Abstract
Motivation
The analysis of mutational signatures is becoming increasingly common in cancer genetics, with emerging implications in cancer evolution, classification, treatment decision and prognosis. Recently, several packages have been developed for mutational signature analysis, with each using different methodology and yielding significantly different results. Because of the non-trivial differences in tools’ refitting results, researchers may desire to survey and compare the available tools, in order to objectively evaluate the results for their specific research question, such as which mutational signatures are prevalent in different cancer types.
Results
Due to the need for effective comparison of refitting mutational signatures, we introduce a user-friendly software that can aggregate and visually present results from different refitting packages.
Availability and implementation
MetaMutationalSigs is implemented using R and python and is available for installation using Docker and available at: https://github.com/EESI/MetaMutationalSigs.
1 Introduction
Mutational signature analysis provides an operative framework to understand the somatic evolution of cancer from normal tissue (Alexandrov et al., 2020; Brunner et al., 2019; Moore et al., 2020; Robinson et al., 2020; Yoshida et al., 2020). From the earliest phases of neoplastic changes, cells may acquire several types of mutations in the form of single nucleotide variants, insertions and deletions, copy number changes and chromosomal aberrations. These mutations are caused by multiple mutational processes operative in cancer leaving behind specific footprints in the DNA that can by captured by mutational signature analysis (Alexandrov et al., 2013, 2020). It is becoming increasingly evident that these mutational signatures are not only important for understanding cancer evolution but also may have therapeutic implications, thus this a very active and important area of research (Alexandrov et al., 2020; Campbell et al., 2017; Chung et al., 2021; Iqbal et al., 2021).
The basic idea behind mutational signatures is that mutational processes create specific patterns of mutations. Thus, it follows that if one can identify these patterns in a given sample then they can essentially detect the corresponding mutational processes. The possible mutations are grouped into six single mutation types based on the base where the mutation was observed. These six single mutation types are C > A, C > G, C > T, T > A, T > C and T > G. Now, these six types of single mutations are further divided based on their context, e.g. one base preceding and one base following the single mutation type, resulting in 4^2 * 6 = 96 mutation types. Alexandrov et al. (2013) first developed and applied this idea to cancer data acquired from many datasets and identified the first iteration of 30 unique single base substitution (SBS) mutational signatures, which are common patterns of occurrences of the 96 mutation types and were compiled into the Catalogue Of Somatic Mutations In Cancer (COSMIC). COSMIC came to be used as the de facto reference for signature refitting, we refer to these V2 signatures as COSMIC Legacy SBS signatures (Campbell et al., 2020; Forbes et al., 2017). The initial study was then expanded to the analysis of data from the Pan-Cancer Analysis of Whole Genomes (PCAWG) project (Campbell et al., 2020), resulting in two additional variant classes, doublet base substitutions (DBS) signatures and insertion/deletion (ID) signatures. With these added to the SBS, the three mutational signatures are in the COSMIC V3 catalog, which are in (Alexandrov et al., 2020).
The mutational signature analysis workflow involves multiple steps that require different amounts of time and processing power. Briefly, preprocessing steps to take Binary Alignment Format (BAM) files, align them to a reference genome and use a variant caller step to output Variant Calling Format (VCF) files, are required from the user. These steps are usually very resource-intensive and thus do not allow for much experimentation on personal computers; the downstream steps of variant filtering and annotation are much faster. The final step, the mutational signature analysis, is the least resource-intensive and, therefore, is easier for users to compare multiple methods on their desktop. Therefore, to facilitate comprehensive mutational signature refitting analyses, we developed the package, MetaMutationalSigs, to analyze the mutational signatures in the VCF files. We developed a wrapper for four typically used refitting packages (Blokzijl et al., 2018; Gori and Baez-Ortega, 2018; Rosenthal et al., 2016; Wang et al., 2020), that have diverse underlying methodologies, including multiple linear regression, non-negative least squares (NNLS), Bayesian inference and simulated annealing (SA), respectively. Here, we have developed a standard format for inputs and outputs for easy interoperability and effective comparison, respectively. With our previous experience in visualization of genomic data (Lan et al., 2014), we have implemented standard visualizations for the results of all mutational signature packages to ensure easy analysis. MetaMutationalSigs software is easy to install and use through Docker.
