids GABA and 5-aminovalerate (5AVA) are common metabolic intermediates in P. putida, appearing as part of arginine, putrescine, and lysine catabolisms (15, 120, 121). Previous work has demonstrated that these C4 and C5 ω-amino acids can cyclize following activation to an acyl-CoA thioester (16, 118, 122). With no functional hydrolase, these lactams would serve as dead-end metabolites, and the reduction in carbon and nitrogen availability might be detrimental to growth. To test whether these lactamase systems serve this purpose, we conducted growth assays of ΔPP_4575-7, ΔPP_2920-2, and ΔoplBA with their lactam substrate’s corresponding ω-amino acid as a nitrogen source (Fig. S7). We observed slight growth lags and decreased maximal optical densities (ODs) in the knockouts versus the wild type, supporting the idea that these lactam hydrolases might exist for this purpose.
In previous work, the lactamase responsible for the hydrolysis of valero- and caprolactam, OplBA, was identified using proteomics data (16). Although the authors did search for the lactam hydrolase with BarSeq experiments using valerolactam as a sole carbon source, there was no significant fitness data for OplBA. Interestingly, we observed significant negative and positive fitness phenotypes for OplBA mutants with valerolactam and caprolactam as the nitrogen sources, respectively (Fig. 6A). The positive fitness data for OplBA mutants on caprolactam is especially surprising, because previous work has shown that oplBA knockouts cannot grow with caprolactam as the sole nitrogen source (16). However, it could be explained by the nature of BarSeq experiments. Library mutants with a functional OplBA hydrolyze the caprolactam to 6-aminocaproic acid (6ACA), which then accumulates due to P. putida’s slow utilization of this nitrogen source (Fig. S2). Then, mutants with the oplBA genes disrupted can consume the freed 6ACA without needing to expend the resources to produce this protein in already nitrogen-limited growth conditions.
The slow utilization of caprolactam, and therefore 6ACA, as a nitrogen source may be because P. putida is unable to utilize the product of 6ACA transamination, adipic acid (123). We suspected that adipic acid was accumulating in the media, which we confirmed with a metabolomics experiment. Caprolactam was used as a sole nitrogen source, and samples of the supernatant were taken at 1, 6, 12, 24, 48, and 72 h. Within 48 h, caprolactam is no longer detectable in any of our samples, while adipic acid slowly accumulates at approximately the same rate that biomass increases (Fig. 6B).
Adjacent to each of these three lactam hydrolases are three transcription factors: oplR (PP_3516), PP_2919, and PP_4579. The fitness data suggest that each of these transcription factors activates expression of its neighboring lactam hydrolase in response to the presence of its lactam substrate. However, we cannot determine from the fitness data if specificity for the different lactams is due to the transcription factor, the lactam hydrolase, or both. To answer this question, we examined the specificity of the lactam hydrolases through a complementation assay and the specificity of the transcription factors using (red fluorescent protein) RFP reporters.
For each of our three hydrolase knockout strains, we attempted to restore growth on its corresponding lactam with plasmid-based expression of the three different lactam hydrolases, resulting in a total of 9 strains tested. If a lactam hydrolase was more promiscuous than its regulator, we would expect it to restore growth in backgrounds with a different lactamase knocked out. However, plasmid-based expression of the lactam hydrolases only restored growth in the expected nitrogen source condition, indicating that the lactam hydrolases are fairly specific (Fig. S8).
The transcription factor oplR has already been identified as the regulator of oplBA and developed into a suite of highly sensitive valero- and caprolactam biosensor plasmids (10). However, although it is strongly suggested, we cannot assume from the fitness data alone that the other adjacent regulators are responsible for inducing lactamase expression. To confirm the role of each transcription factor, we conducted assays of two-plasmid systems in E. coli, as done by Thompson et al. with oplR (10). The transcription factor was expressed under the control of an arabinose inducible promoter on a low copy plasmid (pSC101 origin), while the region 200 bp upstream of the lactamase was cloned 5′ of the RFP gene in a medium-copy-number plasmid (BBR1). We refer to these plasmids as the sensor and reporter plasmid, respectively. The transcription factor induction level and concentration of the expected lactam ligand were then combinatorially varied. RFP, normalized with OD, increased with increasing lactam concentrations, and the dynamic range of the assay changed with transcription factor induction (Fig. S9B). This indicated that the transcription factors do in fact induce expression of their adjacent lactam hydrolases in response to their corresponding lactams. However, total induction and dynamic range were very small, likely because these transcription factors do not function optimally in E. coli. Therefore, we decided to probe the specificity of lactam regulation in P. putida. Unfortunately, our sensor plasmids are not compatible with P. putida. Instead, we opted to test the ligand range of these transcription factors by transforming our reporter plasmids into P. putida and relying on native expression levels of the transcription factors.
Both the butyrolactam and 5-oxoproline systems displayed high inducer specificity, with RFP induction occurring only in the presence of their expected ligand. The dynamic range of both of our reporter plasmid systems was quite wide, and both systems displayed high sensitivity. In particular, the butyrolactam reporter had normalized RFP signal roughly 900 times above zero induction at the lowest tested butyrolactam concentration (50 μM). This system has the potential to be highly effective as a biosensor for detecting butyrolactam production in P. putida. We also observed a response to butyrolactam from the reporter plasmid for OplR. Previous work by Thompson et al. showed that OplR is highly specific to valero- and caprolactam, but the maximum butyrolactam concentration tested was 0.5 mM and the host was E. coli (10). We detected fluorescence at butyrolactam concentrations greater than ∼ 6.25 mM, however, because we are relying on native expression of the transcription factor, we cannot know for certain whether this is due to a response from OplR to the butyrolactam or cross talk between PP_2919 and the oplBA promoter (Fig. S9C).
Polyamines, ω-amino acids, and GABA isomers.
Polyamines are abundant in nearly all forms of life and are involved in numerous cellular functions, including gene expression, stress response, cell growth, and membrane homeostasis (124). Beyond their biological significance, polyamines also have great industrial value, for example, putrescine is a direct nylon precursor, and propanediamine is used in textile finishing and strengthening agents (125, 126). The ω-amino acids, several of which are the degradation products of polyamines, also have both biological and industrial relevance. Many are produced as metabolic intermediates in pathways such as proteinogenic amino-acid degradation, while some, such as GABA, can act as signaling molecules (127, 128). Industrially, the aforementioned 6ACA, 5AVA, and GABA are precursors to several polymers and plastics (118).
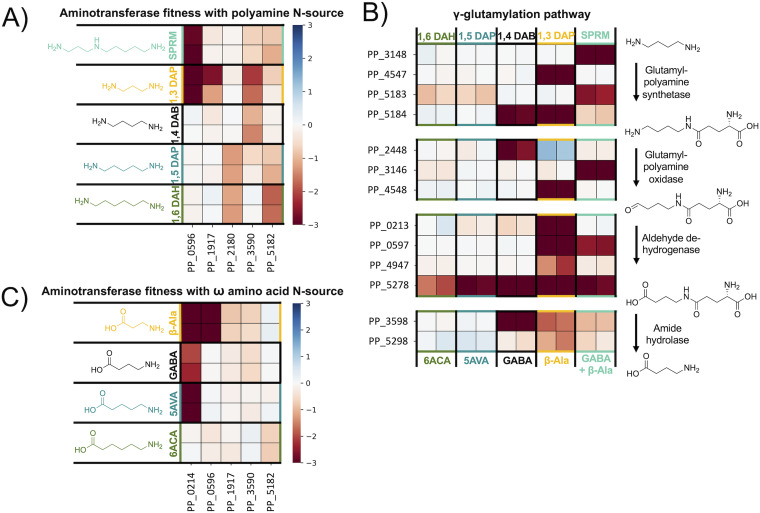
The three transaminases that appear to be most important for the utilization of polyamines, ω-amino acids, and GABA isomers are the pyruvate:alanine aminotransferases PP_0596, PP_2180 (SpuC-I), and PP_5182 (SpuC-II). They have different specific fitness phenotypes on both polyamines and amino acids, however, because the tested polyamines are converted to ω-amino acids, we could not determine if any acted directly on polyamines. Therefore, we purified these enzymes to assay their substrate range in vitro. Surprisingly, all three of the aminotransferases showed some in vitro activity on nearly all of the substrates tested, including substrates on which they had no significant fitness phenotype (Table 4) (Fig. S10).
TABLE 4.
In vitro substrate range of purified pyruvate:alanine aminotransferasesa
| Substrate class | Substrate | % conversion by PP_0596 | % conversion by PP_2180 | % conversion by PP_5182 |
|---|---|---|---|---|
| Polyamines | 1,3 diaminopropane | 65 ± 0.9% | 95 ± 0.7% | 96 ± 3.5% |
| putrescine | 2 ± 0.7% | 109 ± 1.4% | 112 ± 1.8% | |
| cadaverine | 9 ± 4.2% | 97 ± 1.8% | 103 ± 8.5% | |
| 1,6 diaminohexane | 14 ± 1.5% | 85 ± 4% | 88 ± 0.6% | |
| spermidine | 101 ± 1.1% | 105 ± 5.8% | 112 ± 1.2% | |
| ω-amino acids | β-alanine | 51 ± 0.5% | NA | NA |
| γ-aminobutyric acid | 75 ± 2% | 76 ± 1.5% | 83 ± 4.2% | |
| 5-aminovaleric acid | 70 ± 2.5% | 71 ± 1.4% | 73 ± 0.9% | |
| 6-aminocaproic acid | 78 ± 1.9% | 82 ± 0.8% | 84 ± 1% | |
| GABA isomers | dl-2-aminobutyric | 55 ± 1.3% | 95 ± 1.2% | 100 ± 5.6% |
| 3-aminobutyric acid | 87 ± 3.1% | 12 ± 1.6% | 16 ± 3.8% | |
| dl-3-aminoisobutyric | 70 ± 2.4% | 6 ± 1.3% | 12 ± 2.5% | |
| Proteinogenic amino acids | l-isoleucine | NA | NA | 1 ± 0.5% |
| l-leucine | NA | 5 ± 0.7% | NA | |
| l-lysine | 59 ± 49.4% | 61 ± 3.8% | 65 ± 4.9% | |
| l-valine | NA | NA | NA |
Tested substrates were added in excess to pyruvate. Activity of the aminotransferases on the tested substrates was measured indirectly through the quantification of the co-product alanine. Percent conversion is relative to the expected measurement of alanine if the amine acceptor pyruvate was completely transformed. Error represents the standard deviation of three replicates. NA indicates that no activity was statistically detected above controls.
The promiscuity of these enzymes directly contrasts with the specific phenotypes of the fitness data. For example, PP_2180 or PP_5182 have no fitness defect on 1,3-diaminopropane (1,3-DAP), yet both demonstrate greater activity on this substrate than PP_0596, which does have a significant fitness defect. This disparity between the fitness data and biochemical data is best explained by a difference in the promiscuity of these enzymes and their regulators. To investigate our hypothesis, we constructed fluorescent reporter systems.
As previously described in the Lactams section, we used a two-plasmid approach with RFP as an output to test transcription factor and aminotransferase promoter combinations in E. coli. We were able to identify the transcription factors likely responsible for regulation of the aminotransferases PP_0596 and PP_2180 (Fig. S11). Adjacent to PP_0596 is the LysR-type transcription factor PP_0595, which has fitness data that closely mirrors the phenotypes of PP_0596. The two-plasmid system for this combination worked as anticipated, and it appears that PP_0595 activates the promoter in response to both dl-3-aminoisobutyric acid (3AIBA) and β-alanine (Fig. S11). The aminotransferase PP_2180 had opposite fitness data to the adjacent MerR-family transcription factor PP_2181, which is oftentimes characteristic for transcriptional repressors. PP_2180 appears to be part of an operon and using the region upstream of PP_2177 as a promoter results in a response to 1,6-diaminohexane (1,6-DAH) and cadaverine (Fig. S11). The specificity of these transcription factors was much higher than the in vitro substrate specificity of the aminotransferases they regulate, suggesting that the role of PP_0596 and PP_2180 in vivo is determined by regulation.
