Abstract
Objectives.
Plasmodium vivax malaria was thought to be rare in Africans who lack the Duffy blood group antigen expression. However, recent studies indicate that P. vivax can infect Duffy-negative individuals and has penetrated into areas of high Duffy-negativity across Africa. This study compares epidemiological and genetic features of P. vivax between African regions.
Methods.
We utilized a standardized approach to identify and quantify P. vivax from Botswana, Ethiopia, and Sudan, where Duffy-positive and Duffy-negative individuals coexist. We sequenced Duffy Binding Protein (DBP) gene and inferred genetic relationships among all Africa P. vivax.
Results.
Among 1,215 febrile patients, the proportions of Duffy negativity range from 20–36% in East Africa to 84% in Southern Africa. P. vivax prevalence among Duffy-negative populations ranging from averaged 9.2% in Sudan to 86% in Botswana. Parasite density in Duffy-negative is significantly lower than in Duffy-positive infections. P. vivax in Duffy-negative populations were not monophyletic. Duffy-negative and Duffy-positive P. vivax shared similar DBP haplotypes and occurred in multiple well-supported clades.
Conclusions.
Duffy-negative Africans are not resistant to P. vivax and the public health significance should not be neglected. This study highlights need for standardized approach and more resources/training to diagnosis of vivax malaria in Africa.
Keywords: Malaria, Plasmodium vivax, Duffy negatives, Sub-Saharan Africa, Genetic relationships, Molecular epidemiology
Introduction
Plasmodium vivax malaria was previously thought to be rare or absent in African populations who lack the Duffy blood group antigen expression (Miller et al., 1976, Howes et al., 1995). A point mutation (c.1–67T>C; rs2814778) in the GATA-1 transcription factor binding site of the Duffy antigen/receptor for chemokines (DARC) gene promoter alters erythroid expression, eliminating Duffy antigen expression on the surface of the red blood cells (Tournamille et al. 1995, King et al., 2011). However, recent studies reported several cases of P. vivax infection in Duffy-negative people in different parts of Africa (Zimmerman, 2017, Gunalan et al., 2018), including countries where Duffy-negatives are predominant (Brazeau et al., 2018, Mendes et al., 2011, Motshoge et al., 2016, Niangaly et al., 2017, Russo et al., 2017) (Table 1). In addition, 29 African countries including six previously undocumented endemic countries (Benin, Comoros, Mozambique, Senegal, Zambia and Zimbabwe) have reported P. vivax clinical cases, infected vectors or asymptomatic parasitemia (Niang et al., 2018, Oboh et al., 2020, Poirier et al., 2016). These reports indicate that the endemic range of P. vivax has extended beyond East Africa and penetrated into areas of very high Duffy-negativity (Gunalan et al., 2018, Twohig et al., 2019). While P. falciparum is considered to be the deadliest malaria parasite with the most severe clinical outcomes, P. vivax is more widespread and often associated with high levels of morbidity. Compared to P. falciparum, P. vivax has a broader temperature tolerance, an earlier onset of gametocyte development, and can form dormant hypnozoites causing relapse (Livingstone, 1984), enabling P. vivax to spread through the diverse African climate and outcompete P. falciparum (Battle et al., 2019). Primaquine and 8-aminoquinoline are antimalarials effective in clearing hypnozoites and preventing relapses, but they may promote hemolysis in subjects with G6PD deficiency (Baird, 2019). These factors make P. vivax malaria difficult to control and eliminate, highlighting the concern of this ‘new’ P. vivax strains that infect Duffy-negative hosts to spread through much of Africa and result in substantial, negative public health and economic impacts.
Table 1.
Summary of P. vivax infections with available Duffy blood group information in African countries from the literature. nPCR: Nested PCR of P. vivax 18S rRNA gene; qPCR: Quantitative real-time PCR of P. vivax 18S rRNA gene.
| Country | Sample collection year | Symptoms | Sample size | Duffy negative, n (%) | Malaria diagnostic method | Plasmodium spp pos (%) | P. vivax pos (% of P. spp+) | Pv+ in Duffy neg (% of total Pv+) | Reference |
|---|---|---|---|---|---|---|---|---|---|
| East-Southern Africa | |||||||||
| Angola | 2006–07 | No | 898 ^ | * | nPCR | 245/898 (28.9%) | 7/245 (2.8%)1 | 7/7 (100%)* | Mendes C, et al. PLoS NTDs, 2011;5(6): e1192. |
| Ethiopia | 2009 | Yes | 1,931 | 41/205 (20%)2 | nPCR | 205/1,931 (10.6%) | 111/205 (54.1%) | 3/111 (2.7%) | Woldearegai TG, et al. Trans R Soc Trop Med Hyg 2013;107:328–31. |
| Ethiopia | 2013–14 | Yes | 416 | 94/416 (29.7%) | qPCR | 331/416 (79.5%) | 197/331 (59.5%)3 | 2/197 (1%)4 | Lo E, et al. Malaria J, 2015;14:84. |
| Ethiopia | 2013–14 | No | 390 | 139/390 (35.6%) | qPCR | 73/390 (18.7%) | 24/73 (32.9%)5 | 4/24 (16.6%) | Lo E, et al. Malaria J, 2015;14:84. |
| Kenya | 1999–2000 | Yes | 31 ^6 | 31/318 (100%) | microscopy | 31/316 (100%) | 11/31 (35.4%) | 9/11 (81.8%) | Ryan JR, et al. Am J Trop Med Hyg, 2006;75:575–81. |
| Madagascar | 2006–07 | Yes | 1837 | * | nPCR | 183/183 (100%)7 | 183/183 (100%)7 | 17/183 (9.3%)* | Ménard D, et al. PNAS, 2010;107(13):5697–71. |
| Madagascar | 2006–07 | No | 661 ^ | 476/661 (72%) | nPCR§ | 251/661 (38%) | 86/251 (34.3%)8 | 42/86 (48.8%) | Ménard D, et al. PNAS, 2010;107(13):5697–71. |
| Madagascar | 2014 | No | 2,063 | 914/1,878 (48.7%) | nPCR | 285/2,063 (13.8%)9 | 137/285 (48.1%)10 | 44/914 (4.8%) | Howes RE, et al. Am. J. Trop. Med. Hyg., 2018;99(4):995–1002. |
| West-Central Africa | |||||||||
| Benin | 2009–10 | No | 84 ^^ | * | nPCR | 25/84 (29.8%) | 13/25 (52%)11 | 13/13 (100%)* | Poirier P, et al., Malar J, 2016;15:570. |
| Botswana | 2012 | No | 3,62422 | N.A. | nPCR | 179/3,624 (5%) | 169/179 (94.4%) | N.A. | Matshoge T, et al. BMC Inf Dis 2016;16:520. |
| Cameroon | N.A. | Yes | 485 | * | nPCR | 201/485 (41.4%) | 8/201 (4%)12 | 8/8 (100%)* | Ngassa Mbenda HG & Das A. PLoS ONE 2014;9(8):e103262. |
| Cameroon | 2008–09 | No | 269 | * | nPCR | 87/267 (32.3%) | 13/87 (14.9%)13 | 6/13 (46.1%)* | Fru-Cho J, et al. Malaria J, 2014;13:170. |
| Cameroon | 2012–13 | Yes | 484 | 224/228 (98.3%)15 | nPCR | 70/484 (14.4%) | 27/70 (38.6%)15 | 27/27 (100%) | Russo G et al. Malaria J, 2017;16(1):74. |
| Democratic Republic of Congo | 2013–14 | No | 292 ^ | * | nPCR | 194/292 (66.4%) | 14/194 (7.2%) | 14/14 (100%)* | Brazeau NF, et al. Am. J. Trop. Med. Hyg., 2018;99(5):1128–33. |
| Equatorial Guinea | 2005 | No | 97 | * | nPCR | 84/97 (86.6%) | 8/84 (9.5%)16 | 8/8 (100%)* | Mendes C, et al. PLoS NTDs, 2011;5(6):e1192. |
| Mali | 2009–11 | No | 300 ^ | * | qPCR | 135/300 (15%) | 25/135 (18.5%) | 25/25 (100%)* | Niangaly A, et al. Am J Trop Med Hyg 2017;97(3):744–52. |
| Mauritania | 2007–09 | Yes | 277 | 52/258 (20.1%) | qPCR | 110/277 (39.7%) | 110/110 (100%) | 1/110 (0.9%) | Wurtz N et al. Malaria J, 2011;10:336. |
| Nigeria | 2016–17 | Yes | 436 | * | nPCR | 256/436 (58.7%) | 5/256 (1.9%)17 | 5/5 (100%)* | Oboh MA, et al., Malar J, 2018;17:439 and 2020;19:229. |
| Senegal | 2009–2013 | Yes | 263 | N.A. | nPCR | 164/263 (62.3%) | 4/164 (2.4%)24 | N.A. | Niang et al., Malar J, 2015; 14: 281. |
| Senegal | 2010–2011 | No | 48(x4)^18 | 48/48 (100%) | nPCR | 74/192 (38.5%) | 15/74 (20.3%) | 5/5 (100%) | Niang M, et al. Trop Med & Hyg 2018;46:45. |
| Sudan | 2009 | Yes | 126 | * | nPCR | N.A. | 48/126 (38.1%) | 4/48 (8.3%)* | Abdelraheem MH, et al. Trans R Soc Trop Med Hyg 2016;110:258–60 |
| Sudan | 2016 | Yes | 99219 | * | microscopy | 992/992 (100%)19 | 190/992 (19.1%)20 | 34/190 (17.9%)* | Albsheer MMA, et al. Genes, 2019;10:437. |
| Uganda | 2016 | Yes | 49921 | N.A. | nPCR | 499/499 (100%) | 4/499 (0.8%)23 | N.A. | Asua, V. et al. Am J Trop Med Hyg. 2017;97:753–57. |
Duffy-Ag assessed only among Pv pos patients;
only children;
only blood-donors; N.A. not available.
