Abstract
We outline a framework for elucidating tumor genetic complexity through multidimensional protein-protein interaction maps and apply it to enhance the understanding of head-and-neck squamous cell carcinoma. This network uncovers 771 interactions from cancer and non-cancerous cell states including wild-type and mutant protein isoforms. Prioritization of cancer enriched interactions reveals a previously unidentified association of the FGF receptor tyrosine kinase 3 with Daple, a guanine-nucleotide exchange factor, resulting in activation of Gαi and PAK1/2 to promote cancer cell migration. Additionally, we observe mutation enriched interactions between the HER3 receptor tyrosine kinase and PIK3CA that can inform the response to HER3 inhibition in vivo. We anticipate the application of this framework will be valuable to translate genetic alterations into a molecular and clinical understanding of the underlying biology of many disease areas.
One Sentence Summary:
Comparative protein interaction analysis of genes altered in head and neck cancer instructs the selection of therapeutic targets.
Genome sequencing efforts over the past decade have profiled the genetic landscape of thousands of patient tumors and solidified the concept of cancer as a highly heterogeneous disease (1-8). Evidence from these efforts has also revealed that thousands of genes are altered in cancer, presenting an overwhelming degree of complexity that has limited the power of connecting individual alterations with cancer patient phenotypes. To facilitate interpretation, powerful network biology approaches have been developed, in which protein network knowledge is used to aggregate individual tumor mutations and, on the basis of altered networks, predict patient survival and response to therapy (9-19). Such network-based approaches rely strongly on existing databases of molecular interactions. To date, the vast majority of publicly available human protein-protein interactions (PPIs) networks have been populated primarily by systematic efforts either without human cellular context (yeast two-hybrid) (20, 21) or by affinity purification-mass spectrometry (AP-MS) (22-24) in workhorse cell lines that lack cancer context, such as HEK293T embryonic kidney cells. With the growing recognition that such PPIs can vary significantly across cellular contexts (25), the generation and incorporation of physical and functional networks in a cancer context likely represents a critical component to interpret and predict cancer biology and its clinical outcomes (26).
To explore the utility of PPI maps generated in a cancer context, we conducted AP-MS experiments to map protein networks in the context of head and neck squamous cell carcinoma (HNSCC) guided by analyses such as TCGA. HNSCC is a cancer affecting squamous mucosal epithelial cells in the oral cavity, pharynx and larynx, and is estimated to be the sixth most common malignancy worldwide (27). Despite a wealth of data detailing the genetic alterations in this tumor type (28), only two types of targeted therapies are presently available (27). Therefore, HNSCC presents an opportunity to apply emerging, quantitative, systems approaches to both identify new therapeutic targets, as well as to further our understanding of existing targets, such as PIK3CA. PIK3CA is the most commonly mutated oncogene in HNSCC and encodes p110alpha (p110α), the catalytic subunit of phosphatidylinositol 3-kinase (PI3K). A hallmark of numerous tumor types, hyperactivation of PI3K can be directly attributed to either amplification or mutation of the PIK3CA and results in activation of the Akt/mTOR pathway. While the function of the canonical PIK3CA mutations (e.g. E542K, E545K, and H1047R) are well-studied, much remains to be learned about how the numerous non-canonical mutations regulate PIK3CA interactions and function (29-31). Here, we present a comparative AP-MS analysis across 3 cell lines for 31 genes frequently altered in HNSCC, including 16 PIK3CA mutations.
Mapping of the head and neck cancer interactome
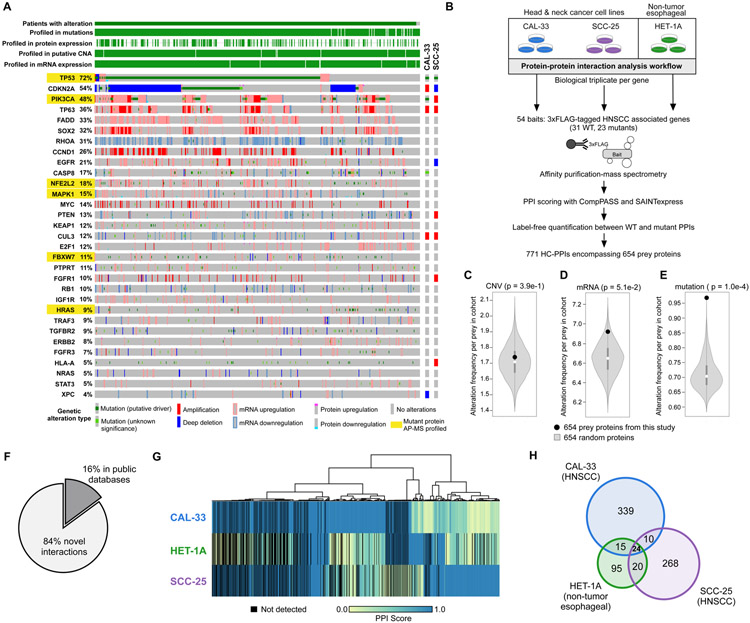
To characterize the protein-protein interaction landscape of HNSCC, we selected proteins based on altered molecular pathways identified from the TCGA analysis of HNSCC tumors (Fig. 1A) (7). Additional proteins were added based on genes with recurrent point mutations or a previously published association with HNSCC (32-35). In total, we selected 33 protein baits, of which 31 were experimentally tractable (Materials and methods and table S1). Importantly, 99% of HNSCC patients harbor an alteration in one or more of these proteins (Fig. 1A).
Figure 1. Experimental design and workflow.
(A) The alteration frequencies from the HNSCC TCGA provisional dataset (n = 530 patients) for the 31 experimentally tractable genes selected as AP-MS baits in this study. Proteins analyzed in this study are listed, along with the percentage of patients with an alteration in that gene/protein. Each patient is represented by a grey box that is colored based on the occurrence and type of alteration(s) observed in that patient. Both the wild-type and mutant protein sequence(s) were analyzed for genes highlighted in yellow. The genetic alteration types in the two cancer cell lines (CAL-33 and SCC-25) are also displayed. (B) The experimental workflow in which each bait was expressed in biological triplicate in 3 cell lines and subjected to AP-MS analysis. (C-E) Permutation test illustrating the frequency of CNVs (C), mRNA alterations (D), or mutations (E), from randomly selected genes in the HNSCC TCGA data. The white circle indicates the median of the random sampling, and the grey bar represents +/− 1 standard deviation. The frequency of alterations found in the prey retrieved in this PPI dataset is indicated in the black circle. (F) Percentage of HC-PPIs identified in a panel of public PPI databases (CORUM, BioPlex 2.0, or BioGRID low throughput and multivalidated, and IMEX (23, 36-38)). (G) Clustering analysis of all HC-PPIs (n = 771) based on their PPI score, which is an average of the confidence scores reported from SAINTexpress and CompPASS score (see Materials and methods for details). A PPI score of 1.0 represents the highest confidence in a PPI. (H) Venn diagram illustrating the overlap in HC-PPIs among the 3 cell lines. For this analysis, only those PPIs passing the HC-PPI filtering criteria by both SAINTexpress and CompPASS were classified as an HC-PPI within an individual cell line.
For those baits with recurrent point mutations, both the wild-type (WT) and mutant forms of the protein were tagged, purified and analyzed. Each bait was expressed as a 3xFLAG-tagged protein under the control of a doxycycline-inducible promoter in biological triplicate in three separate cell lines (Fig. 1B). We selected two HPV-negative HNSCC cell lines (SCC-25 and CAL-33) that harbor many genetic alterations present in the HNSCC patient population (Fig. 1A) and have previously been shown to have RNA profiles highly correlated with those of HNSCC patients (Spearman correlation = 0.66 and 0.69 for CAL-33 and SCC-25, respectively) (7, 33, 35, 39). Additionally, an immortalized non-tumorigenic cell line, HET-1A, was used from a similar anatomical location (esophagus) for comparison. We then used a previously described AP-MS workflow to identify PPIs from these three cell lines (Fig. 1B) (40). We elected to report a conservative and high-confidence protein-protein interaction map by requiring PPIs to pass stringent criteria by two complementary PPI scoring algorithms; SAINTexpress and CompPASS (Materials and methods) (22, 41, 42). Using this workflow, a total of 771 high-confidence PPIs (HC-PPIs) were identified involving 654 proteins (Fig. 1B, fig. S1A-B, data S2 and data S3), for an average of 25 PPIs per bait gene.
We and others have shown that alteration profiles in cancer are organized into molecular networks in which the interaction partners of frequently altered proteins incur a higher rate of alteration than a random selection of genes (9, 10, 43, 44). Thus, we tested whether our HNSCC HC-PPI set was enriched for different types of alterations measured in the HNSCC TCGA cohort (Materials and methods). Our dataset was indeed highly enriched for preys with point mutations; however, this enrichment was not observed for alterations in mRNA expression or for chromosomal rearrangements (Fig. 1C-E). Despite the overall lack of enrichment for generic mRNA alterations, we do find that PPIs from each cell line are significantly enriched in proteins whose mRNA expression profiles are prognostic in TCGA for HNSCC (Fig. S1C).
Of the 771 HC-PPIs detected, the majority (84%) had not been previously reported in public PPI databases (Fig. 1F). This high percentage of novel interactions likely reflects the fact that differences across cellular contexts exist for PPIs and that nearly all systematic PPI analyses to date have been performed in only HEK293T or HeLa cell lines (22-24). This proportion of novel interactions, presumably due to cell type specificity, is also supported by the observation that significant differences in PPIs are observed even across the cell lines in this study (Fig. 1G), with only 24 PPIs being conserved across all cell lines analyzed (Fig. 1H and fig. S1D). Notably, many well-studied cancer proteins are included in the novel interactions. For example, in SCC-25 cells we observed physical interactions between the proto-oncoprotein MYC and each of two DNA repair proteins, PARP1 and TOP1. MYC has previously been shown to regulate PARP1 activity (45). The MYC:PARP1 interaction is supported by previous studies reporting MYC:TOP1 (46) and PARP1:TOP1 interactions (47).
