Abstract
Aetokthonotoxin (AETX) is a cyanobacterial neurotoxin that causes vacuolar myelinopathy, a neurological disease that is particularly deadly to bald eagles in the United States. The recently characterized AETX is structurally unique among cyanotoxins and is composed of a pentabrominated biindole nitrile. Herein we report the discovery of an efficient, five-enzyme biosynthetic pathway that the freshwater cyanobacterium Aetokthonos hydrillicola uses to convert two molecules of tryptophan to AETX. We demonstrate that the biosynthetic pathway follows a convergent route in which two functionalized indole monomers are assembled and then reunited by biaryl coupling catalyzed by the cytochrome P450 AetB. Our results revealed enzymes with novel biochemical functions, including the single-component flavin-dependent tryptophan halogenase AetF and the iron-dependent nitrile synthase AetD.
Aetokthonotoxin (AETX, 1) is a potent neurotoxin produced by the epiphytic cyanobacterium Aetokthonos hydrillicola.1 As the causative agent of vacuolar myelinopathy, AETX (1) is responsible for the mortality of bald eagles in the United States that prey on intoxicated waterbirds and other animals that consume the invasive aquatic plant Hydrilla verticillata carrying toxic A. hydrillicola. Structurally, AETX (1) is an unusually configured biindole alkaloid decorated with five bromine atoms and a nitrile functional group. Although brominated biindoles are widely reported in the literature from marine organisms,2,3 AETX is the first with an N1–C2′ connection. Its unsymmetrical bromination pattern featuring two bromine atoms on the pyrrole ring of indole is an unusual motif for brominated biindoles. Additionally, nitrile-containing molecules are very rare in nature,4 and as such, our knowledge regarding biosynthetic strategies for generating nitrile groups is limited. Intrigued by these unique chemical functionalities in a newly described environmental toxin, we sought to decipher the total biosynthesis of AETX (1) and explore the possibility of rare biosynthetic transformations and promising biocatalysts. Toward this goal, we were particularly interested in studying three aspects of AETX (1) biosynthesis: biaryl coupling, nitrile generation, and polybromination.
Recently, Breinlinger et al. sequenced the genome of A. hydrillicola and identified the putative AETX biosynthetic gene cluster (BGC) (Figure 1a).1 Of the six biosynthetic genes identified, one gene encoding tryptophan halogenase AetF was confirmed to be responsible for the regioselective dibromination of l-tryptophan (4) at the five- and seven-positions of the indole ring. This regioselective bromination catalyzed by AetF is consistent with the bromination pattern in AETX (1) and thus served as initial support that the identified BGC was likely associated with its biosynthesis. Of the remaining genes, denoted aetA–aetE, only three of the translated protein sequences shared strong homology with known enzyme classes: AetA with flavin-dependent tryptophan halogenases, AetB with cytochrome P450s, and AetE with pyridoxal phosphate (PLP)-dependent tryptophanases (Figure 1a). The functions of the remaining two enzymes, AetC and AetD, were unclear based on sequence homology, as the closest AetC homologues were annotated as cyclases and AetD lacked any conserved domains by BLAST analysis. These annotations guided our retrobiosynthetic proposal. We envisaged that the biosynthesis of AETX (1) follows a convergent route reminiscent of efficient synthetic approaches5 in which indole building blocks 2 and 3 would be separately biosynthesized before coupling to form the final biindole product 1 (Figure 1b).