Conventional PCR of genes PvCOI and PvDBP.
2 Pf-Pv co-infections;
Duffy-Ag available only among Plasmodium spp pos;
33 Pf-Pv co-infections;
2 Pf-Pv co-infections;
1 Pf-Pv co-infection;
31 children Duffy neg affected by malaria enrolled in a precedent study (anemia study);
only Pv pos analysed (153 Pv mono-infections and 30 Pf-Pv co-infections);
34 Plasmodium mixed-infections (species not specified);
42 co-infections (25 Pf-Pv, 5 Pf-Pm, 9 Pv-Pm, 1 Pv-Po, 1 Pf-Pv-Pm, 1 Pf-Pv-Pm-Po);
37 co-infections (25 Pf-Pv, 9 Pv-Pm, 1 Pv-Po, 1 Pf-Pv-Pm, 1 Pf-Pv-Pm-Po);
9 Pf-Pv and 1 Pf-Pv-Pm co-infections;
2 Pf-Pv co-infections;
3 Pf-Pv and 1 Pf-Pv-Pm co-infections;
Duffy-Ag assessed among 228 participants (including all those infected);
2 Pf-Pv co-infections;
4 Pf-Pv co-infections;
4 Pf-Pv co-infections;
48 school children followed during 2 years (4 samples for each children, 192 total samples analyzed)
only Plasmodium spp pos sample analysed;
4 Pf-Pv co-infections;
all Plasmodium pos children aged 2 months-10 years;
all children aged 2–12 years;
all 4 were co-infections: 3 Pf-Pv and 1 Pv-Pm;
2 Pf-Pv co-infections
There is a major knowledge gap in the P. vivax invasion mechanisms in Duffy-negative erythrocytes. In P. falciparum, erythrocyte invasion involves multiple interactions between parasite ligands and host receptors, some of which have overlapping and partially redundant roles (Cowman et al. 2017, Kumar and Tolia, 2019). Several established invasion ligands from Erythrocyte Binding Antigens such as EBA-175, EBA-181/JESEBL and EBA-140/BAEBL and Reticulocyte binding homolog proteins such as RH1, RH2a, RH2b, RH4 and RH5 are used by P. falciparum for invasion (Gunalan et al., 2013, Kumar and Tolia, 2019). In P. vivax, only a single P. vivax ligand-receptor interaction has so far been studied in any detail, P. vivax Duffy Binding Protein (PvDBP1). Previous study has shown that mutations in PvDBP1 region II unique to P. vivax in Duffy-negative people in Ethiopia did not lead to binding of Duffy-negative erythrocytes (Gunalan et al., 2016). Salvador (Sal) I P. vivax infects Squirrel monkeys without PvDBP1 binding to Squirrel monkey erythrocytes (Gunalan et al., 2019). Further, EBP/DBP2 region II, a paralog of PvDBP1, was shown to bind to Duffy-positive and Duffy-null human erythrocytes at low frequency (Gunalan et al., 2016, Ntumngia et al., 2016), despite being deleted in Sal-I P. vivax (Hester et al., 2013). Recently, reticulocyte binding protein RBP2b of P. vivax was shown to bind to transferrin receptor in the reticulocytes (Gruszczyk et al., 2018). These findings suggested that there are other Duffy-independent pathways that enable erythrocyte invasion and explain the widespread phenomenon of P. vivax infections in Africa.
Despite the fact that several case reports from almost all countries across the African continent are emerging from various entomological and serological studies, community surveys, and clinical records (Gunalan et al., 2018, Twohig et al., 2019), the documentation of P. vivax infections across Africa is diverse, context-specific, and primarily driven by the specific objectives of isolated clinical or epidemiological activities. The varied diagnostic and methodological approaches used across studies have limited our ability to identify distinct epidemiological characteristics of P. vivax between regions (Table 1). This situation is concerning because there is no comprehensive genetic and epidemiological data of P. vivax in Africa available to National Malaria Programs or World Health Organization to assess impacts and confer control strategies. Therefore, in this study, we utilized a standardized assay to examine the epidemiological attributes of P. vivax in three African countries where Duffy-positive and Duffy-negative individuals coexist. Specifically, we (1) compared the prevalence of Duffy negativity and P. vivax infections among countries; (2) compared P. vivax parasitemia between Duffy-negative and Duffy-positive infections collected from the same area; and (3) inferred the genetic relationships among the African P. vivax isolates. The epidemiological and genetic features of P. vivax from different parts of Africa will fill critical gap in understanding how widespread this phenomenon is impacting malaria control and the important effect of P. vivax as a cause of anemia.
Materials and Methods
Study sites and sample collection
A total of 1,215 febrile patients were collected from seven study sites in three countries including (1) Jimma and Bonga in Ethiopia; (2) Khartoum, River Nile, and New Halfa in Sudan; and (3) Tutume and Kweneng East in Botswana (Figure 1). Finger-prick blood samples were obtained from patients who visited the health facilities. Thick and thin blood smears were prepared for microscopic screening. Three to four blood spots were blotted on Whatman 3MM filter paper from each participant. Parasite DNA was extracted from dried blood spots by the Saponin/Chelex method (Bereczky et al., 2005). Eluted DNA was used for PCR diagnosis, quantification and genotyping of malaria parasites.
Figure 1.
