Abstract
Certolizumab pegol (CZP) is a PEGylated Fc-free tumor necrosis factor (TNF) inhibitor antibody approved for use in the treatment of rheumatoid arthritis (RA), Crohn’s disease, psoriatic arthritis, axial spondyloarthritis and psoriasis. In a clinical trial of patients with severe RA, CZP improved disease symptoms in approximately half of patients. However, variability in CZP efficacy remains a problem for clinicians, thus, the aim of this study was to identify genetic variants predictive of CZP response. We performed a genome-wide association study (GWAS) of 302 RA patients treated with CZP in the REALISTIC trial to identify common single nucleotide polymorphisms (SNPs) associated with treatment response. Whole-exome sequencing was also performed for 74 CZP extreme responders and non-responders within the same population, as well as 1546 population controls. No common SNPs or rare functional variants were significantly associated with CZP response, though a non-significant enrichment in the RA-implicated KCNK5 gene was observed. Two SNPs near spondin-1 and semaphorin-4G approached genome-wide significance. The results of the current study did not provide an unambiguous predictor of CZP response.
Introduction
Rheumatoid arthritis is a chronic, systemic autoimmune disease of unknown etiology, affecting between 0.3 and 1% of the global population. It affects the joints, connective tissues, muscle, tendons, and fibrous tissue leading to reduced quality of life, disability, and early mortality for sufferers [1].
Established treatment approaches, focused on sequential monotherapies and step-up combination therapies, have underserved many patients. Recently, it has become clear that addressing the underlying inflammatory processes early and prompt disease treatment with biological disease-modifying anti-rheumatic drugs (bDMARDs) such as TNF inhibitors (TNFi), is more successful in terms of limiting radiological progression and minimising loss of mobility and function [2]. However, whilst targeting TNF in RA patients represents a significant advance in treatment options, approximately 30–40% of treatment-naïve patients in clinical trials do not respond adequately to current drug treatment [3–5]. Although these reagents are highly beneficial in the majority of patients, their relatively high cost and potential safety liabilities are problematic in prescribing for broad patient populations.
Several factors have been identified in susceptibility to RA, including 2 to 3-fold increased risk in women and 1.3 to 2.4-fold higher risk among smokers. An increase in risk is also seen in individuals positive for anti-citrullinated peptide antibodies [6]. Despite extensive efforts, there are currently no accepted molecular or genetic biomarkers qualified for use in RA diagnosis and therapy. Therefore, current treatment paradigms operate without guarantee of patient benefit.
Until recently, the search for genetic markers associated with RA and anti-TNF response has focused on genes involved in RA susceptibility and disease pathways [7, 8] and genes involved in TNF production and signalling [9]. Over the last decade, a pool of DNA variants proposed as predictors of response to anti-TNFs has been uncovered following pharmacogenetic analyses of RA cohorts [10]. For example, the common TNF single nucleotide polymorphism (SNP) -308G>A (rs1800629), proposed to be associated with RA treatment outcome [11], has been rigorously investigated with often conflicting results [12–14].
More recently, genome-wide array and sequencing approaches have afforded researchers increased scope to search in an unbiased manner for clinically relevant biomarkers associated with RA and response to TNF blockers. However, the first round of these analyses, focused on European or Caucasian patients undergoing therapy with etanercept, infliximab and adalimumab, have generated further ambiguous and conflicting findings [10, 15–18]. The largest genome-wide association study to date, consisting of 2706 patients from 13 European cohorts, including those which had previously indicated SNP associations with RA, showed no association in a meta-analysis [18], possibly due to data or population heterogeneity.
However, eight candidate loci did show replication in Dutch patients from the DREAM cohort (n = 882), with consistent suggestive associations in two further cohorts in a subsequent meta-analysis [10]. Although none of these markers reached genome-wide significance, the directionality of association for three SNPs (rs1568885, rs1813443 and rs4411591) was consistent in all four analyses and may merit further study [10].
Subsequent genome-wide studies and meta-analyses have had limited success at replicating most candidate SNPs [19–21], however associations with GFRA1, MED15, PTPRC, and the PDE3A-SLCO1C1 region have been successfully replicated in at least one independent cohort [22–25]. Recent evidence suggests that some these SNP associations may be drug-specific [24], which could explain the inability of some studies to replicate them. Integrated analysis of genomic and transcriptomic datasets has confirmed the therapy-specific nature of TNFi response [26]. In addition, substantial phenotypic heterogeneity between RA cohorts may make replication of TNFi SNP associations more difficult. Therefore, machine learning approaches capable of abstracting clinical and genetic predictors have been developed in an effort to classify patients according to their likelihood of responding to treatments [27, 28]. Despite the relatively high heritability of TNFi response [29], inclusion of SNPs previously associated with TNFi response provides limited improvement in the accuracy of machine learning models [27].
To date, no GWAS has been reported for certolizumab pegol. We report the first genome-wide study of a North American cohort of RA patients with moderate to severe RA, with the objective of discovering DNA variants associated with CZP response.
Materials and methods
Patients
The study protocol was approved by the Institutional Review Boards at Duke and Columbia University and written informed consent for participation was obtained at study entry for all subjects. North American patients (≥18 years) were selected from the REALISTIC phase IIIb study (NCT00717236). All participants were diagnosed with adult onset RA as defined by 1987 American College of Rheumatology (ACR) criteria for at least 3 months. In addition, they had either not tolerated or responded unsatisfactorily to at least one DMARD prior to study entry [30]. Patients were assessed using a 28 joint count and were defined as having active disease if they had at least five tender and at least four swollen joints, C-reactive protein (CRP) ≥10 mg/l and/or ≥ 28mm/hour erythrocyte sedimentation rate (ESR) and had moderate to severe RA at study entry [30] (Table 1).
Table 1. Characteristics of study sample for GWAS.
| Patient demographics | |
| Self-reported gender, % female | 79.5 |
| Age, mean (S.D) years | 56+/-12 |
| Duration, years | |
| Mean (S.D.) | 9.1+/-9.0 |
| Median (interquartile range) | 6.0 (2.3–13.6) |
| Disease duration <2 years, n (%) | 66 (21.8) |
| Tender joint count, mean (S.D.) | 15.4+/-6.8 |
| DAS 28 (ESR), mean (S.D.) | 6.4+/-1.0 |
| ACR20 at week 6, n (%) | 138 (45.7) |
| ACR20 at week 12, n (%) | 146 (48.3) |
| ACR70 at week 6, n (%) | 17 (5.6) |
| ACR70 at week 12, n (%) | 34 (11.3) |
| Self-reported ethnicity, n (%) | |
| Caucasian | 293 (97.0) |
| CRP, mg/l; Median (interquartile range) | 9.0 (5.0–18.0) |
| ESR, mm/h: Median (interquartile range) | 36.0 (25.0–51.0) |
| Anti-CCP positive at baseline, n (%) | 180 (66.4) |
| RF positive at baseline, n (%) | 208 (74.8) |
| Treatment history | |
| Previous TNF inhibitor use, n (%) | 142 (47.0) |
| Other Medication at baseline, n (%) | |
| Methotrexate | 216 (71.5) |
| Steroids | 186 (61.6) |
| Statins | 68 (22.5) |
| Lefluonamide | 21 (7.0) |
| Azathioprine | 1 (0.03) |
ACR20, 20% improvement in ACR score; ACR70, 70% improvement in ACR score; DAS, Disease Activity Score; CCP, cyclic citrullinated peptide; RF, rheumatoid factor.
GWAS of common SNPs
Genotyping
A total of 2,372,361 SNPs in 413 patients were genotyped using the Illumina Omni2.5M array platform. SNP genotypes were called using the Illumina GenomeStudio Software package Version 2011.1 with genotyping module version 1.9.4 according to the manufacturer’s instructions.
Quality control (QC). A series of QC checks were carried out to ensure sample integrity. Biological sex was assessed for concordance between the genetically-inferred and self-reported gender. Duplicate samples and cryptic relatedness were identified based on genetic data using PLINK [31]. Samples found to be related at the level of first cousins or closer (pairwise estimate of identity-by-descent exceeding 0.125) were considered excessively related and one sample from each pair was removed at random. Quantitative estimates of ancestry (principal components analysis (PCA) implemented in EIGENSTRAT software [32]) were performed using LD pruned SNPs to estimate genomic ancestry and compared with self-reported ethnicity.
Data quality was also evaluated at the marker level to remove low quality genotypes. Two subjects missing >10% of genotype calls and one subject with an anomalously low inbreeding coefficient (F statistic outside of 6 standard deviations from the mean) were excluded. A total of 8629 poorly performing markers missing >10% of genotype calls and 3144 heterozygous haploid genotypes were removed. In addition, rare variants with a minor allele frequency (MAF) of <1% were excluded. No SNPs were found to deviate from Hardy-Weinberg equilibrium when a Bonferonni-adjusted threshold of 2.91x10-8 was applied.
Following QC, data were available for 1,685,541 SNPs in 360 individuals of primarily European ancestry. Of these, 302 individuals treated with CZP were included in the statistical analysis. Disease activity Score (DAS28 ESR) data were not available for all individuals (Table 2) and where data was missing for individual outcome measures, those patients were excluded from subsequent analysis. Only a minor difference in ACR20 response (45.7% vs 48.3%, Table 1) was observed between weeks 6 and 12, therefore only week 6 (ACR20_W6) was used for the analysis.