Figure 1.
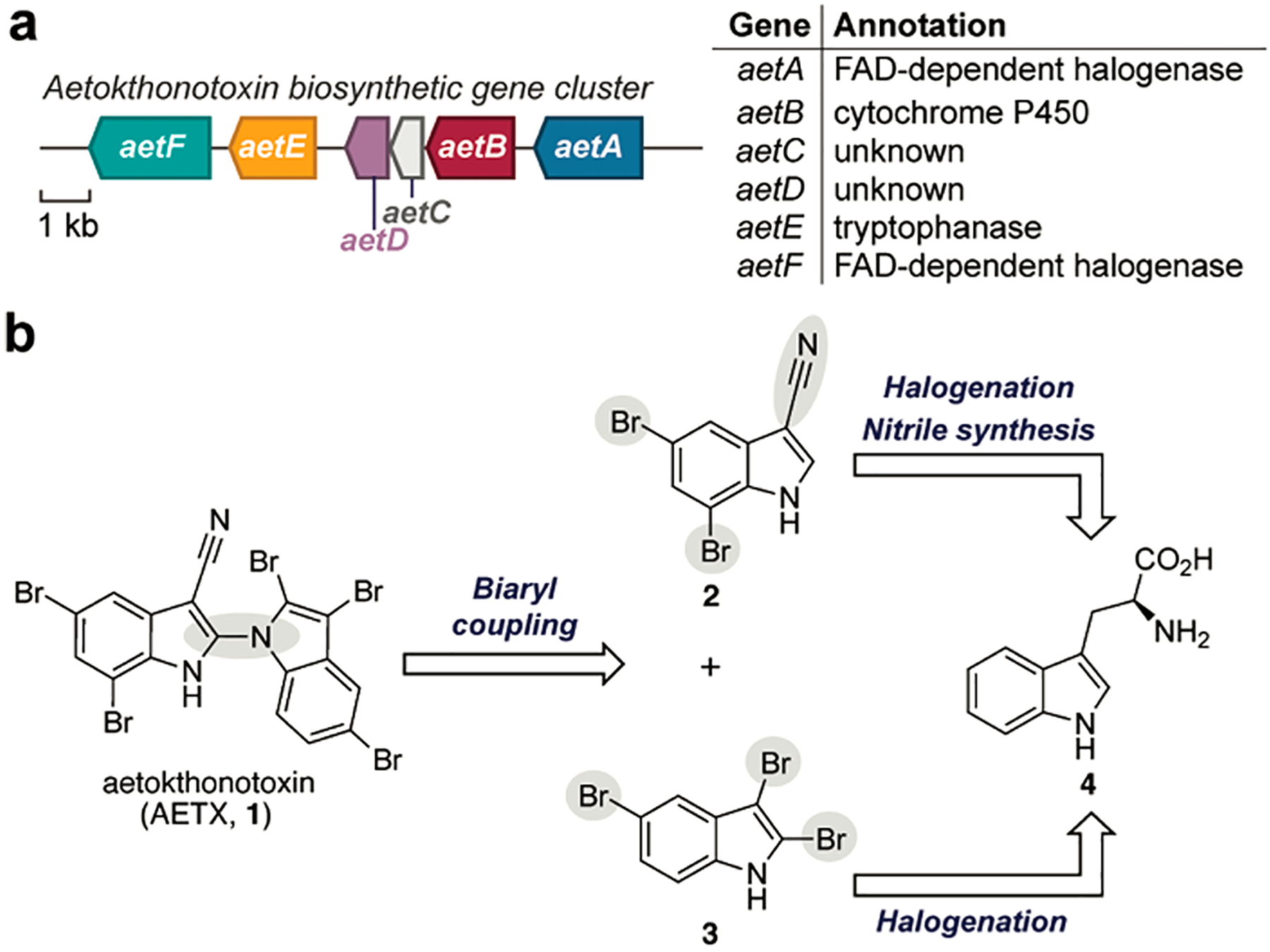
(a) Biosynthetic gene cluster and predicted annotations from A. hydrillicola proposed to be responsible for the biosynthesis of AETX. (b) Retrobiosynthetic proposal for AETX (1) biosynthesis from two tryptophan-derived indoles.
To interrogate our retrobiosynthetic proposal, we opted to take an in vitro approach by overexpressing each of the enzymes in Escherichia coli and functionally characterizing the proteins. AetA, AetC, AetD, and AetF were readily overexpressed in their soluble form and could be purified by Ni-affinity chromatography in sufficient quantities (Figures S1–S3). However, AetB and AetE were insoluble under standard conditions. The solubility of AetB was improved through constructing a maltose-binding protein (MBP)–AetB fusion protein, whereas AetE solubility was enhanced by coexpression in the presence of GroEL and GroES chaperone proteins6 (Figure S3). With all of the purified proteins in hand, we started our journey to deconvolute the biosynthesis of AETX (1).
We began with the previously studied halogenase AetF by incubating the purified protein with l-tryptophan (4) and NADPH to form 5-bromo-l-tryptophan (5) and 5,7-dibromo-l-tryptophan (6), as described1 (Figure 2a). This result is surprising, as all previously reported flavin-dependent tryptophan halogenases, with the exception of engineered halogenase–reductase fusion proteins,7 require a separate flavin reductase as a partner protein to demonstrate activity,8,9 rendering AetF the first example of a naturally occurring single-component, flavin-dependent tryptophan halogenase. The identity of the flavin cofactor in this protein was confirmed to be flavin adenine dinucleotide (FAD) by liquid chromatography–mass spectrometry (LC-MS), a result that is consistent with all other tryptophan halogenases10 (Figure S6). Furthermore, AetF showed a distinct preference for NADPH over NADH as the nicotinamide cofactor in the time-dependent reduction of bound FAD when incubated with l-tryptophan (4) (Figure S7). Intrigued by this unique enzyme and to gain insight into the relationship of AetF relative to other known flavin-dependent monooxygenases (FMOs), we generated a sequence similarity network (SSN)11 consisting of representative FMOs from each functional group, as defined by Huijbers et al. (Figure 2b).12 The FMO groups A–H are differentiated by flavin preference (FAD or flavin adenine mononucleotide (FMN)), reductase requirement (“single-component” enzymes versus “two-component” enzymes that require a reductase partner), and the type of chemical reaction performed. Interestingly, this analysis revealed that AetF does not cluster with any characterized FMO, including other tryptophan halogenases or single-component brominases.13 To further explore AetF in the context of the broader flavin-dependent halogenase family, we generated a second SSN with AetF included in a curated list of 3347 flavin-dependent halogenase sequences compiled in Fisher et al. (Figure S8).14 This expanded comparison also resulted in no AetF clustering, suggesting that the enzyme is unique among known halogenases.
Figure 2.
