Abstract
Background
The massive increase in COVID-19 infection had generated a second wave in India during May–June 2021 with a critical pandemic situation. The Delta variant (B.1.617.2) was a significant factor during the second wave. Conversely, the UK had passed through the crucial phase of the pandemic from November to December 2020 due to B.1.1.7. The study tried to comprehend the pandemic response in the UK and India to the spread of the B.1.1.7 (Alpha, UK) variant and B.1.617.2 (Delta, India) variant.
Methods
This study was performed in three directions to understand the pandemic response of the two emerging variants. First, we served comparative genomics, such as genome sequence submission patterns, mutational landscapes, and structural landscapes of significant mutations (N501Y, D614G, L452R, E484Q, and P681R). Second, we performed evolutionary epidemiology using molecular phylogenetics, scatter plots of the cluster evaluation, country-wise transmission pattern, and frequency pattern. Third, the receptor binding pattern was analyzed using the Wuhan reference strain and the other two variants.
Results
The study analyzed the country-wise and region-wise genome sequences and their submission pattern, molecular phylogenetics, scatter plot of the cluster evaluation, country-wise geographical distribution and transmission pattern, frequency pattern, entropy diversity, and mutational landscape of the two variants. The structural pattern was analyzed in the N501Y, D614G L452R, E484Q, and P681R mutations. The study found increased molecular interactivity between hACE2-RBD binding of B.1.1.7 and B.1.617.2 compared to the Wuhan reference strain. Our receptor binding analysis showed a similar indication pattern for hACE2-RBD of these two variants. However, B.1.617.2 offers slightly better stability in the hACE2-RBD binding pattern through MD simulation than B.1.1.7.
Conclusion
The increased hACE2-RBD binding pattern of B.1.1.7 and B.1.617.2 might help to increase the infectivity compared to the Wuhan reference strain.
Keywords: Comparative genomics, Evolutionary epidemiology, Receptor binding, Variance of UK and India
Abbreviations: BFE, Binding free energy; CDC, Centre for Disease Prevention and Control; ECDC, European Centre for Disease Prevention and Control; GISAID, Global Initiative on Sharing All Influenza Data; hACE2, Human angiotensin-converting enzyme 2; ICU, Intensive Care Unit; MDS, Molecular dynamics simulation; MM/GBSA, Molecular mechanics/generalized born surface area; NAMD, Nanoscale molecular dynamics; PDB, Protein Data Bank; RBD, Receptor-binding domain; RMSF, Root mean square fluctuation; RMSD, Root mean square deviation; S-glycoprotein, Spike glycoprotein; VMD, Visual molecular dynamics; VOC, Variant of concern; VOI, Variant of interest; WHO, World Health Organization; nAb, Neutralizing antibody
Graphical abstract
1. Introduction
India has passed through a massive pandemic. A severe increase in the number of infected cases has been observed since March 2021, which has created the current COVID-19 surge in India. New COVID-19 patients were approximately 273,810 (as of April 19, 2021) in a single day, the maximum number of daily cases recorded from daily data worldwide. This incident set a new record during the pandemic (Mallapaty, 2021). A sudden increase in infection and death cases resulted in the second wave and a massive public health crisis in India. At present, this is an international concern (Rubin et al., 2021; Thiagarajan, 2021). At first, a sudden rise in infection cases was observed in Maharashtra (India)in March 2021. During that period, Indian scientists attempted to determine the biological cause of the spread of the infection. They found that one variant might be responsible for the sudden burst in the cases. A distinct B.1.617.2 variant was identified from India with three significant mutations in spike proteins from the genome sequence (Chakraborty et al., 2021a). Now the variant is circulating throughout the globe, and the dominance of the B.1.617.2 variant is noted in several countries (Bhattacharya et al., 2021a). The identified three important mutations were L452R, E484Q, and P681R (Cherian et al., 2021). Among these three mutations, L452R and E484Q are located in the RBD region. The B.1.617.2 variant was primarily identified in December 2020 in India (WHO, 2020). This B.1.617.2 variant is a significant variant in India and is currently circulating. It is claimed that this variant is associated with more infections and is linked with India's current COVID-19 surge, which is regarded as the second wave of COVID-19 (Vaidyanathan, 2021). B.1.617.2 variant has spread to approximately 50 nations, including Singapore, the UK, the USA, Germany, and Australia.
Due to its high spreading capacity and increased infectivity, the ECDC declared B.1.617.2 as a variant of concern (VOC)(ECDC, 2021a). However, the WHO affirmed B.1.617.2 variant as a variant of interest (VOI) (WHO, 2020). The CDC, USA, described the B.1.617.2 variant as VOI (Centers for Disease Control and Prevention, 2021). Several B.1.617.2 variant sequences were uploaded to the GISAID database from different countries. Most of the sequences were gathered from India and other countries such as the UK, Singapore, and the USA (Rambaut et al., 2020). Three distinct sub-lineages were observed within B.1.617. The first sub-lineage is B.1.617.1, the second sub-lineage is B.1.617.2, and the third sub-lineage is B.1.617.3. Some significant mutations were associated with spike protein alteration within B.1.617.2. The major mutations observed were L452R, E484Q, and P681R (ECDC, 2021a, ECDC, 2021b). L452R is related to a reduction in therapeutic antibodies and is associated with neutralization using convalescent plasma. This mutation is also associated with increased transmissibility (Deng et al., 2021). While, E484Q has been reported to decrease neutralization in convalescent sera (Jangra et al., 2021). Furthermore, the P681R mutation is located outside the RBD, near the furin cleavage site of the S-glycoprotein. This mutation may be associated with cell entrance and infectivity (Afrin et al., 2021; Cherian et al., 2021). Another significant mutation, D614G, was reported by researchers and was found outside the RBD (Zhang et al., 2020). However, more scientific data are needed regarding variant B.1.617.2, such as the transmissibility and reproduction number (R0) variant, the possibility for hospitalization and ICU admission, more structural detail in the mutation site, and receptor binding pattern.
Another more infective and rapidly spreading variant of SARS-CoV-2 is B.1.1.7 (Graham et al., 2021). This variant was first observed in different places in the UK and spread to 122 countries (Andrew Banchich, 2021; Volz et al., 2021). Volz et al. collected whole-genome SARS-CoV-2 sequences from the UK. They identified 9134 VOC sequences from London, 4413 VOC sequences from East England, and 5,609,413 VOC sequences from southeast England. In addition, they identified 52,795 non-VOC genomes. They observed that VOCs had spread widely across England. This variant's reproduction number (R0) is higher than other variants. The study reported that the R0 of this variant was 50%–100% higher (Volz et al., 2021). In addition, another study showed a high proportion of infections (>80%) among patients due to B.1.1.7 (Graham et al., 2021). Grint et al. observed that B.1.1.7 may be responsible for the augmented death rate among patients (Grint et al., 2021). Therefore, due to the high risk, the variant was also declared as VOC by the CDC, USA (Centers for Disease Control and Prevention, 2021), WHO (WHO, 2020), and ECDC (ECDC, 2021c). The ECDC demonstrated that this variant might cause a higher possibility of hospitalization and ICU admission (ECDC, 2021c). The variant has significant mutations. One primary mutation is N501Y, which is located in the RBD. Ostrov also confirmed this significant mutation associated with B.1.1.7 (N501Y), which increased the binding ability between the S-glycoprotein and ACE2 (Angiotensin-Converting Enzyme 2) receptor (Ostrov, 2021). Widera et al. found a high viral load in patients infected with B.1.1.7 variant having the same mutation (Widera et al., 2021). A decreased affinity was observed in B.1.1.7 variant S-glycoprotein and neutralizing mAbs (monoclonal antibodies) due to the mutation (N501Y substitution) (Cheng et al., 2021). The variant has another mutation, P681H, located outside the RBD and near the furin cleavage site (S1/S2 furin cleavage) (Lubinski et al., 2021). The D614G mutation, also observed in S-glycoprotein, was reported in this variant and was related to increased infectivity and virion density (Zhang et al., 2020).
Several single-molecule drugs and several combination therapies were assessed to comprehend the treatment strategies against COVID-19. In this direction, numerous small to major clinical trials were performed, and the significant clinical trials are ACTT-1, ACTT-2, ACTT-3, ACTT-4 study group RECOVERY trials, etc. Several single drug molecules were assessed like ritonavir/lopinavir, hydroxychloroquine, remdesivir, favipiravir, dexamethasone, ivermectin, and heparin. At the same time, several immunotherapeutic drugs were also evaluated, such as interferons, baricitinib, mavrilimumab, and tocilizumab. Some clinicians also used convalescent plasma therapy to treat COVID-19 patients. It has been noted that single dosages therapy or combination therapy of remdesivir, dexamethasone, baricitinib, tofacitinib, tocilizumab, and sarilumab might provide better results against hospitalized COVID-19 patients (Chakraborty et al., 2021b; Alam et al., 2021). Simultaneously, several vaccines have been approved for the immunization against the SARS CoV-2 virus. Among the vaccines, DNA, mRNA, protein, and attenuated vaccines are important platforms for vaccine development. The significant vaccines are Johnson and Johnson, AstraZeneca/Oxford vaccine, Moderna, Pfizer/BioNTech, Sinopharm, Sinovac, COVAXIN, and Covovax (Chakraborty et al., 2021c; Chakraborty et al., 2021d; Chakraborty et al., 2021e). Due to the continuous generation of variants, some major issues are different escape or resistance abilities such as immune escape, nAb escape, partial vaccine escape, and therapeutic resistance. Developing a new drug or vaccine that can protect against these significant variants and their mutations is necessary. Therefore, to understand the significant variants and their major mutations, comparative genomics epidemiological and evolutionary epidemiology properties are necessary for new therapeutic and vaccine development to combat the variants. This study was aimed in three directions to comprehend the biological and epidemiological properties of two emerging variants, namely B.1.1.7 (Alpha, UK origin) and B.1.617.2 (Delta, India origin).The first aim was to comprehend the comparative genomics such as country-wise and region-wise genome sequences and their submission patterns, the mutational landscape of two variants, and the structural landscape of significant mutations such as N501Y, D614G, L452R, E484Q, and P681R of these two variants. The second aim was to evaluate the evolutionary epidemiology, such as molecular phylogenetics, the molecular clock of the evolution, country-wise geographical distribution, and transmission pattern. The third aim was to analyze the receptor binding interaction, such as molecular docking analysis, calculation of binding free energy, and molecular dynamics simulation to understand the hACE2 receptor-RBD interactions.
2. Material and methods
2.1. Data collection
Data was collected for the Wuhan strain, B.1.1.7 and B.1.617.2, from different sources such as NCBI, CDC(USA), (Control and Prevention, 2021), WHO (WHO, 2020), and ECDC (ECDC, 2021c). Relevant keywords were searched in several significant databases such as PubMed (Liu, 2020; Zuo et al., 2021), Web of Science (Farooq et al., 2021), and Google Scholar. Different keywords for database search such as “B.1.1.7,” “B.1.617.2,” “variants of interest (VOI),” “variants of concern (VOC),” “variants of consequence” were used in this study.
The data was also collected using several databases and servers such as Github and Pango lineages (O'Toole and McCrone, 2020), Pango lineages (O'Toole et al., 2020), SARS-CoV-2 resources GISAID (Velazquez et al., 2020), and Nextstrain server (Hadfield et al., 2018; Nextstrain, 2020). The NextStrain server fetches the records from the GISAID server. All the data were obtained from the different servers during the first week of July 2021.The statistical calculation and verification were performed using different advanced statistical tools and techniques wherever it is needed, such as MATLAB, and PAST 4.03 software (Hammer et al., 2001; MathWorks, 1992).
2.2. Comparative genomics, evolutionary epidemiology analysis
2.2.1. Country-wise and region-wise genome sequences analysis of B.1.617.2 and B.1.1.7 and their submission pattern
For analysis of genome sequences and their submission pattern, for these two variants, we used servers such as GitHub and Pango lineages (O'Toole and McCrone, 2020), Pango lineages (O'Toole et al., 2020), and GISAID (Velazquez et al., 2020).
