Abstract
Background:
The observance of non-random space–time groupings of childhood cancer has been a concern of health professionals and the general public for decades. Many childhood cancers are suspected to have initiated in utero; therefore, we examined the spatial–temporal randomness of the birthplace of children who later developed cancer.
Methods:
We performed a space–time cluster analysis using birth addresses of 5,896 cases and 23,369 population-based, age-, sex- and race/ethnicity-matched controls in California from 1997–2007, evaluating 20 types of childhood cancer and three a priori designated subgroups of childhood acute lymphoblastic leukemia (ALL). We analyzed data using a newly designed semiparametric analysis program, ClustR, and a common algorithm, SaTScan.
Results:
We observed evidence for non-random space–time clustering for ALL diagnosed at 2–6 years of age in the South San Francisco Bay Area (ClustR p=0.04, SaTScan p=0.07), and malignant gonadal germ cell tumors in a region of Los Angeles (ClustR p=0.03, SaTScan p=0.06). ClustR did not identify evidence of clustering for other childhood cancers, though SaTScan suggested some clustering for Hodgkin lymphoma (p=0.09), astrocytoma (p=0.06) and retinoblastoma (p=0.06).
Conclusion:
Our study provides evidence that childhood ALL diagnosed at 2–6 years and malignant gonadal germ cell tumors sporadically occurs in non-random space–time clusters. Further research is warranted to identify epidemiologic features that may inform the underlying etiology.
Keywords: acute lymphoblastic leukemia, gonadal germ cell tumors, childhood cancer, space–time clustering, ClustR, SaTScan
Background
Cancer Etiology.
In the United States, cancer is the leading cause of death by disease for children past infancy1. Acute lymphoblastic leukemia (ALL) is the most common form of childhood cancer, and consequently the most commonly researched childhood cancer.
Multiple studies have concluded that ALL is frequently initiated by a genetic translocation in utero 2,3, suggesting the importance of parental and in-utero exposures in the etiology of ALL. A far smaller number of studies have also suggested that testicular cancer, a subset of malignant gonadal germ cell tumors, may have a fetal origin4. Other childhood cancers have not received the same investigative efforts in part due to the rarity and therefore difficulty of obtaining a sufficient number of cases. Therefore more is known about the etiology of ALL than most other childhood cancers, and we use the etiologic understanding of ALL as a framework for our investigations. A number of environmental exposures, such as to pesticides and traffic emissions, before and during pregnancy have been associated with ALL5. Recently, a strong association was reported between in utero cytomegalovirus infection and subsequent development of ALL (odds ratio (OR) = 3.71, 95% confidence interval (CI): 1.71, 8.95), particularly in Hispanic children (OR: 5.90, 95% CI: 1.89, 25.96), further highlighting the etiologic significance of exposures during pregnancy6.
ALL exhibits heterogeneity across demographic groups. Hispanic children experience the highest risk of ALL, while black children experience the lowest (1,7). Additionally, age of diagnosis is associated with specific genetic subtypes of ALL, with diagnosis as an infant (≤ 1 year of age), child (2 to 6 years of age), and adolescent or adult representing three divergent distributions of molecular subtype8,9. The diversity of ALL risk and subtype across demographic groups suggests a variety of etiologic pathways may be at play.
Cluster investigations have a long history in public health. In general most cluster investigations have not identified etiologic agents associated with the putative cluster. Many of these investigations have suffered from a variety of epidemiologic flaws10,11. Childhood leukemia has been a focus of both public health and academic investigations for decades and has been observed to form ‘clusters’ or non-random space–time groupings, which can be leveraged to gain insight about potential causes of the disease. While less studied, other childhood cancers, such as lymphomas and central nervous system (CNS) tumors, have also exhibited these patterns 12,13. Many other childhood cancers, such as malignant gonadal germ cell tumors, have not been examined for clustering due to their rarity. In addition, cluster studies in the past have generally followed public concerns, leading to retrospective or ad hoc cluster analyses rather than agnostic evaluations. Most of these investigations have not uncovered the underlying cause of the cluster, if one did indeed exist. However, some cluster investigations have nominated putative causes such as the cluster in Woburn, Mass, USA, where trichloroethylene (TCE) and other chlorinated compounds were found in the water supply and associated with disease 14. A poignant example of an investigation into birth clustering in Japan led to the independent identification of human t-lymphotrophic virus (HTLV)-induced adult T-cell leukemia15,16. This example highlights the power of combining cluster analysis with molecular investigations. Although we do not have direct evidence for in utero initiation of other childhood cancers beyond childhood ALL we suspect that the in utero and early life time period may be a critical programming window for the initiation of multiple cancers.
Cluster Analysis.
Cluster analysis is a statistical tool that allows researchers to explore non-random groupings of an outcome, such as cancer, across a dimension like the spatial plane without designating a hypothesized association a priori. This analysis is well suited to study diseases that are infectious or may be influenced by environmental factors. For example, cluster findings from the Centers for Disease Control and Prevention linked leukemia and lymphoma to the spread of infectious agents in several communities across the United States17. Spatial cluster analysis, which identifies high incidence areas across the spatial plane, has been a common approach in childhood cancer research13,18–23. Some studies have also employed spatial–temporal cluster analysis, which identifies high incidence areas in both space and time12,24–30. Both types of studies have used a variety of methods, ranging from simple statistical tests to software that employ complex sampling algorithms.
Today, SaTScan, a publicly available software tool developed by Martin Kulldorff, is widely utilized for cluster analysis31. SaTScan employs scan statistics to identify clusters using user-specified models such as the Bernoulli model and can output results for visualization by ArcGIS or Google Maps (SaTScan™ User Guide, 2015). SaTScan has been used in three studies that found significant spatial clusters of pediatric leukemia in Florida, Argentina and Spain20,25,30 and one study that found significant space-time clusters for central nervous system tumors and neuroblastomas in Spain12.
The present study evaluated birth addresses for space–time clustering of 20 different types of childhood cancer in California, the most populous state in the United States with 39 million people as of 2018, and it is the first agnostic statewide childhood cancer cluster analysis conducted for the California population. We employed SaTScan and developed a novel methodology, ClustR, to conduct space–time cluster analysis. ClustR employs a unique statistical methodology, runs in the R framework, and provides more information about the analysis to the user. Furthermore, compared to SaTScan, the tool is faster, easier to implement and interpret, and has trade-offs in sensitivity and specificity across different cluster types 32.
Methods
Data.
