Abstract
To test the possibility that MexX-MexY, a new set of efflux system components, is associated with OprM and contributes to intrinsic resistance in Pseudomonas aeruginosa, we constructed a series of isogenic mutants lacking mexXY and/or mexAB and/or oprM from a laboratory strain PAO1, and examined their susceptibilities to ofloxacin, tetracycline, erythromycin, gentamicin, and streptomycin. Loss of either MexXY or OprM from the MexAB-deficient mutant increased susceptibility to all agents tested, whereas loss of MexXY from the MexAB-OprM-deficient mutant caused no change in susceptibility. Introduction of an OprM expression plasmid decreased the susceptibility of the mexAB-oprM-deficient-/mexXY-maintaining mutant, yet caused no change in the susceptibility of a mexAB-oprM- and mexXY-deficient double mutant. Immunoblot analysis using anti-MexX polyclonal rabbit serum generated against synthetic oligopeptides detected expression of MexX in the PAO1 cells grown in medium containing tetracycline, erythromycin, or gentamicin, although expression of MexX was undetectable in the cells incubated in medium without any agent. These results suggest that MexXY induced by these agents is functionally associated with spontaneously expressed OprM and contributes to the intrinsic resistance to these agents.
A variety of multicomponent efflux systems are coded on the chromosomes of gram-negative bacteria and contribute to intrinsic and acquired resistance against antimicrobial agents, disinfectants, organic solvents, and heavy metals (17–20, 24). Pseudomonas aeruginosa is a clinically significant pathogen exhibiting highly intrinsic resistance to various antimicrobial agents. One of the mechanisms contributing to its intrinsic resistance is a spontaneous expression of the efflux system MexA-MexB-OprM encoded on a mexA-mexB-oprM operon of the chromosome of P. aeruginosa (10). This system energy-dependently extrudes many antimicrobial agents from the cell interior in cooperation with the periplasmic, inner membrane, and outer membrane components organized through the two membranes. While the disruption of each component gene of the mexA-mexB-oprM operon increases the susceptibility to many antimicrobial agents, the disruption of oprM increases the susceptibility to a greater extent than the disruption of mexA or mexB (5, 21, 30, 31). Due to the presence of a weak promoter in the mexB gene upstream of oprM gene, the polar effect from the disruptions of mexA and mexB does not entirely suppress the expression of the oprM gene (31). Thus, OprM can contribute to the intrinsic resistance by cooperation with unknown periplasmic and inner membrane components. Recently, mexG-mexH (GenBank accession no. AF073776 [1]), mexX-mexY (GenBank accession no. AB015853 [16]), and amrA-amrB (GenBank accession no. AF147719 [29]), three new sets of mexA-mexB homologous operons lacking outer membrane component genes, have been discovered independently on the chromosome of P. aeruginosa. However, homology searches of hypothetical closed genome sequence data from the Pseudomonas genome sequencing project (http: //www.pseudomonas.com/) conducted by the BLASTN program (National Center for Biotechnology Information) show the existence of one operon highly homologous to mexGH, mexXY, and amrAB in the whole genome, suggesting that they are the same genes. Thus, we use the nomenclature mexXY for the homologous operon as proposed by Aires et al. (2). Aires et al. reported that MexXY appears to function with OprM in P. aeruginosa, whereas Westbrock-Wadman et al. (29) reported that OprM is unlikely to be the outer membrane component associated with MexXY.