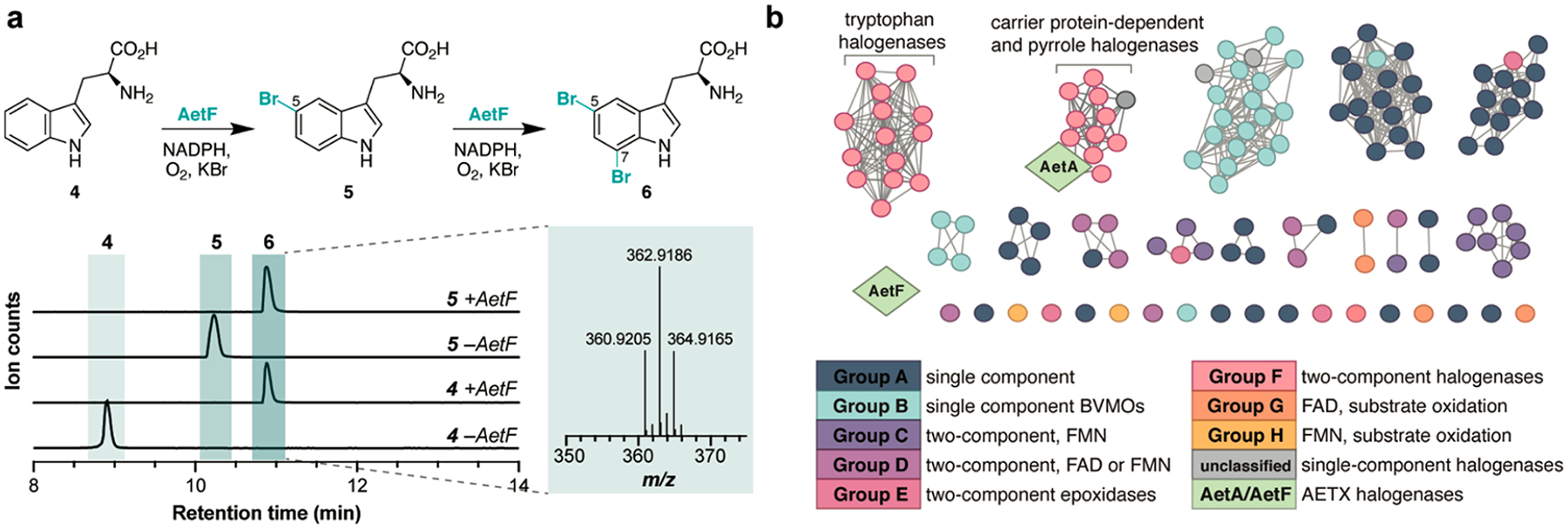
(a) AetF reaction and analysis by LC-MS. Chromatograms shown are extracted ion chromatograms (EICs) for 4 (m/z 205.0972), 5 (m/z 283.0077), and 6 (m/z 362.9162) in positive ionization mode. (b) Sequence similarity network of representative, functionally characterized FMOs and the flavin-dependent halogenases from the AETX (1) biosynthetic gene cluster. See Figure S8 for a comprehensive network analysis with 3347 flavin-dependent halogenase sequences.
Next, we turned our attention to putative cytochrome P450 AetB. Cytochrome P450 enzymes are known to catalyze biaryl coupling reactions,15 and we anticipated that AetB might fulfill such a role in catalyzing the final reaction sequence in AETX (1) biosynthesis. To test this hypothesis, we chemically synthesized the two brominated indole building blocks 5,7-dibromo-indole-3-carbonitrile (2) and 2,3,5-tribromoindole (3) following established literature procedures.16 AetB was then incubated with substrates 2 and 3 along with spinach ferredoxin (Fdx) and spinach ferredoxin reductase (FdR) and NADPH to complement electron transfer. LC-MS analysis of the reaction mixture revealed the formation of a new pentabrominated product only in the full reaction but absent in all negative controls. The identity of the enzymatic product was confirmed as AETX (1) based on coelution with a chemically synthesized standard16 (Figure 3). In our hands, 2 and 3 were the only two monomers coupled by AetB when given a mixture of other indole monomers (e.g., 5-bromoindole (7), 5,7-dibromoindole (S2)), thus corroborating our proposal that indole functionalization takes place prior to the coupling step (Figure S9).
Figure 3.
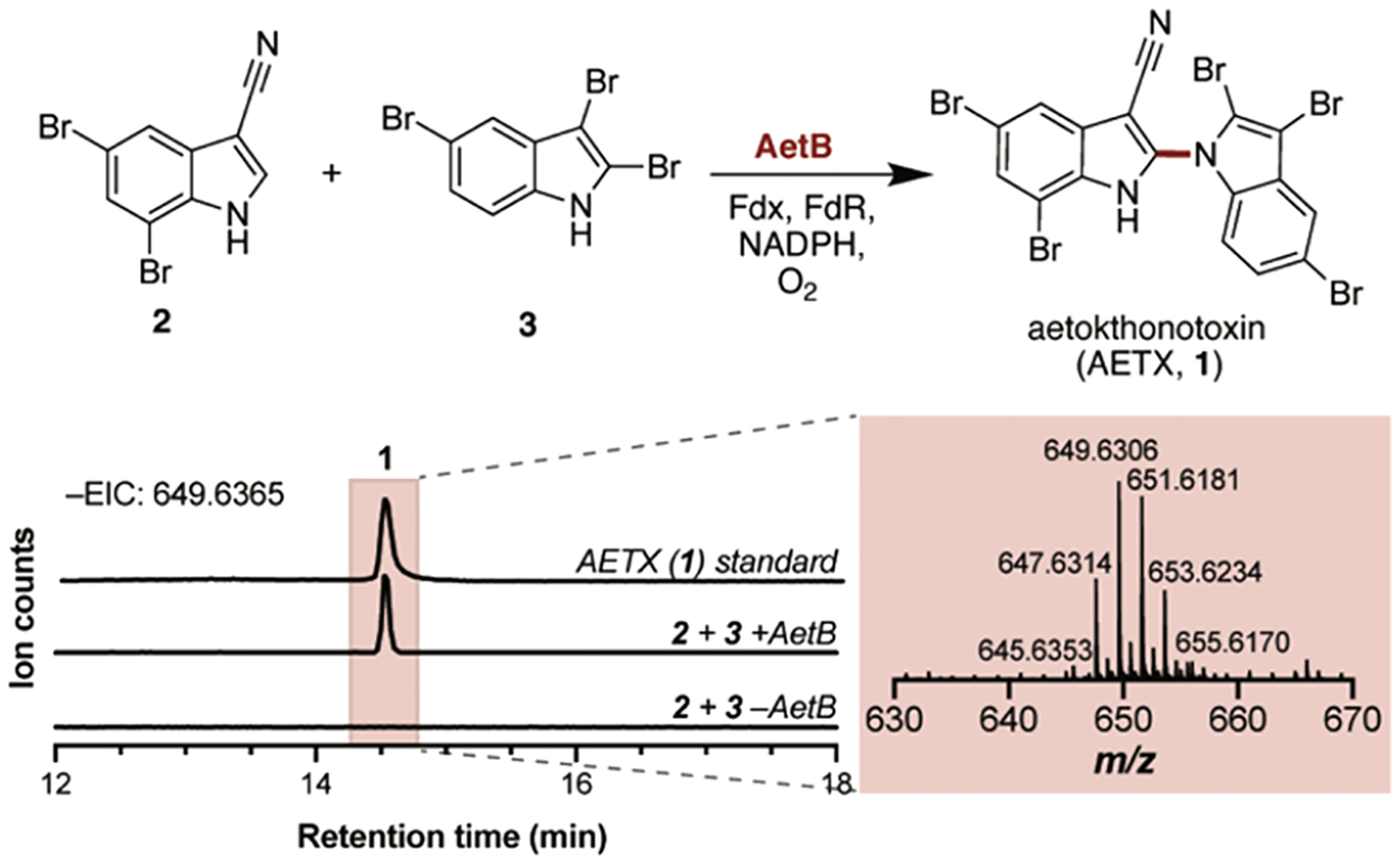
AetB reaction with 2 and 3 yields the final natural product AETX (1). Chromatograms are the EICs of AETX (m/z 649.6365) analyzed in negative ionization mode.