We obtained data from the California Department of Public Health under the Childhood Cancer Record Linkage Project (CCRLP) which has been described in detail elsewhere 33. Briefly, the CCRLP links birth records maintained by the California Department of Public Health with data from the California Cancer Registry. Cases were defined as any child born between 1 January 1978 and 31 December 2009 in the state of California and diagnosed with cancer at the ages of 0–14 years. For each case, up to four controls were randomly selected from the birth records, matched on year and month of birth, sex, and race or ethnicity (non-Hispanic white, non-Hispanic black, Hispanic/Latino, Asian/Pacific Islander, other). Matching varied by year depending on information recorded on the birth certificate. The child was recorded as Hispanic if the mother or father was recorded as Hispanic on the birth certificate. Matching was performed by first matching by child’s recorded race; if child’s race was unavailable maternal race was used, and if if maternal race was unavailable, paternal race was used. This analysis includes birth years 1997-2007 where detailed address information is available. We analyzed all childhood cancers, based on the International Classification of Childhood Cancer (World Health Organization, 2008), with at least 50 cases in the dataset. Analyses were also performed on three subgroups of ALL cases and their respective controls: Hispanic cases, cases diagnosed at 0 to 1 years of age, and cases diagnosed at 2 to 6 years of age. These groups were chosen a priori to investigate possible etiologically relevant subtypes of interest9. These age strata were only applied to ALL; these age and race strata are not explicitly implicated as etiologically relevant in other childhood cancers and, because of lower incidence, sample size limits investigating additional strata in non-ALL cancer groups. Maternal residential addresses from birth records were geocoded using ArcGIS (ESRI v10.2, Redlands, CA) and linked to the 2000 census to obtain neighborhood characteristics at the block group level, including median household income, the proportion of adults who did not complete high school, and population density. The study protocol was approved by the Institutional Review Boards at the California Health and Human Services Agency and the University of California at Berkeley where the statistical analyses took place.
ClustR.
ClustR is a R package designed to conduct cluster analysis using case–control data. ClustR conducts the main analysis by simulating a null distribution of study subjects over the underlying population distribution, comparing case–control data to the generated null distribution, and generating descriptive and inferential statistics. In space–time cluster analysis, the null distribution arises from data where disease status is randomly distributed across space and time with respect to controls. The random distribution is created by scrambling the case control status of the actual data to be analyzed. The two chi-square distributions are compared to identify if any samples from the provided data would be highly unusual for the null distribution of the randomized data. Further detail can be found in 32,which compares the sensitivity and specificity of ClustR to SaTScan across a variety of cluster types. We ran ClustR using five bootstraps of 1000 samples each where samples could have radii of 0.05, 0.1, 0.2, 0.5, and 1 decimal degrees and durations of 75, 175, 275, 375, and 500 days. We allowed ClustR to test many different sample sizes given the variety in sizes and durations of clusters found in previous research. Significant clusters were samples with p-values < 0.05 after adjustment for multiple testing using the Benjamini-Hochberg procedure34. While the utility of p-values in epidemiology is debatable, we chose to include this metric to represent our a priori cutoff of a significant difference, while correcting for multiple testing, between case and control series to aid in interpretation.
SaTScan.
Since ClustR and SaTScan have different strengths and weaknesses32, SaTScan was also employed as a parallel analysis tool. As SatScan is designed for aggregated data, we aggregated datasets by zip code tabulation area (ZCTA, a geographical boundary used by the U.S. Census Bureau which approximates zip codes) and month using total counts of cases and controls. We ran SaTScan under the following settings: type of analysis: retrospective space–time analysis; scanning for: clusters with high or low rates; probability model: the Bernoulli model; time aggregation and precision: month. All other settings were left at default.
We defined a priori that any analysis group that reached the statistical significance at p≤0.05 in either ClustR or SaTScan would be deemed significant.
Results
The final dataset included 5,896 cases and 23,369 controls (Table 1). Individuals removed from the analysis included cases who had been previously diagnosed with another cancer and their matched controls (n = 1,142), and subjects without complete latitude and longitude information for maternal address in birth records (presumably maternal residence during pregnancy and child’s address at birth, n = 587). Finally, we excluded cases of cancers that had fewer than 50 cases in the dataset and their matched controls (n = 2,348). Only 10.1% of cases had fewer than four controls, and none had fewer than two controls. On average, cases had older mothers and fathers, had birth residences in wealthier, more educated, and had a different distribution of fathers’ race, compared to controls. Subjects who were excluded for missing coordinate data tended to have younger mothers and were born in more recent years, compared to those included (eTable 1).
Table 1.
Characteristics of the Study Population, California (1997–2007)
| Characteristic | Cases | Controls | |
|---|---|---|---|
| N | 5896 | 23369 | |
| Birth year | 1997–1999 | 2046 (35%) | 8114 (35%) |
| 2000–2002 | 1718 (29%) | 6805 (29%) | |
| 2003–2005 | 1454 (25%) | 5760 (25%) | |
| 2006–2007 | 678 (12%) | 2690 (12%) | |
| Age at diagnosis | 0–1 | 1853 (31%) | NA |
| 2–6 | 3187 (54%) | NA | |
| >6 | 856 (15%) | NA | |
| Sex | Male | 3278 (56%) | 12979 (56%) |
| Female | 2618 (44%) | 10390 (45%) | |
| Age of mother | Mean (SD) | 28.40 (6.4) | 27.96 (6.3) |
| Age of father | Mean (SD) | 31.21 (7.2) | 30.85 (7.1) |
| Median household income ($) | Mean (SD) | 49828.4 (24708) | 48433.53 (24221) |
| Proportion of Adults without a GED (%) | Mean (SD) | 0.29 (0.2) | 0.30 (0.2) |
| Population density (persons/m2) | Mean (SD) | 0.0038 (0.0040) | 0.0040 (0.0041) |
| Race/ethnicity of mother | White, non-Hispanic | 2097 (36%) | 8078 (35%) |
| Hispanic | 2704 (46%) | 10886 (47%) | |
| Black, non-Hispanic | 288 (5%) | 1158 (5%) | |
| Asian & Pacific Islander, non-Hispanic | 578 (10%) | 2339 (10%) | |
| Alaskan & Native American, non-Hispanic | 25 (1%) | 94 (1%) | |
| Other | 204 (4%) | 814 (4%) | |
| Race/ethnicity of father | White, non-Hispanic | 2016 (34%) | 7587 (33%) |
| Hispanic | 2555 (43%) | 10196 (44%) | |
| Black, non-Hispanic | 335 (6%) | 1300 (6%) | |
| Asian & Pacific Islander, non-Hispanic | 485 (8%) | 2095 (9%) | |
| Alaskan & Native American, non-Hispanic | 18 (1%) | 100 (1%) | |
| Other | 487 (8%) | 2091 (9%) |
Abbreviations: GED=general education diploma, SD=standard deviation.
After exclusions, the types of cancer analyzed in this study included: lymphoid leukemias, acute myeloid leukemia, Hodgkin lymphoma, non-Hodgkin lymphoma, Burkitt lymphoma, miscellaneous lymphoreticular neoplasms, ependymomas and choroid plexus tumor, astrocytomas, intracranial and intraspinal embryonal tumors, other gliomas, neuroblastoma and ganglioneuroblastoma, retinoblastoma, nephroblastoma and other non-epithelial renal tumors, hepatoblastoma, osteosarcomas, rhabdomyosarcomas, other specified soft tissue sarcomas (besides rhabdomyosarcomas and fibrous neoplasms), malignant extracranial and extragonadal germ cell tumors, malignant gonadal germ cell tumors, and malignant melanomas. Demographic characteristics for the individual cancer subsets can be found in the supplementary materials (eTables 2–24).
