Abstract
Background
COVID-19 manifests with respiratory, systemic, and gastrointestinal (GI) symptoms.1, SARS-CoV-2 RNA is detected in respiratory and fecal samples, and recent reports demonstrate viral replication in both the lung and intestinal tissue.2, 3, 4 Although much is known about early fecal RNA shedding, little is known about long-term shedding, especially in those with mild COVID-19. Furthermore, most reports of fecal RNA shedding do not correlate these findings with GI symptoms.5
Methods
We analyzed the dynamics of fecal RNA shedding up to 10 months after COVID-19 diagnosis in 113 individuals with mild to moderate disease. We also correlated shedding with disease symptoms.
Findings
Fecal SARS-CoV-2 RNA is detected in 49.2% [95% confidence interval, 38.2%–60.3%] of participants within the first week after diagnosis. Whereas there was no ongoing oropharyngeal SARS-CoV-2 RNA shedding in subjects at 4 months, 12.7% [8.5%–18.4%] of participants continued to shed SARS-CoV-2 RNA in the feces at 4 months after diagnosis and 3.8% [2.0%–7.3%] shed at 7 months. Finally, we found that GI symptoms (abdominal pain, nausea, vomiting) are associated with fecal shedding of SARS-CoV-2 RNA.
Conclusions
The extended presence of viral RNA in feces, but not in respiratory samples, along with the association of fecal viral RNA shedding with GI symptoms suggest that SARS-CoV-2 infects the GI tract and that this infection can be prolonged in a subset of individuals with COVID-19.
Funding
This research was supported by a Stanford ChemH-IMA grant; fellowships from the AACR and NSF; and NIH R01-AI148623, R01-AI143757, and UL1TR003142.
Keywords: COVID-19, SARS-CoV-2, fecal RNA, gastrointestinal infection, viral shedding
Graphical abstract
Context and significance
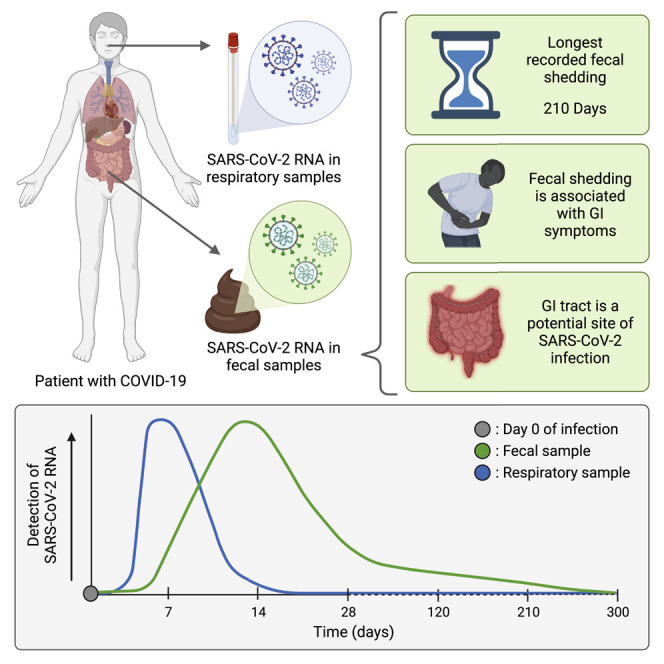
Gastrointestinal symptoms and SARS-CoV-2 RNA shedding in feces point to the gastrointestinal tract as a possible site of infection in COVID-19. Researchers from Stanford University measured the dynamics of fecal viral RNA in patients with mild to moderate COVID-19 followed for 10 months post-diagnosis. The authors found that fecal viral RNA shedding was correlated with gastrointestinal symptoms in patients who had cleared their respiratory infection. They also observed that fecal shedding can continue to 7 months post-diagnosis. In conjunction with recent related findings, this work presents compelling evidence of SARS-CoV-2 infection in the gastrointestinal tract and suggests a possible role for long-term infection of the gastrointestinal tract in syndromes such as “long COVID.”
Natarajan et al. perform a longitudinal study of fecal SARS-CoV-2 RNA shedding in patients with mild to moderate COVID-19, revealing that patients can shed RNA for up to 7 months after infection, that shedding is associated with gastrointestinal symptoms, and that the gastrointestinal tract may be infected even after the respiratory infection has cleared.
Introduction
COVID-19 is a disease with protean manifestations, ranging from respiratory to gastrointestinal to systemic. Although the primary site of SARS-CoV-2 infection is the respiratory tract, the presence of symptoms affecting other organ systems (e.g., abdominal pain, nausea, arthralgia), coupled with in vitro evidence of SARS-CoV-2 infectivity in a variety of other tissues, suggests that SARS-CoV-2 infection can extend beyond the respiratory system. Meta-analyses of studies that focus on hospitalized individuals with COVID-19 estimate the pooled incidence of gastrointestinal (GI) symptoms such as nausea, vomiting, and diarrhea to be between 11% and 18%.1 , 2 , 3 , 6, 7, 8 Additionally, within this moderate- to severe-disease group, SARS-CoV-2 RNA has been detected in 40%–85% of fecal samples (reviewed in Brooks EF and Bhatt AS9), indicating that SARS-CoV-2 viral RNA is found in feces nearly as frequently as in respiratory secretions.10 Patients with moderate to severe COVID-19 have been well studied; by contrast, much less is known about the clearance of SARS-CoV-2 RNA in the feces of patients with mild to moderate disease despite the fact that they make up ∼81% of those who contract COVID-19.11 , 12 Furthermore, most studies are cross-sectional, and the few reported longitudinal studies have focused on the early time period after diagnosis. Thus, a comprehensive understanding of the dynamics of fecal clearance of SARS-CoV-2 RNA in individuals with mild to moderate COVID-19 is both of crucial importance and lacking.