The presence of the gene encoding putative tryptophanase AetE suggests that the transformation of a tryptophan species to an indole may be required in the biosynthesis of AETX (1). On the basis of our retrobiosynthetic scheme, we assessed the reactivity of AetE with 5-bromo-l-tryptophan (5) and confirmed that AetE indeed catalyzes the PLP-dependent conversion of 5 to 5-bromoindole (7) (Figure 4a). The coproduct pyruvate was detected using the derivatizing agent o-(2,3,4,5,6-pentafluorobenzyl)-hydroxylamine (PFBHA) (Figure S10). Next, we interrogated the putative role of AetA in catalyzing the brominations on the pyrrole ring of 5-bromoindole (7). AetA was colorless as purified, suggesting that the required FAD cofactor for catalysis was not bound after purification. This property is common among flavin-dependent halogenases, which are known to bind reduced FAD that is generated by a separate NAD(P)H-dependent reductase.10 The classification of AetA as a canonical flavin-dependent halogenase is corroborated by its placement in our SSN (Figure 2b). AetA consistently clustered with pyrrole halogenases rather than tryptophan or indole halogenases (Figure 2b, Figure S8), and we hypothesized that AetA may perform the role of halogenating the pyrrole ring of indole rather than the benzene ring, as required in indole building block 3. Upon the incubation of AetA in the presence of 7, FAD, NADPH, and flavin reductase SsuE,17 two products were generated, which we characterized as 3,5-dibromoindole (8) and 2,3,5-tribromoindole (3) through comparisons with synthetic standards (Figure 4b). This result suggests that bromination at the three-position of 5-bromoindole (7) occurs first followed by the two-position, which is further confirmed by the direct conversion of 8 to 3 (Figure S5). Activity was not observed when incubated in the presence of l-tryptophan (4), 5-bromo-l-tryptophan (5), or desbromo-AETX (Figure S4), validating the proposed order of enzymatic steps in the biosynthesis of AETX (1). In several bioinformatic databases, AetA is annotated as a tryptophan halogenase; however, the observed lack of activity on tryptophan and derivatives underscores the ongoing discrepancy of the misannotation of enzyme function in public databases.18
Figure 4.
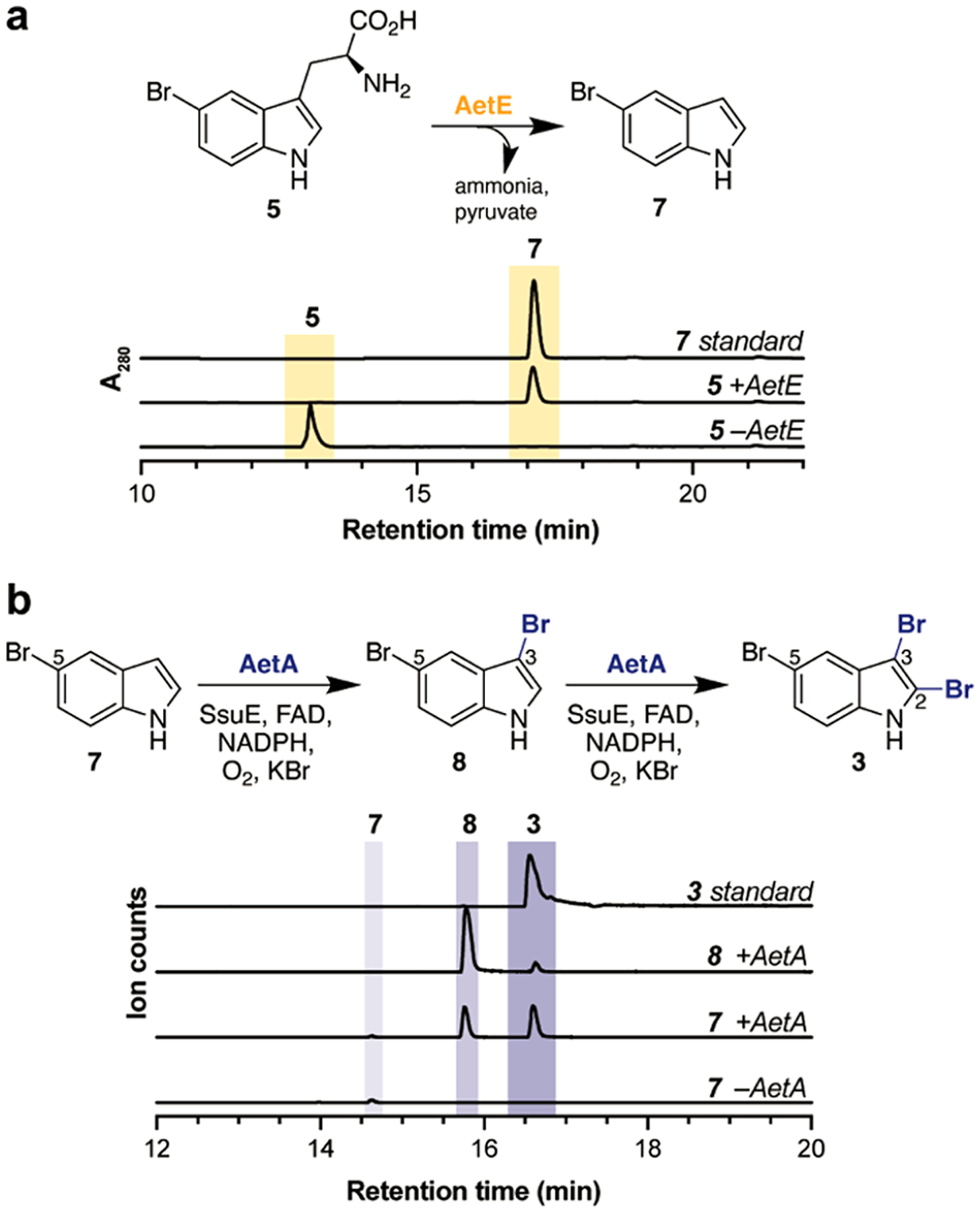
(a) AetE reaction with 5-bromo-l-tryptophan (5). Chromatograms are high-performance liquid chromatography (HPLC) traces monitored at 280 nm. (b) AetA reaction with 5-bromoindole (7) and 3,5-dibromoindole (8). Chromatograms are the EICs of 7 (m/z 193.9611), 8 (m/z 273.8696), and 3 (m/z 351.7801) analyzed in negative ionization mode.
