Abstract
Disruptions to the microbiota can have pathological consequences, which highlights the need to understand the factors that contribute to its stability. Although decades of research have focused on the importance of IgA during pathogenic infection, much of the IgA that is generated in the gut targets the resident commensal microorganisms. Despite this observation, the role of antibodies in regulating microbiota composition remains controversial and poorly understood. Here we propose that antibodies generated in response to microbial colonization of the gut shape the composition of the microbiota to benefit the health of the host through a process that we term antibody-mediated immunoselection (AMIS). Given the exquisite specificity of antibodies and an emerging interest in the use of immunotherapies, we suggest that understanding AMIS of the microbiota will highlight novel uses of antibodies to manipulate microbial communities for therapeutic benefit.
Antibodies in the gut target and control pathogenic infection, but they also target common resident commensal microorganisms. This suggests that immunoglobulins might influence microbiota composition during homeostasis. Although the immune system has evolved many ways to control microorganisms, the antibody response does so in an antigen-specific manner. Recent studies exploiting sequencing innovations together with immunological techniques and ecological theory have begun to demonstrate how host antibody responses might influence microbiota composition and function, a process that we refer to as antibody-mediated immunoselection (AMIS). Immunoselection refers to a process of natural selection within a host organism that is mediated by the immune system to influence microbial fitness, and consequently microbial ecology and evolution. All immune responses can influence microbial fitness, but different individuals do not respond to the same microorganism in the same way. In fact, emerging evidence suggests that even otherwise genetically identical individuals develop largely non-overlapping antibody repertoires1. We use the term AMIS to emphasize the potential importance of variability in immunoselection among individuals in shaping microbiota composition. Indeed, a central tenet of this article is that personalized antibody repertoires result in a dynamic pattern of immunoselection that leads to the establishment of unique microbial communities among individuals. We briefly review the available scientific literature pertaining to the role of adaptive immunity and IgA antibody responses in the gut to support our hypothesis that a process of AMIS shapes microbiota composition. We also describe how the microbiota can modulate this process, how AMIS promotes a ‘healthy’ microbiota, and how this process might be harnessed to treat diseases associated with dysbiosis (that is, unhealthy host–microbiota interactions).
The factors regulating microbiota composition are diverse and complex, and the hypothesis that adaptive immunity participates in this process is controversial2. Much of this debate stems from the difficulties in designing experiments that adequately control for confounding environmental variables that are known to influence microbiota composition. Co-housing of mice has been used in an attempt to address this concern but is insufficient because coprophagy in rodents has a homogenizing effect on microbiota composition. One potential solution to this issue might be to co-house wild-type and mutant animals in partitioned mouse cages that limits the transfer of bedding and faecal material but allows for the exchange of air. Another approach could be to design experiments in such a way that the effects of both the environment and host genetics can be quantified.
Generation of intestinal antibodies
In the gut, naive B cells are found in the immune cell-rich tissues immediately underlying the epithelium (the lamina propria), but they reach their highest concentrations in the specialized gut-associated lymphoid tissues, where they mature into long-lived memory B cells or antibody-secreting plasma cells.
Of the five known antibody isotypes, IgA, IgM and IgG are known to be secreted into the lumen of the gut under steady-state conditions, but IgA is by far the most abundant. Secretory antibodies can exist in polymeric (IgM, IgA) and monomeric forms (IgG). IgM and IgA bind the polymeric immunoglobulin receptor (PIgR), which is expressed on the basolateral surface of gut epithelium, and are transported across the epithelium to the apical surface. Proteolytic cleavage of the antibody-bound portion of PIgR, known as the secretory component, results in the release of secretory IgA and IgM. The secretion of IgG into the gut is mediated by a different transporter known as the Fc neonatal receptor3. Although IgA responses have been the primary focus of research into AMIS in the gut, other antibody classes may also influence this process.
IgA targets the microbiota.
Germ-free mice provide a powerful tool to understand the effects of the microbiota on the host4. It has long been appreciated that the absence of commensal microorganisms is associated with reduced antibody concentrations in the gut. When germ-free mice are colonized with a complex microbiota, plasma cell abundance rapidly increases in the lamina propria, and faecal IgA concentrations markedly increase5–7. Not surprisingly, much of this IgA directly targets the colonizing microorganisms. Various methods have been used to study the coating of particular commensals by IgA. Flow cytometry-based assays suggested that approximately 45% of the microbiota within human faeces is covered with IgA8. Using a simpler system, in which germ-free mice were mono-associated with a single bacterial organism, Morganella morganii, more than 90% of the bacteria were coated with IgA 70-days post-colonization7. Consistent with observations made in humans, animals colonized with a complex microbiota have 10–60% of the bacteria bound by IgA9–13. It seems that more bacteria are targeted by IgA within the small intestine than the colon in both mice and humans12. Thus, a large proportion of commensals within animals and humans are bound by IgA.
So, which bacterial organisms are targeted by IgA, and do they represent a specialized subset? Using high-throughput sequencing technologies, multiple investigations have shown that IgA binds to a distinct group of bacteria within the colon10,12,14,15. A recent study from our laboratory showed that when IgA-bound and IgA-unbound microorganisms are compared to the total community present within the mucosa or lumen, the IgA-bound fraction was more similar to the mucosa community than the faecal community10, suggesting that IgA tends to target organisms that are more closely associated with host tissues. Supporting this, IgA preferentially targets organisms such as segmented filamentous bacteria, flagellated members of the microbiota belonging to Enterobacteriaceae, and mucus-degrading bacteria such as Akkermansia muciniphila, which are all known to localize to host tissue12–14,16. Although this is discussed in more detail later, IgA seems to bind organisms that are associated with inflammation and disease such as members of the Enterobacteriaceae15. Differences in the physical properties of mucus might influence IgA concentrations in the gut17,18. For example, the heavily glycosylated secretory component of dimeric IgA helps to anchor secretory IgA to the mucus lining of the gut19. Thus, the increased antibody binding to mucosa-associated organisms could be due to their localization in a site enriched for IgA antibodies. Alternatively, it may reflect a bias in the immune response towards targeting the tissue-associated microbial community, although there is currently no direct evidence to support this possibility.
