Abstract
Ciliated protists are among the oldest unicellular organisms with a heterotrophic lifestyle and share a common ancestor with Plantae. Unlike any other eukaryotes, there are two distinct nuclei in ciliates with separate germline and somatic cell functions. Here, we assembled a near-complete macronuclear genome of Fabrea salina, which belongs to one of the oldest clades of ciliates. Its extremely minimized genome (18.35 Mb) is the smallest among all free-living heterotrophic eukaryotes and exhibits typical streamlined genomic features, including high gene density, tiny introns, and shrinkage of gene paralogs. Gene families involved in hypersaline stress resistance, DNA replication proteins, and mitochondrial biogenesis are expanded, and the accumulation of phosphatidic acid may play an important role in resistance to high osmotic pressure. We further investigated the morphological and transcriptomic changes in the macronucleus during sexual reproduction and highlighted the potential contribution of macronuclear residuals to this process. We believe that the minimized genome generated in this study provides novel insights into the genome streamlining theory and will be an ideal model to study the evolution of eukaryotic heterotrophs.
Keywords: ciliate, genome streamlining, genome evolution
Introduction
With the evolution of prokaryotes into eukaryotes, the complexity of organisms has increased dramatically, but phenotypic complexity is not proportional to genome size. For example, the prokaryotic genome Sorangium cellulosum (14.78 Mb) (Han et al. 2013) is even larger than that of the unicellular eukaryotic green alga Ostreococcus tauri (12.56 Mb) (Derelle et al. 2006). Genome size evolution is deemed to be shaped by distinct directions, that is, expansion or contraction (Koonin 2009). Studies on the genomes of Prochlorococcus (Garcia-Fernández et al. 2004), SAR11 (Grote et al. 2012), and SAR86 (Molloy 2012) have contributed to the genome streamlining theory, which explains the observed small, compact, and relatively AT-rich genomes. The characteristics of a typical streamlined genome include a small genome size, a low ratio of noncoding DNA and low numbers of paralogs (Giovannoni et al. 2014). In theory, a large effective population size is essential for the efficiency of evolutionary selection, and an oligotrophic marine environment is the base of selective pressure (Giovannoni et al. 2014). Additional studies further revealed the ubiquity of streamlined genomes in bacteria and archaea in the ocean (Getz et al. 2018). However, the status of genome streamlining in eukaryotes is not unclear.
Ciliates are among the most diverse clades of unicellular eukaryotes, with 4,500 recorded free-living species and an estimated number of 27,000–40,000 species (Foissner et al. 2007). Ciliates originated approximately 1,100 million years ago (Fernandes and Schrago 2019), and they disperse virtually overall freshwater, marine, and terrestrial environments. An important feature of ciliates is that each cell harbors two types of nuclei, the diploid germline micronucleus (MIC) and the polyploid somatic macronucleus (MAC). High species and population diversity, as well as unusual genome characteristics, make ciliates good model organisms for studies of genome evolution and environmental adaptation. Previous studies on the ciliate Paramecium tetraurelia revealed that it has experienced three whole-genome duplication events, resulting in a somatic genome of 72.09 Mb (Aury et al. 2006). Additionally, the largest somatic genome (117 Mb) among the sequenced ciliate species was reported in Entodinium caudatum (Park et al. 2021). However, the minimum number of genes in the somatic genome required to maintain heterotrophic free-living life remains unknown.
Fabrea salina is a heterotrichous ciliate that dominates in hypersaline environments such as salt marshes, hypersaline lakes, and solar salterns. It can grow and reproduce at almost all salinities in the range of 35–180 ppt (Hotos 2021), and it is highly adaptable to various environmental conditions, with a short generation period and significant resistance to harsh environments (Guermazi et al. 2008). The body length along the long axis of F. salina is approximately 100–350 μm (fig. 1A). The major morphological characteristic of F. salina is that its oral cavity forms a vortex-like S-shaped feature. Under a suitable growth environment, cells proliferate stably by asexual reproduction. In vegetative cells, most of them contain one ribbon-like MAC, while a few may have multiple MACs (supplementary fig. S1A and B, Supplementary Material online). In this study, we employed both Illumina and Nanopore sequencing technologies to assemble the somatic genome of F. salina and described the characteristics of its extremely streamlined genome. By combining both genome and transcriptome data, we analyzed transcriptional changes in macronuclear residuals during sexual reproduction. In addition, through comparative genomic and transcriptomic analyses, we further systematically explored its genetic basis for adaptation to hypersaline environments.
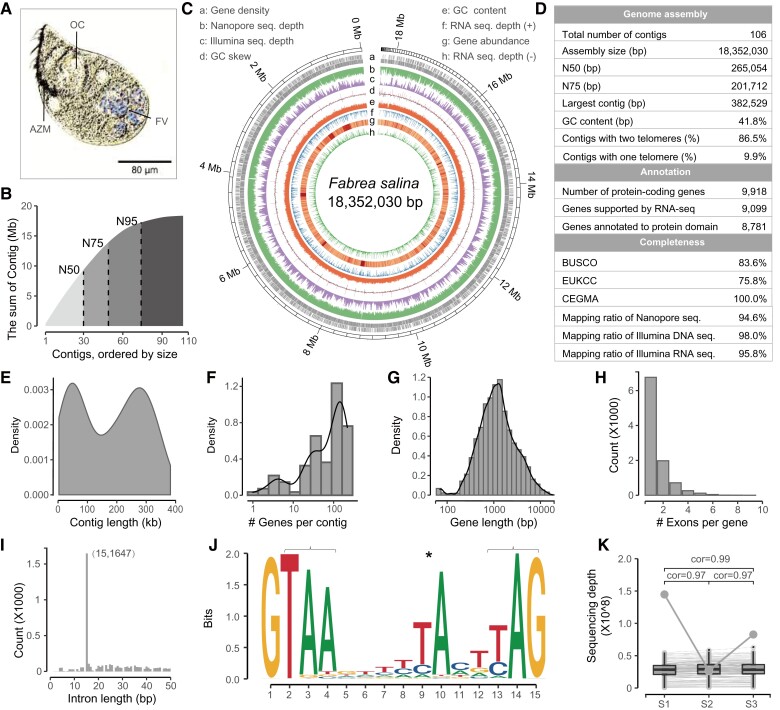
Fig. 1.
Sequencing and assembly of the macronuclear genome of Fabrea salina. (A) Morphology of F. salina in optical microscope. (B) The cumulative distribution of contig length. The N50, N75, and N95 was 265.05, 201.71, and 62.71 kb, respectively. (C) Characteristics of the 106 assembled contigs of F. salina. Tracks a–h represent the distribution of gene density, genome coverage of nanopore reads, genome coverage of Illumina reads, GC skew, GC density, genome coverage of RNA-seq reads in the forward strand (+), gene expression abundance, and genome coverage of RNA-seq reads in the reverse strand (−), respectively, with values calculated in 2 kb sliding windows. (D–I) Statistics on assembly and annotation of the F. salina MAC genome, which shows the distribution of contig length, gene number in each contig, gene length, exon number and intron length. (J) Weblogo plot of 15 bp intron. Asterisk denotes an atypical internal TA dinucleotide, and brackets denote two potential stop codons. (K) Consistency of contig copy number among samples. The bottom value shows the Spearman correlation coefficients between samples. The same contig is connected by a line. The grey dot represents the most variable contig, which is the mitochondrial genome. FV, food vacuoles; OC, oral cavity; AZM, adoral zone of membranelles.
Results and Discussion
Shotgun Sequencing and Assembly of the Macronuclear Genome of F. salina
To address the assembly challenge arising from highly fragmented nanochromosomes of ciliates, we employed both Illumina and Nanopore sequencing technologies to generate short and long reads for F. salina, respectively. Then, we assembled an 18.35 Mb (18,352,030 bp, N50 = 265.05 kb) draft genome based on 185.8 million Illumina paired-end reads (27.86 Gb) and 0.33 million Nanopore long-reads (1.17 Gb) by using a combination of the Flye (Kolmogorov et al. 2019) – Racon (Vaser et al. 2017) – Medaka – Pilon (Walker et al. 2014) pipeline (fig. 1B and C). We found that 97.98 and 94.56% of the Illumina and Nanopore DNA-seq reads, respectively, and 95.82% of 60.5 million RNA-seq reads could be properly aligned to the assembled genome (fig. 1D), indicating its high completeness. Notably, 86 of 106 assembled chromosomes (15.85 Mb) were capped with telomeres. We further used three different approaches (BUSCO [Seppey et al. 2019], EUKCC [Saary et al. 2020] and CEGMA [Parra et al. 2007]) to evaluate its completeness and potential contamination. As shown in figure 1D, the completeness of the assembled F. salina genome was comparable with that of well-studied ciliate model species (supplementary table S1, Supplementary Material online). The GC content distribution of assembled contigs did not show any algal or bacterial contaminants with discordant GC content (supplementary fig. S1C, Supplementary Material online).