Acute Lymphoblastic Leukemia (ALL)
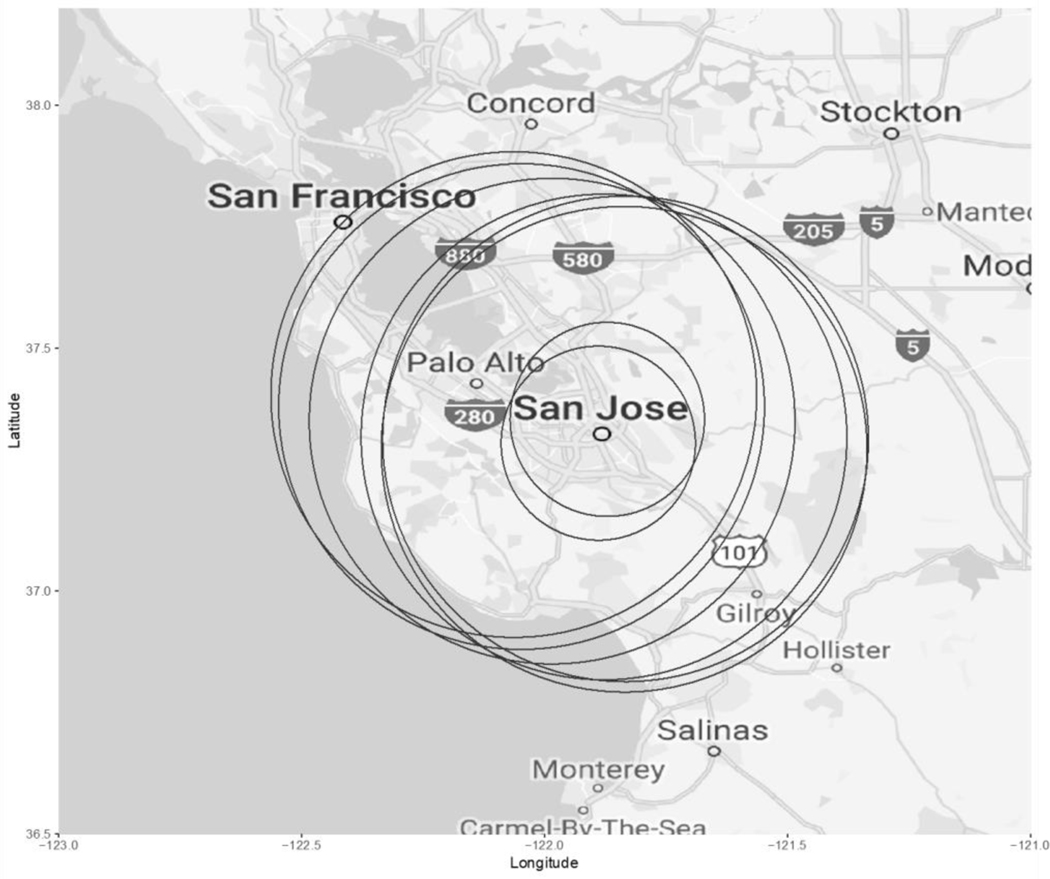
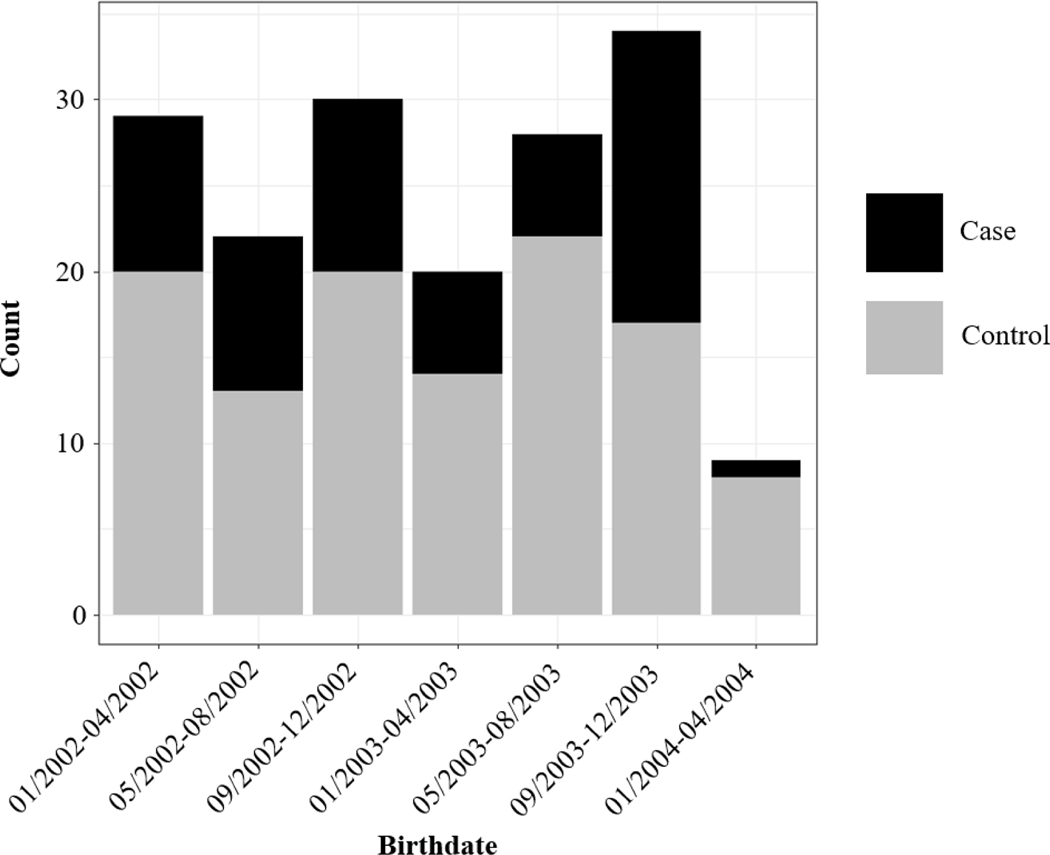
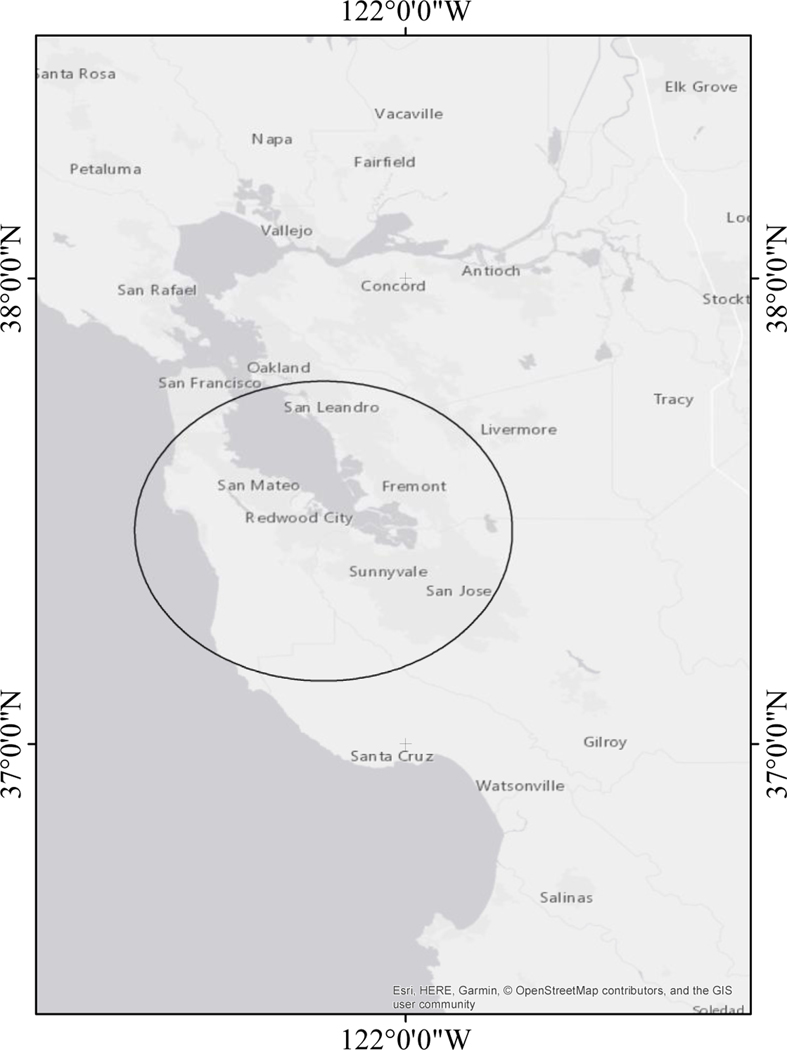
ALL overall demonstrated some marginal clustering in ClustR (p = 0.07), while SaTSCan did not find significant clustering for ALL overall (p = 0.63). Among ALL cases that were diagnosed at the age of 2–6 years and their respective controls (n = 7,113), ClustR identified eight distinct samples which yielded adjusted p-values of 0.04. These samples overlapped spatially in the neighboring regions of San Jose, Union City, and Fremont (Figure 1). Birthdates in the samples ranged from January 2002 to March 2004 (Figure 2). SaTScan identified a cluster in the same region from January 2002 to January 2004 with a p-value of 0.07 (Figure 3). Neither of the other subgroups, defined by age (cases diagnosed ages 0–1 years) and racial/ethnic background (Hispanic), showed any evidence of clustering.
