Summary
Replication complexes of (+)RNA viruses of eukaryotes are associated with specialized membranous domains, termed replication organelles. How these structures develop is poorly understood. In a recent Cell paper, Laufman et al. (2019) reveal that enteroviruses recruit lipid droplets to support lipid synthesis required for the structural development of replication organelles.
(+)RNA viruses are by far the largest group of viruses of eukaryotic hosts. Many of them are associated with human, animal and plant diseases incurring multibillion economic losses and posing global health challenges. (+)RNA viruses actively build a unique membrane environment to harbor their replication complexes, rather than just occupy membranes available before infection. Since infection relies on specific changes in cellular membrane metabolism, targeting infection-specific membrane synthesis and trafficking pathways may open directions for creating safe and effective broad-spectrum antiviral therapeutics.
Enteroviruses, a group of viruses belonging to the Picornaviridae family, include important human pathogens, such as poliovirus, enteroviruses A71 and D68, coxsackieviruses, and rhinoviruses (Laufman et al., 2019). These viruses code for just a handful of proteins and rely on simple but effective mechanisms to trick the highly elaborate cellular controls of membrane homeostasis. In a recent issue of Cell, Laufman et al (2019) provide a comprehensive picture of recruitment of lipid droplets (LDs) that support the development of replication organelles (ROs) of enteroviruses (Laufman et al., 2019).
Enterovirus infection dramatically changes intracellular membrane architecture. Distinct ROs appear early in infection and soon occupy a large volume of the cytoplasm. This rapid expansion of ROs is due to massive activation of structural phospholipid production supported by the virus-induced release of the nuclear depot of CCTα, an enzyme controlling a rate-limiting step in phosphatidylcholine synthesis (Viktorova et al., 2018). At the same time, viral protein 3A coordinates the recruitment of several ER and Golgi factors (e.g. phosphatidylinositol kinase PI4KB, sterol transporter OSBP and ArfGEF GBF1), and triggers the formation of membrane contact sites in which lipids are shuttled between these compartments and ROs. This imparts a specific PI4P- and cholesterol-enriched lipid signature to the ROs and provides an optimal environment for the functioning of the replication machinery (van der Schaar et al., 2016).
LDs are dynamic cellular organelles coordinating synthesis, storage and mobilization of neutral lipids. They are composed of a triglyceride and cholesteryl ester core surrounded by a monolayer of phospholipids derived from the ER membrane. The lipids stored in LDs can be mobilized by a complex of LD-associated lipases, or by a specific branch of autophagy, lipophagy, which delivers the LD-derived lipids to lysosomes. The long chain fatty acids (FAs) released by lipase activity serve as substrates in multiple biosynthesis pathways, including those generating structural phospholipids, or support cellular energy metabolism. Basic LD metabolism is similar in different cell types, yet cell-specific variations in LD biogenesis and utilization have been reported (Olzmann and Carvalho, 2019).
In a series of papers published in the last years, LDs emerged as crucial participants of the enterovirus life cycle. It has been demonstrated that LDs provide the bulk of cholesterol required to maintain the cholesterol-rich environment of the ROs of several rhinoviruses (Roulin et al., 2014). It was also found that the phospholipid synthesis activated upon poliovirus and rhinovirus infection is mostly sustained by FAs released from LDs through LD-associated lipases, including hormone-sensitive lipase (HSL) (Viktorova et al., 2018, Roulin et al., 2014). Furthermore, electron microscopic analysis of rhinovirus-infected cells (Roulin et al., 2014) and whole-cell tomography of coxsackievirus-infected cells (Melia et al., 2019) revealed that ROs form contact sites and develop in close association with LDs.
The work by Laufman and colleagues to the large part confirms and significantly extends these findings. By quantitative analysis of microscopy images, they demonstrated clustering of LDs upon poliovirus infection. LDs were found to often form membrane contact sites with the RO membranes, and sometimes engage a nearby ER membrane, likely facilitating exchange of lipids and proteins among these structures. The authors demonstrated that poliovirus proteins 2B, 2C and their precursor 2BC associate with LDs, and that both 2B and 2C possess amphipathic helix (AH) domains mediating LD targeting. The capacity of 2C to oligomerize, previously known to be important for viral RNA replication, was responsible for clustering of LDs (Figure 1). The development of poliovirus infection was accompanied by displacement of a cellular protein, TIP47, from the surface of LDs, which was recapitulated by ectopic expression of 2BC, and which may contribute to the increased accessibility of the neutral lipid core to lipases. Accordingly, co-IP experiments revealed that viral protein 3A is engaged in interactions with LD-associated lipases, adipose triglyceride lipase (ATGL) and HSL. As 3A localizes to ROs rather than to LDs, these interactions were suggested to promote the recruitment of the lipolysis machinery to the membrane contact sites between LDs and ROs (van der Schaar et al., 2016). In line with these findings, inhibitors of ATGL or HSL activity, but not lipophagy, prevented development of ROs and suppressed viral replication. Importantly, the key findings were replicated with other enteroviruses, such as enterovirus A71 and coxsackievirus B3, indicating shared mechanisms of LD recruitment and lipase activation, which can be targeted for the development of broad-spectrum antivirals.