Then, the MAKER (Holt and Yandell 2011) pipeline was used to annotate the MAC genome. By combining transcriptome-based, homology-based, and ab initio approaches, we finally obtained 9,918 protein-coding gene models, 89 tRNAs, and 76 rRNAs. Among the predicted protein-coding genes, almost all of them (99.9%) were supported by RNA-seq data, and 88.5% had significant hits in the InterPro database by InterProScan (Jones et al. 2014) (fig. 1D). The gene number in each contig correlates with contig length. The median gene length was 1,145 bp, and each chromosome had an average of 95.3 genes (fig. 1F and G). Most genes (68.2%) contained only one exon, while multiexon genes had 5,156 introns in total (fig. 1H). Most introns are 15 bp (fig. 1I), which is similar to other heterotrich ciliates (e.g., Stentor coeruleus [Slabodnick et al. 2017] and Condylostoma magnum [Swart et al. 2016]). We also found that the F. salina genome contained canonical introns with a highly conserved branch point adenosine (fig. 1J), similar to that in S. coeruleus (Slabodnick et al. 2017). We have scanned the nucleotide sequence and confirmed that the F. salina genome was translated with a standard codon (supplementary table S2, Supplementary Material online).
In addition, we uncovered a high consistency of contig copy number in three F. salina individuals based on their sequencing depth distribution (cor = 0.96) (fig. 1K). We observed that the assembled contig with the greatest copy number variation was the mitochondrial genome (supplementary fig. S1D, Supplementary Material online), which was 42,610 bp and contained 2 rRNA genes, 5 tRNA genes, 26 known protein-coding genes, and 20 unclassified open reading frames. Similar to other sequenced ciliate mitochondrial genomes (Huang et al. 2021), the F. salina mitochondrial genome is a linear molecule.
General Characteristics of the Smallest Genome in Free-Living Protozoa
To characterize the structure of the F. salina MAC genome, nine well-studied ciliate genomes were compared with F. salina. The phylogenetic tree showed that S. coeruleus was a closely related species of F. salina (fig. 2A). The length of the basal branch leading to Heterotrichea was shorter than that of the other classes, suggesting that Heterotrichea was a class of ancient ciliates that experienced more conservative evolution (Fernandes and Schrago 2019). The number of chromosomes in F. salina is similar to that of Oligohymenophorea but fewer than that of Spirotrichea characterized by fragmented genomes termed nanochromosomes. Meanwhile, 96.7% of the F. salina genome was covered by genes (fig. 2A), indicating the greatest nucleotide utilization efficiency and the highest gene density. The gene number of F. salina was much lower than that of other free-living ciliates but comparable with that of the parasitic ciliate Ichthyophthirius multifiliis (fig. 2A). Notably, F. salina has the smallest genome, even smaller than I. multifiliis (48.7 Mb) and the symbiotic ciliate Paramecium bursaria (29.2 Mb). Next, we compared the genome size with all published protozoan genomes and found that F. salina has the smallest genome among all free-living protozoa described thus far (fig. 2B). Its size is even smaller than Tieghemostelium lacteum (23.4 Mb) (Narita et al. 2020), parasitic Plasmodium falciparum (23.5 Mbp) (Moser et al. 2020), and parasitic Entamoeba histolytica (20.8 Mb) (Al-Ardi 2021). Considering that parasites and symbionts are dominated by reductive evolution with extensive losses of genes and introns (Wolf and Koonin 2013), their genomes are usually smaller than their free-living relatives. Unexpectedly, free-living F. salina has the smallest genome, suggesting that it may have undergone unusual genome streamlining.
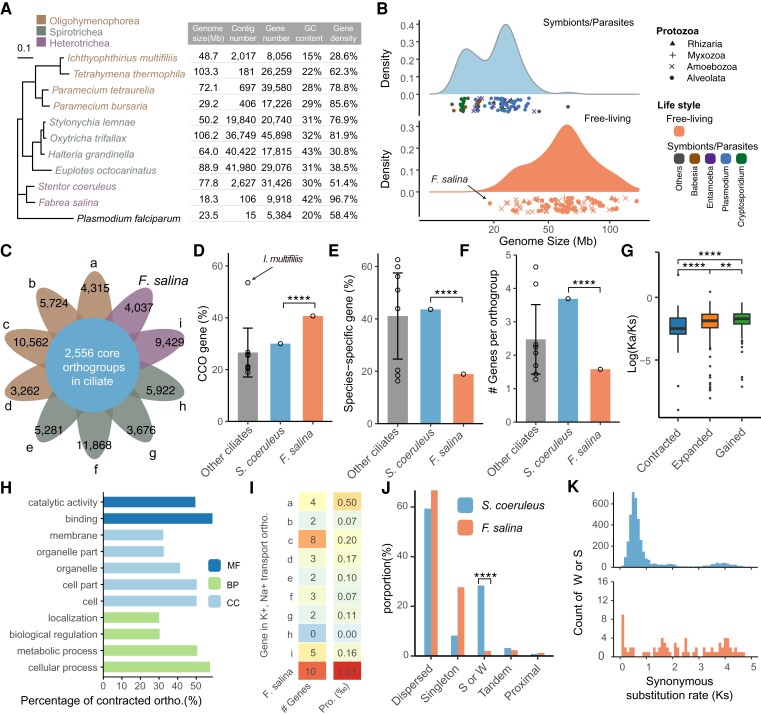
Fig. 2.
Comparative genomic analysis among Fabrea salina and its relatives. (A) Phylogenetic tree of 10 representative ciliates based on the 18S rRNA gene, with Plasmodium falciparum serving as an outgroup. The right panel shows their corresponding genomic features. (B) Comparison of genome size among Protozoa. The top and bottom panels show the genome size distribution of the studied symbionts/parasites and free-living species, respectively. The dot shapes represent different taxonomic classifications, namely rhizaria, myxozoa, amoebozoa, and alveolata. The dot colors represent four main groups of symbiotic or parasitic microorganisms. (C) The Venn diagram represents the number of ciliate core orthogroups present in >7 ciliates. The value in petals denotes the gene number in CCO for each species. The order from a to i represents (a) Ichthyophthirius multifiliis, (b) Tetrahymena thermophila, (c) Paramecium tetraurelia, (d) Paramecium bursaria, (e) Stylonychia lemnae, (f) Oxytricha trifallax, (g) Halteria grandinella, (h) Euplotes octocarinatus, and (i) Stentor coeruleus. (D) The proportion of the CCO gene in each ciliate. The proportions of S. coeruleus and F. salina were 30.00% and 40.70%, respectively. The mean proportion of other ciliates was 26.59% (95% CI: 17.16–36.02%). Each dot in the bar denotes one species. The height of the bar represents the mean value of the remaining eight species, and the error bar represents the 95% confidence interval of the mean. ****P < 0.0001, based on χ2 test. (E) The proportion of species-specific genes in each species. ****P < 0.0001, based on χ2 test. (F) The median gene number in orthology group for each ciliate. (G) Boxplot for the distribution of Kn/Ks. Gained gene families show a significantly higher Kn/Ks ratio than the expanded and contracted gene families. ****P < 0.0001, ***P < 0.001, **P < 0.01, based on a two-tailed Mann–Whitney test. (H) The GO classification of the contracted orthogroups. (I) Gene content of two orthogroups in 10 ciliates. (J) Distribution of gene duplication events in S. coeruleus and F. salina segmental duplicates/whole-genome duplication, S or W. (K) Ks plots for duplicate genes in S. coeruleus (top) and F. salina (bottom). MF, molecular function; BP, biological process; CC, cellular component.