Interestingly, in the few studies that have investigated longitudinal fecal samples, prolonged fecal shedding of SARS-CoV-2 RNA can occur even after respiratory shedding ceases. Indeed, in one notable pediatric case, fecal viral RNA shedding extended beyond 70 days after disease onset.8 If SARS-CoV-2 RNA shedding in the feces is indicative of a GI infection, this suggests that SARS-CoV-2 infection of the GI tract can continue after clearance from the respiratory tract.
While the presence of SARS-CoV-2 RNA in feces is well established, whether live, infectious SARS-CoV2 is commonly shed in stool remains an outstanding question (reviewed in Guo M et al.13). Five studies have reported isolating infectious SARS-CoV-2 from stool samples collected from participants with severe COVID-19,14, 15, 16, 17, 18 whereas others have reported being unable to isolate infectious virions from stool.19 , 20 Therefore, it remains unclear whether the presence of infectious virions of SARS-CoV-2 in the stool is a rare or common phenomenon. However, there is mounting evidence of possible SARS-CoV-2 infection of the GI tract. Specifically, the presence of SARS-CoV-2 RNA,4 , 21, 22, 23 protein antigen,21 , 24 and virions4 , 23 , 25 in GI biopsies all point to a potential infection of the GI tract. Additionally, the presence of a gut immune response26 and inflammation markers such as fecal calprotectin27 , 28 in individuals with COVID-19 provides supporting evidence of a GI infection. Finally, in vitro experiments reveal that SARS-CoV-2 is able to successfully infect enteroid models of the gut29, 30, 31 and intestinal cell lines.32 This phenomenon of possible GI tract involvement is not surprising, as bovine coronavirus (BCoV) and human enteric coronavirus (HECoV-4408), both of the same genus as SARS-CoV-2 (Betacoronaviruses), can infect respiratory and GI tissues.33 Taken together, these data indicate that the GI tract may be an important site of SARS-CoV-2 infection.33
SARS-CoV-2 presence in the GI tract has additional relevance to patient health. The GI tract is a highly immunoactive tissue, and SARS-CoV-2 antigens in this body site may hone a humoral immune response against variants of the SARS-CoV-2 virus.21 Furthermore, prolonged presence of SARS-CoV-2 in the GI tissue may also have an impact on the hitherto mysterious phenomenon of post-acute sequelae of SARS-CoV-2 infection (PASC) or “Long COVID,” where individuals suffer from an unusual constellation of symptoms even after recovery from the respiratory SARS-CoV-2 infection.34 Taken together, it is critical that we understand whether or not the GI tract is infected and the dynamics of the infection in this tissue, from the standpoint of both the acute infection and the long-term sequelae of COVID-19.
Here, we sought to better define the features of SARS-CoV-2 presence in the GI tract and its relevance for short- and long-term human health. We leveraged longitudinal fecal and respiratory samples from individuals enrolled in a randomized controlled study of Peg-interferon lambda-1a (IFN-λ) versus a placebo control for the treatment of mild to moderate COVID-19 (n = 120).35 While the intervention did not shorten the duration of oropharyngeal shedding of SARS-CoV-2 RNA (primary outcome) or disease symptoms (secondary outcome), the study provided a rich, prospectively collected dataset from which to evaluate fecal shedding dynamics and its relation to GI symptoms.
Using fecal samples collected at regular intervals from the time of COVID-19 diagnosis to 10 months after diagnosis, we compared fecal viral RNA shedding with the presence of GI and other symptoms and found that it is positively correlated with GI symptoms. This constitutes the largest longitudinal analysis of paired fecal viral RNA shedding and disease symptomatology data in individuals with mild to moderate COVID-19, and it reveals important information about the pathophysiology of the disease.
Results
Description of study participants and sample collection
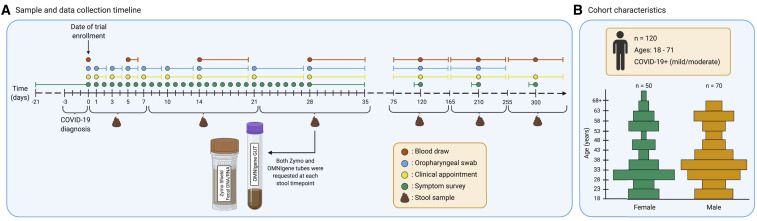
The Peg-IFN-λ clinical trial (NCT04331899) enrolled 120 participants with mild to moderate COVID-19 between 25 April and 17 July 2020.35 Of these, 113 participants collected at least one stool sample at one of the six predefined stool collection time points. These collection time points centered around days 3 (range = 0–7 days), 14 (8–21), 28 (22–35), 120 (75–165), 210 (166–255), and 300 (>255 days) post-enrollment (Figure 1A). Of these 113 participants, 86 provided samples for at least three time points (summarized in Data S1; the overall Data S1 file includes additional data and analysis that informs methods and conclusions in the study and is related to Figures 1, 2 , 3 , and STAR Methods).
Figure 1.