Figure 1.
Map of Samples Identified by ClustR with Clusters of ALL Cases Diagnosed at Ages 2–6 Years, and Born from 2002 to 2004
Figure 2.
Distribution of Birth Dates Among Regions Identified by ClustR with Clusters of ALL Cases Diagnosed at Ages 2–6 Years
Figure 3.
Map of the Most Likely Cluster Identified by SaTScan with Cluster of ALL Cases Diagnosed Ages 2–6 Years
We examined the demographic characteristics of the clusters identified among the ALL cases diagnosed at ages 2 to 6 years and their controls. Although cases and controls were matched on demographic characteristics, creating an artificial similarity between their demographic distributions overall, the controls within clusters are not necessarily those matching the cases within clusters. Nevertheless, within the clusters, we did not observe any differences between cases and controls with respect to demographic characteristics (Table 2). However, in the same area of California before and after the cluster, median income was higher among the cases.
Table 2.
Demographic Characteristics of the Identified Cluster of ALL Cases Diagnosed at the Age of 2–6 years: During, Before and After
| Cluster | Cluster Area Before | Cluster Area After | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Demographics | Cases (n=58) | Controls (n=114) | Cases (n=80) | Controls (n=386) | Cases (n=47) | Controls (n=175) |
| Median income ($) | 69676.1 | 74671.6 | 69822.7 | 62912.9 | 68312.2 | 64803.1 |
| Adults without GED or equivalent (%) | 21% | 19% | 19% | 20% | 19% | 24% |