Effects of antibodies on the microbiota
Antibodies secreted in the gut could directly affect microbial fitness through various effector functions, including agglutination of microbial cells, binding of surface epitopes to block the attachment of microorganisms to the host, opsonization of bacteria to enhance phagocytosis and antigen presentation by dendritic cells in the lamina propria, and neutralization of microbial toxins or other secreted factors. The myriad ways microorganisms have evolved to circumvent this immune defence20 suggests that antibodies are a potent force of immune selection capable of influencing microbial ecology and evolution. Therefore, AMIS is predicted to have a significant effect on microbiota composition in the gut.
Adaptive immune deficiency alters microbiota composition.
The tremendous functional variability of antibody repertoires is generated by the processes of somatic recombination and receptor editing; the addition of non-templated nucleotides between V, D and J gene cassettes, somatic hypermutation (SHM) and class switch recombination (CSR). The absence of key enzymes involved in these processes causes complete loss of T cells and B cells (such as recombinase-activating gene (RAG) deficiency) and IgA deficiency (such as activation-induced cytidine deaminase (AID) deficiency), which has been shown to be associated with changes to the composition of microbial communities in the steady state.
Three recent studies by Dimitriu et al.21, Zhang et al.22 and Kawamoto et al.23 characterized the importance of adaptive immunity in shaping microbiota composition. All of these studies demonstrated that RAG-deficient mice had significantly altered microbiota composition compared with their respective wild-type controls. However, they identified subtly different effects on the microbiota. Kawamoto et al. reported that a lack of adaptive immunity led to a marked reduction in the diversity of microorganisms present in the gut. However, Zhang et al. did not report a difference in diversity between wild-type and RAG-deficient animals, but they did show outgrowth of a mucosa-associated organism, A. muciniphila. Importantly, when RAG-deficient animals were given a bone marrow transplant to restore adaptive immunity, the levels of A. muciniphila were markedly reduced. Dimitriu et al. identified increased variation of the microbial communities between RAG-deficient mice, suggesting that adaptive immunity may contribute to stabilizing inter-individual variation around a ‘core microbiota’. In particular, organisms from the families Helicobacteraceae and Alcaligenaceae were increased in the absence of B cells and T cells. The families Helicobacteraceae and Alcaligenaceae include members such as Helicobacter spp. and Burkholderia spp., respectively, which are known to have pathogenic potential. These studies suggest that adaptive immunity promotes diversity of the microbiota while limiting the levels of bacteria found at the mucosa.
Antibody deficiency alters microbiota composition.
Mouse models of B cell and immunoglobulin deficiency are the most direct way of testing the hypothesis that AMIS influences microbiota composition. Indeed, Ighm−/− mice, which lack B cells but have normal numbers of T cells, had an overall reduced microbial diversity compared with their Ighm+/− littermate controls, similar to what is observed in RAG-deficient animals23. Some of the first evidence that AMIS shapes microbiota composition came from studies of Aicda (the gene that encodes AID)-knockout mice, which cannot undergo class-switching to the IgA isotype and therefore do not have secretory IgA in the gut24,25. Fagarasan et al.25 showed that these animals suffered from a severe lymphoid hyperplasia that was associated with the expansion of anaerobic members of the microbiota. In a follow-up study, secretory IgA-deficient Rag2−/− animals had a similar expansion of anaerobic bacteria, and their abundance could be restored to wild-type levels when bone marrow from AID-sufficient (Aicda+/−) but not Aicda−/− mice was transferred to Rag2−/− hosts26. Finally, using a mouse model in which a knock-in mutation in the Aicda gene results in a polymorphism (AIDG23S) that allows CSR but severely reduces SHM, it was demonstrated that SHM as well as CSR were crucial for controlling microbiota composition27. Supporting these studies, IgA-deficient (Igha−/−) mice have a unique microbiota composition compared with wild-type controls, in which Proteobacteria outgrowth is a typical feature28. Finally, Reikvam et al.29 used 16S ribosomal RNA gene sequencing to profile steady-state microbiota composition across four sites in the gut (faeces, whole caecum, caecal luminal contents and caecal mucosa) between wild-type and PIgR-deficient mice (which are unable to secrete antibodies into the intestinal lumen). Significant differences in microbiota composition were observed across all four sites in PIgR-deficient mice compared with wild-type counterparts with notable decreases in putatively ‘beneficial’ bacteria, such as Bifidobacterium spp., and increases in known pathobionts, such as Helicobacter spp. Together, these studies indicate the importance of isotype switching and antibody secretion into the gut to prevent the outgrowth of potentially harmful organisms, such as Proteobacteria, and the maintenance of advantageous species.
T cell subsets differentially influence AMIS.
T cells have a central role in promoting antibody responses against commensals. Indeed, Cd3e−/− mice (which possess B cells but lack T cells) had significantly reduced microbiota diversity compared with wild-type controls23. B cells can mature into antibody-secreting cells with or without the help of T cells, and these two pathways of terminal B cell differentiation are referred to as T cell-dependent (TD) and T cell-independent (TI) responses (FIG. 1). In general, IgA responses generated in Peyer’s patches involve B2 cell–T cell interactions, which result in the generation of high-affinity and specific IgA responses30. B1 cells are thought to contribute to the majority of TI IgA responses that generate low-affinity polyreactive responses within the lamina propria. B1 cell-derived IgA is considered an ‘innate’ form of IgA because its generation is independent of T cell help, and it is the main contributor to the ‘natural’ antibody pool5,31. The generation of TD and TI IgA responses in the gut has been extensively reviewed elsewhere32–34. However, the relative importance of these two pathways of IgA synthesis on AMIS in the gut has only very recently begun to be appreciated.
Figure 1 |. Role of immune recognition pathways in antibody-mediated immunoselection of the microbiota.