A typical streamlined genome is usually characterized by a smaller genome size, a lower ratio of noncoding DNA, fewer paralogs, and nonessential genes (Giovannoni et al. 2014). To investigate genome streamlined traits in F. salina, orthogroups were determined using OrthoFinder (Emms and Kelly 2019) in nine representative ciliates as well as F. salina. A total of 207,761 genes were assigned to 33,032 orthogroups (fig. 2C). A total of 2,556 orthogroups were found in more than seven ciliates and thus were considered ciliate core orthogroups (CCOs). A total of 4,037 of 9,918 coding genes in F. salina could be classified into CCOs. The ratio of CCOs in F. salina (40.7%) was only lower than that of I. multifiliis (53.5%) but significantly higher than that of its relative S. coeruleus (30.0%) (χ2 test, P < 2.2 × 10−16) (fig. 2D). The proportion of species-specific genes (nonessential genes) showed an opposite trend, in which F. salina was significantly fewer than S. coeruleus (χ2 test, P < 2.2 × 10−16) (fig. 2E). In addition, the gene number of F. salina in each orthogroup was much lower than that of S. coeruleus (fig. 2F), indicating its highly compact genome with limited paralogs.
Next, nonsynonymous/synonymous substitution ratio analysis (Ka/Ks) revealed that gained and expanded gene families showed higher Ka/Ks values than contracted gene families (Mann–Whitney test, P = 2.7 × 10−13 and P = 1.3 × 10−8, respectively) (fig. 2G), suggesting that the gained and expanded gene families may be subjected to more relaxed selective pressure. We performed Gene Ontology (GO) functional enrichment analysis on the contracted genes and found that among 317 contracted orthogroups, most of them were involved in organelle part (32.8%), catalytic activity (49.8%), or metabolic process (50.8%) (fig. 2H). GO:0010766 (negative regulation of sodium ion transport) and GO:1903288 (positive regulation of potassium ion transmembrane transport) were only present in one expanded orthogroup, where F. salina has the largest number of paralogous genes in this orthogroup (fig. 2I and supplementary fig. S2, Supplementary Material online), which may contribute to an increased ability to maintain intracellular Na+ and K+ homeostasis in high salinity environments.
Considering that whole-genome duplication often plays a key role in genome amplification, MCScanX (Wang et al. 2012) was used to detect gene duplication events compared with S. coeruleus, which has an almost 4-fold larger genome and 3-fold more genes. The ratio of segmental or whole-genome duplication (S/W) events in F. salina was significantly lower than that in S. coeruleus (fig. 2J and supplementary table S3, Supplementary Material online). The KS distribution indicated that a recent S/W event occurred in S. coeruleus but was absent in F. salina (fig. 2K).
Comparative Genomics Analysis
To explore how such a streamlined genome adapts to diverse hypersaline environments, comprehensive comparative genomic analyses were performed. First, all protein-coding genes in 10 ciliates were classified as KO by using Kofamscan (Aramaki et al. 2020). Then, the unassigned genes were further annotated using the most frequent KO terms in their orthogroups. As a result, 72.4% of 254,039 protein-coding genes were successfully annotated, and a median of 76.2% of genes was annotated in each ciliate genome. The proportion of annotated genes was 91.46 and 85.38% for I. multifilliis and F. salina, respectively, which was higher than that of any other ciliate (fig. 3A). This indicates the core gene composition in F. salina and I. multifilliis, which is characteristic of streamlined genomes (Giovannoni et al. 2014). Among annotated functions, genetic information processing (37.1%) ranked first, followed by signaling and cellular processes (22.5%) and metabolism (15.7%) (fig. 3B).
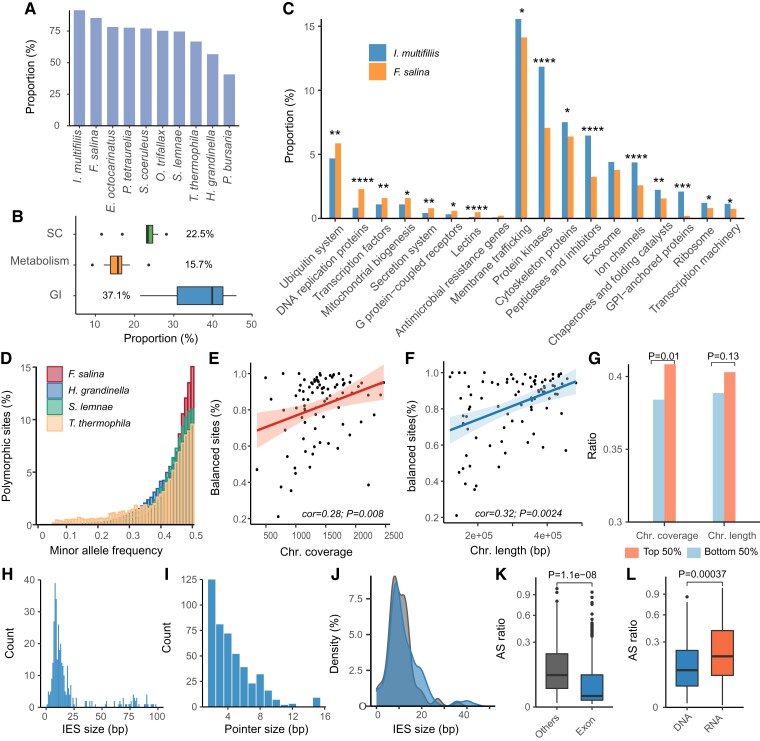
Fig. 3.
Metabolic pathways and macronuclear genomes polymorphisms in ciliates. (A) The proportion of annotated orthogroups based on KEGG. (B) The boxplot shows the proportion of classified genes based on the KEGG BRITE database in 10 ciliates. (C) Significantly differentiated GO terms with adjusted P values <0.05. ****Adj. P < 0.0001, ***adj. P < 0.001, **adj. P < 0.01, *adj. P < 0.05, χ2 test. The adjusted P values were calculated using the Benjamini–Hochberg method. (D) Distribution of MAF for polymorphic sites. The bin size is 0.01. (E and F) Spearman correlation between the proportion of balanced sites (MAF > 0.4) and chromosome coverage (E) and length (F). Each dot denotes one chromosome in F. salina. (G) Characteristics of the distribution of ciliate core orthogroup genes on chromosomes. The P value was determined based on Fisher’s exact test. (H and I) The length distribution of IESs (H) and pointer (I). (J) The length distribution of IESs located in exonic (blue) and other regions (dark gray). The P value was determined based on Mann–Whitney test. (K and L) Distribution of AS ratios in locations (K) and biological processes (L). The P value was determined based on the Mann–Whitney test. GI, genetic information processing; SC, signaling and cellular processes.
Next, F. salina was compared against parasitic I. multifiliis, which had a comparable number of genes. We found that even compared with the parasitic ciliate, F. salina exhibited a significantly lower gene content in the signaling and cellular processes and metabolic pathway (Fisher’s exact test, P < 0.01) (supplementary fig. S3A, Supplementary Material online). Notably, the number of genes related to genetic information processing in F. salina was significantly higher than those in other ciliates (21.43–42.80%) but comparable with that in JCVI-syn3A (Fisher’s exact test, P = 0.698), an artificially designed bacteria with the fewest genes necessary for life (Breuer et al. 2019). This finding suggests that genes in F. salina should be functionally optimized during evolution, with only the most viable and essential genes being retained.
KO-based functional enrichment analysis revealed that genes involved in DNA replication, mitochondrial biogenesis, ubiquitination, transcription, G protein−coupled receptors, and lectins were enriched in F. salina compared with I. multifiliis (Fisher’s exact test, P < 0.05) (fig. 3C). In addition, the gene content related to DNA replication and mitochondrial biogenesis was the highest in F. salina compared with any other ciliate (supplementary fig. S3B, Supplementary Material online), which suggests that a higher proportion of energy and replication-related genes retained in streamlined genomes may have a selective advantage in extreme environments.