Having identified the biosynthetic steps to access 2,3,5-tribromoindole (3), we shifted our attention to the assembly of indole building block 2, 5,7-dibromoindole-3-carbonitrile. This indole species has never been reported in the context of natural products aside from the isolation of a closely related compound, indole-3-carbonitrile, from a halophilic bacterium suspected to belong to the genus Bacillus.19 We speculated that the unannotated AetC and AetD might be involved in the transformation of 6 to 2. The incubation of 6 along with AetC and AetD indeed resulted in the formation of a small amount of 2 (Figure S11). Remarkably, further control experiments confirmed that AetD alone could catalyze this transformation (Figure S11). However, the yield of this enzymatic transformation was very low, suggesting the possible absence of a required cofactor or cosubstrate. Bioinformatic analysis of the AetD amino acid sequence provided limited insight regarding the identity of the required cofactor, as AetD does not show strong homology with any characterized protein. In efforts to reconstitute multiple turnovers and identify the missing reaction component, we screened several common organic cofactors, including PLP, FAD, NADPH, and S-adenosyl-l-methionine (SAM). However, to our dismay, we did not observe any improvement in the enzyme activity. At this point, we screened the first-row transition metals, for example, Mn(II), Fe(II), Fe(III), Co(II), Ni(II), Cu(II), and Zn(II), hypothesizing that a metal cofactor could enable the desired chemistry (Figure S12). To our delight, we observed a significant increase in AetD activity only when the reaction mixture was incubated with Fe(II) (Figure 5). The addition of Fe(III) in the reaction mixture did not influence the activity of AetD, confirming the necessity of reduced iron during catalysis. Nitrile biosynthesis has been studied primarily in plants in the context of the biosynthesis of cyanogenic glycosides.20 In this case, N-hydroxylation and decarboxylation of amino acids afford aldoximes that are converted to the corresponding nitrile through enzymatic dehydration. The only well-studied bacterial nitrile-forming enzyme is ToyM, which catalyzes the ATP-dependent conversion of a carboxyl group to a nitrile via an amide intermediate during the biosynthesis of the antibiotic toyocamycin.21 Although gene candidates have been proposed for nitrile functionalization during the biosynthesis of the enediyne-derived natural product cyanosporaside22 and the PKS-derived macrolide borrelidin,23 the exact reaction sequences are yet to be identified. AetD is fundamentally different from any of these known nitrile synthases, and the characterization of this enzyme presents a new paradigm for nitrile generation in biosynthesis.
Figure 5.