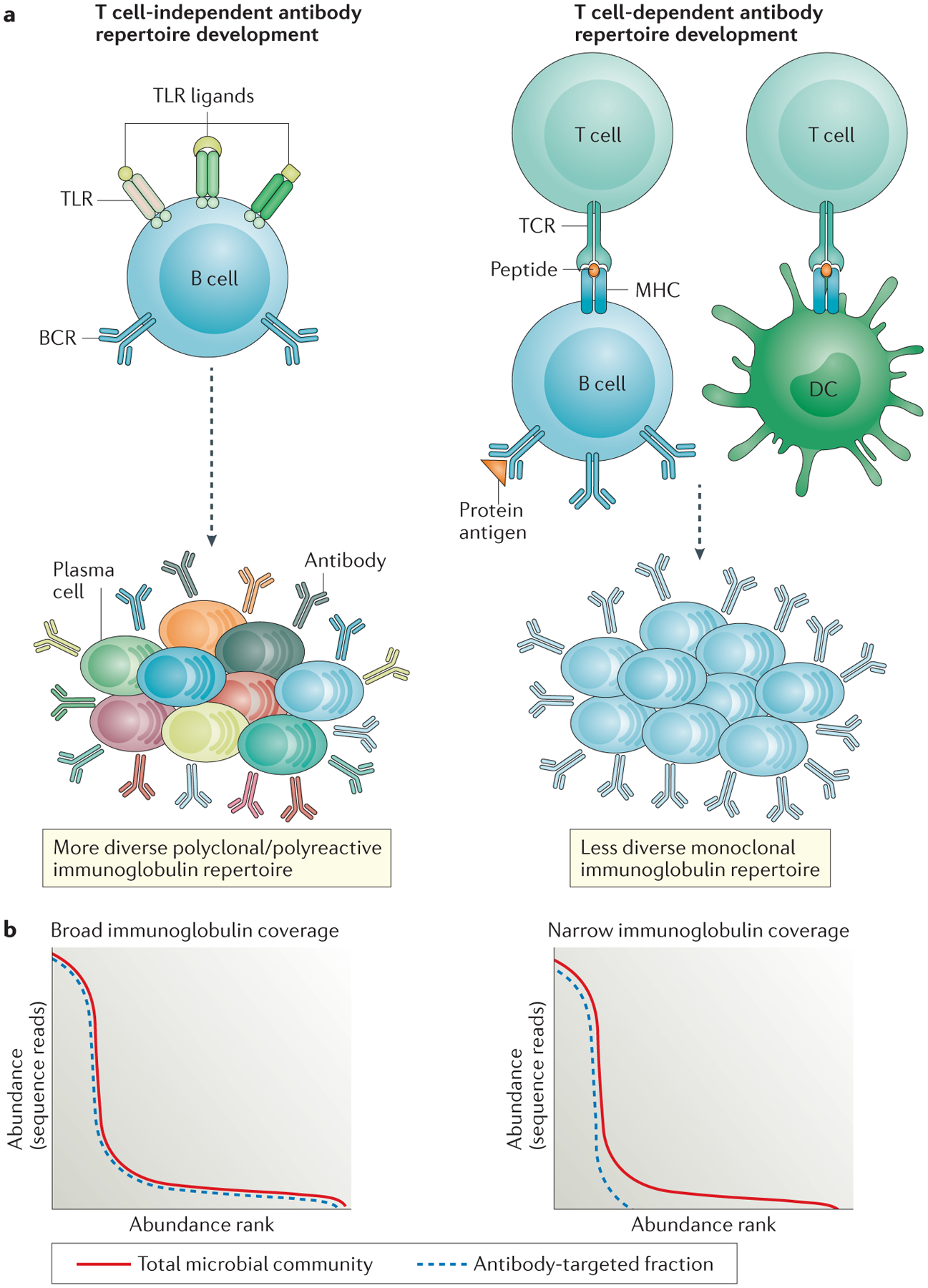
a | Microbial products can differently influence the development of the antibody repertoire in the gut. This schematic shows how Toll-like receptor (TLR)- and MHC class II-mediated signalling influence T cell-independent and T cell-dependent antibody responses, respectively. T cell-independent antibody responses are promoted by the mitogenic activity of TLR engagement on B cells, and this leads to the generation of relatively low-affinity antibodies with polyreactive specificities. Peptide–MHC class II–T cell receptor (TCR) interactions place more stringent selection on maturing B cells, which reduces the overall diversity of antibody repertoires but results in the generation of high-affinity antibodies with high epitope specificity. b | Two models using species rank abundance curves predict the effect on the microbiota of differential immune signalling in T cell-independent and T cell-dependent antibody responses. The red lines in both panels represent the typical distribution of species in microbiota communities from the gut. A small subset of species numerically dominate the community, but most of the community diversity resides in the long-tailed distribution of rare species. Low-affinity polyreactive antibody repertoires are predicted to bind a wider array of species — that is, have greater antibody coverage of the total microbial community (blue dashed line in left panel) — whereas high-affinity monoreactive antibody repertoires are predicted to bind a more limited array of species — that is, have a narrower range of coverage of the total microbial community (blue dashed line in right panel). By focusing the antibody response against the most abundant species, T cell-dependent antibody responses should favour diversity by constraining the growth of the fastest replicators, which maintains niches for rare and more fastidious microbial species. DC, dendritic cell.
Kawamoto et al.23 used adoptive-transfer experiments to show that the quality of the TD IgA response is important for promoting microbiota diversity. Although Cd3e−/− mice had a significantly reduced microbial diversity compared with wild-type mice, the transfer of total naive CD4+ T cells led to an even further decrease in bacterial diversity. However, the transfer of regulatory T (Treg) cells to these mice completely restored the diversity of the microbiota. In addition, the IgA molecules produced in the absence of Treg cells had a low-affinity maturation index, suggesting defective affinity selection within the follicle, but led to an increase in the amount of bacteria that was bound by IgA. We believe these data indicate that forkhead box P3 (FOXP3)+ Treg cells promote the production of more specific IgA molecules that target a narrower range of bacterial species in the gut, in a process similar to that described in FIG. 1. In a subsequent study, Bunker et al.12 compared the composition of IgA-bound bacteria between wild-type and T cell-deficient (Tcrb−/−Tcrd−/−) animals and found that TI and TD IgA responses in the gut targeted distinct subsets of commensal microorganisms. Although wild-type and T cell-deficient mice had similar levels of IgA-coated bacteria throughout the intestine, Mucispirillum spp. and segmented filamentous bacteria were highly enriched within the IgA-coated fraction. Both of these organisms are known to localize close to the mucosa, which might suggest that TD IgA responses are important for mediating IgA against invasive commensals.