Limited Chromosomal Drift During Asexual Reproduction
One of the characteristics of ciliates is that they can reproduce asexually via less accurate amitosis, which may lead to uneven distribution of MAC polyploid chromosomes in progeny cells, known as chromosomal copy drift (CCD) (Zheng et al. 2021). The occurrence of CCD results in a deviation of 1:1 in the allele frequency of the heterozygous sites on the genome. Given that CCD is thought to reduce cell fitness and thus many ciliates cannot be maintained for a long period under laboratory conditions (Bell and Graham 1988; Zheng et al. 2021), we investigated the CCD in the F. salina MAC genome by analyzing the minimum allele frequencies (MAFs) of heterozygous loci on each chromosome. We detected a total of 37,653 heterozygous sites under a strict threshold (minimum frequency >0.05, total coverage of each site >5×, and coverage of the minor allele >5×). The MAF of 15.77% heterozygous loci was <0.4 (referring to unbalanced sites, with a large deviation from 1:1), which was the lowest proportion among the 10 ciliate species (fig. 3D and supplementary fig. S4A, Supplementary Material online), indicating a limited influence of CCD in F. salina. Interestingly, we observed that the overall proportion of balanced sites (MAF > 0.4) in the genome was negatively correlated with chromosome number (supplementary fig. S4B, Supplementary Material online).
Next, we determined the proportion of balanced sites on each chromosome to explore potential factors affecting CCD. The proportion of balanced sites showed a significant positive correlation with the copy number and chromosome length (cor = 0.28 and 0.32, respectively) (fig. 3E and F). Likewise, chromosome length and copy number were positively correlated (cor = 0.37, P = 0.0004) (supplementary fig. S4C, Supplementary Material online), showing that longer contigs tended to have more copies. We hypothesized that more functionally important genes tend to be located on the chromosomes where CCD is less likely to occur to reduce the impact of unequal MAC division. As expected, we found that the CCO genes in F. salina were significantly enriched in the top 50% of contigs with higher copies (Fisher’s exact test, P = 0.01) but not in the top 50% of contigs with longer lengths (Fisher’s exact test, P = 0.13) (fig. 3G). Chromosomes with more copies are more likely to avoid the CCD effect, and similar observations were also made in Oxytricha trifallax (Swart et al. 2013) and Halteria grandinella (Zheng et al. 2021).
Cryptically Excised Internally Eliminated Sequences Increase Transcriptional Diversity During Sexual Reproduction
During sexual reproduction in ciliates, the MAC is diminished and forms a variable number of MAC residuals. Generally, most parental MAC residuals are completely degenerated, while a few of them may be retained and eventually fuse with the new MAC (Gong et al. 2020). However, it is still not clear whether MAC residuals play a role in sexual reproduction. During the formation of new MACs, DNA segments of MICs named internally eliminated sequences (IESs) are spliced (Zheng et al. 2021). Therefore, detecting whether the IESs are absent in the sequence can be used as a signal to identify the origin of the transcriptionally expressed genes.
Given that some IESs may be alternatively retained in MACs, we identified 461 candidates alternatively spliced (AS) IESs with pointer structures. The median AS ratio was 1.39%, which is comparable with observations in P. tetraurelia (Duret et al. 2008) and H. grandinella (Zheng et al. 2021), indicating that cryptically excised IESs are retained in most copies of the polyploid MAC genome, with only a small fraction of the polypoid chromosomes undergoing complete excision of IESs. The median lengths of AS-IESs and pointers were 14 and 4 nt, respectively (fig. 3H and I). However, the pointer sequence did not show any obvious motif and may have undergone a different excision mechanism compared with the typical fully excised IES. We found that 346 IESs were located in the exonic region (exonic IESs), which was longer than those located in other regions (Mann–Whitney test, P = 0.0003) (fig. 3J), with median lengths of 16 and 12 nt, respectively. The AS ratio of exonic IESs was significantly lower than that in other regions (Mann–Whitney test, P < 0.001) (fig. 3K). Among all the IESs, 133 were found in the transcriptome data, and the AS ratio at the transcriptional level was higher than that at the genomic level (Mann–Whitney test, P < 0.001) (fig. 3L). Genes containing exonic IESs have multiple spliced isoforms in the genome, which may have different functions. These results suggest that the IES in the exonic regions greatly increases protein diversity, which may represent a fine regulation of gene content and their related functions in ciliates.
Next, we induced conjugation and sequenced the transcriptome at five consecutive time points (supplementary figs. S5–S8, Supplementary Material online). Eighteen RNA-seq libraries were built and sequenced to generate 3,655 million paired-end reads (54.83 Gb). As shown in Supplementary figure S8, Supplementary Material online, the degradation of the MAC begins, and several areas in the MAC gradually depress and break into a varying number of irregularly shaped pieces. As conjugation proceeds, the MAC residual becomes regular and rounded. The MAC residuals were still present 72 h after conjugation pair (CP) separation (supplementary figs. S7 and S8, Supplementary Material online). Interestingly, all 77 exonic IESs could be detected at all five time points, and samples from different conjugation periods were clearly separated (supplementary fig. S5A, Supplementary Material online), indicating that MAC residuals retained transcriptional activity during sexual reproduction. Next, 33 exonic IESs present in more than five samples were selected to describe the expression pattern in detail (supplementary fig. S6, Supplementary Material online). The results showed that despite being located on the same chromosome, different genes showed opposite directions (supplementary fig. S6, Supplementary Material online), suggesting a dynamic regulation of the ratio between different versions of the chromosome during conjugation. Next, six genes that have a highly variable exonic IES (SD > 2) were identified (supplementary fig. S5B, Supplementary Material online). Fabrea_09599, Fabrea_02192, and Fabrea_00615 have a WD40 repeat domain (Pfam ID: PF00400) that is associated with multiple functions, including RNA processing and control of cell division (Suganuma et al. 2008). Fabrea_00480 was annotated as DIAPH2 (diaphanous 2, K05741) involved in cytoskeletal proteins. Fabrea_09158 was annotated as FRAS1 (extracellular matrix protein FRAS1, K23379), which is associated with a variety of physiological processes, including shape change, proliferation, and motility (Wu 2004). These findings indicate that the cryptically excised IESs in the MAC may play important functions during sexual reproduction.
Genetic Basis for Salinity Tolerance
To better understand its broad salinity adaptation, we analyzed the gene expression profiles of F. salina at different salinity levels: 80‰, 130‰, and 180‰ (fig. 4A). A total of 1,129 differentially expressed genes (DEGs) were found in the DEG analysis of the high and optimum salinity groups, and 1,353 DEGs were detected between optimum and low salinity (fig. 4B). However, only 23.4% of the DEGs were shared in both comparisons, and clustering analysis showed a different expression signature (fig. 4C and E), suggesting that F. salina may have distinct mechanisms to adapt to low and high osmotic pressures. Further analysis confirmed that DEGs of high salinity group were significantly enriched (χ2 test, P = 5.86 × 10−6) in the expanded gene families (fig. 4D and F), most of which were involved in G protein-coupled receptors and chaperones and folding catalysts (fig. 4G). These findings mirror the results of previous studies, which showed that G protein receptors play an important role in the response to abiotic stresses, especially high salinity (Liu et al. 2018). Activation of chaperones and folding catalysts is a key way for organisms to deal with misfolded proteins resulting from the accumulation of reactive oxygen species owing to salt stress (Zhang et al. 2021). Next, we found that the most significant changed DEGs were enriched in the genes specific to the high salinity group (Fisher’s exact test, P = 1.4 × 10−7), rather than those shared with the low salinity group (fig. 4H and 4I). Functional enrichment analysis showed that the GO terms such as biosynthetic process, regulation of cellular process, cellular component organization or biogenesis, were up-regulated in high salinity environments, whereas the GO terms related to carbohydrate derivative binding, ion binding, microtubule-based process were down-regulated (fig. 4J).
