Abstract
Cryptosporidiosis is a major global health problem and a primary cause of diarrhea, particularly in young children in low- and middle-income countries (LMICs). The zoonotic Cryptosporidium parvum and anthroponotic Cryptosporidium hominis cause most human infections. Here, we present a comprehensive whole-genome study of C. hominis, comprising 114 isolates from 16 countries within five continents. We detect two lineages with distinct biology and demography, which diverged circa 500 years ago. We consider these lineages two subspecies and propose the names C. hominis hominis and C. hominis aquapotentis (gp60 subtype IbA10G2). In our study, C. h. hominis is almost exclusively represented by isolates from LMICs in Africa and Asia and appears to have undergone recent population contraction. In contrast, C. h. aquapotentis was found in high-income countries, mainly in Europe, North America, and Oceania, and appears to be expanding. Notably, C. h. aquapotentis is associated with high rates of direct human-to-human transmission, which may explain its success in countries with well-developed environmental sanitation infrastructure. Intriguingly, we detected genomic regions of introgression following secondary contact between the subspecies. This resulted in high diversity and divergence in genomic islands of putative virulence genes, including muc5 (CHUDEA2_430) and a hypothetical protein (CHUDEA6_5270). This diversity is maintained by balancing selection, suggesting a co-evolutionary arms race with the host. Finally, we find that recent gene flow from C. h. aquapotentis to C. h. hominis, likely associated with increased human migration, maybe driving the evolution of more virulent C. hominis variants.
Keywords: Cryptosporidium hominis, speciation, whole genome sequencing, comparative genomics, population structure, population genetics, recombination, gene flow, secondary contact, evolution
Introduction
Cryptosporidiosis is a leading cause of diarrhea in children under five globally (Kotloff et al. 2013; Khalil et al. 2018), resulting in an estimated 48,000 deaths annually. Among parasitic diseases, it is second only to malaria in global health burden, with an overall impact of ∼12.8 million disability adjusted life-years (Khalil et al. 2018). Human cryptosporidiosis is primarily caused by Cryptosporidium parvum, a zoonoses common in young ruminants (Ryan et al. 2016; Santin 2020), and Cryptosporidium hominis, which is anthroponotic and the more prevalent species globally (Razakandrainibe et al. 2018). The disease burden is overwhelmingly skewed to low- and middle-income countries (LMICs) (Yang et al. 2021), particularly in sub-Saharan Africa (Khalil et al. 2018), where C. hominis predominates. However, Cryptosporidium remains a significant public health problem in wealthy countries through large water or foodborne outbreaks and direct transmission in daycare facilities, hospitals, and other institutions (Craun et al. 1998; Putignani and Menichella 2010). Presently, there are no vaccines or effective drugs to treat the infection. Hence, control depends on the prevention of infection, which is driven by a strong understanding of the parasite’s epidemiology.
The global epidemiology of cryptosporidiosis varies by geographic region, socioeconomic status, and a range of risk factors (Nichols et al. 2014; Yang et al. 2021). Understanding of this epidemiology is underpinned by molecular typing, based mainly on the highly polymorphic gp60 gene (Feng et al. 2018). This work has identified numerous genetic variants within each species and indicated a complex population genetic structuring. In C. parvum, population structure varies globally from clonal to epidemic to panmictic, likely due to varying ecological factors (Morrison et al. 2008; Herges et al. 2012; Wang et al. 2014). Genetic variants found exclusively in humans point to an anthroponotic C. parvum lineage (IIc) (King et al. 2017; King et al. 2019), with a recent genomic study recognizing two subspecies, the zoonotic C. parvum parvum and the anthroponotic C. parvum anthroponosum (Nader et al. 2019). Interestingly, this study found that both subspecies occasionally still hybridize and exchange genetic variation. These exchanges overlapped with similar genomic regions undergoing genetic introgression between C. hominis and C. p. anthroponosum, indicating candidate sites underpinning adaptation to human-specific infection (Nader et al. 2019). The signature of introgression can be identified by comparing the DNA sequence (dis)similarity between two or more haplotypes. Briefly, introgressed regions are characterized by high nucleotide similarity, resulting from a relatively recent coalescence of the introgressed sequence. By comparing the sequence variation of three or more haplotypes, the directionality of genetic exchange can be inferred. In addition, the genetic divergence (i.e., number of nucleotide substitutions) can be used to estimate how long ago the sequence was exchanged between the ancestors of the haplotypes. These are the principles of software such as HybridCheck (Ward and van Oosterhout 2016) that enable the study of genetic introgression. Cryptosporidium hominis appears to have a largely clonal population structure, dominated by specific variants in different regions (Yang et al. 2021). The IbA10G2 gp60 subtype, although found in LMICs (Jex and Gasser 2010), is the dominant variant in high-income countries and accounts for up to ∼45% of all gp60-typed human infections (Jex and Gasser 2010). This subtype is linked to most major waterborne outbreaks in wealthy countries for which genetic typing is available (Zhou et al. 2003; Chalmers et al. 2010; Widerström et al. 2014; Segura et al. 2015; Efstratiou et al. 2017). An epidemiological study in the UK that investigated household transmission of Cryptosporidium found that person–person transmission was a key pathway for C. hominis, with 78% of 40 samples subtyped as IbA10G2 (McKerr et al. 2022).
Studies of C. hominis molecular epidemiology pose several essential questions that have major implications for the global control of cryptosporidiosis. Specifically (1) what is the IbA10G2 subtype, and is this gp60-defined subtype reflective of a phylogenomically divergent C. hominis lineage that predominates in wealthy countries; (2) if so, does this lineage undertake reduced levels of genetic recombination with global C. hominis populations; (3) and can signatures within the genome sequences of IbA10G2 typed isolates identify its taxonomic status, reveal the factors underpinning its putatively increased virulence and its dominance in wealthy countries, and identify its influence on global parasite population structure? To address these questions, we performed a global study of 114 C. hominis genome sequences, comparing the IbA10G2 subtype to published genome sequences representing locally acquired infections from 16 countries across five continents. The insights gained from these analyses are particularly relevant for public health; the IbA10G2 subtype has already been identified as an emerging host-adapted, likely more virulent and transmissible population, making it a threat to human health in high-income countries (Li et al. 2013; Feng et al. 2014; Cacciò and Chalmers 2016).
Results
Genomic Evidence of Population Substructuring at the Continental Level
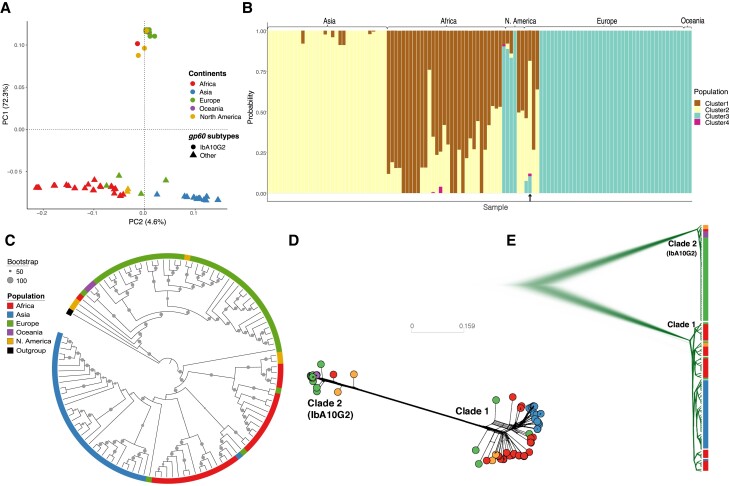
All 114 C. hominis isolates included in this study were confirmed as single variant infections (estimated multiplicity of infection = 1 and Fws > 0.95) (see supplementary table S2, Supplementary Material online). We identified 5,618 biallelic single nucleotide polymorphisms (SNPs) among these samples and used these to explore the population structure of C. hominis. Our analyses identified two major clusters (fig. 1A), separating European, North American, and Oceanian samples from Asian and African samples. We also saw minor clustering separating African from Asian samples. STRUCTURE analysis provided further support for these findings (fig. 1B). Overall, ∼94% of isolates are clustered geographically. Exceptions included a small number of infections (e.g., UKH30 [from the UK]) acquired during international travel. STRUCTURE analysis also identified a fourth cluster, including three African and one European isolate, with unique population ancestry (cluster 4 in fig. 1B).
Fig. 1.