Summary of study protocol and cohort demographics
(A) Sample and data collection timeline represented in days. Day 0 marks the day of enrollment in the trial, within 72 h of a COVID-19 diagnosis. Each sample collection event is marked by a colored dot, where orange represents a blood draw and blue an oropharyngeal (OP) swab. Additionally, clinical appointments and symptom surveys are marked by yellow and green dots, respectively. Some of these events are marked by day ranges to represent collection time frames. The symptom survey at day 0 retrospectively collected symptomatology for 3 weeks prior to enrollment using a single questionnaire. Symptom surveys at time points centered around days 120, 210, and 300 retrospectively collected symptomatology for 1 week prior to the appointment using a single questionnaire at each timepoint. Collection of stool samples and their respective day ranges are marked below the timeline. Subjects were asked to provide samples in the OMNIgene GUT collection tube (OG) and the Zymo DNA/RNA shield fecal collection tube (ZY) at six time points.
(B) Cohort characteristics. 120 participants were enrolled in the clinical trial. Participants had a COVID-19 infection of mild to moderate severity and were between the ages of 18 and 71. The age and sex distributions of the paticipants are reprented here. The x axis separates the groups by self-reported sex, and the y axis lists age in years. Each bar represents a range of 5 years.
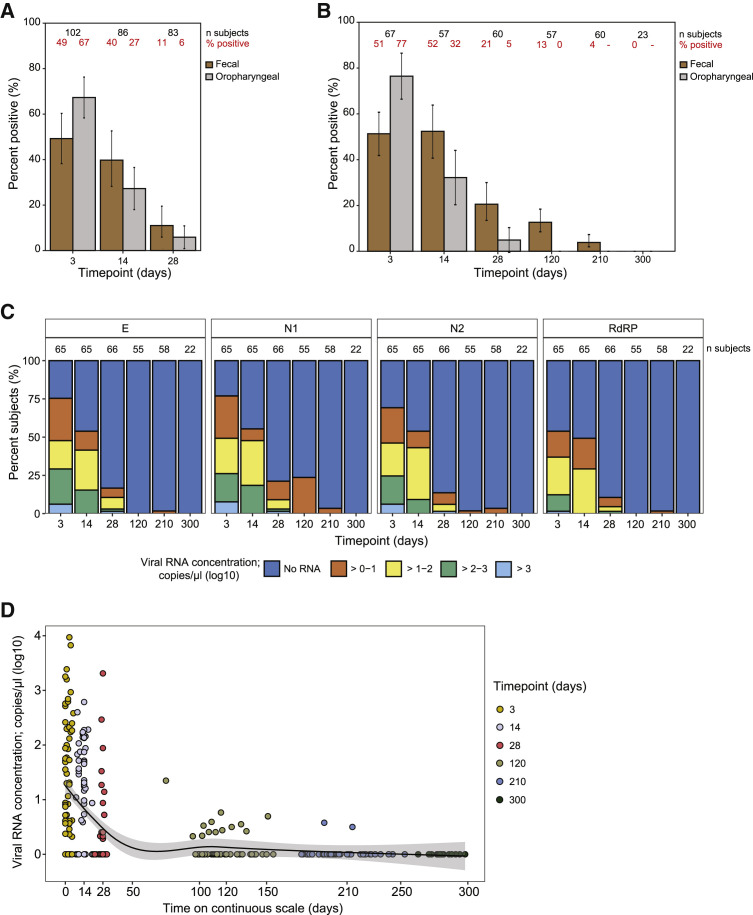
Figure 2.
Fecal and oropharyngeal viral gRNA measurements over time
(A) Summary of viral RNA positivity rates as determined by fecal and OP samples acquired from participants enrolled in the study for a period of around 28 days. The x axis lists time point categories since enrollment as days 3 (range 0–7), 14 (8–21), and 28 (22–35). The y axis lists the percentage of fecal samples (brown bar) and OP samples (gray bar) that tested positive at each of the time points. Fecal positivity rates are evaluated using the logistic GEE model described in the statistical methods section (see STAR Methods), which averages over all of the sample collection methods, gene types, and technical replicates. OP positivity rates are evaluated for the swab taken within 3 days of the fecal sample. Each bar also marks the 95% confidence interval (CI). Number of participants and percent positivity are listed as numbers at the top of the plot in black and red fonts, respectively, and summarized in Data S1.
(B) Same as (A), except restricted to the subset of those who participated in the extended study, and following them through all six time points. As before, the x axis lists time point categories since enrollment: days 3 (range 0–7), 14 (8–21), 28 (22–35), 120 (75–165), 210 (166–255), and 300 (>255 days), and the y axis lists the percentage of participants with positive fecal samples (brown bar) and OP samples (gray bar) at each of the time points, with 95% CI. Number of participants and percent positivity are listed in black and red fonts and summarized in Data S1.
(C) SARS-CoV-2 viral RNA concentration in stool samples collected in the ZY kit from participants (n = 104) with mild to moderate COVID-19 infection over a time period of 300 days from enrollment in the study. Note that the ZY kits had higher overall positivity rates than the OG kits, so positivity rates in this panel tend to be slightly larger than the numbers in the previous two panels, which average over kits and genes. Fecal viral RNA concentration was determined using RT-qPCR with primers/probes targeting the E, N1, N2, and RdRP genes in the SARS-CoV-2 genome, as indicated in the tab at the top of each panel. The x axis lists time point categories since enrollment. The y axis lists the percentage of participants with a given viral RNA concentration, as indicated by the color scheme in the stacked bar plot; dark blue refers to those with no detectable viral RNA, orange to viral RNA concentrations between 0 and one log10 copy/μL, yellow between one and two log10 copies/μL, green between two and three log10 copies/μL, and light blue over three log10 copies/μL. Number of participants per time point is listed above each bar in the stacked bar plot.