Table 2. Summary of data available for individual clinical outcome measuresa.
| ACR20_W6 | RD28_W6 | ΔDAS28_W6 | ΔDAS28_W12 | |
|---|---|---|---|---|
| Total | 362 | 347 | 344 | 339 |
| CZP | 302 | 287 | 287 | 284 |
| Placebo | 60 | 60 | 57 | 55 |
ACR20_W6, 20% reduction in ACR at week 6; RD28_W6, reduction in DAS28 at week 6 (dichotomous phenotype); ΔDAS28_W6, change in DAS28 at week 6; ΔDAS28_W12, change in DAS28 at week 12.
aIncomplete outcome data available for DAS28.
Imputation
After QC, non-ambiguous SNPs, including rare variants with MAF<1%, were used as input for imputation using haplotypes from the 1000 Genomes phase1 integrated reference panel. Prephasing of haplotypes was performed using SHAPEIT software [33]. Imputation of missing and un-genotyped SNPs was then performed on 5Mb segments from each autosome and chromosome X using IMPUTE2 [34] according to recommended best practices. High confidence SNPs with greater than 90% imputation confidence were then merged using GTOOL and converted back to PLINK format for association analysis.
Statistical analysis
Four primary outcomes were tested for association within the study: ACR20 and decrease in DAS28 ESR >1.2 at week 6 (both as dichotomous variables), and change in DAS28 ESR at week 6 and 12 (continuous variables). Testing for association between individual outcome measures and each SNP were carried out using logistic or linear regression, after correcting for gender and genetic ancestry using the top 3 principal component axes with significant Tracy-Widom statistics, explaining 2.78% of the observed variance.
Analysis of candidate SNPs
Using a targeted approach harnessing known GWAS hits in autoimmune disease and genes involved in the TNF pathway, SNPs with higher prior probability of association with CZP response were tested for enrichment of association beyond that expected under the null hypothesis. Of 1618 SNPs previously reported to be associated with auto-immune diseases (SNPs with p< 5x10-8 obtained by searching GWAS Central for the MeSH term “autoimmune disease”), 72 were specifically associated with RA. These SNPs have a higher probability of association with the CZP outcomes than SNPs selected randomly. A total of 1403 out of 1618 SNPs were present on the Omni2.5M chip or were available in the imputed dataset. Of the 72 RA-associated SNPs, 64 were available for testing. To examine the role of genetic variation in genes involved in TNF signaling, a further hypothesis driven approach was carried out using 13303 common SNPs located within 112 genes that were annotated as participating in TNF signaling (hsa04668) according to KEGG pathway ontologies [35]. Significance scores for the subset of 13303 TNF-signaling SNPs were compared against a uniform distribution of p-values that would be expected under the null hypothesis using quantile-quantile plots. A review of the literature was used to identify 94 SNPs previously reported to be associated with TNF inhibitor response. Of these, 86 SNPs were present in the Omni2.5M array or imputed dataset.
Whole-exome sequencing
Patient selection
Extreme non-responders (hereafter referred to as “non-responders”) were defined as ACR20 failures at week 6 and 12, with maximum reduction in DAS 28 ESR at week 12 compared with baseline (BL) of 0.33 and maximum reduction in DAS 28 ESR at week 6 compared with BL of 0.6, to eliminate secondary loss of response. Extreme responders (hereafter referred to as “responders”) were defined as individuals with a positive ACR70 score at 12 weeks. Using these criteria, a total of 81 non-responders and 40 responders were identified (summary shown in Table 3).
Table 3. Characteristics of study sample for whole exome sequencing.
| Patient demographics | Responders | Non-responders |
|---|---|---|
| Self-reported gender, % female | 77.5 | 80.2 |
| Age, mean (S.D) years | 54.5+/-15 | 54.7+/-12 |
| Duration, years | ||
| Mean (S.D.) | 8.9+/-10.4 | 7.5+/-7.8 |
| Median (interquartile range) | 4.7 (1.5–13.3) | 4.0 (1.7–11.0) |
| Disease duration <2 years, n (%) | 13 (32.5) | 21 (25.9) |
| Tender joint count, mean (S.D.) | 14.4+/-7.0 | 15.7+/-7.0 |
| DAS 28 (ESR), mean (S.D.) | 6.3+/-1.0 | 6.3+/-1.1 |
| ACR20 at week 6, n (%) | 34 (85.0) | 0 (0.0) |
| ACR20 at week 12, n (%) | 40 (100) | 0 (0.0) |
| ACR70 at week 6, n (%) | 12 (30.0) | 0 (0.0) |
| ACR70 at week 12, n (%) | 40 (100) | 0 (0.0) |
| Self-reported ethnicity, n (%) | ||
| Caucasian | 30 (75) | 75 (93) |
| CRP, mg/l; Median (interquartile range) | 9 (3.8–21.3) | 7 (4.0–16.0) |
| ESR, mm/h: Median (interquartile range) | 42.0 (26.3–57.8) | 38.0 (27.0–52.0) |
| Anti-CCP positive at baseline, n (%)a | 24 (66.6) | 45 (61.6) |
| RF positive at baseline, n (%)a | 28 (75.7) | 57 (76.0) |
| Treatment history | ||
| Previous TNF inhibitor use, n (%)a | 16 (40.0) | 37 (45.6) |
| Other Medication at baseline, n (%) | ||
| Methotrexate | 32 (80.0) | 59 (72.8) |
| Steroids | 26 (65.0) | 47 (58.0) |
| Statins | 7 (17.5) | 20 (24.7) |
| Lefluonamide | 0 (0.0) | 3 (3.7) |
| Azathioprine | 0 (0.0) | 0 (0.0) |
aIncomplete clinical data.
Sequencing and quality control
Whole-exome sequencing (WES) was performed using the Roche Nimblegen Human Exon v2 capture kit and KAPA library prep kit according to the manufacturer’s instructions. Sequencing was performed using the 100bp paired-end read protocol on the Illumina HiSeq 2000 instrument. After quality filtering the raw sequence data using CASAVA (Illumina, Inc., San Diego, CA, USA), adapter sequences were trimmed and the sequencing reads were aligned to the reference human genome (NCBI37/hg19) using BWA software [36]. Duplicate reads were then removed from the dataset using Picard (Broad Institute, Boston, MA, USA). Variant calling was performed using GATK [37] with local re-alignment around insertion/deletion variants (indels) and base quality recalibration for single-nucleotide variants (SNVs) [38]. Similarly sequenced control samples available at the Duke Center for Human Genome Variation (CHGV) served as the comparison group for a subset of the exome sequencing analyses.
Assesment of population structure
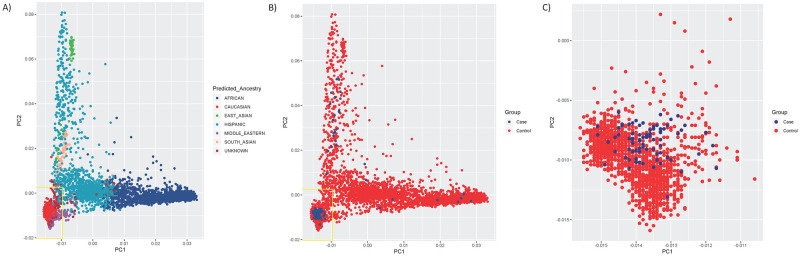
As the majority of subjects were self-described Caucasians, we sought to limit our analyses to that ancestry group in order to minimize the influence of population stratification between cases and controls. Preliminary ancestry predictions were perfomed during bioinfomatic processing using a panel of 12840 high coverage SNPs, based on principal components generated by EIGENSTRAT for cases, controls, and set of 4289 samples with pre-evaluated genetic ancestries. All cases and controls were initially assigned to one of six geographic ancestry groups (Caucasian, African, Hispanic, East Asian, South Asian or Middle Eastern) using a multinomial logistic regression model which provided a probability estimate of a sample belonging to each of the six groups. After restricting the cases and control samples to those predicted to be of Caucasian ancestry, all samples were further required to have EIGENSTRAT PC scores within 6 standard deviations of the mean on PCs 1 and 2. From an initial 121 CZP-treated patients and 2654 CHGV controls, ancestry pruning resulted in 74 CZP-treated patients (19 responders, 55 non-responders) and 1546 controls of European ancestry (Fig 1A and 1B). After removal of ancestry outliers, no major differences in population structure were observed between case-control groups (Fig 1C). Therefore no additional adjustment for genetic ancestry was included in the WES association analysis. The first two principal components accounted for 0.3% of the observed variance. Study characteristics after ancestry pruning are shown in S1 Table.
Fig 1. Scatterplot of Principal Component 1 (PC1) vs Principal Component 2 (PC2) from EIGENSTRAT ancestry analysis.