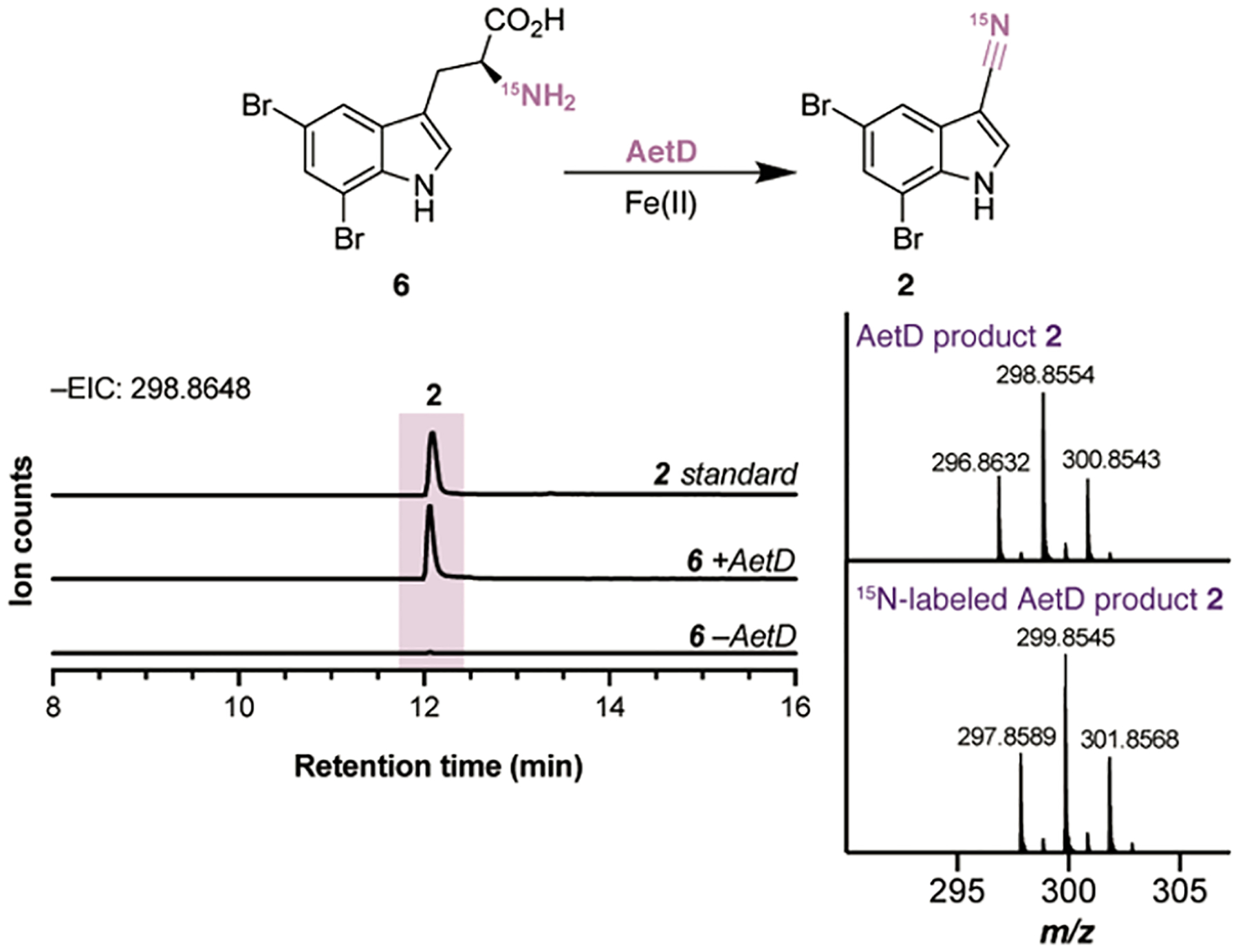
AetD reaction with 5,7-dibromo-l-tryptophan (6) to yield nitrile 2. Chromatograms are EICs of 2 (m/z 298.8648) analyzed in negative ionization mode. MS spectra shown are of the unlabeled and 15N-labeled AetD product 2.
Following the establishment of robust enzyme activity with AetD, we sought to determine the source of the nitrogen atom in the nitrile functional group. We considered several possibilities for sources of the nitrogen donor including ammonium salt, an AetD protein residue, an unknown nitrogenous cosubstrate copurified with the protein, and the α-amino group of substrate 6. The role of ammonium salt was ruled out because no 15N-incorporation was observed when the enzymatic reaction was carried out in the presence of 15N-labeled ammonium chloride. The observation that we were achieving multiple turnovers after supplementation with Fe(II) eliminated the possibility of a protein residue or any unknown nitrogenous cosubstrate. This led us to hypothesize that the α-amino group of 6 is the source of the nitrogen atom in the nitrile, which was confirmed by incubating AetD with enzymatically prepared 15N-labeled 6 (Scheme S1) and analyzing the product by mass spectrometry. A predicted 1 Da increase in the mass of the product was observed when 15N-labeled substrate was used, suggesting an unprecedented intramolecular rearrangement (Figure 5). The nature of the metallocofactor and how this novel rearrangement chemistry occurs in the AetD active site are yet to be unveiled.
In summary, we successfully delineated the five-enzyme biosynthetic pathway of the cyanobacterial toxin AETX (1) from two tryptophan molecules (Scheme 1). The pathway requires the choreographed construction of the two indole monomers 2 and 3 that diverge from the AetF monobrominated intermediate 5 prior to their ultimate reunion in the pentabrominated biindole 1. Our work unveiled enzymes with novel biochemical capabilities that have potential as biocatalysts. Specifically, AetF is a unique single-component, flavin-dependent tryptophan halogenase that does not require an accompanying reductase partner. AetA is also an unusual halogenase that regioselectively brominates the two- and three-positions of 5-bromoindole (7), shedding light on the biosynthesis of 2,3,5-tribromoindole (3), a metabolite previously reported from red algae,24 molluscs,25 and cyanobacteria.2 Above all, the iron-dependent nitrile synthase AetD is a first-of-its-kind enzyme that constructs a nitrile functional group via unprecedented rearrangement chemistry. Many questions remain unanswered regarding its mechanism, and our ongoing studies are directed toward a detailed mechanistic investigation. The role of the remaining enzyme encoded by the BGC, AetC, has not been confirmed, as the in vitro biosynthesis of AETX (1) is achievable without its participation. It is possible that AetC may provide structural or regulatory support for the biosynthesis in vivo. Ultimately, we anticipate that the knowledge gained in this study will enable the identification of related BGCs containing enzymes with similar understudied functionality that may lead to new environmental toxins.