(D) Fecal viral RNA concentration in stool samples collected in the ZY kit from participants (n = 104) with mild to moderate COVID-19 infection and assayed using RT-qPCR detecting the N1 gene (viral RNA concentration in log10 copies per μL) versus time (continuous variable; x axis). Time point categories are indicated by color scheme: yellow for days 3 (range 0–7), lavender for day 14 (8–21), red for day 28 (22–35), gray for day 120 (75–165), light blue for day 210 (166–255), and dark blue for day 300 (>255 days). A smoothed line generated using LOESS regression (span parameter = 0.75) and 95% CI is marked in the scatterplot. Note that all viral RNA concentration measurements are expressed on a logarithmic scale by applying the transformation log10 (viral RNA concentration+1).
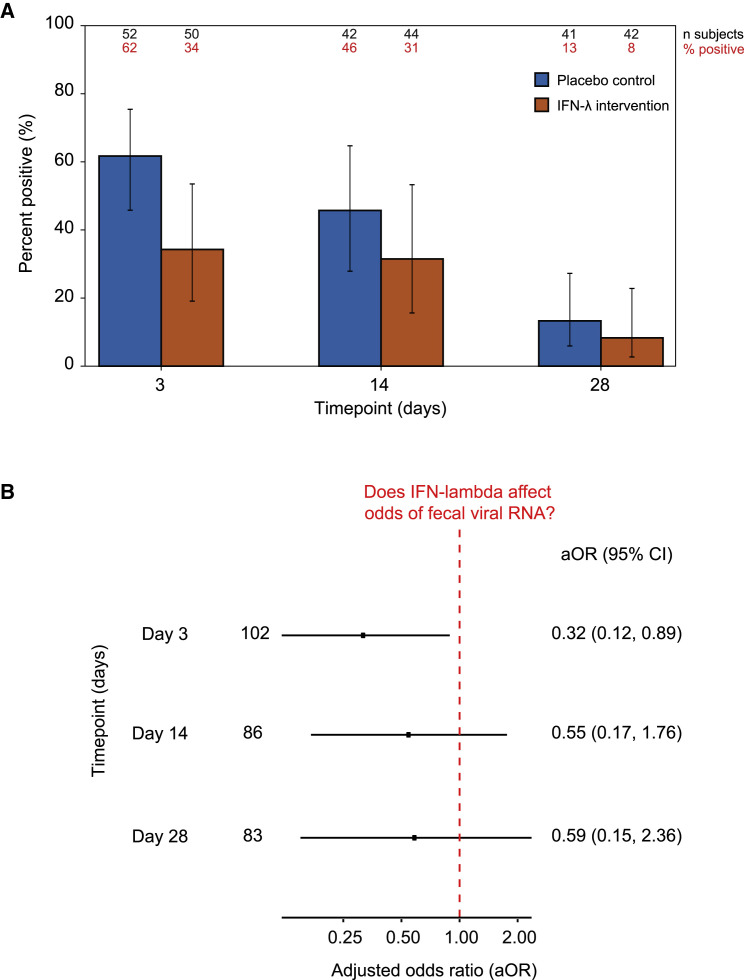
Figure 3.
The effect of IFN-λ on fecal viral RNA shedding
(A) Percentage of participants with detectable fecal SARS-CoV-2 RNA across each of the study arms, as evaluated using the logistic GEE model described in the statistical methods section (STAR Methods). The x axis marks the time point in the study: days 3 (range 0–7), 14 (8–21), and 28 (22–35). The y axis indicates the percentage of participants with detectable fecal SARS-CoV-2 RNA. The blue bar corresponds to participants in the placebo control arm, and the orange bar corresponds to participants in the IFN-λ intervention arm. Each bar also marks the 95% CI. Number of participants and percentage of participants that provided a positive stool sample are listed above each stacked bar in black and red fonts, respectively, and summarized in Data S1.
(B) Odds ratio comparing detectable fecal SARS-CoV-2 RNA shedding in the IFN-λ intervention arm with the placebo arm at each time point in the first month of the study. The x axis marks the odds ratio adjusted for age, sex, collection kit type (OG or ZY), and target gene (E, N1, N2, or RdRP) (aOR). The y axis marks the time point in the study: days 3 (range 0–7), 14 (8–21), and 28 (22–35). The point marks the aOR, flanked by lines denoting the 95% CIs. The red dashed vertical line at aOR = 1.0 indicates no association.
We originally started collecting stool samples in the OMNIGene GUT collection tube (OG), which is extensively used in gut microbiome studies.36 Parallel work from our group10 and one other group37 optimized and benchmarked stool collection and processing methods for the detection of fecal SARS-CoV-2 RNA; our group found that the Zymo DNA/RNA shield fecal collection tube (ZY) performs better than OG in viral RNA preservation. Therefore, starting on 14 May 2020, study participants were asked to provide samples in both the OG and ZY kits. Overall, a total of 326 samples were collected in the OG kit, and 347 in the ZY kit (sample collection compliance is summarized in Data S1 and the STAR Methods). In addition to these stool samples, oropharyngeal (OP) swabs were obtained daily during the initial part of the study, and at each study visit on days 120, 210, and 300; blood samples were drawn at days 0, 5, 14, 28, 120, 210, and 300 (Figure 1A). Clinical specimens were paired with self-reported symptom data collected through questionnaires administered on the day of enrollment and then daily from days 1 through 28, and on days 120, 210, and 300. Additionally, symptoms experienced in the 3 weeks preceding study enrollment were surveyed on the day of enrollment. Finally, long-term follow-up questionnaires on days 120, 210, and 300 collected symptoms occurring in the 7 days leading up to the appointment.