(A) Scatter plot of PCs 1 & 2 prior to ancestry pruning, with subjects colored according to their predicted genetic ancestry. Yellow box indicates the region encompassing 6 s.d on PC1 and PC2. (B) Scatter plot of PCs 1 & 2 prior to ancestry pruning, with subjects colored by case/control status. Yellow box indicates the region encompassing 6 s.d on PC1 and PC2. (C) Scatter plot of PCs 1 & 2 after ancestry pruning and PCA outlier removal.
Variant quality filtering and association testing
Variants were filtered to include only those located in CCDS genes, with a GATK variant quality score in the 99.9 percentile and in the following functional categories: stop-gained, stop-lost, nonsynonymous coding, essential splice site, and coding region indels. Genotypes with ≤3 reads were considered missing for any individual. Variants were excluded if they were missing in ≥10% of cases or controls, or if they showed significant deviation from Hardy-Weinberg equilibrium in controls. In addition, 6044 variants previously identified as being artifacts based on comparison of CHGV controls with publically available data from the Exome Variant Server (EVS; http://evs.gs.washington.edu/EVS/) were removed from the dataset.
Individual variants were tested for association using Fisher’s exact test. In addition, gene-based collapsing tests were performed following the method of Li and Leal [39], with adjustment for differences in coverage at the exon level between cases and controls. Variants qualified for inclusion in these tests if they had a minor allele frequency (MAF) <1% in both the study sample (combined cases and controls) and EVS, and fell into the functional categories above.
There were three major comparisons in the study: responders vs non-responders, responders vs controls + non-responders, and non-responders vs controls + responders. The within-cohort comparison of responders and non-responders benefits from the clarity of the two alternative phenotypes; however, since the patients in these groups were selected from the extremes of the outcome distribution, it is reasonable to also compare each extreme to a population control sample, as the misclassification rate in controls is unlikely to be high and the sample sizes available are much larger. Statistical significance was assessed using Bonferroni correction for the number of tested variants or genes (in the collapsing analysis).
Results
We have carried out a two-stage genetic analysis seeking to identify DNA variants associated with response to CZP in a North American cohort with moderate to severe RA.
Patient population
The patient population was derived from the REALISTIC phase IIIb study [30] and the subjects used for the GWAS are summarized in Table 1. 79.5% of participants in the study were women, with average age at entry of 56 years. Mean duration of disease was 9.1 years with 22% of the population having disease duration of <2 years. Median CRP level at entry was 9.0 mg/l and median ESR was 36.0 mm/h. Mean DAS 28 ESR at entry was 6.4. 60% of the population for which data were available was positive for anti-citrullinated peptide antibodies and 69% positive for rheumatoid factor. Previous exposure to anti-TNF agents had occurred in 47% of patients, while 72% were on methotrexate, 62% on steroids and 23% on statins at baseline. A small number of patients were receiving other DMARDs on study entry (lefluonamide n = 21; azathioprine n = 1).
GWAS
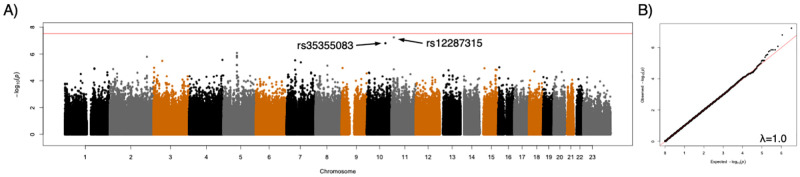
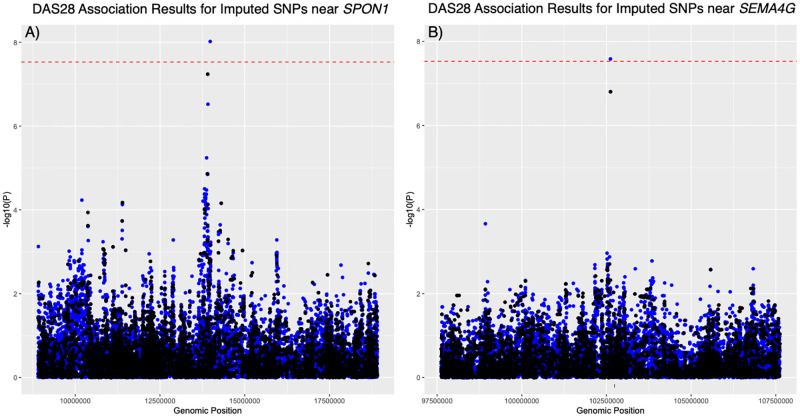
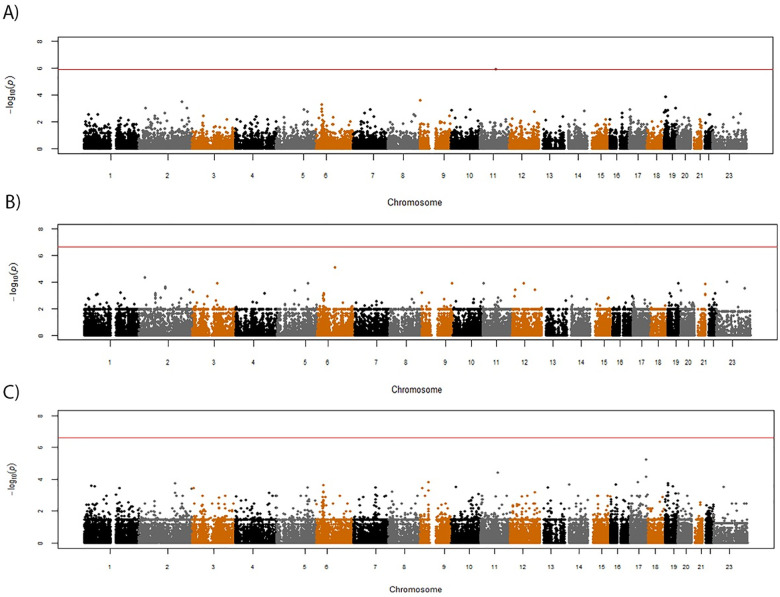
After Bonferroni correction for multiple testing, using categorical or continuous outcome variables (ACR20 and DAS28), we were unable to demonstrate genome-wide significant associations between any genotyped SNP and therapeutic response to CZP for any of the outcome measures. For three of the measures, ACR20 and DAS28 ESR reduction of >1.2 at week 6 (dichotomous variables), and change in DAS28 ESR at week 6 (a continuous variable), there was no evidence of SNPs associated with outcome (S1 Fig). For the change in DAS28 ESR at week 12 outcome measure, two SNPs approaching genome-wide significance were observed (rs12287315 and rs35355083) (Fig 2). Neither of these SNPs were located within genes; rs12287315 (p = 5.78x10-8; β = -1.08) is located 76 kb upstream of the spondin-1 (SPON1) gene and rs35355083 (p = 1.57x10-7; β = -1.14) is located in an intergenic region 110 kb upstream of the semaphorin 4G (SEMA4G) gene. Imputation of ungenotyped SNPs in a 10Mb window around each lead SNP identified two additional SNPs in these regions with stronger associations with DAS28 ESR at week 12 (Fig 3). These included rs78675205 (p = 2.61x10-8; β = -1.21) on chromosome 10, located downstream of the Paired Box 2 (PAX2) gene, and rs72873110 (p = 9.61x10-9; β = -1.13) located on chromosome 11, approximately 3kb upstream of the SPON1 coding region. While the significance scores for these two variants exceeded the original threshold for statistical significance, they were not statistically significant when the association analysis was performed using all common SNPs from a genome-wide imputation and no additional associations were detected outside of these two regions (S2 Fig).
Fig 2. Results of genome-wide association study of certolizumab pegol response, as measured by DAS28 (week 12) scores.
(A) Manhattan plot of significance scores from linear regression analysis. (B) Quantile-quantile plot of observed significance scores vs expected under the null hypothesis. The red line indicates the Bonferroni-adjusted threshold for statistical significance.
Fig 3. Fine mapping of regions with suggestive associations with DAS28 (week 12).
(A) Manhattan plot of significance scores from linear regression analysis of DAS28 for common SNPs in a 10Mb window around the SPON1 locus. (B) Manhattan plot of significance scores from linear regression analysis of DAS28 for common SNPs in a 10Mb window around the SEMA4G locus. SNPs genotyped on the Omni2.5M array are shown in black while imputed SNPs appear in blue. The dotted red line indicates the original Bonferroni threshold for significance.
Candidate SNP and pathway analysis
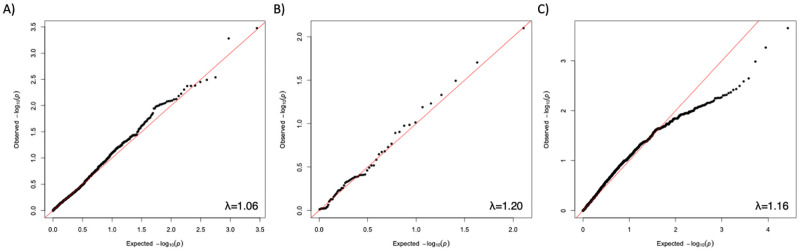
We supplemented our genome-wide search for variants with a targeted analysis focused on SNPs with a higher prior probability of association, through previous documentation of association with auto-immune disease, as well as a sub-group of 64 SNPs associated specifically with RA. Similarly, we generated a candidate list of 13303 common variants observed in 112 genes reported to be involved in TNF biology [35]. No enrichment of association beyond that expected under the null hypothesis was observed using these approaches (Fig 4, S2–S4 Tables).