2 Approach
The two major methods typically used for mutational signature analysis are signature refitting and de-novo signature extraction. Signature refitting methods try to reconstruct the observed mutational pattern in the sample (the frequencies of 96 types of mutations) using linear combinations of known signatures (COSMIC Legacy SBS and COSMIC V3 SBS, ID, DBS, etc.), these methods work quite well on small sample sizes (such as single samples) and are widely used with small datasets (Omichessan et al., 2019). Signature extraction methods infer signatures from a given dataset, and then compare the extracted signatures with known reference signatures. Each extracted signature is assigned to a known signature if their cosine similarity exceeds a set threshold, otherwise signatures with similarity less than the threshold are ignored (Alexandrov et al., 2013). There are a few important caveats to signature extraction as recently discussed in Omichessan et al. (2019): (i) a novel signature can be very similar to several reference signatures and the assignment is not always perfect and (ii) the threshold for assignment plays a crucial role but is not widely agreed upon and using a different threshold can change the assignment (Omichessan et al., 2019).
We chose signature refitting as our primary task because refitting techniques use COSMIC signatures that are well-established, are able to analyze signatures in smaller sets of samples than de-novo techniques and are computationally less intense than de-novo techniques. We implemented high performing packages as identified in Omichessan et al. (2019) that were implemented in R using a common input matrix generated using SigProfilerMatrixGenerator (Bergstrom et al., 2019). While other techniques exist, including convenient web-based tools, such as Mutalisk (Lee et al., 2018), that refits using a maximum likelihood estimation of the signature contributions, and Signal (Degasperi et al., 2020), which uses quadratic programming or SA, there may be a desire to run the mutational signature analysis on local machines, that do not require uploading data to a third party. To address this problem, we implement a software package to compare four mutational signature analyzers—DeconstructSigs (Rosenthal et al., 2016), MutationalPatterns (Blokzijl et al., 2018), Sigfit (Gori and Baez-Ortega, 2018), Sigminer (Wang et al., 2020), which build up on other tools such as Mayakonda et al. (2018) and Huang et al. (2018). DeconstructSigs is the most cited method and uses a multiple linear regression model, with coefficients constrained to positive values, to find contributions of mutational signatures to an overall signature. MutationalPatterns is also popular and uses NNLS optimization to estimate the mutational signature weighting. Sigfit uses Bayesian inference to perform fitting. Sigminer uses an SA method to mutational signature fitting. Between multiple linear regression, optimization via NNLS and SA, and Bayesian estimation, users can survey how a variety of techniques can estimate COSMIC signatures in sample(s).
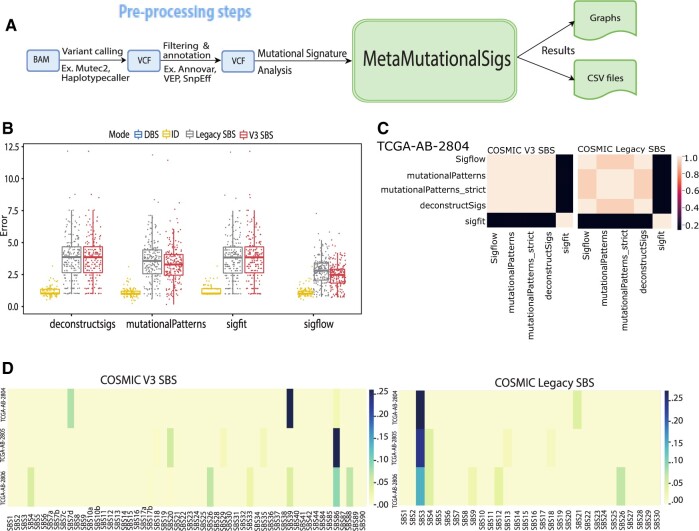
Our package outputs several data files in comma separated values (CSV) format ready for further analysis and visualization using external packages along with visualizations of the signature contributions as described in Table 1. In Figure 1A, we illustrate the workflow of the analysis (including preprocessing steps in blue and our package’s steps in green). In Figure 1B, we compare packages using the root mean squared error (RMSE) between the reconstructed and actual signals for 188 myeloid leukemia (LAML) patients obtained from The Cancer Genome Atlas portal (Weinstein et al., 2013). RMSE is a performance metric commonly used in signal processing (Rosen, 2007). In Figure 1C, we plot heatmaps of distances between methods for predicting signature contributions for SBS V2 (Legacy) and V3. In Figure 1D, we plot the heatmap of SBS contributions to overall signature reconstruction for three of the LAML patient samples.
Table 1.