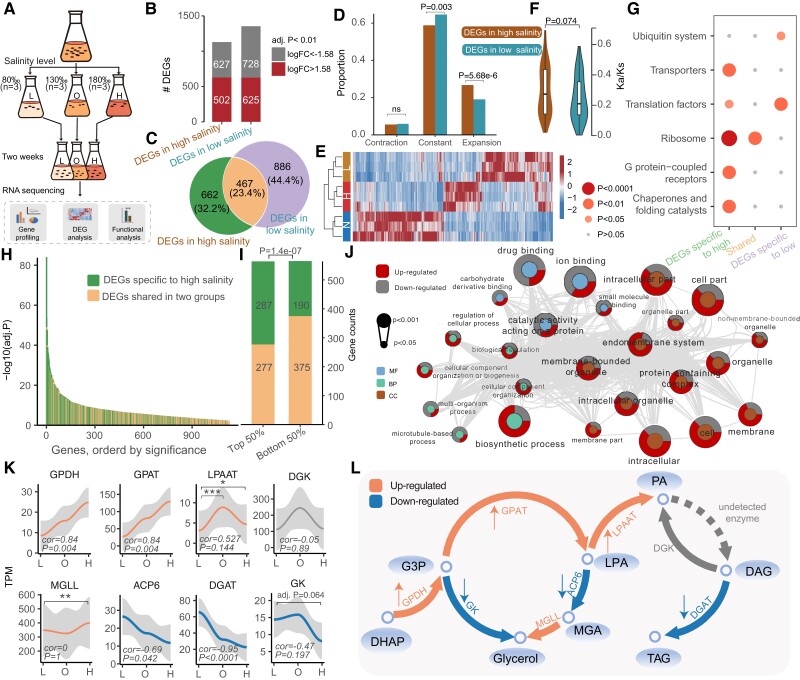
Fig. 4.
Genetic adaptation for salinity tolerance. (A) Flow chart of the experimental design and transcriptome analysis of F. salina under salinity stress. (B) The number of differentially expressed genes in the high and low salinity groups. Both groups were compared against the optimum salinity group. (C) Venn diagram of differentially expressed genes in the two groups. (D) The bar graph indicates the enrichment of DEGs in expanded and contracted gene families. (E) Heatmap for differentially expressed genes. (F) Kn/Ks of differentially expressed genes. P value was determined based on Mann–Whitney test. (G) KEGG enrichment analysis of DEGs in different categories. The color of the dots represents the P value. (H) Differential significance distribution of the DEGs in in high salinity group. (I) Gene counts of the DEGs in the high salinity group. (J) Significant GO terms (P < 0.05) in the high salinity-specific DEGs. The P value in I and J was determined based on Fisher’s exact test. (K) Distribution of gene expression at different salinities. The Spearman correlations and their significance are marked at the bottom of the image. The adjusted P values resulted from DEG analysis and are marked at the top of the image. ****Adj. P < 0.0001, ***adj. P < 0.001, **adj. P < 0.01, *adj. P < 0.05. (L) Schematic diagram of the pathways associated with glycerol and PA synthesis. Orange and blue colors represent up-regulated and down-regulated steps under hypersalinity stress, respectively. GPDH, glycerol 3-phosphate dehydrogenase; GPAT, glycerol-3-phosphate acyltransferase; LPAAT, lysophosphatidic acid acyltransferase; DGK, diacylglycerol kinase; GK, glycerol kinase; MGLL, monoglyceride lipase; ACP6, lysophosphatidic acid phosphatase type 6 precursor; DGK, diacylglycerol kinase; DGAT, diacylglycerol acyltransferase; DHAP; dihydroxyacetone-phosphate; G3P; glycerol-3-phosphate dehydrogenase; MGA, monoacylglycerol; LPA, lysophosphatidic acid; PA, phospholipid acid; DAG, diacylglycerol; TAG, triacylglycerol.
Microorganisms counteract salt stress by the accumulation of compatible solutes that act as osmoprotectants to help cells resist osmotic pressure without altering intracellular enzyme activity (Harding et al. 2016). A large number of osmoprotectants, including proline, glycine betaine (GB), ectoine, and glycerol, have been reported in halophilic and halotolerant organisms (Weinisch et al. 2019). Weinisch et al. (2019) found that GB and ectoine increased linearly with salinity stress in F. salina. Here, we observed an incomplete biosynthetic pathway for GB, with the absence of Ect/5-hydroxyectoine-related genes (ectABCD) in the MAC genome. For betaine aldehyde dehydrogenase (BADH), one of the important gene involved in the biosynthetic pathway of GB, its abundance was negatively correlated with salinity (cor = −0.74, P = 0.023). Interestingly, we found that the expression of enzymes related to glycerol biosynthesis decreased with increasing salinity (fig. 4K and L). The abundance of ACP6 was significantly negatively correlated with salinity (cor = −0.685, P = 0.042). The expression level of GK in the 80‰ salinity group was lower than that in the 180‰ group, although this difference was not statistically significant (adj. P = 0.064). These results suggest that the compatible solutes in F. salina may not be fully self-synthesized and confirmed by findings in the heterotrophic ciliate Schmidingerothrix salinarum, which could directly absorb GB and ectoine from the environment (Weinisch et al. 2018).
As a lipid second messenger, phosphatidic acid (PA) was reported to be involved in intracellular salt stress signaling in eukaryotes (Shen et al. 2019). In F. salina, we found that the expression of GPAT, which acylates G3P to produce LPA, was significantly positively correlated with salinity (cor = 0.84, P = 0.004). The same expression trend was also observed in GPDH (cor = 0.84 and P = 0.004). The expression of another important gene, lysophosphatidic acid acyltransferase (LPAAT), which produces PA with lysophosphatidic acid (LPA) and acyl-CoA as substrates, was significantly higher in the 180‰ and 130‰ groups than in the 80‰ group (adj. P = 0.049 and adj. P = 0.001, respectively) (fig. 4K and L). Such increased expression of the enzymes associated with PA synthesis was also observed in the alga Parachlorella kessleri grown under salt stress conditions (You et al. 2019). We demonstrated that the accumulation of PA may play an important role in resistance to high osmotic pressure.
Conclusion
In this study, we used both Illumina and Nanopore sequencing technologies to assemble a high-quality macronuclear genome of F. salina characterized by an unusually streamlined and compacted genome in free-living protozoa. We revealed the minimal gene content that allows eukaryotes to live freely and heterotrophically, which suggests that genome streamlining is in fact not unique to prokaryotes but has also been adopted by some eukaryotes. Given their dual nuclei system, ciliates represent an ideal model to study genome expansion or contraction events. Our study provides a unique and valuable resource for understanding the genomic conservation and evolutionary history of eukaryotic organisms.
Methods and Materials
Culture of F. salina
Fabrea salina was collected from the BeiDaGang wetland along the Bohai Sea coast at Tianjin (38°57′N, 117°37′E), China, by the Tianjin Academy of Agricultural Sciences and was identified by its morphological features using differential interference contrast microscopy. Cells were incubated in automatically filtered sterile seawater (pH 7.8) at 30 °C and 80 pSU (actual units of salinity) under lighting. Dunaliella salina was supplied as food source. After 7–10 days of culture, the population density reached 100 cells/1 ml of medium. The status and cell numbers of F. salina were checked using an anatomical lens.
Nucleic Acid Isolation and Sequencing
As described previously (He et al. 2019), F. salina cells were isolated through a 15-μm-pore-size nylon filter membrane and washed three times. Then, the cells were starved for 1 day, and 1 × penicillin–streptomycin antibiotics (Invitrogen, Carlsbad, USA) were added to eliminate bacterial contamination. Before extracting nucleic acids, the cells were washed five times with sterile water to remove any bacteria that may be attached to the cell surface. For single-cell genome sequencing, 12 cells were placed in three sterile 0.2-ml tubes as biological replicates, with 3, 4, and 5 cells in each tube. DNA extraction was carried out following the Repli-g single-cell kit manufacturer’s instructions (Qiagen). Then, approximately 12,000 cells were harvested and generated 1µg DNA for Nanopore long-read sequencing. To obtain high-quality genomes, we constructed two types of libraries. The native library was constructed following with native barcoding genomic DNA protocol (with EXP-NBD104, EXP-NBD114, SQK-LSK109, ONT, UK). The polymerase chain reaction (PCR) library was constructed according to PCR barcoding genomic DNA protocol (with SQK-LSK109, EXP-PBC001, ONT, UK) with 12 PCR cycles. The library mean length was 8 kb and was sequenced on R9.4 FlowCells using the MinION sequencer (ONT, UK) for 24 h. ONT Guppy software (v1.8) was used for base calling on fast5 files, and the “passed filter” reads were used for downstream analysis.
In addition, the Illumina paired-end sequencing DNA library was constructed by shearing genomic DNA to 300 bp fragments on a Covaris S220 system (Covaris, Woburn, MA, USA). A Nextera DNA Flex Library Prep kit (Illumina, San Diego, CA, USA) was used following the manufacturer’s protocol. Three DNA libraries were sequenced using the Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA) for 2 × 150 bp reads and generated 27.86 Gb of data.