Fig 4. Quantile-quantile (QQ) plots of results from candidate SNP analysis.
Plot of ACR20 significance scores from (A) 1403 SNPs previously reported to be associated with autoimmune disease, (B) 64 SNPs previously associated with rheumatoid arthritis, and (C) 13303 SNPs occurring in genes involved in TNF biological pathways.
We further tested whether SNPs previously associated with response to TNF inhibitors showed evidence of association in the current study. As shown in Table 4, the previously reported associations with TNF inhibitor response were not observed in our cohort.
Table 4. SNPs previously associated with therapeutic response to TNF inhibitors.
| SNP | PUBMEDID | Region | Nearest Gene | Genotyped/ Imputed | P-value (ACR20 Wk6) | P-value (DAS28 Wk6) | P-value (DAS28 Wk12) | P-value (RD28 Wk 6) |
|---|---|---|---|---|---|---|---|---|
| rs1800896 | 18615156 | 1q32.1 | IL19 | Genotyped | 0.398 | 0.3498 | 0.6376 | 0.1547 |
| rs1800629 | 18615156 | 6p21.33 | TNF | Genotyped | 0.4269 | 0.5255 | 0.3321 | 0.78 |
| rs983332 | 18615156 | 1p22.3 | LMO4 | Genotyped | 0.8091 | 0.7796 | 0.6857 | 0.2909 |
| rs928655 | 18615156 | 1p22.2 | GBP6 | Genotyped | 0.1043 | 0.1377 | 0.1266 | 0.1585 |
| rs13393173 | 18615156 | 2q24.3 | CERS6 | Genotyped | 0.5742 | 0.1443 | 0.1691 | 0.1655 |
| rs437943 | 18615156 | 4p15.1 | ARAP2 | Genotyped | 0.7274 | 0.2707 | 0.429 | 0.3062 |
| rs10945919 | 18615156 | 6q26 | QKI | Genotyped | 0.579 | 0.1387 | 0.0937 | 0.2423 |
| rs854555 | 18615156 | 7q21.3 | PON1 | Genotyped | 0.5385 | 0.557 | 0.9665 | 0.9778 |
| rs854548 | 18615156 | 7q21.3 | PPP1R9A | Genotyped | 0.2678 | 0.6907 | 0.5764 | 0.5743 |
| rs854547 | 18615156 | 7q21.3 | PPP1R9A | Genotyped | 0.3115 | 0.6884 | 0.7034 | 0.6514 |
| rs7046653 | 18615156 | 9p21.2 | MOB3B | Genotyped | 0.8241 | 0.4069 | 0.1583 | 0.3181 |
| rs868856 | 18615156 | 9p21.2 | MOB3B | Genotyped | 0.8859 | 0.5538 | 0.1812 | 0.3844 |
| rs774359 | 18615156 | 9p21.2 | C9ORF72 | Genotyped | 0.8746 | 0.358 | 0.1237 | 0.4198 |
| rs2814707 | 18615156 | 9p21.2 | MOB3B | Genotyped | 0.7717 | 0.4574 | 0.09011 | 0.3859 |
| rs3849942 | 18615156 | 9p21.2 | C9ORF72 | Genotyped | 0.8352 | 0.2486 | 0.06975 | 0.3508 |
| rs6028945 | 18615156 | 20q12 | MAFB | Genotyped | 0.04575 | 0.3062 | 0.681 | 0.4589 |
| rs6138150 | 18615156 | 20p11.21 | CST5 | Genotyped | 0.7479 | 0.4045 | 0.6091 | 0.3948 |
| rs6071980 | 18615156 | 20q12 | MAFB | Genotyped | 0.005448 | 0.2701 | 0.8796 | 0.157 |
| rs2372536 | 20847201 | 2q35 | ATIC | Imputed | 0.4728 | 0.3225 | 0.8358 | 0.8671 |
| rs1127354 | 20847201 | 20p13 | ITPA | Imputed | 0.8614 | 0.573 | 0.04869 | 0.4101 |
| rs1801133 | 20847201 | 1p36.22 | MTHFR | Genotyped | 0.9395 | 0.8252 | 0.5648 | 0.4344 |
| rs10919563 | 20847201 | 1q32.1 | PTPRC | Imputed | 0.6454 | 0.04959 | 0.1749 | 0.008771 |
| rs12081765 | 21061259 | 1q23.3 | RXRG | Genotyped | 0.08769 | 0.4604 | 0.9424 | 0.3248 |
| rs1532269 | 21061259 | 5p13.3 | PDZD2 | Imputed | 0.06234 | 0.1323 | 0.6655 | 0.6793 |
| rs17301249 | 21061259 | 6q23.2 | EYA4 | Imputed | 0.2642 | 0.7368 | 0.8703 | 0.5782 |
| rs7305646 | 21061259 | 12p12.3 | LMO3 | Genotyped | 0.9131 | 0.7942 | 0.6261 | 0.8969 |
| rs4694890 | 21061259 | 4p11 | TEC | Genotyped | 0.9198 | 0.7517 | 0.04464 | 0.3293 |
| rs1350948 | 21061259 | 11p14.3 | CCDC179 | Genotyped | 0.7382 | 0.907 | 0.6072 | 0.9468 |
| rs4411591 | 23233654 | 18p11.31 | LOC100130480 | Imputed | 0.9059 | 0.2898 | 0.1311 | 0.5541 |
| rs7767069 | 23233654 | 6q12 | LOC102723883 | Imputed | 0.1295 | 0.1288 | 0.7267 | 0.494 |
| rs4651370 | 23233654 | 1q31.1 | PLA2G4A | Imputed | 0.1486 | 0.6554 | 0.6513 | 0.2614 |
| rs1813443 | 23233654 | 11q22.1 | CNTN5 | Imputed | 0.1955 | 0.01845 | 0.05309 | 0.06672 |
| rs1447722 | 23233654 | 3q23 | CLSTN2 | Imputed | 0.6619 | 0.2028 | 0.114 | 0.8809 |
| rs1568885 | 23233654 | 7p21.3 | ETV1 | Imputed | 0.5729 | 0.8817 | 0.8937 | 0.7815 |
| rs12142623 | 23233654 | 1q31.1 | PLA2G4A | Imputed | 0.3175 | 0.6917 | 0.6671 | 0.4263 |
| rs2378945 | 23233654 | 14q12 | NUBPL | Imputed | 0.1607 | 0.4346 | 0.7431 | 0.1113 |
| rs10520789 | 22569225 | 15q26.2 | NR2F2 | Imputed | 0.4158 | 0.5916 | 0.9036 | 0.3318 |
| rs11870477 | 22569225 | 17q24.3 | KCNJ16 | Genotyped | 0.6034 | 0.3105 | 0.9661 | 0.5014 |
| rs16973982 | 22569225 | 15q26.2 | NR2F2 | Imputed | 0.2349 | 0.8241 | 0.6655 | 0.6806 |
| rs12001550 | 22569225 | 9q33.1 | TLR4 | Imputed | 0.2199 | 0.6921 | 0.216 | 0.1803 |
| rs885814 | 22569225 | 1p36.12 | ALPL | Genotyped | 0.4722 | 0.8331 | 0.39 | 0.6981 |
| rs2293137 | 22569225 | 3p13 | FOXP1 | Imputed | 0.9456 | 0.7823 | 0.6059 | 0.5943 |
| rs885813 | 22569225 | 1p36.12 | ALPL | Genotyped | 0.2844 | 0.4211 | 0.6313 | 0.1756 |
| rs1875620 | 22569225 | 9q22.1 | C9ORF47 | Genotyped | 0.3423 | 0.2853 | 0.4925 | 0.7107 |
| rs11525966 | 22569225 | 9q22.1 | C9ORF47 | Imputed | 0.2175 | 0.3616 | 0.751 | 0.6859 |
| rs960902 | 22569225 | 2p22.2 | QPCT | Genotyped | 0.6766 | 0.9054 | 0.4399 | 0.7923 |
| rs1539909 | 22569225 | 18q22.3 | CBLN2 | Genotyped | 0.6148 | 0.4174 | 0.8415 | 0.5087 |
| rs11124586 | 22569225 | 2p22.2 | CDC42EP3 | Genotyped | 0.7767 | 0.8684 | 0.747 | 0.2929 |
| rs17679567 | 22569225 | 16q23.1 | CNTNAP4 | Genotyped | 0.5491 | 0.6337 | 0.6196 | 0.9315 |
| rs1827596 | 22569225 | 2q14.3 | CNTNAP5 | Imputed | 0.9189 | 0.7095 | 0.1244 | 0.9499 |
| rs4412918 | 22569225 | 15q21.3 | PRTG | Imputed | 0.2184 | 0.6313 | 0.6727 | 0.7144 |
| rs17002731 | 22569225 | 4q21.1 | CXCL13 | Genotyped | 0.9293 | 0.1418 | 0.8304 | 0.07673 |
| rs1835353 | 22569225 | 2q14.3 | CNTNAP5 | Genotyped | 0.5036 | 0.406 | 0.1828 | 0.4105 |
| rs6427528 | 23555300 | 1q23.3 | CD84 | Imputed | 0.7892 | 0.5482 | 0.2071 | 0.779 |
| rs1503860 | 23555300 | 1q23.3 | CD84 | Imputed | 0.8815 | 0.5997 | 0.1289 | 0.5712 |
| rs12570744 | 23555300 | 10p14 | LINP1 | Genotyped | 0.2621 | 0.4243 | 0.4004 | 0.08183 |
| rs7141276 | 23555300 | 14q13.1 | SNX6 | Imputed | 0.3357 | 0.4961 | 0.4497 | 0.2217 |
| rs10833455 | 23555300 | 11p15.1 | NELL1 | Imputed | 0.7821 | 0.3469 | 0.6566 | 0.8805 |
| rs10833456 | 23555300 | 11p15.1 | NELL1 | Imputed | 0.6108 | 0.255 | 0.719 | 0.9805 |
| rs7932820 | 23555300 | 11p15.1 | NELL1 | Imputed | 0.6176 | 0.3293 | 0.5869 | 0.8421 |