Among the participants who returned at least one stool sample, the median age was 36 years (IQR = 29–51 years), 46 (41%) were female, and 72 (65%) were Hispanic (Figure 1B and Table 1 ). We describe the overall cohort, as well as two subsets: those reporting gastrointestinal (GI) symptoms (n = 54, 49%) at the first time point and those reporting no GI symptoms (i.e., exclusively respiratory symptoms or no symptoms at all) at that time point. Participants with GI symptoms at baseline are more likely to also experience a constellation of other symptoms, including myalgias (participants with GI symptoms, 78%; without GI symptoms, 30%; standard difference, −1.09), chills (59%, 21%, −0.84), decreased smell (63%, 30%, −0.7), headache (70%, 42%, −0.59), and joint pain (46%, 19%, −0.6). A comparison of those with and without GI symptoms, in terms of age, sex, ethnicity, and clinical measures at enrollment, including temperature, blood oxygen saturation, white blood cell count, blood alanine aminotransferase (ALT) concentration, and SARS-CoV-2 IgG seropositivity reveal no large differences and are presented in Table 1.
Table 1.
Cohort demographics and associated metadata
| Overall | GI symptoms at enrollment |
Standardized difference | ||
|---|---|---|---|---|
| Yes | No | |||
| n | 111 | 54 | 57 | |
| Age, median (IQR) | 36 (29–51) | 36 (29–49) | 37 (30–53) | 0.05 |
| Female, n (%) | 46 (41%) | 26 (48%) | 20 (35%) | −0.27 |
| BMI (kg/m2), median (IQR) | 27.7 (24.8–31.8) | 28.2 (25.0–32.1) | 27.4 (24.7–30.5) | −0.25 |
| Race/Ethnicity, n (%) | ||||
| Hispanic | 72 (65%) | 38 (70%) | 34 (60%) | −0.22 |
| White | 28 (25%) | 12 (22%) | 16 (28%) | 0.13 |
| Asian | 4 (4%) | 3 (6%) | 1 (2%) | −0.2 |
| Unknown | 6 (5%) | 1 (2%) | 5 (9%) | 0.31 |
| Symptomatology | ||||
| Asymptomatic at enrollment, n (%) | 8 (7%) | 0 (0%) | 8 (14%) | 0.56 |
| Duration of symptoms in days prior to randomization, median (IQR) | 5 (4–7) | 6 (5–8) | 5 (3–7) | −0.61 |
| GI symptoms at enrollment | ||||
| Any GI symptom | 54 (49%) | 54 (100%) | 0 (0%) | |
| Abdominal pain | 13.0 (12%) | 13.0 (24%) | 0 (0%) | −0.8 |
| Diarrhea | 29.0 (26%) | 29.0 (54%) | 0 (0%) | −1.53 |
| Nausea | 31.0 (28%) | 31.0 (57%) | 0 (0%) | −0.8 |
| Vomiting | 5.0 (5%) | 5.0 (9%) | 0 (0%) | −0.45 |
| Other symptoms at enrollment | ||||
| Body aches (myalgias) | 59.0 (53%) | 42.0 (78%) | 17.0 (30%) | −1.09 |
| Chest pain/pressure | 21.0 (19%) | 15.0 (28%) | 6.0 (11%) | −0.45 |
| Chills | 44.0 (40%) | 32.0 (59%) | 12.0 (21%) | −0.84 |
| Cough | 62.0 (56%) | 38.0 (70%) | 24.0 (42%) | −0.59 |
| Decreased smell | 51.0 (46%) | 34.0 (63%) | 17.0 (30%) | −0.7 |
| Fatigue | 68.0 (61%) | 43.0 (80%) | 25.0 (44%) | −0.78 |
| Fever (>99.5°F) | 10 (9%) | 4 (7%) | 6 (11%) | 0.11 |
| Headache | 62.0 (56%) | 38.0 (70%) | 24.0 (42%) | −0.59 |
| Joint pain | 36.0 (32%) | 25.0 (46%) | 11.0 (19%) | −0.6 |
| Shortness of breath | 28.0 (25%) | 17.0 (32%) | 11.0 (19%) | −0.28 |
| Sore throat | 43.0 (39%) | 27.0 (50%) | 16.0 (28%) | −0.46 |
| Rash | 6.0 (5%) | 4.0 (7%) | 2.0 (4%) | −0.17 |
| Runny nose | 24.0 (22%) | 16.0 (30%) | 8.0 (14%) | −0.38 |
| Laboratory values at enrollment, median (IQR) | ||||
| Absolute lymphocyte count (cells/μL) | 1.5 (1.2–2.2) | 1.4 (1.1–1.9) | 1.6 (1.2–2.3) | 0.33 |
| Alanine aminotransferase (IU/L) | 30.0 (22.0–48.5) | 31.5 (22.0–47.8) | 28.0 (22.0–50.0) | 0.07 |
| Aspartate aminotransferase (IU/L) | 30.0 (25.0–39.0) | 32.5 (26.0–41.0) | 29.0 (24.0–34.0) | −0.03 |
| Seropositivity at enrollment, n (%) | 46 (41%) | 22 (41%) | 24 (42%) | 0.03 |
| White blood cell count (cells/μL) | 5.5 (4.2–7.1) | 5.4 (3.8–7.1) | 5.8 (4.7–7.1) | 0.18 |
Longitudinal dynamics of SARS-CoV-2 RNA in stool