Global population structure of Cryptosporidium hominis isolates illustrating their substructing and diversification. (A) PCA of isolates based on the filtered set of whole-genome SNPs, highlighting three clusters of isolates which are predominately based on continents of origin. Isolates were color coded with their continent of origin. Isolates associated with gp60 subtype IbA10G2 were represented with solid circles while non-IbA10G2 with solid triangles. (B) Structure plot illustrating population genetic ancestry and the admixed nature of the C. hominis isolates. The plot was obtained for an optimum value of K = 4. The black arrow (bottom) indicates the highly admixed isolate (UK_UKH4), which includes all four ancestries. (C) Maximum likelihood-based phylogenetic tree. (D) Splitstree and (E) Densitree are also demonstrating two major clades. Cryptosporidium hominis hominis (clade 1) includes isolates associated with other gp60 subtypes, whereas Cryptosporidium hominis aquapotentis (clade 2) includes isolates associated with gp60 subtype IbA10G2.
Genomic Evidence of Population Diversification
Maximum likelihood (ML) analysis (fig. 1C) identified two major clades, one corresponding to Asia and Africa (clade 1) and the other to Europe, North America, and Oceania (clade 2). All isolates within clade 2 had gp60 subtype IbA10G2, and no IbA10G2 isolates clustered with clade 1. The two clades were estimated to have diverged 488 (84–2,199; 95% highest posterior density [HPD]) years ago. This divergence is supported by SplitTree network (fig. 1D) and DensiTree (fig. 1E) analyses. Considering the stability of these two genomically distinct lineages and evidence below of their reproductive isolation, we propose their recognition as separate subspecies. We propose clade 1, which comprises infections observed in LMICs, mostly from Africa and Asia, be recognized as C. h. hominis, referring to the fact that this subspecies represents the majority population. We propose clade 2 (IbA10G2 subtype) includes isolates from high-income countries, namely Europe, North America, and Oceania, be named C. hominis aquapotentis (strong water), as it predominates in countries with long-standing high sanitation and water quality indices.
Demographic Histories
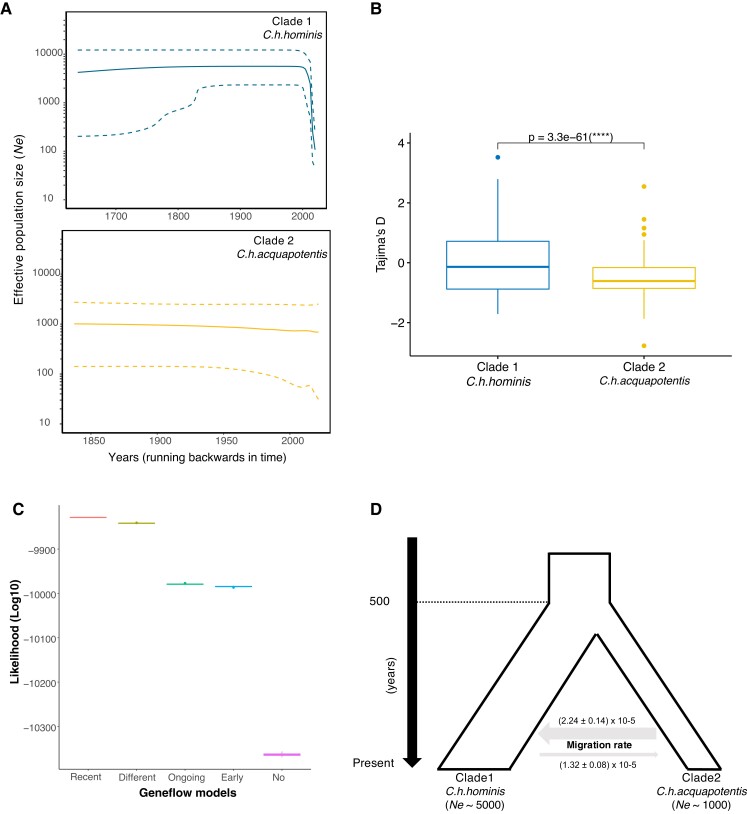
To understand the demographic histories and estimate the change in the effective population size (Ne) through time, we constructed a Bayesian skyline plot (BSP) (Drummond et al. 2005) for C. h. hominis (clade 1) and C. h. aquapotentis (clade 2, gp60 subtype IbA10G2 IbA10G2). Parasite isolates from low-income countries (C. h. hominis, clade 1) experienced a marked population contraction recently from Ne = ∼5000 to Ne = ∼100 (fig. 2A), which is supported by a higher proportion of positive Tajima’s D values (fig. 2B). In contrast, the isolates from high-income countries (C. h. aquapotentis (clade 2), gp60 subtype IbA10G2) had a stable effective population size (Ne = ∼1000) and a higher proportion of negative Tajima’s D values (fig. 2C). The distribution of Tajima’s D in C. h. aquapotentis is significantly smaller than zero (one-sided t-test: t = −28.883, df = 681, P value < 2.2×10−16), which is consistent with recent population expansion. BSP analysis of C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) suggests a stable Ne, whereas the overall negative value of Tajima’s D analysis is consistent with a recent population size expansion or selective sweep. BSP analyses are based on coalescent theory, whereas Tajima’s D compares the constitution of polymorphisms, that is, the mean number of pairwise differences and the number of segregating sites. The latter is more sensitive to recent demographic events. Altogether, our analyses thus imply that after a relatively stable Ne, the population has started to expand only very recently, or that it has been affected by a recent selective sweep. Simulation-based projections of the evolution of the overall C. hominis population finds it is being shaped by “recent gene flow”, likely indicating recent secondary contact (fig. 2C and D) between the two subspecies ∼165 (24–647; 5–95% confidence interval [CI]) years ago.
Fig. 2.
Demographic histories and population size and secondary contact between Cryptosporidium hominis hominis (clade 1) and Cryptosporidium hominis aquapotentis (clade 2). (A) BSPs depicting change in Ne (effective population size) through time, for both the clades. The central dark line and the upper and lower dashed lines on Y-axis are mean estimates and 95% HPD intervals of Ne, respectively. X-axis is time in years, running backwards. (B) Boxplot showing significant difference (two-sided t-test) in Tajima’s D values between C. h. hominis (clade 1) and C. h. aquapotentis (clade 2). (C) Higher likelihood (log10) for “recent gene flow” model. Comparing likelihood distributions of gene flow models and observed significant difference (one-way ANOVA test, F = 2629761, df = 4, P value < 2 × 10−16). Further, post hoc Tukey–HSD test revealed difference in likelihood between all the models (P value < 1 × 10−16). (D) Graphical representation of demographic history of C. hominis, illustrating recent secondary contact and migration rates between the two clades (mean ± SE).
We found evidence of two selective sweeps on chromosome 6 in C. h. hominis (clade 1) based on composite likelihood ratio (CLR) statistic using SweeD (Pavlidis et al. 2013; supplementary fig. S1, Supplementary Material online). However, we do not see any other hallmarks of a selective sweep in these regions based on π or Tajima’s D (see Supplementary fig. S2, Supplementary Material online). Possibly, the selective sweeps were incomplete or occurred in the distant past, eroding their genomic signatures. Nucleotide diversity and Tajima’s D are simple statistical approaches to identify selective sweep, but they are also affected by population size changes. In contrast, SweeD analyses site frequency spectra (SFS) and uses complex statistical approaches such as likelihood-based methods to identify selective sweeps in whole-genome data. However, the likelihood of selective sweep identified in our analysis is low which might be the reason that nucleotide diversity and Tajima’s D methods did not identify any statistically significant sweeps. Nevertheless, this may have contributed to the recent decline in effective population size of C. h. hominis (clade 1) (fig. 2A); genetic variation could have been lost during the selective sweep not only in the affected chromosomal regions but throughout the genome in this largely clonally reproducing organisms, as the selectively favored variant replaced other existing variants.
Linkage, Recombination, Introgression, and Gene Flow between Subspecies
Distinct Patterns of Decay of Linkage Disequilibria
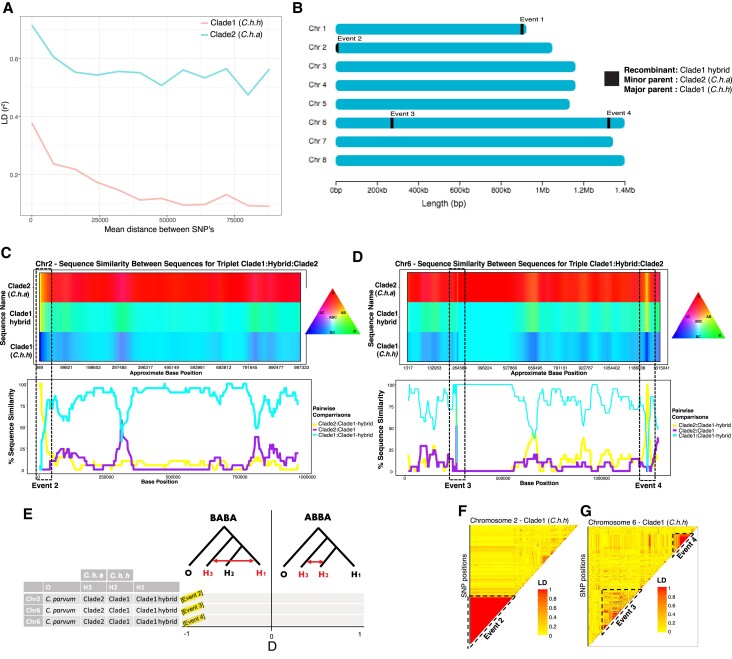
We performed independent linkage analyses for each subspecies to infer the recombination rate within parasite populations from low- and high-income countries (fig. 3A). Cryptosporidium hominis hominis (clade 1) had more rapid linkage disequilibrium (LD) decay than C. h. aquapotentis (clade 2, gp60 subtype IbA10G2), consistent with genetic exchanges through gene flow and recombination. In contrast, the strong LD in C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) supports our hypothesis of a recent population expansion (see below).