| rs8009551 | 23555300 | 14q13.1 | SNX6 | Genotyped | 0.2048 | 0.4228 | 0.4865 | 0.1568 |
| rs4336372 | 23555300 | 5q35.2 | DRD1 | Genotyped | 0.9886 | 0.8532 | 0.01123 | 0.8786 |
| rs10265155 | 23555300 | 7q21.11 | MAGI2 | Genotyped | 0.04158 | 0.04413 | 0.5201 | 0.3038 |
| rs1990099 | 23555300 | 7q21.11 | MAGI2 | Imputed | 0.2394 | 0.03951 | 0.6735 | 0.2659 |
| rs284515 | 26776603 | 6q15 | MAP3K7 | Imputed | 0.1955 | 0.8197 | 0.09406 | 0.4495 |
| rs75908454 | 26776603 | 6q27 | WDR27 | Imputed | 0.21 | 0.08461 | 0.891 | 0.8489 |
| rs1679568 | 26776603 | 10q25.3 | GFRA1 | Imputed | 0.5555 | 0.6671 | 1 | 0.9108 |
| rs284511 | 26776603 | 6q15 | MAP3K7 | Imputed | 0.8658 | 0.1713 | 0.8586 | 0.03014 |
| rs6941263 | 25896534 | 6q21 | ARMC2 | Imputed | 0.359 | 0.2057 | 0.3872 | 0.132 |
| rs113878252 | 25896534 | 22q11.21 | MED15 | Imputed | 0.1638 | 0.2092 | 0.9521 | 0.3039 |
| rs6065221 | 25896534 | 20q12 | MAFB | Genotyped | 0.7959 | 0.3276 | 0.2482 | 0.5876 |
| rs7195994 | 30166627 | 16q12.2 | FTO | Imputed | 0.9607 | 0.8829 | 0.7041 | 0.2118 |
| rs10739537 | 30166627 | 9q33.1 | DBC1 | Imputed | 0.4139 | 0.4443 | 0.6199 | 0.4562 |
| rs948138 | 30166627 | 11q22.2 | MMP20 | Genotyped | 0.3166 | 0.4068 | 0.7111 | 0.7946 |
| rs11599217 | 30166627 | 10q26.2 | DOCK1 | Imputed | 0.8675 | 0.8999 | 0.9294 | 0.9163 |
| rs2187874 | 30166627 | 4p16.3 | ZNF595 | Imputed | 0.1811 | 0.5109 | 0.5323 | 0.2259 |
| rs150537045 | 30166627 | 8q23.3 | CSMD3 | Imputed | 0.5835 | 0.4199 | 0.3298 | 0.7194 |
| rs76668869 | 30166627 | 1p31.1 | ADGRL2 | Imputed | 0.4255 | 0.164 | 0.7019 | 0.05351 |
| rs2295463 | 30166627 | 14q13.2 | KIAA0391 | Imputed | 0.4716 | 0.6512 | 0.3877 | 0.8045 |
| rs34619498 | 30166627 | 4q24 | EMCN | Imputed | 0.2414 | 0.2441 | 0.2936 | 0.6553 |
| rs140142800 | 30166627 | 10q21.2 | RHOBTB1 | Imputed | 0.8841 | 0.9169 | 0.4225 | 0.664 |
| rs78368496 | 30166627 | 11p15.1 | NAV2 | Imputed | 0.3588 | 0.7284 | 0.4222 | 0.9777 |
| rs147859879 | 30166627 | 5p15.33 | MRPL36 | Imputed | 0.6686 | 0.3167 | 0.557 | 0.12 |
| rs337527 | 30166627 | 9q22.33 | GABBR2 | Imputed | 0.2747 | 0.441 | 0.1885 | 0.8569 |
| rs11045392 | 22569225 | 12p12.2 | PDE3A | Genotyped | 0.5394 | 0.01316 | 0.06091 | 0.2323 |
| rs3794271 | 22569225 | 12p12.2 | SLCO1C1 | Genotyped | 0.4297 | 0.006361 | 0.04621 | 0.1448 |
Exome sequencing analysis
In a complementary analysis, we screened for rare variants by WES of 19 extreme responders and 55 non-responders. Additionally, patients from these extremes of the response distribution were compared with a large sample of more than 1500 ethnically matched control samples sequenced in the same laboratory using identical data processing methods. S5 Table shows the sample size and the number of variants tested in each comparison.
For all comparisons, the single-variant tests did not reveal any variant showing significant association with responder or non-responder status (Fig 5). The p-value distributions were non-uniform and tended to be less significant than expected under the null, probably owing to both the small sample sizes and the relatively low allele frequency distribution of the variants tested. For gene-based collapsing tests, the results were similar (Fig 6), with no individual gene showing significant association with therapeutic response after correcting for multiple testing. The top ten SNPs and genes most significantly associated with CZP response are shown in Tables 5 and 6.
Fig 5. Manhattan plots of significance scores from single-variant analysis of exome sequencing data of patients selected from extremes of the CZP response distribution.
In panel A, 19 CZP super-responders are compared to 55 non-responders. Panel B shows FET significance scores for 19 CZP super-responders compared to sequence data from a larger group of 1546 ethnically matched population controls and 55 non-responders, while panel C shows results of analysis of 55 CZP non-responders versus 1546 ethnically matched population controls and 19 super-responders. Horizontal red lines in figures A-C indicate the Bonferroni-corrected threshold for statistical significance.
Fig 6. Results of gene-based collapsing analysis of CZP responder and non-responder exome sequencing data.
(A) QQ plot of observed vs null FET significance values for CZP super-responders vs non-responders. (B) CZP super-responders are compared to sequence data from 1546 population controls and 55 non-responders, while C shows QQ plot of CZP non-responders compared to 1546 population controls and 19 super-responders. In all three analyses, no individual genes exceeded what was expected under the null hypothesis.
Table 5. Top ten most strongly associated individual variants from whole exome sequencing.
| Variant | Gene | P-value | |
|---|---|---|---|
| Responders vs. Non-responders + Controls | 6_80228535_G | LCA5 | 8.541E-006 |
| 2_26689619_A | OTOF | 5.046E-005 | |
| X_49082499_T | CACNA1F | 1.03E-004 | |
| 3_113376202_C | KIAA2018 | 1.34E-004 | |
| 9_136305552_A | ADAMTS13 | 1.34E-004 | |
| 11_3661588_3661588_1bp_INS | ART5 | 1.346E-004 | |
| 12_53448166_T | TENC1 | 1.362E-004 | |
| 19_47910643_T | MEIS3 | 1.362E-004 | |
| 5_140237030_G | PCDHA10 | 1.365E-004 | |
| 21_47836571_T | PCNT | 1.469E-004 | |
| Non-responders vs. Responders + Controls | 17_73520485_C | TSEN54 | 6.133E-006 |
| 11_76062206_C | PRKRIR | 3.811E-005 | |
| 17_72240177_G | TTYH2 | 7.369E-005 | |
| 9_35663069_T | ARHGEF39 | 1.487E-004 | |
| 17_33690160_T | SLFN11 | 1.487E-004 | |
| 19_15587345_C | PGLYRP | 1.852E-004 | |
| 2_167055393_C | SCN9A | 1.877E-004 | |
| 16_24564879_24564882_4bp_DEL | RBBP6 | 2.213E-004 | |
| 14_20711681_A | OR11H4 | 2.256E-004 | |
| 6_32797809_T | TAP2 | 2.35E-004 | |
| Responders vs. Non-responders | 11_71249125_71249145_21bp_INS | KRTAP5-8 | 1.257E-006 |
| 19_7528734_G | ARHGEF18 | 1.362E-004 | |
| 9_712156_G | KANK1 | 2.466E-004 | |
| 2_196720705_C | DNAH7 | 3.06E-004 | |
| 6_25914853_A | SLC17A2 | 4.977E-004 | |
| 19_51530741_G | KLK11 | 9.068E-004 | |
| 2_220072431_A | ZFAND2B | 9.082E-004 | |
| 2_32713706_T | BIRC6 | 9.897E-004 | |
| 6_26017542_C | HIST1H1A | 0.0011 | |
| 10_88930249_T | FAM35A | 0.0012 |
Table 6. Top ten most strongly associated genes in collapsing tests of multiple rare variants.