| Population density (persons/m2) | 0.00396 | 0.00345 | 0.00483 | 0.00395 | 0.00413 | 0.00479 |
| White, non-Hispanic | 29% | 38% | 40% | 39% | 19% | 27% |
| Black, non-Hispanic | 5% | 3% | 6% | 4% | 4% | 2% |
| Hispanic | 36% | 33% | 29% | 33% | 43% | 35% |
| Asian & Pacific Islander, non-Hispanic | 28% | 25% | 25% | 22% | 21% | 18% |
| Alaskan & Native American, non-Hispanic | 0% | 0% | 0% | 0% | 0% | 0% |
| Other | 2% | 2% | 0% | 2% | 13% | 17% |
Abbreviations: GED (general education diploma)
Malignant gonadal germ cell tumors
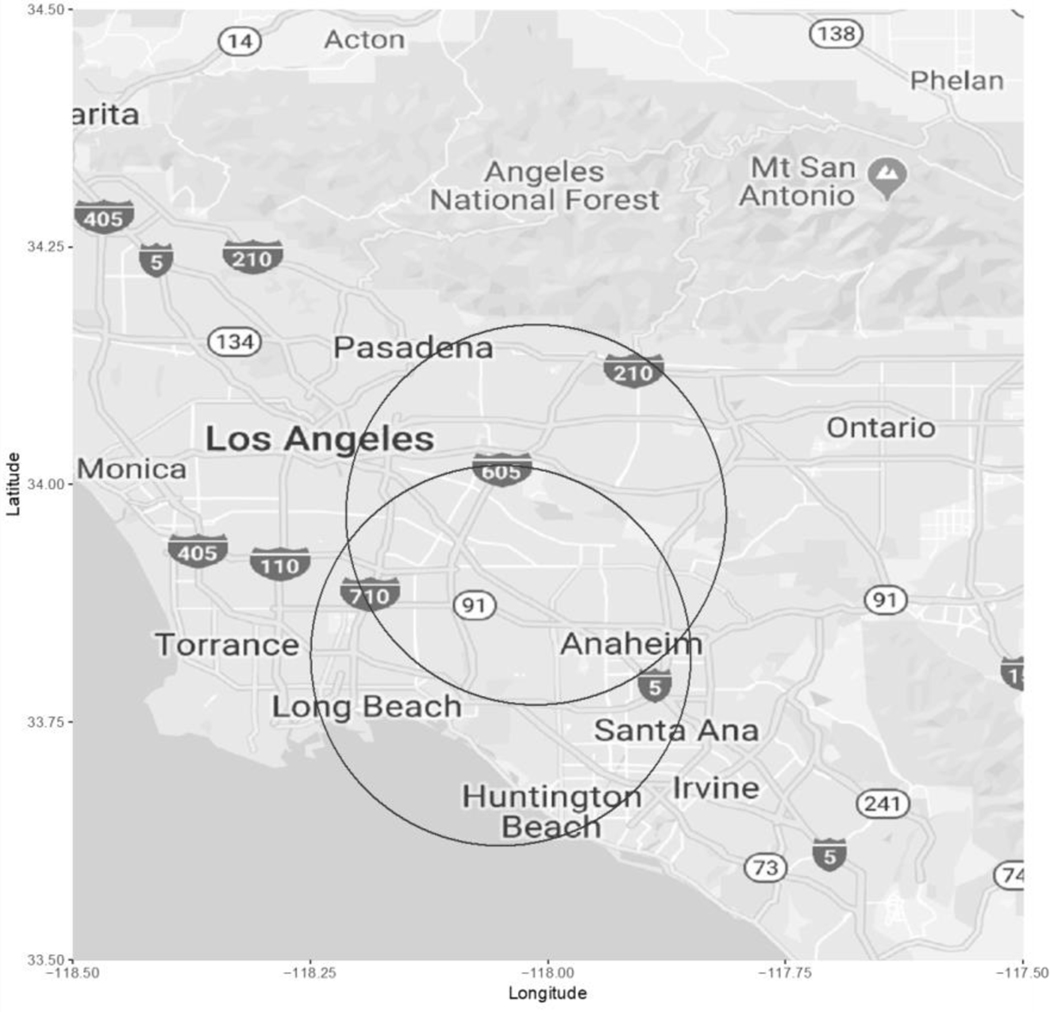
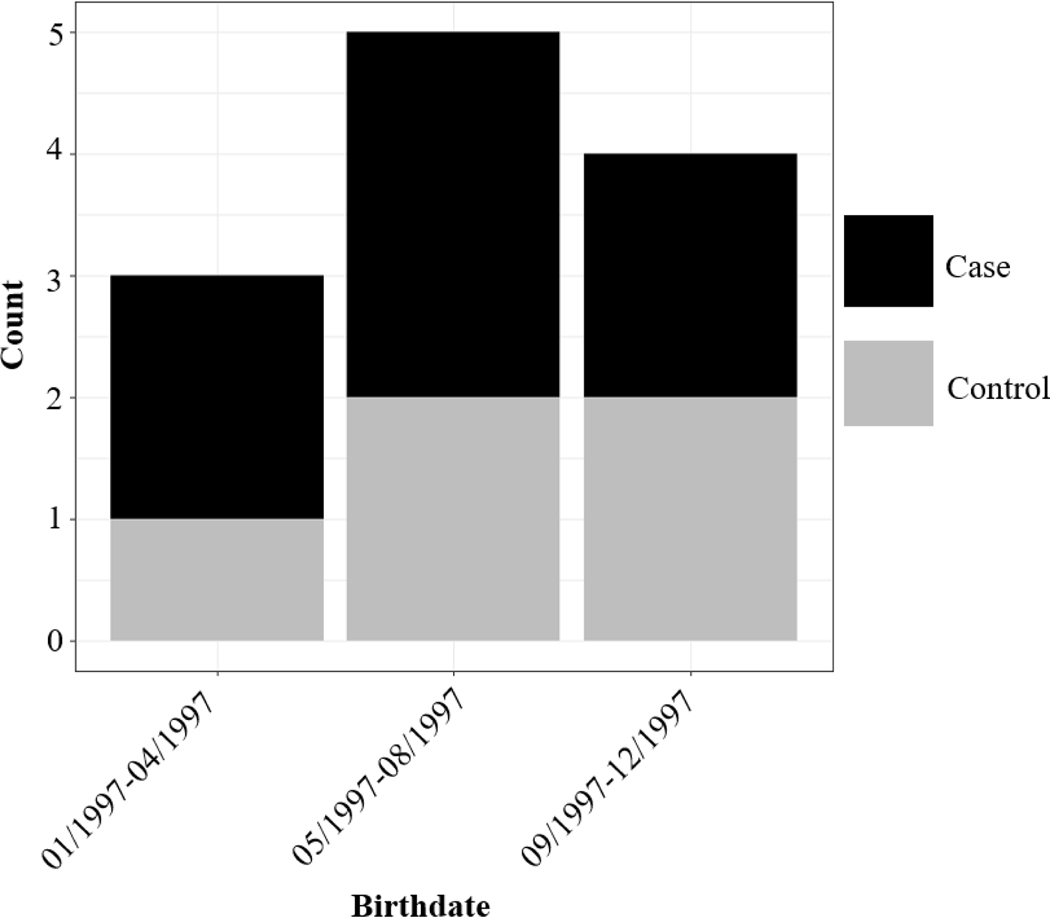
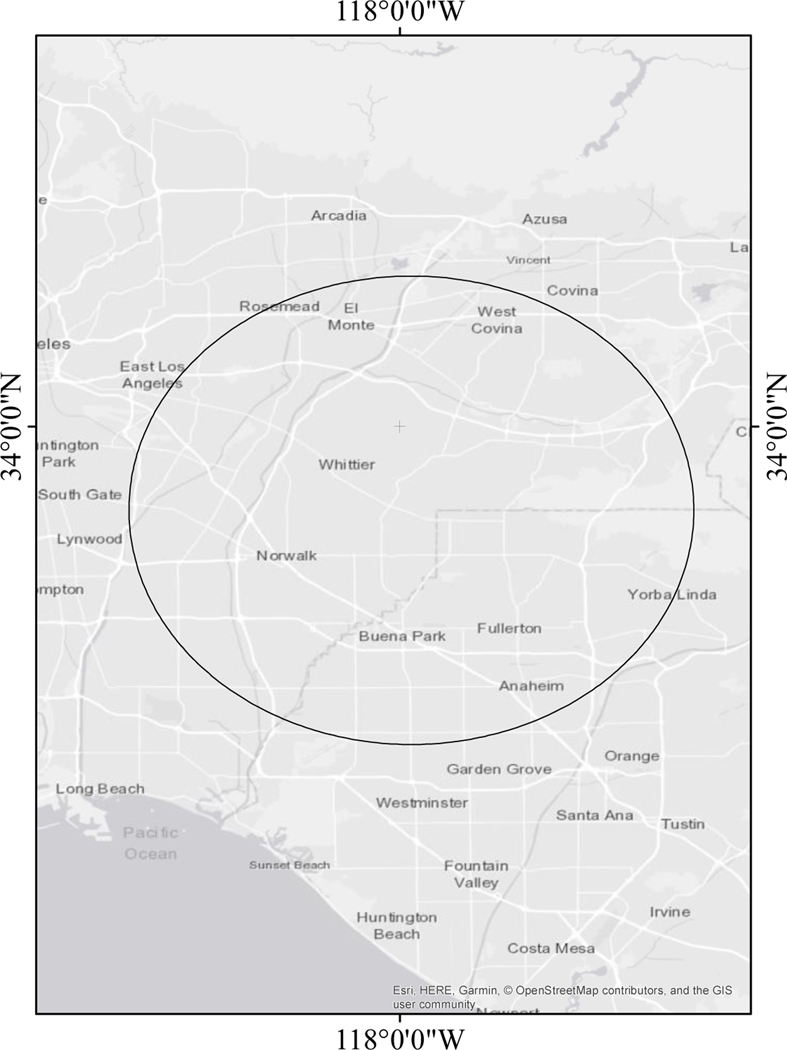
In the germ cell tumor subset, where 30.6% cases had tumors of the ovaries and 69.4% cases had tumors of the testis, ClustR identified two distinct samples with adjusted p-values of 0.03. These samples overlapped spatially in central Los Angeles (Figure 4). Birthdates in the clusters ranged from January 1997 to December 1997 (Figure 5). The most likely cluster identified by SaTScan was in the same region from April 1997 to December 1997 with a p-value of 0.06 (Figure 6).
Figure 4.
Map of Samples Identified by ClustR with Clusters of Malignant Gonadal Germ Cell Tumors
Figure 5.
Distribution of Birth Dates Among Regions Identified by ClustR with Clusters of Malignant Gonadal Germ Cell Tumors
Figure 6.
Map of the Most Likely Cluster Identified by SaTScan for Malignant Gonadal Germ Cell Tumors in 1997.