Fig. 3.
Analyses of recombination and gene flow between Cryptosporidium hominis hominis (clade 1) and Cryptosporidium hominis aquapotentis (clade 2). (A) LD decay plot showing rapid decay of linkage between SNPs in C. h. hominis (clade 1) compared with C. h. aquapotentis (clade 2). (B) Graphical representation of recombinant breakpoint positions detected by RDP4 program between C. h. hominis (clade 1) and C. h. aquapotentis (clade 2). (C,D) HybridCheck plots representing genomic signature of introgression in chromosomes 2 and 6, respectively. Analysis for chromosome 1 was excluded due to unknown parental sequences. The plots were generated for random set of triplets that includes recombinant (hybrid), minor (donor) and major (recipient) parental sequence, as detected by RDP4 program. Introgressed blocks (recombinant breakpoints) were illustrated with dashed boxes, showing high similarity between the recombinant (C. h. hominis hybrid isolates) and minor parent (C. h. aquapotentis isolates). The top panel illustrates the visualization of sequence similarity between sequences within the triplet, using RBG color triangle. The two sequences are colored same (yellow, purple, or turquoise) if they share polymorphism. (E) Gene flow analyses with ABBA–BABA test, representing D statistics for the random sets of triplets (as used in c,d) along with Cryptosporidium parvum as an outgroup. D statistic values close to −1 at all three recombinant events, suggesting gene flow between H1 and H3. (f,g) Pairwise LD of SNPs in chromosomes 2 and 6 of C. h. hominis showing red blocks of high linkage between SNPs in introgressed events 2–4.
Recombination and Regions of Secondary Contact
Using Recombination Detection Program version 4 (RDP4) (Martin et al. 2015), we identified significant recombination events between the two clades in chromosomes 1 (event 1), 2 (event 2), and 6 (events 3 and 4) (fig. 3B and Supplementary material table S3, Supplementary Material online). This indicated secondary contacts between C. h. hominis (clade 1) and C. h. aquapotentis (clade 2, gp60 subtype IbA10G2), which resulted in rare genetic exchanges between these otherwise diverged clades. We also undertook additional analyses of the highly admixed European isolate (UK_UKH4 of gp60 subtype IaA14R3) that showed unique ancestry (fig. 1B) and clustered with low-income countries (C. h. hominis [clade 1]) (see supplementary text, Supplementary Material online).
Signature of Introgression and Gene Flow between the Clades
We analysed the recombination events in more detail to better understand the implications of genetic introgression between the two subspecies. Determining the signature of genetic introgression is crucial as these regions can also be responsible for increasing genetic diversity and providing novel substrate for natural selection in host–parasite coevolution (Van Oosterhout 2021). We used HybridCheck (Ward and van Oosterhout 2016) to perform introgression analyses for recombinant events 2–4 (event 1 was excluded due to a missing parental sequence). We randomly selected a triplet (recombinant, minor parent, and major parental lines) from the RDP output (supplementary table S3, Supplementary Material online), which revealed a clear signature of introgression (fig. 3C and D), also supported by an ABBA–BABA test (fig. 3E). Additionally, we calculated the pairwise r2 between SNPs within chromosomes of C. h. hominis (clade 1) to assess linkage among SNPs in the introgressed regions (fig. 3F and G). Large blocks of high LD that encompass the introgressed regions suggested each had been exchanged as a single event between C. h. hominis (clade 1) and C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) ∼165 (24–647; 5–95% CI) years ago.
Could Recent Introgression Increase Virulence?
Our analyses suggest C. h. hominis (clade 1) and C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) have diverged and largely reproductively isolated for ∼500 years. Noting that the latter subspecies has come to dominate infections within high-income countries (Guo et al. 2015) is more virulent (Cama et al. 2008), appears better able to transmit through direct human-to-human contact (McKerr et al. 2022), and possibly at a lower infectious dose (Segura et al. 2015); C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) may owe its success to being better adapted to human infection. If this is the case, it is possible that recent introgression between these two subspecies could select for more virulent and transmissible C. hominis subspecies in low-income countries, particularly noting the recent genetic bottlenecking we have observed in C. h. hominis (clade 1) within the last decades.
Identification of Potential Virulence Genes
To explore this, we first predicted candidate virulence genes in C. hominis likely to be involved in the host–parasite interaction and engaged in a coevolutionary arms race with the host. Broadly, such genes are under continuous adaptation while interacting with hosts (Van Oosterhout 2021). During coevolution, evolutionary forces act on virulence genes to create genetic variation through mutation, recombination, and gene flow, and this variation is molded by natural selection (and genetic drift). To identify putative virulence genes in C. hominis, we selected the top 5% most highly polymorphic genes based on nucleotide diversity (π). These were filtered for the top 25% of genes under balancing selection, based ranked Tajima’s D. Finally, these genes were filtered by selecting the top 50% of genes with the highest proportion of nonsynonymous mutations, based on the ranked Ka/Ks ratio. Using this approach, we identified 24 highly polymorphic genes (supplementary table S5, Supplementary Material online) that are rapidly mutating at the protein level and under selective pressure. These genes were significantly more polymorphic (two-sided t-test: t = −2.9062, df = 23, P value = 0.007957), under stronger balancing selection (two-sided t-test: t = −10.393, df = 23, P value = 2.533 × 10−10) and positive selection (two-sided t-test: t = −2.4736, df = 23, P value = 0.021) compared with all remaining polymorphic genes. Moreover, these genes are enriched in recombination regions (χ2 = 225.04, df = 1, P value = 0.00049), showing that gene flow within C. hominis is inclined toward elevated levels of genetic variation. Overall, these genes are enriched for extracellular (χ2 = 13.608, df = 1, P value = 0.00349) and signal peptide proteins (χ2 = 6.69, df = 1, P value = 0.015), which is consistent with their potential relevance for host–parasite interaction. Together, these results provided the evidence of genomic islands of putative virulence genes (GIPVs: fig. 4) that are likely to be in a coevolutionary arms race with the host, undergoing frequent recombination and accumulating beneficial polymorphisms that are under selection and that this increased population diversity.
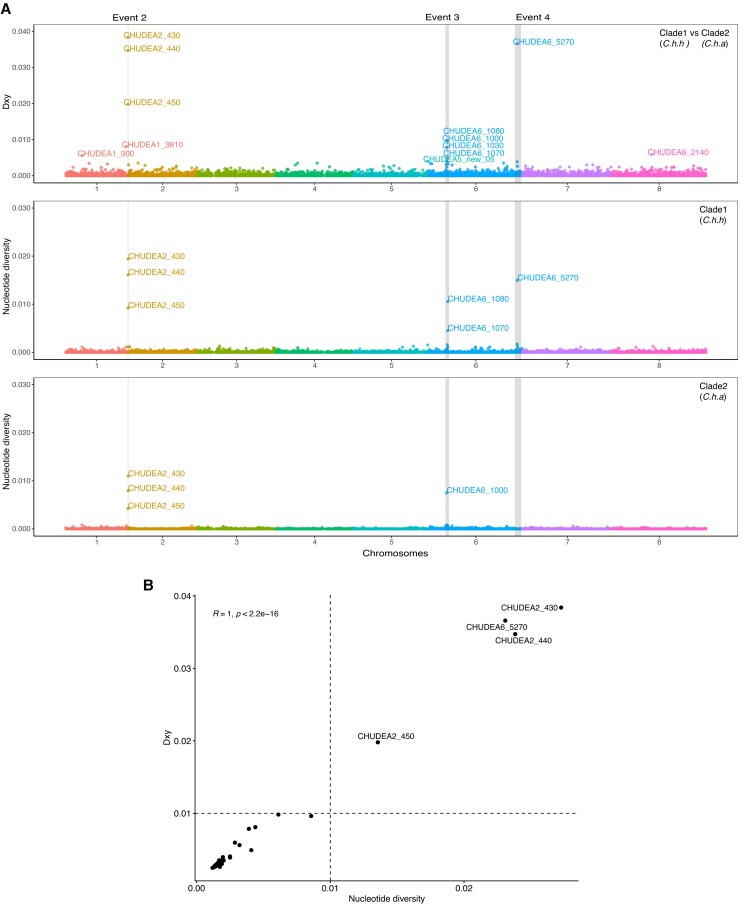
Fig. 4.