A total of 673 stool samples collected from 113 participants over a period of 10 months were processed as per a recently optimized and benchmarked protocol10 outlined in the STAR Methods and summarized in Figure S1. Briefly, RNA was extracted from each of these stool samples and assayed for four target genes in the SARS-CoV-2 genomic RNA (gRNA) encoding the envelope protein (E), nucleocapsid protein (N1 and N2), and RNA-dependent RNA polymerase (RdRP) in technical duplicate, using RT-qPCR. We also assayed 278 of the 673 RNA samples, derived predominantly from samples collected in the first month of the study for the N1 and E gene, using multiplexed droplet digital PCR (ddPCR) assays because ddPCR is more robust to the presence of PCR inhibitors than RT-qPCR.38 We found the measurement of the N1 and E genes using ddPCR to be concordant with one another (Figure S2) and thus assayed the remainder of the samples (n = 395) only for the N1 gene. In total, 5,384 RT-qPCR assays and 951 ddPCR assays measuring the concentration of fecal SARS-CoV-2 gRNA were carried out. This dataset was then analyzed as summarized in the STAR Methods. SARS-CoV-2 viral RNA concentrations estimated by RT-qPCR and ddPCR targeting the N1 gene were found to be concordant (Figure S3; ZY, Pearson’s correlation, R = 0.98, p < 0.0001; OG, Pearson’s correlation, R = 0.9, p < 0.0001). Given the relative concordance between the RT-qPCR and ddPCR results, and the fact that that we had a richer dataset across four target genes in duplicate reactions using RT-qPCR, we decided to carry out the rest of our analyses using the RT-qPCR results alone; where relevant, associated analyses using ddPCR-derived viral RNA concentrations are included in Data S1 and are referenced below. We applied a logistic regression model that averaged RT-qPCR-derived viral RNA concentrations over all four target genes and both sample collection kits with fixed effects to correct for systematic differences. The model uses a generalized estimating equations (GEE) approach and is described in the STAR Methods; it was used in all our primary analyses except where noted.
In study participants with uncomplicated COVID-19, the GEE model that considers RT-qPCR-derived viral RNA concentrations across all four target genes in the gRNA shows that 49% (95% confidence interval = 38%–60%) of participants (n = 102) were positive for fecal SARS-CoV-2 RNA at the first time point around day 3 (Figure 2A). The proportion of participants with fecal shedding of SARS-CoV-2 RNA gradually declined to 40% (95% confidence interval = 28%–53%, n = 86) on day 14 and 11.0% (6%–20%, 83) on day 28. To determine whether fecal SARS-CoV-2 RNA shedding continues after oropharyngeal shedding ceases, we compared the presence of SARS-CoV-2 RNA in fecal samples to that in OP samples from the same participant.35 At 4 months (120 days) post-enrollment, all participants (n = 57) who provided paired fecal and OP samples tested negative for SARS-CoV-2 RNA in their OP samples but 12.7% (95% confidence interval = 8.5%–18.4%) of their fecal samples were positive for SARS-CoV-2 RNA (Figure 2B). OP samples were not tested beyond the 4-month time point. However, at 7 months (210 days) post-enrollment, 3.8% (2.0%–7.3%) of the participants’ fecal samples were positive for SARS-CoV-2 RNA. Among the 23 fecal samples collected at 10 months (300 days), none were positive for SARS-CoV-2 RNA. It should be noted that the presence of viral RNA in the feces at the later time points could be the consequence of prolonged infection and viral RNA shedding or the consequence of a re-infection.
We then calculated the absolute concentrations of fecal SARS-CoV-2 RNA using RT-qPCR of samples collected in the ZY kit (Figure 2C; corresponding data from samples collected in the OG kit are presented in Figure S4). In samples collected around day 3, between 54% and 77% of the participants shed viral RNA in their stool, depending on the gene targeted in the assay. At the first time point, looking at viral RNA concentrations derived from measuring the N1 gene, the gene that yielded the most number of SARS-CoV-2-positive fecal samples at this time point, we find that positive stool samples had between 0.32 and 3.97 log10 copies of viral RNA per microliter of eluate. We found that these viral RNA concentration data were concordant when measured using an orthogonal assay using ddPCR (Figure S5). Finally, to understand the temporal dynamics of shedding, we treated time since enrollment in the study as a continuous variable (Figure 2D), and we observed a decline in fecal viral gRNA concentration over the first month post-enrollment, with a few individuals demonstrating extended shedding versus evidence of a possible re-infection at the 4- and 7-month time points.