Scheme 1.
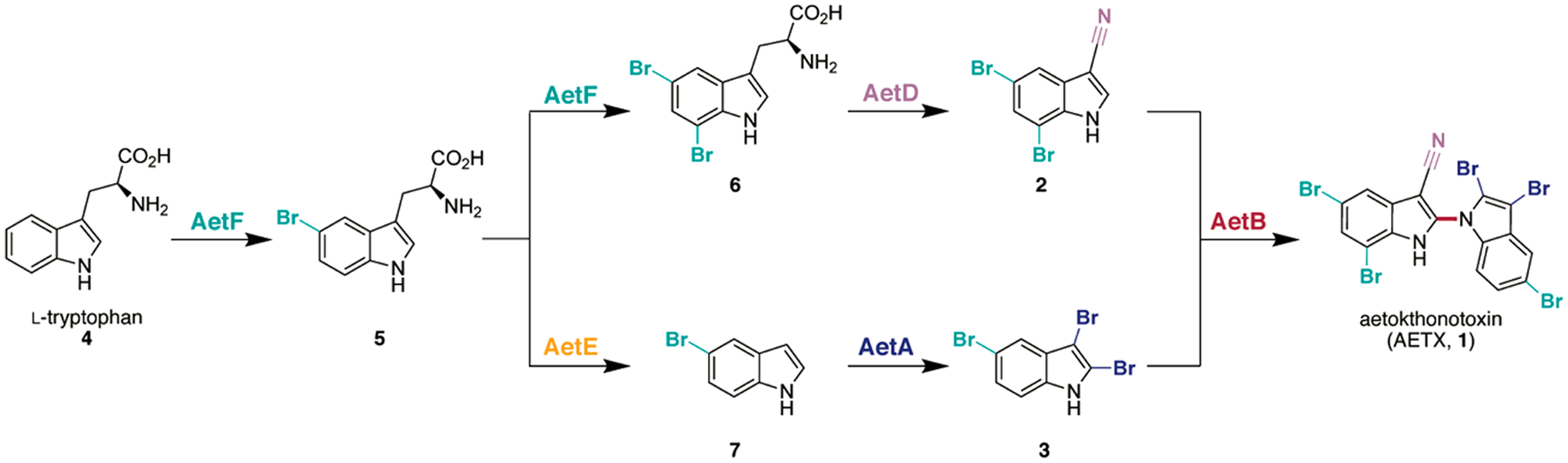
Elucidated Scheme for the Biosynthesis of Aetokthonotoxin (1)
Supplementary Material
ACKNOWLEDGMENTS
Funding was generously provided by the National Institute of Environmental Health Sciences (F32-ES033540 to A.L.L. and R01-ES030316 to B.S.M.), the National Science Foundation (OCE-1837116 to B.S.M.), and a postdoctoral fellowship from the Swiss National Science Foundation to R.J.B.S.
ABBREVIATIONS
- AETX
aetokthonotoxin
- ATP
adenosine triphosphate
- BGC
biosynthetic gene cluster
- EIC
extracted ion chromatogram
- FAD
flavin adenine dinucleotide
- FdR
spinach ferredoxin reductase
- Fdx
spinach ferredoxin
- FMN
flavin mononucleotide
- FMO
flavin-dependent monooxygenase
- HPLC
high-performance liquid chromatography
- LC-MS
liquid chromatography–mass spectrometry
- MBP
maltose-binding protein
- NADH
nicotinamide adenine dinucleotide hydride
- NADPH
nicotinamide adenine dinucleotide phosphate hydride
- PLP
pyridoxal 5′-phosphate
- PKS
polyketide synthase
- SSN
sequence similarity network
- SAM
S-adenosyl-l-methionine
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.1c12778.
General procedures, experimental details, and NMR spectra (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/jacs.1c12778
The authors declare no competing financial interest.
Contributor Information
Sanjoy Adak, Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California at San Diego, La Jolla, California 92093, United States.
April L. Lukowski, Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California at San Diego, La Jolla, California 92093, United States.
Rebecca J. B. Schäfer, Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California at San Diego, La Jolla, California 92093, United States
Bradley S. Moore, Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography and Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California at San Diego, La Jolla, California 92093, United States.