To test the possibility that MexXY is associated with OprM and contributes to intrinsic resistance, we constructed a series of isogenic mutants lacking mexXY and/or mexAB and/or oprM from laboratory strain PAO1 and compared their susceptibilities to antimicrobial agents. We also showed that the expression of MexXY is induced by exposure to several kinds of antimicrobial agents in PAO1.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids used in this study are listed in Table 1. Bacterial cells were grown on Mueller-Hinton II agar (MHA) (Becton Dickinson Microbiology Systems, Cockeysville, Md.) or in Mueller-Hinton broth (MHB) (Becton Dickinson Microbiology Systems) at 37°C. Minimal agar medium (4) was used for selection of P. aeruginosa. The following antibiotics were added to media at the indicated concentrations: carbenicillin, 100 μg/ml for Escherichia coli and 200 μg/ml for P. aeruginosa; and chloramphenicol, 30 μg/ml for E. coli and MexAB-OprM-deficient P. aeruginosa and 300 μg/ml for MexAB-OprM-producing P. aeruginosa. MHA was supplemented with 10% (wt/vol) sucrose as required. Gentamicin-ofloxacin-resistant mutants were isolated by plating PAO1 on MHA containing 4 μg of gentamicin and 1 μg of ofloxacin per ml. They were obtained at a frequency of 8 × 10−8. One clone was selected and designated N135.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| P. aeruginosa strains | ||
| PAO1 | Prototroph | |
| OCR1 | MexAB-OprM-overproducing nalB mutant | 13 |
| KG2225 | mexA::Ω of PAO1 | Unpublished datab |
| KG2239 | ΔmexR-mexA-mexB-oprM of PAO1 | 6 |
| N101 | ΔmexXY of PAO1 | This study |
| N102 | ΔmexXY of KG2225 | This study |
| N103 | ΔmexXY of KG2239, ΔmexRAB-oprM of PAO1 | This study |
| N126 | ΔmexAB of OCR1 | Submittedc |
| N128 | ΔmexXY of a MexXY-overproducing mutant of N126 called N127 | This study |
| N135 | MexXY-overproducing mutant of PAO1 | This study |
| N136 | ΔmexXY of N135 | This study |
| Plasmids | ||
| pMT5059 | pBend2 derivative carrying the multiple-cloning site and NotI site; Cbr | 27 |
| pMT5071 | pMOB3 derivative carrying Ω-Cm instead of Cm; Cmr | 28 |
| pNS001 | pMT5059 with 1.2-kb PCR fragment with 3′ flanking region of mexY in SpeI-HindIII; Cbr | This study |
| pNS002 | pNS001 with 1.0-kb PCR fragment with 5′ flanking region of mexX in MluI-NheI; Cbr | This study |
| pNS003 | pNS002 with Mob cassette in NotI; Cbr Cmr | This study |
| pAK1900 | E. coli-P. aeruginosa shuttle cloning vector; Cbr | 22 |
| pKMM128 | pAK1900 derivative carrying the partial mexR, XbaI linker, and oprM gene on a 4.3-kb fragment; Cbr | 7 |
Abbreviations: Cbr, carbenicillin resistant; Cmr, chloramphenicol resistant.
K. Okamoto, N. Gotoh, H. Tsujimoto, and T. Nishino.
Masuda et al.
Susceptibility testing.
MICs were determined by the usual twofold agar dilution technique with MHA with an inoculum size of 104 cells. All antimicrobial agents used in this study were obtained from commercial sources.
Molecular biology techniques.
Chromosomal DNA and plasmids were isolated using a DNeasy tissue kit and QIAfilter plasmid kit (Qiagen K.K., Tokyo, Japan), respectively. PCRs were performed with a Perkin-Elmer 480 thermal cycler using PfuTurbo DNA polymerase (Stratagene, La Jolla, Calif.). The thermal cycle profile for amplification of the mexX-mexY-flanking regions was 1 min at 96°C, 1 min at 59°C, 1 min at 72°C, and 30 cycles. The thermal cycle profile for amplification of the mexX-mexY region was 1 min at 96°C, 1 min at 68°C, 10 min at 72°C, and 30 cycles. Restriction endonucleases, alkaline phosphatase, and the DNA ligation kit were obtained from Takara Shuzo Co., Ltd., Kyoto, Japan. Restriction fragments were isolated, as required, from agarose gels using TaKaRa RECOCHIP (Takara). All molecular biology techniques were carried out according to the manufacturer's instructions or as described by Sambrook et al. (25). Transformation of E. coli (6) and P. aeruginosa (9) with plasmid DNA was performed as described previously.
Deletion of mexXY by gene replacement.