Utilization of polyamines as a nitrogen source can proceed through two pathways, direct transamination by an aminotransferase or the γ-glutamylation pathway. However, because the product of the γ-glutamylation pathway is an ω-amino acid that is also then a substrate for an aminotransferase, we cannot say with certainty which pathways are used based on fitness data alone. In 1,6-DAH and cadaverine nitrogen source conditions, we see mild fitness defects (< −1.0) for the two aminotransferases PP_5182 and PP_2180. As previously discussed, both aminotransferases act on these substrates in vitro and the regulator PP_2181 responds specifically to 1,6-DAH and cadaverine. This suggests that transamination, not γ-glutamylation, is the preferred method of utilization for longer chain polyamines. In-frame deletions of either aminotransferase alone does not effectively abolish growth on 1,6-DAH and cadaverine, likely because they are functionally redundant. Strains with both PP_5182 and PP_2180 deleted show almost no growth with cadaverine and 1,6-DAH as sole nitrogen sources, although we did observe very slight growth in the double knockout strain after 72 h (Fig. S12). This could be due to the background activity of another aminotransferase or the γ-glutamylation pathway.
Conversely, the γ-glutamylation pathway appears to be equally if not more important than direct transamination for the utilization of putrescine, 1,3-diaminopropane (1,3-DAP), and spermidine, with each condition resulting in significant fitness phenotypes for all steps of the pathway (Fig. 7B). It is possible that putrescine, 1,3-DAP, and spermidine may also be directly deaminated by aminotransferases, although this is less likely for putrescine, which results in no significant fitness phenotypes for any aminotransferases.
FIG 7.
Heatmaps of polyamine and ω-amino acid catabolism in P. putida. For each enzyme class shown, HMMER was used to identify all putative genes present in P. putida corresponding to that pFam in P. putida. Data was filtered to find genes in each class with fitness values >–1.0 and t > |5| in the nitrogen sources shown. (A) Heatmap with fitness values (n = 2) for genes putatively involved in polyamine transamination in P. putida KT2440. (B) Heatmap with fitness values (n = 2) for genes putatively involved in polyamine γ-glutamylation in P. putida KT2440. (C) Heatmap with fitness values (n = 2) for genes putatively involved in omega-amino acid transamination in P. putida KT2440. Spermidine (SPRM) = teal; 1,3 diaminopropane (1,3 DAP) = yellow; β-alanine = yellow; putrescine/1,4-diaminobutane (1,4 DAB) = black; γ-aminobutyric acid (GABA) = black; cadaverine/1,5-diaminopentane (1,5 DAP) = blue; 5-aminovalerate (5AVA) = blue; 1,6-diaminohexane (1,6 DAH) = green; 6-aminocaproic acid (6ACA) = green.
The ω-amino acids, whether provided directly or released from polyamine catabolism, are degraded by transamination to an aldehyde then oxidation to a dicarboxylic acid. The most poorly utilized ω-amino acid we tested is 6ACA, which typically results in little to no visible growth until close to 72 h. This is likely due to a combination of regulatory issues and the accumulation of its product adipic acid, which P. putida is unable to utilize further (Fig. 6B). The spuC paralogs PP_5182 and PP_2180 are likely involved in 6ACA catabolism, despite their weak (> −1) phenotypes. If the aminotransferases for which we tested the in vitro substrate range are just as promiscuous in vivo and regulatory issues do contribute to the slow growth rate of 6ACA, we would expect that plasmid-based expression of these aminotransferases would improve growth. To test this theory, we provided functional gene copies to knockout mutants of PP_0596, PP_5182, and PP_2180 by introducing the arabinose-inducible pBADT plasmid harboring the corresponding gene (Fig. S13). Surprisingly, the control strain, wild-type P. putida harboring pBADT-RFP, was unable to grow under these culture conditions. We hypothesize that the burden of plasmid maintenance and insufficient kanamycin-resistance protein levels lead to early stage cell death when 6ACA is used as the sole source of nitrogen. However, our functionally complemented strains all demonstrated growth on 6ACA, indicating that that regulation contributes to the slow utilization of 6ACA and expressing any of the three aminotransferases with known activity on 6ACA independently of native regulation improves growth.
As previously discussed, the transcription factor PP_0595 is specific for β-alanine, and the main substrate of the aminotransferase it regulates, PP_0596, appears to be β-alanine in vivo. We also found PP_0596 necessary for utilization of uracil, 3ABA, and 3AIBA (Fig. S14). Both of these genes have a very strong fitness defect in the β-alanine nitrogen source condition (−7.8). Located in the same operon is the methylmalonate-semialdehyde dehydrogenase PP_0597 (−5.9), which catalyzes the conversion of malonate-semialdehyde to acetyl-CoA. The requirement for PP_0596, davT, and gltBD in the β-alanine condition may indicate a higher-order metabolic pathway for efficient metabolism of β-alanine. In the β-alanine condition, DavT may catalyze the reverse reaction (succinate-semialdehyde and glutamate to 4-ABA and 2-oxoglutarate) to reduce intracellular alanine concentrations produced by PP_0596. This is supported by the requirement of DavD (PP_0213, succinate/glutarate dehydrogenase) (Fig. 2B). Alternatively, DavT and PP_0596 may both have activity on β-alanine, and a knockout in either gene could result in detrimental loss of flux to malonate-semialdehyde. This peculiarity of β-alanine metabolism requires further investigation (Fig. S4).
In P. putida, GABA can serve as the sole source of both carbon and nitrogen (129). Because GABA is abundantly present in root exudates, P. putida possesses sensitive and specific receptors that detect GABA and facilitate chemotaxis (129, 130). Unlike E. coli, the genes for the degradation and transport of GABA are not clustered in a single operon (68, 131). Furthermore, the observed fitness defects are mostly unspecific, which makes it more difficult to identify distinct phenotypes. However, the main aminotransferase acting on GABA appears to be DavT (PP_0214; −2.15) , producing succinate-semialdehyde that is then converted to succinate by DavD (68). The fitness defect for davD (−0.45) is not significant (|t | < 5) suggesting that additional dehydrogenases such as PP_2488 (0.0) or PP_3151 (−0.2) catalyze this reaction. Moreover, our biochemical assay revealed that at least three other aminotransferases (PP_0596, PP_2180, PP_5182) accept GABA as a substrate (Table 4). Although the second aminotransferase (PP_3590) identified by BarSeq has an insignificant phenotype in the GABA condition (−0.55; |t | = 3.7), it exhibits a significant fitness defect in the dl-2-aminobutyric acid condition (−0.95).
2-aminobutyrate (2-ABA) is an unnatural amino acid that is used in the production of pharmaceuticals, and has been a target molecule for biological production in E. coli (132, 133). The only known usage of 2-ABA in biological systems is the substitution of l-cysteine during glutathione synthesis to form ophthalmic acid (134). Biosynthetically, it can occur during isoleucine synthesis via threonine by IlvE transamination of 2-oxobutyrate (133). Due to the essentiality of ilvE in minimal media conditions, we are not able to confirm this in P. putida using BarSeq. However, we were able to identify the d-amino acid oxidoreductase PP_5270 (−2.75) which acts on the alanine produced during pyruvate transaminase activity. Although there is no significant phenotype for PP_0596 in this condition, we have shown that PP_0596 can transaminate 2-ABA (Table 4). Furthermore, the transamination product 2-oxobutyrate is an intermediate in isoleucine biosynthesis. Because of this, there appears to be a requirement for the downstream steps of isoleucine biosynthesis. Instead of producing isoleucine, its precursor (S)-3-methyl-2-oxopentanoate might be directly funneled into the isoleucine degradation pathway via the branched-chain ɑ-ketoacid dehydrogenase (BCKD) complex (−1.54). This decarboxylative route is further supported by the requirement of the methylcitrate cycle (PP_2334-6; −1.5). These genes are important for the degradation of propionyl-CoA, which can only be produced from 2-ketobutyrate via a decarboxylative reaction (135).
The last GABA isomer we tested was 3ABA, previously described in the Purines and Pyrimidines section. 3ABA is one of several GABA isomers that can prime plant immunity, and it is applied agriculturally to prevent crop blight (136). Although the most effective route for 3ABA degradation seems to be the pyruvate transaminase, PP_0596 (−5.25), it appears that the cotranscribed gene, PP_0597 (malonyl semialdehyde dehydrogenase), is deleterious during growth on 3ABA (+2.85). The dehydrogenase PP_0597 is not the correct enzyme for further processing of the downstream product acetoacetate, and is likely a waste of resources. Furthermore, the reaction catalyzed by PP_0597 is identical to that of PP_4667, and it is possible that expression of PP_0597 results in a metabolic disturbance in valine metabolism.
Future directions.
In this work, hundreds of genes critical for growth of P. putida KT2440 on diverse nitrogen sources were identified. Not only is the generated wealth of data useful to those studying P. putida KT2440, but it could also be applied to infer possible gene functions of homologs in closely related species. Furthermore, this work provides a roadmap for the characterization of other microbes using functional genomics. Deeper analysis of related gammaproteobacteria through similar omics methodologies could enable investigations into the evolution and diversification of these metabolic phenotypes. The Pseudomonas genus alone contains a litany of species with interesting metabolisms. Further interrogation of this genus through BarSeq or multi-omics could shine a light on the evolutionary and ecological importance of these metabolisms.
The majority of our functional genomics data validates what has already been predicted by major databases such as KEGG or SEED. We and others have confirmed the reliability of TnSeq data as a guide for successful metabolic engineering (11, 14, 16, 28). These data provide a level of in vivo evidence for gene function that is distinct from prior mutational interrogations. We contend that these data are highly predictive and can be used in conjunction with existing databases (BioCyc, SEED, KEGG) to inform genetic, biochemical, and bioengineering studies. A process for integrating the acquired information from these and other TnSeq studies into public-facing databases would be desirable.
The identification of genetic interactions through t-SNE and correlation analyses could also enable more detailed interrogations of their translated proteins. With the recent publication of Alphafold2 and RoseTTA fold, it is now possible to elucidate structures and protein-protein interfaces using purely in silico approaches (137, 138). Using computer-generated protein structures and docking simulations, one could propose and test more detailed hypotheses regarding structure-function relationships. This could further deepen our understanding of the molecular mechanisms that enable these diverse phenotypes.
MATERIALS AND METHODS
Media, chemicals, and culture conditions.
General E. coli cultures were grown in lysogeny broth (LB) Miller medium (BD Biosciences, USA) at 37°C while P. putida was grown at 30°C. When indicated, P. putida and E. coli were grown on modified MOPS minimal medium, which is comprised of 32.5 μM CaCl2, 0.29 mM K2SO4, 1.32 mM K2HPO4, 8 μM FeCl2, 40 mM MOPS, 4 mM tricine, 0.01 mM FeSO4, 9.52 mM NH4Cl, 0.52 mM MgCl2, 50 mM NaCl, 0.03 μM (NH4)6Mo7O24, 4 μM H3BO3, 0.3 μM CoCl2, 0.1 μM CuSO4, 0.8 μM MnCl2, and 0.1 μM ZnSO4 (103). For most experiments, nitrogen-free MOPS was used, in which case the NH4CL was omitted and the nitrogen source of interest was added at concentrations ranging from 4 to 10 mM. Cultures were supplemented with kanamycin (50 mg/L, Sigma-Aldrich, USA), gentamicin (30 mg/L, Fisher Scientific, USA), or carbenicillin (100 mg/L, Sigma-Aldrich, USA), when indicated. All other compounds were purchased through Sigma-Aldrich (Sigma-Aldrich, USA).
Strains, plasmids, and primers.
The strains and plasmids used in this work are listed in Table 5, and plasmids used in this work are listed in Table 6. Primers used in this study can be found in Table 7. All strains and plasmids created in this work are available through the public instance of the JBEI registry (https://public-registry.jbei.org/folders/715). Device Editor and Vector Editor software were used to design the plasmids, and primers used for the construction of plasmids were designed using j5 software (139–141). All primers were purchased from Integrated DNA Technologies (IDT, Coralville, IA). Plasmids were assembled via Gibson Assembly using standard protocols and isolated with the Qiaprep Spin Miniprep kit (Qiagen, USA) (142).