We examined the demographic characteristics of this cluster of malignant gonadal germ cell tumor cases, despite the small sample size and limited statistical power (Table 3). We observed that cases lived in higher population density areas and were wealthier and more educated than controls, though these differences were imprecise and not conclusive of an actual difference. By contrast, in the same area of California after the cluster, cases lived in far less wealthy neighborhoods than controls. Since the cluster occurred at the beginning of the study period, we did not have data on the area before the cluster. Finally, over 57.1% of cases in the clusters had tumors of the ovaries and 42.9% of cases had tumors of the testis.
Table 3.
Demographic Characteristics of the Identified Cluster of Malignant Gonadal Germ Cell Tumors: During, Before and After
| Cluster | Cluster Area Before | Cluster Area After | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Demographics | Cases (n=7) | Controls (n=5) | Cases (n=0) | Controls (n=0) | Cases (n=12) | Controls (n=49) |
| Median income ($) | 63492.29 | 56976.00 | NA | NA | 49112.86 | 70477.00 |
| Adults without GED or equivalent (%) | 35% | 29% | NA | NA | 36% | 27% |
| Population density (persons/m2) | 0.00665 | 0.00363 | NA | NA | 0.00413 | 0.00316 |
| White, non-Hispanic | 14% | 20% | NA | NA | 8% | 0% |
| Black, non-Hispanic | 0% | 0% | NA | NA | 0% | 2% |
| Hispanic | 71% | 80% | NA | NA | 83% | 76% |
| Asian & Pacific Islander, non-Hispanic | 14% | 0.0% | NA | NA | 8% | 8% |
| Alaskan & Native American, non-Hispanic | 0% | 0% | NA | NA | 0% | 0% |
| Other | 0% | 0% | NA | NA | 0% | 14% |
Abbreviations: GED (general education diploma)
Other cancers.
ClustR did not identify any other significant clusters with adjusted p < 0.05 (Table 4). However, for both astrocytomas and ependymomas/choroid plexus tumors, ClustR identified suggestive clustering at the p=0.10 level. SaTScan and ClustR generally obtained similar results, including that SaTScan also suggested clustering for astrocytomas with p=0.06. However, SaTScan also indicated some clustering for Hodgkin lymphoma (p=0.09) and retinoblastoma (p=0.06), while it did not support clustering for ependymomas/choroid plexus tumors.
Table 4.
Overview of Results of ClustR Analyses on All Types of Cancer
| ICCC Code | Cancer | Cases | Controls | Adjusted p for ClustR’s Most Significant Sample | Count of Samples with p < 0.05 | Count of Samples with p < 0.10 | p for SaTScan’s MLC |
|---|---|---|---|---|---|---|---|
| 11 | Lymphoid leukemias | 2014 | 7980 | 0.07 | 0 | 7 | 0.63 |
| 11 | Lymphoid leukemias, Diagnosed Age 0 to 1 | 324 | 1285 | 0.96 | 0 | 0 | 0.66 |
| 11 | Lymphoid leukemias, Diagnosed Age 2 to 6 | 1444 | 5720 | 0.04 | 12 | 17 | 0.07 |
| 11 | Lymphoid leukemias, Hispanic | 1050 | 4125 | 1.00 | 0 | 0 | 0.26 |
| 12 | Acute myeloid leukemia | 321 | 1290 | 0.99 | 0 | 0 | 0.15 |
| 21 | Hodgkin lymphoma | 85 | 332 | 0.96 | 0 | 0 | 0.09 |
| 22 | Non-Hodgkin lymphoma (except Burkitt lymphoma) | 166 | 651 | 0.29 | 0 | 0 | 0.28 |
| 23 | Burkitt lymphoma | 76 | 307 | 0.79 | 0 | 0 | 0.49 |
| 24 | Miscellaneous lymphoreticular neoplasms | 73 | 293 | 0.91 | 0 | 0 | 1.00 |
| 31 | Ependymomas and choroid plexus tumor | 146 | 575 | 0.10 | 0 | 5 | 0.67 |
| 32 | Astrocytomas | 535 | 2123 | 0.10 | 0 | 31 | 0.06 |
| 33 | Intracranial and intraspinal embryonal tumors | 333 | 1316 | 0.99 | 0 | 0 | 0.32 |
| 34 | Other gliomas (besides ICCCMDG codes 31–33) | 159 | 637 | 0.62 | 0 | 0 | 0.26 |
| 41 | Neuroblastoma and ganglioneuroblastoma | 529 | 2096 | 0.70 | 0 | 0 | 0.81 |
| 50 | Retinoblastoma | 293 | 1142 | 0.14 | 0 | 0 | 0.06 |
| 61 | Nephroblastoma and other nonepithelial renal tumors | 415 | 1649 | 0.32 | 0 | 0 | 0.52 |
| 71 | Hepatoblastoma | 143 | 568 | 0.98 | 0 | 0 | 0.91 |
| 81 | Osteosarcomas | 72 | 283 | 0.98 | 0 | 0 | 0.47 |
| 91 | Rhabdomyosarcomas | 242 | 967 | 0.97 | 0 | 0 | 0.63 |
| 94 | Other specified soft tissue sarcomas (besides ICCCMDG codes 91–93) | 85 | 334 | 0.80 | 0 | 0 | 0.74 |
| 102 | Malignant extracranial and extragonadal germ cell tumors | 87 | 337 | 0.96 | 0 | 0 | 1.00 |
| 103 | Malignant gonadal germ cell tumors | 72 | 291 | 0.03 | 5 | 5 | 0.06 |
| 114 | Malignant melanomas | 50 | 198 | 0.62 | 0 | 0 | 0.31 |
Abbreviations: MLC (most likely cluster) ICCC (International Classification of Childhood Cancer)
Discussion
We have found evidence of non random space-time groupings of childhood ALL and malignant gonadal germ cell tumors, and suggestive evidence in Hodgkin lymphoma, astrocytoma and retinoblastoma. To our knowledge this study is the largest cluster analysis of childhood cancer birth residences ever conducted. A driver of this analysis is the underlying hypothesis of a prenatal origin of childhood cancer, a theory that is well described for ALL2,35. We chose to analyze birth addresses to focus on the prenatal and directly postnatal exposure window, and for direct comparability to the control series.
Regarding ALL, ClustR, and SaTScan did not detect any potential clusters among the vast majority of ALL cases, ALL cases diagnosed at the age of ≤ 1 year, or among Hispanic ALL cases. However, both programs identified an area with higher-than-expected number of births of children who went on to develop ALL at the age of 2–6 years in the southern region of the San Francisco Bay Area during 2002–2004. We a priori designated the 2–6 year age range for stratified analysis due to the high proportion of TEL-AML1 (ETV6-RUNX1) and high hyperdiploid subtype that present during this age window. TEL-AML1 and high hyperdiploid ALLs are the most common subtypes of childhood ALL, share an early pre-B cell phenotype (CD10+, CD19+), and are suspected to have a distinct etiology from other subtypes of ALL36. Unfortunately, we do not have detailed cytogenetic information on the tumor types for any of our subjects in this analysis. Our results are consistent with a finding from a study using Swiss registry data that observed clustering of individuals with ALL with the TEL-AML1 subtype24. Our analysis provides further evidence that TEL-AML1 leukemias may be non-randomly distributed in space and time.