Enteroviruses engage lipid droplets to support the development of the replication organelles.
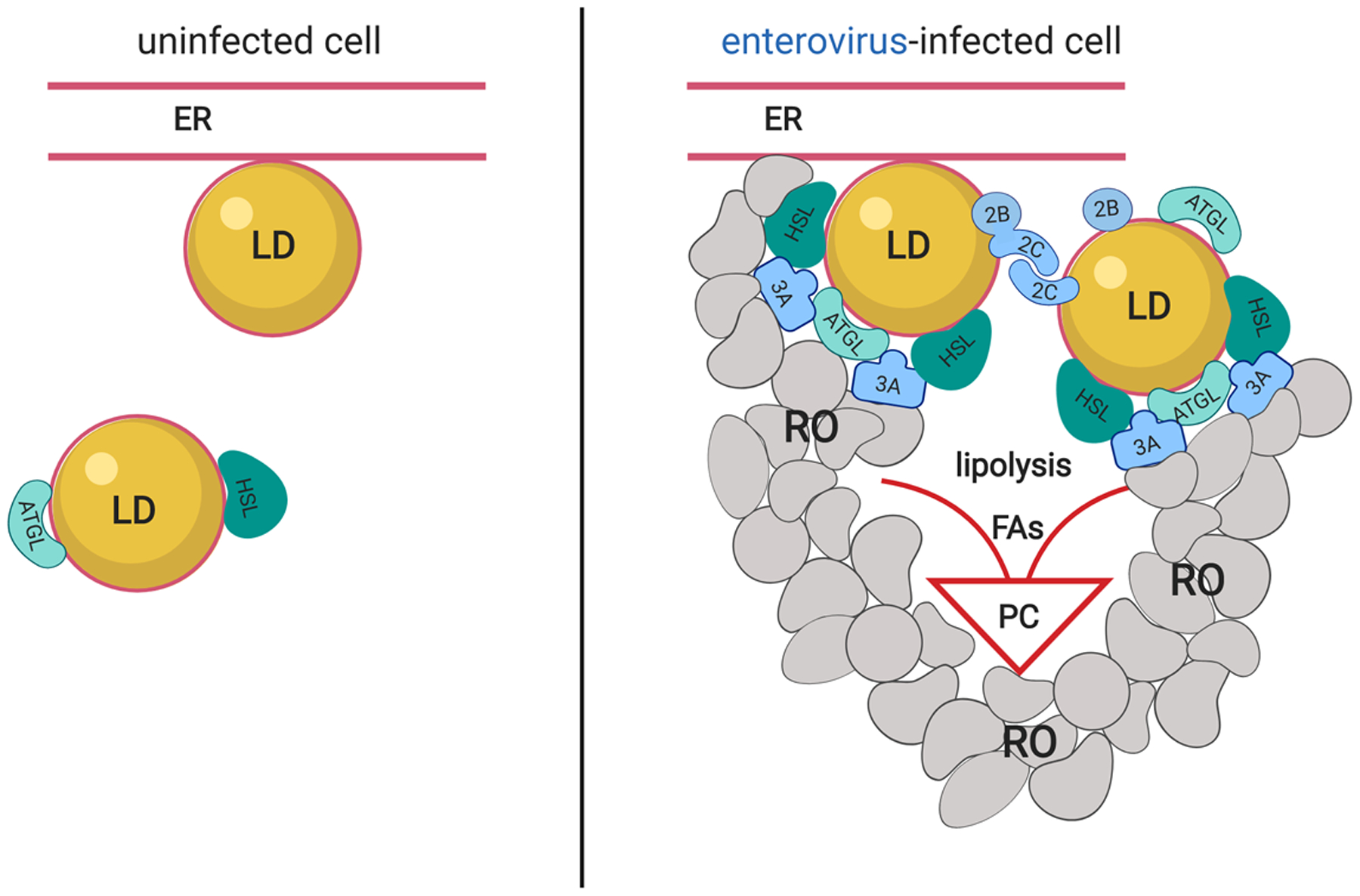
Lipid droplets (LDs) are formed in association with the ER membrane and support basic cellular metabolism in uninfected cells. Neutral lipids are hydrolyzed by LD-associated lipases, including ATGL and HSL. Upon enterovirus infection viral proteins 2B, 2C and precursor 2BC associate with LDs, and oligomerization of 2C results in LD clustering. Viral protein 3A, localized on the membranes of replication organelles (RO), facilitates recruitment of lipases HSL and ATGL to LDs, likely by stimulating establishment of membrane contact sites between RO, LDs and ER, and/or interactions with the lipases. Increased lipase activity releases long chain fatty acids (FAs) promoting synthesis of structural phospholipids (PC), leading to massive expansion of the membranes of ROs.