T follicular helper (TFH) cells are one of the most important T cell subtypes involved in TD IgA induction35. Using programmed cell death 1 (Pdcd1)-knockout animals that have an abnormally high abundance of TFH cells, Fagarasan and colleagues explored how TFH cells influence the structure of the microbiota9. Pdcd1−/− mice had undetectable levels of Bifidobacteria spp. and Bacteriodes spp. and markedly higher levels of Enterobacteriaceae compared with wild-type controls. Thus, a deficiency in Pdcd1 leads to the loss of bacterial members that are associated with a homeostatic environment and the outgrowth of organisms that are associated with pathology. Although these mice had higher levels of free IgA in the gut, the IgA bound to significantly fewer luminal bacteria. Pdcd1−/− mice had increased TFH cells and a higher turnover of IgA-producing B cells. Collectively, these results suggest that altering the contribution of TD and TI IgA responses in the gut has important consequences for microbiota composition and host health.
Maternal antibodies define the initial stages of host–microbiota symbiosis.
Maternal antibodies that are passed in milk from a mother to her offspring may also influence microbiota composition. Maternal antibodies control bacterial growth in the neonatal gut and delay the development of antibody responses in the offspring36. Indeed, most of the IgA that binds to faecal bacteria during the first few weeks of life comprises passively transferred maternal antibodies. During lactation, plasma cells dramatically increase in number in mammary glands, and secretory IgA antibodies are delivered into the milk duct via PIgR-mediated transport. CC-chemokine receptor 10 (CCR10), which is important for plasma cell recruitment to the gut, is also involved in plasma cell recruitment to the mammary glands, suggesting a common route of migration of plasma cells between the gut and mammary tissues37. Indeed, a recent study showed that the IgA-producing plasma cell pool in mammary glands has tremendous overlap with the gut-associated IgA-producing plasma cell repertoires, which suggests that most of the antibodies that are passively transferred in mother’s milk are derived from maternal B cell responses generated against gut antigens38. Current research is focused on understanding how maternal antibody transfer influences microbiota community assembly. A recent study demonstrated that maternal antibodies may influence AMIS in the newborn gut39. Using a simple but elegant breeding strategy to manipulate maternal IgA transfer, Koch et al.40 crossed Pigr+/− females (that are PIgR sufficient) with Pigr−/− males, and Pigr−/− females with Pigr+/− males. Pigr+/− and Pigr−/− offspring passively received IgA antibodies from the milk of Pigr+/− mothers but not Pigr−/− mothers. The authors observed markedly different microbiota composition in weanling Pigr+/− animals that did and did not receive maternal antibodies. Moreover, animals that received maternal antibodies were less likely to suffer systemic invasion by an opportunistic pathogen and had lower colitis scores in the dextran sodium sulfate (DSS) model of microbiota-induced inflammatory bowel disease. Maternal transmission of IgG can also function to maintain homeostasis within the infant gut by dampening immune responses40.
These studies provide evidence supporting the hypothesis that maternal antibody transfer represents a vertically transmissible programme of AMIS in the gut. By facilitating the engraftment of microorganisms that are favoured in the maternal gut, this phenomenon could in part account for the high degree of similarity in the microbiota between mother and offspring. Vertical transmission of AMIS may favour the establishment of a beneficial microbiota that has been ‘pre-screened’ by maternal antibodies. Lack of this selective screen could increase the chances of a harmful microbiota developing. This might explain why formula-fed infants have been shown to have a higher incidence of juvenile inflammatory bowel disease and enteric infection than breast-fed infants41,42.
Immune recognition pathways in AMIS
Commensal bacteria have many ligands and antigens that are recognized by the immune system. Two immune recognition pathways that may have important roles in coordinating antibody responses in the gut and controlling microbiota composition are the Toll-like receptor (TLR) pathway and the antigen presentation pathway, which is mediated by MHC molecules.
Commensal ligands can induce AMIS through TLRs.
TLRs are generally thought to elicit inflammatory responses against pathogenic infection, but a seminal study using mice that lack the TLR signalling molecule MYD88 (myeloid differentiation primary response 88) identified that these animals developed more severe colitis43. This was a surprising result given that deletion of Myd88 should reduce inflammation and therefore theoretically ameliorate intestinal autoimmunity. The observation that Myd88−/− animals developed enhanced inflammation suggested a role for TLRs in the maintenance of tolerance. Since then several studies have demonstrated that part of this regulatory role involves the induction of antibodies.
B cells express TLRs, and therefore B cell migration and antibody production can be directly influenced by microbial ligands. For example, TLR ligands present in the serum are important for the recruitment of B1 cells from the peritoneum into the gut44. A role for TLR-mediated recognition of microbial ligands by B cells has recently been evaluated by deleting Myd88 from B cells (B-Myd88−/− mice)45. These mice are more susceptible to systemic bacterial invasion in response to DSS treatment compared with wild-type animals, and antibiotic treatment ameliorates disease. Sequencing of the bacterial 16S rRNA gene from the livers of B-Myd88−/− mice implied an overgrowth of members of the phylum Proteobacteria. These data suggest that loss of B cell-intrinsic TLR signalling altered the microbiota, resulting in a more pro-inflammatory composition that may have contributed to the observed patterns of disease. However, the adaptor protein MYD88 is also important for the TACI (transmembrane activator and calcium-modulating cyclophilin ligand interactor) signalling pathway in B cells, which promotes CSR and TI antibody responses46. This was not addressed in this study so it is not known how much of the observed effect by Kirkland et al.45 was due to TLR or TACI deficiency. Also, although it was not shown that the dysbiosis observed in B-Myd88−/− animals was due to a deficiency in B1 cell recruitment to the gut, a study by Ha et al.44 supports this conclusion.
Results from experiments in our laboratory demonstrated that animals with a T cell-specific deficiency of Myd88 (termed T-Myd88−/− mice) had a marked reduction in TFH cell numbers in the gut10. This led to a loss of germinal centre B cell responses and a failure to target commensal bacteria with IgA. The loss of IgA binding of bacteria in these models correlated with the overgrowth of mucosa-associated bacteria including Desulfovibrionaceae, Mucispirillum spp. and Ruminococcus spp. These observations further support a role for TD AMIS in controlling communities at the mucosa. Interestingly, the communities found in T-Myd88−/− animals had greater inter-individual variability, suggesting that AMIS functions to constrain and stabilize the resident bacterial communities. The percentage of germinal centre B cells within these animals positively correlated with increased phylogenetic diversity, further suggesting that AMIS also functions to diversify the microbiota.