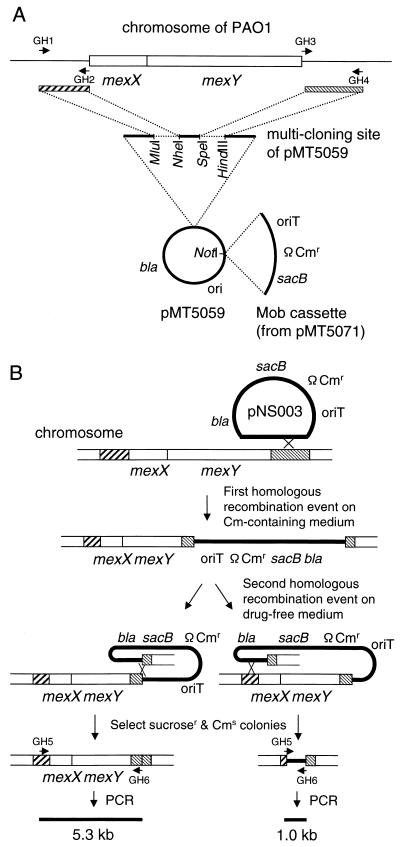
To construct a series of isogenic mutants lacking the mexX-mexY region, PCR primers for amplification of the mexX-mexY region and its flanking regions were synthesized based on nucleotide sequences of the Pseudomonas genome sequencing project database (Fig. 1A). After amplifying a 1.2-kb region downstream of mexY on PAO1 chromosomal DNA as a template using GH3 (5′-TGTACTAGTTGATGCCCCTAGCGAAACTCTC-3′) and GH4 (5′-TTTAAGCTTGACCTACAGGACGCTGCTG-3′), a primer pair containing a newly added cutting site (underlined) for restriction nucleases, the region was ligated into the SpeI-HindIII site in a multicloning site of pMT5059 (27) to yield pNS001. Next, a 1.0-kb region upstream of the mexX gene was amplified by PCR using the primer pair GH1 (5′-TGTACGCGTATTCGGAACAAGGCGTCTGC-3′) and GH2 (5′-TTCTGCTAGCGATGTGCATGGGTGTCCCTC-3′), and this region was ligated into the MluI-NheI site of pNS001 to yield pNS002. (A 24-bp sequence derived from a multicloning site of pMT5059 was still left between the two DNA fragments inserted on pNS002.) After addition of a NotI-flanked Mob cassette from pMT5071 (28), the resulting plasmid pNS003 was mobilized from an E. coli strain S17-1 (26) to the P. aeruginosa strains to introduce deletion of the mexX-mexY region into the recipient chromosomes by allelic exchange (Fig. 1B). The cell mixture was mated on MHA at 37°C for 3 h and then suspended in saline. Aliquots of the suspensions were plated on minimal agar plates supplemented with 30 to 300 μg of chloramphenicol per ml and incubated at 30°C for 2 days to isolate chloramphenicol-resistant pNS003 integrants. The obtained transconjugants were subsequently grown on drug-free MHA overnight for a second allelic exchange and then plated on MHA supplemented with 10% (wt/vol) sucrose. Chloramphenicol-susceptible and sucrose-resistant clones were selected and subjected to PCR analysis using the primer pair GH5 (5′-TCGCACTTGAGGTAGAGGATCTCCAGCACC-3′) and GH6 (5′-TCCTCACCGATCTGTCGAGCCTCTACTACG-3′). The sizes of the amplified DNA fragments obtained using these primers were 5.3 kb for the wild-type strain and 1 kb for the mexXY-deficient strain (data not shown). Thus, we constructed mutants lacking the mexXY region from MexAB-OprM-producing strain PAO1, MexAB-deficient- and OprM-producing mutant KG2225, and MexAB-OprM-deficient mutant KG2239, as described above, and we designated them N101, N102, and N103, respectively.
FIG. 1.
Schematic representation of procedure for markerless deletion of the chromosomal mexXY gene. (A) Construction of plasmid for deletion of mexXY. The striped boxes represent the mexXY-flanking regions amplified by PCR. (B) Procedure for deletion of chromosomal mexXY. The thin line represents the plasmid sequences. The thick line represents the P. aeruginosa chromosome. The straight lines at the bottom represent the size of PCR products.
Production of polyclonal antisera specific to MexA and MexX.