Population genetic analyses of GIPVs. (A) Population genetic and divergence analyses of introgressed regions. X-axis represents genomic positions of eight chromosomes highlighted with different colors. Population divergence (Dxy) between Cryptosporidium hominis hominis (clade 1) and Cryptosporidium hominis aquapotentis (clade 2) for each gene were plotted on Y-axis (top panel). Nucleotide diversity (π) for C. h. hominis (middle panel) and C. h. aquapotentis (bottom panel) for each gene, was also plotted on Y-axis, respectively. The breakpoints of four recombination events (event 1–4) were indicated by gray vertical boxes. Event 1 was undetected in C. h. aquapotentis. (B) Correlation between π and Dxy were plotted to identify polymorphic and potential virulence genes.
Subspecific Divergence of Putative Virulence Genes
We investigated whether any virulence genes predicted above were highly diverged between C. h. hominis (clade 1) and C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) and under diversifying selection between the two subspecies. To do this, we calculated absolute divergence (Dxy), representing the average proportion of differences between all pairs of sequences between C. h. hominis (clade 1) and C. h. aquapotentis (clade 2, gp60 subtype IbA10G2), revealing differential gene flow during their reproductive isolation (fig. 4A). We then calculated the correlation between diversity (π) and divergence (Dxy) for each gene (fig. 4B). This approach identified four clear outlier genes, which were the most divergent, diverse, and rapidly mutating at the protein level. These genes encoded three mucins (CHUDEA2_430, CHUDEA2_440, and CHUDEA2_450) arrayed in a cluster on chromosome 2, and a hypothetical protein (CHUDEA6_5270) found on chromosome 6 (fig. 4B, supplementary table S5, Supplementary Material online). The gp60 gene ranked sixth in the list of virulence genes but does not sit as a clear outlier from the other genes identified in our assessment. Two of these genes, CHUDEA2_430 (LRT, P value = 0.005) and CHUDEA6_5270 (LRT, P value = 0.017), are under statistically significant diversifying selection between C. h. hominis (clade 1) and C. h. aquapotentis (clade 2, gp60 subtype IbA10G2).
Lastly, we looked at each codon position within each of the four outlier genes to detect codon specific diversifying selection, which might be overlooked in the overall gene. We detected episodic diversifying selection (LRT P value < 0.05) at codon positions 92, 111, and 138 of CHUDEA2_430 and 97 and 105 of CHUDEA6_5270 using the mixed effects model of evolution method implemented in Datamonkey (Pond and Frost 2005). These sites represent putative codons harboring genetic polymorphisms that experience periods of strong diversifying selection. This strongly suggests CHUDEA2_430 and CHUDEA6_5270 are likely candidate virulence factors participating in a coevolutionary arms race and contributing to the divergence of C. h. hominis (clade 1) and C. h. aquapotentis (clade 2, gp60 subtype IbA10G2).
Recent Introgression of Putative Virulence Genes
Finally, we asked whether the recent recombination events in chromosomes 2 and 6 between C. h. hominis (clade 1) and C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) might include genes associated with increased virulence, indicating increasing virulence of C. hominis globally within the past decades, possibly linked to increased human migration. These introgressed regions had significantly elevated levels of nucleotide diversity (two-sided t-test: t = −3.0026, df = 27, P value = 0.0057), divergence (two-sided t-test = −3.0043, df = 27, P value = 0.0057) and balancing selection (two-sided t-test: t = −3.0125, df = 27, P value = 0.0055). Interestingly, we observed a pattern of high diversity and divergence (fig. 4A), particularly for genes within the recombinant blocks (fig. 3B–E). The 38 (= top 1%) most divergent genes between C. h. hominis (clade 1) and C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) were enriched in recombinant regions (χ2 = 287.08, df = 1, P value = 0.00049), with 37% present in these regions (supplementary table S6, Supplementary Material online), compared to 0.9% of the other 1,645 divergent genes. These results suggest the genes undergoing frequent recombination accumulated beneficial polymorphisms that were maintained by balancing selection and that this increased population diversity.
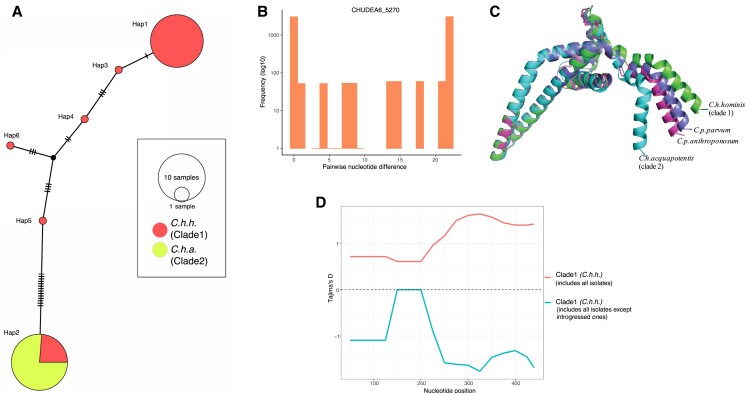
Notably, the introgressed regions between C. h. hominis (clade 1) and C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) included CHUDEA2_430 (muc5) for event 2 (chromosome 2), CHUDEA6_1080 (gp60) for event 3 (chromosome 6), and CHUDEA6_5270 (a hypothetical protein) for event 4 (chromosome 6). The introgression at CHUDEA6_5270 is particularly intriguing as it sheds further light on the evolution of C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) and C. h. hominis (clade 1), revealing how recent recombination events could be driving the virulence evolution of C. h. hominis (clade 1). We identified two major CHUDEA6_5270 (hypothetical gene) haplotypes; Hap1 representing most C. h. hominis (clade 1) isolates, and Hap2 represents all C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) plus a small subset of C. h. hominis (clade 1) (fig. 5A), as well as many haplotypes associated only with C. h. hominis (clade 1). No mutations were observed in Hap2, which is consistent with C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) having evolved recently, being an estimated 392 years (29–1699 years; 5–95% CI) old, assuming a mutation rate of u = 10−8 and 48 h cell division time. Hap1 and Hap2 are diverged by 22 SNPs (fig. 5B), which is unlikely to represent standing genetic variation, given that these are 3-fold higher than the mean deviation among all other CHUDEA6_5270 haplotypes identified here. Instead, we propose CHUDEA6_5270-Hap2 might be an introgressed variant from a highly diverged, unsampled (sub)species that diverged from C. hominis around 2.36 million generations ago (assuming a mutation rate of u = 10−8), which is equal to 12,742 (8,901–17,497 years; 5–95% CI) years (assuming 48 h/replication). The emergence of CHUDEA6_5270-Hap2 in some C. h. hominis (clade 1) isolates (i.e., the red section of Hap2 in fig. 5A) strongly implies that C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) has introgressed (ca. 400 years ago) into C. h. hominis (clade 1) leading to evolution under balancing selection (fig. 5D).
Fig. 5.
Illustrating diversifying selection between Cryptosporidium hominis subspecies and host adaptation at CHUDEA6_5270 (hypothetical gene). (A) Haplotype network analyses illustrating haplotype diversification between Cryptosporidium hominis hominis (clade 1) and Cryptosporidium hominis aquapotentis (clade 2). (B) Pairwise nucleotide divergence shows bimodal distribution, which, theoretically, can be explained both by balancing selection (Lighten et al. 2017), as well as by genetic introgression. (C) Comparison of predicted models of protein structure of CHUDEA6_5270 gene between Cryptosporidium species and subtypes demonstrates variation towards C-terminal region. (D) Introgressed isolates driving balancing selection at gene CHUDEA6_5270 in C. h. hominis. Red line represents balancing selection (positive Tajima’s D) in C. h. hominis that also includes introgressed isolates. Blue line represents purifying selection (negative Tajima’s D) in C. h. hominis after excluding introgressed isolates.
We modeled the 3D protein structure of CHUDEA6_5270-Hap1 and Hap2 and compared these to similarly predicted 3D protein structures for the orthologous genes from C. p. parvum and C. parvum anthropanosum (fig. 5C). This modeling identified several conserved alpha-helices that appear to form a coiled-coil domain. CHUDEA6_5270 from C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) encodes mutations near the C-terminal end, resulting in a notable kink that deviates from the other structures. This structural variation overlaps with an increase in Tajima’s D values toward the C-terminus of the protein, suggesting the region is under balancing selection (fig. 5D).