| Gene | P-value | |
|---|---|---|
| Responders vs. Non-responders + Controls | C11ORF93 | 3.637E-004 |
| CUL2 | 6.964E-004 | |
| PPME1 | 7.715E-004 | |
| EOGT | 8.394E-004 | |
| CROT | 8.583E-004 | |
| PIK3AP1 | 9.998E-004 | |
| OR5AU1 | 9.998E-004 | |
| SAMD10 | 0.0013 | |
| CLRN3 | 0.0016 | |
| KIAA1107 | 0.0016 | |
| Non-responders vs. Responders + Controls | KIAA1522 | 1.01E-004 |
| BTN1A1 | 1.703E-004 | |
| POF1B | 2.297E-004 | |
| IFNA4 | 3.292E-004 | |
| GGN | 3.786E-004 | |
| GREB1 | 3.887E-004 | |
| TSEN54 | 4.814E-004 | |
| EIF3B | 6.756E-004 | |
| ACTL9 | 6.779E-004 | |
| OR11H4 | 0.0011 | |
| Responders vs. Non-responders | CROT | 0.0034 |
| ADAMTS2 | 0.014 | |
| RBMXL3 | 0.014 | |
| ADAMTS16 | 0.0149 | |
| CLRN3 | 0.0149 | |
| CNGB1 | 0.0149 | |
| DUPD1 | 0.0149 | |
| FARSB | 0.0149 | |
| KCNK5 | 0.0149 | |
| KDM3B | 0.0149 |
Interestingly, among the top ten most highly associated results in the gene-based analysis was the potassium channel, subfamily K, member 5 (KCNK5) gene, which is known to be a critical factor in T cell activation [40]. In addition, KCNK5 expression has previously been reported to be strongly correlated with RA disease severity [41] and up-regulation of KCNK5 expression was found to be predictive of treatment failure in RA patients receiving the anti-IL-6 therapy tocilizumab [41], which inhibits RA inflammation through a different biological pathway to that targeted by anti-TNF therapies. In our collapsing analysis, non-synonymous variants in the KCNK5 gene were present in 16% (3/16) of CZP extreme responders, while no patients (0/55) from the CZP non-responder group were found to have any predicted functional variants in the KCNK5 gene.
Discussion
We have completed an exploratory pharmacogenetic analysis of a cohort of 302 RA patients of Western European descent treated with CZP. A two-stage analytical approach was employed, using high content beadchip genotyping analysis to examine the role of common human genetic variation, followed by a second stage consisting of WES of extreme responders and non-responders, to investigate the association of rare genetic variants with CZP treatment response.
In the first stage of the analysis examining common variation, we were unable to demonstrate robust associations with any of the clinical outcome measures. Two novel candidates (rs35355083 near SEMA4G and rs12287315 near SPON1), showed strong trends toward association with change in DAS28 ESR at week 12, but did not meet genome-wide statistical significance. These variants have not been associated with response to TNF inhibitors previously, however a low-frequency variant located in the FAR1-SPON1 intergenic region was previously reported to be associated with Etanercept response [42]. Imputation of ungenotyped SNPs near these suggestive associations revealed two additional variants with stronger associations with DAS28 ESR at week 12. While the p-values for these associations exceeded the original threshold for statistical significance, they were not found to be significant when the association analysis was performed using all common variants from a genome-wide imputation.
Semaphorins are a diverse group of proteins involved in regulation of cell movement and migration via interaction with cognate plexin or neuropilin receptors [43]. Recent studies have indicated that they play a role in many aspects of the immune system, including innate immunity and cell trafficking [44]. In the mouse, SEMA4G is required for cerebellar development [45], however, a clear role in auto-immune disease has yet to emerge.
Spondin 1(SPON1) is an extracellular matrix protein involved in regulation of neuronal outgrowth and inhibition of angiogenesis. It has been shown to bind to the extracellular domain of amyloid precursor protein and impairs cleavage by the beta secretase BACE 1, reducing beta-amyloid production [46]. Its expression has been reported to be elevated in osteoarthritis lesions in both humans and rodents, and its activation of TGF-beta in situ may promote osteoarthritis pathogenesis [47].
The involvement of these genes in immune signaling, as well as the reported role of spondin 1 in cartilage homeostasis [47], suggests that these associations may be biologically relevant. However, their failure to show significant association with CZP response after correction for multiple testing should prompt caution with regard to interpretation of the findings. As with all such candidates, replication of these SNPs in additional cohorts and biological validation will be required.
Whilst the genotyping stage of the analysis was intended to screen for common variants associated with CZP response, for complex diseases such as RA, such approaches typically require large cohorts and/or large effect sizes. One particularly compelling reason to search for rare variants in the study of therapeutic response to drugs is that drug response may be correlated with the underlying genetic cause of disease, and it is expected that much of the risk for human disease will be found in the rare-variant spectrum [48]. To address the risk of overlooking low frequency variants that contribute to CZP response, we implemented a WES approach in the second stage of the study. This approach enables identification of rare variants with larger effect sizes, despite the small sample sizes available. In this case, sequencing focused on 74 patients from the REALISTIC trial with extreme responses to CZP treatment; including 55 non-responders and 19 super-responders. Neither the single-variant tests nor the gene-based collapsing method identified any gene or variant that was significantly associated with CZP response. A modest, but non-significant, enrichment of variation was identified in the KCNK5 gene in our collapsing analysis. The previous implication of this gene in RA disease progression, as well as the specific observation that increases in KCNK5 expression are correlated with the failure of antibody-based therapies targeting similar inflammatory pathways involved in RA disease pathology, may suggest that additional investigation of KCNK5 as a modifier of RA therapies is merited.
With the caveat that KCNK5 and the two SNPs in SEMA4G and SPON1 may be worthy of follow up, we have failed to demonstrate the presence of any variants with a clinically robust association with response to the anti-TNF agent CZP. While the lack of statistically significant associations could result from the limited size of our study, this finding is also consistent with many previous candidate gene and genome-wide efforts to discover genetic determinants of response to anti-TNF agents. Researchers have either been unable to discover or replicate initial findings in clinically relevant cohorts, or the statistical performance of the candidates has not been sufficiently compelling to justify follow up or clinical implementation. A post hoc analysis of the estimated statistical power for the GWAS component of our study indicated it had over 80% power to detect common variants with a MAF of 25% and a relative risk of 2.5, but was only sufficiently powered to detect more rare variants with a MAF of 5% when the relative risk was over 5 (S3A Fig). Similarly, the WES portion of our study was likely to be underpowered to detect enrichments of rare variants with moderate effect sizes (RR<6.0; S3B Fig).
RA is a heterogeneous disease with complex genetic and environmental etiology. Although we have focused our analysis on a largely homogenous Caucasian cohort of 302 patients, the study population comprised patients with differing clinical characteristics at entry. We have accommodated variability in disease severity and duration, and use of other medications in the analysis but were not able to adjust for the variability in biology underpinning disease heterogeneity and response/non-response. In addition, the development of antibodies against TNF inhibitors has recently been recognized a significant contributor to treatment failure [49, 50]. Our study did not account for the induction of antibodies against CZP. It therefore remains a possibility that heritable biomarkers predictive of risk do exist for sub-populations within diverse RA populations such as this one, but that their penetrance is diluted to an extent whereby they are not detectable.
Further complexity is added by the routine use of composite outcome measures such as DAS28 and ACR20, the primary endpoints used in this clinical study. They are comprised of a mixture of objective physical measurements, laboratory biomarker data, and subjective patient assessments. Robust associations may be discernible with a more refined focus on a single objective endpoint, although it should be noted that this may be misleading in a mixed patient population.
The goal of personalizing RA therapy is to identify patients where the likelihood of a beneficial outcome is optimized and the risk of adverse events is reduced or eliminated. Many studies with conflicting and ambiguous outcomes have been published investigating the pharmacogenomics of etanercept, infliximab and adalimumab in RA populations, and to date there have been no predictive anti-TNF genomic biomarkers established for clinical use. This report, the first pharmacogenomic analysis of CZP response, is in accord with these earlier efforts, and has been unable to demonstrate compelling evidence for the existence of genomic biomarkers of value in targeting this anti-TNF to a responsive patient sub-population.
In diseases such as RA, where there are complex, polygenic contributions to pathology and response to therapeutic intervention, it remains a possibility that combinations of clinical, environmental, and molecular markers will hold more promise for managing therapy.
Supporting information
Manhattan and quantile-quantile plots of significance scores for association with (A) ACR20 week 6 response, (B) logistic regression of reduction in DAS28 ESR week 6, and (C) linear regression of DAS28 ESR at week 6. The red line indicates the Bonferroni-adjusted threshold for statistical significance. Genomic inflation factor (λ) for each analysis is indicated in the bottom right of quantile-quantile plots.