2.2.2. Molecular phylogenetics, the molecular clock of evolution, country-wise transmission prototype and geographical positioning B.1.617.2 and B.1.1.7
Here, we used servers and databases like Nextstrain (Hadfield et al., 2018; Rambaut et al., 2020) and GISAID (Velazquez et al., 2020) to analyze the molecular phylogenetics, the molecular clock of evolution, country-wise geographical distribution, and transmission pattern, and entropy diversity of these two variants. The Nextstrain server is used to evaluate a pathogen's real-time evolution and is a very efficient tool for phylodynamics analysis (Hadfield et al., 2018). At the same time, GISAID is one of the largest genome sequences servers. This is one of the significant resources for analyzing SARS-CoV-2 variants during pandemics, especially for VOC and VOI. Several scientists have used the GISAID to analyze the millions of sequences in different variants such as B.1.1.7 (Alpha; originated from the UK), B.1.617.2 (Delta; originated from India), P1 (Gamma; originated from Brazil), B.1.351 (Beta; originated from South Africa) (Kalia et al., 2021; Zelenova et al., 2021; Khare et al., 2021). The classification methodology as proposed by Rambaut et al. (2020) was followed.
2.3. Analysis of the structural landscape of significant mutations such as N501Y, D614G L452R, E484Q, and P681R, which are found in these two variants
The COVID-3D server was used for the structural analysis of significant mutations (N501Y, D614G L452R, E484Q, and P681R) in two emerging variants (Portelli et al., 2020). Subsequently, the GISAID database (Velazquez et al., 2020) was used for significant mutational landscape analysis. COVID-3D server is used to analyze and visualize the structural landscape of genetic variation (Portelli et al., 2020).
2.4. Molecular docking analysis
In this analysis, the binding interaction between human ACE2 (hACE2) and RBD of SARS-CoV-2 in the Wuhan strain (PDB ID: 6VW1) and more infective variants such as B.1.1.7 (PDB ID: 7BWJ) and B.1.617.2 were predicted. The Wuhan strain was used for molecular docking experiments, and the infective variants were used for comparative analysis. Using UCSF chimera software, the PDB files were pre-processed and downloaded from the RCSB protein data bank (Kouranov et al., 2006). The S-glycoprotein 3D structure of B.1.617.2 was not found in the RCSB PDB. Therefore, the generated RDD of the B.1.617.2 variant by introducing two significant mutations in control RBD, 452R and E484Q. The molecular docking was performed using the HDOCK server and visualized and edited the PDB structure using PyMOL software. The HDOCK server predicts the binding complexes between two molecules, such as protein-protein docking, using a unique hybrid algorithm. The hybrid algorithm was designed using a template-based method and an ab initio free docking modeling server (Yan et al., 2020; De Vries et al., 2010; Yan et al., 2017).
2.5. Binding free energy (BFE) calculation
In this study, the HawkDock online server was employed to evaluate the BFE of our docked complexes using MM/GBSA approaches (Weng et al., 2019; Chen et al., 2016;). For MM/GBSA analysis, the binding free energy (ΔGbind) of the system can be defined as a change in free energy, which is expressed as
| (1) |
In the above, ΔEMM depicts the changes in the MM energy of the system, ΔGsolv indicates changes in solvation free energy, and TΔS denotes the total system binding entropy. Further equations calculate how the ΔEMM energy and ΔGsolv solvation free energy are obtained from the system. The equation is
| (2) |
Where ΔEinternal is the change in internal MM energy, ΔEelectrostatic is the change in electrostatic energy, and ΔEvdw is the change in van der Waals energy.
| (3) |
The equation represents deviations of solvation free energy combined with non-electrostatic solvation (ΔGSA) and energy electrostatic solvation (ΔGGB).
2.6. Molecular dynamics simulation (MDS)
MDS is an efficient method for identifying and predicting biological or chemical processes at the atomic level (Cao et al., 2017). To generate the MDS analysis, the NAMD 2.14b2_win64-multicore software package was used for three docked complexes (Kalé et al., 1999; Phillips et al., 2005; Phillips et al., 2020; Melo et al., 2018) and optimized the parameters using CHARMM22 and CHARM36 all-atom force field (MacKerell Jr et al., 2000). Protein topologies were prepared using the psf module, water solvation was prepared around the complex molecules, and equilibrated TIP3P (transferable intermolecular potential 3P) water was added as solvation. After that, the system was placed at least 1.0 nm from the box edge, and 56 sodium ions and 30 Cl ions were included in the solvation molecule, which was used to neutralize the solvated protein structure. The energy minimization of the system uses the NAMD minimizer of 1000 steps using an algorithm called the steepest descent algorithm. The system equilibration was then achieved in two-step: NVT ensembles and NPT ensembles for 1000 ps. MD simulations of the equilibrated system were 20 ns (Timestep) at 2 fs speed. The simulation trajectories were analyzed with VMD 1.9.3 graphic visualization software and evaluated with some other types of secondary analysis such as bond angle, dihedral, etc. (Humphrey et al., 1996).
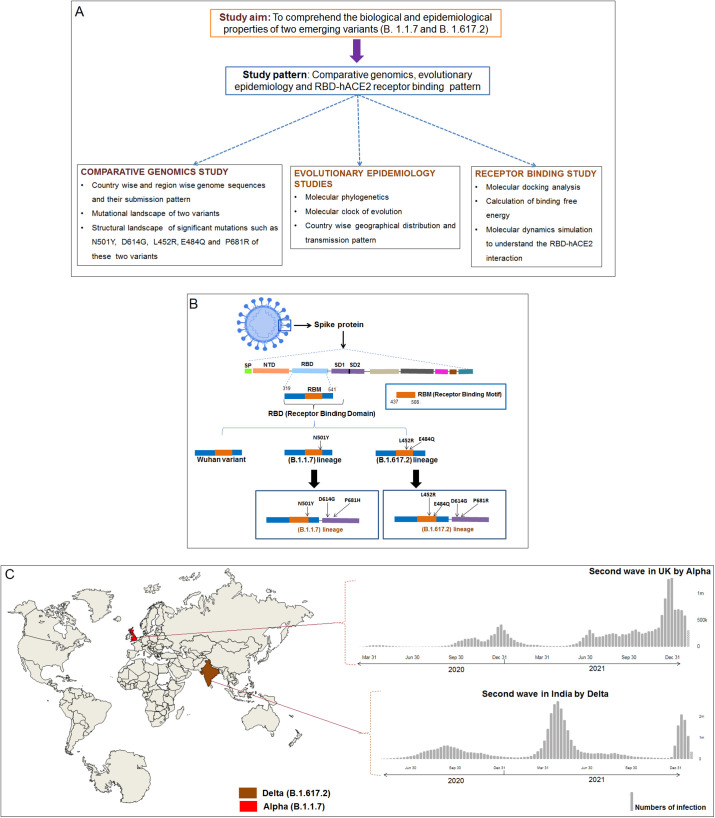
The entire study methodology is represented through a flowchart to understand the overall study process (Fig. 1A). A schematic diagram of two variants along with the Wuhan strain is illustrated in Fig. 1B. Another schematic diagram of these two variants and their pandemic response is depicted in Fig. 1C.
Fig. 1.
The flowchart of the study methodology and schematic diagram of the S-protein of three strains (one control and two variants). (A) Flowchart of our study methodology that describes the workflow in three directions which are comparative genomics, evolutionary epidemiology, and receptor binding. (B) The schematic diagram shows the features of RBD of one control (Wuhan strain) and two variants (B.1.617.2 and B.1.1.7). The diagram shows significant mutations in B.1.1.7 (N501Y, D614G, and P681R) and B.1.617.2 (N501Y, D614G, L452R, E484Q, and P681R) (C). Two variants (B.1.1.7 and B.1.617.2) and their pandemic response in their country of origin. B.1.1.7 (Alpha) originated in the UK, and it is responsible for the second wave generation in the UK in the second half of 2020. Similarly, B.1.617.2 (Delta) is originated in India, and the variant is responsible for the generation second wave in India during the first half of 2021.
3. Results
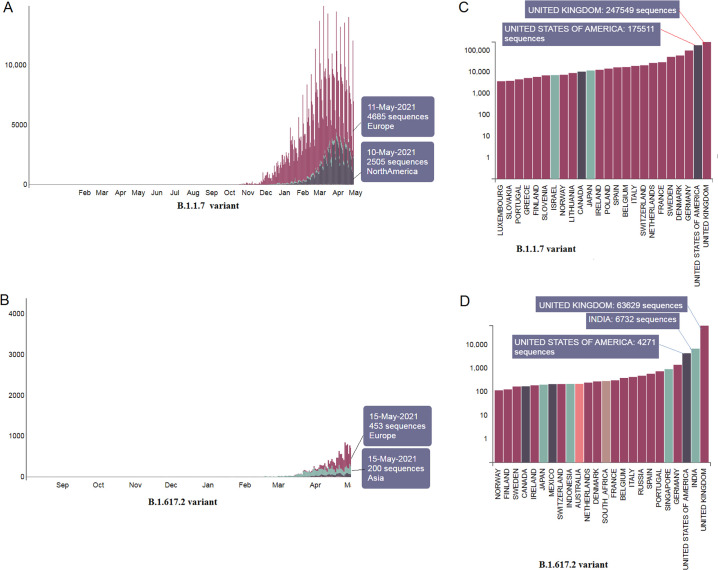
3.1. Country-wise and region-wise submitted genome sequences number and pattern of these two variants
The PANGO lineage data shows these two variants' region-wise submitted genome sequence pattern. Data indicates that more sequences were submitted for the B.1.1.7 (Fig. 2A) than the B.1.617.2 variant (Fig. 2B). The country-wise submission patterns of these variants were also mapped. Furthermore, genome sequences for the B.1.1.7 (Fig. 2C) compared to the B.1.617.2 variant were also found (Fig. 2D). We observed the highest number of genome sequences for B.1.1.7, submitted from various places in the UK (247,549 nos), and the second-highest number of genome sequences for this variant was introduced from the USA (175,511 nos). The genome summation pattern of the B.1.1.7 variant indicates that this variant was transmitted to different parts of the world from the UK.
Fig. 2.
Country-wise and region-wise submitted genome sequences pattern of two variants. (A) Region-wise submitted genome sequences pattern of B.1.1.7. (B) Region-wise submitted genome sequences pattern of B.1.617.2. (C) Country-wise submitted genome sequences number of B.1.1.7. It shows the maximum sequences were deposited from the UK. The Scatter plot of the cluster shows the dots of B.1.617.2, located in the upper position of the regression line. (D) Country-wise submitted genome sequences number of B.1.617.2. It also shows the maximum sequences were deposited from India.
For the B.1.617.2 variant, 6732 genome sequences were found from India, 63,629 genome sequences from the UK, and 4271 genome sequences from the USA. The study shows that the B.1.1.7 variant is widely spread and well-studied. However, this B.1.617.2 variant has just started to spread from India to other countries. Probably, this is the time to alert other countries for the spared of the B.1.617.2 variant.
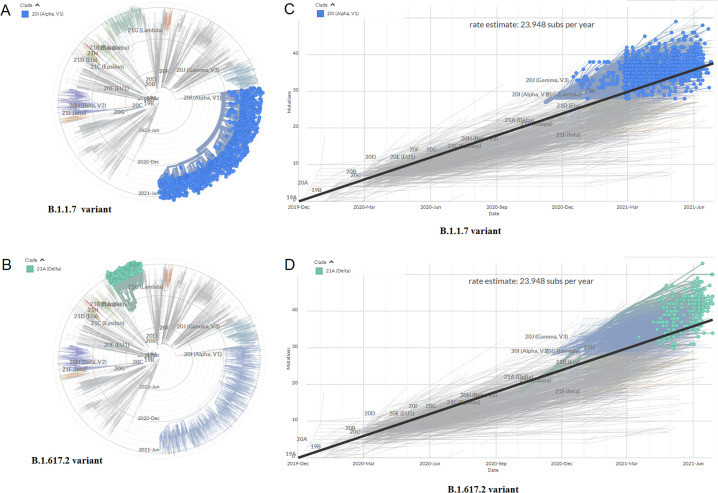
3.2. Molecular phylogenetics of these two variants
Molecular phylogenetic analysis was performed using the genome sequences of these two variants. A radial phylogenetic tree of the B.1.1.7 variant was developed using 890 sequences between September 2020 and May 2021 (Fig. 3A). A radial phylogenetic tree of the B.1.617.2 variant used 66 sequences between January 2021 and May 2021 (Fig. 3B).
Fig. 3.
Evaluation of molecular phylogenetics and scatter plot of a cluster of two variants. (A) Radial phylogenetic tree of the B.1.1.7 using 890 sequences. (B) Radial phylogenetic tree of the B.1.617.2 using 66 sequences. (C) Scatter plot of a cluster of the B.1.1.7 using 890 sequences. The Scatter plot of the cluster shows the dots of B.1.1.7, located in the upper position of the regression line.