Finally, the effects on the microbiota have also been well studied in mice that lack TLR5, which is activated by bacterial flagellin. Loss of TLR5 alters microbiota composition47 and leads to the production of more gut IgA than wild-type controls16. However, the IgA produced in Tlr5−/− animals fails to target commensal flagellin, which implies that defects in TLR5-mediated IgA responses against the microbiota might contribute to the observed community differences between Tlr5−/− and wild-type animals. Interestingly, meta-transcriptomic analysis of the microbial communities from Tlr5−/− and wild-type animals showed that flagella-related gene expression was completely absent in wild-type animals but highly expressed in the microbiota of Tlr5−/− mice. Moreover, IgA produced in TLR5-sufficient animals directly downregulated the expression of flagellin and immobilized bacteria in vitro. Thus, sensing of flagellar ligands by TLR5 promotes an IgA response that induces the downregulation of epitopes that prevent invasion and movement of commensal bacteria. This is consistent with earlier findings showing a role for IgA in regulating epitope expression by commensal microorganisms48.
Despite the aforementioned evidence, it is still unclear as to what extent TLR signalling influences microbiota composition in the gut. Ubeda et al.1 compared the microbiota composition of wild-type and TLR-deficient littermates from several mouse models of TLR deficiency (that is, Myd88−/−, Tlr2−/−, Tlr4−/−, Tlr5−/− or Tlr9−/− animals) and found no detectable effect of host genotype on microbiota composition, and any compositional differences appeared to be solely due to cage and litter effects. This is in stark contrast to many experiments demonstrating differences in some of these deficiency models. The authors suggest that discrepancies between their study and earlier work could be due to differences between mouse colonies reared at different institutions. Alternatively, it is possible that the homogenizing effect of coprophagy during cohousing from birth masked the effect of the host genotype in this study. Clearly, more work is needed to determine whether TLR-mediated immune responses control microbiota composition.
Variable antigen presentation influences IgA responses and microbiota composition.
MHC genes are some of the most polymorphic loci in vertebrates, and thus there is marked inter-individual variation with respect to the set of MHC molecules that are expressed. We recently explored whether natural variation in MHC genes among individuals can drive differences in microbial community structure11. In this study, we used three different lines of MHC-congenic animals that have the same genetic background but express different types of MHC molecules and had differing microbiota composition. Differences in the expressed MHC genes led to unique IgA repertoires in each MHC-congenic line, resulting in differential AMIS of the microbiota. Thus, the unique MHC repertoire of individuals might influence, in part, the unique community of commensal organisms found in the gut. This is one of the first studies to demonstrate that naturally occurring genetic variation relevant to IgA responses in the gut can influence microbiota composition and potentially contribute to the development of a microbiota that is unique to the individual.
How AMIS promotes a healthy gut
The antibody response benefits host physiology in many ways. Broadly, antibody responses in the gut promote health by limiting bacterial dissemination, regulating bacterial virulence factor expression and managing microbiota composition to maintain benign communities. It is not clear what constitutes a ‘benign’ microbial community, and this is an ongoing focus of research in many laboratories. This is complicated by the fact that some commensal microorganisms may have evolved ways to circumvent or exploit antibody responses to facilitate their own colonization or inhibit that of others (FIG. 2). However, numerous studies have now clearly demonstrated that defective or variable antibody responses result in alterations to microbiota composition that can influence an individual’s susceptibility to disease.
Figure 2 |. Antibody–microorganism interactions that influence antibody-mediated immune-selection of the microbiota.
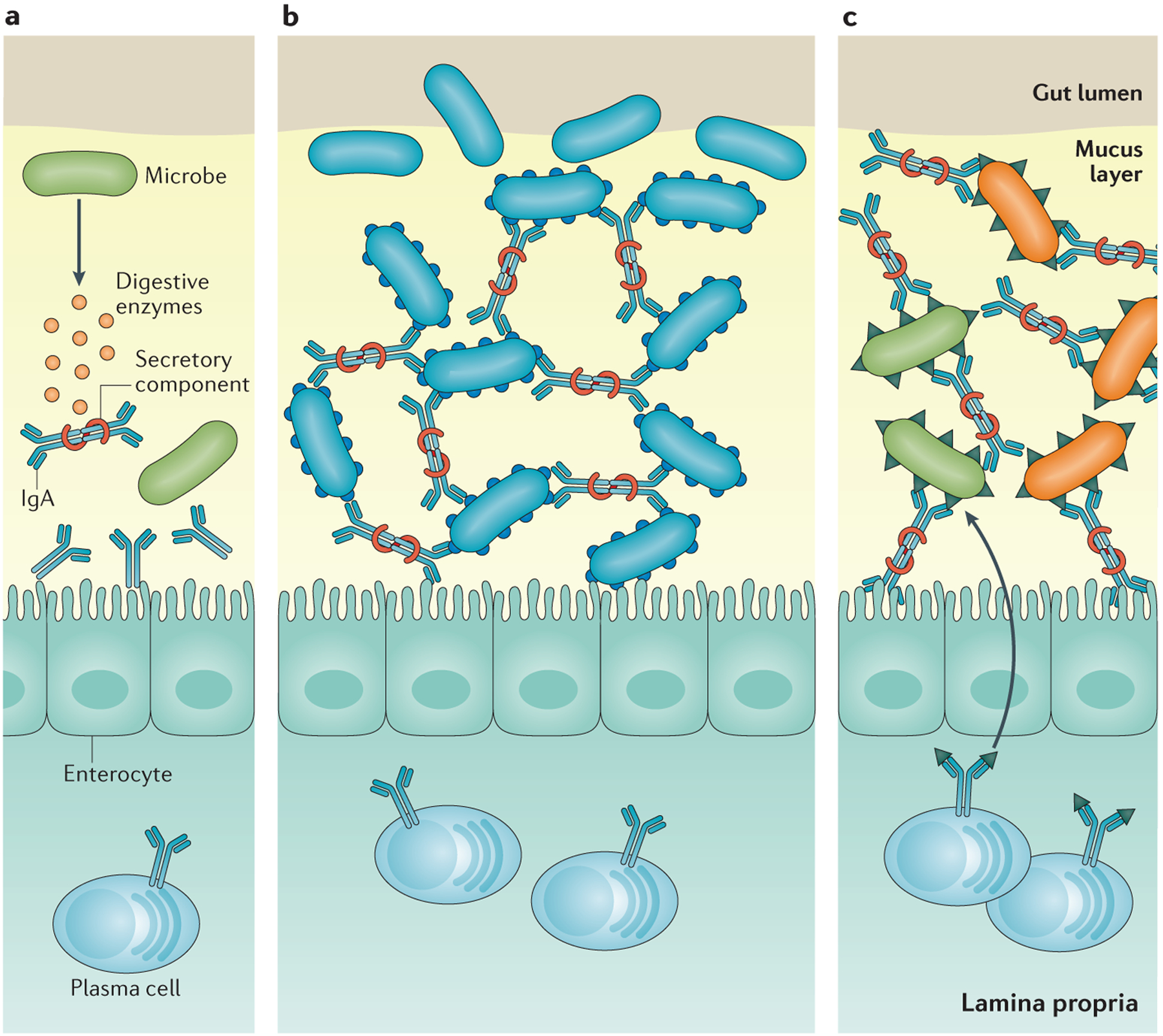
a | Some microorganisms can secrete enzymes that degrade the secretory component of secretory antibodies, which disrupts the stability of the antibody molecule, rendering it ineffective. b | Some microorganisms may express surface epitopes that bind secretory IgA and facilitate colonization of the mucus lining in the gut. c | Some microorganisms express surface epitopes that are very similar to those on other microorganisms. Antibodies generated against one microorganism can lead to the generation of antibody specificities that cross-react with similar epitopes found on other microorganisms.