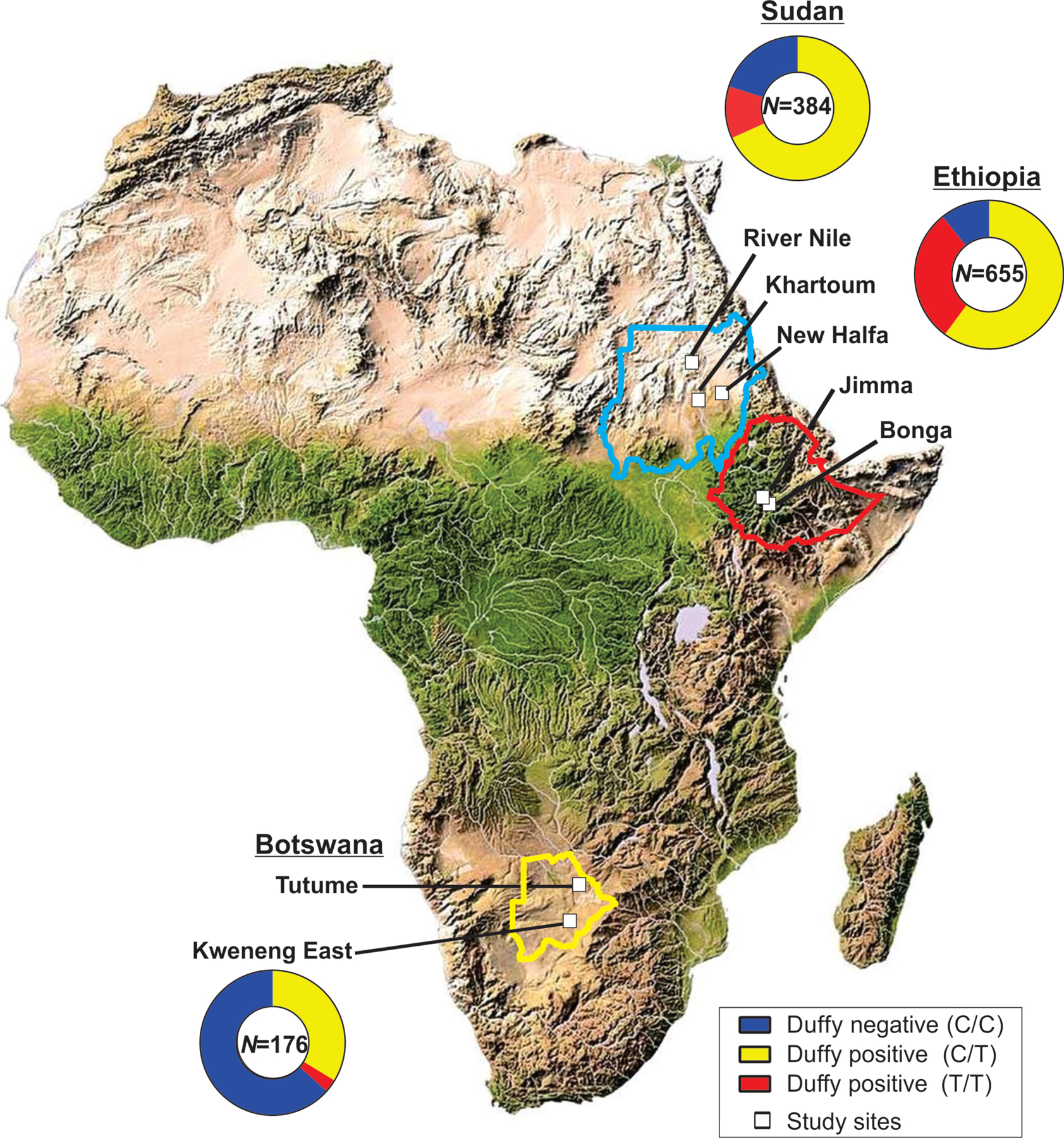
Map showing the distribution of study sites and the Duffy status of febrile patients included in the present study.
Molecular screening of P. vivax
Parasite gene copy number was estimated using the SYBR Green detection method (Lo et al., 2015) using P. vivax-specific primers that targeted the 18S rRNA genes (detail in Supplementary File 1). Each assay included positive controls of P. vivax Pakchong (MRA-342G) and Nicaragua (MRA-340G) isolates, in addition to negative controls. A standard curve was produced from a ten-fold dilution series of the P. vivax control plasmid to determine the amplification efficiency (E). Melting curve analyses were performed to confirm the specificity of gene amplifications. The mean threshold cycle (Ct) and standard error were calculated from three independent assays of each sample. The amount of parasite density in a sample was calculated using the follow equation: Parasite densitysample = 2 E×(40-Ctsample). The differences in the log-transformed parasite density between samples among the study sites were assessed for significance by one-tailed t-tests.
Duffy blood group genotyping
For all febrile patients, we first employed qPCR-based TaqMan assay to examine the point mutation (c.1–67T>C; rs2814778) of the DARC gene (Supplementary File 1). A no-template control was used in each assay. The Fy genotypes were determined by the allelic discrimination plot based on the fluorescent signal emitted from the allele-specific probes. For P. vivax positive samples, a 1,100-bp fragment of the DARC gene was further amplified using published primers (Menard et al., 2010). PCR products were sequenced to confirm the Fy genotypes.
Phylogenetic analyses of P. vivax from Duffy negative and Duffy positive samples
We amplified and obtained PvDBP sequences of 4 Duffy-positive and 4 Duffy-negative P. vivax samples from Botswana, 107 Duffy-positive and 9 Duffy-negative P. vivax samples from Ethiopia, and 53 Duffy-positive and 16 Duffy-negative P. vivax samples from Sudan (Genbank accession number: MZ062224-MZ062409). These sequences were aligned with 36 previously published P. vivax isolates from other parts of Africa including Uganda (n=31), Madagascar (n=4), and Mauritania (n=1; Supplementary File 2). Duffy status of the published sequences are unknown. The DBP sequence of Sal-1 (NC_009911.1) and EBP sequence of P. cynomolgi (Y11396.1) were used as outgroups. Phylogenetic trees were reconstructed using the maximum likelihood method implemented in RAxML v8.0 with 500 bootstrap replicates to assess clade support (details in Supplementary File 1). We further examined the nucleotide and haplotype diversity of PvDBP sequences in Duffy-negative and Duffy-positive samples using DnaSP v6.12.03.
Results and Discussion
Contrasting proportion of Duffy-negatives and P. vivax prevalence
Duffy genotyping shows different proportions of Duffy-negative among febrile patients in Botswana, Ethiopia, and Sudan (Figure 1). In Botswana, the proportion of Duffy-negative was 83.5% (147/176) among febrile patients (Figure 1). In Kweneng East, our qPCR analyses indicated that 3% (9 out of 301) of the febrile patients were P. vivax positive. Among them, eight were Duffy-negative (C/C) and one was Duffy-positive (T/C) (Table 2; Supplementary File 3). In Tutume, 6.8% (12/176) of the febrile patients were detected with P. vivax and 10 of them were Duffy-negative. Vivax malaria was first reported in asymptomatic children in a survey during the 2012–2013 transmission season [10]. The average rate of asymptomatic P. vivax cases was 4.7%, but with large variation among districts. Compared to other parts of Botswana, Tutume and Kweneng East accounted for most of the P. vivax cases, with previously reported rates of 16.9% (54/320) and 13.6% (93/686), respectively (Motshoge et al., 2016).
Table 2.