To further understand the context of this space–time cluster, we examined aspects of the population and time period within and surrounding this cluster. We did not find any strong demographic trends, though this finding may be expected given the matching on several demographics. We did observe that both the cases and controls had a lower median income during the cluster window, which may be due to economic issues at the time. Intriguingly, the cluster occurred at a time surrounding the “dotcom crash” in which the U.S. economy crashed after a boom in technology and business following the birth of the internet. The southern San Francisco Bay Area, which includes Silicon Valley, was especially heavily impacted by this crash 37. Impacts of the crash and the period preceding it, including increased maternal stress, heavy industrial growth (and decline) especially related to semiconductor manufacturing, and population mixing may be possible contributors to this spike in ALL cases, which require further investigation beyond the scope of this current analysis38.
Due to the diverse population of California, this study provides unparalleled power to examine Hispanic ethnicity in space–time grouping of ALL, which is particularly important given that Hispanics have the highest risk of ALL39; however, we found no ethnic differences in our cluster analyses.
We observed a significant space–time cluster of malignant gonadal germ cell tumors in Southern California. These tumors are more common in males, though female cases were overrepresented within the identified cluster. Previous studies suggest a trend towards increasing incidence over time in males but not females40. Little is known about the etiology of malignant gonadal germ cell tumors, and our study appears to be the first to observe a non-random space–time clustering. When examining demographic patterns in and around the cluster window, we found that cases were wealthier than controls during the cluster window but not after. Further research with higher power is needed to explore this relationship and how it may relate to the space–time clustering of these tumors and their possible environmental risk factors.
There was also some indication for clustering in Hodgkin lymphoma, astrocytoma, and retinoblastoma, although none of them met our predefined significance cutoff of p=0.05 in either ClustR or SaTScan.
The key strength of this study comes from the population-based case and control selection with minimal selection bias. The controls serve as a proxy of the underlying population base from which the cases arose. Many case–control studies of childhood cancer suffer from strong selection biases in control selection that damage the inference of spatial analyses due to residual confounding; this study is not prone to such bias. A matched study design, often used, prevents the identification of clusters due to the demographic features used for matching. Furthermore, our sample size is unusually large compared to previous studies, particularly for childhood cancer, allowing us to examine cancers which have not been previously analyzed for clustering. In addition to the data, the methods are a strength of this study. Two different methods, SaTScan and ClustR, are employed and can serve to corroborate one another. ClustR and SaTScan have different strengths and weaknesses in terms of sensitivity32 and indeed the two programs disagreed on the results of four of the 23 subsets. Nevertheless, this study serves as a proof of concept for ClustR, establishing it as a viable R-based cluster analysis tool that is flexible and transparent. The study itself provides an example of how ClustR can be leveraged and interpreted in the context of a study, which may aid other researchers conducting cluster analysis.
This study also has several limitations due to both data issues and the methods used. We have limited verification of our data originating from the birth and California cancer registry. Inclusion criteria may have induced slight selection bias, as 587 observations were removed due to missing latitude and longitude. A disproportionate percentage of subjects missing data were controls, and the observations with missing data tended to come from areas with lower socioeconomic status. For purposes of evaluating potential bias, median household income was aggregated by zip code tabulation area so that addresses that could not be geocoded could be evaluated. The median household income for observations included in the spatial analysis was $47,092, while the median household income for observations removed due to missing data was $42,510 (p < 0.05), indicating that the individuals excluded from the analysis were born in zip codes with lower median income than individuals included in the analysis. Thus, bias could be induced if controls from poorer areas were not included, causing an artificially high rate of cases in those areas. If there had been any positive findings in areas with low median income, a concern would be that controls may be missing from that region due to data issues, rather than an actual shortage of controls. However, there is no difference in the median household income of the case and control series overall (Table 1), indicating that the effect of this bias is likely minimal. Additionally, we have analyzed birth addresses that capture the in utero exposure window but also may be the source of some misclassification due to residential mobility during pregnancy, this misclassification is likely non-differential unless residential mobility during pregnancy is associated with the development of childhood cancer. Previous research studying residential mobility of childhood ALL in California found no differences between cases and controls41. Furthermore, misclassification of disease status may result from a child that moved out of the state prior to diagnosis. We estimate that approximately 10%−20% of the birth cohort may have moved out of California during the study window; the result would only create controls that may indeed have a cancer diagnosis. Due to the rarity of childhood cancer and the resulting bias towards the null we believe the impact of this misclassification to be minimal. ClustR and SaTScan only sampled cylinders of data, which may not accurately capture certain disease patterns. For example, an infectious disease may be best represented with a conical shape in space time, as disease starts in a small region and expands outwards with time. Furthermore, a study by Tango & Takahashi demonstrated that SaTScan has trouble detecting spatially non-circular clusters42. Therefore, some non-cylindrical clusters may have been missed by both programs.
In conclusion, the present study provides evidence that childhood ALL diagnosed at 2–6 years of age and malignant gonadal germ cell tumors may occur in non-random space–time clusters at the birth location, potentially suggesting that risk factors of these cancers may also cluster across space and time. Further research into these epidemiologic features is warranted.
Supplementary Material
Acknowledgments
Financial Support:
The research was supported by a grant from Alex’s Lemonade Stand Foundation. The collection of cancer incidence data used in this study was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885; the National Cancer Institute’s Surveillance, Epidemiology and End Results Program under contract HHSN261201000140C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute; and the Centers for Disease Control and Prevention’s National Program of Cancer Registries, under agreement U58DP003862-01 awarded to the California Department of Public Health. The ideas and opinions expressed herein are those of the author(s) and endorsement by the State of California, Department of Public Health, the National Cancer Institute, and the Centers for Disease Control and Prevention or their Contractors and Subcontractors is not intended nor should be inferred.