TABLE 5.
Strains used in this study
| Strain | Description | Reference | JBEI part ID |
|---|---|---|---|
| E. coli XL1 Blue | Agilent | ||
| E. coli BL21(DE3) | NEB | ||
| P. putida KT2440 | Wild type | ATCC 47054 | |
| ΔPP_0596 | Strain with complete internal in-frame deletion of PP_0596 | This work. | JPUB_019820 |
| ΔPP_2180 | Strain with complete internal in-frame deletion of PP_2180 | This work. | JPUB_019821 |
| ΔPP_5182 | Strain with complete internal in-frame deletion of PP_5182 | This work. | JPUB_019822 |
| ΔPP_4575-7 | Strain with complete internal in-frame deletion of PP_4575-7 | This work. | JPUB_019823 |
| ΔPP_2920-2 | Strain with complete internal in-frame deletion of PP_2920-2 | This work. | JPUB_019824 |
| ΔPP_3514-5 | Strain with complete internal in-frame deletion of PP_3514-5 | (16) |
TABLE 6.
Plasmids used in this study
| Plasmid | Description | Reference | JBEI part ID |
|---|---|---|---|
| pMQ30 | Suicide vector for allelic replacement with Gmr, SacB | (156) | |
| pMQ30 ΔPP_0596 | Suicide vector for in-frame deletion of PP_0596 | This work. | JPUB_019825 |
| pMQ30 ΔPP_2180 | Suicide vector for in-frame deletion of PP_2180 | This work. | JPUB_019827 |
| pMQ30 ΔPP_5182 | Suicide vector for in-frame deletion of PP_5182 | This work. | JPUB_019829 |
| pMQ30 ΔPP_4575-7 | Suicide vector for in-frame deletion of PP_4575-7 | This work. | JPUB_019861 |
| pMQ30 ΔPP_2920-2 | Suicide vector for in-frame deletion of PP_2920-2 | This work. | JPUB_019831 |
| pMQ30 ΔPP_3514-5 | Suicide vector for in-frame deletion of PP_3514-5 | (16) | |
| pET28a | Protein expression vector with ColE1 origin, N-terminal 6xHis-tag and T7lac promoter | (157) | |
| pET28a PP_0596 | Protein expression vector for Ni-NTA purification of PP_0596 | This work. | JPUB_019857 |
| pET28a PP_2180 | Protein expression vector for Ni-NTA purification of PP_2180 | This work. | JPUB_019859 |
| pET28a PP_5182 | Protein expression vector for Ni-NTA purification of PP_5182 | This work. | JPUB_019833 |
| pBADT | Broad host-range vector with BBR1 origin, arabinose inducible promoter, and kanamycin resistance marker | (158) | |
| pBADT PP_0596 | Complementation vector for PP_0596 | This work. | JPUB_019835 |
| pBADT PP_2180 | Complementation vector for PP_2180 | This work. | JPUB_019837 |
| pBADT PP_5182 | Complementation vector for PP_5182 | This work. | JPUB_019839 |
| pBADT PP_4575-7 | Complementation vector for PP_4575-7 | This work. | JPUB_019841 |
| pBADT PP_2920-2 | Complementation vector for PP_2920-2 | This work. | JPUB_019843 |
| pBADT PP_3514-5 | Complementation vector for PP_3514-5 | (16) | |
| pBBR1k | pBADT derived broad host-range expression vector with araC and PBAD removed | (10) | |
| pBBR1k PP_2920p-RFP | Expression vector with region 200 bp upstream of PP_2920 used as promoter for RFP | This work. | JPUB_019855 |
| pBBR1k PP_4578p-RFP | Expression vector with region 200 bp upstream of PP_4578 used as promoter for RFP | This work. | JPUB_019853 |
| pBBR1k PP_3515p-RFP | Expression vector with region 200 bp upstream of PP_3515 used as promoter for RFP | (10) | |
| pBBR1k PP_0596p-RFP | Expression vector with region 200 bp upstream of PP_0596 used as promoter for RFP | This work. | JPUB_019845 |
| pBBR1k PP_2177p-RFP | Expression vector with region 200 bp upstream of PP_2177 used as promoter for RFP | This work. | JPUB_019847 |
| pBbS8a | pSC101 origin with carbenicillin resistance and arabinose inducible promoter | (159) | |
| pBbS8a PP_0595 | pBbS8a expressing the transcription factor PP_0595 | This work. | JPUB_019851 |
| pBbS8a PP_2181 | pBbS8a expressing the transcription factor PP_2181 | This work. | JPUB_019849 |
TABLE 7.
Primers used in this study
| Primer name | Sequence (5′ to 3′) | Use |
|---|---|---|
| pBbS8a_fwd | ggatccaaactcgagtaaggatctcc | pBbS8a backbone plasmid construction |
| pBbS8a_rev | atgtatatctccttcttaaaagatcttttgaattcc | pBbS8a backbone plasmid construction |
| pBbS8a-PP_2181 PP_2181_fwd | ggaattcaaaagatcttttaagaaggagatatacatatgaaagtgcacgaagaaatcgaaggcc | pBbS8a-PP_2181 construction |
| pBbS8a-PP_2181 PP_2181_rev | ggagatccttactcgagtttggatcctcagctgaacacccaaagcacgcg | pBbS8a-PP_2181 construction |
| pBbS8a-PP_0596 PP_0596_fwd | ggaattcaaaagatcttttaagaaggagatatacatatgagccgacgccccgatcc | pBbS8a-PP_0596 construction |
| pBbS8a-PP_0596 PP_0596_rev | ggagatccttactcgagtttggatcctcaacgtgtcgcagccaggc | pBbS8a-PP_0596 construction |
| BBR1k_fwd | tttaagaaggagatatacatatggcgag | BBR1k backbone plasmid construction |
| BBR1k_rev | gacgtcggaattgccagctg | BBR1k backbone plasmid construction |
| BBR1k-PP_2177p-RFP PP_2177p_fwd | gcgccccagctggcaattccgacgtcgtcgggctggaggattcagc | BBR1k-PP_2177p-RFP construction |
| BBR1k-PP_2177p-RFP PP_2177p_rev | cgctactcgccatatgtatatctccttcttaaacgtttcggactgccgtgaaatttttag | BBR1k-PP_2177p-RFP construction |
| BBR1k-PP_0596p-RFP PP_0596p_fwd | gcgccccagctggcaattccgacgtcactcgacgacgctgcgatag | BBR1k-PP_0596p-RFP construction |
| BBR1k-PP_0596p-RFP PP_0596p_rev | cgctactcgccatatgtatatctccttcttaaacggcggttcctcggaagacg | BBR1k-PP_0596p-RFP construction |
| BBR1k-PP_4578p-RFP PP_4578p_fwd | gcgccccagctggcaattccgacgtctttctgccgtcaggcggaagct | BBR1k-PP_4578p-RFP construction |
| BBR1k-PP_4578p-RFP PP_4578p_rev | accaacaagccgaggcatcctttaagaaggagatatacatatggcgagtagcg | BBR1k-PP_4578p-RFP construction |
| BBR1k-PP_2920p-RFP PP_2920p_fwd |
gcgccccagctggcaattccgacgtcaggtggatcgccgcttgc | BBR1k-PP_2920p-RFP construction |
| BBR1k-PP_2920p-RFP PP_2920p_rev |
gtgccaggagacgcggtctttaagaaggagatatacatatggcgagtagcga | BBR1k-PP_2920p-RFP construction |
| pET28_fwd | atgagatccggctgctaacaaagccc | pET28 backbone plasmid construction |
| pET28_rev | tgctagccatatggctgccgcg | pET28 backbone plasmid construction |
| pET28-PP_2180 PP_2180_fwd |
gccgcgcggcagccatatggctagcagtgaacagaattcgcagacccttgcc | pET28-PP_2180 construction |
| pET28-PP_2180 PP_2180_rev |
cgggctttgttagcagccggatctcattaccgaacagcctcataggtcaggtcc | pET28-PP_2180 construction |
| pET28-PP_5182 PP_5182_fwd |
gccgcgcggcagccatatggctagcagcgtcaacaacccgcaaaccc | pET28-PP_5182 construction |
| pET28-PP_5182 PP_5182_rev |
cgggctttgttagcagccggatctcattattgaatcgcctcaagggtcaggtcc | pET28-PP_5182 construction |
| pET28-PP_0596 PP_0596_fwd |
gccgcgcggcagccatatggctagcaacatgcccgaaactggtcctgcc | pET28-PP_0596 construction |
| pET28-PP_0596 PP_0596_rev |
cgggctttgttagcagccggatctcatcagtcgatcaggttcagggtttcgc | pET28-PP_0596 construction |
| pBADT_fwd | aggatccaaactcgagtaaggatctcc | pBADT backbone plasmid construction |
| pBADT_rev | atgtatatctccttcttaaaagatcttttgaattcc | pBADT backbone plasmid construction |
| pBADT-PP_5182 PP_5182_fwd |
tcaaaagatcttttaagaaggagatatacatatgagcgtcaacaacccg | pBADT-PP_5182 construction |
| pBADT-PP_5182 PP_5182_rev |
ggagatccttactcgagtttggatccttattgaatcgcctcaagggtcagg | pBADT-PP_5182 construction |
| pBADT-PP_0596 PP_0596_fwd |
tcaaaagatcttttaagaaggagatatacatatgaacatgcccgaaactggtcctgc | pBADT-PP_0596 construction |
| pBADT-PP_0596 PP_0596_rev |
ggagatccttactcgagtttggatcctcagtcgatcaggttcagggtttcgc | pBADT-PP_0596 construction |
| pBADT-PP_2920-2 PP_2920-2_fwd |
tcaaaagatcttttaagaaggagatatacatatgcaagcagtggatttcaactcgg | pBADT-PP_2920-2 construction |
| pBADT-PP_2920-2 PP_2920-2_rev |
ggagatccttactcgagtttggatcctcagctgaagagcctggccagg | pBADT-PP_2920-2 construction |
| pBADT-PP_4575-7 PP_4575-7_fwd |
tcaaaagatcttttaagaaggagatatacatatgcacaacgacaagggagac | pBADT-PP_4575-7 construction |
| pBADT-PP_4575-7 PP_4575-7_rev |
ggagatccttactcgagtttggatccttacgaccaacgccccagg | pBADT-PP_4575-7 construction |
| pBADT-PP_2180 PP_2180_fwd |
tcaaaagatcttttaagaaggagatatacatatgagtgaacagaattcgcagaccc | pBADT-PP_2180 construction |
| pBADT-PP_2180 PP_2180_rev |
ggagatccttactcgagtttggatccttaccgaacagcctcataggtcaggtcc | pBADT-PP_2180 construction |
| pMQ30_fwd | tctaagaaaccattattatcatgacattaacc | pMQ30 backbone plasmid construction |
| pMQ30_rev | aaaactgtattataagtaaatgcatgtatactaaac | pMQ30 backbone plasmid construction |
| PP_4575-7_up_fwd | gtttagtatacatgcatttacttataatacagttttgcgctaatccccgaagactggc | pMQ30_PP_4575-7 construction |
| PP_4575-7_up_rev | taagactgctggccctttcgcgg | pMQ30_PP_4575-7 construction |
| PP_4575-7_down_fwd | cgcccgcgaaagggccagcagtcttacatatgcctaccttaccgcgcctgc | pMQ30_PP_4575-7 construction |
| PP_4575-7_down_rev | ggttaatgtcatgataataatggtttcttagagcaactgacggtgttgtggttcgc | pMQ30_PP_4575-7 construction |
| PP_0596_up_fwd | gtttagtatacatgcatttacttataatacagttttcaccaggttggggcgccg | pMQ30_PP_0596 construction |
| PP_0596_up_rev | atcggcggttcctcggaagacg | pMQ30_PP_0596 construction |
| PP_0596_down_fwd | taatcgtcttccgaggaaccgccgatgtgatctttccgatattgatgccaggcg | pMQ30_PP_0596 construction |
| PP_0596_down_rev | ggttaatgtcatgataataatggtttcttagacagcgccttgatctgtggctcc | pMQ30_PP_0596 construction |
| PP_2920-2_up_fwd | gtttagtatacatgcatttacttataatacagttttcatggaagtaattgcgcaccg | pMQ30_PP_2920-2 construction |
| PP_2920-2_up_rev | catgaccgcgtctcctggc | pMQ30_PP_2920-2 construction |
| PP_2920-2_down_fwd | cacaagtgccaggagacgcggtcatgtgagcatcatgttttcgctgtagccg | pMQ30_PP_2920-2 construction |
| PP_2920-2_down_rev | ggttaatgtcatgataataatggtttcttagacccgcccaggccgagctg | pMQ30_PP_2920-2 construction |
| PP_5182_up_fwd | gtttagtatacatgcatttacttataatacagttttcgcccccgagatggagttctacc | pMQ30_PP_5182 construction |
| PP_5182_up_rev | ggcggtccaataacggagaagcacat | pMQ30_PP_5182 construction |
| PP_5182_down_fwd | ggcggtccaataacggagaagcacatgtaagcgtttgctaggctagcgt | pMQ30_PP_5182 construction |
| PP_5182_down_rev | ccggtactttctacgacatggtcgccatctaagaaaccattattatcatgacattaacc | pMQ30_PP_5182 construction |
| PP_2180_up_fwd | gtttagtatacatgcatttacttataatacagttttaaacaaataccaggagcaactggacttcatgca | pMQ30_PP_2180 construction |
| PP_2180_up_rev | acagctaaccaggccctctgctgttacattgacttcactccatcaagttgcgcagc | pMQ30_PP_2180 construction |