Comparison of P. vivax infection rate in Duffy-negative populations across different study sites in Botswana, Ethiopia, and Sudan based on febrile patient samples collected in this study.
| Region | Country | Study site | Collection period | Type of collection | Total samples | Infection rate of P. vivax | Duffy-negative among P. vivax infections |
|---|---|---|---|---|---|---|---|
| Southern Africa | |||||||
| Botswana | Tutume | 2017 – 2018 | Symptomatic | 176 | 12 (6.8%) | 10 (83.3%) | |
| Kweneng East | 2017 – 2018 | Symptomatic | 301 | 9 (3%) | 8 (88.9%) | ||
| East Africa | |||||||
| Ethiopia | Jimma | April - October 2017 | Symptomatic | 358 | 134 (37.4%) | 16 (11.9%) | |
| Bonga | October – November 2019 | Symptomatic | 297 | 76 (25.6%) | 8 (10.5%) | ||
| Sudan | River Nile | August 2018 – February 2019 | Symptomatic | 213 | 52 (24.4%) | 2 (3.8%) | |
| Khartoum | August 2018 – February 2019 | Symptomatic | 525 | 42 (8%) | 4 (9.5%) | ||
| New Halfa | August 2018 – February 2019 | Symptomatic | 93 | 7 (7.5%) | 1 (14.3%) | ||
In Ethiopia, the proportion of Duffy-negative was 35.9% (235/655) among febrile patients (Figure 1), similar to our earlier finding in Asendabo indicating that 35.1% (137/390) of the general population was Duffy-negative (Lo et al., 2015). Among the 358 febrile patient samples collected in Jimma, 36% (129/358) were Duffy-negatives (Figure 1) and 37.4% (134/358) were detected with P. vivax (Table 2). About 11.9% (16/134) of the confirmed P. vivax infections were from Duffy-negatives. Likewise, in Bonga, 30.3% (125/413) of the febrile patients were detected with P. vivax and 3.2% (4/125) were from Duffy-negatives (Table 2). For these 20 Duffy-negatives P. vivax infections, microscopy, nested and quantitative PCRs indicated that 16 were single infections and four were mixed with P. falciparum. Vivax malaria is a significant problem in Ethiopia (Lo et al., 2015, Woldearegai et al., 2013). Our previous study has shown that the asymptomatic prevalence of P. vivax is 5.9% (23/390) in Asendabo and Duffy-negatives accounted for 8.7% (2/23) of the P. vivax infections (Lo et al., 2015). A lower proportion of Duffy-negativity in febrile patients and the general population in Ethiopia, as compared to Botswana, is consistent with the ethnic diversity and complex admixture history in East Africa (Hollfelder et al., 2017, Pickrell et al., 2014).
In Sudan, the proportion of Duffy-negative was 20% (77/384) among febrile patients (Figure 1). Over a 6-month collection period between 2018 and 2019, 101 out of 831 febrile patients were confirmed as P. vivax positive by qPCR assays (Table 2). Further testing revealed that 4 of the 101 P. vivax samples were mixed with P. falciparum. The highest rate of P. vivax infection was observed in River Nile, of which 24.4% (52/213) of the febrile patients were confirmed with P. vivax and Duffy-negatives accounted for 3.8% (2/52) of these infections (Table 2). In Khartoum, 8% (42/525) of the febrile patients were P. vivax-positive and Duffy-negatives accounted for 9.5% (4/42) of these infections. In New Halfa, despite a smaller sample size, 7.5% (7/93) of the febrile patients was P. vivax-positive and Duffy-negatives accounted for 14.3% (1/7) of these infections (Table 2). Across the country, there has been an increase in P. vivax detection and reports in recent years (Albsheer et al., 2019) and our findings indicated that the infection rate in Duffy-negative individuals varies among study sites.
Historical movement and genetic admixture explain distribution of Duffy-negative people in Africa
Historical human movement and human genetics are highly relevant to the distribution of Duffy-negative people and P. vivax in Africa. Recent genome-wide studies of African populations have refined earlier models of the continent’s history and its impact on genetic diversity of its inhabitants (Choudhury et al., 2020). Our data showing a Duffy negative rate of 83.5% among febrile patients in Botswana (Figure 1) is consistent with the Bantu expansion and admixture theories (Choudhury et al., 2020, Grollemunda et al., 2015). The Bantu expansion and population admixture are two main historical events that shape the present distribution and genetic make-up of ethnic groups across Africa. The Bantu and Khoisan are two major ethnic groups in West-Central and Southern Africa, with the Bantu heartland in the region between southern Nigeria and Cameroon where malaria transmission was and still is endemic (Grollemunda et al., 2015). A component of Bantu ancestry (likely Duffy-negative) was found in the Southern African Khoisan, which were originally and mostly Duffy-positive ancestors (Hamblin et al., 2002, Petersen et al., 2013). The Duffy-negative allele from Bantu of West-Central Africa may have reached south of the continent within the last 750 years and mixed with the indigenous Khoisan, resulting in a variable Khoisan ancestry (Busby et al., 2016, Schuster et al., 2010).
While the direction of the Bantu expansion is still in debate, there is evidence showing that the Bantu migrated towards East Africa where other ethnic groups such as the Cushitic and Nilotic dominated, potentially around 2,000 years ago (Pickrell et al., 2014). Our data showing a Duffy negative rate of 20–36% in Southwestern Ethiopia and East Sudan (Figure 1) is consistent with the complex admixture history. The Ethiopian and Sudanese population, with an admixture of several Eurasian ancestries and some Nilotic and Semitic-Cushitic components, migrated south after the Bantu expansion 2–5 thousand years (Hollfelder et al., 2017, Pickrell et al., 2014). Many population groups in Sudan are dominated by Nilotic and Eurasian admixtures with minimal West African component. One such exception is the Afro-Asiatic speaking Hausa population in the Middle Eastern Sudan, which have migrated from West Africa within the past 300 years (Hollfelder et al., 2017). These migrations could have spread P. vivax from West-Central to other parts of Africa.
Low parasitemia in symptomatic Duffy-negative P. vivax infections and implications on invasion mechanism
In Botswana, Ethiopia, and Sudan, Duffy-positive and Duffy-negative individuals coexist. P. vivax parasite density in Duffy-negative infected individuals is significantly lower than the Duffy-positive infected individuals, regardless of geographical differences (Figure 2). Duffy-positive individuals with heterozygous C/T and homozygous T/T were not significantly different in parasitemia. Both genotypes showed significantly higher parasitemia than Duffy negative C/C. The Duffy-negative P. vivax samples in Ethiopia and Sudan showed a greater range of parasitemia variation than those in Botswana. This may be due to differences in sample size (Figure 2; Supplementary File 3). In very few cases the asexual parasites were detected by microscopy in Duffy-negative individuals. For example, among the 20 P. vivax infections identified in Duffy-negative patients from Ethiopia, only four were microscopic-positive and they all showed a relatively higher parasitemia compared to the submicroscopic infections. The Duffy-negative individuals who were infected with P. vivax were mostly submicrocopic and exhibited fever at the time of sample collection. Without highly sensitive diagnostic tools and vigorous on-site training and screening of P. vivax in different parts of Africa, the public health burden, economic impact, and severity associated with vivax malaria could have been vastly underestimated. The clinical spectrum of P. vivax malaria ranges from asymptomatic parasitemia and uncomplicated febrile illness to severe and fatal malaria (Naing et al., 2014). Moreover, P. vivax can cause anemia during chronic undetected infections (Niangaly et al., 2017). Other severe clinical manifestations include multiorgan dysfunction associated with anemia and thrombocytopenia, spontaneous abortions, premature and low birth weight in pregnant women (Naing et al., 2014). These clinical features have mostly been described for Duffy-positive populations. It is unclear if the spectrum of clinical symptoms is different in Duffy-negative patients in Africa.
Figure 2.