(TIF)
Manhattan and quantile-quantile plots of significance scores for association with (A) ACR20 week 6 response, (B) logistic regression of reduction in DAS28 ESR week 6, (C) linear regression of DAS28 ESR at week 6, and (D) linear regression of DAS28 ESR at week 12. The red line indicates the Bonferroni-adjusted threshold for statistical significance. Genomic inflation factor (λ) for each analysis is indicated in the bottom right of quantile-quantile plots.
(TIF)
Graphs of statistical power estimated across a range of minor allele frequencies and effect sizes for (A) the GWAS of common SNPs and ACR20 response, and (B) collapsing analysis of rare variants identified by WES for non-responders vs population controls and super-responders. For rare variant collapsing analysis, cMAF indicates the cumulative minor allele frequency summed across multiple rare variants. All power calculations were performed using CaTS.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data can be found here: https://www.gwascentral.org/study/HGVST432.
Funding Statement
This study was funded by UCB Pharma. DBG received partial salary support and funding for the research materials described in our study. The funder also provided support in the form of salaries for authors IRW, CP, CC, and DM, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Woolf AD, Pfleger B. Burden of major musculoskeletal conditions. Bull World Health Organ. 2003;81(9):646–56. Epub 2004/01/09. . [PMC free article] [PubMed] [Google Scholar]
- 2.Singh JA, Furst DE, Bharat A, Curtis JR, Kavanaugh AF, Kremer JM, et al. 2012 update of the 2008 American College of Rheumatology recommendations for the use of disease-modifying antirheumatic drugs and biologic agents in the treatment of rheumatoid arthritis. Arthritis Care Res (Hoboken). 2012;64(5):625–39. Epub 2012/04/05. doi: 10.1002/acr.21641 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maini R, St Clair EW, Breedveld F, Furst D, Kalden J, Weisman M, et al. Infliximab (chimeric anti-tumour necrosis factor alpha monoclonal antibody) versus placebo in rheumatoid arthritis patients receiving concomitant methotrexate: a randomised phase III trial. ATTRACT Study Group. Lancet. 1999;354(9194):1932–9. Epub 2000/01/06. doi: 10.1016/s0140-6736(99)05246-0 . [DOI] [PubMed] [Google Scholar]
- 4.Lipsky PE, van der Heijde DM, St Clair EW, Furst DE, Breedveld FC, Kalden JR, et al. Infliximab and methotrexate in the treatment of rheumatoid arthritis. Anti-Tumor Necrosis Factor Trial in Rheumatoid Arthritis with Concomitant Therapy Study Group. N Engl J Med. 2000;343(22):1594–602. Epub 2000/11/30. doi: 10.1056/NEJM200011303432202 . [DOI] [PubMed] [Google Scholar]
- 5.Weinblatt ME, Keystone EC, Furst DE, Moreland LW, Weisman MH, Birbara CA, et al. Adalimumab, a fully human anti-tumor necrosis factor alpha monoclonal antibody, for the treatment of rheumatoid arthritis in patients taking concomitant methotrexate: the ARMADA trial. Arthritis Rheum. 2003;48(1):35–45. Epub 2003/01/16. doi: 10.1002/art.10697 . [DOI] [PubMed] [Google Scholar]
- 6.Silman AJ, Hochberg MC. Epidemiology of the rheumatic diseases. 2nd ed. Oxford; New York: Oxford University Press; 2001. xii, 382 p. p. [Google Scholar]
- 7.Barton A, Worthington J. Genetic susceptibility to rheumatoid arthritis: an emerging picture. Arthritis Rheum. 2009;61(10):1441–6. Epub 2009/10/01. doi: 10.1002/art.24672 . [DOI] [PubMed] [Google Scholar]
- 8.Raychaudhuri S. Recent advances in the genetics of rheumatoid arthritis. Curr Opin Rheumatol. 2010;22(2):109–18. Epub 2010/01/16. doi: 10.1097/BOR.0b013e328336474d . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowes JD, Potter C, Gibbons LJ, Hyrich K, Plant D, Morgan AW, et al. Investigation of genetic variants within candidate genes of the TNFRSF1B signalling pathway on the response to anti-TNF agents in a UK cohort of rheumatoid arthritis patients. Pharmacogenet Genomics. 2009;19(4):319–23. Epub 2009/03/06. doi: 10.1097/FPC.0b013e328328d51f . [DOI] [PubMed] [Google Scholar]
- 10.Umicevic Mirkov M, Cui J, Vermeulen SH, Stahl EA, Toonen EJ, Makkinje RR, et al. Genome-wide association analysis of anti-TNF drug response in patients with rheumatoid arthritis. Ann Rheum Dis. 2013;72(8):1375–81. Epub 2012/12/13. doi: 10.1136/annrheumdis-2012-202405 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuchacovich M, Soto L, Edwardes M, Gutierrez M, Llanos C, Pacheco D, et al. Tumour necrosis factor (TNF)alpha -308 G/G promoter polymorphism and TNFalpha levels correlate with a better response to adalimumab in patients with rheumatoid arthritis. Scand J Rheumatol. 2006;35(6):435–40. Epub 2007/03/09. doi: 10.1080/03009740600904284 . [DOI] [PubMed] [Google Scholar]
- 12.Lee YH, Rho YH, Choi SJ, Ji JD, Song GG. Association of TNF-alpha -308 G/A polymorphism with responsiveness to TNF-alpha-blockers in rheumatoid arthritis: a meta-analysis. Rheumatol Int. 2006;27(2):157–61. Epub 2006/08/16. doi: 10.1007/s00296-006-0175-7 . [DOI] [PubMed] [Google Scholar]
- 13.O’Rielly DD, Roslin NM, Beyene J, Pope A, Rahman P. TNF-alpha-308 G/A polymorphism and responsiveness to TNF-alpha blockade therapy in moderate to severe rheumatoid arthritis: a systematic review and meta-analysis. Pharmacogenomics J. 2009;9(3):161–7. Epub 2009/04/15. doi: 10.1038/tpj.2009.7 . [DOI] [PubMed] [Google Scholar]
- 14.Pavy S, Toonen EJ, Miceli-Richard C, Barrera P, van Riel PL, Criswell LA, et al. Tumour necrosis factor alpha -308G->A polymorphism is not associated with response to TNFalpha blockers in Caucasian patients with rheumatoid arthritis: systematic review and meta-analysis. Ann Rheum Dis. 2010;69(6):1022–8. Epub 2009/12/08. doi: 10.1136/ard.2009.117622 . [DOI] [PubMed] [Google Scholar]
- 15.Liu C, Batliwalla F, Li W, Lee A, Roubenoff R, Beckman E, et al. Genome-wide association scan identifies candidate polymorphisms associated with differential response to anti-TNF treatment in rheumatoid arthritis. Mol Med. 2008;14(9–10):575–81. Epub 2008/07/11. doi: 10.2119/2008-00056.Liu . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plant D, Bowes J, Potter C, Hyrich KL, Morgan AW, Wilson AG, et al. Genome-wide association study of genetic predictors of anti-tumor necrosis factor treatment efficacy in rheumatoid arthritis identifies associations with polymorphisms at seven loci. Arthritis Rheum. 2011;63(3):645–53. Epub 2010/11/10. doi: 10.1002/art.30130 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krintel SB, Palermo G, Johansen JS, Germer S, Essioux L, Benayed R, et al. Investigation of single nucleotide polymorphisms and biological pathways associated with response to TNFalpha inhibitors in patients with rheumatoid arthritis. Pharmacogenet Genomics. 2012;22(8):577–89. Epub 2012/05/10. doi: 10.1097/FPC.0b013e3283544043 . [DOI] [PubMed] [Google Scholar]
- 18.Cui J, Stahl EA, Saevarsdottir S, Miceli C, Diogo D, Trynka G, et al. Genome-wide association study and gene expression analysis identifies CD84 as a predictor of response to etanercept therapy in rheumatoid arthritis. PLoS Genet. 2013;9(3):e1003394. Epub 2013/04/05. doi: 10.1371/journal.pgen.1003394 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suarez-Gestal M, Perez-Pampin E, Calaza M, Gomez-Reino JJ, Gonzalez A. Lack of replication of genetic predictors for the rheumatoid arthritis response to anti-TNF treatments: a prospective case-only study. Arthritis Res Ther. 2010;12(2):R72. Epub 2010/04/29. doi: 10.1186/ar2990 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marquez A, Ferreiro-Iglesias A, Davila-Fajardo CL, Montes A, Pascual-Salcedo D, Perez-Pampin E, et al. Lack of validation of genetic variants associated with anti-tumor necrosis factor therapy response in rheumatoid arthritis: a genome-wide association study replication and meta-analysis. Arthritis Res Ther. 2014;16(2):R66. Epub 2014/03/13. doi: 10.1186/ar4504 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Rodriguez R, Perez-Pampin E, Marquez A, Blanco FJ, Joven B, Carreira P, et al. Validation study of genetic biomarkers of response to TNF inhibitors in rheumatoid arthritis. PLoS One. 2018;13(5):e0196793. Epub 2018/05/08. doi: 10.1371/journal.pone.0196793 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honne K, Hallgrimsdottir I, Wu C, Sebro R, Jewell NP, Sakurai T, et al. A longitudinal genome-wide association study of anti-tumor necrosis factor response among Japanese patients with rheumatoid arthritis. Arthritis Res Ther. 2016;18:12. Epub 2016/01/19. doi: 10.1186/s13075-016-0920-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Julia A, Fernandez-Nebro A, Blanco F, Ortiz A, Canete JD, Maymo J, et al. A genome-wide association study identifies a new locus associated with the response to anti-TNF therapy in rheumatoid arthritis. Pharmacogenomics J. 2016;16(2):147–50. Epub 2015/04/22. doi: 10.1038/tpj.2015.31 . [DOI] [PubMed] [Google Scholar]
