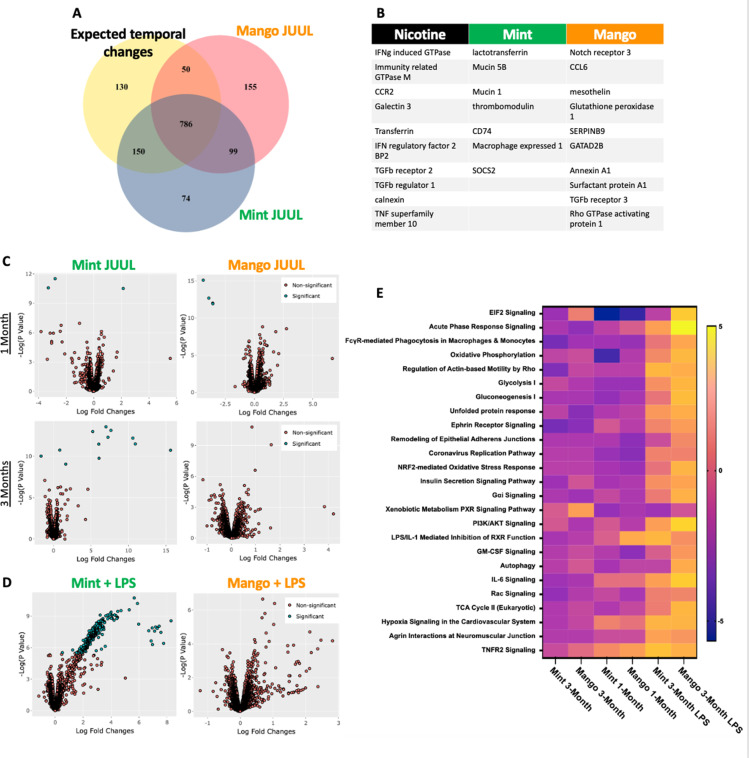
Figure 6. Unique RNAseq signatures in the lungs exposed to different flavors of JUUL aerosols.
(A) Venn diagram of gene expression unique to JUUL Mango (155) and JUUL Mint (Alhaddad et al., 2014b). Gene expression changes common to both aerosols (99) suggest that they are due to chemicals found in both of the flavored e-liquids (nicotinic salts, propylene glycol, glycerin, benzoic acid, etc). (B) Greatest gene expression changes associated with nicotine and flavorant chemicals within aerosols. (C) Volcano plots demonstrating gene expression changes specific for each flavor after sub-acute exposure (1 month; top row) and 3 months of exposure (bottom row). (D) Gene expression changes associated with inhalation of JUUL aerosols with different flavors in the setting of inflammatory challenge with inhaled LPS.(E) Heat map of the pathways most notably impacted by 1- and 3-month daily exposures to JUUL Mint and Mango, as well as in the setting of LPS challenge at 3 months. IFNg: interferon gamma; CCR: C-C motif chemokine related; IFN: interferon; BP: binding protein; TGFb: transforming growth factor beta; TNF: tumor necrosis factor; SOCS: suppressor of cytokine signaling; CCL: C-C motif ligand.