Although gRNA is regularly used as an indicator of SARS-CoV-2 infection, this biomolecule does not mark an active infection, because non-infective viral particles can also harbor gRNA. Subgenomic RNA (sgRNA) is a possible indicator of an actively replicating virus, although there is ongoing debate about its specificity. Hence, we quantified sgRNA as previously described;39 23.8% (95% confidence interval = 15.2%–35.3%) of participants had detectable sgRNA (0.8–5.69 log10 copies of viral sgRNA per μL of eluate) in the first time point after diagnosis (Figure S6). This is in comparison with the 49.2% (38.2%–60.3%) of participants who had detectable gRNA in the first time point after diagnosis. Although there were samples that tested positive for gRNA that did not test positive for sgRNA, there were no samples where sgRNA was detected but gRNA was not. Finally, at the fourth time point, SARS-CoV-2 sgRNA had almost totally cleared, with 0.7% (0.2–3.0%) of samples remaining positive for sgRNA.
Impact of interferon lambda on fecal shedding of SARS-CoV-2 RNA
As samples from this study were collected from individuals on a randomized controlled trial of Peg-IFN-λ , we carried out an exploratory analysis to determine whether this intervention affected fecal SARS-CoV-2 RNA clearance in the first month after treatment. We found that there was no significant difference in the percentage of participants who shed SARS-CoV-2 RNA in their feces between the two arms of the study at the first three time points (Figure 3A). We went on to calculate the odds ratio adjusted for age, sex, collection kit type, and target gene (adjusted odds ratio, aOR) that a person who received the IFN-λ intervention would also be shedding viral RNA in stool at the first three time points (Figure 3B). At the first time point, around 3 days after enrollment in the study, we find that receiving the IFN-λ intervention was associated with lower odds of shedding viral RNA in stool (aOR = 0.32, 95% confidence interval = 0.12–0.89). While the association between exposure to IFN-λ and lower odds of fecal viral RNA shedding was intriguing and suggested that exposure to the intervention on day 1 might decrease short-term fecal viral RNA shedding, this association failed to replicate upon execution of several sensitivity analyses (Figure S7 and Data S1). In summary, in the current study, we did not observe a robust effect of a single 180-μg subcutaneous dose of IFN-λ on fecal SARS-CoV-2 RNA shedding.
Subjects with detectable fecal SARS-CoV-2 RNA also manifest GI symptoms
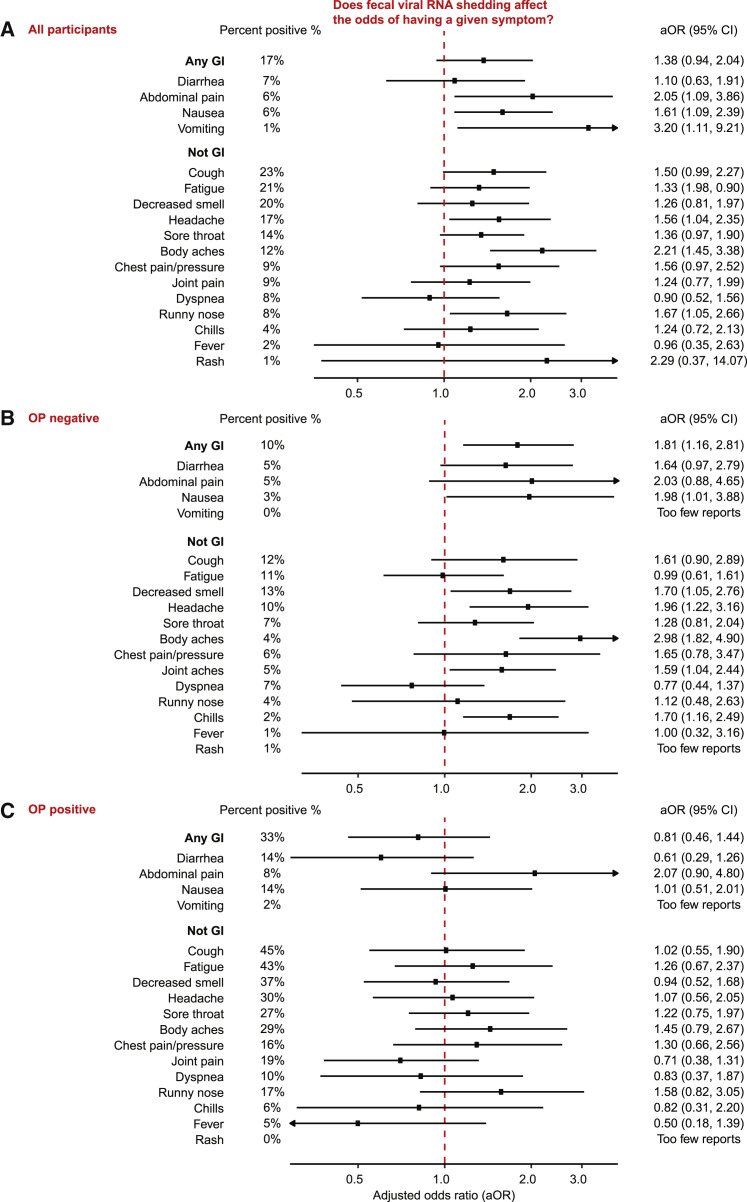
In limited recent studies, the presence of fecal SARS-CoV-2 RNA has been linked to the presence of GI symptoms. However, these studies are mostly cross-sectional in nature, collect symptomatology data retrospectively and do not use a uniform, benchmarked methodology for quantification of SARS-CoV-2 RNA in stool. To address the question of whether fecal viral RNA shedding is associated with GI symptoms, we collected comprehensive longitudinal symptomatology data, including information on GI symptoms from study participants in this interventional trial and compared these with absolute viral RNA concentrations measured in their feces (Figure 4A). Across the first month of the study, we found that participants who shed viral RNA in their stool were more likely to report nausea (aOR = 1.61, 95% confidence interval = 1.09–2.39), vomiting (3.20, 1.11–9.21), and abdominal pain (2.05, 1.09–3.86); no association was observed between viral RNA shedding and diarrhea (1.10, 0.63–1.91) or when considering any GI symptom (1.38, 0.94–2.04). Respiratory and systemic symptoms including runny nose (1.67, 1.05–2.66), headaches (1.56, 1.04–2.35), and body aches (2.21, 1.45–3.38) were also associated with the presence of fecal SARS-CoV-2 RNA. These results taken together, fecal SARS-CoV-2 RNA shedding is positively associated with most GI symptoms and with specific systemic and respiratory symptoms.