Footnotes
Conflicts of Interest: None declared.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2.Wiemels JL, Cazzaniga G, Daniotti M, Eden OB, Addison GM, Masera G, Saha V, Biondi A, Greaves MF. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet 1999;354(9189):1499–503. [DOI] [PubMed] [Google Scholar]
- 3.Ford AM, Bennett CA, Price CM, Bruin MC, Van Wering ER, Greaves M. Fetal origins of the TEL-AML1 fusion gene in identical twins with leukemia. Proc Natl Acad Sci U S A 1998;95(8):4584–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGlynn KA, Cook MB. Etiologic factors in testicular germ-cell tumors. Future Oncol 2009;5(9):1389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitehead TP, Metayer C, Wiemels JL, Singer AW, Miller MD. Childhood Leukemia and Primary Prevention. Curr Probl Pediatr Adolesc Health Care 2016;46(10):317–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francis SS, Wallace AD, Wendt GA, Li L, Liu F, Riley LW, Kogan S, Walsh KM, de Smith AJ, Dahl GV, Ma X, Delwart E, Metayer C, Wiemels JL. In utero cytomegalovirus infection and development of childhood acute lymphoblastic leukemia. Blood 2017;129(12):1680–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dores GM, Devesa SS, Curtis RE, Linet MS, Morton LM. Acute leukemia incidence and patient survival among children and adults in the United States, 2001–2007. Blood 2012;119(1):34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greaves M. Childhood leukaemia. BMJ 2002;324(7332):283–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greaves M Infection, immune responses and the aetiology of childhood leukaemia. Nat Rev Cancer 2006;6(3):193–203. [DOI] [PubMed] [Google Scholar]
- 10.Goodman M, Naiman JS, Goodman D, LaKind JS. Cancer clusters in the USA: what do the last twenty years of state and federal investigations tell us? Crit Rev Toxicol 2012;42(6):474–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodman M, LaKind JS, Fagliano JA, Lash TL, Wiemels JL, Winn DM, Patel C, Van Eenwyk J, Kohler BA, Schisterman EF, Albert P, Mattison DR. Cancer cluster investigations: review of the past and proposals for the future. Int J Environ Res Public Health 2014;11(2):1479–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ortega-Garcia JA, Lopez-Hernandez FA, Fuster-Soler JL, Martinez-Lage JF. Space-time clustering in childhood nervous system tumors in the Region of Murcia, Spain, 1998–2009. Childs Nerv Syst 2011;27(11):1903–11. [DOI] [PubMed] [Google Scholar]
- 13.Ye X, Torabi M, Lix LM, Mahmud SM. Time and spatial trends in lymphoid leukemia and lymphoma incidence and survival among children and adolescents in Manitoba, Canada: 1984–2013. PLoS One 2017;12(4):e0175701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costas K, Knorr RS, Condon SK. A case-control study of childhood leukemia in Woburn, Massachusetts: the relationship between leukemia incidence and exposure to public drinking water. Sci Total Environ 2002;300(1–3):23–35. [DOI] [PubMed] [Google Scholar]
- 15.Uchiyama T, Yodoi J, Sagawa K, Takatsuki K, Uchino H. Adult T-cell leukemia: clinical and hematologic features of 16 cases. Blood 1977;50(3):481–92. [PubMed] [Google Scholar]
- 16.Yoshida M, Miyoshi I, Hinuma Y. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc Natl Acad Sci U S A 1982;79(6):2031–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heath CW Jr., Community clusters of childhood leukemia and lymphoma: evidence of infection? Am J Epidemiol 2005;162(9):817–22. [DOI] [PubMed] [Google Scholar]
- 18.Kohli S, Noorlind Brage H, Lofman O. Childhood leukaemia in areas with different radon levels: a spatial and temporal analysis using GIS. J Epidemiol Community Health 2000;54(11):822–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hjalmars U, Kulldorff M, Gustafsson G, Nagarwalla N. Childhood leukaemia in Sweden: using GIS and a spatial scan statistic for cluster detection. Stat Med 1996;15(7–9):707–15. [DOI] [PubMed] [Google Scholar]
- 20.Carceles-Alvarez A, Ortega-Garcia JA, Lopez-Hernandez FA, Orozco-Llamas M, Espinosa-Lopez B, Tobarra-Sanchez E, Alvarez L. Spatial clustering of childhood leukaemia with the integration of the Paediatric Environmental History. Environ Res 2017;156:605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramis R, Gomez-Barroso D, Tamayo I, Garcia-Perez J, Morales A, Pardo Romaguera E, Lopez-Abente G. Spatial analysis of childhood cancer: a case/control study. PLoS One 2015;10(5):e0127273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosavi-Jarrahi A, Moini M, Mohagheghi MA, Alebouyeh M, Yazdizadeh B, Shahabian A, Nahvijo A, Alizadeh R. Clustering of childhood cancer in the inner city of Tehran metropolitan area: A GIS-based analysis. International Journal of Hygiene and Environmental Health 2007;210(2):113–119. [DOI] [PubMed] [Google Scholar]
- 23.Francis SS, Selvin S, Yang W, Buffler PA, Wiemels JL. Unusual space-time patterning of the Fallon, Nevada leukemia cluster: Evidence of an infectious etiology. Chem Biol Interact 2012;196(3):102–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreis C, Grotzer M, Hengartner H, Spycher BD, Swiss Paediatric Oncology G, Swiss National Cohort Study G. Space-time clustering of childhood cancers in Switzerland: A nationwide study. Int J Cancer 2016;138(9):2127–35. [DOI] [PubMed] [Google Scholar]
- 25.Agost L. Analysis of spatial-temporal clusters of childhood cancer incidence in the province of Cordoba, Argentina (2004–2013). Arch Argent Pediatr 2016;114(6):534–543. [DOI] [PubMed] [Google Scholar]
- 26.Gustafsson B, Carstensen J. Evidence of space-time clustering of childhood acute lymphoblastic leukaemia in Sweden. Br J Cancer 1999;79(3–4):655–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gustafsson B, Carstensen J. Space-time clustering of childhood lymphatic leukaemias and non-Hodgkin’s lymphomas in Sweden. Eur J Epidemiol 2000;16(12):1111–6. [DOI] [PubMed] [Google Scholar]
- 28.McNally RJ, Alexander FE, Birch JM. Space-time clustering analyses of childhood acute lymphoblastic leukaemia by immunophenotype. Br J Cancer 2002;87(5):513–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilman EA, Knox EG. Childhood cancers: space-time distribution in Britain. J Epidemiol Community Health 1995;49(2):158–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amin R, Burns JJ. Clusters of adolescent and young adult thyroid cancer in Florida counties. Biomed Res Int 2014;2014:832573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulldorff M, Huang L, Konty K. A scan statistic for continuous data based on the normal probability model. Int J Health Geogr 2009;8:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Enders C HR, Selvin S, Metayer C, Francis S. ClustR: A Space-Time Cluster Analysis R Package for Individual-level Data. Epidemiology 2019;31:XXX. [DOI] [PubMed] [Google Scholar]
- 33.Wang R, Metayer C, Morimoto L, Wiemels JL, Yang J, DeWan AT, Kang A, Ma X. Parental Age and Risk of Pediatric Cancer in the Offspring: A Population-Based Record-Linkage Study in California. Am J Epidemiol 2017;186(7):843–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med 1990;9(7):811–8. [DOI] [PubMed] [Google Scholar]
- 35.McHale CM, Wiemels JL, Zhang L, Ma X, Buffler PA, Guo W, Loh ML, Smith MT. Prenatal origin of TEL-AML1-positive acute lymphoblastic leukemia in children born in California. Genes Chromosomes Cancer 2003;37(1):36–43. [DOI] [PubMed] [Google Scholar]
- 36.Inaba H, Greaves M, Mullighan CG. Acute lymphoblastic leukaemia. Lancet 2013;381(9881):1943–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quigley JM. Froth in the Silicon Valley Housing Market? The Economists’ voice 2006(5). [Google Scholar]
- 38.Mann A, Luo T. Crash and reboot: Silicon Valley high-tech employment and wages, 2000–08. Monthly Labor Review 2010:59–73. [Google Scholar]
- 39.Barrington-Trimis JL, Cockburn M, Metayer C, Gauderman WJ, Wiemels J, McKean-Cowdin R. Rising rates of acute lymphoblastic leukemia in Hispanic children: trends in incidence from 1992 to 2011. Blood 2015;125(19):3033–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stang A, Trabert B, Wentzensen N, Cook MB, Rusner C, Oosterhuis JW, McGlynn KA. Gonadal and extragonadal germ cell tumours in the United States, 1973–2007. Int J Androl 2012;35(4):616–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Urayama KY, Von Behren J, Reynolds P, Hertz A, Does M, Buffler PA. Factors associated with residential mobility in children with leukemia: implications for assigning exposures. Ann Epidemiol 2009;19(11):834–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tango T, Takahashi K. A flexible spatial scan statistic with a restricted likelihood ratio for detecting disease clusters. Stat Med 2012;31(30):4207–18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.