Similarly, purification of tagged KEAP1 in SCC-25 cells revealed an interaction with AJUBA, a scaffolding protein involved in the regulation of numerous cellular processes, including negative regulation of Wnt/β-catenin signaling (48). Until recently, AJUBA was not associated with HNSCC, however tumor genome analysis revealed it is inactivated in 7% of HPV-negative tumors (7). The KEAP1:AJUBA interaction was further supported by our identification of a physical connection in HET-1A cells between KEAP1 and SQSTM1, a known AJUBA interactor (49-52).
A statistical approach to evaluate cell-type specificity of interactions
To identify interactions with relevance to cancer biology, we sought to compare PPIs across cell lines and prioritize those that are seemingly cancer-enriched; i.e. those that exist in both CAL-33 and SCC-25, the two HNSCC cancer cell lines, but absent in the HET-1A non-tumorigenic cell line. However, a simple overlap analysis of the sets of HC-PPIs identified by each cell line does not faithfully represent whether a PPI is shared. For example, a PPI might erroneously appear to be specific for a single cell line when it passes the threshold for HC-PPIs in that cell line (i.e., a true positive) while falling slightly below the threshold (i.e., false negative) in a second. Accordingly, we developed a method for calculating differential interaction scores (DIS) for each PPI, with associated Bayesian false discovery rates (BFDR). This method is based on the SAINTexpress score (41), which reports on the probability of a PPI in a single cell line given the AP-MS data. Here, quantitative SAINTexpress probabilities were combined across the three cell lines to generate the DIS (Materials and methods), allowing for the identification of PPIs that are enriched in either the two cancer cell lines or the non-cancerous cells.
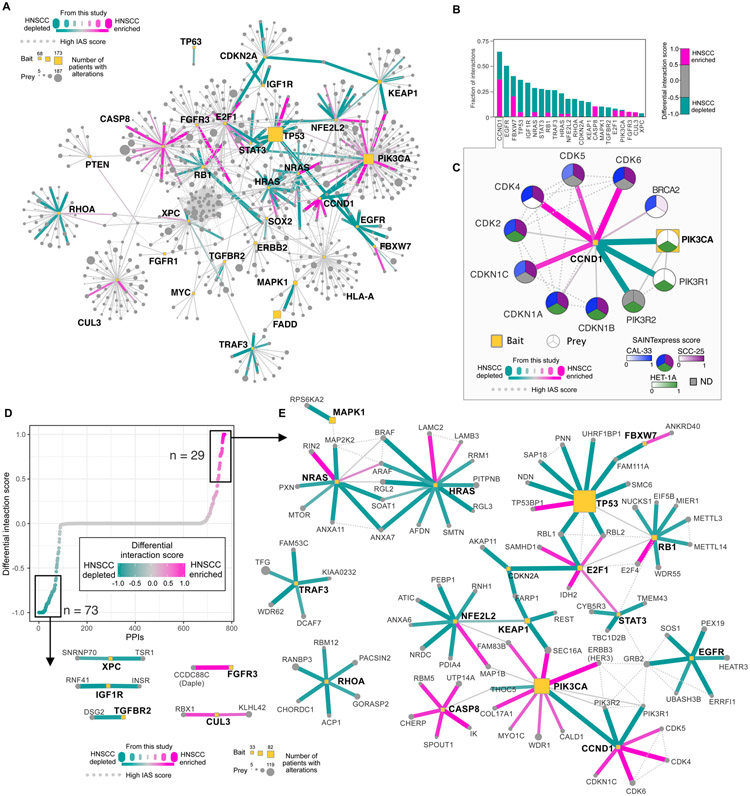
Application of the DIS method to our HC-PPIs identified interactions enriched in HNSCC cells as well as those enriched in the HET-1A non-tumorigenic background (Fig. 2A-B and data S4). For example, the interaction profile for cyclin D1 was dramatically rewired between HNSCC and HET-1A (Fig. 2C). Cyclin D1, encoded by the CCND1 gene, is one of the most commonly altered oncogenes in HNSCC, being amplified in 31% of HPV-negative HNSCC tumors (7). Cyclin D1 interacts with the cyclin-dependent kinase inhibitors CDKN1A (p21) and CDKN1B (p27) in all three cell lines, but preferentially interacts with multiple cyclin-dependent kinases (CDKs) only in HNSCC cells. This interaction preference was not unexpected, as CCND1:CDK4/6 interactions are known to be essential for cell proliferation and thus can contribute to uncontrolled cell cycle progression in cancer cells (53). Consistent with these findings, we observe the HNSCC cell lines have increased growth rates when compared to HET-1A (Fig. S2A).
Figure 2. Differential interaction analysis of the HNSCC enriched and depleted interactome.
(A) Interactome of the union of all HC-PPIs detected across all cell lines. Edges are colored based on their differential interaction score (DIS), with pink edges representing PPIs that are enriched in HNSCC (both SCC-25 and CAL-33) as compared to HET-1A cells, and teal lines representing PPIs that are depleted from HNSCC cell lines. IAS connections represent physical protein-protein association derived from prior studies (55) (see Material and methods). (B) For baits with ∣DIS∣ > 0.5, the fraction of PPIs for that bait having HNSCC-enriched PPIs with DIS > 0.5, or HNSCC-depleted DIS < −0.5. (C) CCND1 interactome. Here the SAINTexpress score, used for calculation of the DIS, is displayed for each cell line within the prey node, ND indicates not detected. (D) DIS for the entire interactome represented in panel A ranked by DIS. (E) Subnetwork of the interactome of the HNSCC-enriched and -depleted interactions.
We also found a previously uncharacterized interaction of cyclin D1 with components of the PI3K complex (PIK3CA, PIK3R1/2) exclusively detected by AP-MS in HET-1A cells, an interaction that was further validated by a proximity ligation assay (PLA) in HET-1A cells expressing FLAG-tagged CCND1 (Fig. S2B-D). Lastly, we evaluated the cellular localization of the interaction, as cyclin D1 is usually associated with the nucleus, while PI3K is primarily associated with cytoplasmic and plasma membrane localization. We observed the interaction to be 80% cytoplasmic in localization (Fig. S2E), indicating a non-canonical localization of CCND1, which has been observed previously in certain cell types (54). The cell type enrichment of this particular interaction, along with several others, is further supported by targeted proteomic analysis (Fig. S3A). Lastly, we evaluated the relationship between cell-type PPI enrichment and both bait expression levels or prey expression levels (Fig. S3B-D). In general, we find virtually no correlation between these factors, suggesting that a diversity of factors likely influence PPI cell-type specificity.
Identification of a novel FGFR3:Daple interaction that regulates Gαi-mediated migratory signaling
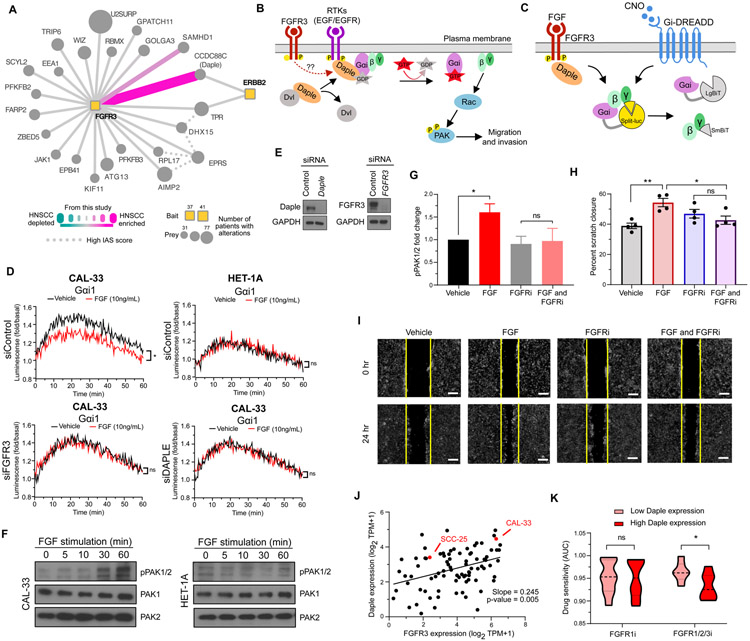
To uncover cancer-enriched interactions, we ranked PPIs by their DIS (Fig. 2D), focusing on those PPIs with greatest enrichment (DIS > 0.5) or depletion (DIS < −0.5) in the HNSCC cell lines (Fig. 2E). This analysis prioritized a previously unknown interaction between FGFR3 and CCDC88C, which was strongly observed in both CAL-33 and SCC-25 cells but not in HET-1A (Fig. 3A). FGFR3 is a receptor tyrosine kinase (RTK) that recognizes fibroblast growth factor (FGF) and mediates cellular proliferation, survival and differentiation. Meanwhile, CCDC88C, also known as Daple, is a 228-kDa scaffolding protein with roles in mediating both canonical and non-canonical Wnt signaling (56-59). Daple regulates Wnt through its interaction with the protein Disheveled (Dvl) (56), and it can also interact with RTKs, including EGFR and ERBB2 (HER2) (57), leading to its phosphorylation and dissociation from Disheveled (57). Upon this dissociation, Daple translocates from the cytoplasm to the plasma membrane where it functions as a guanine nucleotide exchange factor (GEF) to activate G proteins (Gαi) and promote Akt signaling, cell migration, and invasion (Fig. 3B) (60). We detected the previously characterized ERBB2:Daple interaction (57) in CAL-33 cells, in addition to the FGFR3:Daple interaction, which we hypothesized may function to promote Gαi activation in an FGFR3-dependent manner.
Figure 3. An HNSCC-enriched FGFR3:Daple interaction mediates activation of cell migratory proteins.