Summary of result files
| File name | Format | Description |
|---|---|---|
| Heatmap_contributions_all_sigs_[signature_version].svg | svg | Contributions from all signatures of the [signature_version] to the overall signature |
| Heatmap_[signature_version].svg | svg | Heatmap of cosine similarity between the predicted contributions by different tools for [signature_version] |
| [signature_version]_bar_charts.html | html | Bar charts of signature contributions per sample and per tool for [signature_version] |
| rmse_box_plot.svg | svg | Box plot of RMSE between the reconstructed signal (from the reference signatures) and the overall signature |
| [tool_name]\[signature_version]_sample_error.csv | csv | Data about the difference between reconstructed and signal for each signature of [signature_version] for each [tool_name] for each sample. This is used to create rmse_box_plot.svg |
| [tool_name]\[signature_version]_contribution.csv | csv | Data about the contribution of each signature of [signature_version] for each [tool_name] for each sample. This is used to create the Heatmap_contributions_all_sigs_[singature_version].svg, Heatmap_[signature_version].svg and [signature_version]_bar_charts.html |
Note: Signature version corresponds to COSMIC Legacy or V3. Tool_name corresponds MutationalPatterns, Sigfit, Sigflow and DeconstructSigs.
Fig. 1.
Workflow and results for MetaMutationalSigs. (A) The workflow for mutational signature analysis starts with preprocessing steps (shown in blue boxes) required by the user to conduct variant calling, filtering and annotation on a BAM file of a sequenced genome or exome. Our tool, MetaMutationalSigs, conducts the steps shown in green boxes and analyzes the signatures found in a VCF. (B) RMSE between the reconstructed signal (from the reference signatures) and the overall signature, plotted as dots for each of the 188 patient samples in the Acute Myeloid Leukemia (TCGA, GDAC Firehose Legacy, Study ID: laml_tcga) dataset, for each tool (lower values are better) and for signature type (V2 and V3 SBS and IDs; no tool predicted DBS for the samples used). While RMSE does not change for DeconstructSigs and Sigfit, the RMSE significantly drops for MutationalPatterns and Sigflow with COSMIC v3. (C) Heatmap of cosine similarity between the predicted contributions of COSMIC v3 SBS versus COSMIC Legacy SBS signatures by different tools for the same TCGA patient sample. With the legacy signatures, tools are generally less in agreement in their resulting signature contributions, while with COSMIC v3 signatures, the standard use tools are all in agreement with each other. Sigflow had the lowest RMSE and was selected for analysis in (D). (D) Heatmaps of Sigflow analysis of COSMIC v3 SBS versus COSMIC Legacy SBS mutational signature contributions using whole-exome sequence data from three TCGA patients with acute myeloid leukemia. Here, each row is a patient sample. Left. COSMIC v3 SBS refitting provides different dominant signature contributions, TCGA-AB-2804: unknown etiology, TCGA-AB-2805 and TCGA-AB-2806: unknown chemotherapy and different DNA mismatch repair signatures, SBS20 and 26, respectively. COSMIC Legacy SBS refitting provides signature 3 (failure of double-strand break-repair by homologous recombination) as the dominant signature for all samples. The COSMIC v3 SBS refitting reveals multiple mutational processes may be playing a role in the overall signature contribution than is found with the COSMIC Legacy SBS refitting
3 Discussion
The massive increase in the number of software packages has made managing dependencies quite burdensome, coupled with incompatible data formats for signature matrices can make mutational signature refitting results difficult and hard to compare. Our package, MetaMutationalSigs, provides a simplified approach for performing the setup related tasks so that more focus can be placed on the analysis. Investigators should keep in mind that refitting approaches need a priori knowledge about the samples and each package for effective interpretation (Maura et al., 2019), and the results should not be used as-is without an assessment of the cell biology and genomics.
Future work for this project would focus on expanding the tool to work with more packages and keep the reference signatures updated as new versions are released. Due to the open-source nature of the project, we also welcome additional feature requests using the project link on GitHub https://github.com/EESI/MetaMutationalSigs.
Funding
P.P. and G.R. were supported by the NSF [awards #1936791 and #1919691], and P.P. was also supported by the Fox Chase Cancer Center Risk Assessment Program Funds. S.A. was supported by the DOD W81XWH-18-1-0148 award and a CEP award from the Yale Head and Neck Cancer NIH SPORE.
Conflict of Interest: S.A. performs collaborative research (with no funding) with Caris Life Sciences, Foundation Medicine, Inc., Ambry Genetics and Invitae Corporation. S.A. has several patents and/or pending patents related to cancer diagnostics/treatment. All other authors declare no competing interests.