(d) Scatter plot of a cluster of the B.1.617.2 using 66 sequences. The Scatter plot of the cluster shows the dots of B.1.617.2, located in the upper position of the regression line.
This study used the Nextstrain server and GISAID to access the molecular phylogenetics, the molecular clock of evolution, country-wise geographical distribution, transmission pattern, etc., two variants. Researchers have frequently applied these two servers to understand the emergence and evolution of SARS-CoV-2 variants (Islam et al., 2021; Alai et al., 2021).
3.3. Molecular clock of the evolution of these two variants
The molecular clock estimation was performed and depicted as scatter plots, showing the cluster genome samples of the sampling date and estimated mutation with regression lines. The advanced tools estimated the substitution rate (23.948 substitutions per year). The molecular clock of the B.1.1.7 variant using 1132 sequences (Fig. 3C) and the B.1.617.2 variant using 235 sequences was developed (Fig. 3D). The molecular clock was generated using a linear regression model. This model shows the sample values scatter as plots that are spread around the regression line. The plot illustrates the general patterns of the molecular clock and the mutations of these two variants through the plotted points.
3.4. Speard and country-wise transmission prototype of these two variants
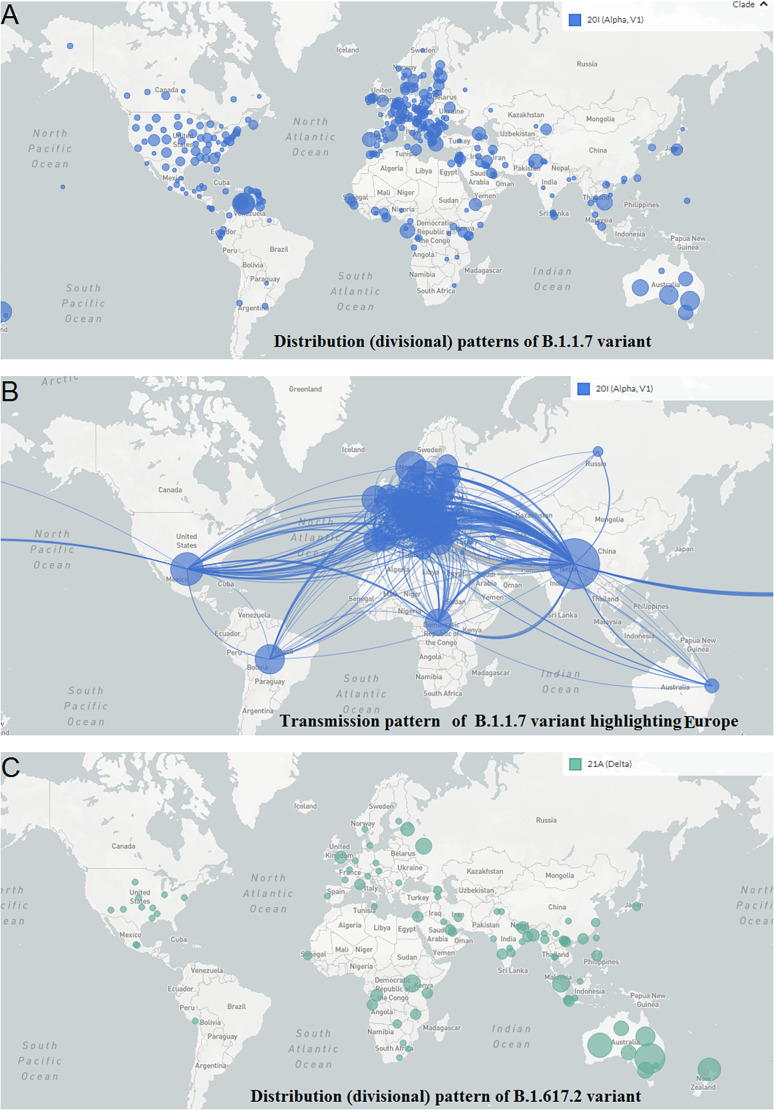
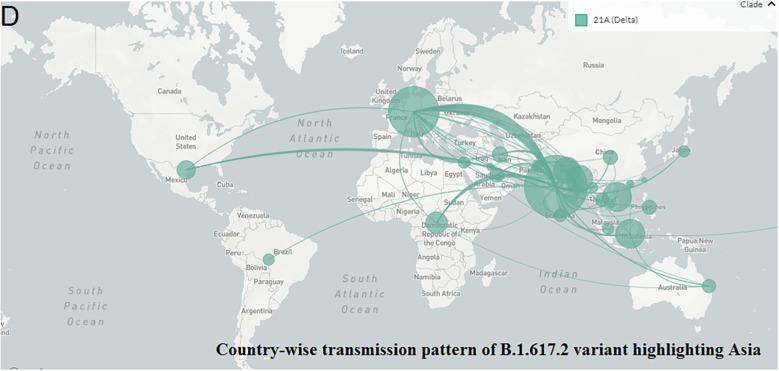
The geographical distribution (divisional) and country-wise transmission prototype of B.1.1.7 were evaluated. The geographical spread and country-wise transmission prototype highlighting B.1.1.7 in Europe are represented in Fig. 4A and B, respectively. This lineage emerged in the UK. From the transmission model, it appears that the variant is transmitted all over the countries of the European Union, different parts of the USA, and Canada. It is also widespread in several parts of South Africa, Latin America, and Asia. It also got transmitted to India, Sri Lanka, Australia, and the Philippines. The geographical distribution (divisional) and country-wise transmission pattern of the B.1.617.2 variant is also depicted. The geographical spread and country-wise transmission pattern highlighting B.1.617.2 in Asia are represented in Fig. 4C and D, respectively. The lineage was originated in India and transmitted to several regions of the UK, the USA, and some parts of South Africa. This variant also got transmitted to Australia, Malaysia, and Thailand.
Fig. 4.
Geographical distribution and country-wise transmission pattern of the two variants. (A) Geographical distribution of the B.1.1.7. (B) Country-wise transmission pattern of the B.1.1.7. (C) Geographical distribution of the B.1.617.2. (D) Country-wise transmission pattern of the B.1.617.2.
3.5. Entropy diversity of each nucleic acid position throughout the genome, S-glycoprotein, and RBD region of these two variants
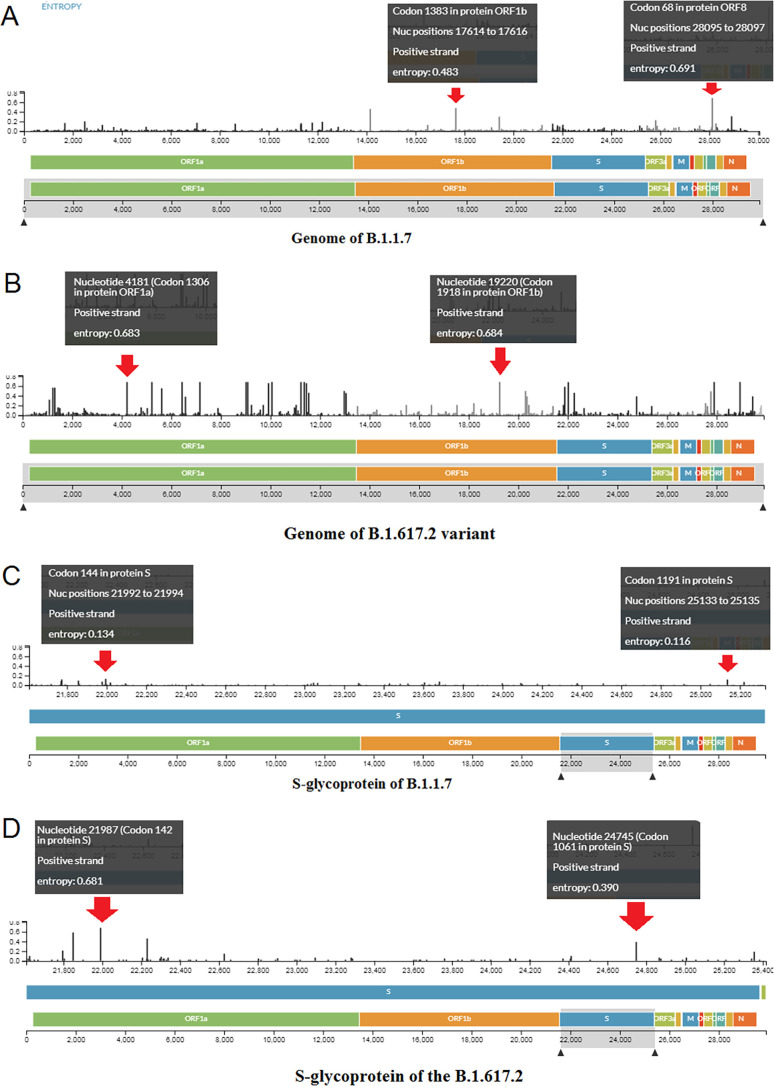
The entropy diversity of each nucleic acid position throughout the genome of the B.1.1.7 variant in a frame was evaluated (Fig. 5A ). The entropy diversity of each nucleic acid position all over the genome of the Delta variant in a frame was also recorded (Fig. 5B).
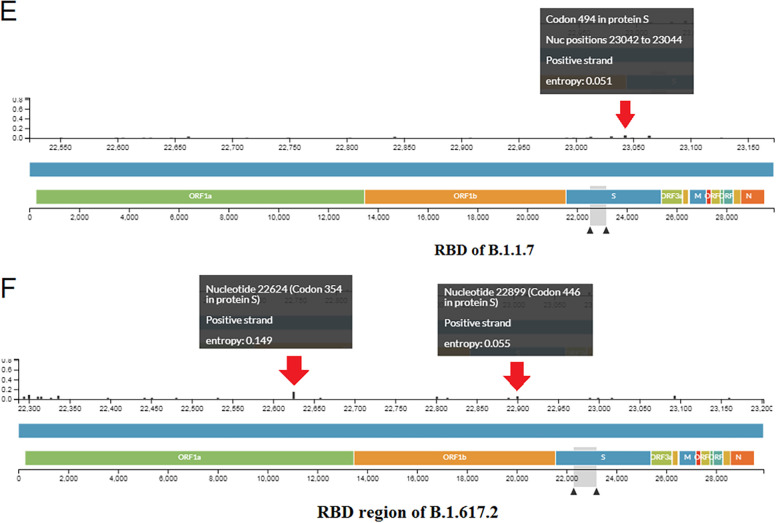
Fig. 5.
Frequencies of two variants (A) Entropy diversity of each nucleic acid position throughout the genome of the B.1.1.7.(B) Entropy diversity of each nucleic acid position throughout the genome of the B.1.617.2. (C)The entropy diversity of each nucleic acid position of S-glycoprotein of B.1.1.7.(D) The entropy diversity of each nucleic acid position of S-glycoprotein of B.1.617.2. (E) The entropy diversity of each nucleic acid position of the RBD region of the B.1.1.7. (F) The entropy diversity of each nucleic acid position of the RBD region of the B.1.617.2.
Furthermore, the entropy diversity of each nucleic acid position of the S-glycoprotein of the B.1.1.7 variant was developed, shown in Fig. 5C. The entropy diversity of each nucleic acid position of the S-glycoprotein of the B.1.617.2 variant was also evaluated (Fig. 5D).
Finally, we evaluated the entropy diversity of each nucleic acid position in the RBD region of the B.1.1.7 variant (Fig. 5E). We also evaluated the entropy diversity of each nucleic acid position of the RBD region of the B.1.617.2 variant (Fig. 5F).
We found the maximum entropy of each nucleic acid position throughout the genome, S-glycoprotein, and RBD region of B.1.617.2, compared to B.1.1.7.