(A) Differential scoring analysis of the FGFR3 interactome highlights CCDC88C (Daple) as an HNSCC-enriched interaction partner to both FGFR3 and ERBB2 (HER2). (B) Activation of RTKs can disrupt the interaction between Disheveled (Dvl) and Daple, allowing Daple to function as a GEF for Gαi. GTP binding causes dissociation of the G protein, leaving Gβγ subunits free to activate migratory signaling through Rac and PAK. (C) NanoBiT biosensor measures Gαi activation through dissociation of the luciferase split between Gα and Gβγ. CNO mediates canonical GPCR signaling through the synthetic Gαi-coupled DREADD receptor. FGF mediates HNSCC-specific signaling through FGFR3 and Daple. (D) Luminescence was measured in CAL-33 and HET-1A cells transfected with Gαi NanoBiT and siRNA (control, FGFR3, or Daple) and stimulated with FGF (10ng/mL) (*P < 0.05 when compared with the vehicle-treated group). (E) Immunoblot analysis of CAL-33 subject to siRNA knockdown. (F) PAK1/2 autophosphorylation measured by immunoblot analysis over a time course of FGF stimulation (0, 5, 10, 30, 60 minutes) in CAL-33 and HET-1A cells. (G) PAK1/2 autophosphorylation measured by immunoblot analysis in CAL-33 cells stimulated with FGF (10ng/mL) and/or treated with 0.5μM of the pan FGFR inhibitor Infigratinib (*P < 0.05 when compared with the vehicle-treated group). (H-I) A vertical scratch was introduced to fibronectin-plated CAL-33 cells and cells were stimulated with FGF (10ng/mL) and/or treated with 0.5μM of Infigratinib. Replicate scratch closures were quantified (H, (*P < 0.05, **P < 0.01 when compared with the vehicle-treated group)), and (I) images were taken at 0 and 24 hours after FGF stimulation (scale bar = 250μm). (J) Daple and FGFR3 expression are plotted for all upper airway and esophageal cell lines in DepMap (62), with the two cancer cell lines used in this study highlighted in red. (K) The sensitivity of cell lines with high or low Daple expression to either a FGFR1 inhibitor (sorafinib), or a FGFR1/2/3 inhibitor (AZD4547) as quantified by area under the curve (AUC) (*P < 0.05). Cell lines were selected from panel J, and for those with corresponding drug sensitivity data the top 5 Daple expressing cells (High Daple) or the bottom 5 Daple expressing cells (Low Daple), were used.
To test this idea, we used a split luciferase assay (Gαi NanoBiT) in which signal is lost upon activation of Gαi and dissociation from Gβγ (Fig. 3C). As a control, we first transfected an engineered Designer Receptor Exclusively Activated by Designer Drugs (DREADD) receptor and stimulated the resulting cell population with the DREADD ligand, clozapine-N-oxide (CNO). We observed robust Gαi activation and corresponding loss of luciferase signal in both CAL-33 and HET-1A cell lines (Fig. S3A). Next, we observed that in the CAL-33 cells, where we had detected the interaction between FGFR3 and Daple, FGF stimulation similarly induced Gαi activation; however, no such activation occurred in HET-1A cells (Fig. 3D). Using siRNA knockdowns, we found that Gαi activation in CAL-33 cells was dependent on both FGFR3 and Daple (Fig. 3D-E and fig. S3B). FGF also rapidly induced ERK phosphorylation in both CAL-33 and HET-1A cells, in line with canonical RTK signaling (Fig. S3C). FGF-mediated Gαi activation in CAL-33 cells results in downstream phosphorylation of PAK1/2, an event not observed in HET-1A (Fig. 3F), and this increased phosphorylation was dependent upon both FGFR3 and Daple (Fig. S3D-E). PAK1/2 activity is known to promote cell migration and invasion and is associated with aggressive tumor behavior and poor patient prognosis in HNSCC (61). Thus, we also evaluated whether FGF stimulation promoted cell migration and, indeed, a statistically significant increase was observed (Fig. S3F-G). Importantly, the FGF-induced migration was not blocked by mitomycin C treatment, suggesting that the effects of FGF promoted cell migration, and were independent of growth factor-stimulated proliferation.
Next, we evaluated if this pathway could be inhibited by the FGFR inhibitor, Infigratinib. Indeed, we find that Infigratinib prevented PAK1/2 phosphorylation upon FGF stimulation (Fig. 3G and fig. S3H), and likewise also prevented cell migration (Fig. 3H-I) in CAL-33 cells. To determine if these observations may be more broadly applicable in more cell types, we first looked at FGFR3 and Daple expression in all upper airway and esophageal cell lines using DepMap (62). We find that both cancer cell lines in which we detect the FGFR3:Daple interaction have above average Daple expression (Fig. 3J). Stratification of cell lines by high and low Daple expression revealed that cell lines with high Daple were more sensitive to a pan FGFR inhibitor (FGFR1/2/3) than those with low Daple expression (Fig. 3K). Importantly, there was no difference in sensitivity to inhibition of FGFR1 among high and low Daple expressing cells, suggesting that the inhibition of FGFR3 may be particularly important for cells with high Daple expression. In total, these results support a mechanism for regulating Gαi activity via FGFR3 and Daple, resulting in increased PAK1/2 activation and cell migration, signaling that can be effectively inhibited using FGFR inhibitors.
Quantitative analysis of the mutant PIK3CA interactome
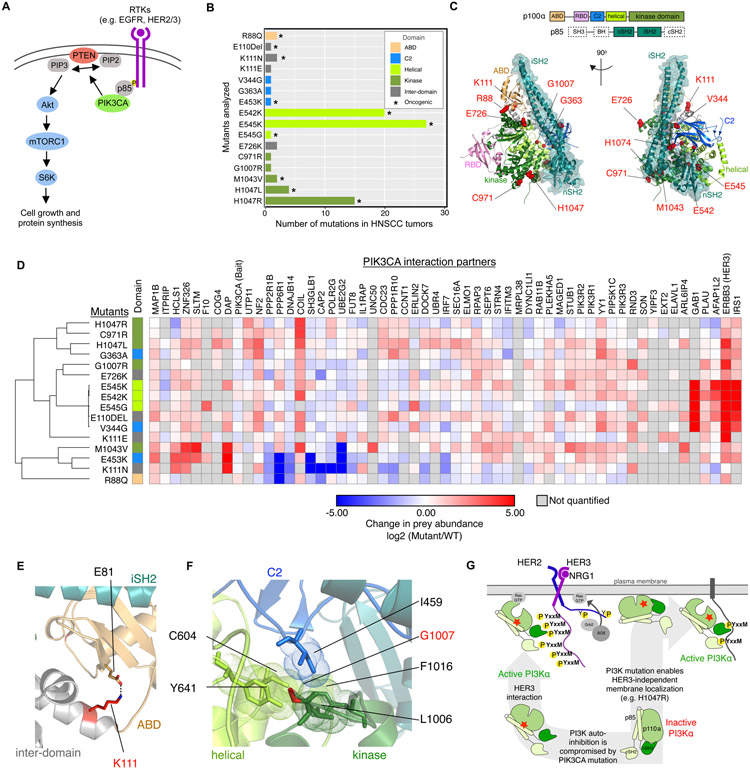
Phosphatidylinositol 3-kinase (PI3K) is a multiprotein kinase complex consisting of a p110alpha (p110α) catalytic subunit that is encoded by the PIK3CA gene, and a p85 regulatory subunit. Engagement of the SH2 domains of p85 with phosphorylated YxxM motifs is essential for PI3K signaling by releasing p110α autoinhibition and mediating recruitment of PI3K to the plasma membrane (63). Upon activation, PI3K is a potent mediator of cellular signaling, interacting with both intracellular small GTPases (e.g., RAS proteins) as well as receptor kinases (e.g., EGFR) to regulate downstream signaling primarily through the Akt/mTOR pathway (Fig. 4A). We selected 16 different PIK3CA mutations observed in HNSCC patients and quantitatively assessed the effects of these mutations on p110α interaction partners (Fig. 4B, Fig. S5 and data S5). Importantly, these mutations were distributed across multiple domains within the p110α protein (Fig. 4C).
Figure 4. PIK3CA mutant interactome.
(A) Overview of the PIK3CA signaling pathway, which is often stimulated by RTKs that interact with PIK3CA to stimulate RAS/Raf-mediated or Akt/mTORC1-mediated downstream signaling. (B) Analyzed PIK3CA mutants and their frequency in HNSCC tumors from TCGA. Asterisk (*) denotes mutations annotated as oncogenic in OncoKB (65). Graph bars corresponding to each mutation were color-coded to indicate their localization within the p110α domain (as indicated in the legend in top right corner). (C) Selected PIK3CA mutations were mapped on the structure of PI3K (PDB: 4L23) (66) by highlighting the mutated residues as red spheres. (D) Quantification of PPIs for all PIK3CA HC-PPIs detected in the SCC-25 cell line (all cell lines displayed in Figure S4A). (E) Cartoon representation of a zoomed-in view of PI3K illustrating a salt bridge formed between K11 and E81 (PDB: 4L23). (F) A zoomed-in view depicting interactions made by G1007 in PI3K (PDB: 4L23). (G) Cartoon representation of different mutation induced PI3K activation mechanisms and their respective HER3 binding preferences.
Examining the protein-protein interaction profiles of WT PIK3CA and the corresponding mutants in SCC-25 cells revealed a high similarity in interaction patterns for five of the PIK3CA mutants (E110DEL, V344G, E542K, E545G, and E545K) (Fig. 4D), driven by a strong increase in interaction of these mutants with three proteins: ERBB3 (HER3), GAB1, and IRS1. All of these prey proteins contain multiple YxxM motifs, representing consensus binding sites for the two SH2 domains (nSH2 and cSH2) located in the PI3K p85 regulatory subunit (64). The interaction between phosphorylated YxxM tyrosine and the SH2 domain serves to release the PI3KCA autoinhibition and recruit it to the plasma membrane to enable PIP2 phosphorylation. The helical domain mutants (E545K, E545G, E542K) are poised to disrupt the interaction of p110α with its auto-inhibitory p85 subunits, making the p85 nSH2 domain more readily available for interaction with phosphorylated YxxM motifs. Outside of this primary cluster of mutations, we also observed other mutation sites (e.g., K111E and G1007R) with a strong increase in HER3 binding. In these cases also, mutations are expected to compromise the p85-imposed inhibition of the p110α catalytic module, either by disruption of the ABD domain relative to the inhibitory iSH2 module of p85 (K111E, Fig. 4E), or by disruption of a hydrophobic cluster coordinating amino acids from multiple p110α domains (G1007R, Fig. 4F).