To obtain antibodies specific to MexA and MexX, the oligopeptides (C)YQIDPATYEADYQSA (amino acids 92 to 106 of MexA) and (C)EDSPTPLTRVEQID (amino acids 197 to 210 of MexX) were synthesized and conjugated to keyhole limpet hemocyanin by Chiron Technologies Pty., Ltd. (Clayton Victoria, Australia). A cysteine residue was added to each N terminus for conjugation. Rabbit antiserum raised against each antigen was prepared by Takara Shuzo Co., Ltd. (Otsu, Japan).
Isolation of total and outer membranes, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and immunoblot assay.
Exponentially growing cells in MHB were harvested as described previously (14). MHB was supplemented with tetracycline, erythromycin, or gentamicin as required. Total membranes (6) and outer membranes (14) were prepared as described previously. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrophoretic transfer were performed as described previously (15). Production levels of MexA, MexX, and OprM were tested by immunoblot assay using rabbit polyclonal antisera for MexA and MexX (see above), and murine monoclonal antibody for OprM (TM001 [6]). Binding of the primary antibodies was detected as described previously (6) using alkaline phosphatase-conjugated goat antibodies to rabbit or mouse immunoglobulins (Biosource International, Camarillo, Calif.) as the secondary antibodies and an alkaline phosphatase conjugate substrate kit (Bio-Rad Laboratories, Hercules, Calif.) for color development.
RESULTS
Functional association of MexXY with OprM.
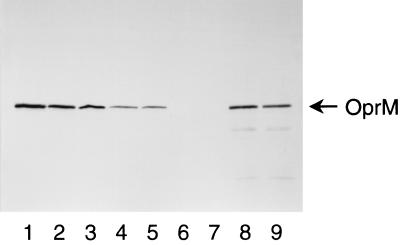
Western immunoblot analysis using the previously described murine monoclonal antibodies (6) showed that KG2225 produced a decreased level of OprM (Fig. 2, lane 4) and no detectable amounts of MexA or MexB (data not shown). These results are consistent with the previous report (31), which demonstrated the presence of a weak promoter upstream from oprM. Table 2 shows the MICs for the constructed mutants and their parent strains. KG2225 showed lower susceptibility to ofloxacin, tetracycline, erythromycin, and gentamicin than the mexAB-oprM deletion mutant KG2239. Deletion of mexXY (refer to N102) from KG2225 increased the susceptibility to these agents up to the same level of susceptibility demonstrated by KG2239, whereas deletion of mexXY (refer to N103) from KG2239 caused no significant change in susceptibility. Furthermore, introduction of the OprM expression plasmid pKMM128 (7) into KG2239 (MexAB− OprM− MexXY+) induced the production of OprM (Fig. 2, lane 8) and increased resistance to ofloxacin, tetracycline, erythromycin, gentamicin, and streptomycin (Table 2). In contrast, introduction of the plasmid into N103 (MexAB− OprM− MexXY−) caused no significant change in susceptibility to these agents (Table 2) in spite of production of OprM (Fig. 2, lane 9). These results suggest that MexXY does not contribute to the resistance without OprM. Deletion of mexXY from PAO1 (refer to N101) caused increases in susceptibility to tetracycline, erythromycin, gentamicin, and streptomycin (but not to ofloxacin), suggesting that MexXY contributes to the intrinsic resistance by functional association with OprM. Furthermore, the fact that OprM did not cause resistance without both MexAB and MexXY (refer to N103 and N102; N103/pAK1900 and N103/pKMM128) suggests that OprM does not associate with periplasmic and inner membrane components other than MexAB and MexXY in the wild strain.
FIG. 2.
Western immunoblot analysis with a monoclonal antibody to OprM. Each lane contains 20 μg of outer membrane protein. Lanes: 1, PAO1; 2, N101; 3, N126; 4, KG2225; 5, N102; 6, KG2239; 7, N103; 8, KG2239/pKMM128; 9, N103/pKMM128.
TABLE 2.