3.6. Mutational landscape of S-glycoprotein of these two variants
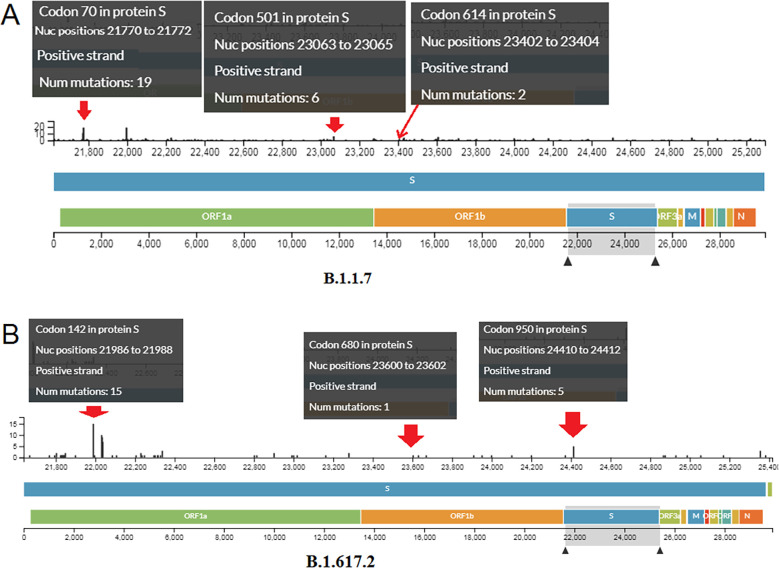
We evaluated the mutational landscape of all mutations in these two significant variants. Based on the results, we found several mutations in a different positions in the S-glycoprotein of the B.1.1.7 variant (Fig. 6A). We found nineteen mutations in codon 70, six in codon 501, and two in codon 614 in this variant. Several mutations at different positions in the S-glycoprotein of the B.1.617.2 variant were also observed (Fig. 6B). We also observed fifteen mutations in codon 142, one mutation in codon 452, one mutation in codon 680, and five mutations in codon 950 in this variant.
Fig. 6.
Mutational landscape of S-glycoprotein of two variants. (A) Mutations in a different particular position in the S-glycoprotein of the B.1.1.7. (B) Mutations in a different position in the S-glycoprotein of the B.1.617.2.
3.7. The structural landscape of significant mutations (N501Y, D614G L452R, E484Q, and P681R) found in these two variants
The structural landscape of mutation analysis revealed significant mutations, such as N501Y, D614G L452R, E484Q, and P681R, which are frequently found in these two variants.
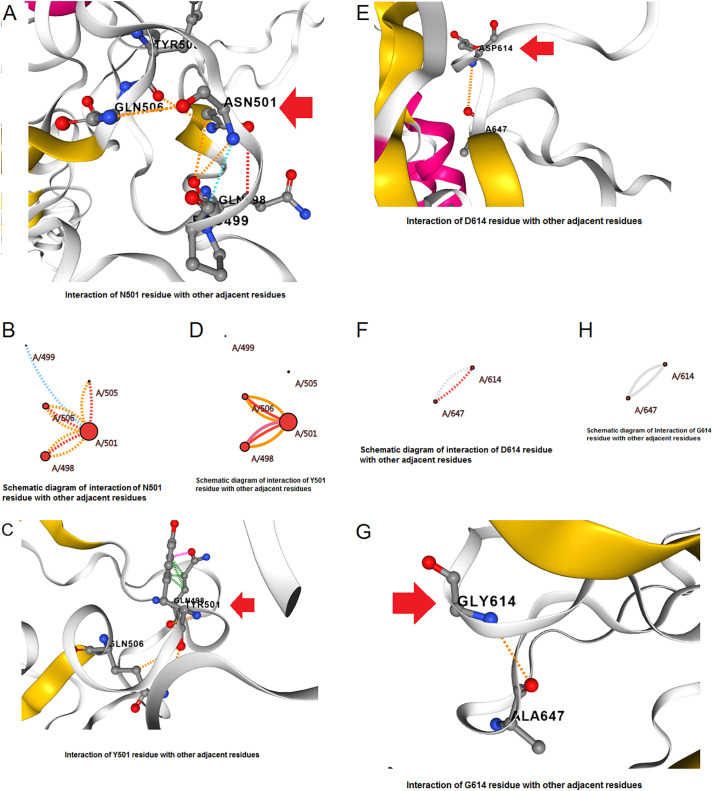
We evaluated the N501Y mutation in the UK variant (Alpha). Within the N501Y structure, the amino acid was altered from Asn501→Tyr. The structural analysis of N501Y showed different forms of interactions with other residues such as Q506 etc. In this variant mutation, the interactivity among the other residues in the wild form (N501) is shown in Fig. 7A . Again, we have shown a schematic interaction demonstrating the interaction of the wild residue via different bond formations with the nearby residues. The schematic interaction pattern among the residues in the wild form (N501) is shown in Fig. 7B and displays diverse types of interaction with different types of bonds with other nearby residues. Likewise, we have shown the interaction of the mutant-type residue and its molecular interaction with the other residues. The mutant-type residue (Y501) and its molecular interaction among the other nearby residues are shown in Fig. 7C. The schematic diagram shows the mutant-type residue (Y501) and its interaction among the other nearby residues (Fig. 7D). It extensively shows the different types of interaction with different types of bond formation among other nearby residues.
Fig. 7.
The structural landscape of significant mutations of the B.1.1.7. (A) Molecular association between the residues in the wild-type N501Y mutation. (B) The schematic diagram shows the molecular association between the residues in the wild-type N501Y mutation. (C) Molecular association between the residues in the mutant type N501Y mutation. (D) The schematic diagram shows the molecular association between the residues in the mutant type N501Y mutation. (E) Molecular association between the residues in the wild-type D614G mutation. (F) The schematic diagram shows the molecular association between the residues of wild type D614G mutation. (G) Molecular association between the residues in the mutant type D614G mutation. (H) The schematic diagram shows the molecular association between the residues of mutant type D614G mutation.
Again, like the previous point AA mutation, we also evaluated the D614G mutation, which was identified as the B.1.1.7 variant. In the D614G structure, the amino acid changed from Asp614→Gly. The structural evaluation of the D614G mutation was performed, showing different forms of interactions. In this variant mutation, the molecular association of the wild type (D614) and the surrounding residues is shown in Fig. 7E. At the same time, a schematic interrelation diagram has been shown to depict the different bond formation and interaction between the wild type residue (D614) and the adjacent residues (Fig. 7F). Similarly, the mutant type residue (G614) and its interactions between the neighboring residues are shown in Fig. 7G. An interrelation diagram has also been depicted to show the interaction and bond formation between the mutant residue (G614) and adjacent residues (Fig. 7H).
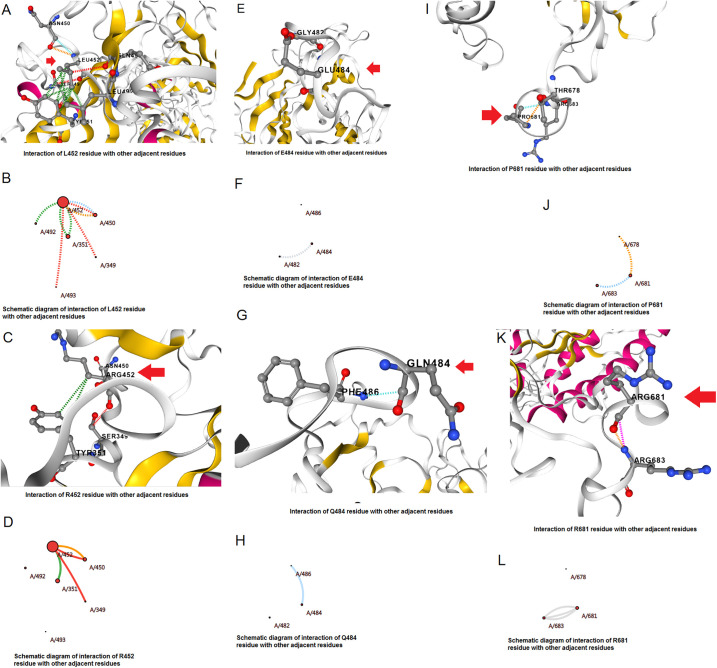
Furthermore, the L452R mutation was evaluated, which was identified in B.1.617.2 variant. In the L452R structure, the amino acid was altered from Leu452→Arg. Structural analysis of the L452R mutation showed different forms of interactions. In this variant, the interrelation between the wild-type residue (L452) and adjacent residues is shown in Fig. 8A . Similarly, we have drawn a schematic interaction diagram between the wild-type residue (L452) and nearby residues, and a schematic diagram for interrelation is shown in Fig. 8B. Again, we have shown the molecular association between the mutant type residue (R452) and surrounding residues (Fig. 8C). To make the interaction clearer, we depicted a schematic interaction diagram and a schematic interrelation between the mutant type residue (R452) and adjacent residues (Fig. 8D).
Fig. 8.
The structural landscape of significant mutations of the B.1.617.2. (A) Molecular association between the residues in the wild-type L452R mutation. (B) The schematic diagram shows the molecular association between the residues in the wild-type L452R mutation. (C) Molecular association between the residues in the mutant type L452R mutation. (D) The schematic diagram shows the molecular association between the residues of mutant type L452R mutation. (E) Molecular association between the residues in the wild-type E484Q mutation. (F) The schematic diagram shows the molecular association between the residues in the wild-type E484Q mutation. (G) Molecular association between the residues in the mutant type E484Q mutation. (H) The schematic diagram shows the molecular association between the residues of mutant type E484Q mutation. (I) Molecular association between the residues in the wild-type P681R mutation. (J) The schematic diagram shows the molecular association between the residues in the wild-type P681R mutation. (K) Molecular association between the residues in the mutant type P681R mutation. (L) The schematic diagram shows the molecular association between the residues of mutant type P681R mutation.
We next evaluated the E484Q mutation, which was identified in B.1.617.2 variant. In the E484Q structure, the amino acid changed from Glu484→Gln. Structural analysis of the E484Q mutation showed different interactions between the residues. In this AA variant (E484Q), the interaction between the wild type (E484) is shown in Fig. 8E. We have drawn a schematic representation to show the interaction and the bond formation between the wild type (E484) and nearby residues (Fig. 8F). Simultaneously, we have also shown the interrelation between mutant type residue (Q484) and adjacent residues in Fig. 8G. A schematic representation was developed to show the molecular association among the mutant type residue (Q484) and adjacent residues, and a schematic interaction figure is shown in Fig. 8H.
Finally, we evaluated the P681R mutation identified in the B.1.617.2 variant. In the P681R structure, the amino acid was altered from Pro681→Arg. The structural analysis of P681R showed different forms of interactions between the residues. In variant P681R, the wild-type residue (P681) and its interrelation with other residues are shown in Fig. 8I. Furthermore, a schematic diagram illustrates the molecular association among the wild-type residue (P681) and neighboring residues (Fig. 8J). The interactivity between the residue of the mutant type (R681) and others is shown in Fig. 8K. Simultaneously, a schematic diagram was drawn to visualize the graphical interaction between the mutant type residue (R681) and other residues (Fig. 8L).
COVID-3D server is noteworthy severer to analyze the mutational landscape. Presently, many researchers are using the COVID-3D server to understand the mutational landscape of SARS-CoV-2 variants. Jacob et al. have used the COVID-3D server in their mutational assessment of the variants and apprehend the destabilizing or stabilizing properties (Jacob et al., 2020). We have also used previously to understand the mutational residue characteristics of the SARS-CoV-2 variants using the COVID-3D server (Chakraborty et al., 2021f).
3.8. Molecular docking analysis
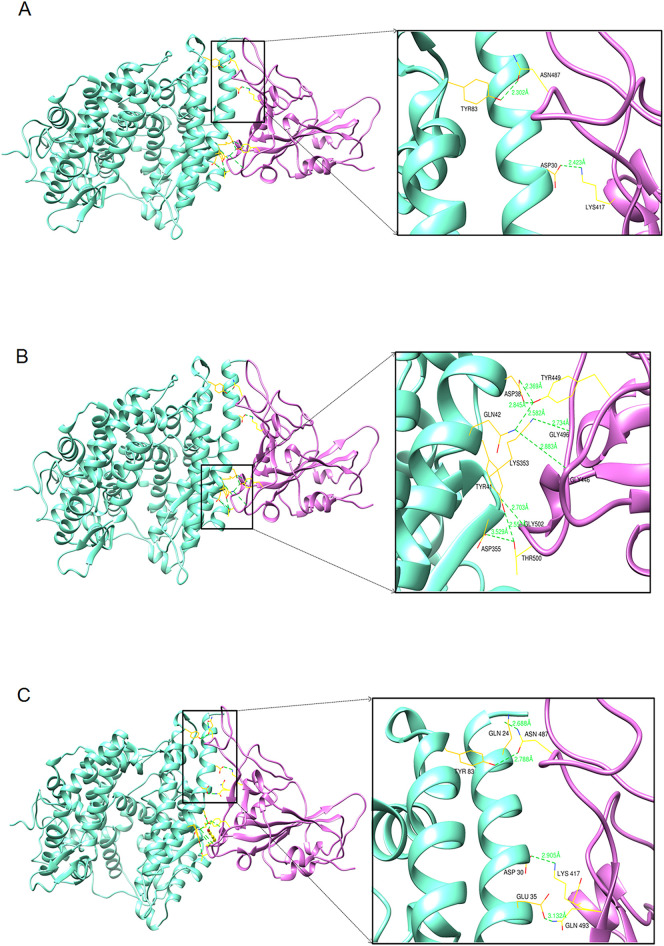
The results of the molecular docking analysis of the Wuhan strain are illustrated in Fig. 9A and B. Fig. 9A represents the top view (zoom) of the interaction between hACE2 and RBD residues of the Wuhan strain. Fig. 9B depicts the bottom view (zoom) of the interaction between hACE2 and RBD residues of the Wuhan strain.