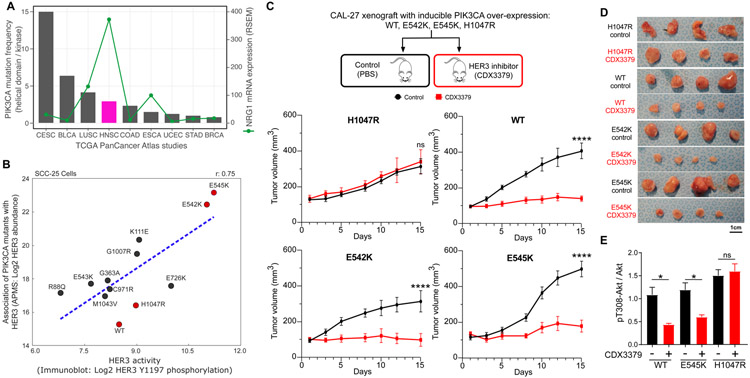
HNSCC tumors display a high preference for PIK3CA helical mutations as compared to kinase domain mutations (Fig. 5A). Our results suggest that PIK3CA mutations which disrupt the autoinhibition of PI3K (e.g. helical domain mutations) may rely on upstream signals that present multiple phosphorylated YxxM sites for signaling (Fig. 4G). In HNSCC, this signal is likely to be an activated HER3 receptor. In support of this synergy, HNSCC tumors have the highest mRNA levels for the HER3-activating ligand, neuregulin-1 (NRG1), across TCGA PanCancer studies with PIK3CA mutations (Fig. 5A). In vitro analysis in the SCC-25 mutant PIK3CA cell lines also demonstrates a strong positive correlation between the interaction of individual PIK3CA mutants with HER3, as measured by AP-MS, and HER3 activation, as measured by immunoblotting of Y1197 phosphorylation (r = 0.75, Fig. 5B and fig. S6A). Furthermore, the increased interaction between helical domain PIK3CA mutants and HER3 is conserved across several additional cell lines tested (Fig. S6B-D). In contrast, kinase domain mutations are known to innately associate with the membrane and therefore to be less dependent on other proteins for membrane recruitment (67). Indeed, we observed low levels of HER3 interaction and phosphorylation (Fig. 4D, Fig. 5B, fig. S6B-D) with the H1047R mutant, suggesting that it might drive oncogenesis in a HER3-independent fashion. We therefore hypothesized that HNSCC tumors harboring PIK3CA helical domain mutations may be selectively sensitive to HER3 inhibitor treatment, while the kinase domain mutation (H1047R) may confer resistance to HER3-inhibition in vivo.
Figure 5. In vivo targeting of HER3 in the context of different PIK3CA mutants.
(A) Bar chart representing the ratio of helical domain (E545 and E542) mutations as compared to kinase domain mutations (H1047) across TCGA PanCancer Altas studies represented in cBioPortal (68). Line graph showing the mRNA expression (RSEM) for NRG1 across the same studies. (B) Correlation of Log2 HER3 interaction levels from AP-MS experiments and Log2 HER3 Y1197 phosphorylation levels from immunoblot analysis. All values are normalized by FLAG-PIK3CA levels in their respective experiments. Mutations marked in red were selected for in vivo experiments. (C-D) CAL-27 cells expressing inducible PIK3CA variants were transplanted into athymic nude mice. Mice were fed with doxycycline to induce PIK3CA expression. When tumor volumes reached approximately 100 mm3, mice were treated with vehicle (PBS) or CDX3379 (10mg/kg, twice a week) for approximately 15 days, as indicated. Shown are (C) tumor growth curves, (D) representative tumor images, and (C) last day tumor volume (****P < 0.0001 when compared with the control-treated group). (E) Quantification of immunoblot analysis of signaling events in the same CAL-27 cells in vitro. PIK3CA variant expression was induced by doxycycline (1μg/ml in culture medium), cells were treated with CDX3379 (1μg/ml, 1hr), and lysates were analyzed by immunoblotting as indicated. Densitometry analysis of western blots was performed using ImageJ. Data are represented as mean ± SEM, n= 3 in each group. (*P < 0.05 when compared with the control-treated group).
To test this hypothesis, we generated isogenic CAL-27 cell lines overexpressing either WT, E542K, E545K, or H1047R mutant isoforms of PIK3CA - CAL-27 cells were used, as they are diploid for WT PIK3CA. We injected these engineered lines into the flanks of athymic nude mice (Materials and methods), and then treated the mice with either saline (control) or the HER3 monoclonal inhibitor CDX3379 over the course of 15 days. Tumor size was monitored and as expected, tumors harboring the H1047R mutant were resistant to CDX3379. Intriguingly, both the helical domain mutants, E542K and E545K and wild-type PIK3CA were sensitive to CDX3379 treatment, resulting in almost complete inhibition of tumor growth (Fig. 5C-D and fig. S6E). From these results, we hypothesize that even in the presence of wild-type PIK3CA, low levels of HER3 binding and HER3 phosphorylation are sufficient and represent an essential mechanism for recruitment of PIK3CA to the membrane and subsequent PI3K activation.These results underscore that only HNSCC patients carrying PI3KCA variants that are still dependent on association with HER3 (wild-type and helical domain mutants) will benefit from HER3-targeted therapeutics.
To further investigate the mechanisms regulating these in vivo phenotypes, we assessed the levels of phosphorylated Akt (pAkt), a downstream mediator of PI3K signaling, in CAL-27 cells. For mutants in which CDX3379 treatment inhibited tumor growth in vivo, in vitro treatment resulted in significant downregulation of pAkt levels, whereas no such decrease was observed for the CDX3379-resistant H1047R-expressing cells (Fig. 5E and fig. S6F).
Discussion
In order to truly understand the complexities of the cell, it has been hypothesized that one would need a list of the protein machines, or complexes, that carry out all of its functions (69). Obtaining such a map would be instrumental in not only understanding how a healthy cell functions but how mutations impact on these machines, and the pathways in which they function (70), in disease states like cancer. Using this premise as a motivation, in this study, we examined the physical landscape of protein-protein interactions targeting genes genetically linked to HNSCC, revealing hundreds of novel PPIs. We observe that these interactions are highly specific to the cell line of study and that mutations in key cancer genes impact on PPI interactions in ways that can provide important mechanistic insight and inform response to targeted treatments. In support of previous observations (25) and our accompanying manuscript (Kim et al., submitted), these results suggest the exciting premise that there remains a vast network of PPIs left to discover beyond the thousands annotated from HEK293T and HeLa cells (22-24). We anticipate that developments in high-throughput protein complex determination, such as co-elution (71), proximity-labeling (72, 73), and cross-linking MS (74), will enable the rapid advancement of systematic PPI mapping in a diverse array of cancer cell contexts.
An important goal of cancer therapy is to identify drug targets that are applicable across many patients and that achieve high specificity for cancer cells among a heterogeneous tumor cell population. In the context of PPIs, this goal requires moving beyond simply cataloging protein-protein interactions towards robust comparative analysis of PPIs across cellular contexts. For this purpose, we have created and demonstrated the value of a differential interaction score (DIS) to statistically compare PPIs across contexts, which will aid in not only understanding the underlying biology behind HNSCC, but other cancers and disease in general, as evidenced by our application of this approach for breast cancer (Kim et al., accompanying manuscript). While the DIS revealed a subset of interactions to be cancer enriched in the three cell lines used in our study, future analysis in additional cell lines would be beneficial to further support these observations.
One interaction uncovered by our DIS approach is a connection between the FGFR3 receptor tyrosine kinase and the guanine-nucleotide exchange factor, Daple, which was seen exclusively in the cancer cell lines. Our findings build upon previous work by demonstrating that FGF stimulation can activate Gαi in a Daple- and FGFR3-dependent manner, which results in activation of PAK1/2 kinases and cell motility. PAK1 expression is highly correlated with aggressive tumor behavior and poor patient prognosis in HNSCC (61, 75). Our work becomes increasingly important as FGFR inhibitors progress towards the clinic. Phase II clinical trials with rogaratinib, an FGFR inhibitor, are underway for HNSCC patients with FGFR1/2/3 mRNA overexpression (NCT03088059), after phase I trials demonstrated a 67% objective response rate for solid tumors with FGFR mRNA overexpression (76). Additionally, a complete response was observed in a metastatic HNSCC tumor with multiple FGFR amplifications, including FGFR3, when treated with a pan-FGFR inhibitor (77). Further work may determine if the FGFR3:Daple interaction results in frequent coupling of FGFR and PAK1/2 activity in HNSCC patients and if other cancer types exploit this signaling mechanism. More direct studies are necessary to determine the extent to which FGFR and PAK1/2 activity contribute to clinical outcomes, as PAK1/2 activity could serve as an additional biomarker of patients benefiting from FGFR targeted therapy.
Our results pertaining to PIK3CA also highlight that oncogenic mechanisms of individual mutations in cancer genes can be reflected in their differences in the corresponding PPIs, and that these differences can be exploited for therapeutic benefit. We postulate that the mechanism for the selectivity we uncovered using PIK3CA mutants lies in the dependence on HER3 signaling that the helical domain mutations maintain. These features of PI3K mutants seemingly contradict previous studies showing that addition of the phosphorylated YxxM motif-containing peptides increases in vitro catalytic activity of the H1047R mutant but not the helical domain mutants (78). However, we hypothesize that phosphorylated RTK tails are necessary not for activation of the helical domain PI3K mutants, but for their recruitment to the plasma membrane where they need to interact with RasGTP for full activation (67). This strong dependence renders cells with such mutations sensitive to HER3 inhibition. Most importantly, we also identify a number of other PI3K mutants that share HER3 binding features with the helical domain mutants, and we predict that their oncogenic potential will also be HER3-dependent (Fig. 4G). Our data also indicate that upstream PI3K activators with a high density of tyrosine phosphorylation sites represented by the YxxM consensus motifs, such as HER3 and IRS1/2, will be particularly efficient in synergizing with the PI3K helical domain mutants.