| PP_2180_down_fwd | taacagcagagggcctgg | pMQ30_PP_2180 construction |
| PP_2180_down_rev | ggttaatgtcatgataataatggtttcttagaccaccgccgggtccatctc | pMQ30_PP_2180 construction |
| PP_0596_KOCON_fwd | ggatgtcgaaatcgctgact | Knockout confirmation for ΔPP_0596 |
| PP_0596_KOCON_rev | gcgaaatcttggcttcgttc | Knockout confirmation for ΔPP_0596 |
| PP_2180_KOCON_fwd | cgagttccaggtcatggataa | Knockout confirmation for ΔPP_2180 |
| PP_2180_KOCON_rev | gttcgacctgggacagaaa | Knockout confirmation for ΔPP_2180 |
| PP_5182_KOCON_fwd | gcagtacctgggcaagaaa | Knockout confirmation for ΔPP_5182 |
| PP_5182_KOCON_rev | cttgatccctgtctgcttctc | Knockout confirmation for ΔPP_5182 |
| PP_4575-7_KOCON_fwd | tatgtgtggatccagcgtttc | Knockout confirmation for ΔPP_4575-7 |
| PP_4575-7_KOCON_rev | ttcttcgcctcgggtatct | Knockout confirmation for ΔPP_4575-7 |
| PP_2920-2_KOCON_fwd | gaactttggcgcacctacta | Knockout confirmation for ΔPP_2920-2 |
| PP_2920-2_KOCON_rev | ggagatgttcaggtggatcg | Knockout confirmation for ΔPP_2920-2 |
| pMQ30_seq_fwd | ttgtctcatgagcggatacatatt | Forward sequencing of inserts in pMQ30 plasmids |
| pMQ30_seq_rev | tcaccgtcatcaccgaaac | Reverse sequencing of inserts in pMQ30 plasmids |
| RFPseq_up_rev | ggaaggacagtttcaggtagt | Sequencing region upstream of RFP in reporter plasmids |
| pBbS8a_seq_rev | ccttactcgagtttggat | Reverse sequencing of inserts in pBbS8a plasmids |
| pBADT_seq_fwd | atgccatagcatttttatcc | Forward sequencing of inserts in pBADT and pBbS8a plasmids |
| pBADT_seq_rev | gccctaggtataaacgcagaaa | Reverse sequencing of inserts in pBADT plasmids |
| pET28a_seq_fwd | taatacgactcactataggg | Forward sequencing of inserts in pET28a plasmids |
| pET28a_seq_rev | gctagttattgctcagcgg | Reverse sequencing of inserts in pET28a plasmids |
Plate-based growth assays.
Growth studies of bacterial strains were conducted using microplate reader kinetic assays as described previously (10). Overnight cultures were washed three times with nitrogen-free MOPS minimal medium and used to inoculate 48-well plates at a ratio of 1:100 (Falcon, 353072). Each well contained 500 μL of MOPS medium with 10 mM the tested nitrogen source. Plates were sealed with a gas-permeable microplate adhesive film (VWR, USA), and then optical density and fluorescence were monitored for 24 to 72 h in an Biotek Synergy H1M plate reader (BioTek, USA) at 30°C with fast continuous shaking. Optical density was measured at 600 nm. RFP was measured with an excitation wavelength of 535 nm, an emission of 620 nm, and a gain of 100.
BarSeq assays.
BarSeq experiments utilized the P. putida library JBEI-1 and were completed as previously described (15). Aliquots (2 mL) of libraries of JBEI-1 were thawed on ice, added to 25 mL of LB medium with kanamycin, and then grown at 30°C to an OD600 of 0.5. Then, three 1-mL aliquots were removed, pelleted, and stored at −80°C as zero time points. The libraries were washed three times in MOPS minimal medium with no nitrogen source, and then used to inoculate each experiment at a ratio of 1:100. Experiments were conducted in 24-well plates; each well contained 2 mL of nitrogen-free MOPS minimal medium with 10 mM each tested nitrogen source. Plates were grown at 30°C with shaking at 200 rpm, and 1 mL samples were collected after 24 to 72 h, depending on when cultures appeared sufficiently turbid for DNA extraction. Samples were pelleted and stored at −80°C until DNA extraction, which was done with a DNeasy UltraClean Microbial kit (Qiagen, Germany). BarSeq analysis was performed as previously described (19, 37). Strain fitness is defined as the normalized log2 ratio of the barcode reads in the experimental sample to the barcode reads in the time zero sample. The fitness of a gene is defined as the weighted average of the strain fitness for insertions in the central 10% to 90% of the gene. The gene fitness values are normalized such that the typical gene has a fitness of zero. The primary statistic t value represents the form of fitness divided by the estimated variance across different mutants of the same gene. Statistic t values of >|4| were considered significant. A more detailed explanation of fitness score calculations can be found in Wetmore et al. (143). All experiments described here passed the quality testing metrics described previously. Experiments were conducted in biological duplicates, and the fitness data are publicly available at http://fit.genomics.lbl.gov.
Protein production and purification.
The protocol for the ω-amino acid aminotransferases purification is a modified version of the previously described procedure by Yuzawa et al. (144). Briefly, E. coli BL21(DE3) harboring the pET28a vectors for PP_0596, PP_2180, or PP_5182 were cultured in LB medium with kanamycin at 37°C until OD600 reached 0.4. Protein expression was induced with 250 μM IPTG and cultures were grown at 18°C for 24 h. Cells were harvested by centrifugation for 20 min at 5,000 g and the pellet was resuspended in 30 mL wash buffer (50 mM sodium phosphate pH 7.6, 300 mM NaCl, 10 mM imidazole, 4°C). The cells were disrupted by sonication (8 × 30 seconds) and cell debris was removed by three subsequent centrifugations (15 min, 8,000 g, 4°C). The soluble fraction was mixed with 4 mL Nickel-NTA agarose beads (Thermo Fisher Scientific) for 1 h at 4°C. The mixture was applied to a Nickel-NTA column and washed three times with wash buffer. The protein was eluted with 12 mL elution buffer (150 mM sodium phosphate buffer pH 7.6, 50 mM NaCl, 150 mM imidazole, 4°C) and buffer was exchanged with stock buffer (100 mM sodium phosphate pH 7.5, 0.5 mM DTT, 10% glycerol, 4°C) by dialysis (SnakeSkin Dialysis Tubing, 10k MWCO, Thermo Fisher Scientific). Protein was concentrated in 30k MWCO Amicon Ultra-15 centrifugal filters (MilliporeSigma) and stored at −80°C.
In vitro pyruvate transamination and product quantification.
Substrate specificity of the purified aminotransferases was determined by in vitro l-alanine production. The reaction was carried out in 100 mM sodium phosphate buffer pH 9 with 500 μM pyruvate, 1 mM PLP, 5 μM enzyme and 5 mM substrate. After addition of the substrate, the reaction mixture was immediately incubated at 30°C for 30 min. The reaction was stopped by boiling the mixture for 10 min at 100°C. Alanine concentrations were determined enzymatically using the Alanine assay kit (Cell Biolabs, Inc.) following the manufacturer’s instructions.
Caprolactam degradation assay and LC-ESI-QTOF-MS analysis.
To investigate the degradation of caprolactam in P. putida KT2440, wild-type cells were grown in modified MOPS minimal media with either 10 mM ammonium chloride or caprolactam as the sole source of nitrogen. Cells were cultured as triplicates in 25 mL medium in shake flasks at 30°C and 200 rpm. Samples were collected and OD600 was measured after 1, 6, 12, 24, 48, and 72 h. For metabolomics analysis, cell culture samples were centrifuged at 16,000 g for 1 min, and 300 μL supernatant was mixed with −80°C cold methanol. After that the mixture was filtered by centrifugation (30 min, 14,000 g, 4°C) using 3 kDa Amicon Ultra-0.5 centrifugal filters (MilliporeSigma) and stored at −80°C for further analysis.
For the measurement of caprolactam, liquid chromatographic separation was conducted with a Kinetex HILIC column (100-mm length, 4.6-mm internal diameter, 2.6-μm particle size; Phenomenex, Torrance, CA) using a 1260 HPLC system (Agilent Technologies, Santa Clara, CA, USA). The injection volume for each measurement was 2 μL. The sample tray and column compartment were set to 6°C and 20°C, respectively. The mobile phase was composed of 10 mM ammonium formate and 0.2% formic acid in water (solvent A) and 10 mM ammonium formate and 0.2% formic acid in 90% acetonitrile and 10% water (solvent B). Reagent grade and source are as follows: Ammonium formate (Analytical grade) and formic acid (98% to 100% chemical purity): Sigma-Aldrich, St. Louis, MO, USA; Acetonitrile (LC-MS grade) and water (LC-MS grade): Honeywell Burdick & Jackson, CA, USA. Analytes were separated with the following gradient: 90% to 70%B in 4 min, held at 70%B for 1.5 min, 70% to 40%B in 0.5 min, held at 40%B for 2.5 min, 40% to 90%B in 0.5 min, held at 90%B for 2 min. The flow rate was varied as follows: held at 0.6 mL/min for 6.5 min, linearly increased from 0.6 mL/min to 1 mL/min in 0.5 min, and held at 1 mL/min for 4 min. The total run time was 11 min.
The 1260 HPLC system was coupled to an Agilent Technologies 6520 QTOF-MS system (for quadrupole time-of-flight mass spectrometric detection). The LC column effluent was delivered to the 6520 QTOF-MS system’s electrospray ionization (ESI) ion source via a 1:4 post-column split ratio. ESI was used to facilitate the production of gas-phase [M+H]+ ions in the positive ion mode. The capillary voltage was set to 3500 V. Fragmentor, skimmer, and OCT 1 RF voltages were set to 100 V, 50 V, and 250 V, respectively. Drying gas temperature, drying gas flow rate, and nebulizer were set to 350°C, 12 L/min, and 25 lb/In2, respectively. The instrument was tuned for a range of m/z 50 to 1,700 with the Agilent ESI-Low TOF tuning mix. High mass accuracy was achieved via reference mass correction with purine and HP-0921 reference solutions, which were purchased from Agilent Technologies. Data was acquired within the 50 to 1,100 m/z range. Analytes were quantified via a six-point calibration curve following 2-fold serial dilutions from 25 μM to 0.78125 μM. MassHunter Workstation (Agilent Technologies), MassHunter Qualitative Analysis and MassHunter Profinder (Agilent Technologies) software were used for data acquisition and processing.