Susceptibility of constructed mutants and their parent strains to antimicrobial agents
| Strain | Genotypea
|
MIC (μg/ml) ofb:
|
||||||
|---|---|---|---|---|---|---|---|---|
| AB | XY | M | OFX | TEC | ERY | GEN | STR | |
| PAO1 | + | + | + | 0.5 | 16 | 256 | 1 | 16 |
| N101 | + | − | + | 0.25 | 2 | 64 | 0.13 | 2 |
| KG2225 | − | + | + | 0.13 | 2 | 64 | 1 | 4,096 |
| N102 | − | − | + | 0.03 | 0.5 | 32 | 0.13 | 512 |
| KG2239 | − | + | − | 0.03 | 0.5 | 16 | 0.25 | 2 |
| N103 | − | − | − | 0.03 | 0.25 | 16 | 0.25 | 2 |
| KG2239/pAK1900 | − | + | − | 0.03 | 0.25 | 16 | 0.13 | 2 |
| KG2239/pKMM128 | − | + | + | 0.13 | 8 | 64 | 1 | 16 |
| N103/pAK1900 | − | − | − | 0.03 | 0.13 | 16 | 0.13 | 2 |
| N103/pKMM128 | − | − | + | 0.03 | 0.13 | 16 | 0.13 | 2 |
| N126 | − | + | + | 0.13 | 16 | 128 | 1 | 8 |
| N128 | − | − | + | 0.03 | 0.5 | 32 | 0.13 | 1 |
AB, mexAB possessing (+) or -deficient (−); XY, mexXY possessing (+) or deficient (−); M, oprM (+) or deficient (−).
Abbreviations: OFX, ofloxacin; TEC, tetracycline; ERY, erythromycin; GEN, gentamicin; STR, streptomycin.
Inducible expression of MexX in PAO1.
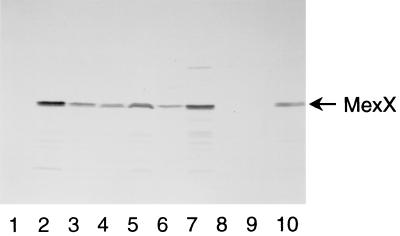
To confirm the expression of MexXY in wild-type strains, we prepared rabbit antiserum specific to MexX. For this purpose, a synthetic oligopeptide based on the deduced amino acid sequences of this protein was synthesized and used to immunize rabbits. The increase in susceptibility to tetracycline, erythromycin, and gentamicin caused by the deletion of mexXY from PAO1 suggests that MexXY is expressed in PAO1. However, an immunoblot assay using the antiserum as the primary antibody showed no production of MexX in PAO1 (Fig. 3, lane 1). To elucidate the discrepancy between the contribution of MexXY on intrinsic resistance and absence of MexX in PAO1, we examined the expression of MexX in PAO1 exposed to antimicrobial agents. MHB containing subinhibitory concentrations of antimicrobial agents and approximately 5 × 105 CFU of PAO1 per ml were incubated for 18 h at 37°C with shaking, and then total membranes were prepared. The exposure to subinhibitory concentrations of tetracycline, erythromycin, and gentamicin induced the production of MexX in PAO1 (Fig. 3, lanes 2 to 4). The amounts of MexA and OprM in the presence of these agents were comparable to those in the absence of the agents (data not shown). Production of MexX was also induced by subinhibitory concentrations of tetracycline in KG2225 and KG2239 (Fig. 3, lanes 5 and 6) but not in N101, N102, and N103 (data not shown). These results suggest that the MexXY induced by these agents is associated with spontaneously expressed OprM and contributes to the intrinsic resistance to these agents.
FIG. 3.
Detection of MexX with antiserum directed against synthetic oligopeptide containing part of the amino acid sequence of MexX. Each lane contains 25 μg of total membrane protein. PAO1 (lanes 1 to 4, and 9), KG2225 (lane 5), KG2239 (lane 6), N135 (lane 7), N136 (lane 8), and N126 (lane 10) were grown in MHB (lanes 1, 7, and 8) or MHB containing 8 (lane 2), 1 (lane 5), or 0.25 (lane 6) μg of tetracycline per ml, 128 μg of erythromycin per ml (lane 3), 0.5 μg of gentamicin per ml (lane 4), or 1 (lane 9) or 0.25 (lane 10) μg of ofloxacin per ml. The MICs of ofloxacin for PAO1 and N126 in MHB were 2 and 0.5 μg/ml, respectively.
MexXY-OprM is the primary system for intrinsic resistance to tetracycline and erythromycin.