REFERENCES
- (1).Breinlinger S; Phillips TJ; Haram BN; Mareš J; Martínez Yerena JAM; Hrouzek P; Sobotka R; Henderson WM; Schmieder P; Williams SM; Lauderdale JD; Wilde HD; Gerrin W; Kust A; Washington JW; Wagner C; Geier B; Liebeke M; Enke H; Niedermeyer THJ; Wilde SB Hunting the eagle killer: A cyanobacterial neurotoxin causes vacuolar myelinopathy. Science 2021, 371, eaax9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Norton RS; Wells RJ A series of chiral polybrominated biindoles from the marine blue-green alga Rivularia firma. Application of carbon-13 NMR spin-lattice relaxation data and carbon-13-proton coupling constants to structure elucidation. J. Am. Chem. Soc 1982, 104, 3628–3635. [Google Scholar]
- (3).Jennings LK; Khan NMD; Kaur N; Rodrigues D; Morrow C; Boyd A; Thomas OP Brominated bisindole alkaloids from the celtic sea sponge Spongosorites calcicola. Molecules 2019, 24, 3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Fleming FF Nitrile-containing natural products. Nat. Prod. Rep 1999, 16, 597–606. [Google Scholar]
- (5).Gao Y; Ma D In pursuit of synthetic efficiency: Convergent approaches. Acc. Chem. Res 2021, 54, 569–582. [DOI] [PubMed] [Google Scholar]
- (6).Nishihara K; Kanemori M; Kitagawa M; Yanagi H; Yura T Chaperone coexpression plasmids: differential and synergistic roles of DnaK-DnaJ-GrpE and GroEL-GroES in assisting folding of an allergen of Japanese cedar pollen, Cryj2, in Escherichia coli. Appl. Environ. Microbiol 1998, 64, 1694–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Andorfer MC; Belsare KD; Girlich AM; Lewis JC Aromatic halogenation by using bifunctional flavin reductase–halogenase fusion enzymes. Chembiochem 2017, 18, 2099–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Andorfer MC; Lewis JC Understanding and improving the activity of flavin-dependent halogenases via random and targeted mutagenesis. Annu. Rev. Biochem 2018, 87, 159–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Agarwal V; Miles ZD; Winter JM; Eustáquio AS; El Gamal AA; Moore BS Enzymatic halogenation and dehalogenation reactions: Pervasive and mechanistically diverse. Chem. Rev 2017, 117, 5619–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Heine T; van Berkel WJH; Gassner G; van Pée KH; Tischler D Two-component FAD-dependent monooxygenases: Current knowledge and biotechnological opportunities. Biology 2018, 7, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Gerlt JA; Bouvier JT; Davidson DB; Imker HJ; Sadkhin B; Slater DR; Whalen KL Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim Biophysic Acta 2015, 1854, 1019–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Huijbers MME; Montersino S; Westphal AH; Tischler D; van Berkel WJH Flavin dependent monooxygenases. Arch. Biochem. Biophys 2014, 544, 2–17. [DOI] [PubMed] [Google Scholar]
- (13).Agarwal V; El Gamal AA; Yamanaka K; Poth D; Kersten RD; Schorn M; Allen EE; Moore BS Biosynthesis of polybrominated aromatic organic compounds by marine bacteria. Nat. Chem. Biol 2014, 10, 640–647. [DOI] [PMC free article] [PubMed] [Google Scholar]; Agarwal V; Blanton JM; Podell S; Taton A; Schorn M; Busch J; Lin Z; Schmidt EW; Jensen PR; Paul VJ; Biggs JS; Golden JW; Allen EE; Moore BS Metagenomic discovery of polybrominated diphenyl ether biosynthesis by marine sponges. Nat. Chem. Biol 2017, 13, 537–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Fisher BF; Snodgrass HM; Jones KA; Andorfer MC; Lewis JC Site-selective C–H halogenation using flavin-dependent halogenases identified via family-wide activity profiling. ACS Cent. Sci 2019, 5, 1844–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Aldemir H; Richarz R; Gulder TA The biocatalytic repertoire of natural biaryl formation. Angew. Chem., Int. Ed 2014, 53, 8286–8293. [DOI] [PubMed] [Google Scholar]
- (16).Ricardo MG; Schwark M; Llanes D; Niedermeyer THJ; Westermann B Total synthesis of aetokthonotoxin, the cyanobacterial neurotoxin causing vacuolar myelinopathy. Chem.—Eur. J 2021, 27, 12032–12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Eichhorn E; van der Ploeg JR; Leisinger T Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. J. Biol. Chem 1999, 274, 26639–26646. [DOI] [PubMed] [Google Scholar]
- (18).Schnoes AM; Brown SD; Dodevski I; Babbitt PC Annotation error in public databases: Misannotation of molecular function in enzyme superfamilies. PLoS Comput. Biol 2009, 5, e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Fu X; Schmitz FJ; Tanner RS Chemical constituents of halophilic facultatively anaerobic bacteria, 1. J. Nat. Prod 1995, 58, 1950–1954. [DOI] [PubMed] [Google Scholar]
- (20).Knaggs AR The biosynthesis of shikimate metabolites. Nat. Prod. Rep 2003, 20, 119–136. [DOI] [PubMed] [Google Scholar]
- (21).Nelp MT; Bandarian V A single enzyme transforms a carboxylic acid into a nitrile through an amide intermediate. Angew. Chem., Int. Ed 2015, 54, 10627–10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Lane AL; Nam S-J; Fukuda T; Yamanaka K; Kauffman CA; Jensen PR; Fenical W; Moore BS Structures and comparative characterization of biosynthetic gene clusters for cyanosporasides, enediyne-derived natural products from marine actinomycetes. J. Am. Chem. Soc 2013, 135, 4171–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Olano C; Moss SJ; Braña AF; Sheridan RM; Math V; Weston AJ; Méndez C; Leadlay PF; Wilkinson B; Salas JA Biosynthesis of the angiogenesis inhibitor borrelidin by Streptomyces parvulus Tü4055: insights into nitrile formation. Mol. Microbiol 2004, 52, 1745–1756. [DOI] [PubMed] [Google Scholar]
- (24).Sridevi KV; Venkatesham U; Vijender Reddy A; Venkateswarlu Y Chemical constituents of the red alga Nitophyllum marginata. Biochem. Syst. Ecol 2003, 31, 335–337. [Google Scholar]
- (25).Rahelivao MP; Gruner M; Andriamanantoanina H; Andriamihaja B; Bauer I; Knölker H-J Red algae (Rhodophyta) from the coast of Madagascar: Preliminary bioactivity studies and isolation of natural products. Mar. Drugs 2015, 13, 4197–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