Contributor Information
Palash Pandey, Ecological and Evolutionary Signal-Processing and Informatics Laboratory, Department of Electrical and Computer Engineering, College of Engineering, Drexel University, Philadelphia, PA 19104, USA; Cancer Prevention and Control Program, Fox Chase Cancer Center, Philadelphia, PA 19111, USA.
Sanjeevani Arora, Cancer Prevention and Control Program, Fox Chase Cancer Center, Philadelphia, PA 19111, USA; Department of Radiation Oncology, Fox Chase Cancer Center, Philadelphia, PA 19111, USA.
Gail L Rosen, Ecological and Evolutionary Signal-Processing and Informatics Laboratory, Department of Electrical and Computer Engineering, College of Engineering, Drexel University, Philadelphia, PA 19104, USA.
References
- Alexandrov L.B. et al. (2013) Deciphering signatures of mutational processes operative in human cancer. Cell Rep., 3, 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov L.B. et al. ; PCAWG Consortium. (2020) The repertoire of mutational signatures in human cancer. Nature, 578, 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom E.N. et al. (2019) SigProfilerMatrixGenerator: a tool for visualizing and exploring patterns of small mutational events. BMC Genomics, 20, 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokzijl F. et al. (2018) MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med., 10, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner S.F. et al. (2019) Somatic mutations and clonal dynamics in healthy and cirrhotic human liver. Nature, 574, 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell B.B. et al. (2017) Comprehensive analysis of hypermutation in human cancer. Cell, 171, 1042–1056.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P.J. et al. (2020) Pan-Cancer analysis of whole genomes. Nature, 578, 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J. et al. (2021) DNA polymerase and mismatch repair exert distinct microsatellite instability signatures in normal and malignant human cells. Cancer Disc., 11, 1176–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degasperi A. et al. (2020) A practical framework and online tool for mutational signature analyses show intertissue variation and driver dependencies. Nat. Cancer, 1, 249–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes S.A. et al. (2017) COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res., 45, D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gori K., Baez-Ortega A. (2018) Sigfit: Flexible Bayesian Inference of Mutational Signatures. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. doi: 10.1101/372896 [DOI] [Google Scholar]
- Huang X. et al. (2018) Detecting presence of mutational signatures in cancer with confidence. Bioinformatics, 34, 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal W. et al. (2021) RRM2B is frequently amplified across multiple tumor types: implications for DNA repair, cellular survival, and cancer therapy. Front. Genet., 12, 628758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y. et al. (2014) POGO-DB—a database of pairwise-comparisons of genomes and conserved orthologous genes. Nucleic Acids Res., 42, D625–D632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. et al. (2018) Mutalisk: a web-based somatic MUTation AnaLyIS toolKit for genomic, transcriptional and epigenomic signatures. Nucleic Acids Res., 46, W102–W108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maura F. et al. (2019) A practical guide for mutational signature analysis in hematological malignancies. Nat. Commun., 10, 2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayakonda A. et al. (2018) Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res., 28, 1747–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L. et al. (2020) The mutational landscape of normal human endometrial epithelium. Nature, 580, 640–646. [DOI] [PubMed] [Google Scholar]
- Omichessan H. et al. (2019) Computational tools to detect signatures of mutational processes in DNA from tumours: a review and empirical comparison of performance. PLoS One, 14, e0221235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson P.S. et al. (2020) Elevated Somatic Mutation Burdens in Normal Human Cells due to Defective DNA Polymerases. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. https://doi.org/10.1101/2020.06.23.167668 [Google Scholar]
- Rosen G. (2007) Comparison of autoregressive measures for DNA sequence similarity. In: IEEE International Workshop on Genomic Signal Processing and Statistics, pp. 1–4, https://doi.org/10.1109/GENSIPS.2007.4365814.
- Rosenthal R. et al. (2016) DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol., 17, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. et al. (2020) Copy Number Signature Analyses in Prostate Cancer Reveal Distinct Etiologies and Clinical Outcomes. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. https://doi.org/10.1101/2020.04.27.20082404 [Google Scholar]
- Weinstein J.N. et al. ; Cancer Genome Atlas Research Network. (2013) The Cancer Genome Atlas Pan-Cancer Analysis project. Nat. Genet., 45, 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K. et al. (2020) Tobacco smoking and somatic mutations in human bronchial epithelium. Nature, 578, 266–272. [DOI] [PMC free article] [PubMed] [Google Scholar]