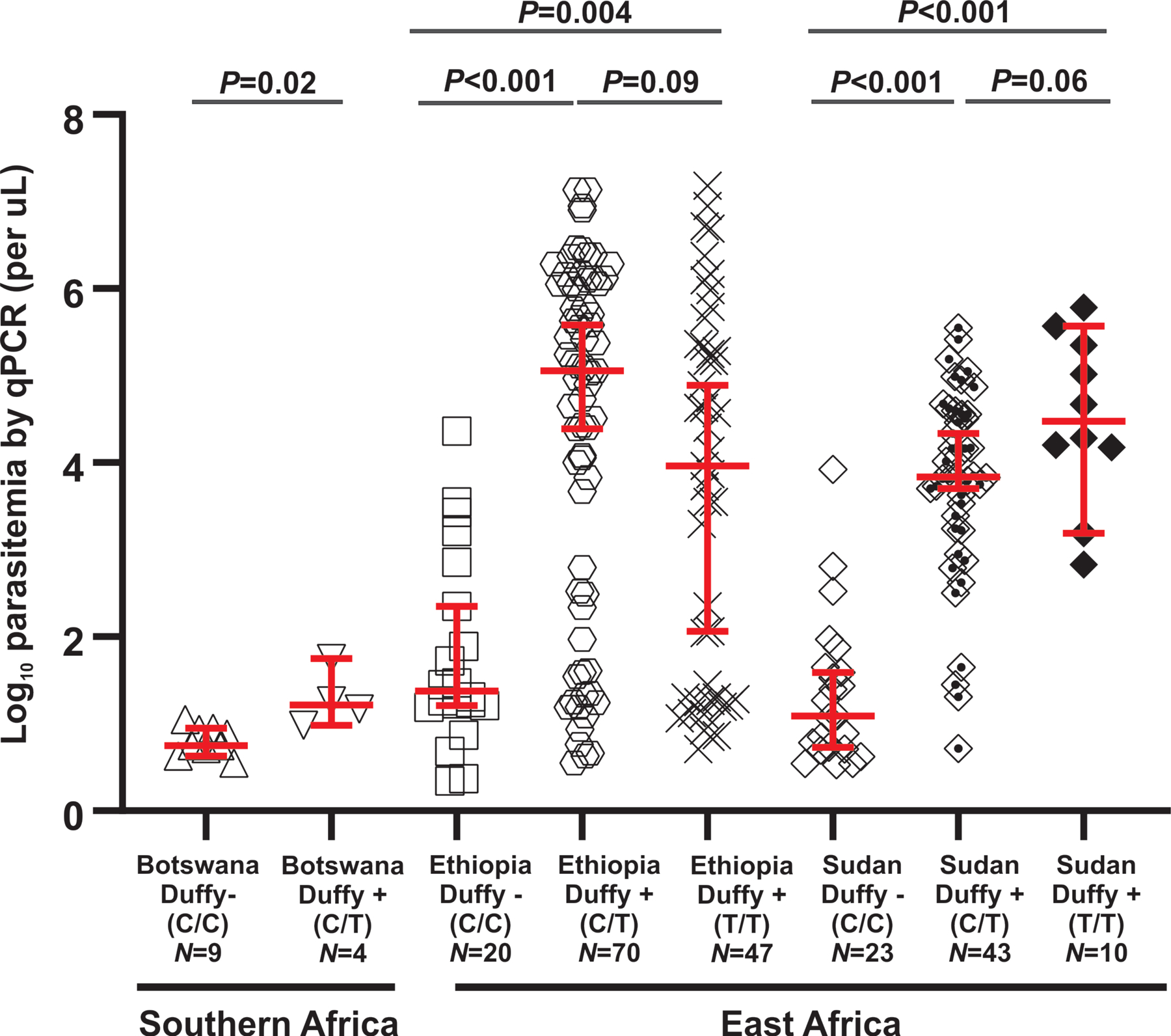
Comparison of P. vivax parasitemia based on quantitative PCR assays between Duffy negative and Duffy positive symptomatic infections among different geographical regions in Africa. Variations in parasitemia among samples were presented as boxplots showing the median and interquartile range values.
Low parasitemia observed in Duffy-negative individuals might suggest a low invasion capability of P. vivax in Duffy-negative individuals. Recent study has shown that mutations in PvDBP1 region II unique to P. vivax in Duffy-negative people in Ethiopia did not lead to binding of Duffy-negative erythrocytes (Gunalana et al., 2016). Also, Sal-I P. vivax infects Squirrel monkeys without PvDBP1 binding to Squirrel monkey erythrocytes (Gunalana et al., 2019). These findings suggested that there are other Duffy-independent pathways that enable erythrocyte invasion. For example, EBP/DBP2 region II has shown to bind to Duffy-positive and Duffy-negative human erythrocytes at low frequency (Gunalana et al., 2016, Ntumngia et al., 2016). CD71 (Transferrin Receptor 1, TfR1) has been shown to bind readily to the reticulocyte binding proteins (PvRBP2b) based on in vitro experiments (Chan et al., 2020, Gruszczyk et al., 2018). Given reticulocytes constitute only a small fraction of all red blood cells, invasion via this RBP2b-TfR1 pathway may result in only a small number of infected erythrocytes and this may explain the considerably low parasitemia observed in Duffy-negative P. vivax infections (Figure 2). Further, recent transcriptomic study has also indicated that genes belonging to tryptophan-rich antigen and merozoite surface protein families were highly expressed in the Saimiri-infected P. vivax, of which erythrocytes did not bind to DBP1 from the Belem isolate of P. vivax (Gunalana et al., 2019). There is growing evidence that members of the tryptophan-rich antigen gene family are involved in erythrocyte invasion (Zeeshan et al., 2015). Various other invasion ligands may also mediate the recognition and invasion to reticulocytes, providing a potential mechanism for variations in reticulocyte preference (Baquero et al., 2017, Moreno-Pérez et al., 2017). Successful schizont development has been shown to be associated with increased younger reticulocytes in the Indian P. vivax isolates (Lim et al., 2016). The low prevalence of schizonts in peripheral blood has led to the hypothesis that P. vivax could be sequestering in reticulocyte-rich zones such as the bone marrow (Mayor and Alano, 2015), resulting in lower detectable parasitemia. Future studies should clarify the expression and role of various P. vivax ligand proteins and their respective receptors in Duffy-negative erythrocyte invasion.
Genetic relationships and origin hypotheses of P. vivax in Duffy-negative Africans
Maximum likelihood analyses of the African P. vivax isolates based on PvDBP indicated that P. vivax from Duffy-negative individuals were not monophyletic but found in multiple well-supported clades (clades I-III in Figure 3). These clades did not show clear geographical boundary but a mixture of P. vivax from different African countries. For instance, Duffy-negative P. vivax from Botswana, Ethiopia, and Sudan were closely related to Duffy-positive P. vivax from the same area, as well as to P. vivax from neighboring Uganda (clade II; bootstrap 91%). The Duffy-negative P. vivax were clustered together with the Duffy-positive ones without genetic distinction. The present data may imply that Duffy-negative and Duffy-positive individuals shared similar P. vivax strains possibly by the same ancestral origin or through recent transmission. The evolution of PvDBP region II could be also driven by functional selection rather than by geographical isolation. Interestingly, Duffy-negative P. vivax samples from Ethiopia and Sudan showed a higher nucleotide and haplotype diversity than the Duffy-positive ones, despite a smaller sample size (Table 3). Among all geographical isolates, P. vivax from Uganda and Madagascar had the highest level of genetic variation, though Duffy status of these samples are unclear (Table 3). These findings offered a hypothesis on the origin of Duffy-negative P. vivax, but PvDBP could be biased by selection or has limited resolution. Extensive phylogenetic analyses using whole genome sequences of Duffy-negative P. vivax from West-Central, Southern, and East Africa, together with the existing data of the P. vivax-like isolates in African apes are needed to adjudicate these origin hypotheses.
Figure 3.