Clinical inhibition of HER3 in HNSCC patients is currently being pursued in phase II clinical trials with the monoclonal antibody CDX3379 (NCT03254927) (79). This drug locks the HER3 extracellular domain in an inactive configuration (80) and prevents not only dimerization with co-activating RTKs (e.g., HER2) but also activation of HER3 by neuregulins (e.g. NRG1). These properties make HER3 a particularly promising target, as NRG1 is expressed at higher levels in HNSCC than in any other tumor type (81). The results presented here further suggest that HER3 inhibitors present an opportunity to potently target specific PIK3CA mutant tumors, a utility that had not been evaluated previously. This is important, as PIK3CA is one of the most commonly mutated oncogenes in HNSCC (7), yet direct targeting of PIK3CA in the clinic has been limited by toxicity (82), likely due to its pleiotropic roles in cancer and maintenance of normal cell states. In light of our findings, patient pre-selection, such as exclusion of PIK3CA H1047R mutation carriers and inclusion of those harboring helical domain mutants, may be a valuable consideration as future phases of clinical trials proceed.
In summary, this study, along with the accompanying manuscripts (Kim et al., accompanying manuscript and Zheng et al., accompanying manuscript), outlines a framework for elucidating genetic complexity through multidimensional maps of cancer cell biology and demonstrates that such maps can reveal mechanisms of cancer pathogenesis, instructs the selection of therapeutic targets, and informs which point mutations in the tumor are most likely to respond to treatment (26). As such, we anticipate the generation and incorporation of cancer-specific physical and functional networks may represent a critical component to interpret and predict cancer biology and its clinical outcomes. Finally, the framework described here applies not only to cancer but to many other genetically defined disease areas as well.
Supplementary Material
Key Resources
| Reagent or resource | Source | Identifier |
|---|---|---|
| Cell lines | ATCC | HEK293T, HET-1A |
| Cell lines | Thomas Carey (University of Michigan) | SCC-25 |
| Cell lines | Gerard Milano (University of Nice, Nice, France) | CAL-33 |
| NanoBiT G-protein dissociation assay | Dr. Asuka Inoue (Tohoku University, Japan) | NanoBiT plasmids (pCAGGS) include Gαi1-LgBiT, Gβ1-native, and SmBiT-Gγ2 (CAAX C68S mutant). Gαi-DREADD (pcDNA3.1) |
| Antibodies | ||
| RSK1/2/3 antibody | Cell Signaling Technology | 9355 |
| ERK1/2 | Cell Signaling Technology | 4695 |
| phospho-PAK1(S199/204)/PAK2(S192/197) | Cell Signaling Technology | 2605 |
| PAK1 | Cell Signaling Technology | 2602 |
| PAK2 | Cell Signaling Technology | 2608 |
| pERK | Cell Signaling Technology | 9106 |
| FGFR3 | OriGene | TA801078 |
| Daple | Millipore EMD | ABS515 |
| GAPDH | Cell Signaling Technology | 2118 |
| secondary goat anti-rabbit HRP | Southern Biotech | 4010-05 |
| P-HER3-Y1197 | Cell Signaling Technology | 4561 |
| HER3 | Cell Signaling Technology | 12708 |
| goat anti-mouse HRP | Southern Biotech | 1010-05 |
| anti-B-tubulin | Abcam | ab6276 |
| ERK | Cell Signaling Technology | 9102 |
| Deposited data | ||
| Unprocessed peptide files | This paper | PRIDE ProteomeXchange: PXD019469 |
| Raw data | This paper | PRIDE ProteomeXchange: PXD019469 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tris | G-Biosciences | RC108 |
| Acetonitrile, HPLC grade (ACN) | Thermo Fisher Scientific | A955-4 |
| cOmplete protease inhibitor cocktail tablets mini, EDTA-free | Roche | 11846 170 001 |
| Dithiothreitol (DTT) | Sigma-Aldrich | 43819 |
| Formic acid (FA) | Thermo Fisher Scientific | 28905 |
| Iodoacetamide (IAA) | Acros Organic | 122270250 |
| Sequencing-grade modified trypsin | Promega | V5111 |
| Benzonase | Sigma | E1014-25KU |
| Trifluoroacetic acid (TFA) | Thermo Fisher Scientific | 28904 |
| Urea | Sigma-Aldrich | U5378-1kg |
| Fetal bovine serum (FBS) | Gibco | A3160502 |
| DMEM | Corning | MT10013CV |
| DMEM/F12 | Corning | MT10092CV |
| Water, HPLC grade | Sigma-Aldrich | 270733-4 L |
| Igepal (NP-40) | Sigma-Aldrich | I3021 |
| Minimal Essential Media | Corning | 10-009-CV |
| Opti-MEM | Thermo Fisher Scientific | 31985062 |
| BEGM™ (Lonza) | Lonza | CC-3170 |
| 1% Penicillin-Streptomycin | Corning | MT30002Cl |
| Paraformaldehyde, 4% solution in PBS | Thermo Scientific | MFCD00133991 |
| PolyJet | SignaGen | SL100688 |
| Lipofectamine 3000 | ThermoFisher Scientific | L3000008 |
| hydrocortisone | Sigma | H6909-10ML |
| Rapigest | Waters | 186001861 |
| 3x Flag Peptide | Sigma | F4799-4MG |
| Anti-Flag M2 Magnetic Beads | Sigma | M8823-5ML |
| Lipofectamine RNAiMAX | Thermo Fisher Scientific | 100014472 |
| siFGFR3 | Sigma Aldrich | SIHK0780, SIHK0781, SIHK0782 |
| native coelenterazine | Biotium | 10110-1 |
| pooled siControl | Dharmacon | D-001810-10-20 |
| siDaple | Dharmacon | L-033364-01-0005 |
| 10μM clozapine-N-oxide | Cayman Chemical | NC1044836 |
| 5μM native coelenterazine | Biotium | 10110-1 |
| RPS6KA1 siRNA pool | OriGene | SR304161 |
| non-targeting control siRNA | Dharmacon | D-001810-10 |
| Triton X-100 | Thermo Scientific | 9002-93-1 |
| Software and Algorithms | ||
| artMS | Bioconductor | https://www.bioconductor.org/packages/release/bioc/html/artMS.html |
| MSstats | Bioconductor | https://bioconductor.org/packages/release/bioc/html/MSstats.html |
| Skyline | MacCoss Lab | https://skyline.ms/project/home/begin.view? |
| The R Project for Statistical Computing | R Core Team, 2019. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. | http://www.r-project.org/index.html |
| Morpheus | Broad Institute | https://software.broadinstitute.org/morpheus |
| MaxQuant (version 1.6.2.10) | Jurgen Cox Lab | https://www.maxquant.org/ |
| InstantClue | http://www.instantclue.uni-koeln.de/ | |
| CompPASS (version 0.0.0.9000) | github | https://github.com/dnusinow/cRomppass/blob/master/R/comppass.R |
| SAINTexpress (version 3.6.1) | Sourceforge | https://sourceforge.net/projects/saint-apms/files/ |
| Other | ||
| 1.9μM C18 particles | Dr. Maisch | R119.aq.0001 |
| picotip column | New Objective | PF360-75-10-N-5 |
| C18 Stage tips | Rainin | 17014047 |
| Orbitrap Elite mass spectrometer | Thermo Fisher Scientific | IQLAAEGAAPFADBMAZQ |
| Q-Exactive Plus mass spectrometer | Thermo Fisher Scientific | IQLAAEGAAPFALGMBDK |
Acknowledgments:
Funding: NIH U54 CA209891 to JRG, JSG, TI, and NJK. George and Judy Marcus Award in Precision Medicine Innovation to JRG and NJK. NIH R35CA231998 and an American Cancer Society Clinical Research Professorship to JRG. NIH R01DE028289 to JRG and DEJ. NIH R01DE026644, R01DE026870, and R01CA247551 to JSG. NIH U24 CA184427 and NIH P41 GM103504 to TI. NIH R50 CA243885 to JFK. NIH R01 GM109176 to NJu. NIH F32CA239333 to MB. Martha and Bruce Atwater Breast Cancer Research Program, UCSF Prostate Cancer Program, and Benioff Initiative for Prostate Cancer Research to MK. Data for this study were acquired at the Center for Advanced Light Microscopy- Nikon Imaging Center at UCSF on instruments obtained using grants from the UCSF Program for Breakthrough Biomedical Research funded in part by the Sandler Foundation and the UCSF Research Resource Fund Award.
Footnotes
Competing interests: JSG is a Member of the Board of Scientific Advisors for Vividion, Oncoceutics Pharmaceuticals, and Domain Pharmaceuticals. TI is the co-founder of Data4Cure, Inc. with an equity interest, and he has a funded sponsored research agreement from Ideaya BioSciences, Inc., with an equity stake. The lab of NJK has received research support from Vir Biotechnology and F. Hoffmann-La Roche. Nevan Krogan has consulting agreements with the Icahn School of Medicine at Mount Sinai, New York, Maze Therapeutics and Interline Therapeutics, is a shareholder of Tenaya Therapeutics and has received stocks from Maze Therapeutics and Interline Therapeutics. NJu is a member of the SAB of Turning Point Therapeutics, SUDO Biosciences and Type6 Therapeutics, a shareholder of Turning Point Therapeutics and stock option holder of SUDO Biosciences and Type6 Therapeutics.
Data and materials availability: Further information and requests for resources and reagents should be directed and will be fulfilled by NJK (nevan.krogan@ucsf.edu). All raw MS data files and search results are available from the Pride partner ProteomeXchange repository under the PXD019469 identifier, and the PPI network is viewable on NEDx with the DOI 10.18119/N9R010.