The LC-ESI-TOF-MS method for adipic acid has been described previously (145).
t-stochastic neighbor embedding of BarSeq results.
The data used for the t-SNE visualization was filtered by t-scores and fitness scores (|t | > 5 and |fitness| > 1). This resulted in a file containing fitness scores for 615 significantly affected genes across 129 sole-nitrogen source growth assays in 51 different conditions, and 19 amino acid dropout conditions. 2-ABA was excluded from the analysis due to the large number of significantly affected genes in this condition. For each gene, the “significant condition” was defined as the condition that caused the largest change in fitness score. The fitness values were then used as the input into the t-SNE module supplied in the python package Scikit-Learn (19, 146). Clusters (n = 100) were defined using the Ward-clustering module in Scikit-Learn. The most frequently occurring “significant condition” was used to name each cluster. The Jupyter notebook used in these analyses can be found at: https://github.com/mschmidt75/ppnitrogentsne.git
Bioinformatic analyses.
All statistical analyses were carried out using either the Python Scipy or Numpy libraries (147, 148). To identify the number of aminotransferases, the HMMER version 3.3.2 (http://hmmer.org/) was used to scan the P. putida KT2440 genome against a profile database file consisting out of the Pfam HMM files of PF00155, PF00202, PF01063, and PF00266. The E-value cutoff was set to 1e-20, and significant hits annotated as transcriptional regulators were excluded from the final data set. HMMER was also used to identify possible genes of the γ-glutamyl cycle, with the Pfam HMM files of PF00120, PF01266, PF00171, and PF07722 corresponding to the glutamyl-polyamine synthetase, the glutamyl-polyamine oxidase, the aldehyde dehydrogenase, and the amide hydrolase.
The databases MiST 3.0 and TransportDB 2.0 were used to extract transcription factors and transport associated proteins from the BarSeq data set (17, 18). EggNOGmapper was used to generate the clusters of orthologous groups (COGs) for P. putida KT2440 (149, 150). Additionally, manual analysis of the data and proposals of metabolic pathways relied heavily on BioCyc and PaperBlast (151, 152).
ACKNOWLEDGMENTS
We thank Morgan Price and Megan Garber for their assistance in analyzing BarSeq data, Alberto Nava for his advice on manifold learning methods, Elias Englund for his help on protein purification, and Namil Lee for his advice on gene annotations. Mitchell Thompson is a Simons Foundation Awardee of the Life Sciences Research Foundation. The laboratory of LMB is partially funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy within the Cluster of Excellence FSC 2186 “The Fuel Science Center.” This work was part of the DOE Joint BioEnergy Institute (https://www.jbei.org) supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, supported by the U.S. Department of Energy, Energy Efficiency and Renewable Energy, Bioenergy Technologies Office, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy. The views and opinions of the authors expressed herein do not necessarily state or reflect those of the United States Government or any agency thereof. Neither the United States Government nor any agency thereof, nor any of their employees, makes any warranty, expressed or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe privately owned rights.
Conceptualization: M.S., A.N.P., M.R.I., M.G.T.; methodology: M.S., A.N.P., M.R.I., M.G.T.; investigation: M.S., A.N.P., M.R.I., E.E.K.B., M.G.T., R.K.; writing – original draft: M.S., A.N.P., M.R.I.; writing – review and editing: all authors; resources and supervision: M.G.T., L.M.B., A.M.D., P.M.S., and J.D.K.
J.D.K. has financial interests in Amyris, Ansa Biotechnologies, Apertor Pharma, Berkeley Yeast, Demetrix, Lygos, Napigen, ResVita Bio, and Zero Acre Farms.
Footnotes
[This article was published on 14 March 2022 with a different version of Fig. 5. A revised version of Fig. 5 was posted on 22 March 2022.]
Supplemental material is available online only.
Contributor Information
Jay D. Keasling, Email: jdkeasling@lbl.gov.
Ning-Yi Zhou, Shanghai Jiao Tong University.
REFERENCES
- 1.López-Farfán D, Reyes-Darias JA, Matilla MA, Krell T. 2019. concentration dependent effect of plant root exudates on the chemosensory systems of pseudomonas putida KT2440. Front Microbiol 10:78. 10.3389/fmicb.2019.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernández M, Morel B, Corral-Lugo A, Krell T. 2016. Identification of a chemoreceptor that specifically mediates chemotaxis toward metabolizable purine derivatives. Mol Microbiol 99:34–42. 10.1111/mmi.13215. [DOI] [PubMed] [Google Scholar]
- 3.Neal AL, Ahmad S, Gordon-Weeks R, Ton J. 2012. Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS One 7:e35498. 10.1371/journal.pone.0035498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neal AL, Ton J. 2013. Systemic defense priming by Pseudomonas putida KT2440 in maize depends on benzoxazinoid exudation from the roots. Plant Signal Behav 8:e22655. 10.4161/psb.22655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hervás AB, Canosa I, Santero E. 2008. Transcriptome analysis of Pseudomonas putida in response to nitrogen availability. J Bacteriol 190:416–420. 10.1128/JB.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poblete-Castro I, Escapa IF, Jäger C, Puchalka J, Lam CMC, Schomburg D, Prieto MA, Martins dos Santos VAP. 2012. The metabolic response of P putida KT2442 producing high levels of polyhydroxyalkanoate under single- and multiple-nutrient-limited growth: highlights from a multi-level omics approach. Microb Cell Fact 11:34. 10.1186/1475-2859-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mozejko-Ciesielska J, Dabrowska D, Szalewska-Palasz A, Ciesielski S. 2017. Medium-chain-length polyhydroxyalkanoates synthesis by Pseudomonas putida KT2440 relA/spoT mutant: bioprocess characterization and transcriptome analysis. AMB Express 7:92. 10.1186/s13568-017-0396-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galman JL, Slabu I, Weise NJ, Iglesias C, Parmeggiani F, Lloyd RC, Turner NJ. 2017. Biocatalytic transamination with near-stoichiometric inexpensive amine donors mediated by bifunctional mono- and di-amine transaminases. Green Chem 19:361–366. 10.1039/C6GC02102F. [DOI] [Google Scholar]
- 9.Han T, Kim GB, Lee SY. 2020. Glutaric acid production by systems metabolic engineering of an l-lysine-overproducing Corynebacterium glutamicum. Proc Natl Acad Sci USA 117:30328–30334. 10.1073/pnas.2017483117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson MG, Pearson AN, Barajas JF, Cruz-Morales P, Sedaghatian N, Costello Z, Garber ME, Incha MR, Valencia LE, Baidoo EEK, Martin HG, Mukhopadhyay A, Keasling JD. 2020. Identification, characterization, and application of a highly sensitive lactam biosensor from pseudomonas putida. ACS Synth Biol 9:53–62. 10.1021/acssynbio.9b00292. [DOI] [PubMed] [Google Scholar]
- 11.Banerjee D, Eng T, Lau AK, Sasaki Y, Wang B, Chen Y, Prahl J-P, Singan VR, Herbert RA, Liu Y, Tanjore D, Petzold CJ, Keasling JD, Mukhopadhyay A. 2020. Genome-scale metabolic rewiring improves titers rates and yields of the non-native product indigoidine at scale. Nat Commun 11:5385. 10.1038/s41467-020-19171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nogales J, Mueller J, Gudmundsson S, Canalejo FJ, Duque E, Monk J, Feist AM, Ramos JL, Niu W, Palsson BO. 2020. High-quality genome-scale metabolic modelling of Pseudomonas putida highlights its broad metabolic capabilities. Environ Microbiol 22:255–269. 10.1111/1462-2920.14843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson MG, Incha MR, Pearson AN, Schmidt M, Sharpless WA, Eiben CB, Cruz-Morales P, Blake-Hedges JM, Liu Y, Adams CA, Haushalter RW, Krishna RN, Lichtner P, Blank LM, Mukhopadhyay A, Deutschbauer AM, Shih PM, Keasling JD. 2020. Fatty acid and alcohol metabolism in Pseudomonas putida: functional analysis using random barcode transposon sequencing. Appl Environ Microbiol 86. 10.1128/AEM.01665-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Incha MR, Thompson MG, Blake-Hedges JM, Liu Y, Pearson AN, Schmidt M, Gin JW, Petzold CJ, Deutschbauer AM, Keasling JD. 2020. Leveraging host metabolism for bisdemethoxycurcumin production in Pseudomonas putida. Metab Eng Commun 10:e00119. 10.1016/j.mec.2019.e00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson MG, Blake-Hedges JM, Cruz-Morales P, Barajas JF, Curran SC, Eiben CB, Harris NC, Benites VT, Gin JW, Sharpless WA, Twigg FF, Skyrud W, Krishna RN, Pereira JH, Baidoo EEK, Petzold CJ, Adams PD, Arkin AP, Deutschbauer AM, Keasling JD. 2019. Massively parallel fitness profiling reveals multiple novel enzymes in Pseudomonas putida lysine metabolism. mBio 10. 10.1128/mBio.02577-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson MG, Valencia LE, Blake-Hedges JM, Cruz-Morales P, Velasquez AE, Pearson AN, Sermeno LN, Sharpless WA, Benites VT, Chen Y, Baidoo EEK, Petzold CJ, Deutschbauer AM, Keasling JD. 2019. Omics-driven identification and elimination of valerolactam catabolism in Pseudomonas putida KT2440 for increased product titer. Metab Eng Commun 9:e00098. 10.1016/j.mec.2019.e00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gumerov VM, Ortega DR, Adebali O, Ulrich LE, Zhulin IB. 2020. MiST 3.0: an updated microbial signal transduction database with an emphasis on chemosensory systems. Nucleic Acids Res 48:D459–D464. 10.1093/nar/gkz988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elbourne LDH, Tetu SG, Hassan KA, Paulsen IT. 2017. TransportDB 2.0: a database for exploring membrane transporters in sequenced genomes from all domains of life. Nucleic Acids Res 45:D320–D324. 10.1093/nar/gkw1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Maaten L, Hinton G. 2008. Visualizing data using t-SNE. J Machine Learning Res 9:2579–2609. [Google Scholar]
- 20.Price MN, Wetmore KM, Waters RJ, Callaghan M, Ray J, Liu H, Kuehl JV, Melnyk RA, Lamson JS, Suh Y, Carlson HK, Esquivel Z, Sadeeshkumar H, Chakraborty R, Zane GM, Rubin BE, Wall JD, Visel A, Bristow J, Blow MJ, Arkin AP, Deutschbauer AM. 2018. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557:503–509. 10.1038/s41586-018-0124-0. [DOI] [PubMed] [Google Scholar]
- 21.Sonawane AM, Röhm KH. 2004. A functional gltB gene is essential for utilization of acidic amino acids and expression of periplasmic glutaminase/asparaginase (PGA) by Pseudomonas putida KT2440. Mol Genet Genomics 271:33–39. 10.1007/s00438-003-0951-y. [DOI] [PubMed] [Google Scholar]
- 22.Eberl L, Ammendola A, Rothballer MH, Givskov M, Sternberg C, Kilstrup M, Schleifer KH, Molin S. 2000. Inactivation of gltB abolishes expression of the assimilatory nitrate reductase gene (nasB) in Pseudomonas putida KT2442. J Bacteriol 182:3368–3376. 10.1128/JB.182.12.3368-3376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reitzer L. 2003. Nitrogen assimilation and global regulation in Escherichia coli. Annu Rev Microbiol 57:155–176. 10.1146/annurev.micro.57.030502.090820. [DOI] [PubMed] [Google Scholar]