Fig. 9.
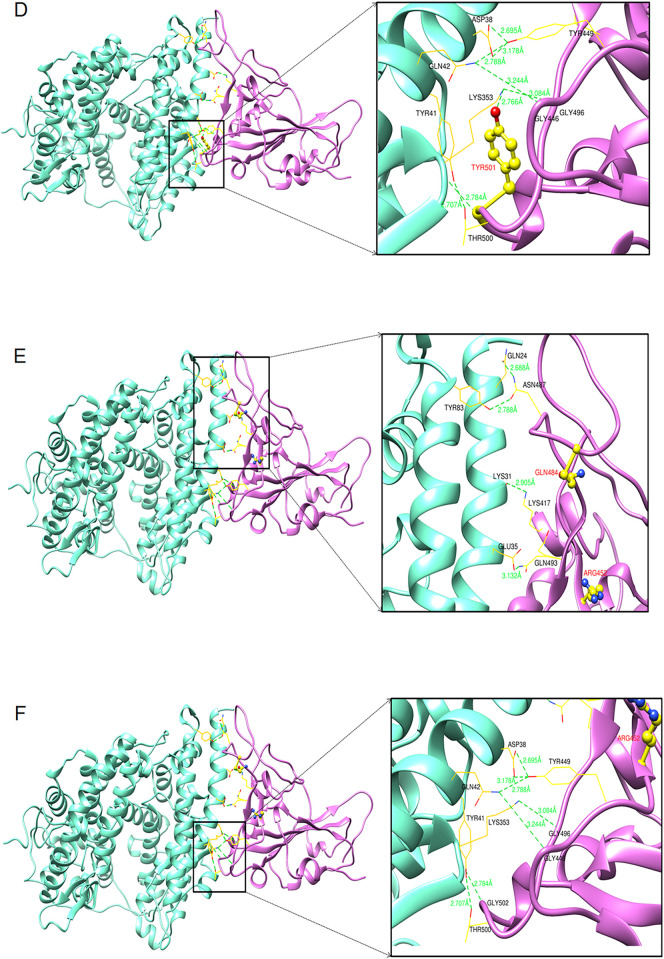
The diagram shows the protein-protein docking complexes of hACE2 and RBD. The hACE2 is colored by cyan, and its interacting RBD domains are colored by purple. Comparison of the binding interaction of residues of RBD and hACE2 of Wuhan strain and other two variants. (A) Top zoom view of the interaction between RBD and hACE2 residues of the Wuhan strain. (B) Bottom zoom view of the interaction between RBD and hACE2 residues of the Wuhan strain. (C) Top zoom view interaction between RBD and hACE2 residues of B.1.1.7 variant. (D) The bottom zoom view of the interaction between RBD and hACE2 residues of B.1.1.7 variant and yellow color stick represent the mutation of N501Y. (E) The top zoom view of the interaction between RBD and hACE2 residues of B.1.617.2 variant and yellow color stick represent the mutation of L452R and E484Q. (F) Bottom zoom view of the interaction between RBD and hACE2 residues of B.1.617 variant.
The molecular docking analysis of the B.1.1.7 variant is shown in Fig. 9C and D. Fig. 9C shows the top view (zoom) of the interaction between hACE2 residues and RBD residues of the B.1.1.7 variant. Similarly, Fig. 9D illustrates the bottom view (zoom) of the interaction between hACE2 and RBD residues of B.1.1.7.
The results of the molecular docking evaluation of the B.1.617.2 variant are shown in Fig. 9E and F. Fig. 9E illustrates the top view (zoom) of the interaction between hACE2 residues and RBD residues of the B.1.617.2 variant. Simultaneously, Fig. 9F shows the bottom view (zoom) of the interaction between RBD and hACE2 residuesof the Delta variant.
The molecular docking study shows the comparative hydrogen bonds formed during the interaction of hACE2 and RBD residues of the three variants (Wuhan strain, Alpha variant, and Delta variant) and is presented in Table 1 . The study showed that the molecular interactivity of the residues of hACE2 and RBD of B.1.1.7 formed more hydrogen bonds during the interaction (12 numbers) compared to the B.1.617.2 variant (11 numbers).
Table 1.
Comparative analysis of hydrogen bonds formed during the interaction of hACE2 and RBD residues of one control and two variants.
| Sl. no. | Variant | Number of hydrogen bonds (H-bonds) | Residues involve in hydrogen bond (ACE2_Spike protein) |
Hydrogen bond length |
|---|---|---|---|---|
| 1 | Wuhan strain | 9 | TYR83-ASN487 ASP30-LYS417 ASP38-TYR449 GLN42-TYR449 GLN42-GLy446 TYR41-THR500 LYS353-GLY496 LYS353-GLY502 ASP355-THR500 |
2.302 Å 2.423 Å 2.369 Å 2.582 Å 2.885 Å 2.553 Å 2.734 Å 2.703 Å 3.529 Å |
| 2 | B.1.1.7 (UK) variant | 11 | GLN24-ASN487 TYR83-ASN487 ASP30-LYS417 GLU35-GLN493 ASP38-TYR449 GLN42-TYR449 GLN42-GLy446 TYR41-THR500 LYS353-GLY496 LYS353-GLY502 LYS353-TYR501 |
2.688 Å 2.788 Å 2.905 Å 3.132 Å 2.695 Å 2.778 Å 3.244 Å 2.707 Å 3.084 Å 2.784 Å 2.766 Å |
| 3 | B.1.617 (Indian) variant | 10 | GLN24-ASN487 TYR83-ASN487 ASP30-LYS417 GLU35-GLN493 ASP38-TYR449 GLN42-TYR449 GLN42-GLy446 TYR41-THR500 LYS353-GLY496 LYS353-GLY502 |
2.688 Å 2.788 Å 2.905 Å 3.132 Å 2.695 Å 2.788 Å 3.244 Å 2.707 Å 3.084 Å 2.784 Å |
Molecular docking performed by the HDOCK server revealed that B.1.617.2 has a higher binding affinity (Fig. 1B), which was further analyzed using the MM/GBSA technique. The study found increased molecular interactivity between hACE2-RBD binding of B.1.1.7 and B.1.617.2 compared to the Wuhan reference strain.
In this study, the HDOCK server was used to dock the RBD and hACE2 residues of B.1.1.7. and B.1.617.2. HDOCK server is used to perform protein-protein docking very frequently by researchers. Recently, Calcagnile et al. have used the HDOCK server in their molecular docking study to understand the hACE2 gene polymorphism, which can interfere with the interaction landscape between the SARS-CoV-2 spike and hACE2 (Calcagnile et al., 2021). Bhattacharya et al. have used the HDOCK server in several studies to perform the protein-protein interaction (Bhattacharya et al., 2020a; Bhattacharya et al., 2020b; Bhattacharya et al., 2021b; Bhattacharya et al., 2022a).
3.9. BFE calculation
The HawkDock is a server for online docking online. A freely available docking server uses the ATTRACT docking algorithm to calculate the BFE using the MM/GBSA technique. The server describes the free energy of binding of hACE2 and RBD of the three strains. The predicted negative score of binding free energy indicates that the binding affinity of the hACE2 and RBD residues is vital for these two variants. The comparative free energies of the binding complexes of the three variants' hACE2 residues and RBD residues are shown in Table 2 . It was noted that the lowest negative binding free energy for B.1.1.7 during the molecular association of the hACE2 residues and RBD residues is ΔGbind = −64.27 and ΔEvdw = −104.01, respectively. Data also revealed an increased molecular association between hACE2-RBD binding of these two variants compared to the Wuhan reference strain.
Table 2.
Comparative evaluation of binding free energy using MM/GBSA method. Here we used one control and two variants of the RBD-hACE2 complex.
| Energy components (kcal/mol) | Wuhan spike_ variant_hACE2 |
B.1.617 spike_ variant_hACE2 |
B.1.1.7 spike_ variant_hACE2 |
|---|---|---|---|
| Binding free energy (ΔGbind) | −59.71 | −64 | −64.27 |
| Van der Waal energy (ΔEvdw) | −99.84 | −99.6 | −104.01 |
| Electrostatic energy (ΔEelectrostatic) | −662.07 | −1084.18 | −672.78 |
| Electrostatic solvation energy (ΔGGB) | 715.29 | 1132.7 | 725.93 |
| Non-electrostatic solvation energy(ΔGSA) | −13.09 | −12.93 | −13.41 |
In this study, we used the HawkDock server for Binding free energy (BFE) calculation to understand the binding affinity of the hACE2 residues and RBD residues of B.1.1.7 and B.1.617.2 variants. Several scientists have used the HawkDock server to calculate BFE. Akachar et al. have used the HawkDock server to understand the binding efficiency of newly designed peptides inhibitors against spike RBD SARS-CoV-2 (Akachar et al., 2020). Parate et al. (2021) have applied the HawkDock server to illustrate the BFE of the binding interaction of the inhibitory protein of Raf Kinase (Parate et al., 2021). Tallei et al. used the HawkDock server to evaluate the BFE to understand the attachment of a fruit-derived small peptide. It has been noted that the small peptide can restrain the attachment of variants of SARS-CoV-2 RBD to hACE2 (Tallei et al., 2022).
3.10. Molecular dynamics simulation
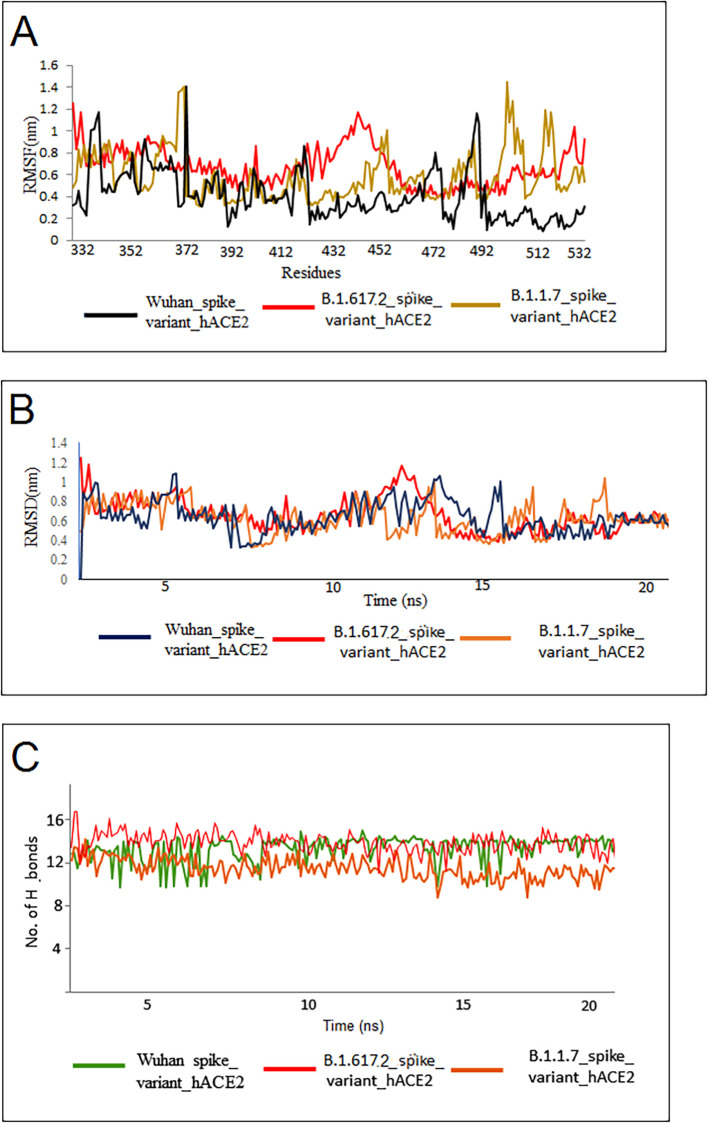
The RMSF, RMSD, and the quantity of hydrogen bonding were estimated using the MDS trajectories of all three variants. The Wuhan strain RBD-hACE2 complex and the developed and equilibrated RBDs of these two variants were simulated for 20 ns. The average RMSF of the Wuhan RBD-hACE2 complex was 1.5 ± 0.85 nm, and the average RMSF of B.1.1.7 variant RBD-hACE2 complex protein was 1.68 ± 0.99 nm, the average RMSF of B.1.617.2 variant RBD-hACE2 complex was 1.26 ± 0.4 nm (Fig. 10A). During the evaluation process of RMSD, it was observed that all these complexes showed a parallel deviation of about 1.1 nm from the initial point to the endpoint (20 ns of simulations). The results of RMSD of RBD-hACE2 of Wuhan strain (1.2 ± 0.34 nm), RBD-hACE2 of B.1.17 variant (1.0 ± 0.25 nm), and RBD-hACE2 of B.1.617.2 variant (1.1 ± 0.12 nm), are shown in Fig. 10B. After analyzing the deviations and fluctuations within the complex, the hydrogen bond count was evaluated for the RBD-hACE2 complex for all three variants. We observed that the Wuhan strain formed a hydrogen bond between RBD-hACE2 (10 ± 3), followed by the B.1.617.2 variant (13 ± 2), the highest number of hydrogen bonds present in this analysis, and the B.1.1.7 variant formed 11 ± 4 hydrogen bonds from all the three MDS analyses, respectively, (Fig. 10C). A similar hydrogen bond pattern was noted between the RBD-hACE2 formations in these two variants.