Paradoxically, PAO1 lacking mexXY (refer to N101) and PAO1 lacking mexAB (refer to KG2225) showed eight- and fourfold increases in susceptibility to tetracycline and erythromycin, respectively (Table 2), while deletion of either mexXY or mexAB should cause a twofold increase in susceptibility if the two systems contribute equally to intrinsic resistance to these agents. To elucidate the reason for this discrepancy, we tested the susceptibility of N126 (N. Masuda, E. Sakagawa, S. Ohya, N. Gotoh, T. Tsujimoto, and T. Nishino, submitted for publication), which is a markerless mexAB-deficient mutant of a MexAB-OprM-overproducing mutant of PAO1 called OCR1, because the decreased level of OprM (Fig. 2, lane 4) in KG2225 might affect the susceptibility. The amount of OprM produced in N126 was lower than that in OCR1 but almost the same as that in PAO1 (Masuda et al., submitted) (Fig. 2, lane 3). N126 had almost the same susceptibility to tetracycline and erythromycin as PAO1, whereas N101 and KG2225 showed susceptibilities to these agents four to eight times lower than that of PAO1 (Table 2). In addition, N128, an N126-derived-mexXY-deficient strain, showed a susceptibility to tetracycline and erythromycin lower than those of N126 and N101 and almost equal to those of N102 and N103. These results suggest that the decreased level of OprM diminishes the efflux activity of MexXY-OprM in KG2225 and increases the susceptibility of KG2225 to tetracycline and erythromycin. The data also suggest that MexXY-OprM is the primary system to extrude tetracycline and erythromycin in the wild-type strain and that MexAB-OprM supports this process as a supplementary system.
MexXY-OprM is a compensatory system to extrude ofloxacin.
To confirm the involvement of MexXY-OprM in quinolone-resistance, we isolated a gentamicin-ofloxacin-resistant mutant of PAO1 and designated it N135. N135 showed a decrease in susceptibility to quinolones and aminoglycosides (Table 3) and produced MexX constitutively (Fig. 3, lane 7). Deletion of mexXY from N135 (refer to N136) increased the susceptibility to quinolones and aminoglycosides until it reached the levels of susceptibility of N101 (Table 3) and caused deficiency in MexX (Fig. 3, lane 8). These results suggest that MexXY-OprM also extrudes quinolones. Deletion of mexXY from PAO1 caused no significant change in susceptibility to ofloxacin, whereas overexpression of MexXY in N135 caused a significant increase in the susceptibility to ofloxacin. This discrepancy, which was also observed in the previous report (2), stands to reason when we note that no MexX was induced in PAO1 by exposure to ofloxacin (Fig. 3, lane 9). Thus, MexXY-OprM does not contribute to intrinsic resistance to ofloxacin in spite of the potency of MexXY-OprM in extruding ofloxacin. In contrast, ofloxacin induced the production of MexX in N126 lacking MexAB (Fig. 3, lane 10). These results suggest that MexAB-OprM is the primary system to extrude ofloxacin in the wild-type strain and that MexXY-OprM is a compensatory system to extrude ofloxacin in the mutant lacking MexAB.
TABLE 3.
Susceptibilities of ofloxacin-gentamicin-resistant mutant and its mexXY-deficient mutant
| Antimicrobial agent | MIC (μg/ml) for:
|
|||
|---|---|---|---|---|
| PAO1 | N135 | N101 | N136 | |
| Ofloxacin | 0.25 | 2 | 0.25 | 0.25 |
| Ciprofloxacin | 0.06 | 0.25 | 0.03 | 0.03 |
| Sparfloxacin | 0.5 | 2 | 0.25 | 0.13 |
| Streptomycin | 16 | 128 | 2 | 1 |
| Gentamicin | 2 | 8 | 0.13 | 0.13 |
| Amikacin | 2 | 32 | 0.5 | 0.5 |
DISCUSSION
AcrA-AcrB of E. coli are encoded on an operon having no outer membrane component gene, and they express their activity in association with the outer membrane protein TolC, which is encoded on a different region on the chromosome (3). In the present study, our constructed mutants revealed that MexXY needs the outer membrane component OprM to contribute to the intrinsic resistance to antimicrobial agents. This finding is consistent with the result reported by Aires et al. (2), who did not mention the inducible expression of MexXY. In sum, these results support the concept that an outer membrane component is also essential to the extrusion activities of multicomponent efflux systems.