Antibody responses limit the inflammatory potential of gut-resident microorganisms by prohibiting their dissemination into the systemic compartment. The first study of Pigr−/− mice reported that these animals have a leaky gut phenotype with markedly elevated serum levels of Escherichia coli-targeted IgG antibodies49. This implies aberrant activation of systemic immunity and outgrowth of potentially disease-promoting members of the Enterobacteriaceae. Supporting this idea, Reikvam et al.29 demonstrated that the altered microbiota community observed in Pigr−/− animals was associated with a more severe colitis following DSS treatment compared with wild-type controls and could be ameliorated by antibiotic treatment. Increased dissemination of bacteria into the systemic compartment has also been observed in mouse models of B cell deficiency27 and defective TD IgA responses11, and in Aicda−/− mice50. Collectively, these studies indicate that deficient antibody responses in the gut result in bacterial invasion of host tissues.
Another beneficial role of antibody responses in the gut is the regulation of microbial virulence factor expression. Antibodies can protect host tissues by directly neutralizing microbial toxins in the gut or by downregulating the expression of pro-inflammatory epitopes. For example, J chain-deficient mice that are unable to make secretory IgA cannot neutralize cholera toxin and are significantly more susceptible to diarrhoeal disease51. In another study, IgA responses in the gut reduced the expression of inflammation-promoting surface proteins in the commensal microorganism Bacteroides thetaiotamicron48. Antibodies can also directly influence the expression of virulence factors that promote attachment and invasion of the gut epithelium. IgA responses against Salmonella surface antigens have been shown to limit attachment and SPI-1-mediated invasion of gut epithelial cells52,53. More recently, it was shown that lower levels of flagellin-specific antibody in Tlr5−/− mice increase flagella expression by various commensal Proteobacteria species, leading to increased motility and penetration of flagellated bacteria into the systemic compartment16. IgG responses in the gut have also recently been shown to directly limit virulence factors expressed by Citrobacter rodentium that facilitate attachment of the bacterium to the gut epithelium54. These studies demonstrate that antibody responses in the gut limit infection by enteric pathogens by blocking the effects of virulence factors that promote pathogen colonization. They also demonstrate that antibodies reduce the inflammatory potential of normally occurring commensal species by regulating the expression of pro-inflammatory surface epitopes.
Differences in the quality of the IgA response and their effect on microbiota composition can have important implications for health. As mentioned above, we showed that deficiencies in TFH cell development and IgA responses (in T-Myd88−/− animals) resulted in altered IgA targeting of commensals and was associated with the establishment of a more pro-inflammatory microbiota10. In a second study, we showed that the naturally occurring polymorphisms in MHC genes resulted in differences in microbiota composition between different lines of MHC-congenic mice, and this influenced colonization resistance against an enteric pathogen11. Both of these studies used microbiota transplantations in germ-free mice to demonstrate that defects or naturally occurring variability in AMIS among individuals lead to the establishment of unique microbial communities that can influence host susceptibility to inflammatory and infectious disease in the gut.
An important question is whether antibody responses generated against the microbiota can protect hosts from more dangerous bacterial species through the generation of cross-reactive antibodies. Nunez and colleagues recently addressed this question and demonstrated that the generation of serum IgG antibodies against a widely conserved epitope (murein lipoprotein) from commensal Gram-negative bacteria in the gut conferred protection against systemic invasion by these commensal species as well as against systemic infection by the Gram-negative pathogen Salmonella enterica subsp. enterica serovar Typhimurium, which also expresses this epitope50. The mechanisms by which the microbiota can influence AMIS are summarized in FIG. 2.
Implications for AMIS in immunotherapy
Currently, there is great interest in the potential of harnessing the immune response to treat a variety of human diseases (such applications are termed immunotherapies). Given the potential importance of antibodies for selection of the microbiota together with the significance of maintaining an appropriate bacterial community structure, we propose that future endeavours could be directed at developing immunotherapies that specifically function to manipulate the microbiota (FIG. 3). The most obvious immunotherapy is direct modulation of the antibody response in the gut through oral vaccination. Oral vaccines could be designed to limit the growth of pathogens or to favour colonization by microbial species that are antagonistic to a pathogen. Alternatively, monoclonal antibodies could be generated that target pathogen virulence factors like flagella or type I pili and could be used therapeutically to treat immune-compromised patients that are unable to mount an effective antibody response to these antigens. The microbiota is known to generate unique metabolites and secrete molecules that positively influence intestinal homeostasis. It is possible that the body generates antibodies to these molecules that interfere with their function or availability. Therefore, blocking antibodies against these novel molecules might also prove to be a useful therapy. As we continue to better understand the function of different organisms within the gut, commensal-targeted antibodies might be used as biomarkers for susceptibility to or diagnosis of certain diseases, similarly to the use of anti-Saccharomyces cerevisiae antibodies (ASCAs) in the diagnosis of Crohn disease55. A detailed understanding of the nature of antigen selection on developing B cell repertoires is necessary to assess the feasibility of exploiting these potential therapies, and should remain a focus in the IgA field.
