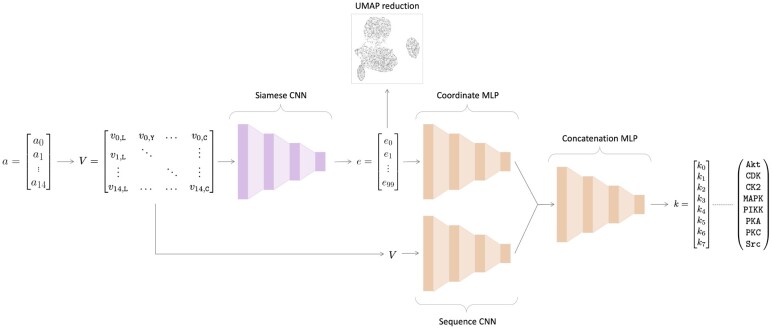
Fig. 2.
EMBER model architecture. Here, the previously trained Siamese neural network is colored pink, and the classifier architecture is colored orange. The 15 amino acid-length motif, a, is converted into a one-hot encoded matrix, V. The one-hot encoded matrix is then fed into a single twin from the Siamese neural network. The 100-dimensional embedding, e, is output by the Siamese neural network. Here, we reduce e to a 2D space for illustrative purposes using UMAP. Then, e is fed into a MLP alongside V, which is fed into a CNN. Then, the last layers of the separate neural networks are concatenated, followed by a series of fully connected layers. The final output is a vector, k, of length eight, where each value corresponds to the probability that the motif a was phosphorylated by one of the kinase families indicated in k