Fig. 10.
The MDS evaluation of the three complexes. (A) The RMSF plot of one control and two variants. The RMSF plot reveals the flexibility of three complexes. The mutated residues show lesser fluctuations and are comparable in all three structures. (B) The RMSD plot of one control and two variants. The RMSD plot represents similar deviations from the simulated structure. (C) MDS analysis shows hydrogen bonds of one control and two variants. The number of hydrogen bonds (H-bonds) count indicates that mutated variants have more H-bonds than the Wuhan strain. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Here, we used the NAMD server to perform the MD simulation and illustrate the molecular dynamics of B.1.617.2 RBD-hACE2 and B.1.1.7 RBD-hACE2. Several researchers are using the NAMD server to perform the MD simulation during the simulation performance. Baral et al. have also performed an MD simulation of B.1.617.2 RBD-neutralizing Abs and RBD-ACE2 to understand the RBD-neutralizing Abs interactions and RBD-ACE2 interactions of the Delta variant using the NAMD server (Baral et al., 2021). Chakraborty et al. have used the MD simulation to access the molecular interaction prototype of Spike RBD and hACE2 of SARS-CoV-2 using the NAMD server (Chakraborty et al., 2021h).
4. Discussion
Several variants have been generated from time to time and circulated worldwide. The significant variants are B.1.1.7 (Alpha; originated from the UK), B.1.617.2 (Delta; originated from India), P1 (Gamma; originated from Brazil), and B.1.351 (Beta; originated from South Africa). However, several other variants have been reported in other regions (Chakraborty et al., 2021a; Chakraborty et al., 2021f; Chakraborty et al., 2021g; Chakraborty et al., 2021h; Bhattacharya et al., 2021b).The recent one is Omicron which was first observed in South Africa (Mohapatra et al., 2022). However, among these variants, two variants (Delta and Alpha) have a significant role at a certain time point of the pandemic period and increased the infection, which generated a new wave in both the countries (UK and India) (Fig. 1c). B.1.1.7 was noted to increase SARS-CoV-2 infection in the UK during 2020 September/October, which is circulating in different places in the country (Wilton et al., 2021). It also got transferred to other countries like the USA (Galloway et al., 2021). The mutated variants may show significant differences in epidemiological patterns, such as reinfection possibility, severity, and transmissibility. The variant was observed with some significant mutations in spike protein (Cherian et al., 2021).Comparative genomics assessment of these two variants has revealed specific unique mutations, such as N501Y, D614G L452R, E484Q, and P681R in the S-glycoprotein. Several scientists have illustrated several mutations from time to time and described their significance in the pandemic situation (Harvey et al., 2021; Mohammadi et al., 2021). N501Y, D614G L452R, and E484Q mutations are reported as significant mutations in different variants in SARS-CoV-2. Some mutations are responsible for immune escape, antibody escape, and partial vaccine escape (Chakraborty et al., 2022a: Chakraborty et al., 2022b). One of the significant mutations is D614G which is responsible for infectivity and re-infectivity. The mutation also increases the ACE2 receptor binding capacity of S-glycoprotein and also augments the fitness of SARS-CoV-2 variants (Bhattacharya et al., 2021a: Plante et al., 2021). D614G is found in all VOIs and VOCs, and the mutation might be the outcome of a positive selection (Chakraborty et al., 2021j).
Our analyses focused on country-wise and region-wise genome sequences and their submission patterns, molecular phylogenetics, scatter plots of the cluster evaluation, country-wise geographical distribution and transmission pattern, frequencies, and entropy diversity mutational landscape of B.1.1.7 and B.1.617.2. Finally, we evaluated the comparative receptor binding (hACE2) pattern with the Wuhan strain, VOC B.1.1.7 variant (RBD N501Y mutation), and VOI B.1.617.2 variant of the spike protein (RBD double mutations L452R, E484Q). Our study illustrated a similar binding pattern of these two variants, which might play a crucial role in the entry of these viruses into the cell and thereby infection. A stable binding pattern was noted for the binding of the B.1.617.2 variant, which may help explain the more infectious property of the variant. Therefore, this study will help understand the variants' infectivity and their public health importance.
Molecular dynamics (MD) simulation patterns and structural mutational landscape data help understand the variants' infectivity (Khan et al., 2021). Our previous study analyzed the MD simulation of the interaction of human ACE2 and the RBD region of S-glycoprotein with the Wuhan strain (Chakraborty et al., 2021k). In this study, the MD simulation of the human ACE2 and the RBD of the Wuhan strain showed a very similar pattern as observed previously (Chakraborty et al., 2021k). The MD simulation pattern of the B.1.617.2 variant showed a more stable binding than the Alpha variant. The RMSF results showed that the variant was more stable in most residues; however, some fluctuations were noted from 412 to 472.
Furthermore, the RMSD showed more or less stability throughout the simulation. However, one small peak was noted between 13 and 14 ns. The Delta variant showed a few more H-bonds than the Alpha variant and Wuhan strain during MD simulation, and this may provide a more stable binding in the B.1.617.2 variant. Simultaneously, these variants formed 11H-bonds during residue interactions with RBD and receptor binding (hACE2). This variant formed one more hydrogen bond during the interaction than the Wuhan strain (10H-bonds). Similarly, the B.1.1.7 variant formed one more hydrogen bond during the interaction (12H-bonds) than the B.1.617.2 variant. The free energy calculations confirmed the increased binding pattern of ACE2 and spike RBD to these two variants. Our comparative binding study of RBD and receptor binding (hACE2) will help comprehend all binding properties and further understand epidemiological patterns such as transmissibility and reinfection possibility.
The L452R mutation, found in the B.1.617.2 variant, is linked with increased transmissibility. This mutation was detected in other variants, B.1.429/ B.1.427 (VOI), which is also related to augmented transmissibility. This mutation shows a moderate reduction in neutralization in post-vaccination sera and the generation of a few monoclonal antibodies (Centers for Disease Control and Prevention, 2021; World Health Organization, 2021). We also analyzed the genome submission pattern of the B.1.617.2 variant from the GISAID server. The increased sequence submission pattern of the Delta variant suggests that this variant is more dominant and has a higher growth rate than other circulating variants in India (Rambaut et al., 2020: O'Toole et al., 2020;). The variant with increased transmissibility might be one of the significant factors contributing to the second wave generation in India, along with other factors (Chakraborty et al., 2021i).
Molecular recognition of proteins is essential in molecular biology, which helps administer protein-ligand interactions. Therefore, the entropy landscape of a protein is an important factor in understanding the receptor-ligand binding properties of a protein (Caro et al., 2017). Researchers are trying to understand the entropy of the RNA genome. It has been observed that conformational entropy is useful to develop the RNA secondary structures from the RNA sequences (Garcia-Martin and Clote, 2015; Manzourolajdad and Arnold, 2015). Our previous studies have described the entropy at different positions of the nucleotides in the wild strain of SARS-CoV-2 and recent variants like Omicron (Chakraborty et al., 2021f; Bhattacharya et al., 2022b). In this study, we found more entropy at different positions in the B.1.617.2 variant nucleotides compared to B.1.1.7. Like, RNA entropy, the entropy of a protein also helps to understand several protein characteristics like protein-ligand interaction. Consecutively, higher entropy was noted in various places in the nucleotides of S-glycoprotein in the B.1.617.2 variant. More entropy or higher entropy-enthalpy in the protein helps to interact with ligand molecules, and the whole entropy is linked with protein binding (Du et al., 2016; Fenley et al., 2012). The augmented entropy condition of the S-glycoprotein of the B.1.617.2 variant may support the binding of ACE2 and spike RBD, and this property supports the infective nature of this variant.
This study also performed a comparative genomics analysis of these two variants. The distribution pattern and country-wise genome sequence submission pattern in the depository server showed the further spread of these two significant variants worldwide. This evolutionary epidemiology study directs phylogenetic analysis associated with the additional adaptation of these two variants. It may help in the future to continue evidence-based research on the evolutionary changes of these two variants, which may be substantial for public health measures. Recently, Day et al. described the evolutionary epidemiology of SARS-CoV-2, where they described the importance of genome-wide analysis of mutational events and genome-wide analysis of entropy calculations (Day et al., 2020). We can also understand the individual variants shared mutations with other variants using bioinformatics and molecular phylogenetics. This will help us understand the epidemiological characteristics and activity of circulating variants and guide us to formulate strategies for pandemic response (Bauer et al., 2020).
5. Limitation of the study
The study has performed a comprehensive analysis of comparative genomics, evolutionary epidemiology, and RBD-hACE2 receptor binding pattern of two significant variants, Alpha and Delta. Moreover, the study revealed the pandemic response in UK and India due to the variants. The study was performed through computational biology approaches. In this study, we have performed molecular dynamics (MD) simulations. We could not achieve very high resolution (200 ns or above) due to resource invalidity, which is a limitation of our study.
However, to curtail the COVID-19 pandemic, any kind of data produced, even through bioinformatics methods for Alpha and Delta variants, is necessary for future researchers and society. These two variants have been marked as VOI by WHO and made the pandemic more serious in UK and India. From this point of view, our data is very significant and highly beneficial for society. However, all the data needs to be validated through in vitro and in vivo methods, another limitation of the study. We urgently urge future researchers to validate our data through in vitro and in vivo analysis to end the pandemic crisis and prepare for a future pandemic.
6. Future perspectives
This study of the evolutionary epidemiology of these two SARS-CoV-2 variants will help understand the emergence and their geographic spread. It can also be related to the epidemiological pattern and infectivity. The evolution pattern of these two variants might explain the dynamic behavior patterns of these two new emerging variants. With the help of these primary data, one can comprehend the epidemiological dynamics, transmission rates, asymptomatic infections, disease progression, and virulence patterns. The data will help better understand future researchers to illustrate the evolutionary mechanism and develop an epidemiological model in the future.
Conversely, the genome sequences of B.1.1.7 and B.1.617.2 were deposited in more than 100 countries. Therefore, these variants have already spread in countries other than the country of origin, which is a severe concern. More data are urgently required to understand the epidemiological characteristics, including reinfection possibility, severity, transmissibility, activity against the nAb, nAb escape phenomena, and partial vaccine escape event of B.1.1.7 and B.1.617.2 variant. Understanding the molecular mechanism might help to fight against the pandemic in a stipulated time.