- 24.Venturi V. 2003. Control of rpoS transcription in Escherichia coli and Pseudomonas: why so different? Mol Microbiol 49–:1–9. 10.1046/j.1365-2958.2003.03547.x. [DOI] [PubMed] [Google Scholar]
- 25.Lapouge K, Schubert M, Allain FH-T, Haas D. 2008. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol 67:241–253. 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 26.Martínez-Gil M, Ramos-González MI, Espinosa-Urgel M. 2014. Roles of cyclic Di-GMP and the Gac system in transcriptional control of the genes coding for the Pseudomonas putida adhesins LapA and LapF. J Bacteriol 196:1484–1495. 10.1128/JB.01287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bentley GJ, Narayanan N, Jha RK, Salvachúa D, Elmore JR, Peabody GL, Black BA, Ramirez K, De Capite A, Michener WE, Werner AZ, Klingeman DM, Schindel HS, Nelson R, Foust L, Guss AM, Dale T, Johnson CW, Beckham GT. 2020. Engineering glucose metabolism for enhanced muconic acid production in Pseudomonas putida KT2440. Metab Eng 59:64–75. 10.1016/j.ymben.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Eng T, Banerjee D, Lau AK, Bowden E, Herbert RA, Trinh J, Prahl J-P, Deutschbauer A, Tanjore D, Mukhopadhyay A. 2021. Engineering Pseudomonas putida for efficient aromatic conversion to bioproduct using high throughput screening in a bioreactor. Metab Eng 66:229–238. 10.1016/j.ymben.2021.04.015. [DOI] [PubMed] [Google Scholar]
- 29.Demling P, Ankenbauer A, Klein B, Noack S, Tiso T, Takors R, Blank LM. 2021. Pseudomonas putida KT2440 endures temporary oxygen limitations. Biotechnol Bioeng 118:4735–4750. 10.1002/bit.27938. [DOI] [PubMed] [Google Scholar]
- 30.Merrick MJ, Edwards RA. 1995. Nitrogen control in bacteria. Microbiol Rev 59:604–622. 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pino C, Olmo-Mira F, Cabello P, Martínez-Luque M, Castillo F, Roldán MD, Moreno-Vivián C. 2006. The assimilatory nitrate reduction system of the phototrophic bacterium Rhodobacter capsulatus E1F1. Biochem Soc Trans 34:127–129. 10.1042/BST0340127. [DOI] [PubMed] [Google Scholar]
- 32.Luque-Almagro VM, Gates AJ, Moreno-Vivián C, Ferguson SJ, Richardson DJ, Roldán MD. 2011. Bacterial nitrate assimilation: gene distribution and regulation. Biochem Soc Trans 39:1838–1843. 10.1042/BST20110688. [DOI] [PubMed] [Google Scholar]
- 33.Caballero A, Esteve-Núñez A, Zylstra GJ, Ramos JL. 2005. Assimilation of nitrogen from nitrite and trinitrotoluene in Pseudomonas putida JLR11. J Bacteriol 187:396–399. 10.1128/JB.187.1.396-399.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romeo A, Sonnleitner E, Sorger-Domenigg T, Nakano M, Eisenhaber B, Bläsi U. 2012. Transcriptional regulation of nitrate assimilation in Pseudomonas aeruginosa occurs via transcriptional antitermination within the nirBD-PA1779-cobA operon. Microbiology (Reading) 158:1543–1552. 10.1099/mic.0.053850-0. [DOI] [PubMed] [Google Scholar]
- 35.Wang B, Rensing C, Pierson LS, Zhao H, Kennedy C. 2014. Translational coupling of nasST expression in Azotobacter vinelandii prevents overexpression of the nasT gene. FEMS Microbiol Lett 361:123–130. 10.1111/1574-6968.12621. [DOI] [PubMed] [Google Scholar]
- 36.Maeda S, Okamura M, Kobayashi M, Omata T. 1998. Nitrite-specific active transport system of the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol 180:6761–6763. 10.1128/JB.180.24.6761-6763.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao S-H, Fan L, Yuan Z, Bond PL. 2015. The concentration-determined and population-specific antimicrobial effects of free nitrous acid on Pseudomonas aeruginosa PAO1. Appl Microbiol Biotechnol 99:2305–2312. 10.1007/s00253-014-6211-8. [DOI] [PubMed] [Google Scholar]
- 38.Kusumawardhani H, van Dijk D, Hosseini R, de Winde JH. 2020. Novel toxin-antitoxin module SlvT-SlvA regulates megaplasmid stability and incites solvent tolerance in Pseudomonas putida S12. Appl Environ Microbiol 86. 10.1128/AEM.00686-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skjerning RB, Senissar M, Winther KS, Gerdes K, Brodersen DE. 2019. The RES domain toxins of RES-Xre toxin-antitoxin modules induce cell stasis by degrading NAD+. Mol Microbiol 111:221–236. 10.1111/mmi.14150. [DOI] [PubMed] [Google Scholar]
- 40.Fuhrer T, Fischer E, Sauer U. 2005. Experimental identification and quantification of glucose metabolism in seven bacterial species. J Bacteriol 187:1581–1590. 10.1128/JB.187.5.1581-1590.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikel PI, Fuhrer T, Chavarría M, Sánchez-Pascuala A, Sauer U, de Lorenzo V. 2021. Reconfiguration of metabolic fluxes in Pseudomonas putida as a response to sub-lethal oxidative stress. ISME J 15:1751–1766. 10.1038/s41396-020-00884-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.del Castillo T, Ramos JL, Rodríguez-Herva JJ, Fuhrer T, Sauer U, Duque E. 2007. Convergent peripheral pathways catalyze initial glucose catabolism in Pseudomonas putida: genomic and flux analysis. J Bacteriol 189:5142–5152. 10.1128/JB.00203-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roon RJ, Levenberg B. 1968. An adenosine triphosphate-dependent, avidin-sensitive enzymatic cleavage of urea in yeast and green algae. J Biol Chem 243:5213–5215. [PubMed] [Google Scholar]
- 44.Roon RJ, Levenberg B. 1972. Urea amidolyase. I. Properties of the enzyme from Candida utilis. J Biol Chem 247:4107–4113. [PubMed] [Google Scholar]
- 45.Mackay EM, Pateman JA. 1982. The regulation of urease activity in Aspergillus nidulans. Biochem Genet 20:763–776. 10.1007/BF00483972. [DOI] [PubMed] [Google Scholar]
- 46.Kanamori T, Kanou N, Atomi H, Imanaka T. 2004. Enzymatic characterization of a prokaryotic urea carboxylase. J Bacteriol 186:2532–2539. 10.1128/JB.186.9.2532-2539.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott E, Peter F, Sanders J. 2007. Biomass in the manufacture of industrial products–the use of proteins and amino acids. Appl Microbiol Biotechnol 75:751–762. 10.1007/s00253-007-0932-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hermann T. 2003. Industrial production of amino acids by coryneform bacteria. J Biotechnol 104:155–172. 10.1016/s0168-1656(03)00149-4. [DOI] [PubMed] [Google Scholar]
- 49.Nijkamp K, Westerhof RGM, Ballerstedt H, de Bont JAM, Wery J. 2007. Optimization of the solvent-tolerant Pseudomonas putida S12 as host for the production of p-coumarate from glucose. Appl Microbiol Biotechnol 74:617–624. 10.1007/s00253-006-0703-0. [DOI] [PubMed] [Google Scholar]
- 50.Craig JW, Chang F-Y, Kim JH, Obiajulu SC, Brady SF. 2010. Expanding small-molecule functional metagenomics through parallel screening of broad-host-range cosmid environmental DNA libraries in diverse proteobacteria. Appl Environ Microbiol 76:1633–1641. 10.1128/AEM.02169-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verhoef S, Ballerstedt H, Volkers RJM, de Winde JH, Ruijssenaars HJ. 2010. Comparative transcriptomics and proteomics of p-hydroxybenzoate producing Pseudomonas putida S12: novel responses and implications for strain improvement. Appl Microbiol Biotechnol 87:679–690. 10.1007/s00253-010-2626-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xing X, Jiang P. April 2011. Recombinant bacteria for producing deoxyviolacein and uses thereof. US patent 20110183384A1.
- 53.Schmitz S, Nies S, Wierckx N, Blank LM, Rosenbaum MA. 2015. Engineering mediator-based electroactivity in the obligate aerobic bacterium Pseudomonas putida KT2440. Front Microbiol 6:284. 10.3389/fmicb.2015.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Molina-Santiago C, Cordero BF, Daddaoua A, Udaondo Z, Manzano J, Valdivia M, Segura A, Ramos J-L, Duque E. 2016. Pseudomonas putida as a platform for the synthesis of aromatic compounds. Microbiology (Reading) 162:1535–1543. 10.1099/mic.0.000333. [DOI] [PubMed] [Google Scholar]
- 55.Aboulmagd E, Voss I, Oppermann-Sanio FB, Steinbüchel A. 2001. Heterologous expression of cyanophycin synthetase and cyanophycin synthesis in the industrial relevant bacteria Corynebacterium glutamicum and Ralstonia eutropha and in Pseudomonas putida. Biomacromolecules 2:1338–1342. 10.1021/bm010075a. [DOI] [PubMed] [Google Scholar]
- 56.Voss I, Diniz SC, Aboulmagd E, Steinbüchel A. 2004. Identification of the Anabaena sp. strain PCC7120 cyanophycin synthetase as suitable enzyme for production of cyanophycin in gram-negative bacteria like Pseudomonas putida and Ralstonia eutropha. Biomacromolecules 5:1588–1595. 10.1021/bm049861g. [DOI] [PubMed] [Google Scholar]
- 57.Wiefel L, Bröker A, Steinbüchel A. 2011. Synthesis of a citrulline-rich cyanophycin by use of Pseudomonas putida ATCC 4359. Appl Microbiol Biotechnol 90:1755–1762. 10.1007/s00253-011-3224-4. [DOI] [PubMed] [Google Scholar]
- 58.Loeschcke A, Thies S. 2015. Pseudomonas putida-a versatile host for the production of natural products. Appl Microbiol Biotechnol 99:6197–6214. 10.1007/s00253-015-6745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Radkov AD, Moe LA. 2013. Amino acid racemization in Pseudomonas putida KT2440. J Bacteriol 195:5016–5024. 10.1128/JB.00761-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mercenier A, Simon JP, Haas D, Stalon V. 1980. Catabolism of L-arginine by Pseudomonas aeruginosa. J Gen Microbiol 116:381–389. 10.1099/00221287-116-2-381. [DOI] [PubMed] [Google Scholar]
- 61.Stalon V, Vander Wauven C, Momin P, Legrain C. 1987. Catabolism of arginine, citrulline and ornithine by Pseudomonas and related bacteria. J Gen Microbiol 133:2487–2495. 10.1099/00221287-133-9-2487. [DOI] [PubMed] [Google Scholar]
- 62.Li C, Lu C-D. 2009. Arginine racemization by coupled catabolic and anabolic dehydrogenases. Proc Natl Acad Sci USA 106:906–911. 10.1073/pnas.0808269106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patil MD, Rathod VP, Bihade UR, Banerjee UC. 2019. Purification and characterization of arginine deiminase from Pseudomonas putida: Structural insights of the differential affinities of l-arginine analogues. J Biosci Bioeng 127:129–137. 10.1016/j.jbiosc.2018.07.021. [DOI] [PubMed] [Google Scholar]
- 64.Vander Wauven C, Jann A, Haas D, Leisinger T, Stalon V. 1988. N2-succinylornithine in ornithine catabolism of Pseudomonas aeruginosa. Arch Microbiol 150:400–404. 10.1007/BF00408314. [DOI] [PubMed] [Google Scholar]
- 65.Tricot C, Vander Wauven C, Wattiez R, Falmagne P, Stalon V. 1994. Purification and properties of a succinyltransferase from Pseudomonas aeruginosa specific for both arginine and ornithine. Eur J Biochem 224:853–861. 10.1111/j.1432-1033.1994.00853.x. [DOI] [PubMed] [Google Scholar]
- 66.Radkov AD, Moe LA. 2018. A broad spectrum racemase in Pseudomonas putida KT2440 plays a key role in amino acid catabolism. Front Microbiol 9. 10.3389/fmicb.2018.01343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang YF, Adams E. 1971. Induction of separate catabolic pathways for L- and D-lysine in Pseudomonas putida. Biochem Biophys Res Commun 45:570–577. 10.1016/0006-291x(71)90455-4. [DOI] [PubMed] [Google Scholar]