References and Notes
- 1.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, Beltran H, Abida W, Bradley RK, Vinson J, Cao X, Vats P, Kunju LP, Hussain M, Feng FY, Tomlins SA, Cooney KA, Smith DC, Brennan C, Siddiqui J, Mehra R, Chen Y, Rathkopf DE, Morris MJ, Solomon SB, Durack JC, Reuter VE, Gopalan A, Gao J, Loda M, Lis RT, Bowden M, Balk SP, Gaviola G, Sougnez C, Gupta M, Yu EY, Mostaghel EA, Cheng HH, Mulcahy H, True LD, Plymate SR, Dvinge H, Ferraldeschi R, Flohr P, Miranda S, Zafeiriou Z, Tunariu N, Mateo J, Perez-Lopez R, Demichelis F, Robinson BD, Sboner A, Schiffman M, Nanus DM, Tagawa ST, Sigaras A, Eng KW, Elemento O, Sboner A, Heath EI, Scher HI, Pienta KJ, Kantoff P, de Bono JS, Rubin MA, Nelson PS, Garraway LA, Sawyers CL, Chinnaiyan AM, Integrative Clinical Genomics of Advanced Prostate Cancer. Cell. 162, 454 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research, Network, Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 455, 1061–1068 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research, Network, Integrated genomic analyses of ovarian carcinoma. Nature. 474, 609–615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas, Network, Comprehensive molecular portraits of human breast tumours. Nature. 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, Chang DK, Cowley MJ, Gardiner BB, Song S, Harliwong I, Idrisoglu S, Nourse C, Nourbakhsh E, Manning S, Wani S, Gongora M, Pajic M, Scarlett CJ, Gill AJ, Pinho AV, Rooman I, Anderson M, Holmes O, Leonard C, Taylor D, Wood S, Xu Q, Nones K, Fink JL, Christ A, Bruxner T, Cloonan N, Kolle G, Newell F, Pinese M, Mead RS, Humphris JL, Kaplan W, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chou A, Chin VT, Chantrill LA, Mawson A, Samra JS, Kench JG, Lovell JA, Daly RJ, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N, Australian Pancreatic Cancer Genome, Initiative, Kakkar N, Zhao F, Wu YQ, Wang M, Muzny DM, Fisher WE, Brunicardi FC, Hodges SE, Reid JG, Drummond J, Chang K, Han Y, Lewis LR, Dinh H, Buhay CJ, Beck T, Timms L, Sam M, Begley K, Brown A, Pai D, Panchal A, Buchner N, De Borja R, Denroche RE, Yung CK, Serra S, Onetto N, Mukhopadhyay D, Tsao MS, Shaw PA, Petersen GM, Gallinger S, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Schulick RD, Wolfgang CL, Morgan RA, Lawlor RT, Capelli P, Corbo V, Scardoni M, Tortora G, Tempero MA, Mann KM, Jenkins NA, Perez-Mancera PA, Adams DJ, Largaespada DA, Wessels LF, Rust AG, Stein LD, Tuveson DA, Copeland NG, Musgrove EA, Scarpa A, Eshleman JR, Hudson TJ, Sutherland RL, Wheeler DA, Pearson JV, McPherson JD, Gibbs RA, Grimmond SM, Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 491, 399–405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, Yates LR, Papaemmanuil E, Beare D, Butler A, Cheverton A, Gamble J, Hinton J, Jia M, Jayakumar A, Jones D, Latimer C, Lau KW, McLaren S, McBride DJ, Menzies A, Mudie L, Raine K, Rad R, Chapman MS, Teague J, Easton D, Langerod A, Oslo Breast Cancer, Consortium, Lee MT, Shen CY, Tee BT, Huimin BW, Broeks A, Vargas AC, Turashvili G, Martens J, Fatima A, Miron P, Chin SF, Thomas G, Boyault S, Mariani O, Lakhani SR, van de Vijver M, van ’t Veer L, Foekens J, Desmedt C, Sotiriou C, Tutt A, Caldas C, Reis-Filho JS, Aparicio SA, Salomon AV, Borresen-Dale AL, Richardson AL, Campbell PJ, Futreal PA, Stratton MR, The landscape of cancer genes and mutational processes in breast cancer. Nature. 486, 400–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas, Network, Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 517, 576–582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, Akbani R, Bowlby R, Wong CK, Wiznerowicz M, Sanchez-Vega F, Robertson AG, Schneider BG, Lawrence MS, Noushmehr H, Malta TM, Cancer Genome Atlas Network, Stuart JM, Benz CC, Laird PW, Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell. 173, 291–304.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofree M, Shen JP, Carter H, Gross A, Ideker T, Network-based stratification of tumor mutations. Nat. Methods 10, 1108–1115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leiserson MDM, Vandin F, Wu H-T, Dobson JR, Eldridge JV, Thomas JL, Papoutsaki A, Kim Y, Niu B, McLellan M, Lawrence MS, Gonzalez-Perez A, Tamborero D, Cheng Y, Ryslik GA, Lopez-Bigas N, Getz G, Ding L, Raphael BJ, Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat. Genet 47, 106–114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.T. M. Consequences, Consortium, Pathway Analysis working group of the International Cancer Genome, Pathway and network analysis of cancer genomes. Nat. Methods 12, 615–621 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerami E, Demir E, Schultz N, Taylor BS, Sander C, Automated network analysis identifies core pathways in glioblastoma. PLoS One. 5, e8918 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paull EO, Carlin DE, Niepel M, Sorger PK, Haussler D, Stuart JM, Discovering causal pathways linking genomic events to transcriptional states using Tied Diffusion Through Interacting Events (TieDIE). Bioinformatics. 29, 2757–2764 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T, Wernersson R, Hansen RB, Horn H, Mercer J, Slodkowicz G, Workman CT, Rigina O, Rapacki K, Stærfeldt HH, Brunak S, Jensen TS, Lage K, A scored human protein–protein interaction network to catalyze genomic interpretation. Nat. Methods (2016) (available at http://www.nature.com/doifinder/10.1038/nmeth.4083). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horn H, Lawrence MS, Chouinard CR, Shrestha Y, Hu JX, Worstell E, Shea E, Ilic N, Kim E, Kamburov A, Kashani A, Hahn WC, Campbell JD, Boehm JS, Getz G, Lage K, NetSig: network-based discovery from cancer genomes. Nat. Methods 15, 61–66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drier Y, Sheffer M, Domany E, Pathway-based personalized analysis of cancer. Proc. Natl. Acad. Sci. U. S. A 110, 6388–6393 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akavia UD, Litvin O, Kim J, Sanchez-Garcia F, Kotliar D, Causton HC, Pochanard P, Mozes E, Garraway LA, Pe’er D, An integrated approach to uncover drivers of cancer. Cell. 143, 1005–1017 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paczkowska M, Barenboim J, Sintupisut N, Fox NS, Zhu H, Abd-Rabbo D, Mee MW, Boutros PC, PCAWG Drivers and Functional Interpretation Working Group, Reimand J, PCAWG Consortium, Integrative pathway enrichment analysis of multivariate omics data. Nat. Commun 11, 735 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reyna MA, Haan D, Paczkowska M, Verbeke LPC, Vazquez M, Kahraman A, Pulido-Tamayo S, Barenboim J, Wadi L, Dhingra P, Shrestha R, Getz G, Lawrence MS, Pedersen JS, Rubin MA, Wheeler DA, Brunak S, Izarzugaza JMG, Khurana E, Marchal K, von Mering C, Sahinalp SC, Valencia A, PCAWG Drivers and Functional Interpretation Working Group, Reimand J, Stuart JM, Raphael BJ, PCAWG Consortium, Pathway and network analysis of more than 2500 whole cancer genomes. Nat. Commun 11, 729 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rolland T, Taşan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, Kamburov A, Ghiassian SD, Yang X, Ghamsari L, Balcha D, Begg BE, Braun P, Brehme M, Broly MP, Carvunis A-R, Convery-Zupan D, Corominas R, Coulombe-Huntington J, Dann E, Dreze M, Dricot A, Fan C, Franzosa E, Gebreab F, Gutierrez BJ, Hardy MF, Jin M, Kang S, Kiros R, Lin GN, Luck K, MacWilliams A, Menche J, Murray RR, Palagi A, Poulin MM, Rambout X, Rasla J, Reichert P, Romero V, Ruyssinck E, Sahalie JM, Scholz A, Shah AA, Sharma A, Shen Y, Spirohn K, Tam S, Tejeda AO, Trigg SA, Twizere J-C, Vega K, Walsh J, Cusick ME, Xia Y, Barabási A-L, Iakoucheva LM, Aloy P, De Las Rivas J, Tavernier J, Calderwood MA, Hill DE, Hao T, Roth FP, Vidal M, A Proteome-Scale Map of the Human Interactome Network. Cell. 159, 1212–1226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luck K, Kim D-K, Lambourne L, Spirohn K, Begg BE, Bian W, Brignall R, Cafarelli T, Campos-Laborie FJ, Charloteaux B, Choi D, Coté AG, Daley M, Deimling S, Desbuleux A, Dricot A, Gebbia M, Hardy MF, Kishore N, Knapp JJ, Kovács IA, Lemmens I, Mee MW, Mellor JC, Pollis C, Pons C, Richardson AD, Schlabach S, Teeking B, Yadav A, Babor M, Balcha D, Basha O, Bowman-Colin C, Chin S-F, Choi SG, Colabella C, Coppin G, D’Amata C, De Ridder D, De Rouck S, Duran-Frigola M, Ennajdaoui H, Goebels F, Goehring L, Gopal A, Haddad G, Hatchi E, Helmy M, Jacob Y, Kassa Y, Landini S, Li R, van Lieshout N, MacWilliams A, Markey D, Paulson JN, Rangarajan S, Rasla J, Rayhan A, Rolland T, San-Miguel A, Shen Y, Sheykhkarimli D, Sheynkman GM, Simonovsky E, Taşan M, Tejeda A, Tropepe V, Twizere J-C, Wang Y, Weatheritt RJ, Weile J, Xia Y, Yang X, Yeger-Lotem E, Zhong Q, Aloy P, Bader GD, De Las Rivas J, Gaudet S, Hao T, Rak J, Tavernier J, Hill DE, Vidal M, Roth FP, Calderwood MA, A reference map of the human binary protein interactome. Nature. 