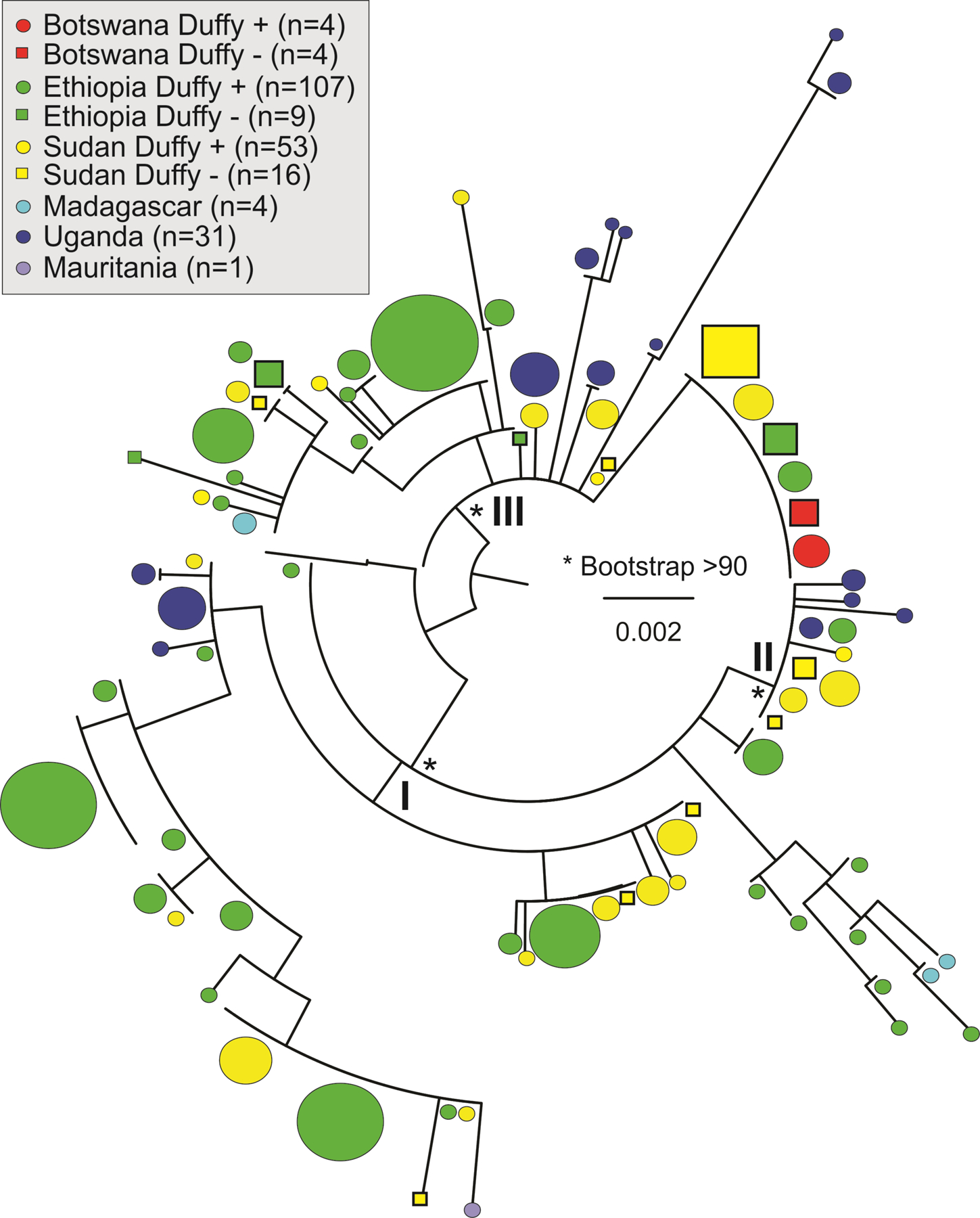
Phylogeny based on PvDBP sequences showing multiple source/origin of Duffy negative P. vivax in Africa. The reference P. vivax strain PVP01 isolated from an Indonesian patient was used as an outgroup. The size of the symbol indicates sample size of each PvDBP haplotype. No clear differentiation was observed between the Duffy negative and Duffy positive P. vivax but nested within one another, suggestive of similar DBP haplotypes.
Table 3.
Nucleotide and haplotype diversity of PvDBP gene sequences between Duffy-negative and Duffy-positive samples from different African countries.
| Region | Country | Duffy | Total samples | Number of polymorphic sites | Nucleotide diversity (SD) | Number of haplotypes | Haplotype diversity (SD) |
|---|---|---|---|---|---|---|---|
| Central Africa | |||||||
| Botswana | Duffy-positive | 4 | 0 | 0 | 1 | 0 | |
| Duffy-negative | 4 | 0 | 0 | 1 | 0 | ||
| East Africa | |||||||
| Ethiopia | Duffy-positive | 107 | 9 | 1.51×10−3 (1.5×10−4) | 11 | 0.762 (0.031) | |
| Duffy-negative | 9 | 9 | 4.18×10−3 (1.0×10−4) | 4 | 0.694 (0.147) | ||
| Sudan | Duffy-positive | 53 | 4 | 3.03×10−3 (2.5×10−4) | 6 | 0.720 (0.039) | |
| Duffy-negative | 16 | 17 | 5.59×10−3 (8.3×10−4) | 8 | 0.758 (0.110) | ||
| Uganda | - | 31 | 28 | 6.51×10−3 (7.7×10−4) | 17 | 0.933 (0.027) | |
| Madagascar | - | 4 | 6 | 7.08×10−3 (2.0×10−4) | 3 | 0.833 (0.222) | |
Previous studies indicated that P. vivax in Southeast Asia and South America evolved in a clade of parasites that infect African monkeys (Loy et al., 2018). Plasmodium vivax in African apes might present a substantial parasite reservoir from which Duffy-positive and Duffy-negative human infections arose from. There are two hypotheses concerning the origin of P. vivax in Duffy-negative Africans (Figure 4). The first hypothesis posits that the ancestral P. vivax infected all African primates including apes and Duffy-positive humans (Liu et al., 2014) (Figure 4A). One of these ancestral lineages evolved to a Duffy-independent pathway and subsequently spread to different parts of Africa via human migration (Choudhury et al., 2020, Grollemunda et al., 2015). The geographical overlap between apes and humans, e. g, in Cameroon and the Democratic Republic of Congo suggest a West-Central African origin of P. vivax in Duffy-negatives (Liu et al., 2014). The second hypothesis posits that the ancestral P. vivax infected only non-human primates in Africa until some of the lineages crossed the species barrier and gave rise to the parasite population currently infecting Duffy-positive humans (Prugnolle et al., 2013). It is possible that Duffy-negative P. vivax observed today across Africa represent separate lineages that were derived multiple times independently from Duffy-positive individuals (Figure 4B). Previous phylogenies based on nuclear genes and partial mitochondrial genomes revealed incongruent genetic relationships (Liu et al., 2014, Prugnolle et al., 2013), possibly due to incomplete lineage sorting or lack of phylogenetic signal (Maddison and Knowles, 2006). Moreover, no African P. vivax isolates from Duffy-positive and Duffy-negative individuals were included. Future studies should employ genome-based phylogenetic approach and molecular dating analyses to clarify the origin of P. vivax in Africa.
Figure 4.
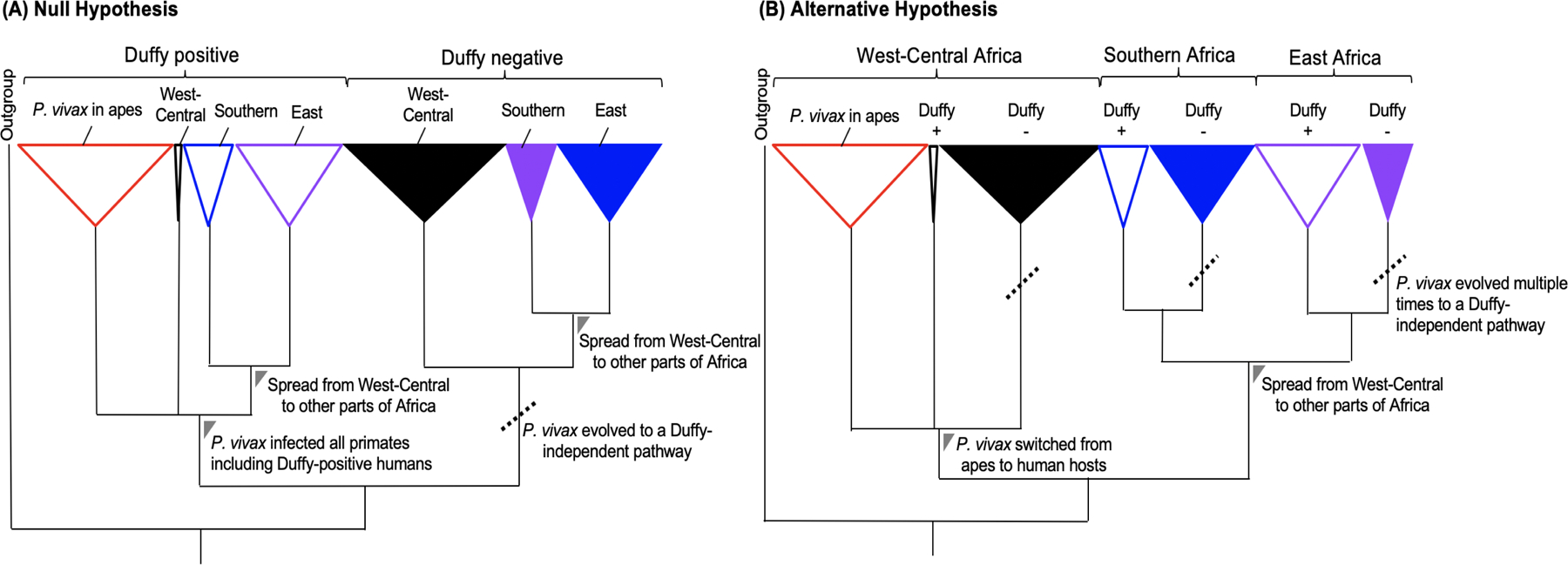
Hypothetical models illustrating the genetic origin of P. vivax in Duffy-negative Africans in a phylogenetic context. (A) The null hypothesis posits that the ancestral P. vivax infected all African primates including apes and Duffy-positive humans forming a monophyletic clade. One of these ancestral lineages evolved to a Duffy-independent pathway (dotted line) and subsequently spread to different parts of Africa via human migration. (B) An alternative hypothesis posits that the ancestral P. vivax infected only non-human primates in Africa until some of the lineages crossed the species barrier and gave rise to the parasite population currently infecting Duffy-positive humans. It is possible that Duffy-negative P. vivax observed today across Africa represent separate lineages that were derived multiple times independently from Duffy-positive individuals (dotted line) forming separate monophyletic clades.