This fundamental work highlights the ingenuity of enteroviruses and their ability to profoundly manipulate cellular metabolism. Additionally, it raises many questions and opens perspectives for research. Many cellular LD metabolism proteins cycle on and off the LDs upon different stimuli, and their recruitment is often also attributed to the AH domains which may sense changes in lipid packaging at the surface of LDs. The AH in 2C belongs to the apolipoprotein class of AHs and strongly resembles that in NS5A in hepatitis C virus (HCV), which is also involved in associating with LDs, whereas that in 2B groups to the lytic polypeptide class of AHs, implying different modes of interactions with LDs. It is also not clear if 2B and 2C are recruited simultaneously to the same LD, or if they may be acting sequentially, thus promoting the transition of LDs from a quiescent to activated state. Furthermore, high resolution microscopy is needed to distinguish if 2B and 2C localize at the LD phospholipid monolayer or at ER(-derived) membranes that surround the LDs, as has been observed for a subpopulation of HCV NS5A (Lee et al., 2019).
The observation that 3A interacts with ATGL and HSL is intriguing. This finding implies that 3A not only has a central role in the formation of membrane contact sites between ROs and the Golgi (van der Schaar et al., 2016), but also between ROs, LDs and ER. How 3A interacts with ATGL and HSL is unclear. These lipases co-precipitated with ectopically expressed 3A but no evidence for a direct interaction has yet been presented. This small viral protein interacts with several cellular partners, including GBF1, a guanine nucleotide exchange factor for small GTPAses Arf, containing a lipid binding domain that can mediate association with LDs. Hence, it cannot be excluded that 3A indirectly pulled down lipase-containing LDs through an interaction with GBF1. More insight into the nature of the interaction of 3A with LD-associated proteins is awaited.
Enterovirus proteins 2B, 2C and 3A have been implicated in several membrane-modifying processes and to interact with multiple host factors. The identification of their role in LD recruitment by Laufman and colleagues underscores the multi-functionality of enterovirus proteins and stimulates re-evaluation of the prevalent view that all viral proteins are more or less present at the same time at the same place, due to their polyprotein-based expression strategy. Clearly, the viral proteins associated with LDs represent a distinct population, raising a question of how they are delivered to LDs. One possibility is the engagement of an existing GBF1/Arf controlled cellular pathway of recruitment of some proteins to LDs. Viral protein 3A specifically recruits GBF1 to ROs, thus providing the possibility to organize local trafficking and LD targeting of viral proteins (van der Schaar et al., 2016). Viral exploitation of this pathway has been observed in Dengue virus-infected cells, where structural proteins are delivered to LDs in a GBF1-dependent manner (Iglesias et al., 2015). However, since individually expressed viral proteins could target to LDs, a passive lateral diffusion in ER or RO membranes juxtaposed to LDs cannot be excluded, similar to the mechanism proposed for LD-associated replication and assembly complexes of HCV (Lee et al., 2019). Still, the data with expression of individual proteins should be interpreted with caution because such systems do not recapitulate all the complexity of the changes of the membrane organization in infected cells.