Figure 3 |. Potential immunotherapies to modulate antibody-mediated immunoselection in the gut.
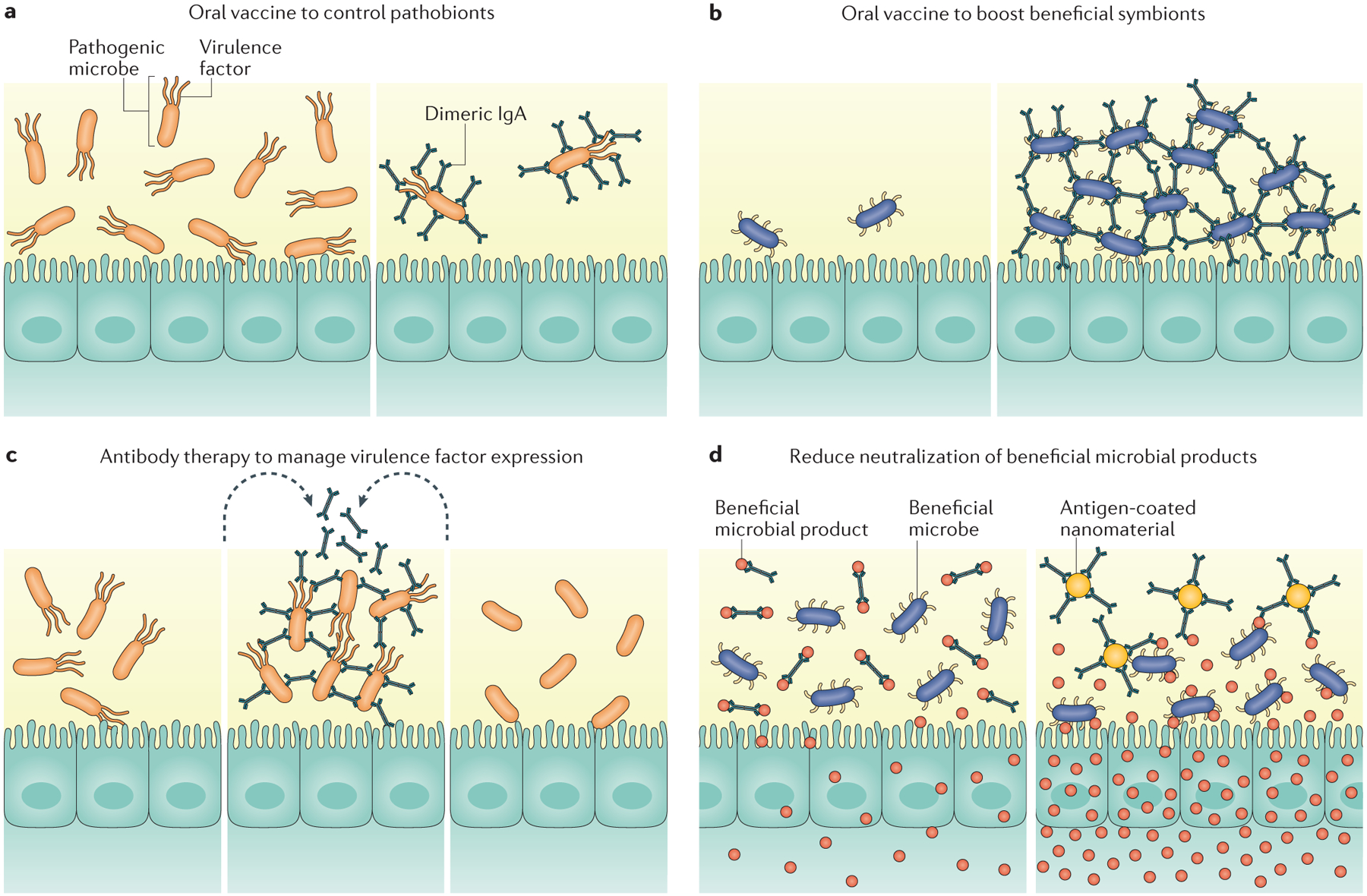
Manipulating antibody-mediated immune-selection (AMIS) to establish a healthy gut can be achieved by enhancing or limiting antibody responses to desired antigens. Oral vaccination could be used to specifically limit pathobiont expansion (part a) or to promote biofilm-formation by beneficial species (part b). Exogenous antigens could be delivered into the gut to manage virulence factor expression by pathobionts (part c). Sequestration of antibodies against beneficial microbial products could increase their availability to the host or to other beneficial microorganisms (part d).
Collectively, we believe these studies support an important role for AMIS of the microbiota to promote diversity while also constraining fluctuations that might result in loss of beneficial microorganisms. AMIS of the microbiota also functions to prune the expression of potentially harmful epitopes of commensals and prevent lethal dissemination by controlling mucosa-associated communities. Thus, we believe AMIS of the microbiota is a critical host mechanism, which mediates symbiosis that could be exploited therapeutically to improve health.
Acknowledgements
The authors would like to thank members of the Round laboratory for their critical review of the manuscript. Support for the laboratory comes from University of Utah’s seed grant program, NIH innovator award DP2AT008746-01, Edward Mallinckrodt Jr. Foundation, Pew Scholars Program, NSF CAREER award (IOS-1253278), Packard Fellowship in Science and Engineering and NIAID K22 (AI95375) and NIAID R21 AI109122 to J.L.R.
Footnotes
Competing interests statement
The authors declare no competing interests.
Contributor Information
Jason L. Kubinak, Department of Biology, University of Texas at Arlington, 501 S. Nedderman Drive, Arlington, Texas 76019, USA.
June L. Round, Department of Pathology (Microbiology and Immunology Division), 15 North Medical Drive East, Salt Lake City, Utah 84112, USA.