Conclusions
With the increasing number of P. vivax cases reported in Duffy-negative individuals as well as across the continent, vivax malaria is no longer a rare but a growing and possibly widespread phenomenon in Africa. To the best of our knowledge, this paper is the first using a standardized approach to characterize and compare the epidemiological and genetic features of Duffy-negative P. vivax from different parts of Africa. The generally low parasitemia observed in the Duffy-negative infections may suggest a less efficient but continuously evolving invasion mechanism that allows a greater negative public health impact in Africa in coming years. The genetic relatedness based on PvDBP sequences suggested similar strains shared between Duffy-negative and Duffy-positive populations, though the transmission capability of P. vivax in Duffy-negative individuals is still unclear. Further investigations are needed to unveil the invasion and transmission mechanisms of these infections. These data would help predict the scale of disease spread and improve existing malaria control measures, beyond P. falciparum in Africa. On the public health front-end, there should be more resources and training allocated to diagnosis and treatment of vivax malaria, given its unique ability in causing relapse and other longer-term health problems such as anemia in asymptomatic infections. Duffy-negative Africans are not resistant to P. vivax infection and the public health significance of vivax malaria in Africa should no longer be neglected.
Supplementary Material
Supplementary File 1. Detailed protocols of P. vivax screening, Duffy genotyping, and PvDBP sequencing.
Supplementary File 2. Genbank accession number and geographical location of PvDBP sequences included in the present study.
Supplementary Fie 3. Duffy genotype and parasite density based on quantitative PCR assay of Duffy negative P. vivax samples.
Acknowledgements
We thank the students and staffs from Jimma University, University of Khartoum, University of Botswana, and University of North Carolina at Charlotte for their technical assistance in sample collection and processing; the communities and hospitals for their support and willingness to participate in this research.
Funding
This research was funded by National Institutes of Health (R15 AI138002). GMP was supported also by Penn Center for AIDS Research (grant # P30 AI045008).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Scientific and ethical clearance was given by the institutional scientific and ethical review boards of Jimma University (Ethiopia), the Ethics Committee of the Institute of Endemic Diseases, University of Khartoum (Reference number: 9/2016), the Health Research and Development Division of the Botswana Ministry of Health and Wellness (Reference number: HPDME: 13/18/1), and University of North Carolina at Charlotte (USA). Written informed consent/assent was obtained from all consenting heads of households, parents/guardians (for minors under 18 years old), and each individual who participated in this study.
References
- 1.Albsheer M, Pestana K, Ahmed S, Elfaki M, Gamil E, Ahmed SM, et al. Distribution of Duffy Phenotypes among Plasmodium vivax Infections in Sudan. Genes 2019;10:437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baird JK. 8-Aminoquinoline therapy for latent malaria. Clin Microbiol Rev 2019;32:e00011–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baquero LA, Moreno-Pérez DA, Garzón-Ospina D, Forero-Rodríguez J, Ortiz-Suárez HD, Patarroyo MA. PvGAMA reticulocyte binding activity: predicting conserved functional regions by natural selection analysis. Parasit Vectors 2017;10: 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battle KE, Lucas TCD, Nguyen M, Howes RE, Nandi AK, Twohig KA, et al. Mapping the global endemicity and clinical burden of Plasmodium vivax, 2000–17: a spatial and temporal modelling study. Lancet 2019;394:P332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bereczky S, Martensson A, Gil JP, Farnert A. Short report: Rapid DNA extraction from archive blood spots on filter paper for genotyping of Plasmodium falciparum. Am J Trop Med Hyg 2005;72:249–51. [PubMed] [Google Scholar]
- 6.Brazeau NF, Whitesell AN, Doctor SM, Keeler C, Mwandagalirwa MK, Tshefu AK, et al. Plasmodium vivax Infections in Duffy-negative individuals in the Democratic Republic of the Congo. Am J Trop Med Hyg 2018;99:1128–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busby GBJ, Band G, Le QS, Jallow M, Bougama E, Mangano VD, et al. Malaria genomic epidemiology network. Admixture into and within sub-Saharan Africa. eLife 2016;5:e15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan LJ, Dietrich MH, Nguitragool W, Tham WH. Plasmodium vivax Reticulocyte Binding Proteins for invasion into reticulocytes. Cell Microbiol 2020;22:e13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choudhury A, Aron S, Botigué LR, Sengupta D, Botha G, et al. High-depth African genomes inform human migration and health. Nature 2020;586:741–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cowman A, Tonkin CJ, Tham WH, Duraisingh MT. The molecular basis of erythrocyte invasion by malaria parasites. Cell Host Microbe 2017;22:232–245. [DOI] [PubMed] [Google Scholar]
- 11.Grollemunda R, Branforda S, Bostoenb K, Meadea A, Vendittia C, Page M. Bantu expansion shows that habitat alters the route and pace of human dispersals. Proc Natl Acad Sci USA 2015;112:13296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruszczyk J, Kanjee U, Chan LJ, Menant S, Malleret B, et al. Transferrin receptor 1 is a reticulocyte-specific receptor for Plasmodium vivax. Science 2018;359:48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunalan K, Gao X, Yap SS, Huang X, Preiser PR. The role of the reticulocyte-binding-like protein homologues of Plasmodium in erythrocyte sensing and invasion. Cell Microbiol 2013;15:35–44. [DOI] [PubMed] [Google Scholar]
- 14.Gunalana K, Lo E, Hostetler JB, Yewhalaw D, Mu J, Neafsey DE, Yan G, Miller LH. Role of Plasmodium vivax Duffy-binding protein 1 in invasion of Duffy-null Africans. Proc Natl Acad Sci USA 2016;113:6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunalan K, Niangaly A, Thera MA, Doumbo OK, Miller LH. Plasmodium vivax Infections of Duffy-negative erythrocytes: Historically undetected or a recent adaptation? Trends Parasitol 2018;34:420–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunalan K, Sá JM, Barros RRM, Anzick SL, Caleon RL, Mershon JP, et al. Transcriptome profiling of Plasmodium vivax in Samira monkeys identifies potential ligands for invasion. Proc Natl Acad Sci USA 2019;116: 7053–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamblin MT, Thompson EE, Di Rienzo A. Complex signatures of natural selection at the Duffy blood group locus. Am J Hum Genet 2002;70:369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollfelder N, Schlebusch CM, Günther T, Babiker H, Hassan HY, Jakobsson M. Northeast African genomic variation shaped by the continuity of indigenous groups and Eurasian migrations. PLoS Genet 2017;13:e1006976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hester J, Chan ER, Menard D, Mercereau-Puijaion O, Barnwell J, Zimmerman PA, Serre D. De novo assembly of a field isolate genome reveals novel Plasmodium vivax