For the single-cell transcriptome, whole-transcriptome amplification was performed using a SMART-Seq2 v4 Ultra Low Input RNA kit (NEB, USA) following the manufacturer’s protocols. Sequencing libraries were prepared with the Illumina Nextera XT kit and sequenced using an Illumina HiSeq 2500 platform. High-throughput sequencing for three samples produced 54.83 Gb of 2 × 150 pair-end data in total.
Genome Assembly
First, Nanopore long reads were trimmed with porechop (version 0.2.4) (https://github.com/rrwick/Porechop) to remove adapters from read ends and split sequences with internal adapters. The processed reads were assembled to draft the genome by Flye (version 2.8.1) with the default parameters (Kolmogorov et al. 2019). Subsequently, a combination of polishing methods was used, including Racon (Vaser et al. 2017), Medaka (version 1.3) (https://github.com/nanoporetech/medaka) and Pilon (version 1.23) polishing with Illumina sequencing reads (Walker et al. 2014). A total of 185,798,514 Illumina paired-end reads were used to correct the draft genome. Finally, we obtained a high-quality F. salina MAC genome (18.3 Mb) with an N50 of 265.05 kb and a complete mitochondrial genome (42,610 bp) (see the Supplementary Materials for more details).
Assessment of Genome Assembly
First, the integrity of core eukaryotic genes in the MAC genome was assessed using CEGMA (version 2.5) (Parra et al. 2007) with default parameters. Two hundred of 248 core eukaryotic orthologous groups (KOGs) were identified. Then, a relaxed restriction of CEGMA search was performed on the remaining genes following the published approach of S. coeruleus (Slabodnick et al. 2017). Hmmscan (HMMER, version 3.3.2, http://hmmer.org/) was used to assign the Pfam domain in KOGs to the predicted genes in F. salina. With this analysis, 46 additional core eukaryotic genes were determined under the full-sequence e-value <1 × 10−3. Finally, the updated HMMs for two undetected KOGs (KOG0563 and KOG2531) in the EggNOG database were downloaded and reassigned to genes by hmmscan. Three genes were assigned to them. Above all, 248 core eukaryotic genes were found in our assembled genome. This result is similar to the completeness rate of fully well-studied ciliate genomes, such as S. coeruleus, Tetrahymena thermophila, P. bursaria, or O. trifallax, which commonly contain 220–243 KOGs.
Additionally, EukCC (version 4.0.6) (Saary et al. 2020) and BUSCO (version 0.2) (Seppey et al. 2019) were used to assess the completeness and contamination based on the predicted gene amino acid sequence in F. salina with default parameters. The results showed that the completeness was 75.76% and 83.6% in BUSCO and EukCC, respectively. The contamination was 3.03% and 6.4% in BUSCO and EukCC, respectively. The other nine published ciliate genomes were evaluated using BUSCO with the same parameters.
Detection of Telomeres
To identify the telomere in the MAC genome of F. salina, 200 bp sequences from both ends of assembled contigs were extracted and analyzed by a custom R script that recognized the telomere repeat 5-CACCCTAA-3. Briefly, the sequences of contig ends were cut into 8 nt kmers, and kmers with significantly higher frequencies were assembled and manually validated.
Gene Prediction and Annotation
The MAKER pipeline (version 2.31.11) (Holt and Yandell 2011) was used to predict gene models. The pipeline includes Augustus (Nachtweide and Stanke 2019), SNAP (Korf 2004), and GeneMark-ES (Borodovsky and Lomsadze 2011) to identify repeats, align ESTs and proteins to the genome, produce ab initio gene predictions, and then combine these data into gene annotations with evidence-based quality values. Considering the short intron length (15–16 nt), the source code (filename: intronmodel.cc, types.cc, extrinsic.cc, GI.pm, protein.pm, altest.pm, est.pm) in Augustus was modified to change the minimum length limit to 9 nt for introns (the default is 39 or 20). Then, the source code was recompiled using these updated settings.
The transcriptome evidence for gene prediction came from two sources. The first was a published F. salina transcriptome assembly (MMETSP1345) containing 10,034 assembled transcripts produced by the Marine Microbial Eukaryotic Transcriptome Sequencing Project (MMETSP) (Keeling et al. 2014). The second was generated in this study, which contained 9.07 Gb of PE150 transcriptome sequencing data. After quality filtration and adapter removal, processed reads were assembled using Trinity (version 2.11.0) with default parameters. A total of 20,356 transcripts were obtained. Cd-hit (version 4.8.1) (Li and Godzik 2006) was used to eliminate redundancy at a threshold of 0.95 similarity (-c 0.95). For homology evidence, amino acid sequences in nine published ciliates were collected as cross-species protein homology evidence.
InterProScan (version 5.48-83.0) (Jones et al. 2014) was used to identify conserved functional motifs and protein domains. The predicted proteins were aligned with parameters (-iprlookup -goterms) to the following databases: PROSITE, HAMAP, PFAM, PRINTS, PRODOM, SMART, TIGRFAMS, PIRSF, SUPERFAMILY, CATH-GENE3D, and PANTHER. The results showed that 8,781 of 9,918 genes contained motifs or protein domains in the F. salina assembled genomes. Additionally, eggNOG-mapper (Huerta-Cepas et al. 2019) was used based on the webserver http://eggnog-mapper.embl.de/. A total of 5,995 genes were assigned to GO terms, KEGG KO, or EC numbers. For noncoding RNA prediction, rRNA and tRNA were predicted by cmscan based on the Rfam (version 14.4) (Kalvari et al. 2018) database. tRNAscan-SE (version 2.0.7) (Chan and Lowe 2019) was used to identify tRNA with default parameters.
Codetta (version 2.0) was used to predict the genetic code from nucleotide sequence data (Shulgina and Eddy 2021).
Phylogenetic Analysis
The F. salina 18S rRNA gene was identified by Rfam. 18S rRNA genes of the following nine ciliates and an outgroup were downloaded from NCBI: H. grandinella (MF002432.1), Stylonychia lemnae (AM233915.1), O. trifallax (FJ545743.1), Euplotes octocarinatus (LT623905.1), T. thermophila (M10932.1), I. multifiliis (U17354.1), P. tetraurelia (AB252009.1), S. coeruleus (JQ282899.1), P. bursaria (MG589318.1), and the outgroup Plasmodium falciparum (MF155937.1). Then, multiple sequence alignment and maximum likelihood tree construction were performed in MEGAX (Kumar et al. 2018). Gene density was defined as the ratio of the total length of protein-coding genes to the genome length.
Heterozygous Loci Detection
First, the genome sequencing data of nine published ciliates were downloaded from public databases (supplementary table S4, Supplementary Material online). Then, the sequence reads were aligned to the reference genome using Bowtie2 (Langmead and Salzberg 2012) (version 2.3.2) with default parameters. Filtering was performed using Bcftools (Danecek et al. 2021) (version 1.8) (-e “QUAL < 10 || DP < 5 || AD < 5 || AD/DP < 0.05” –SnpGap 5).
Orthology Group Analysis
The protein sequences of 10 species were downloaded from NCBI and other databases: H. grandinella (GCA_006369765), S. lemnae (GCA_000751175), O. trifallax (http://oxy.ciliate.org/index.php/home/downloads), E. octocarinatus (http://ciliates.ihb.ac.cn/database/download/#eo), T. thermophila (http://ciliate.org), I. multifiliis (GCA_000220395), P. tetraurelia (GCF_000165425), S. coeruleus (GCA_001970955), and P. bursaria (https://ngdc.cncb.ac.cn/gwh/Assembly/138/show; GWHAAFB00000000). OrthoFinder (version 2.5.2) (Emms and Kelly 2019) was used to infer orthogroups based on the MCL clustering algorithm with default parameters. A total of 207,761 (84.20%) of 246,731 genes were assigned to 33,032 orthogroups. Among them, 2556 orthogroups were considered CCOs, as they were detected in >7 species. A total of 60,623 genes (24.57%) were in 15,978 species-specific orthogroups.