A similar pattern is observed for CHUDEA2_430 (see supplementary fig. S6, Supplementary Material online). However, given that both the subspecies are interspersed in the haplotype network, indicating against a specific directionality of introgression. This may indicate that CHUDEA_430 diverged with the divergence of the subspecies and continued to diversify. We were not able to generate a robust 3D structural model for CHUDEA2_430 or its C. parvum orthologs. We noted the gene encodes a large intrinsically disordered region (supplementary fig. S7, Supplementary Material online) which is a consistent feature of mucin proteins (Carmicheal et al. 2020), whose structural confirmation is influenced by post-translation glycosylation (Perez-Vilar and Hill 1999). Noting this, we did identify eight novel glycosylation sites in C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) CHUDEA2_430 haplotypes not found in C. h. hominis (clade 1) (supplementary fig. S6, Supplementary Material online). Whether these impact interaction with host proteins is not known, but this would be consistent with glycosylated proteins in other pathogens (Lin et al. 2020).
Discussion
In this study, we examined the evolutionary genomics of a major human parasite, C. hominis, studying whole-genome sequence data from 114 isolates from 16 countries across five continents. We posed three questions, and we are able to answer these as follows: (1) the gp60-defined subtype IbA10G2 is reflective of a phylogenomically divergent C. hominis lineage that predominates in wealthy countries; (2) this lineage has experienced significantly reduced levels of genetic recombination with other global C. hominis populations, and we could identify only four genetic exchanges between these otherwise diverged clades; (3) the distinct evolutionary trajectory of the IbA10G2 subtype, characterized by rapid population expansion, warrants a distinct taxonomic status as subspecies. We propose the name C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) to reflect its adaptation to “strong water”, that is, high sanitation and water quality indices.
These two subspecies are estimated to have been reproductively isolated for ∼ 488 (84–2,199) years, except for the more recent genetic exchange at four genetic loci. The reproductive isolation coincides with improvements to sanitation in Europe. It is possible that C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) evolved specializations making it better suited to human infection allowing it to be more successful through direct transmission supported by a lower infectious dose. Such adaptations would have allowed C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) to become dominant in higher-income countries where sanitation has reduced the level of environmental transmission. Indeed, epidemiological investigations in the UK identified direct person-to-person transmission as a key pathway for C. hominis with C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) predominating (McKerr et al. 2022). We hypothesize this may have resulted in its rapid population expansion over the last ∼500 years and (partial) reproductive isolation. In contrast, C. h. hominis (clade 1) experienced recent reduction in effective population size (Ne) within the past few decades. We detected two signatures of recent selective sweep in this subspecies, and we propose that this may have eroded some of the genetic variation, resulting in the marked drop in Ne = 5000 to Ne = 100 in the past. This would have resulted in significant genetic drift and random allele frequency changes, which could have increased the divergence between C. h. hominis (clade 1) and C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) further.
Despite increased migration and international traveling in the past decades, our study suggests that there has been relatively little movement between continents for this parasite. This is in contrast to reports for C. parvum.Corsi et al. (2021) recently found a higher proportion of admixture and gene flow between C. parvum populations and no evidence of population structuring by geographic region. Despite the strong population substructuring in C. hominis, we found evidence of potential recombination and gene flow between the geographic populations and subspecies. We further investigated and identified the introgressed regions where we detected significant gene flow between the low- and high-income countries. Simulation-based analyses indicated this was most likely explained by “recent gene flow” (ca. 165 years ago). This would appear to be a secondary contact between the two subspecies after recent globalization, illustrating higher migration rate from high-income to low-income countries, which facilitated gene flow, recombination, population admixture, and selective sweep.
Genetic exchanges between C. h. hominis (clade 1) and C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) are rare compared with those within C. p. parvum (Corsi et al. 2021), and their frequency might be more comparable with the rate of sequence exchange between C. p. parvum and C. p. anthroponosum (Nader et al. 2019). However, whole-genome analyses of more C. hominis isolates may detect other recombination events in addition to the four events detected in our study. In addition, further studies may be able to discover the unknown parental sequences associated with recombination event 1 in our study. Without this parental sequence, we were unable to reconstruct the evolution of this introgressed sequence on chromosome 1. Yet, this event may be a key player in the evolution of the lineage. We encourage future whole-genome studies on C. hominis, believing this may shed further light on the incipient speciation of C. h. aquapotentis (clade 2, gp60 subtype IbA10G2).
Although our data set comprises samples across five continents, we only studied C. hominis in 16 countries in total, which means that we could have missed local gene flow and patterns of population substructuring within continents. Although the marked biological differences between C. parvum and C. hominis have been well established (Abrahamsen et al. 2004), recent population genomic research is demonstrating that also within these species, the population genetics and evolutionary genetics of their subspecies are remarkably distinct. Large data sets and comparative population genomic and phylogenomic analyses (across Cryptosporidium species) are warranted to examine the evolutionary genomics of these parasites in more detail.
Finally, we have discovered GIPVs contributing to population diversification between C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) and C. h. hominis (clade 1). These islands have experienced relatively elevated recombination rate which has enriched nucleotide variation under balancing selection and the acquisition of nonsynonymous SNPs, consistent with virulence factors driving host–parasite interactions. Intriguingly, the most significant signals within these analyses are driven by gp60, a hypothetical protein (CHUDEA6_5270) and a cluster of mucin-like genes (CHUDEA2_430, CHUDEA2_440, and CHUDEA2_450) found on chromosome 2. These genes are consistently identified as being under selection in the evolution of the C. hominis subspecies here and in similar observations made of C. hominis in Africa (Tichkule et al. 2021). Their orthologs are associated with recombination between human-specific C. p. anthroponosum relative to the zoonotic C. parvum parvum and appear to have driven a convergence of the former with C. hominis (Nader et al. 2019).
CHUDEA2_430 (muc5) and hypothetical protein CHUDEA6_5270 are the most notable, displaying significant diversifying selection between the two subspecies. Broadly, mucins mediate cell–cell interactions (O'Connor et al. 2009) and modulate the infectivity of Cryptosporidium sporozoites and merozoites and oocyst production (Cevallos et al. 2000; O'Connor et al. 2009). In C. parvum, MUC5 is involved in host-cell invasion and an important determinant of host adaptation (O'Connor et al. 2009) and highly expressed in the first 2 h of infection in vitro (Lippuner et al. 2018). MUC5 may also play a role in tethering the sporozoite to the oocyst wall (Chatterjee et al. 2010). Our analyses suggest C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) and C. h. hominis (clade 1) muc5 haplotypes diverged before or with the subspecies and subsequently diversified, which is consistent with prior observations implicating CHUDEA2_430 in the emergence of C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) (Bouzid et al. 2013; Feng et al. 2018). This appears to have resulted in the acquisition of novel glycosylation sites within C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) muc5 haplotypes. We cannot determine the functional consequence of these sites but note that glycosylation sites often mediate the specificity of mucin interactions with host proteins in a variety of pathogens (Lin et al. 2020). In contrast, CHUDEA6_5270 displays a clear signal for the recent introgression of a novel C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) haplotype into C. h. hominis (clade 1) after the divergence of these subspecies. This haplotype has notable, structurally relevant, mutations. Identifying the function of this gene, its potential role in infection, and the relevance of the structural variation we have inferred here should be considered a major research priority.
In conclusion, this work represents the first large-scale population genomic study in any Cryptosporidium species, inferring the global population structure and evolutionary history of C. hominis. We propose recognition of two distinct subspecies, C. h. hominis (clade 1) and C. h. aquapotentis (clade 2, gp60 subtype IbA10G2), with distinct demographic histories that have diverged circa 500 years ago. Although the subspecies differ in their global distribution, their gene pools are not completely isolated, and rare genetic exchanges have occurred in the recent past. We contend that many of the genes, CHUDEA2_430 and CHUDEA6_5270 in particular, in these introgression regions are involved in infection and that their evolution in humans may be driving greater human specificity, virulence, and transmissibility. It appears C. h. aquapotentis (clade 2, gp60 subtype IbA10G2) is playing a key role in this process, which is supported by previous observations based on multilocus typing (Li et al. 2013). This illustrates how human-mediated gene flow is involved in parasite evolution and genomic architecture, and how it could affect virulence evolution. In addition, it shows that the GIPVs that result from population admixture in an anthroponotic species are under selection and involved in the evolutionary arms race.
Materials and Methods
Parasite Isolates
The C. hominis isolates newly sequenced for this study (n = 34) were archived stool samples collected at the Cryptosporidium Reference Unit in the UK. The species was determined by species-specific real-time PCR targeting the A135 gene (Robinson et al. 2020) and subtyped by PCR and sequencing of the gp60 gene (Chalmers et al. 2019). Supplementary table S1, Supplementary Material online provides information about these isolates. Isolates were selected to mainly represent the dominant variant, IbA10G2, as defined by gp60 sequencing.