erythrocyte invasion genes. PLoS Negl Trop Dis 2013;7:e2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howes RRE, Patil APAAP, Piel FBF, Nyangiri OAOOA, Kabaria CWCCW, Gething PW, et al. The global distribution of the Duffy blood group. Nat Commun 2011;2:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King CL, Adams JH, Xianli J, Grimberg BT, McHenry AM, Greenberg LJ, Siddiqui A, Howes RE, da Silva-Nunes M, Ferreira MU, Zimmerman PA. Fy(a)/Fy(b) antigen polymorphism in human erythrocyte Duffy antigen affects susceptibility to Plasmodium vivax malaria. Proc Natl Acad Sci USA 2011;108:20113–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar H, Tolia NH. Getting in: The structural biology of malaria invasion. PLoS Pathog 2019; 15: e1007943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim C, Pereira L, Saliba KS, Mascarenhas A, Maki JN, Chery L, Gomes E, et al. Reticulocyte preference and stage development of Plasmodium vivax isolates. J Infect Dis 2016;214:1081–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu W, Li Y, Shaw KS, Learn GH, Plenderleith LJ, Malenke JA, Sundararaman SA, et al. African origin of the malaria parasite Plasmodium vivax. Nat Commun 2014;5:3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livingstone FB. The Duffy blood groups, vivax malaria, and malaria selection in human populations: a review. Hum Biol 1984;56:413–25. [PubMed] [Google Scholar]
- 26.Lo EYY, Delenasaw Y, Zhong D, Zemene E, Degefa T, Tushune K, et al. Molecular epidemiology of Plasmodium vivax and Plasmodium falciparum malaria among Duffy-positive and Duffy-negative populations in Ethiopia. Malar J 2015;14:84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loy DE, Plenderleith LJ, Sundararaman SA, Liu W, Gruszczyk J, Chen YJ, et al. Evolutionary history of human Plasmodium vivax revealed by genome-wide analyses of related ape parasites. Proc Natl Acad Sci USA 2018;115:E8450–8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maddison WP, Knowles LL. Inferring phylogeny despite incomplete lineage sorting. Syst Biol 2006;55:21–30. [DOI] [PubMed] [Google Scholar]
- 29.Mayor A, Alano P. Bone marrow reticulocytes: a Plasmodium vivax affair? Blood 2015;125:1203–1205. [DOI] [PubMed] [Google Scholar]
- 30.Menard D, Barnadas C, Bouchier C, Henry-Halldin C, Gray LR, Ratsimbasoa A, et al. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci USA 2010;107:5967–5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendes C, Dias F, Figueiredo J, Mora VG, Cano J, de Sousa B, et al. Duffy negative antigen is no longer a barrier to Plasmodium vivax--molecular evidences from the African West Coast (Angola and Equatorial Guinea). PLoS Negl Trop Dis 2011;5:e1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med 1976;295:302–304. [DOI] [PubMed] [Google Scholar]
- 33.Moreno-Pérez DA, Baquero LA, Chitiva-Ardila DM, Patarroyo MA. Characterising PvRBSA: an exclusive protein from Plasmodium species infecting reticulocytes. Parasit Vectors 2017;10:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Motshoge T, Ababio GK, Aleksenko L, Read J, Peloewetse E, Loeto M, et al. Molecular evidence of high rates of asymptomatic P. vivax infection and very low P. falciparum malaria in Botswana. BMC Infect Dis 2016;16:520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naing C, Whittaker MA, Nyunt Wai V, Mak JW. Is Plasmodium vivax Malaria a Severe Malaria? A Systematic Review and Meta-Analysis. PLoS Negl Trop Dis 2014;8:e3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niang M, Sane R, Sow A, Sadio BD, Chy S, Legrand E, Faye O, et al. Asymptomatic Plasmodium vivax infections among Duffy-negative population in Kedougou, Senegal. Trop Med Health 2018;46:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niangaly A, Gunalan K, Ouattara A, Coulibaly D, MS a J, Adams M, Travassos MA, et al. Plasmodium vivax infections over 3 years in Duffy blood group negative Malians in Bandiagara, Mali. Am J Trop Med Hyg 2017;97:744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ntumngia FB, Thomson-Luque R, Torres Lde M, Gunalan K, Carvalho LH, Adams JH. A novel erythrocyte binding protein of Plasmodium vivax suggests an alternate invasion pathway into Duffy-positive reticulocytes. MBio 2016;7:e01261–01216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oboh MA, Singh US, Ndiaye D, Badiane AS, Ali NA, Bharti PK, Das A. Presence of additional Plasmodium vivax malaria in Duffy negative individuals from Southwestern Nigeria. Malar J 2020;19:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petersen DC, Libiger O, Tindall EA, Hardie R-A, Hannick LI, Glashoff RH, et al. Complex patterns of genomic admixture within Southern Africa. PLoS Genet 2013;9:e1003309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pickrell JK, Patterson N, Loh PR, Lipson M, Berger B, Stoneking M, Pakendorf B, Reich D. Ancient west Eurasian ancestry in southern and eastern Africa. Proc Natl Acad Sci USA 2014;111:2632–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poirier P, Doderer-Lang C, Atchade PS, Lemoine JP, de l’Isle MC, Abou-Bacar A, et al. The hide and seek of Plasmodium vivax in West Africa: report from a large-scale study in Beninese asymptomatic subjects. Malar J 2016;15:570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prugnolle F, Rougeron V, Becquarta P, Berryd A, Makangab B, Raholaa N, et al. Diversity, host switching and evolution of Plasmodium vivax infecting African great apes. Proc Natl Acad Sci USA 2013;110:8123–8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russo G, Faggiono G, Paganoti GM, Djeunang Dongho GB, Pomponi A, De Santis R, et al. Molecular evidence of Plasmodium vivax infection in Duffy negative symptomatic individuals from Dschang, West Cameroon. Malar J 2017;16:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuster SC, Miller W, Ratan A, Tomsho LP, et al. Complete Khoisan and Bantu genomes from southern Africa. Nature 2010;463:943–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tournamille C, Colin Y, Cartron JP, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet 1995;10:224–228. [DOI] [PubMed] [Google Scholar]
- 47.Twohig KA, Pfeffer DA, Baird JK, Price RN, Zimmerman PA, Hay SI, et al. Growing evidence of Plasmodium vivax across malaria endemic Africa. PLoS Negl Trop Dis 2019;13: e0007140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woldearegai TG, Kremsner PG, Kun JF, Mordmuller B. Plasmodium vivax malaria in Duffy-negative individuals from Ethiopia. Trans R Soc Trop Med Hyg 2013;107:328–31. [DOI] [PubMed] [Google Scholar]
- 49.Zeeshan M, Tyagi RK, Tuagi K, Alam MS, Sharma YD. Host-parasite interaction: selective Pv-fam-a family proteins of Plasmodium vivax bind to a restricted number of human erythrocyte receptors. J Infect Dis 2015;211:1111–20. [DOI] [PubMed] [Google Scholar]
- 50.Zimmerman PA. Plasmodium vivax Infection in Duffy-Negative People in Africa. Am J Trop Med Hyg 2017;97:636–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1. Detailed protocols of P. vivax screening, Duffy genotyping, and PvDBP sequencing.
Supplementary File 2. Genbank accession number and geographical location of PvDBP sequences included in the present study.
Supplementary Fie 3. Duffy genotype and parasite density based on quantitative PCR assay of Duffy negative P. vivax samples.