DupliPHY (Ames and Lovell 2015) was used to determine gene families with rapid expansions, contractions, gain and/or loss among F. salina and the related ciliates. Among the 5,173 orthogroups in F. salina, we detected 317 contraction events (involving 580 genes) and 252 expansion events (involving 1129 genes). The rate of nonsynonymous substitutions (Kn) and the rate of synonymous substitutions (Ks) for the genes in contraction and expansion orthogroups were calculated based on ParaAT (version 1.0) (with parameters “-m clustalw2 -p proc -f axt”) and KaKs_Calculator (version 2.0) (Wang et al. 2010) (with parameters “-m YN”). A two-tailed Mann–Whitney test was used to determine significance.
To detect gene duplication events in F. salina and S. coeruleus, MCScanX (Wang et al. 2012) was used with default parameters. Genes were classified as singletons, dispersed duplicates, proximal duplicates, tandem duplicates, and WGD/segmental duplicates depending on their copy number and genomic distribution.
MAC Morphological and Transcriptomic Changes During Conjugation
We used a combination of low temperature (4 and 0 °C), starvation, and darkness to induce conjugation. Due to the adaptability of F. salina to stress conditions, only a small number of CPs were obtained after conjugation induction. Thus, we selected the CPs with capillary glass pipettes and inspected their nuclear changes. First, we observed that both CP cells were in the same cell cycle by staining with Hoechst 33342. Then, we selected 100 CPs and transferred them to PCR tubes for continuous observation. Cells were selected before CP separation and 12, 24, 48, and 72 h after CP separation. For CP, one cell was stained with Hoechst 33342 to observe nuclear morphology, and the other cell was used for transcriptome sequencing.
Growth Rate at Different Salinity Levels
We set up 13 consecutive salinity gradients between 60‰ and 180‰, with three parallel experiments for each salinity. The volume of each cell culture flask was controlled at 50 ml, and the number of cells was controlled at 200. Dunaliella salina was provided as food and incubated for 2 weeks. During the incubation, sterile seawater or sterile distilled water of varying salinity was added daily to maintain the salinity of the culture. As a result, the maximal growth rate was at a salinity of 130‰. Therefore, we collected F. salina samples at three salinities (80‰, 130‰, and 180‰) with three replicates of each salinity for transcriptome sequencing. Gene expression was quantified using HISAT2 (version 2.0.5) and StringTie (version 1.3.4) (Pertea et al. 2016).
Alternatively Spliced IES Detection
The software ADFinder (Zheng et al. 2020) was used to detect the alternatively spliced IESs in the MAC genome without sequence data of the MIC genome. For the polyploid MAC genome, different versions of the same chromosome may exist with IESs deleted (excised) and IESs retained (nonexcised). As recommended, Bowtie2 (version 2.3.2) software (-local, -k 5, -ma 3) was used to align our RNA-seq and DNA-seq reads to the MAC genome. Then, ADFinder was used to identify the position and pointer sequence of alternatively spliced IESs with default parameters.
Functional Enrichment Analysis
First, all protein-coding genes in ten ciliates were classified into KO by Kofamscan (version 1.3.0) (Aramaki et al. 2020) with the parameter “-E 0.05”. Second, the unassigned genes were further annotated using the most frequent KOs in their orthogroups. Then, all genes were classified into three protein families (metabolism, genetic information processing, and signaling and cellular processes) and 52 sublevel protein families based on the KEGG BRITE database (https://www.genome.jp/kegg/brite.html). As a result, 72.4% of 254,039 protein-coding genes were successfully annotated, and a median of 76.2% of genes was annotated in each ciliate. Then, the function enricher in the R package “clusterProfiler” (version 3.12.0) (https://github.com/YuLab-SMU/clusterProfiler) was used to perform enrichment analysis.
Then WEGO (Ye et al. 2018) (version 2.0) (https://wego.genomics.cn/) was used to perform GO classification and Fisher’s exact test was used to calculate P values. Cytoscape (https://cytoscape.org/) was used to visualize the network.
Salinity Stress-Related Genes
The following genes involved in salinity tolerance were selected: Fabrea_04266 (ACP6, lysophosphatidic acid phosphatase type 6, K14395), Fabrea_06374 (BADH, betaine aldehyde dehydrogenase, K00130), Fabrea_06811 (DGK, diacylglycerol kinase [ATP], K00901), Fabrea_09038 (GK, glycerol kinase, K00864), Fabrea_05028 (GPAT, glycerol-3-phosphate O-acyltransferase, K13506), Fabrea_08063 (GPDH, glycerol-3-phosphate dehydrogenase, K00057), Fabrea_08457 (LPAAT, lysophosphatidic acid acyltransferase/lysophosphatidylinositol acyltransferase, K13523), Fabrea_08078 (MGLL, acylglycerol lipase, K01054), and Fabrea_02753 (DGAT, diacylglycerol O-acyltransferase, K11155). Then, we combined differential expression analysis and Spearman correlation analysis to assess the relationship between gene expression and salinity. Differential expression analysis was performed in the R package DESeq2 (version 1.24.0) (Love et al. 2014). Correlation analysis of gene expression and salinity was calculated using the function cor.test(method = “spearman”) in R.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
F.Z. and M.M. conceived the project. B.Z., T.Z., and F.Z. performed data analysis. L.H. and H.Q. performed the experiments and generated sequencing data. B.Z., F.Z., and M.M. wrote the manuscript with the contribution of all authors. All authors read and approved the final manuscript.
Data availability
All data generated or analyzed during this study are included in this published article, its supplementary information files and publicly available repositories. Reads for the genome assemblies have been deposited to NCBI GenBank (PRJNA822294). The gene training model for Fabrea salina in Augustus and orthogroup results have been deposited to GitHub (https://github.com/callAgene/Supplementary-Materials-for-Fabrea-salina).
Supplementary Material
Funding
This work was supported by grants from the National Natural Science Foundation of China (32070432, 32025009, and 31672279) and the Fundamental Research Funds for the Central Universities.
References
- Al-Ardi MH. 2021. Illumination on the structure and characteristics of entamoeba histolytica genome. Preprints 2021030070. [Google Scholar]
- Ames RM, Lovell SC. 2015. DupliPHY-Web: a web server for DupliPHY and DupliPHY-ML. Bioinformatics 31:416–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, Goto S, Ogata H, Valencia A. 2020. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36:2251–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aury J-M, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Ségurens B, Daubin V, Anthouard V, Aiach N, et al. . 2006. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444:171–178. [DOI] [PubMed] [Google Scholar]
- Bell G, Graham B. 1988. Sex and death in Protozoa: the history of obsession. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Borodovsky M, Lomsadze A. 2011. Eukaryotic gene prediction using GeneMark.hmm-E and GeneMark-ES. Curr Protoc Bioinformatics Chapter 4, Unit 4, 6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer M, Earnest TM, Merryman C, Wise KS, Sun L, Lynott MR, Hutchison CA, Smith HO, Lapek JD, Gonzalez DJ, et al. . 2019. Essential metabolism for a minimal cell. Elife 8:e36842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. . 2021. Twelve years of SAMtools and BCFtools. Gigascience 10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle E, Ferraz C, Rombauts S, Rouzé P, Worden AZ, Robbens S, Partensky F, Degroeve S, Echeynié S, Cooke R, et al. . 2006. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci U S A. 103:11647–11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Cohen J, Jubin C, Dessen P, Goût J-F, Mousset S, Aury J-M, Jaillon O, Noël B, Arnaiz O, et al. . 2008. Analysis of sequence variability in the macronuclear DNA of Paramecium tetraurelia: a somatic view of the germline. Genome Res. 18:585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes NM, Schrago CG. 2019. A multigene timescale and diversification dynamics of Ciliophora evolution. Mol Phylogenet Evol. 139:106521. [DOI] [PubMed] [Google Scholar]
- Foissner W, Chao A, Katz LA. 2007. Diversity and geographic distribution of ciliates (Protista: Ciliophora). In: Foissner W, editor. Protist diversity and geographical distribution. Dordrecht: Springer. p. 111–129. [Google Scholar]