References
- 1.Lindner C et al. Age, microbiota, and T cells shape diverse individual IgA repertoires in the intestine. J. Exp. Med 209, 365–377 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ubeda C et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J. Exp. Med 209, 1445–1456 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshida M et al. Human neonatal Fc receptor mediates transport of IgG into luminal secretions for delivery of antigens to mucosal dendritic cells. Immunity 20, 769–783 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Smith K, McCoy KD & Macpherson AJ Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol 19, 59–69 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Macpherson AJ et al. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 288, 2222–2226 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Benveniste J, Lespinats G, Adam C & Salomon JC Immunoglobulins in intact, immunized, and contaminated axenic mice: study of serum IgA. J. Immunol 107, 1647–1655 (1971). [PubMed] [Google Scholar]
- 7.Shroff KE, Meslin K & Cebra JJ Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect. Immun 63, 3904–3913 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Waaij LA, Mesander G, Limburg PC & van der Waaij D Direct flow cytometry of anaerobic bacteria in human feces. Cytometry 16, 270–279 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Kawamoto S et al. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science 336, 485–489 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Kubinak JL et al. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe 17, 153–163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kubinak JL et al. MHC variation sculpts individualized microbial communities that control susceptibility to enteric infection. Nat. Commun 6, 8642 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bunker JJ et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity 43, 541–553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Planer JD et al. Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature 534, 263–266 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palm NW et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kau AL et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci. Transl Med 7, 276ra24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cullender TC et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe 14, 571–581 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rogier EW, Frantz AL, Bruno ME & Kaetzel CS Secretory IgA is concentrated in the outer layer of colonic mucus along with gut bacteria. Pathogens 3, 390–403 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ermund A, Gustafsson JK, Hansson GC & Keita AV Mucus properties and goblet cell quantification in mouse, rat and human ileal Peyer’s patches. PLoS ONE 8, e83688 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phalipon A et al. Secretory component: a new role in secretory IgA-mediated immune exclusion in vivo. Immunity 17, 107–115 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Finlay BB & McFadden G Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124, 767–782 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Dimitriu PA et al. Temporal stability of the mouse gut microbiota in relation to innate and adaptive immunity. Environ. Microbiol. Rep 5, 200–210 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Sparks JB, Karyala SV, Settlage R & Luo XM Host adaptive immunity alters gut microbiota. ISME J 9, 770–781 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawamoto S et al. Foxp3+ T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity 41, 152–165 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Fagarasan S, Kinoshita K, Muramatsu M, Ikuta K & Honjo T In situ class switching and differentiation to IgA-producing cells in the gut lamina propria. Nature 413, 639–643 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Fagarasan S et al. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science 298, 1424–1427 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Suzuki K et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl Acad. Sci. USA 101, 1981–1986 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei M et al. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat. Immunol 12, 264–270 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Mirpuri J et al. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes 5, 28–39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reikvam DH et al. Epithelial-microbial crosstalk in polymeric Ig receptor deficient mice. Eur. J. Immunol 42, 2959–2970 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Kroese FG et al. Many of the IgA producing plasma cells in murine gut are derived from self-replenishing precursors in the peritoneal cavity. Int. Immunol 1, 75–84 (1989). [DOI] [PubMed] [Google Scholar]
- 31.Suzuki K, Ha SA, Tsuji M & Fagarasan S Intestinal IgA synthesis: a primitive form of adaptive immunity that regulates microbial communities in the gut. Semin. Immunol 19, 127–135 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Fagarasan S, Kawamoto S, Kanagawa O & Suzuki K Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu. Rev. Immunol 28, 243–273 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Fagarasan S & Honjo T Intestinal IgA synthesis: regulation of front-line body defences. Nat. Rev. Immunol 3, 63–72 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Suzuki K, Kawamoto S, Maruya M & Fagarasan S GALT: organization and dynamics leading to IgA synthesis. Adv. Immunol 107, 153–185 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Crotty S Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol 29, 621–663 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Shulzhenko N et al. Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat. Med 17, 1585–1593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morteau O et al. An indispensable role for the chemokine receptor CCR10 in IgA antibody-secreting cell accumulation. J. Immunol 181, 6309–6315 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindner C et al. Diversification of memory B cells drives the continuous adaptation of secretory antibodies to gut microbiota. Nat. Immunol 16, 880–888 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Rogier EW et al. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc. Natl Acad. Sci. USA 111, 3074–3079 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koch MA et al. Maternal IgG and IgA antibodies dampen mucosal T helper cell responses in early life. Cell 165, 827–841 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mikhailov TA & Furner SE Breastfeeding and genetic factors in the etiology of inflammatory bowel disease in children. World J. Gastroenterol 15, 270–279 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanson LA & Winberg J Breast milk and defence against infection in the newborn. Arch. Dis. Child 47, 845–848 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S & Medzhitov R Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Ha SA et al. Regulation of B1 cell migration by signals through Toll-like receptors. J. Exp. Med 203, 2541–2550 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirkland D et al. B cell-intrinsic MyD88 signaling prevents the lethal dissemination of commensal bacteria during colonic damage. Immunity 36, 228–238 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He B et al. The transmembrane activator TACI triggers immunoglobulin class switching by activating B cells through the adaptor MyD88. Nat. Immunol 11, 836–845 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vijay-Kumar M et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peterson DA, McNulty NP, Guruge JL & Gordon JI IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2, 328–339 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Johansen FE et al. Absence of epithelial immunoglobulin A transport, with increased mucosal leakiness, in polymeric immunoglobulin receptor/secretory component-deficient mice. J. Exp. Med 190, 915–922 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeng MY et al. Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity 44, 647–658 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lycke N, Erlandsson L, Ekman L, Schon K & Leanderson T Lack of J chain inhibits the transport of gut IgA and abrogates the development of intestinal antitoxic protection. J. Immunol 163, 913–919 (1999). [PubMed] [Google Scholar]
- 52.Forbes SJ, Eschmann M & Mantis NJ Inhibition of Salmonella enterica serovar typhimurium motility and entry into epithelial cells by a protective antilipopolysaccharide monoclonal immunoglobulin A antibody. Infect. Immun 76, 4137–4144 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mantis NJ & Forbes SJ Secretory IgA: arresting microbial pathogens at epithelial borders. Immunol. Invest 39, 383–406 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kamada N et al. Humoral immunity in the gut selectively targets phenotypically virulent attaching- and-effacing bacteria for intraluminal elimination. Cell Host Microbe 17, 617–627 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruemmele FM et al. Diagnostic accuracy of serological assays in pediatric inflammatory bowel disease. Gastroenterology 115, 822–829 (1998). [DOI] [PubMed] [Google Scholar]