7. Conclusion
In conclusion, we found the increased molecular interactivity between hACE2-RBD binding of these two variants compared to the Wuhan reference strain. The augmented receptor binding pattern of B.1.1.7 and B.1.617.2 might help to increase the infectivity compared to the Wuhan reference strain. Therefore, these two variants are accountable for the second wave generation in the UK and India. Some researchers have attempted to identify different properties, such as transmissibility and reinfection possibility of the newly emerged.B.1.1.7 variant. Our study has provided a better understanding of molecular phylogenetics, the cluster of evaluation, and these two variants' country-wise geographical distribution patterns. Furthermore, we have attempted to understand comparative genomics and comprehend the different mutations related to viral replication rate in terms of infectivity. We also attempted to understand the significant mutations in their structural and functional landscape. In the future, researchers might develop intervention strategies for these essential mutations to stop the spread of these variants, which has public health implications to control the wave of the pandemic period. Finally, we explored the binding association of the RBD and hACE2 of these new variants compared to the Wuhan strain. Consequently, the structural study of significant mutations will provide a strong foundation for structure-based drug design using these mutations against emerging variants to fight against the COVID-19 pandemic.
Informed consent statement
Not applicable.
Data availability statement
Not applicable.
Author contributions
CC: Concept development, Data collection, analysis, review, writing the draft, and editing the final manuscript. ARS: discussion, editing, and reviewing the final manuscript. MB, BM, SSN: Some part of the analysis and scientific discussion of the final manuscript. SSL: Validation and supervised the study.
Declaration of Competing Interest
The authors have no conflict of interest to declare.
References
- Afrin S.Z., Paul S.K., Begum J.A., Nasreen S.A., Ahmed S., Ahmad F.U., Aziz M.A., Parvin R., Aung M.S., Kobayashi N. Extensive genetic diversity with novel mutations in spike glycoprotein of SARS-CoV-2, Bangladesh in late 2020. New Microbes New Infect. 2021:100889. doi: 10.1016/j.nmni.2021.100889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akachar J., Bouricha E.M., Hakmi M., Belyamani L., El Jaoudi R., Ibrahimi A. Identifying epitopes for cluster of differentiation and design of new peptides inhibitors against human SARS-CoV-2 spike RBD by an in-silico approach. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e05739. e05739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alai S., Gujar N., Joshi M., Gautam M., Gairola S. Pan-India novel coronavirus SARS-CoV-2 genomics and global diversity analysis in spike protein. Heliyon. 2021;7 doi: 10.1016/j.heliyon.2021.e06564. e06564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S., Kamal T.B., Sarker M.M.R., Zhou J.R., Rahman S.A., Mohamed I.N. Therapeutic effectiveness and safety of repurposing drugs for the treatment of COVID-19: position standing in 2021. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.659577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew Banchich Á.O.T. Lineage B.1.1.7. PANGO Lineages. 2021. https://cov-lineages.org/global_report_B.1.1.7.html Accessed on September 15, 2021.
- Baral P., Bhattarai N., Hossen M.L., Stebliankin V., Gerstman B.S., Narasimhan G., Chapagain P.P. Mutation-induced changes in the receptor-binding interface of the SARS-CoV-2 Delta variant B. 1.617. 2 and implications for immune evasion. Biochem. Biophys. Res. Commun. 2021;574:14–19. doi: 10.1016/j.bbrc.2021.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D.C., Tay A.P., Wilson L.O., Reti D., Hosking C., McAuley A.J., Pharo E., Todd S., Stevens V., Neave M.J. Supporting pandemic response using genomics and bioinformatics: a case study on the emergent SARS-CoV-2 outbreak. Transbound. Emerg. Dis. 2020;67:1453–1462. doi: 10.1111/tbed.13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M., Sharma A.R., Mallick B., Sharma G., Lee S.S., Chakraborty C. Immunoinformatics approach to understand molecular interaction between multi-epitopic regions of SARS-CoV-2 spike-protein with TLR4/MD-2 complex. Infect. Genet. Evol. 2020;85:104587. doi: 10.1016/j.meegid.2020.104587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M., Sharma A.R., Patra P., Ghosh P., Sharma G., Patra B.C., Saha R.P., Lee S.S., Chakraborty C. A SARS-CoV-2 vaccine candidate: In-silico cloning and validation. Inform. Med. Unlocked. 2020;20:100394. doi: 10.1016/j.imu.2020.100394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M., Chatterjee S., Sharma A.R., Agoramoorthy G., Chakraborty C. D614G mutation and SARS-CoV-2: impact on S-protein structure, function, infectivity, and immunity. Appl. Microbiol. Biotechnol. 2021;105:9035–9045. doi: 10.1007/s00253-021-11676-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M., Sharma A.R., Ghosh P., Lee S.S., Chakraborty C. A next-generation vaccine candidate using alternative epitopes to protect against Wuhan and all significant mutant variants of SARS-CoV-2: an immunoinformatics approach. Aging Dis. 2021;12:2173. doi: 10.14336/AD.2021.0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M., Sharma A.R., Ghosh P., Patra P., Mallick B., Patra B.C., Lee S.S., Chakraborty C. TN strain proteome mediated therapeutic target mapping and multi-epitopic peptide-based vaccine development for Mycobacterium leprae. Infect. Genet. Evol. 2022:105245. doi: 10.1016/j.meegid.2022.105245. [DOI] [PubMed] [Google Scholar]
- Bhattacharya M., Sharma A.R., Dhama K., Agoramoorthy G., Chakraborty C. Omicron variant (B. 1.1. 529) of SARS-CoV-2: understanding mutations in the genome, S-glycoprotein, and antibody-binding regions. GeroScience. 2022:1–19. doi: 10.1007/s11357-022-00532-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcagnile M., Forgez P., Iannelli A., Bucci C., Alifano M., Alifano P. Molecular docking simulation reveals ACE2 polymorphisms that may increase the affinity of ACE2 with the SARS-CoV-2 Spike protein. Biochimie. 2021;180:143–148. doi: 10.1016/j.biochi.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L.-R., Zhang C.-Y., Zhang D.-L., Chu H.-Y., Zhang Y.-B., Li G.-H. Recent developments in using molecular dynamics simulation techniques to study biomolecules. Acta Phys. -Chim. Sin. 2017;33:1354–1365. [Google Scholar]
- Caro J.A., Harpole K.W., Kasinath V., Lim J., Granja J., Valentine K.G., Sharp K.A., Wand A.J. Entropy in molecular recognition by proteins. Proc. Natl. Acad. Sci. 2017;114:6563–6568. doi: 10.1073/pnas.1621154114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention, (2021). SARS-CoV-2 variant classifications and definitions. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html. Accessed, 25 August, 2021.
- Chakraborty C., Bhattacharya M., Sharma A.R. 2021. Present Variants of Concern and Variants of Interest of Severe Acute Respiratory Syndrome Coronavirus 2: Their Significant Mutations in S-Glycoprotein, Infectivity, Re-infectivity, Immune Escape and Vaccines Activity. Wiley Online Library. e2270. [DOI] [Google Scholar]
- Chakraborty C., Sharma A.R., Bhattacharya M., Agoramoorthy G., Lee S.S. The drug repurposing for COVID-19 clinical trials provide very effective therapeutic combinations: lessons learned from major clinical studies. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.704205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C., Sharma A.R., Bhattacharya M., Lee S.S. Lessons learned from cutting-edge immunoinformatics on next-generation COVID-19 vaccine research. Int. J. Pept. Res. Ther. 2021;27:2303–2311. doi: 10.1007/s10989-021-10254-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C., Sharma A.R., Bhattacharya M., Agoramoorthy G., Lee S.S. Asian-origin approved COVID-19 vaccines and current status of COVID-19 vaccination program in Asia: a critical analysis. Vaccines. 2021;9:600. doi: 10.3390/vaccines9060600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C., Sharma A.R., Bhattacharya M., Sharma G., Saha R.P., Lee S.S. Ongoing clinical trials of vaccines to fight against COVID-19 pandemic. Immune Netw. 2021;21(1) doi: 10.4110/in.2021.21.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C., Sharma A.R., Bhattacharya M., Agoramoorthy G., Lee S.S. Evolution, mode of transmission, and mutational landscape of newly emerging SARS-CoV-2 variants. Mbio. 2021;12 doi: 10.1128/mBio.01140-21. e01140–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C., Bhattacharya M., Sharma A.R., Lee S.S., Agoramoorthy G. SARS-CoV-2 Brazil variants in Latin America: more serious research urgently needed on public health and vaccine protection. Ann. Med. Surg. 2021;66:102428. doi: 10.1016/j.amsu.2021.102428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C., Ranjan A., Bhattacharya M., Agoramoorthy G., Lee S.-S. A paradigm shift in the combination changes of SARS-CoV-2 variants and increased spread of delta variant (B.1.617.2) across the world. Aging Dis. 2021 doi: 10.14336/AD.2021.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C., Ranjan A., Bhattacharya M., Agoramoorthy G., Lee S.-S. The current second wave and COVID-19 vaccination status in India. Brain Behav. Immun. 2021;96:1–4. doi: 10.1016/j.bbi.2021.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C., Saha A., Sharma A.R., Bhattacharya M., Lee S.S., Agoramoorthy G. D614G mutation eventuates in all VOI and VOC in SARS-CoV-2: is it part of the positive selection pioneered by Darwin? Mol. Ther. Nucleic Acids. 2021;26:237–241. doi: 10.1016/j.omtn.2021.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C., Sharma A., Mallick B., Bhattacharya M., Sharma G., Lee S. Evaluation of molecular interaction, physicochemical parameters and conserved pattern of SARS-CoV-2 Spike RBD and hACE2: in silico and molecular dynamics approach. Eur. Rev. Med. Pharmacol. Sci. 2021;25:1708–1723. doi: 10.26355/eurrev_202102_24881. [DOI] [PubMed] [Google Scholar]
- Chakraborty C., Bhattacharya M., Sharma A.R. Emerging mutations in the SARS-CoV-2 variants and their role in antibody escape to small molecule-based therapeutic resistance. Curr. Opin. Pharmacol. 2022;62:64–73. doi: 10.1016/j.coph.2021.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C., Sharma A.R., Bhattacharya M., Lee S.S. A detailed overview of immune escape, antibody escape, partial vaccine escape of SARS-CoV-2 and their emerging variants with escape mutations. Front. Immunol. 2022:53. doi: 10.3389/fimmu.2022.801522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Liu H., Sun H., Pan P., Li Y., Li D., Hou T. Assessing the performance of the MM/PBSA and MM/GBSA methods. 6. Capability to predict protein–protein binding free energies and re-rank binding poses generated by protein–protein docking. Phys. Chem. Chem. Phys. 2016;18:22129–22139. doi: 10.1039/c6cp03670h. [DOI] [PubMed] [Google Scholar]
- Cheng L., Song S., Zhou B., Ge X., Yu J., Zhang M., Ju B., Zhang Z. Impact of the N501Y substitution of SARS-CoV-2 Spike on neutralizing monoclonal antibodies targeting diverse epitopes. Virol. J. 2021;18:1–6. doi: 10.1186/s12985-021-01554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherian S., Potdar V., Jadhav S., Yadav P., Gupta N., Das M., Rakshit P., Singh S., Abraham P., Panda S., Team, NIC SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms. 2021;9:1542. doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day T., Gandon S., Lion S., Otto S.P. On the evolutionary epidemiology of SARS-CoV-2. Curr. Biol. 2020;30:R849–R857. doi: 10.1016/j.cub.2020.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries S.J., Van D.M., Bonvin A.M. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010;5(5):883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- Deng X., Garcia-Knight M.A., Khalid M.M., Servellita V., Wang C., Morris M.K., Sotomayor-González A., Glasner D.R., Reyes K.R., Gliwa A.S. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. medRxiv. 2021 doi: 10.1101/2021.03.07.21252647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Li Y., Xia Y.-L., Ai S.-M., Liang J., Sang P., Ji X.-L., Liu S.-Q. Insights into protein–ligand interactions: mechanisms, models, and methods. Int. J. Mol. Sci. 2016;17:144. doi: 10.3390/ijms17020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECDC . ECDC; Stockholm: 2021. Emergence of SARS-CoV-2 B.1.617 Variantsin India and Situation in the EU/EEA; pp. 1–12. May 11 2021. [Google Scholar]