MexB and MexY belong to the resistance-nodulation-division family. Inducible expression of resistance-nodulation-division efflux pumps has been reported in E. coli and Neisseria gonorrhoeae. Ma et al. (12) have shown that transcription of acrAB is elevated by decanoate, ethanol, and NaCl in E. coli, and Rouquette et al. (23) have shown that production of MtrC is induced by Triton X-100 and nonoxynol-9 in N. gonorrhoeae. Inducible expression of major facilitator family transporters by their substrates has also been reported. Firstly, Ahmed et al. (1) reported that bmr transcription is activated by rhodamine 6G and tetraphenylphosphonium in Bacillus subtilis. Secondly, Lomovskaya et al. (11) reported that transcription of emrAB is induced by weakly acidic compounds such as 2,4-dinitrophenol, salicylic acid, and nalidixic acid in E. coli. Thirdly, Grkovic et al. (8) reported that transcription of Staphylococcus aureus qacA is induced by substrates of QacA such as ethidium bromide, benzalkonium chloride, and proflavin. However, our report here is the first to show inducible expression of multidrug efflux pump by clinically used antimicrobial agents such as erythromycin.
Westbrock-Wadman et al. (29) also isolated spontaneous mexXY (amrAB)-overexpressing mutants by plating PAO1 on MHA containing tobramycin. They reported that their mutants (i) were unable to hydrolyze urea, (ii) displayed impaired growth in culture with rich media, (iii) showed dramatic decreases in the amount of OprM compared to PAO1, and (iv) did not show differences in the MICs of quinolones. MexXY-overproducing mutant N135 showed a slight decrease in the growth rate, but it was able to hydrolyze urea and it expressed almost same amount of OprM that was expressed by PAO1 (unpublished data). Since the mutants isolated by Westbrock-Wadman et al. possess diverse changes in phenotype, some change(s) apart from MexXY-overexpression might affect their susceptibility data.
Homology searches have predicted the existence of more than 10 mexAB-homologue operons on the genome in P. aeruginosa. Although the functions and expressions of these homologues are unknown, at least two efflux systems, MexAB-OprM and MexXY-OprM, are known to contribute to the intrinsic resistance of P. aeruginosa to several groups of antimicrobial agents.
ACKNOWLEDGMENTS
This research was partially supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan and by a grant from the Ministry of Health and Welfare of Japan.
We are grateful to K. Okamoto for providing KG2225.
REFERENCES
- 1.Ahmed M, Borsch C M, Taylor S S, Vazquez-Laslop N, Neyfakh A A. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J Biol Chem. 1994;269:28506–28513. [PubMed] [Google Scholar]
- 2.Aires J R, Köhler T, Nikaido H, Plésiat P. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother. 1999;43:2624–2628. doi: 10.1128/aac.43.11.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fralick J A. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukuoka T, Masuda N, Takenouchi T, Sekine N, Iijima M, Ohya S. Increase in susceptibility of Pseudomonas aeruginosa to carbapenem antibiotics in low-amino-acid media. Antimicrob Agents Chemother. 1991;35:529–532. doi: 10.1128/aac.35.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gotoh N, Tsujimoto H, Poole K, Yamagishi J, Nishino T. The outer membrane protein OprM of Pseudomonas aeruginosa is encoded by oprK of the mexA-mexB-oprK multidrug resistance operon. Antimicrob Agents Chemother. 1995;39:2567–2569. doi: 10.1128/aac.39.11.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gotoh N, Tsujimoto H, Tsuda M, Okamoto K, Nomura A, Wada T, Nakahashi M, Nishino T. Characterization of the MexC-MexD-OprJ multidrug efflux system in ΔmexA-mexB-oprM mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:1938–1943. doi: 10.1128/aac.42.8.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gotoh N, Tsujimoto H, Nomura A, Okamoto K, Tsuda M, Nishino T. Functional replacement of OprJ by OprM in the MexCD-OprJ multidrug efflux system of Pseudomonas aeruginosa. FEMS Microbiol Lett. 1998;165:21–27. doi: 10.1111/j.1574-6968.1998.tb13122.x. [DOI] [PubMed] [Google Scholar]