580, 402–408 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, Dong R, Guarani V, Vaites LP, Ordureau A, Rad R, Erickson BK, Wühr M, Chick J, Zhai B, Kolippakkam D, Mintseris J, Obar RA, Harris T, Artavanis-Tsakonas S, Sowa ME, De Camilli Pietro, Paulo JA, Harper JW, Gygi SP, The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell. 162, 425–440 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP, Parzen H, Szpyt J, Tam S, Zarraga G, Pontano-Vaites L, Swarup S, White AE, Schweppe DK, Rad R, Erickson BK, Obar RA, Guruharsha KG, Li K, Artavanis-Tsakonas S, Gygi SP, Harper JW, Architecture of the human interactome defines protein communities and disease networks. Nature. 545, 505–509 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, Hyman AA, Mann M, A Human Interactome in Three Quantitative Dimensions Organized by Stoichiometries and Abundances. Cell. 163, 712–723 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Huttlin EL, Bruckner RJ, Navarrete-Perea J, Cannon JR, Baltier K, Gebreab F, Gygi MP, Thornock A, Zarraga G, Tam S, Szpyt J, Panov A, Parzen H, Fu S, Golbazi A, Maenpaa E, Stricker K, Thakurta SG, Rad R, Pan J, Nusinow DP, Paulo JA, Schweppe DK, Vaites LP, Wade Harper J, Gygi SP, Dual Proteome-scale Networks Reveal Cell-specific Remodeling of the Human Interactome. bioRxiv (2020), p. 2020.01.19.905109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krogan NJ, Lippman S, Agard DA, Ashworth A, Ideker T, The cancer cell map initiative: defining the hallmark networks of cancer. Mol. Cell 58, 690–698 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riaz N, Morris LG, Lee W, Chan TA, Unraveling the molecular genetics of head and neck cancer through genome-wide approaches. Genes Dis. 1, 75–86 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Network, Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 517, 576–582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dogruluk T, Tsang YH, Espitia M, Chen F, Chen T, Chong Z, Appadurai V, Dogruluk A, Eterovic AK, Bonnen PE, Creighton CJ, Chen K, Mills GB, Scott KL, Identification of Variant-Specific Functions of PIK3CA by Rapid Phenotyping of Rare Mutations. Cancer Res. 75, 5341–5354 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rudd ML, Price JC, Fogoros S, Godwin AK, Sgroi DC, Merino MJ, Bell DW, A unique spectrum of somatic PIK3CA (p110alpha) mutations within primary endometrial carcinomas. Clin. Cancer Res 17, 1331–1340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lui VWY, Hedberg ML, Li H, Vangara BS, Pendleton K, Zeng Y, Lu Y, Zhang Q, Du Y, Gilbert BR, Freilino M, Sauerwein S, Peyser ND, Xiao D, Diergaarde B, Wang L, Chiosea S, Seethala R, Johnson JT, Kim S, Duvvuri U, Ferris RL, Romkes M, Nukui T, Kwok-Shing Ng P, Garraway LA, Hammerman PS, Mills GB, Grandis JR, Frequent Mutation of the PI3K Pathway in Head and Neck Cancer Defines Predictive Biomarkers. Cancer Discov. 3, 761–769 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, Shefler E, Ramos AH, Stojanov P, Carter SL, Voet D, Cortés ML, Auclair D, Berger MF, Saksena G, Guiducci C, Onofrio RC, Parkin M, Romkes M, Weissfeld JL, Seethala RR, Wang L, Rangel-Escareño C, Fernandez-Lopez JC, Hidalgo-Miranda A, Melendez-Zajgla J, Winckler W, Ardlie K, Gabriel SB, Meyerson M, Lander ES, Getz G, Golub TR, Garraway LA, Grandis JR, The mutational landscape of head and neck squamous cell carcinoma. Science. 333, 1157–1160 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin D, Abba MC, Molinolo AA, Vitale-Cross L, Wang Z, Zaida M, Delic NC, Samuels Y, Lyons JG, Gutkind JS, The head and neck cancer cell oncogenome: a platform for the development of precision molecular therapies. Oncotarget. 5, 8906–8923 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molinolo AA, Amornphimoltham P, Squarize CH, Castilho RM, Patel V, Gutkind JS, Dysregulated molecular networks in head and neck carcinogenesis. Oral Oncol. 45, 324–334 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Wawrose JS, Gooding WE, Garraway LA, Lui VWY, Peyser ND, Grandis JR, Genomic Analysis of Head and Neck Squamous Cell Carcinoma Cell Lines and Human Tumors: A Rational Approach to Preclinical Model Selection. Mol. Cancer Res 12, 571–582 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orchard S, Kerrien S, Abbani S, Aranda B, Bhate J, Bidwell S, Bridge A, Briganti L, Brinkman FSL, Cesareni G, Chatr-aryamontri A, Chautard E, Chen C, Dumousseau M, Goll J, Hancock REW, Hannick LI, Jurisica I, Khadake J, Lynn DJ, Mahadevan U, Perfetto L, Raghunath A, Ricard-Blum S, Roechert B, Salwinski L, Stümpflen V, Tyers M, Uetz P, Xenarios I, Hermjakob H, Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nature Methods. 9 (2012), pp. 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oughtred R, Rust J, Chang C, Breitkreutz B-J, Stark C, Willems A, Boucher L, Leung G, Kolas N, Zhang F, Dolma S, Coulombe-Huntington J, Chatr-Aryamontri A, Dolinski K, Tyers M, The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 30, 187–200 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giurgiu M, Reinhard J, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Ruepp A, CORUM: the comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res. 47, D559–D563 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu K, Chen B, Aran D, Charalel J, Yau C, Wolf DM, van ’t Veer LJ, Butte AJ, Goldstein T, Sirota M, Comprehensive transcriptomic analysis of cell lines as models of primary tumors across 22 tumor types. Nat. Commun 10, 3574 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jäger S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, Hernandez H, Jang GM, Roth SL, Akiva E, Marlett J, Stephens M, D’Orso I, Fernandes J, Fahey M, Mahon C, O’Donoghue AJ, Todorovic A, Morris JH, Maltby DA, Alber T, Cagney G, Bushman FD, Young JA, Chanda SK, Sundquist WI, Kortemme T, Hernandez RD, Craik CS, Burlingame A, Sali A, Frankel AD, Krogan NJ, Global landscape of HIV-human protein complexes. Nature. 481, 365–370 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teo G, Liu G, Zhang J, Nesvizhskii AI, Gingras A-C, Choi H, SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J. Proteomics 100, 37–43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sowa ME, Bennett EJ, Gygi SP, Harper JW, Defining the Human Deubiquitinating Enzyme Interaction Landscape. Cell. 138, 389–403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouhaddou M, Eckhardt M, Chi Naing ZZ, Kim M, Ideker T, Krogan NJ, Mapping the protein-protein and genetic interactions of cancer to guide precision medicine. Curr. Opin. Genet. Dev 54, 110–117 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eckhardt M, Zhang W, Gross AM, Von Dollen J, Johnson JR, Franks-Skiba KE, Swaney DL, Johnson TL, Jang GM, Shah PS, Brand TM, Archambault J, Kreisberg JF, Grandis JR, Ideker T, Krogan NJ, Multiple Routes to Oncogenesis Are Promoted by the Human Papillomavirus-Host Protein Network. Cancer Discov. 8, 1474–1489 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pyndiah S, Tanida S, Ahmed KM, Cassimere EK, Choe C, Sakamuro D, c-MYC Suppresses BIN1 to Release Poly(ADP-Ribose) Polymerase 1: A Mechanism by Which Cancer Cells Acquire Cisplatin Resistance. Sci. Signal 4, ra19–ra19 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Kalkat M, Resetca D, Lourenco C, Chan P-K, Wei Y, Shiah Y-J, Vitkin N, Tong Y, Sunnerhagen M, Done SJ, Boutros PC, Raught B, Penn LZ, MYC Protein Interactome Profiling Reveals Functionally Distinct Regions that Cooperate to Drive Tumorigenesis. Mol. Cell 72, 836–848.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Czubaty A, Girstun A, Kowalska-Loth B, Trzcińska AM, Purta E, Winczura A, Grajkowski W, Staroń K, Proteomic analysis of complexes formed by human topoisomerase I. Biochim. Biophys. Acta 1749, 133–141 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Haraguchi K, Ohsugi M, Abe Y, Semba K, Akiyama T, Yamamoto T, Ajuba negatively regulates the Wnt signaling pathway by promoting GSK-3beta-mediated phosphorylation of beta-catenin. Oncogene. 27, 274–284 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Feng Y, Longmore GD, The LIM protein Ajuba influences interleukin-1-induced NF-kappaB activation by affecting the assembly and activity of the protein kinase Czeta/p62/TRAF6 signaling complex. Mol. Cell. Biol 25, 4010–4022 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan W, Tang Z, Chen D, Moughon D, Ding X, Chen S, Zhu M, Zhong Q, Keap1 facilitates p62-mediated ubiquitin aggregate clearance via autophagy. Autophagy. 6, 614–621 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lau A, Wang X-J, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD, A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol. Cell. Biol 30, 3275–3285 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Copple IM, Lister A, Obeng AD, Kitteringham NR, Jenkins RE, Layfield R, Foster BJ, Goldring CE, Park BK, Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1-Nrf2 cell defense pathway. J. Biol. Chem 285, 16782–16788 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hamilton E, Infante JR, Targeting CDK4/6 in patients with cancer. Cancer Treat. Rev 45, 129–138 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Fusté NP, Castelblanco E, Felip I, Santacana M, Fernández-Hernández R, Gatius S, Pedraza N, Pallarés J, Cemeli T, Valls J, Tarres M, Ferrezuelo F, Dolcet X, Matias-Guiu X, Garí E, Characterization of cytoplasmic cyclin D1 as a marker of invasiveness in cancer. Oncotarget. 7, 26979–26991 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng F, Tutuncuoglu B, Ono K, Swaney DL, Kim M, Silva E, Liu S, Park J, Kratz A, Yu MK, Chen K, Chen J, Churas C, Pillich R, Patel D, Licon K, Kriesberg JF, Fraley S, Pratt D, Krogan NJ, Ideker T, Convergence of cancer mutation on a hierarchy of protein systems. submitted. [Google Scholar]