- 24.Ferreiro-Iglesias A, Montes A, Perez-Pampin E, Canete JD, Raya E, Magro-Checa C, et al. Evaluation of 12 GWAS-drawn SNPs as biomarkers of rheumatoid arthritis response to TNF inhibitors. A potential SNP association with response to etanercept. PLoS One. 2019;14(2):e0213073. Epub 2019/03/01. doi: 10.1371/journal.pone.0213073 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Acosta-Colman I, Palau N, Tornero J, Fernandez-Nebro A, Blanco F, Gonzalez-Alvaro I, et al. GWAS replication study confirms the association of PDE3A-SLCO1C1 with anti-TNF therapy response in rheumatoid arthritis. Pharmacogenomics. 2013;14(7):727–34. Epub 2013/05/09. doi: 10.2217/pgs.13.60 . [DOI] [PubMed] [Google Scholar]
- 26.Aterido A, Canete JD, Tornero J, Blanco F, Fernandez-Gutierrez B, Perez C, et al. A Combined Transcriptomic and Genomic Analysis Identifies a Gene Signature Associated With the Response to Anti-TNF Therapy in Rheumatoid Arthritis. Front Immunol. 2019;10:1459. Epub 2019/07/18. doi: 10.3389/fimmu.2019.01459 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan Y, Zhang H, Quang D, Wang Z, Parker SCJ, Pappas DA, et al. Machine Learning to Predict Anti-Tumor Necrosis Factor Drug Responses of Rheumatoid Arthritis Patients by Integrating Clinical and Genetic Markers. Arthritis Rheumatol. 2019;71(12):1987–96. Epub 2019/07/26. doi: 10.1002/art.41056 . [DOI] [PubMed] [Google Scholar]
- 28.Koo BS, Eun S, Shin K, Yoon H, Hong C, Kim DH, et al. Machine learning model for identifying important clinical features for predicting remission in patients with rheumatoid arthritis treated with biologics. Arthritis Res Ther. 2021;23(1):178. Epub 2021/07/08. doi: 10.1186/s13075-021-02567-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Umicevic Mirkov M, Janss L, Vermeulen SH, van de Laar MA, van Riel PL, Guchelaar HJ, et al. Estimation of heritability of different outcomes for genetic studies of TNFi response in patients with rheumatoid arthritis. Ann Rheum Dis. 2015;74(12):2183–7. Epub 2014/08/13. doi: 10.1136/annrheumdis-2014-205541 . [DOI] [PubMed] [Google Scholar]
- 30.Weinblatt ME, Fleischmann R, Huizinga TW, Emery P, Pope J, Massarotti EM, et al. Efficacy and safety of certolizumab pegol in a broad population of patients with active rheumatoid arthritis: results from the REALISTIC phase IIIb study. Rheumatology (Oxford). 2012;51(12):2204–14. Epub 2012/08/28. doi: 10.1093/rheumatology/kes150 . [DOI] [PubMed] [Google Scholar]
- 31.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. Epub 2007/08/19. doi: 10.1086/519795 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–9. Epub 2006/07/25. doi: 10.1038/ng1847 . [DOI] [PubMed] [Google Scholar]
- 33.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9(2):179–81. Epub 2011/12/06. doi: 10.1038/nmeth.1785 . [DOI] [PubMed] [Google Scholar]
- 34.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. Epub 2009/06/23. doi: 10.1371/journal.pgen.1000529 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109–14. Epub 2011/11/15. doi: 10.1093/nar/gkr988 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. Epub 2009/05/20. doi: 10.1093/bioinformatics/btp324 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. Epub 2010/07/21. doi: 10.1101/gr.107524.110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–8. Epub 2011/04/12. doi: 10.1038/ng.806 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83(3):311–21. Epub 2008/08/12. doi: 10.1016/j.ajhg.2008.06.024 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meuth SG, Bittner S, Meuth P, Simon OJ, Budde T, Wiendl H. TWIK-related acid-sensitive K+ channel 1 (TASK1) and TASK3 critically influence T lymphocyte effector functions. J Biol Chem. 2008;283(21):14559–70. Epub 2008/04/01. doi: 10.1074/jbc.M800637200 . [DOI] [PubMed] [Google Scholar]
- 41.Bittner S, Bobak N, Feuchtenberger M, Herrmann AM, Gobel K, Kinne RW, et al. Expression of K2P5.1 potassium channels on CD4+ T lymphocytes correlates with disease activity in rheumatoid arthritis patients. Arthritis Res Ther. 2011;13(1):R21. Epub 2011/02/15. doi: 10.1186/ar3245 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Massey J, Plant D, Hyrich K, Morgan AW, Wilson AG, Spiliopoulou A, et al. Genome-wide association study of response to tumour necrosis factor inhibitor therapy in rheumatoid arthritis. Pharmacogenomics J. 2018;18(5):657–64. Epub 2018/09/01. doi: 10.1038/s41397-018-0040-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takamatsu H, Kumanogoh A. Diverse roles for semaphorin-plexin signaling in the immune system. Trends Immunol. 2012;33(3):127–35. Epub 2012/02/14. doi: 10.1016/j.it.2012.01.008 . [DOI] [PubMed] [Google Scholar]
- 44.Suzuki K, Kumanogoh A, Kikutani H. Semaphorins and their receptors in immune cell interactions. Nat Immunol. 2008;9(1):17–23. Epub 2007/12/19. doi: 10.1038/ni1553 . [DOI] [PubMed] [Google Scholar]
- 45.Maier V, Jolicoeur C, Rayburn H, Takegahara N, Kumanogoh A, Kikutani H, et al. Semaphorin 4C and 4G are ligands of Plexin-B2 required in cerebellar development. Mol Cell Neurosci. 2011;46(2):419–31. Epub 2010/12/03. doi: 10.1016/j.mcn.2010.11.005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho A, Sudhof TC. Binding of F-spondin to amyloid-beta precursor protein: a candidate amyloid-beta precursor protein ligand that modulates amyloid-beta precursor protein cleavage. Proc Natl Acad Sci U S A. 2004;101(8):2548–53. Epub 2004/02/26. doi: 10.1073/pnas.0308655100 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Attur MG, Palmer GD, Al-Mussawir HE, Dave M, Teixeira CC, Rifkin DB, et al. F-spondin, a neuroregulatory protein, is up-regulated in osteoarthritis and regulates cartilage metabolism via TGF-beta activation. FASEB J. 2009;23(1):79–89. Epub 2008/09/11. doi: 10.1096/fj.08-114363 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40(6):695–701. Epub 2008/05/30. doi: 10.1038/ng.f.136 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mok CC, Tsai WC, Chen DY, Wei JC. Immunogenicity of anti-TNF biologic agents in the treatment of rheumatoid arthritis. Expert Opin Biol Ther. 2016;16(2):201–11. Epub 2015/11/13. doi: 10.1517/14712598.2016.1118457 . [DOI] [PubMed] [Google Scholar]
- 50.Balsa A, Sanmarti R, Rosas J, Martin V, Cabez A, Gomez S, et al. Drug immunogenicity in patients with inflammatory arthritis and secondary failure to tumour necrosis factor inhibitor therapies: the REASON study. Rheumatology (Oxford). 2018;57(4):688–93. Epub 2018/01/25. doi: 10.1093/rheumatology/kex474 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Manhattan and quantile-quantile plots of significance scores for association with (A) ACR20 week 6 response, (B) logistic regression of reduction in DAS28 ESR week 6, and (C) linear regression of DAS28 ESR at week 6. The red line indicates the Bonferroni-adjusted threshold for statistical significance. Genomic inflation factor (λ) for each analysis is indicated in the bottom right of quantile-quantile plots.
(TIF)
Manhattan and quantile-quantile plots of significance scores for association with (A) ACR20 week 6 response, (B) logistic regression of reduction in DAS28 ESR week 6, (C) linear regression of DAS28 ESR at week 6, and (D) linear regression of DAS28 ESR at week 12. The red line indicates the Bonferroni-adjusted threshold for statistical significance. Genomic inflation factor (λ) for each analysis is indicated in the bottom right of quantile-quantile plots.
(TIF)
Graphs of statistical power estimated across a range of minor allele frequencies and effect sizes for (A) the GWAS of common SNPs and ACR20 response, and (B) collapsing analysis of rare variants identified by WES for non-responders vs population controls and super-responders. For rare variant collapsing analysis, cMAF indicates the cumulative minor allele frequency summed across multiple rare variants. All power calculations were performed using CaTS.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data can be found here: https://www.gwascentral.org/study/HGVST432.