It has long been assumed that ROs are mandatory for viral genome replication. However, recent data demonstrate that inhibition of the structural development of enterovirus ROs does not significantly affect the first cycle of replication (Melia et al., 2017, Viktorova et al., 2018). In contrast, Laufman and colleagues observed that inhibition of lipases not only strongly interfered with RO formation but also reduced virus replication. These different results may reflect the specifics of the experimental systems used. Furthermore, they suggest that during infection in a natural host, enteroviruses may demonstrate different sensitivity to manipulation of membrane metabolism in different cell types and/or under different conditions, which may have important implications for therapeutic approaches. The dependence on LD metabolism seems to be an evolutionarily conserved feature of diverse RNA viruses. Picornaviruses, flaviviruses, and even dsRNA rotaviruses, a group of important enteric pathogens, depend on them for one or another step in their life cycle (Cheung et al., 2010, Iglesias et al., 2015, Laufman et al., 2019, Lee et al., 2019, Melia et al., 2019, Viktorova et al., 2018). Thus, understanding the mechanism of LD functioning in normal and infected cells may open unexpected perspectives to control diverse viral pathogens by broad-range antiviral therapy, and the work by Laufman and colleagues is an important step in this direction.
References
- CHEUNG W, GILL M, ESPOSITO A, KAMINSKI CF, COUROUSSE N, CHWETZOFF S, TRUGNAN G, KESHAVAN N, LEVER A & DESSELBERGER U 2010. Rotaviruses Associate with Cellular Lipid Droplet Components To Replicate in Viroplasms, and Compounds Disrupting or Blocking Lipid Droplets Inhibit Viroplasm Formation and Viral Replication. Journal of Virology, 84, 6782–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IGLESIAS NG, MONDOTTE JA, BYK LA, DE MAIO FA, SAMSA MM, ALVAREZ C & GAMARNIK AV 2015. Dengue Virus Uses a Non-Canonical Function of the Host GBF1-Arf-COPI System for Capsid Protein Accumulation on Lipid Droplets. Traffic, 16, 962–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAUFMAN O, PERRINO J & ANDINO R 2019. Viral Generated Inter-Organelle Contacts Redirect Lipid Flux for Genome Replication. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE JY, CORTESE M, HASELMANN U, TABATA K, ROMERO-BREY I, FUNAYA C, SCHIEBER NL, QIANG Y, BARTENSCHLAGER M, KALLIS S, RITTER C, ROHR K, SCHWAB Y, RUGGIERI A & BARTENSCHLAGER R 2019. Spatiotemporal Coupling of the Hepatitis C Virus Replication Cycle by Creating a Lipid Droplet- Proximal Membranous Replication Compartment. Cell Rep, 27, 3602–3617 e5. [DOI] [PubMed] [Google Scholar]
- MELIA CE, PEDDIE CJ, DE JONG AWM, SNIJDER EJ, COLLINSON LM, KOSTER AJ, VAN DER SCHAAR HM, VAN KUPPEVELD FJM & BARCENA M 2019. Origins of Enterovirus Replication Organelles Established by Whole-Cell Electron Microscopy. MBio, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MELIA CE, VAN DER SCHAAR HM, LYOO H, LIMPENS RWAL, FENG Q, WAHEDI M, OVERHEUL GJ, VAN RIJ RP, SNIJDER EJ, KOSTER AJ, BARCENA M & VAN KUPPEVELD FJM 2017. Escaping Host Factor PI4KB Inhibition: Enterovirus Genomic RNA Replication in the Absence of Replication Organelles. Cell Reports, 21, 587–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OLZMANN JA & CARVALHO P 2019. Dynamics and functions of lipid droplets. Nature Reviews Molecular Cell Biology, 20, 137–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROULIN PS, LOTZERICH M, TORTA F, TANNER LB, VAN KUPPEVELD FJM, WENK MR & GREBER UF 2014. Rhinovirus Uses a Phosphatidylinositol 4-Phosphate/Cholesterol Counter-Current for the Formation of Replication Compartments at the ER-Golgi Interface. Cell Host & Microbe, 16, 677–690. [DOI] [PubMed] [Google Scholar]
- VAN DER SCHAAR HM, DOROBANTU CM, ALBULESCU L, STRATING JRPM & VAN KUPPEVELD FJM 2016. Fat(al) attraction: Picornaviruses Usurp Lipid Transfer at Membrane Contact Sites to Create Replication Organelles. Trends in Microbiology, 24, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VIKTOROVA EG, NCHOUTMBOUBE JA, FORD-SILTZ LA, IVERSON E & BELOV GA 2018. Phospholipid synthesis fueled by lipid droplets drives the structural development of poliovirus replication organelles. PLoS Pathog, 14, e1007280. [DOI] [PMC free article] [PubMed] [Google Scholar]