- 56.Oshita A, Kishida S, Kobayashi H, Michiue T, Asahara T, Asashima M, Kikuchi A, Identification and characterization of a novel Dvl-binding protein that suppresses Wnt signalling pathway. Genes Cells. 8, 1005–1017 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Aznar N, Ear J, Dunkel Y, Sun N, Satterfield K, He F, Kalogriopoulos NA, Lopez-Sanchez I, Ghassemian M, Sahoo D, Kufareva I, Ghosh P, Convergence of Wnt, growth factor, and heterotrimeric G protein signals on the guanine nucleotide exchange factor Daple. Sci. Signal 11 (2018), doi: 10.1126/scisignal.aao4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aznar N, Sun N, Dunkel Y, Ear J, Buschman MD, Ghosh P, A Daple-Akt feed-forward loop enhances noncanonical Wnt signals by compartmentalizing β-catenin. Mol. Biol. Cell 28, 3709–3723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ishida-Takagishi M, Enomoto A, Asai N, Ushida K, Watanabe T, Hashimoto T, Kato T, Weng L, Matsumoto S, Asai M, Murakumo Y, Kaibuchi K, Kikuchi A, Takahashi M, The Dishevelled-associating protein Daple controls the non-canonical Wnt/Rac pathway and cell motility. Nat. Commun 3, 859 (2012). [DOI] [PubMed] [Google Scholar]
- 60.Aznar N, Midde KK, Dunkel Y, Lopez-Sanchez I, Pavlova Y, Marivin A, Barbazán J, Murray F, Nitsche U, Janssen K-P, Willert K, Goel A, Abal M, Garcia-Marcos M, Ghosh P, Daple is a novel non-receptor GEF required for trimeric G protein activation in Wnt signaling. Elife. 4, e07091 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park J, Kim J-M, Park JK, Huang S, Kwak SY, Ryu KA, Kong G, Park J, Koo BS, Association of p21-activated kinase-1 activity with aggressive tumor behavior and poor prognosis of head and neck cancer. Head Neck. 37, 953–963 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, Meyers RM, Ali L, Goodale A, Lee Y, Jiang G, Hsiao J, Gerath WFJ, Howell S, Merkel E, Ghandi M, Garraway LA, Root DE, Golub TR, Boehm JS, Hahn WC, Defining a Cancer Dependency Map. Cell. 170, 564–576.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dornan GL, Burke JE, Molecular Mechanisms of Human Disease Mediated by Oncogenic and Primary Immunodeficiency Mutations in Class IA Phosphoinositide 3-Kinases. Front. Immunol 9, 575 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Songyang Z, Shoelson SE, McGlade J, Olivier P, Pawson T, Bustelo XR, Barbacid M, Sabe H, Hanafusa H, Yi T, Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol. Cell. Biol 14, 2777–2785 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH, Chang MT, Chandarlapaty S, Traina TA, Paik PK, Ho AL, Hantash FM, Grupe A, Baxi SS, Callahan MK, Snyder A, Chi P, Danila D, Gounder M, Harding JJ, Hellmann MD, Iyer G, Janjigian Y, Kaley T, Levine DA, Lowery M, Omuro A, Postow MA, Rathkopf D, Shoushtari AN, Shukla N, Voss M, Paraiso E, Zehir A, Berger MF, Taylor BS, Saltz LB, Riely GJ, Ladanyi M, Hyman DM, Baselga J, Sabbatini P, Solit DB, Schultz N, OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. 2017 (2017), doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao Y, Zhang X, Chen Y, Lu S, Peng Y, Wang X, Guo C, Zhou A, Zhang J, Luo Y, Shen Q, Ding J, Meng L, Zhang J, Crystal Structures of PI3Kα Complexed with PI103 and Its Derivatives: New Directions for Inhibitors Design. ACS Med. Chem. Lett 5, 138–142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao L, Vogt PK, Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc. Natl. Acad. Sci. U. S. A 105, 2652–2657 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N, Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 6, l1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alberts B, The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 92, 291–294 (1998). [DOI] [PubMed] [Google Scholar]
- 70.Hanahan D, Weinberg RA, The hallmarks of cancer. Cell. 100, 57–70 (2000). [DOI] [PubMed] [Google Scholar]
- 71.Salas D, Stacey RG, Akinlaja M, Foster LJ, Next-generation Interactomics: Considerations for the Use of Co-elution to Measure Protein Interaction Networks. Mol. Cell. Proteomics 19, 1–10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lobingier BT, Hüttenhain R, Eichel K, Miller KB, Ting AY, von Zastrow M, Krogan NJ, An Approach to Spatiotemporally Resolve Protein Interaction Networks in Living Cells. Cell. 169, 350–360.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Samavarchi-Tehrani P, Samson R, Gingras A-C, Proximity dependent biotinylation: key enzymes and adaptation to proteomics approaches. Mol. Cell. Proteomics (2020), doi: 10.1074/mcp.R120.001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klykov O, Steigenberger B, Pektaş S, Fasci D, Heck AJR, Scheltema RA, Efficient and robust proteome-wide approaches for cross-linking mass spectrometry. Nat. Protoc (2018), doi: 10.1038/s41596-018-0074-x. [DOI] [PubMed] [Google Scholar]
- 75.Parvathy M, Sreeja S, Kumar R, Pillai MR, Potential role of p21 Activated Kinase 1 (PAK1) in the invasion and motility of oral cancer cells. BMC Cancer. 16 Suppl 1, 293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schuler M, Cho BC, Sayehli CM, Navarro A, Soo RA, Richly H, Cassier PA, Tai D, Penel N, Nogova L, Park SH, Schostak M, Gajate P, Cathomas R, Rajagopalan P, Grevel J, Bender S, Boix O, Nogai H, Ocker M, Ellinghaus P, Joerger M, Rogaratinib in patients with advanced cancers selected by FGFR mRNA expression: a phase 1 dose-escalation and dose-expansion study. Lancet Oncol. 20, 1454–1466 (2019). [DOI] [PubMed] [Google Scholar]
- 77.Dumbrava EI, Alfattal R, Miller VA, Tsimberidou AM, Complete Response to a Fibroblast Growth Factor Receptor Inhibitor in a Patient With Head and Neck Squamous Cell Carcinoma Harboring FGF Amplifications. JCO Precis Oncol. 2 (2018), doi: 10.1200/PO.18.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carson JD, Van Aller G, Lehr R, Sinnamon RH, Kirkpatrick RB, Auger KR, Dhanak D, Copeland RA, Gontarek RR, Tummino PJ, Luo L, Effects of oncogenic p110alpha subunit mutations on the lipid kinase activity of phosphoinositide 3-kinase. Biochem. J 409, 519–524 (2008). [DOI] [PubMed] [Google Scholar]
- 79.Duvvuri U, George J, Kim S, Alvarado D, Neumeister VM, Chenna A, Gedrich R, Hawthorne T, LaVallee T, Grandis JR, Bauman JE, Molecular and Clinical Activity of CDX-3379, an Anti-ErbB3 Monoclonal Antibody, in Head and Neck Squamous Cell Carcinoma Patients. Clin. Cancer Res 25, 5752–5758 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee S, Greenlee EB, Amick JR, Ligon GF, Lillquist JS, Natoli EJ Jr, Hadari Y, Alvarado D, Schlessinger J, Inhibition of ErbB3 by a monoclonal antibody that locks the extracellular domain in an inactive configuration. Proc. Natl. Acad. Sci. U. S. A 112, 13225–13230 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alvarado D, Ligon GF, Lillquist JS, Seibel SB, Wallweber G, Neumeister VM, Rimm DL, McMahon G, LaVallee TM, ErbB activation signatures as potential biomarkers for anti-ErbB3 treatment in HNSCC. PLoS One. 12, e0181356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Janku F, Yap TA, Meric-Bernstam F, Targeting the PI3K pathway in cancer: are we making headway? Nat. Rev. Clin. Oncol 15, 273–291 (2018). [DOI] [PubMed] [Google Scholar]
- 83.Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, Sanli K, von Feilitzen K, Oksvold P, Lundberg E, Hober S, Nilsson P, Mattsson J, Schwenk JM, Brunnström H, Glimelius B, Sjöblom T, Edqvist P-H, Djureinovic D, Micke P, Lindskog C, Mardinoglu A, Ponten F, A pathology atlas of the human cancer transcriptome. Science. 357 (2017), doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 84.Cox J, Mann M, MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 85.Choi M, Chang C-Y, Clough T, Broudy D, Killeen T, MacLean B, Vitek O, MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics. 30, 2524–2526 (2014). [DOI] [PubMed] [Google Scholar]
- 86.Vizcaíno JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Ríos D, Dianes JA, Sun Z, Farrah T, Bandeira N, Binz P-A, Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkley RJ, Kraus H-J, Albar JP, Martinez-Bartolomé S, Apweiler R, Omenn GS, Martens L, Jones AR, Hermjakob H, ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol 32, 223–226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaíno JA, The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ, Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 26, 966–968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Inoue A, Raimondi F, Kadji FMN, Singh G, Kishi T, Uwamizu A, Ono Y, Shinjo Y, Ishida S, Arang N, Kawakami K, Gutkind JS, Aoki J, Russell RB, Illuminating G-Protein-Coupling Selectivity of GPCRs. Cell. 177, 1933–1947.e25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nolte H, MacVicar TD, Tellkamp F, Krüger M, Instant Clue: A Software Suite for Interactive Data Visualization and Analysis. Sci. Rep 8, 12648 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.