- 68.Espinosa-Urgel M, Ramos JL. 2001. Expression of a Pseudomonas putida aminotransferase involved in lysine catabolism is induced in the rhizosphere. Appl Environ Microbiol 67:5219–5224. 10.1128/AEM.67.11.5219-5224.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Revelles O, Espinosa-Urgel M, Molin S, Ramos JL. 2004. The davDT operon of Pseudomonas putida, involved in lysine catabolism, is induced in response to the pathway intermediate delta-aminovaleric acid. J Bacteriol 186:3439–3446. 10.1128/JB.186.11.3439-3446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Revelles O, Espinosa-Urgel M, Fuhrer T, Sauer U, Ramos JL. 2005. Multiple and interconnected pathways for L-lysine catabolism in Pseudomonas putida KT2440. J Bacteriol 187:7500–7510. 10.1128/JB.187.21.7500-7510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Revelles O, Wittich R-M, Ramos JL. 2007. Identification of the initial steps in D-lysine catabolism in Pseudomonas putida. J Bacteriol 189:2787–2792. 10.1128/JB.01538-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Radkov AD, Moe LA. 2014. Bacterial synthesis of D-amino acids. Appl Microbiol Biotechnol 98:5363–5374. 10.1007/s00253-014-5726-3. [DOI] [PubMed] [Google Scholar]
- 73.Duque E, Daddaoua A, Cordero BF, De la Torre J, Antonia Molina-Henares M, Ramos J-L. 2017. Identification and elucidation of in vivo function of two alanine racemases from Pseudomonas putida KT2440. Environ Microbiol Rep 9:581–588. 10.1111/1758-2229.12576. [DOI] [PubMed] [Google Scholar]
- 74.Boulette ML, Baynham PJ, Jorth PA, Kukavica-Ibrulj I, Longoria A, Barrera K, Levesque RC, Whiteley M. 2009. Characterization of alanine catabolism in Pseudomonas aeruginosa and its importance for proliferation in vivo. J Bacteriol 191:6329–6334. 10.1128/JB.00817-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhi J, Mathew E, Freundlich M. 1999. Lrp binds to two regions in the dadAX promoter region of Escherichia coli to repress and activate transcription directly. Mol Microbiol 32:29–40. 10.1046/j.1365-2958.1999.01314.x. [DOI] [PubMed] [Google Scholar]
- 76.Schwanemann T, Otto M, Wierckx N, Wynands B. 2020. Pseudomonas as versatile aromatics cell factory. Biotechnol J 15:e1900569. 10.1002/biot.201900569. [DOI] [PubMed] [Google Scholar]
- 77.Whitaker RJ, Gaines CG, Jensen RA. 1982. A multispecific quintet of aromatic aminotransferases that overlap different biochemical pathways in Pseudomonas aeruginosa. J Biol Chem 257:13550–13556. [PubMed] [Google Scholar]
- 78.Molina-Henares MA, García-Salamanca A, Molina-Henares AJ, de la Torre J, Herrera MC, Ramos JL, Duque E. 2009. Functional analysis of aromatic biosynthetic pathways in Pseudomonas putida KT2440. Microb Biotechnol 2:91–100. 10.1111/j.1751-7915.2008.00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pittard J, Yang J. 2008. Biosynthesis of the aromatic amino acids. Ecosal Plus 3. 10.1128/ecosalplus.3.6.1.8. [DOI] [PubMed] [Google Scholar]
- 80.Nogales J, Palsson BØ, Thiele I. 2008. A genome-scale metabolic reconstruction of Pseudomonas putida KT2440: iJN746 as a cell factory. BMC Syst Biol 2:79. 10.1186/1752-0509-2-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leprince A, de Lorenzo V, Völler P, van Passel MWJ, Martins dos Santos VAP. 2012. Random and cyclical deletion of large DNA segments in the genome of Pseudomonas putida. Environ Microbiol 14:1444–1453. 10.1111/j.1462-2920.2012.02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gadilohar BL, Shankarling GS. 2017. Choline based ionic liquids and their applications in organic transformation. J Mol Liq 227:234–261. 10.1016/j.molliq.2016.11.136. [DOI] [Google Scholar]
- 83.Wood JM. 2011. Bacterial osmoregulation: a paradigm for the study of cellular homeostasis. Annu Rev Microbiol 65:215–238. 10.1146/annurev-micro-090110-102815. [DOI] [PubMed] [Google Scholar]
- 84.Wargo MJ. 2013. Homeostasis and catabolism of choline and glycine betaine: lessons from Pseudomonas aeruginosa. Appl Environ Microbiol 79:2112–2120. 10.1128/AEM.03565-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Geiger O, López-Lara IM, Sohlenkamp C. 2013. Phosphatidylcholine biosynthesis and function in bacteria. Biochim Biophys Acta 1831:503–513. 10.1016/j.bbalip.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 86.Belda E, van Heck RGA, José Lopez-Sanchez M, Cruveiller S, Barbe V, Fraser C, Klenk H-P, Petersen J, Morgat A, Nikel PI, Vallenet D, Rouy Z, Sekowska A, Martins Dos Santos VAP, de Lorenzo V, Danchin A, Médigue C. 2016. The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environ Microbiol 18:3403–3424. 10.1111/1462-2920.13230. [DOI] [PubMed] [Google Scholar]
- 87.Leys D, Basran J, Scrutton NS. 2003. Channelling and formation of “active” formaldehyde in dimethylglycine oxidase. EMBO J 22:4038–4048. 10.1093/emboj/cdg395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wargo MJ, Hogan DA. 2009. Identification of genes required for Pseudomonas aeruginosa carnitine catabolism. Microbiology (Reading) 155:2411–2419. 10.1099/mic.0.028787-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bastard K, Smith AAT, Vergne-Vaxelaire C, Perret A, Zaparucha A, De Melo-Minardi R, Mariage A, Boutard M, Debard A, Lechaplais C, Pelle C, Pellouin V, Perchat N, Petit J-L, Kreimeyer A, Medigue C, Weissenbach J, Artiguenave F, De Berardinis V, Vallenet D, Salanoubat M. 2014. Revealing the hidden functional diversity of an enzyme family. Nat Chem Biol 10:42–49. 10.1038/nchembio.1387. [DOI] [PubMed] [Google Scholar]
- 90.Meadows JA, Wargo MJ. 2018. Transcriptional regulation of carnitine catabolism in Pseudomonas aeruginosa by CdhR. mSphere 3. 10.1128/mSphere.00480-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee S, Takahashi Y, Oura H, Suzuki-Minakuchi C, Okada K, Yamane H, Nomura N, Nojiri H. 2016. Effects of carbazole-degradative plasmid pCAR1 on biofilm morphology in Pseudomonas putida KT2440. Environ Microbiol Rep 8:261–271. 10.1111/1758-2229.12376. [DOI] [PubMed] [Google Scholar]
- 92.Nuccio ML, Russell BL, Nolte KD, Rathinasabapathi B, Gage DA, Hanson AD. 1998. The endogenous choline supply limits glycine betaine synthesis in transgenic tobacco expressing choline monooxygenase. Plant J 16:487–496. 10.1046/j.1365-313x.1998.00316.x. [DOI] [PubMed] [Google Scholar]
- 93.Rontein D, Nishida I, Tashiro G, Yoshioka K, Wu WI, Voelker DR, Basset G, Hanson AD. 2001. Plants synthesize ethanolamine by direct decarboxylation of serine using a pyridoxal phosphate enzyme. J Biol Chem 276:35523–35529. 10.1074/jbc.M106038200. [DOI] [PubMed] [Google Scholar]
- 94.Lundgren BR, Sarwar Z, Pinto A, Ganley JG, Nomura CT. 2016. Ethanolamine catabolism in Pseudomonas aeruginosa PAO1 is regulated by the enhancer-binding protein EatR (PA4021) and the alternative sigma factor RpoN. J Bacteriol 198:2318–2329. 10.1128/JB.00357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roof DM, Roth JR. 1988. Ethanolamine utilization in Salmonella typhimurium. J Bacteriol 170:3855–3863. 10.1128/jb.170.9.3855-3863.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chang GW, Chang JT. 1975. Evidence for the B12-dependent enzyme ethanolamine deaminase in Salmonella. Nature 254:150–151. 10.1038/254150a0. [DOI] [PubMed] [Google Scholar]
- 97.Scarlett FA, Turner JM. 1976. Microbial metabolism of amino alcohols. Ethanolamine catabolism mediated by coenzyme B12-dependent ethanolamine ammonia-lyase in Escherichia coli and Klebsiella aerogenes. J Gen Microbiol 95:173–176. 10.1099/00221287-95-1-173. [DOI] [PubMed] [Google Scholar]
- 98.Cameron B, Briggs K, Pridmore S, Brefort G, Crouzet J. 1989. Cloning and analysis of genes involved in coenzyme B12 biosynthesis in Pseudomonas denitrificans. J Bacteriol 171:547–557. 10.1128/jb.171.1.547-557.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.LaBauve AE, Wargo MJ. 2012. Growth and laboratory maintenance of Pseudomonas aeruginosa. Curr Protoc Microbiol Chapter 6:Unit 6E.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Heller K, Mann BJ, Kadner RJ. 1985. Cloning and expression of the gene for the vitamin B12 receptor protein in the outer membrane of Escherichia coli. J Bacteriol 161:896–903. 10.1128/jb.161.3.896-903.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cadieux N, Bradbeer C, Reeger-Schneider E, Köster W, Mohanty AK, Wiener MC, Kadner RJ. 2002. Identification of the periplasmic cobalamin-binding protein BtuF of Escherichia coli. J Bacteriol 184:706–717. 10.1128/JB.184.3.706-717.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Warren MJ, Raux E, Schubert HL, Escalante-Semerena JC. 2002. The biosynthesis of adenosylcobalamin (vitamin B12). Nat Prod Rep 19:390–412. 10.1039/b108967f. [DOI] [PubMed] [Google Scholar]
- 103.Martens JH, Barg H, Warren MJ, Jahn D. 2002. Microbial production of vitamin B12. Appl Microbiol Biotechnol 58:275–285. 10.1007/s00253-001-0902-7. [DOI] [PubMed] [Google Scholar]
- 104.Zayas CL, Escalante-Semerena JC. 2007. Reassessment of the late steps of coenzyme B12 synthesis in Salmonella enterica: evidence that dephosphorylation of adenosylcobalamin-5’-phosphate by the CobC phosphatase is the last step of the pathway. J Bacteriol 189:2210–2218. 10.1128/JB.01665-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mori K, Bando R, Hieda N, Toraya T. 2004. Identification of a reactivating factor for adenosylcobalamin-dependent ethanolamine ammonia lyase. J Bacteriol 186:6845–6854. 10.1128/JB.186.20.6845-6854.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vogels GD, Van der Drift C. 1976. Degradation of purines and pyrimidines by microorganisms. Bacteriol Rev 40:403–468. 10.1128/br.40.2.403-468.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tipton PA. 2006. Urate to allantoin, specifically (S)-allantoin. Nat Chem Biol 2:124–125. 10.1038/nchembio0306-124. [DOI] [PubMed] [Google Scholar]
- 108.Ramazzina I, Folli C, Secchi A, Berni R, Percudani R. 2006. Completing the uric acid degradation pathway through phylogenetic comparison of whole genomes. Nat Chem Biol 2:144–148. 10.1038/nchembio768. [DOI] [PubMed] [Google Scholar]
- 109.Hassan KA, Liu Q, Henderson PJF, Paulsen IT. 2015. Homologs of the Acinetobacter baumannii AceI transporter represent a new family of bacterial multidrug efflux systems. mBio 6. 10.1128/mBio.01982-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Juttukonda LJ, Green ER, Lonergan ZR, Heffern MC, Chang CJ, Skaar EP. 2019. Acinetobacter baumannii OxyR regulates the transcriptional response to hydrogen peroxide. Infect Immun 87. 10.1128/IAI.00413-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zimmer DP, Soupene E, Lee HL, Wendisch VF, Khodursky AB, Peter BJ, Bender RA, Kustu S. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc Natl Acad Sci USA 97:14674–14679. 10.1073/pnas.97.26.14674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ware E. 1950. The chemistry of the hydantoins. Chem Rev 46:403–470. 10.1021/cr60145a001. [DOI] [PubMed] [Google Scholar]