Processing of Fecal Samples for Whole-Genome Sequencing
Stool samples were processed as previously described (Hadfield et al. 2015). Briefly, saturated salt-flotation was used to obtain a partially purified suspension of oocysts starting from 1 to 2 ml of each fecal sample. Oocysts were further purified from the suspension by immunomagnetic separation (IMS), using the Isolate IMS kit (TCS Biosciences, Botolph Claydon, UK). IMS-purified oocysts were treated with bleach and washed three times with nuclease-free water by centrifugation at 1,100 × g for 5 min. The pellets were suspended in 200 μl of nuclease-free water for DNA extraction.
DNA Preparation and Whole-Genome Sequencing
Genomic DNA was extracted from purified Cryptosporidium oocysts by first performing eight cycles of freezing in liquid nitrogen for 1 min and thawing at 95°C for 1 min, and then using the QIAamp DNA extraction kit (Qiagen, Manchester, UK) according to the manufacturer’s instructions. The genomic DNA was eluted in 50 μl nuclease-free water, and the concentration was measured using the Qubit dsDNA HS Assay Kit with the Qubit 1.0 fluorometer (Invitrogen, Paisley, UK), according to the manufacturer’s instructions.
Whole-genome amplification (WGA) was performed using the Repli-g Midi kit (Qiagen, Milan, Italy), according to the manufacturer’s instructions. Briefly, 5 μl of genomic DNA (containing 1–10 ng of DNA) were mixed with 5 μl of denaturing solution and incubated at room temperature for 3 min. Next, 10 μl of stop solution were added to stabilize denatured DNA fragments. The reaction mixture was completed with 29 μl of buffer and 1 μl of phi29 polymerase and allowed to proceed for 16 h at 30°C. The reaction was stopped by heating at 63°C for 5 min. WGA products were visualized by electrophoresis on a 0.7% agarose gel, purified, and quantified by Qubit as described previously.
For next-generation sequencing (NGS) experiments, about 1 μg of purified WGA product was used to generate Illumina TruSeq 2× 150 bp paired-end libraries (average insert size: 500 bp), which were sequenced on an Illumina HiSeq 4000 platform (Illumina, SanDiego, CA). Library preparation and NGS experiments were performed by a commercial company (GATC, Germany).
Whole-Genome Global Data Set
To perform a global comparative genomics of C. hominis, we supplemented our newly sequenced genome data set by downloading all available published C. hominis genome sequences till date (July 25, 2021), from the sequence read archive of NCBI and from the EMBL’s European Nucleotide Archive (see supplementary table S1, Supplementary Material online). Collectively, these data represented 114 genome sequences of locally acquired infections from 16 countries across five continents.
Data Preprocessing and Variant Calling
Raw reads of the 114 C. hominis isolates were trimmed to remove adapter sequences and filtered for low-quality bases using Trimmomatic v.0.36 (Bolger et al. 2014). The filtered reads were aligned to C. hominis UdeA01 reference genome (Heiges et al. 2006; Isaza et al. 2015) using the maximal exact matches algorithm implemented in Burrows-Wheeler Alignment tool v.0.7 (Li and Durbin 2009) with default settings. PCR duplicates were then marked using Picard MarkDuplicates (https://broadinstitute.github.io/picard/) followed by Genome Analysis Toolkit’s (GATK) indel realignment and base quality score recalibration using default parameters (McKenna et al. 2010). Sequence variants (SNPs) were called from the aligned reads of each isolate using the HaplotypeCaller method in the GATK v.3.7.0 (McKenna et al. 2010) as per GATK’s best practices pipeline (Van der Auwera et al. 2013). SNPs were removed if quality depth <2.0, Fisher strand >60.0, mapping quality <40.0, mapping quality rank-sum test <−12.5, read position rank-sum test <−8.0, Strand odds ratio >4.0. All identified SNPs were combined in one file, and each isolate was genotyped using the GenotypeGVCFs tool (GATK v.3.7.0) (McKenna et al. 2010). To maximize the quality, SNPs were further filtered based on the following criteria and included in the downstream process: biallelic SNPs, quality score >30, allele depth >5, minor allele frequency >0.05, and missing ratio <0.5. Each of the 114 whole-genome sequences assessed here had >80% coverage of the C. hominis reference genome to at least the 5-fold depth. Each of the 114 whole-genome sequences assessed here had at least ∼80% coverage of the C. hominis reference genome to at least the 5-fold depth where 103 of 114 has >80% genome coverage and at least 10× coverage. Mean coverage of all isolates is 158× (Quartile1 = 117×, Quartile3 = 229×) (supplementary table S1, Supplementary Material online).
Population Genetic Structure based on Whole-Genome SNPs
The filtered biallelic SNPs were used for population structure, phylogenetic, and clustering analyses. Multiplicity of infections in each sample was estimated using estMOI (Assefa et al. 2014) and MOIMIX (https://github.com/bahlolab/moimix). MOIMIX calculates Fws statistic (Manske et al. 2012), a fixation index that is used to assess within-host genetic differentiation. An isolate with a single infection is expected to have Fws 0.95–1.00. The R package SNPRelate v.1.18 (Zheng et al. 2012) was used for principal component analysis (PCA). seqVCF2GDS function in SNPRelate R package is used to first convert VCF file into genomic data structure (GDS) file format to store SNP genotypes in an array-oriented matrix format. A genetic covariance matrix is then calculated from genotypes using SNPRelate’s function snpgdsPCA, along with the correlation coefficients between samples and genotypes for each SNP. An ML phylogenetic tree was constructed by IQ-TREE (Nguyen et al. 2014) with 1000 bootstraps and visualized in iTOL v.3 (Letunic and Bork 2016); the sister species C. parvum was used as an outgroup. We also constructed a consensus of 107 trees using DensiTree 2 (Bouckaert and Heled 2014) in BEAST v.2 (Bouckaert et al. 2014). BEAST v.2 (Bouckaert et al. 2014) was also used to estimate the divergence time between the populations by using 95% HPD; and SpeedDate (https://github.com/vanOosterhoutLab/SpeedDate.jl) to estimate the coalescence times between sequences by using 5–95% CI. We used a mutation rate of 10−8 and a generation time of 48 h/replication (Nader et al. 2019) to date the coalescence times between sequences. A neighbor-net algorithm-based network was generated using SplitsTree5 (Huson and Bryant 2006). Genetic structure was analysed by STRUCTURE v.2.3 software (Pritchard et al. 2000) for population number (K) ranging 2–10 and plotted by using plotSTR R package (https://github.com/DrewWham/Genetic-Structure-Tools). The optimal population genetic cluster value K was estimated by using CLUMPAK (Kopelman et al. 2015).
Population Demographic History and Divergence Time Estimation
We used Bayesian Markov chain Monte Carlo (MCMC) method implemented in Beast v.2 program (Bouckaert et al. 2014) to estimate the effective population size (Ne) of the C. hominis population. The nucleotide substitution model of HKY was selected. A strict molecular clock model and a Bayesian skyline coalescent tree prior were used with 109 generations of MCMC chain and 10% burn-ins. Tracer v.1.7 (Rambaut et al. 2018) was used to assess chain convergence and effective sample size >200 and to construct the demographic history over time, that is, BSP. SweeD (Pavlidis et al. 2013) was used to detect windows of selective sweeps from genome-wide SNP data set by using CLR statistic that identifies signature of SFS, with a grid size of 1000.
Demographic histories and migration rates between the C. hominis populations were estimated by using fastsimcoal2 (Excofffier et al. 2021) by using a mutation rate of 10−8 and a generation time of 48 h/replication (Nader et al. 2019). We first inferred the best parameters and the likelihoods for each of the demographic models—no gene flow, ongoing gene flow, early gene flow, recent gene flow, and different gene flow, since the time of divergence (∼500 years) by running 100 independent iterations with 300,000 coalescent simulations and 60 optimization cycles. The demographic model with the highest likelihood (log10) was then selected to run parameter estimation with block-bootstrapping of 100 replicates.
Linkage, Recombination, and Gene Flow Analyses
We inferred the rate of decay of LD by calculating the squared correlation of the coefficient (r2) between SNPs within 50 kb using VCFtools (Danecek et al. 2011). LD blocks were also determined by calculating pairwise r2 between SNPs within chromosomes of each population. Recombination events were identified using the RDP4 (Martin et al. 2015) using the RDP (Martin and Rybicki 2000), GENECONV (Sawyer 1999), BootScan (Salminen et al. 1995), MaxChi (Smith 1992), and Chimera (Posada and Crandall 2001) methods. Events were considered significant if at least three methods predicted their occurrence at a probability value, P ≤ 10−5. Recombination events with undetermined parental sequences were excluded from further HybridCheck analyses. Statistically significant recombination events were visualized and analysed using HybridCheck (Ward and van Oosterhout 2016) to determine the sequence similarity between the isolates involved in the events. HybridCheck program was also used to calculate the D statistic and estimate the gene flow between the populations.