- 8.Grkovic S, Brown M H, Roberts N J, Paulsen I T, Skurray R A. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J Biol Chem. 1998;273:18665–18673. doi: 10.1074/jbc.273.29.18665. [DOI] [PubMed] [Google Scholar]
- 9.Hosaka M, Gotoh N, Nishino T. Purification of a 54-kilodalton protein (OprJ) produced in NfxB mutants of Pseudomonas aeruginosa and production of a monoclonal antibody specific to OprJ. Antimicrob Agents Chemother. 1995;39:1731–1735. doi: 10.1128/aac.39.8.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X-Z, Nikaido H, Poole K. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lomovskaya O, Lewis K, Matin A. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J Bacteriol. 1995;177:2328–2334. doi: 10.1128/jb.177.9.2328-2334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 13.Masuda N, Ohya S. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:1847–1851. doi: 10.1128/aac.36.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masuda N, Sakagawa E, Ohya S. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:645–649. doi: 10.1128/AAC.39.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masuda N, Gotoh N, Ohya S, Nishino T. Quantitative correlation between susceptibility and OprJ production in NfxB mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1996;40:909–913. doi: 10.1128/aac.40.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mine T, Morita Y, Kataoka A, Mizushima T, Tsuchiya T. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:415–417. doi: 10.1128/aac.43.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moken M C, McMurry L M, Levy S B. Selection of multiple-antibiotic-resistant (mar) mutants of Escherichia coli by using the disinfectant pine oil: roles of the mar and acrAB loci. Antimicrob Agents Chemother. 1997;41:2770–2772. doi: 10.1128/aac.41.12.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikaido H. Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin Infect Dis. 1998;27:S32–S41. doi: 10.1086/514920. [DOI] [PubMed] [Google Scholar]
- 20.Nikaido H. Multiple antibiotic resistance and efflux. Curr Opin Microbiol. 1998;1:516–523. doi: 10.1016/s1369-5274(98)80083-0. [DOI] [PubMed] [Google Scholar]
- 21.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poole K, Tetro K, Zhao Q, Neshat S, Heinrichs D, Bianco N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob Agents Chemother. 1996;40:2021–2028. doi: 10.1128/aac.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rouquette C, Harmon J B, Shafer W M. Induction of the mtrCDE-encoded efflux pump system of Neisseria gonorrhoeae requires MtrA, an AraC-like protein. Mol Microbiol. 1999;33:651–658. doi: 10.1046/j.1365-2958.1999.01517.x. [DOI] [PubMed] [Google Scholar]
- 24.Saier M H, Jr, Paulsen I T, Sliwinski M K, Pao S S, Skurray R A, Nikaido H. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 1998;12:265–274. doi: 10.1096/fasebj.12.3.265. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory Manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–790. [Google Scholar]
- 27.Tsuda M, Miyazaki H, Nakazawa T. Genetic and physical mapping of genes involved in pyoverdine production in Pseudomonas aeruginosa PAO. J Bacteriol. 1995;177:423–431. doi: 10.1128/jb.177.2.423-431.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuda M. Use of a transposon-encoded site-specific resolution system for construction of large and defined deletion mutations in bacterial chromosome. Gene. 1998;207:33–41. doi: 10.1016/s0378-1119(97)00601-x. [DOI] [PubMed] [Google Scholar]
- 29.Westbrock-Wadman S, Sherman D R, Hickey M J, Coulter S N, Zhu Y Q, Warrener P, Nguyen L Y, Shawar R M, Folger K R, Stover C K. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob Agents Chemother. 1999;43:2975–2983. doi: 10.1128/aac.43.12.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoneyama H, Ocaktan A, Tsuda M, Nakae T. The role of the mex-gene products in antibiotic extrusion in Pseudomonas aeruginosa. Biochem Biophys Res Commun. 1997;233:611–618. doi: 10.1006/bbrc.1997.6506. [DOI] [PubMed] [Google Scholar]
- 31.Zhao Q, Li X-Z, Srikumar R, Poole K. Contribution of outer membrane efflux protein OprM to antibiotic resistance in Pseudomonas aeruginosa independent of MexAB. Antimicrob Agents Chemother. 1998;42:1682–1688. doi: 10.1128/aac.42.7.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]