- 113.Shorvon S. 1994. Status epilepticus. Cambridge University Press, Cambridge, UK. [Google Scholar]
- 114.Drauz K, Grayson I, Kleemann A, Krimmer H-P, Leuchtenberger W, Weckbecker C. 2007. Amino acids. Ullmann’s encyclopedia of industrial chemistry. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany. [Google Scholar]
- 115.Ogawa J, Kaimura T, Yamada H, Shimizu S. 1994. Evaluation of pyrimidine- and hydantoin-degrading enzyme activities in aerobic bacteria. FEMS Microbiol Lett 122:55–60. 10.1111/j.1574-6968.1994.tb07143.x. [DOI] [Google Scholar]
- 116.Solem E. 1974. The absolute configuration of beta-aminoisobutyric acid formed by degradation of thymine in man. Clin Chim Acta 53:183–190. 10.1016/0009-8981(74)90097-7. [DOI] [PubMed] [Google Scholar]
- 117.Massey LK, Sokatch JR, Conrad RS. 1976. Branched-chain amino acid catabolism in bacteria. Bacteriol Rev 40:42–54. 10.1128/br.40.1.42-54.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang J, Barajas JF, Burdu M, Wang G, Baidoo EE, Keasling JD. 2017. Application of an Acyl-CoA Ligase from Streptomyces aizunensis for Lactam Biosynthesis. ACS Synth Biol 6:884–890. 10.1021/acssynbio.6b00372. [DOI] [PubMed] [Google Scholar]
- 119.Niehaus TD, Elbadawi-Sidhu M, de Crécy-Lagard V, Fiehn O, Hanson AD. 2017. Discovery of a widespread prokaryotic 5-oxoprolinase that was hiding in plain sight. J Biol Chem 292:16360–16367. 10.1074/jbc.M117.805028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Miller DL, Rodwell VW. 1971. Metabolism of basic amino acids in Pseudomonas putida. Intermediates in L-arginine catabolism. J Biol Chem 246:5053–5058. [PubMed] [Google Scholar]
- 121.Bandounas L, Ballerstedt H, de Winde JH, Ruijssenaars HJ. 2011. Redundancy in putrescine catabolism in solvent tolerant Pseudomonas putida S12. J Biotechnol 154:1–10. 10.1016/j.jbiotec.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 122.Chae TU, Ko Y-S, Hwang K-S, Lee SY. 2017. Metabolic engineering of Escherichia coli for the production of four-, five- and six-carbon lactams. Metab Eng 41:82–91. 10.1016/j.ymben.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 123.Tiso T, Narancic T, Wei R, Pollet E, Beagan N, Schröder K, Honak A, Jiang M, Kenny ST, Wierckx N, Perrin R, Avérous L, Zimmermann W, O'Connor K, Blank LM. 2021. Towards bio-upcycling of polyethylene terephthalate. Metab Eng 66:167–178. 10.1016/j.ymben.2021.03.011. [DOI] [PubMed] [Google Scholar]
- 124.Tabor CW, Tabor H. 1985. Polyamines in microorganisms. Microbiol Rev 49:81–99. 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gaymans RJ, Van Utteren TEC, Van Den Berg JWA, Schuyer J. 1977. Preparation and some properties of nylon 46. J Polym Sci Polym Chem Ed 15:537–545. 10.1002/pol.1977.170150303. [DOI] [Google Scholar]
- 126.Xu F, Zhong L, Xu Y, Zhang C, Zhang F, Zhang G. 2019. Highly efficient flame-retardant and soft cotton fabric prepared by a novel reactive flame retardant. Cellulose 26:4225–4240. 10.1007/s10570-019-02374-4. [DOI] [Google Scholar]
- 127.Roberts MR. 2007. Does GABA act as a signal in plants?: hints from molecular studies. Plant Signal Behav 2:408–409. 10.4161/psb.2.5.4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bouché N, Fromm H. 2004. GABA in plants: just a metabolite? Trends Plant Sci 9:110–115. 10.1016/j.tplants.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 129.Reyes-Darias JA, García V, Rico-Jiménez M, Corral-Lugo A, Lesouhaitier O, Juárez-Hernández D, Yang Y, Bi S, Feuilloley M, Muñoz-Rojas J, Sourjik V, Krell T. 2015. Specific gamma-aminobutyrate chemotaxis in pseudomonads with different lifestyle. Mol Microbiol 97:488–501. 10.1111/mmi.13045. [DOI] [PubMed] [Google Scholar]
- 130.Kinnersley AM, Turano FJ. 2000. Gamma aminobutyric acid (GABA) and plant responses to stress. CRC Crit Rev Plant Sci 19:479–509. 10.1080/07352680091139277. [DOI] [Google Scholar]
- 131.Bartsch K, von Johnn-Marteville A, Schulz A. 1990. Molecular analysis of two genes of the Escherichia coli gab cluster: nucleotide sequence of the glutamate:succinic semialdehyde transaminase gene (gabT) and characterization of the succinic semialdehyde dehydrogenase gene (gabD). J Bacteriol 172:7035–7042. 10.1128/jb.172.12.7035-7042.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Weber N, Hatsch A, Labagnere L, Heider H. 2017. Production of (S)-2-aminobutyric acid and (S)-2-aminobutanol in Saccharomyces cerevisiae. Microb Cell Fact 16:51. 10.1186/s12934-017-0667-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang K, Li H, Cho KM, Liao JC. 2010. Expanding metabolism for total biosynthesis of the nonnatural amino acid L-homoalanine. Proc Natl Acad Sci USA 107:6234–6239. 10.1073/pnas.0912903107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Soga T, Baran R, Suematsu M, Ueno Y, Ikeda S, Sakurakawa T, Kakazu Y, Ishikawa T, Robert M, Nishioka T, Tomita M. 2006. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J Biol Chem 281:16768–16776. 10.1074/jbc.M601876200. [DOI] [PubMed] [Google Scholar]
- 135.Ewering C, Heuser F, Benölken JK, Brämer CO, Steinbüchel A. 2006. Metabolic engineering of strains of Ralstonia eutropha and Pseudomonas putida for biotechnological production of 2-methylcitric acid. Metab Eng 8:587–602. 10.1016/j.ymben.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 136.Cohen YR. 2002. β-Aminobutyric acid-induced resistance against plant pathogens. Plant Dis 86:448–457. 10.1094/PDIS.2002.86.5.448. [DOI] [PubMed] [Google Scholar]
- 137.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, Wang J, Cong Q, Kinch LN, Schaeffer RD, Millán C, Park H, Adams C, Glassman CR, DeGiovanni A, Pereira JH, Rodrigues AV, van Dijk AA, Ebrecht AC, Opperman DJ, Sagmeister T, Buhlheller C, Pavkov-Keller T, Rathinaswamy MK, Dalwadi U, Yip CK, Burke JE, Garcia KC, Grishin NV, Adams PD, Read RJ, Baker D. 2021. Accurate prediction of protein structures and interactions using a three-track neural network. Science 373:871–876. 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ham TS, Dmytriv Z, Plahar H, Chen J, Hillson NJ, Keasling JD. 2012. Design, implementation and practice of JBEI-ICE: an open source biological part registry platform and tools. Nucleic Acids Res 40:e141. 10.1093/nar/gks531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chen J, Densmore D, Ham TS, Keasling JD, Hillson NJ. 2012. DeviceEditor visual biological CAD canvas. J Biol Eng 6:1. 10.1186/1754-1611-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hillson NJ, Rosengarten RD, Keasling JD. 2012. j5 DNA assembly design automation software. ACS Synth Biol 1:14–21. 10.1021/sb2000116. [DOI] [PubMed] [Google Scholar]
- 142.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 143.Wetmore KM, Price MN, Waters RJ, Lamson JS, He J, Hoover CA, Blow MJ, Bristow J, Butland G, Arkin AP, Deutschbauer A. 2015. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. mBio 6:e00306-15–e00315. 10.1128/mBio.00306-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yuzawa S, Eng CH, Katz L, Keasling JD. 2013. Broad substrate specificity of the loading didomain of the lipomycin polyketide synthase. Biochemistry 52:3791–3793. 10.1021/bi400520t. [DOI] [PubMed] [Google Scholar]
- 145.Eudes A, Berthomieu R, Hao Z, Zhao N, Benites VT, Baidoo EEK, Loqué D. 2018. Production of muconic acid in plants. Metab Eng 46:13–19. 10.1016/j.ymben.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 146.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay É. 2011. Scikit-learn: machine learning in Python. J Machine Learning Res 12:2825–2830. [Google Scholar]
- 147.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, van der Walt SJ, Brett M, Wilson J, Millman KJ, Mayorov N, Nelson ARJ, Jones E, Kern R, Larson E, Carey CJ, Polat İ, Feng Y, Moore EW, VanderPlas J, Laxalde D, Perktold J, Cimrman R, Henriksen I, Quintero EA, Harris CR, Archibald AM, Ribeiro AH, Pedregosa F, van Mulbregt P, SciPy 1.0 Contributors . 2020. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 17:261–272. 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Harris CR, Millman KJ, van der Walt SJ, Gommers R, Virtanen P, Cournapeau D, Wieser E, Taylor J, Berg S, Smith NJ, Kern R, Picus M, Hoyer S, van Kerkwijk MH, Brett M, Haldane A, Del Río JF, Wiebe M, Peterson P, Gérard-Marchant P, Sheppard K, Reddy T, Weckesser W, Abbasi H, Gohlke C, Oliphant TE. 2020. Array programming with NumPy. Nature 585:357–362. 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, von Mering C, Bork P. 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47:D309–D314. 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. 2021. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol 38:5825–5829. 10.1093/molbev/msab293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Karp PD, Billington R, Caspi R, Fulcher CA, Latendresse M, Kothari A, Keseler IM, Krummenacker M, Midford PE, Ong Q, Ong WK, Paley SM, Subhraveti P. 2019. The BioCyc collection of microbial genomes and metabolic pathways. Brief Bioinform 20:1085–1093. 10.1093/bib/bbx085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Price MN, Arkin AP. 2017. PaperBLAST: text mining papers for information about homologs. mSystems 2. 10.1128/mSystems.00039-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637. 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 154.Galperin MY, Wolf YI, Makarova KS, Vera Alvarez R, Landsman D, Koonin EV. 2021. COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res 49:D274–D281. 10.1093/nar/gkaa1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nicholas DJD, Nason A. 1954. Molybdenum and nitrate reductase II. Molybdenum as a constituent of nitrate reductase. J Biological Chemistry 207:353–360. 10.1016/S0021-9258(18)71275-1. [DOI] [PubMed] [Google Scholar]
- 156.Shanks RMQ, Kadouri DE, MacEachran DP, O'Toole GA. 2009. New yeast recombineering tools for bacteria. Plasmid 62:88–97. 10.1016/j.plasmid.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rosenberg AH, Lade BN, Chui DS, Lin SW, Dunn JJ, Studier FW. 1987. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene 56:125–135. 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- 158.Bi C, Su P, Müller J, Yeh Y-C, Chhabra SR, Beller HR, Singer SW, Hillson NJ. 2013. Development of a broad-host synthetic biology toolbox for Ralstonia eutropha and its application to engineering hydrocarbon biofuel production. Microb Cell Fact 12:107. 10.1186/1475-2859-12-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lee TS, Krupa RA, Zhang F, Hajimorad M, Holtz WJ, Prasad N, Lee SK, Keasling JD. 2011. BglBrick vectors and datasheets: A synthetic biology platform for gene expression. J Biol Eng 5:12. 10.1186/1754-1611-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S15. Download aem.02430-21-s0001.pdf, PDF file, 3.2 MB (3.2MB, pdf)