Population Genetic and Genomic Analyses of Coding Region
Tajima’s D, nucleotide diversity (π), Dxy, and Fst were calculated using the PopGenome R package (Pfeifer et al. 2014). Nonsynonymous (Ka) and synonymous (Ks) mutation rates were calculated by using Ka/Ks_Calculator (Zhang et al. 2006). Protein localization (extracellular) was predicted using WoLF PSORT (Horton et al. 2007) and information regarding predicted protein targeting (signaling peptides) genes were obtained from CryptoDB (Heiges et al. 2006). POPART program was used to generate haplotype networks (Leigh and Bryant 2015). AlphaFold was used to predict the protein structures (Jumper et al. 2021). Glycosylation sites were predicted by using NetNGlyc 4.0 Server (Gupta and Brunak 2002). The intrinsically disordered region in proteins were predicted using IUPred2A (Mészáros et al. 2018). All statistical tests and results were performed and plotted in R (v.3.6.1).
Supplementary Material
Acknowledgments
S.T. acknowledges the Australian Society for Parasitology (ASP) for a student conference travel grant, and JD Smyth Postgraduate Travel Award for a Network Researcher Exchange, Training and Travel Award; and Walter and Eliza Hall Research Institute (WEHI), Australia, for the top-up scholarship. S.T. also acknowledges Namrata Srivastava (Monash University) for assisting with statistical data analysis. This work was supported by the Australian National Health and Medical Research Council (APP1194330) and Victorian State Government Operational Infrastructure Support and Australian Government National Health and Medical Research Council Independent Research Institute Infrastructure Support Scheme to A.R.J. This work was supported by the Interreg 2 Seas program 2014–2020 co-funded by the European Regional Development Fund under subsidy contract no. 2S05–043 H4DC to K.M.T. This work was supported by the European Union's Horizon 2020 research and innovation program, project “Collaborative management platform for detection and analyses of (re-) emerging and foodborne outbreaks in Europe” (COMPARE, www.compare-europe.eu), grant agreement no. 643476 to S.M.C. This work was supported by Walter and Eliza Hall International PhD Scholarship and Melbourne Research Scholarship (MRS) to S.T. This work was supported by the Earth and Life Systems Alliance (ELSA) of the Norwich Research Park (NRP) to C.V.O.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Data Availability
The raw data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession numbers
PRJEB15112, PRJNA610731, PRJNA610732, PRJNA610735, PRJNA610737, PRJNA610738, PRJNA610739, PRJNA610740, PRJNA610741, PRJNA610742, PRJNA610743, PRJNA610744, PRJNA610745, PRJNA610746, PRJNA610747, and PRJNA610748.
Author Contributions
A.R.J., S.M.C., C.V.O., and S.T. conceived the study. A.R.J., S.M.C., C.V.O., and S.T. designed the analyses. S.M.C., R.M.C., G.R., D.E., and K.M.T. were involved in acquisition of data. S.T. performed the bioinformatics associated evolutionary genetic and genomic analyses. A.R.J., S.M.C., C.V.O., and S.T. wrote the manuscript. All authors read and approved the submission of the manuscript for the publication.
References
- Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, et al. . 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304:441–445. [DOI] [PubMed] [Google Scholar]
- Assefa SA, Preston MD, Campino S, Ocholla H, Sutherland CJ, Clark TG. 2014. estMOI: estimating multiplicity of infection using parasite deep sequencing data. Bioinformatics 30:1292–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert RR, Heled J. 2014. DensiTree 2: seeing trees through the forest. bioRxiv 012401. [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 10:e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzid M, Hunter PR, Chalmers RM, Tyler KM. 2013. Cryptosporidium pathogenicity and virulence. Clin Microbiol Rev. 26:115–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacciò SM, Chalmers RM. 2016. Human cryptosporidiosis in Europe. Clin Microbiol Infect. 22:471–480. [DOI] [PubMed] [Google Scholar]
- Cama VA, Bern C, Roberts J, Cabrera L, Sterling CR, Ortega Y, Gilman RH, Xiao L. 2008. Cryptosporidium species and subtypes and clinical manifestations in children, Peru. Emerg Infect Dis. 14:1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmicheal J, Atri P, Sharma S, Kumar S, Chirravuri Venkata R, Kulkarni P, Salgia R, Ghersi D, Kaur S, Batra SK. 2020. Presence and structure-activity relationship of intrinsically disordered regions across mucins. FASEB J. 34:1939–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevallos AM, Bhat N, Verdon R, Hamer DH, Stein B, Tzipori S, Pereira ME, Keusch GT, Ward HD. 2000. Mediation of Cryptosporidium parvum infection in vitro by mucin-like glycoproteins defined by a neutralizing monoclonal antibody. Infect Immun. 68:5167–5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers RM, Robinson G, Elwin K, Elson R. 2019. Analysis of the Cryptosporidium spp. and gp60 subtypes linked to human outbreaks of cryptosporidiosis in England and Wales, 2009 to 2017. Parasit Vectors 12:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers RM, Robinson G, Elwin K, Hadfield SJ, Thomas E, Watkins J, Casemore D, Kay D. 2010. Detection of Cryptosporidium species and sources of contamination with Cryptosporidium hominis during a waterborne outbreak in north west Wales. J Water Health 8:311–325. [DOI] [PubMed] [Google Scholar]
- Chatterjee A, Banerjee S, Steffen M, O'Connor RM, Ward HD, Robbins PW, Samuelson J. 2010. Evidence for mucin-like glycoproteins that tether sporozoites of Cryptosporidium parvum to the inner surface of the oocyst wall. Eukaryot Cell 9:84–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi GI, Tichkule S, Sannella AR, Vatta P, Asnicar F, Segata N, Jex AR, Oosterhout C, Cacciò SM. 2021. Evolutionary epidemiology of a zoonosis. bioRxiv 2021.2010.2015.464618. [Google Scholar]
- Craun GF, Hubbs SA, Frost F, Calderon RL, Via SH. 1998. Waterborne outbreaks of cryptosporidiosis. J AWWA 90:81–91. [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. . 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 22:1185–1192. [DOI] [PubMed] [Google Scholar]
- Efstratiou A, Ongerth JE, Karanis P. 2017. Waterborne transmission of protozoan parasites: review of worldwide outbreaks-an update 2011–2016. Water Res. 114:14–22. [DOI] [PubMed] [Google Scholar]
- Excofffier L, Marchi N, Marques DA, Matthey-Doret R, Gouy A, Sousa VC. 2021. fastsimcoal2: demographic inference under complex evolutionary scenarios. Bioinformatics. 37:4882–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Ryan UM, Xiao L. 2018. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 34:997–1011. [DOI] [PubMed] [Google Scholar]
- Feng Y, Tiao N, Li N, Hlavsa M, Xiao L. 2014. Multilocus sequence typing of an emerging Cryptosporidium hominis subtype in the United States. J Clin Microbiol. 52:524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Tang K, Rowe LA, Li N, Roellig DM, Knipe K, Frace M, Yang C, Feng Y, Xiao L. 2015. Comparative genomic analysis reveals occurrence of genetic recombination in virulent Cryptosporidium hominis subtypes and telomeric gene duplications in Cryptosporidium parvum. BMC Genomics 16:320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Brunak S. 2002. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac Symp Biocomput. 7:310–322. [PubMed] [Google Scholar]
- Hadfield SJ, Pachebat JA, Swain MT, Robinson G, Cameron SJ, Alexander J, Hegarty MJ, Elwin K, Chalmers RM. 2015. Generation of whole genome sequences of new Cryptosporidium hominis and Cryptosporidium parvum isolates directly from stool samples. BMC Genomics 16:650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiges M, Wang H, Robinson E, Aurrecoechea C, Gao X, Kaluskar N, Rhodes P, Wang S, He CZ, Su Y, et al. . 2006. CryptoDB: a Cryptosporidium bioinformatics resource update. Nucleic Acids Res. 34:D419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herges GR, Widmer G, Clark ME, Khan E, Giddings CW, Brewer M, McEvoy JM. 2012. Evidence that Cryptosporidium parvum populations are panmictic and unstructured in the Upper Midwest of the United States. Appl Environ Microbiol. 78:8096–8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park K-J, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. 2007. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35:W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 23:254–267. [DOI] [PubMed] [Google Scholar]