- ECDC . European Centre for Disease Prevention and Control; 2021. SARS-CoV-2 Variants of concern as of May 11 2021.https://www.ecdc.europa.eu/en/covid-19/variants-concern (Accessed on September 15, 2021) [Google Scholar]
- ECDC . European Centre for Disease Prevention and Control; 2021. SARS-CoV-2 Variants of Concern Pose a Higher Risk for Hospitalisation and Intensive Care Admission.https://www.ecdc.europa.eu/en/news-events/sars-cov-2-variants-concern-pose-higher-risk-hospitalisation-and-intensive-care (Accessed on September 15, 2021) [Google Scholar]
- Farooq R.K., Rehman S.U., Ashiq M., Siddique N., Ahmad S. Bibliometric analysis of coronavirus disease (COVID-19) literature published in web of science 2019–2020. J. Fam. Community Med. 2021;28:1–7. doi: 10.4103/jfcm.JFCM_332_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenley A.T., Muddana H.S., Gilson M.K. Entropy–enthalpy transduction caused by conformational shifts can obscure the forces driving protein–ligand binding. Proc. Natl. Acad. Sci. 2012;109:20006–20011. doi: 10.1073/pnas.1213180109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway S.E., Paul P., MacCannell D.R., Johansson M.A., Brooks J.T., MacNeil A., Slayton R.B., Tong S., Silk B.J., Armstrong G.L., Biggerstaff M. Emergence of SARS-CoV-2 b. 1.1. 7 lineage—united states, December 29, 2020–January 12, 2021. Morb. Mortal. Wkly Rep. 2021;70:95. doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martin J.A., Clote P. RNA thermodynamic structural entropy. PLoS One. 2015;10(11) doi: 10.1371/journal.pone.0137859. e0137859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham M.S., Sudre C.H., May A., Antonelli M., Murray B., Varsavsky T., Kläser K., Canas L.S., Molteni E., Modat M. Changes in symptomatology, reinfection, and transmissibility associated with the SARS-CoV-2 variant B. 1.1. 7: an ecological study. Lancet Public Health. 2021;6:e335–e345. doi: 10.1016/S2468-2667(21)00055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grint D.J., Wing K., Williamson E., McDonald H.I., Bhaskaran K., Evans D., Evans S.J., Walker A.J., Hickman G., Nightingale E. Case fatality risk of the SARS-CoV-2 variant of concern B. 1.1. 7 in England, November 16 to February 5. Eurosurveillance. 2021;26:2100256. doi: 10.2807/1560-7917.ES.2021.26.11.2100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Ø., Harper D.A., Ryan P.D. PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001;4:9. [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Islam O.K., Al-Emran H.M., Hasan M.S., Anwar A., Jahid M.I.K., Hossain M.A. Emergence of European and North American mutant variants of SARS-CoV-2 in South-East Asia. Transbound. Emerg. Dis. 2021;68:824–832. doi: 10.1111/tbed.13748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J.J., Vasudevan K., Pragasam A.K., Gunasekaran K., Veeraraghavan B., Mutreja A. Evolutionary tracking of SARS-CoV-2 genetic variants highlights an intricate balance of stabilizing and destabilizing mutations. Mbio. 2020;12 doi: 10.1128/mBio.01188-21. e01188–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra S., Ye C., Rathnasinghe R., Stadlbauer D., Alshammary H., Amoako A.A., Awawda M.H., Beach K.F., Bermúdez-González M.C., Chernet R.L. SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe. 2021;2:e283–e284. doi: 10.1016/S2666-5247(21)00068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalé L., Skeel R., Bhandarkar M., Brunner R., Gursoy A., Krawetz N., Phillips J., Shinozaki A., Varadarajan K., Schulten K. NAMD2: greater scalability for parallel molecular dynamics. J. Comput. Phys. 1999;151:283–312. [Google Scholar]
- Kalia K., Saberwal G., Sharma G. The lag in SARS-CoV-2 genome submissions to GISAID. Nat. Biotechnol. 2021;39:1058–1060. doi: 10.1038/s41587-021-01040-0. [DOI] [PubMed] [Google Scholar]
- Khan A., Zia T., Suleman M., Khan T., Ali S.S., Abbasi A.A., Mohammad A., Wei D.Q. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J. Cell. Physiol. 2021 doi: 10.1002/jcp.30367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S., Gurry C., Freitas L., Schultz M.B., Bach G., Diallo A., Akite N., Ho J., Lee R.T., Yeo W., Team, G.C.C GISAID’s role in pandemic response. China CDC Wkly. 2021;3:1049. doi: 10.46234/ccdcw2021.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A., Xie L., de la Cruz J., Chen L., Westbrook J., Bourne P.E., Berman H.M. The RCSB PDB information portal for structural genomics. Nucleic Acids Res. 2006;34:D302–D305. doi: 10.1093/nar/gkj120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A. Two weeks of “COVID-19” search on PubMed. Gov. Acta Bio Med. Atenei Parmensis. 2020;91 doi: 10.23750/abm.v91i4.11016. e2020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubinski B., Tang T., Daniel S., Jaimes J.A., Whittaker G. Functional evaluation of proteolytic activation for the SARS-CoV-2 variant B. 1.1. 7: role of the P681H mutation. bioRxiv. 2021 doi: 10.1101/2021.04.06.438731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell A.D., Jr., Banavali N., Foloppe N. Development and current status of the CHARMM force field for nucleic acids. Biopolymers Orig. Res. Biomol. 2000;56:257–265. doi: 10.1002/1097-0282(2000)56:4<257::AID-BIP10029>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Mallapaty S. India’s massive COVID surge puzzles scientists. Nature. 2021;592:667–668. doi: 10.1038/d41586-021-01059-y. [DOI] [PubMed] [Google Scholar]
- Manzourolajdad A., Arnold J. Secondary structural entropy in RNA switch (riboswitch) identification. BMC Bioinforma. 2015;16(1):1–77. doi: 10.1186/s12859-015-0523-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MathWorks, Inc . 1992. MATLAB, High-Performance Numeric Computation and Visualization Software: User's Guide: for UNIX Workstations. MathWorks. [Google Scholar]
- Melo M.C., Bernardi R.C., Rudack T., Scheurer M., Riplinger C., Phillips J.C., Maia J.D., Rocha G.B., Ribeiro J.V., Stone J.E., Neese F. Vol. 15. 2018. NAMD Goes Quantum: An Integrative Suite for Hybrid Simulations; pp. 351–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M., Shayestehpour M., Mirzaei H. The impact of spike mutated variants of SARS-CoV2 [alpha, beta, gamma, delta, and lambda] on the efficacy of subunit recombinant vaccines. Braz. J. Infect. Dis. 2021;25 doi: 10.1016/j.bjid.2021.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra R.K., Tiwari R., Sarangi A.K., Islam D.R., Chakraborty C., Dhama K. COMMENTARY omicron (B. 1.1. 529) variant of SARS-CoV-2–concerns, challenges and recent updates. J. Med. Virol. 2022 doi: 10.1002/jmv.27633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nextstrain Genomic epidemiology of novel coronavirus-global subsampling. Nextstrain. Org. 2020;1 https://nextstrain.org/ncov/global?dmax=2020-04-08 (Accessed on September 15, 2021) [Google Scholar]
- Ostrov D.A. Structural consequences of variation in SARS-CoV-2 B. 1.1. 7. J. Cell. Immunol. 2021;3:103. doi: 10.33696/immunology.3.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole Á., McCrone J. GitHub; 2020. Software Package for Assigning SARS-CoV-2 Genome Sequences to Global Lineages.https://github.com/hCoV-2019/pangolin (Accessed on September 15, 2021) [Google Scholar]
- O’Toole A., Scher E., Underwood A., Jackson B., Hill V., McCrone J., Ruis C., Abu-Dahab K., Taylor B., Yeats C. Pangolin: lineage assignment in an emerging pandemic as an epidemiological tool. Virus Evol. 2020;7 doi: 10.1093/ve/veab064. veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parate S., Rampogu S., Lee G., Hong J.C., Lee K.W. Exploring the binding interaction of Raf kinase inhibitory protein with the N-terminal of C-Raf through molecular docking and molecular dynamics simulation. Front. Mol. Biosci. 2021;8:496. doi: 10.3389/fmolb.2021.655035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kale L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J.C., Hardy D.J., Maia J.D., Stone J.E., Ribeiro J.V., Bernardi R.C., Buch R., Fiorin G., Hénin J., Jiang W., McGreevy R. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020;153(4) doi: 10.1063/5.0014475. 044130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., Mirchandani D. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portelli S., Olshansky M., Rodrigues C.H., D’Souza E.N., Myung Y., Silk M., Alavi A., Pires D.E., Ascher D.B. Exploring the structural distribution of genetic variation in SARS-CoV-2 with the COVID-3D online resource. Nat. Genet. 2020;52:999–1001. doi: 10.1038/s41588-020-0693-3. [DOI] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin E.J., Baden L.R., Udwadia Z.F., Morrissey S. Audio interview: India’s Covid-19 crisis. N. Engl. J. Med. 2021;384 doi: 10.1056/NEJMe2107728. e84. [DOI] [PubMed] [Google Scholar]
- Tallei T.E., Adam A.A., Elseehy M.M., El-Shehawi A.M., Mahmoud E.A., Tania A.D., Niode N.J., Kusumawaty D., Rahimah S., Effendi Y., Idroes R. Fruit bromelain-derived peptide potentially restrains the attachment of SARS-CoV-2 variants to hACE2: a pharmacoinformatics approach. Molecules. 2022;27:260. doi: 10.3390/molecules27010260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiagarajan K. Why is India having a covid-19 surge? Brit. Med. J. Publ. Group. 2021;374 doi: 10.1136/bmj.n1124. n2005. [DOI] [PubMed] [Google Scholar]
- Vaidyanathan G. Coronavirus variants are spreading in India-what scientists know so far. Nature. 2021;593:321–322. doi: 10.1038/d41586-021-01274-7. [DOI] [PubMed] [Google Scholar]
- Velazquez A., Bustria M., Ouyang Y., Moshiri N. An analysis of clinical and geographical metadata of over 75,000 records in the GISAID COVID-19 database. medRxiv. 2020 doi: 10.1101/2020.09.22.20199497. [DOI] [Google Scholar]
- Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O’Toole Á. Assessing transmissibility of SARS-CoV-2 lineage B. 1.1. 7 in England. Nature. 2021;593:266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- Weng G., Wang E., Wang Z., Liu H., Zhu F., Li D., Hou T. HawkDock: a web server to predict and analyze the protein–protein complex based on computational docking and MM/GBSA. Nucleic Acids Res. 2019;47:W322–W330. doi: 10.1093/nar/gkz397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . Vol. 579. WHO; Geneva: 2020. Weekly Epidemiological Update–October 27; p. 580. [Google Scholar]
- Widera M., Mühlemann B., Corman V.M., Toptan T., Beheim-Schwarzbach J., Kohmer N., Schneider J., Berger A., Veith T., Pallas C. Surveillance of SARS-CoV-2 in Frankfurt am Main from October to December 2020 reveals high viral diversity including spike mutation N501Y in B. 1.1. 70 and B. 1.1. 7. Microorganisms. 2021;9:748. doi: 10.3390/microorganisms9040748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton T., Bujaki E., Klapsa D., Majumdar M., Zambon M., Fritzsche M., Mate R., Martin J. Rapid increase of SARS-CoV-2 variant B. 1.1. 7 detected in sewage samples from England between October 2020 and January 2021. Msystems. 2021;6 doi: 10.1128/mSystems.00353-21. e00353–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, 2021. COVID-19 weekly epidemiological update, edition 56, 7 September 2021. https://apps.who.int/iris/handle/10665/345454. Accessed, 5 March, 2022.
- Yan Y., Zhang D., Zhou P., Li B., Huang S.-Y. HDOCK: a web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45(W1):W365–W373. doi: 10.1093/nar/gkx407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Tao H., He J., Huang S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020;15:1829–1852. doi: 10.1038/s41596-020-0312-x. [DOI] [PubMed] [Google Scholar]
- Zelenova M., Ivanova A., Semyonov S., Gankin Y. Analysis of 329,942 SARS-CoV-2 records retrieved from GISAID database. Comput. Biol. Med. 2021;139:104981. doi: 10.1016/j.compbiomed.2021.104981. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang L., Jackson C.B., Mou H., Ojha A., Peng H., Quinlan B.D., Rangarajan E.S., Pan A., Vanderheiden A., Suthar M.S. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020;11:1–9. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X., Chen Y., Ohno-Machado L., Xu H. How do we share data in COVID-19 research? A systematic review of COVID-19 datasets in PubMed Central Articles. Brief. Bioinform. 2021;22:800–811. doi: 10.1093/bib/bbaa331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.