- Garcia-Fernández JM, de Marsac NT, Diez J. 2004. Streamlined regulation and gene loss as adaptive mechanisms in Prochlorococcus for optimized nitrogen utilization in oligotrophic environments. Microbiol Mol Biol Rev. 68:630–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getz EW, Tithi SS, Zhang L, Aylward FO. 2018. Parallel evolution of genome streamlining and cellular bioenergetics across the marine radiation of a bacterial phylum. mBio 9:e01089-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Cameron Thrash J, Temperton B. 2014. Implications of streamlining theory for microbial ecology. ISME J. 8:1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong R, Jiang Y, Vallesi A, Gao Y, Gao F. 2020. Conjugation in Euplotes raikovi (Protista, Ciliophora): new insights into nuclear events and macronuclear development from micronucleate and amicronucleate cells. Microorganisms 8:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote J, Thrash JC, Huggett MJ, Landry ZC, Carini P, Giovannoni SJ, Rappé MS. 2012. Streamlining and core genome conservation among highly divergent members of the SAR11 clade. mBio 3:e00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guermazi W, Elloumi J, Ayadi H, Bouain A, Aleya L. 2008. Rearing of Fabrea salina Henneguy (Ciliophora, Heterotrichida) with three unicellular feeds. C R Biol. 331:56–63. [DOI] [PubMed] [Google Scholar]
- Han K, Li Z-f, Peng R, Zhu L-p, Zhou T, Wang L-g, Li S-g, Zhang X-b, Hu W, Wu Z-h, et al. . 2013. Extraordinary expansion of a Sorangium cellulosum genome from an alkaline milieu. Sci Rep. 3:2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding T, Brown MW, Simpson AGB, Roger AJ. 2016. Osmoadaptative sstrategy and its molecular signature in obligately halophilic heterotrophic protists. Genome Biol Evol. 8:2241–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Wang J, Fan X, Liu X, Shi W, Huang N, Zhao F, Miao M. 2019. Genetic basis for the establishment of endosymbiosis in Paramecium. ISME J. 13:1360–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotos GN. 2021. A preliminary survey on the planktonic biota in a hypersaline pond of Messolonghi Saltworks (W. Greece). Diversity 13:270. [Google Scholar]
- Huang N, Chen S, He M, Song Q, Hou L, Zhao Y, Zhao S, Miao M. 2021. Molecular evolutionary analyses of euplotes species living in freshwater and marine habitats: a mitogenomic perspective. Front Mar Sci 8:265. [Google Scholar]
- Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, et al. . 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47:D309–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. . 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I, Nawrocki EP, Argasinska J, Quinones-Olvera N, Finn RD, Bateman A, Petrov AI. 2018. Non-coding RNA analysis using the Rfam database. Curr Protoc Bioinformatics 62:e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ., Burki F, Wilcox HM, Allam B, Allen EE, Amaral-Zettler LA, Armbrust EV, Archibald JM, Bharti AK, Bell CJ, et al. . 2014. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12:e1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 37:540–546. [DOI] [PubMed] [Google Scholar]
- Koonin EV. 2009. Evolution of genome architecture. Int J Biochem Cell Biol. 41:298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 5:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. [DOI] [PubMed] [Google Scholar]
- Liu C, Xu Y, Feng Y, Long D, Cao B, Xiang Z, Zhao A. 2018. Ectopic expression of mulberry G-proteins alters drought and salt stress tolerance in tobacco. Int J Mol Sci. 20:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy S. 2012. Marine microbiology: SAR86: streamlined for success. Nat Rev Microbiol. 10:82. [DOI] [PubMed] [Google Scholar]
- Moser KA, Drábek EF, Dwivedi A, Stucke EM, Crabtree J, Dara A, Shah Z, Adams M, Li T, Rodrigues PT, et al. . 2020. Strains used in whole organism Plasmodium falciparum vaccine trials differ in genome structure, sequence, and immunogenic potential. Genome Med. 12:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtweide S, Stanke M. 2019. Multi-genome annotation with AUGUSTUS. Methods Mol Biol. 1962:139–160. [DOI] [PubMed] [Google Scholar]
- Narita TB, Kawabe Y, Kin K, Gibbs RA, Kuspa A, Muzny DM, Richards S, Strassmann JE, Sucgang R, Worley KC, et al. . 2020. Loss of the Polyketide Synthase StlB Results in Stalk Cell Overproduction in Polysphondylium violaceum. Genome Biol Evol. 12:674–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T, Wijeratne S, Meulia T, Firkins JL, Yu Z. 2021. The macronuclear genome of anaerobic ciliate Entodinium caudatum reveals its biological features adapted to the distinct rumen environment. Genomics 113:1416–1427. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23:1061–1067. [DOI] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 11:1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saary P, Mitchell AL, Finn RD. 2020. Estimating the quality of eukaryotic genomes recovered from metagenomic analysis with EukCC. Genome Biol. 21:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppey M, Manni M, Zdobnov EM. 2019. BUSCO: assessing genome assembly and annotation completeness. Methods Mol Biol. 1962:227–245. [DOI] [PubMed] [Google Scholar]
- Shen L, Zhuang B, Wu Q, Zhang H, Nie J, Jing W, Yang L, Zhang W. 2019. Phosphatidic acid promotes the activation and plasma membrane localization of MKK7 and MKK9 in response to salt stress. Plant Sci. 287:110190. [DOI] [PubMed] [Google Scholar]
- Shulgina Y, Eddy SR. 2021. A computational screen for alternative genetic codes in over 250,000 genomes. Elife 10:e71402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabodnick MM., Ruby JG, Reiff SB, Swart EC, Gosai S, Prabakaran S, Witkowska E, Larue GE, Fisher S, Freeman RM, et al. . 2017. The macronuclear genome of Stentor coeruleus reveals tiny introns in a giant cell. Curr Biol. 27:569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Pattenden SG, Workman JL. 2008. Diverse functions of WD40 repeat proteins in histone recognition. Genes Dev. 22:1265–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart EC, Bracht JR, Magrini V, Minx P, Chen X, Zhou Y, Khurana JS, Goldman AD, Nowacki M, Schotanus K, et al. . 2013. The Oxytricha trifallax macronuclear genome: a complex eukaryotic genome with 16,000 tiny chromosomes. PLoS Biol. 11:e1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart EC, Serra V, Petroni G, Nowacki M. 2016. Genetic codes with no dedicated stop codon: context-dependent translation termination. Cell 166:691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaser R, Sović I, Nagarajan N, Šikić M. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ., Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. . 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, Lee T-h, Jin H, Marler B, Guo H, et al. . 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. 2010. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics 8:77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinisch L, Kirchner I, Grimm M, Kühner S, Pierik AJ, Rosselló-Móra R, Filker S. 2019. Correction to: glycine betaine and ectoine are the major compatible solutes used by four different halophilic heterotrophic ciliates. Microb Ecol. 77:332. [DOI] [PubMed] [Google Scholar]
- Weinisch L, Kühner S, Roth R, Grimm M, Roth T, Netz DJA, Pierik AJ, Filker S, Sourjik V. 2018. Identification of osmoadaptive strategies in the halophile, heterotrophic ciliate Schmidingerothrix salinarum. PLoS Biol. 16:e2003892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Koonin EV. 2013. Genome reduction as the dominant mode of evolution. Bioessays 35:829–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. 2004. The PINCH-ILK-parvin complexes: assembly, functions and regulation. Biochim Biophys Acta 1692:55–62. [DOI] [PubMed] [Google Scholar]
- Ye J, Zhang Y, Cui H, Liu J, Wu Y, Cheng Y, Xu H, Huang X, Li S, Zhou A, et al. . 2018. WEGO 2.0: a web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 46:W71–W75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Zhang Q, Peng Z, Miao X. 2019. Lipid droplets mediate salt stress tolerance in Parachlorella kessleri. Plant Physiol. 181:510–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu D, Zhu D, Liu N, Yan Y. 2021. Endoplasmic reticulum subproteome analysis reveals underlying defense mechanisms of wheat seedling leaves under salt stress. Int J Mol Sci. 22:4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Chen J, Doak TG, Song W, Yan Y. 2020. ADFinder: accurate detection of programmed DNA elimination using NGS high-throughput sequencing data. Bioinformatics 36:3632–3636. [DOI] [PubMed] [Google Scholar]
- Zheng W, Wang C, Lynch M, Gao S. 2021. The compact macronuclear genome of the ciliate Halteria grandinella: aA transcriptome-like genome with 23,000 nanochromosomes. mBio 12:e01964-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article, its supplementary information files and publicly available repositories. Reads for the genome assemblies have been deposited to NCBI GenBank (PRJNA822294). The gene training model for Fabrea salina in Augustus and orthogroup results have been deposited to GitHub (https://github.com/callAgene/Supplementary-Materials-for-Fabrea-salina).