- Isaza JP, Galván AL, Polanco V, Huang B, Matveyev AV, Serrano MG, Manque P, Buck GA, Alzate JF. 2015. Revisiting the reference genomes of human pathogenic Cryptosporidium species: reannotation of C. parvum Iowa and a new C. hominis reference. Sci Rep. 5:16324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jex AR, Gasser RB. 2010. Genetic richness and diversity in Cryptosporidium hominis and C. parvum reveals major knowledge gaps and a need for the application of “next generation” technologies–research review. Biotechnol Adv. 28:17–26. [DOI] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. . 2021. Highly accurate protein structure prediction with AlphaFold. Nature. 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil IA, Troeger C, Rao PC, Blacker BF, Brown A, Brewer TG, Colombara DV, De Hostos EL, Engmann C, Guerrant RL, et al. . 2018. Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: a meta-analyses study. Lancet Glob Health 6:e758–e768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King P, Robinson G, Elwin K, Tyler KM, Hunter PR, Chalmers RM. 2017. Prevalence and epidemiology of human Cryptosporidium parvum IIc infections in England and Wales. Lancet 389:S56. [Google Scholar]
- King P, Tyler KM, Hunter PR. 2019. Anthroponotic transmission of Cryptosporidium parvum predominates in countries with poorer sanitation: a systematic review and meta-analysis. Parasit Vectors 12:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. 2015. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour. 15:1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, et al. . 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. [DOI] [PubMed] [Google Scholar]
- Leigh JW, Bryant D. 2015. popart: full-feature software for haplotype network construction. Methods Ecol Evolut. 6:1110–1116. [Google Scholar]
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44:W242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Xiao L, Cama VA, Ortega Y, Gilman RH, Guo M, Feng Y. 2013. Genetic recombination and Cryptosporidium hominis virulent subtype IbA10G2. Emerg Infect Dis. 19:1573–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lighten J, Papadopulos AST, Mohammed RS, Ward BJ, Paterson I G, Baillie L, Bradbury IR, Hendry AP, Bentzen P, van Oosterhout C. 2017. Evolutionary genetics of immunological supertypes reveals two faces of the Red Queen. Nat Commun. 8:1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, Qing X, Liao J, Zhuo K. 2020. Role of protein glycosylation in host-pathogen interaction. Cells 9:1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippuner C, Ramakrishnan C, Basso WU, Schmid MW, Okoniewski M, Smith NC, Hässig M, Deplazes P, Hehl AB. 2018. RNA-Seq analysis during the life cycle of Cryptosporidium parvum reveals significant differential gene expression between proliferating stages in the intestine and infectious sporozoites. Int J Parasitol. 48:413–422. [DOI] [PubMed] [Google Scholar]
- Manske M, Miotto O, Campino S, Auburn S, Almagro-Garcia J, Maslen G, O'Brien J, Djimde A, Doumbo O, Zongo I, et al. . 2012. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature 487:375–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 1:vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Rybicki E. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562–563. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKerr C, Chalmers RM, Elwin K, Ayres H, Vivancos R, O’Brien SJ, Christley RM. 2022. Cross-sectional household transmission study of Cryptosporidium shows that C. hominis infections are a key risk factor for spread. BMC Infect Dis. 22:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mészáros B, Erdős G, Dosztányi Z. 2018. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 46:W329–W337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison LJ, Mallon ME, Smith HV, MacLeod A, Xiao L, Tait A. 2008. The population structure of the Cryptosporidium parvum population in Scotland: a complex picture. Infect Genet Evol. 8:121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader JL, Mathers TC, Ward BJ, Pachebat JA, Swain MT, Robinson G, Chalmers RM, Hunter PR, van Oosterhout C, Tyler KM. 2019. Evolutionary genomics of anthroponosis in Cryptosporidium. Nat Microbiol. 4:826–836. [DOI] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2014. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols GL, Chalmers RM, Hadfield SJ. 2014. Molecular epidemiology of human cryptosporidiosis. In: Cacciò SM, Widmer G, editors. Cryptosporidium: parasite and disease. Springer. p. 81–147. [Google Scholar]
- O'Connor RM, Burns PB, Ha-Ngoc T, Scarpato K, Khan W, Kang G, Ward H. 2009. Polymorphic mucin antigens CpMuc4 and CpMuc5 are integral to Cryptosporidium parvum infection in vitro. Eukaryotic Cell 8:461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Živkovic D, Stamatakis A, Alachiotis N. 2013. SweeD: likelihood-based detection of selective sweeps in thousands of genomes. Mol Biol Evol. 30:2224–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Vilar J, Hill RL. 1999. The structure and assembly of secreted mucins. J Biol Chem. 274:31751–31754. [DOI] [PubMed] [Google Scholar]
- Pfeifer B, Wittelsbürger U, Ramos-Onsins SE, Lercher MJ. 2014. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol Biol Evol. 31:1929–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SL, Frost SD. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci USA. 98:13757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putignani L, Menichella D. 2010. Global distribution, public health and clinical impact of the protozoan pathogen Cryptosporidium. Interdiscip Perspect Infect Dis. 2010:753512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol. 67:901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razakandrainibe R, Diawara EHI, Costa D, Le Goff L, Lemeteil D, Ballet JJ, Gargala G, Favennec L. 2018. Common occurrence of Cryptosporidium hominis in asymptomatic and symptomatic calves in France. PLoS Negl Trop Dis. 12:e0006355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G, Elwin K, Chalmers RM. 2020. Cryptosporidium diagnostic assays: molecular detection. Methods Mol Biol. 2052:11–22. [DOI] [PubMed] [Google Scholar]
- Ryan U, Zahedi A, Paparini A. 2016. Cryptosporidium in humans and animals—a one health approach to prophylaxis. Parasite Immunol. 38:535–547. [DOI] [PubMed] [Google Scholar]
- Salminen MO, Carr JK, Burke DS, McCutchan FE. 1995. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res Hum Retroviruses 11:1423–1425. [DOI] [PubMed] [Google Scholar]
- Santin M. 2020. Cryptosporidium and Giardia in Ruminants. Vet Clin North Am Food Anim Pract. 36:223–238. [DOI] [PubMed] [Google Scholar]
- Sawyer S. 1999. GENECONV: a computer package for the statistical detection of gene conversion. http://www.math.wustl.edu/∼sawyer.
- Segura R, Prim N, Montemayor M, Valls ME, Muñoz C. 2015. Predominant virulent IbA10G2 subtype of Cryptosporidium hominis in human isolates in Barcelona: a five-year study. PLoS One 10:e0121753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM. 1992. Analyzing the mosaic structure of genes. J Mol Evol. 34:126–129. [DOI] [PubMed] [Google Scholar]
- Tichkule S, Jex AR, van Oosterhout C, Sannella AR, Krumkamp R, Aldrich C, Maiga-Ascofare O, Dekker D, Lamshöft M, Mbwana J, et al. . 2021. Comparative genomics revealed adaptive admixture in Cryptosporidium hominis in Africa. Microb Genom. 7:000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. . 2013. From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinform. 43:11.10.11– 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oosterhout C. 2021. Mitigating the threat of emerging infectious diseases; a coevolutionary perspective. Virulence 12:1288–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Zhang L, Axen C, Bjorkman C, Jian F, Amer S, Liu A, Feng Y, Li G, Lv C, et al. . 2014. Cryptosporidium parvum IId family: clonal population and dispersal from Western Asia to other geographical regions. Sci Rep. 4:4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward BJ, van Oosterhout C. 2016. HYBRIDCHECK: software for the rapid detection, visualization and dating of recombinant regions in genome sequence data. Mol Ecol Resour. 16:534–539. [DOI] [PubMed] [Google Scholar]
- Widerström M, Schönning C, Lilja M, Lebbad M, Ljung T, Allestam G, Ferm M, Björkholm B, Hansen A, Hiltula J. 2014. Large outbreak of Cryptosporidium hominis infection transmitted through the public water supply, Sweden. Emerg Infect Dis. 20:581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Guo Y, Xiao L, Feng Y. 2021. Molecular epidemiology of human cryptosporidiosis in low-and middle-income countries. Clin Microbiol Rev. 34:e00087-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li J, Zhao XQ, Wang J, Wong GK, Yu J. 2006. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genom Proteom Bioinform. 4:259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28:3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Singh A, Jiang J, Xiao L. 2003. Molecular surveillance of Cryptosporidium spp. in raw wastewater in Milwaukee: implications for understanding outbreak occurrence and transmission dynamics. J Clin Microbiol. 41:5254–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession numbers
PRJEB15112, PRJNA610731, PRJNA610732, PRJNA610735, PRJNA610737, PRJNA610738, PRJNA610739, PRJNA610740, PRJNA610741, PRJNA610742, PRJNA610743, PRJNA610744, PRJNA610745, PRJNA610746, PRJNA610747, and PRJNA610748.