Figure 4.
Association between fecal viral RNA shedding and symptoms
We present these results in the overall population, as well as stratified by the presence and absence of ongoing viral RNA shedding from the oropharynx (OP).
(A) Summary of the association between viral RNA shedding and report of a given symptom in all participants. Shedding and symptom data from up to day 28 were included in this analysis. Adjusted odds ratios (aOR) for this association were evaluated using the logistic GEE model described in the statistical methods section (STAR Methods), which averages over collection kits (OG and ZY), target genes (E, N1, N2, and RdRP) and technical replicates and is adjusted for age, sex, collection kit, and target gene. The x axis indicates the aOR for the presence of a given symptom. The y axis lists symptoms divided into those associated with the GI tract and those not associated with the GI tract. The odds ratio for each symptom is indicated by the circle, and associated bars represent the 95% CI. The red dashed vertical line at aOR = 1.0 indicates no association. The percentage of surveys reporting each symptom is provided to the left of these bars. aOR and the 95% CIs are listed to the right of the bars. Analyses where sample size was insufficient are listed as “Too few reports.”
(B and C) Identical data to (A) where (B) lists participants with negative paired OP swabs for SARS-CoV-2 RNA, and (C) lists participants with positive paired OP swabs for SARS-CoV-2 RNA.
To determine whether the observed association between symptoms and fecal shedding was independent of respiratory shedding, we next divided the data into two subsets based on whether or not the participant was shedding virus in the oropharynx at the time the fecal sample was taken; specifically, we looked at participants whose OP swabs were collected within 3 days of the stool sample and (1) did not have any detectable SARS-CoV-2 RNA (n = 69; Figure 4B) or (2) had detectable SARS-CoV-2 RNA (n = 54; Figure 4C). Participants who were shedding viral RNA from the oropharynx had higher rates of almost all COVID-19-related symptoms, and we found no significant association between fecal shedding and symptoms for this subgroup. By contrast, participants who were not shedding viral RNA from the oropharynx had far lower rates of COVID-19-related symptoms in general. However, we found many significant associations between fecal shedding and symptoms in this subgroup. This is consistent with an interpretation that patients with an active infection of the respiratory system can experience an array of COVID-19-related symptoms independent of whether or not they are shedding viral RNA in their feces, but that patients whose respiratory infection has cleared could still be experiencing an active infection of the GI tract, which itself is associated with many different COVID-19-related symptoms. Taken together, these data suggest that fecal shedding of SARS-CoV-2 RNA is a possible indicator of an ongoing GI infection, and that this infection is accompanied by GI and other systemic symptoms.
Discussion
Severe SARS-CoV-2 infections can lead to a life-threatening hypoxemic respiratory failure. Therefore, much of the initial investigation of COVID-19 focused on the respiratory infection and related manifestations of the disease. This may be why, two years into the pandemic, we still do not definitively know whether SARS-CoV-2 infects the GI tract of humans. However, we know that SARS-CoV-2 can infect intestinal cells in vitro, both in cell lines32 and in human tissue-derived intestinal organoids.29, 30, 31 Additionally, the largest autopsy series of patients with COVID-19 to date recently demonstrated consistent evidence of infection of the small intestine by SARS-CoV-2; they also recovered live virus from these intestinal biopsies.4 This evidence suggests that SARS-CoV-2 can infect the GI tract, and perhaps when it does, it induces the GI symptoms observed in individuals with COVID-19. This postulated GI-tropism of SARS-CoV-2 is in keeping with the fact that other Betacoronaviruses that infect mammals can cause GI diseases. For example, BCoV causes severe GI diseases such as calf diarrhea and winter dysentery in cows., 40 What we have lacked in trying to understand whether the GI tract is commonly infected in COVID-19 is longitudinal samples that demonstrate prolonged shedding of fecal viral RNA after respiratory shedding has stopped. We have also lacked data that would enable us to clearly investigate whether or not there is a link between fecal viral RNA shedding and GI symptoms, both during and after respiratory infection by SARS-CoV-2.
To address this gap, we leveraged one of the largest collections of longitudinal fecal samples from patients with mild to moderate COVID-19 to investigate fecal viral RNA shedding and its relationship to both OP viral RNA shedding and COVID-19 symptoms. Among the 113 participants who provided stool samples in this study, 49.2% (95% confidence interval = 38.2–60.3%) shed viral RNA in their feces within 6 days after their COVID-19 diagnosis. The fact that only a subset of individuals with COVID-19 exhibited fecal viral RNA shedding may be a consequence of a broad, nearly 1-week, window for the first sample collection from the time of diagnosis; alternatively, this may also be the result of physiological and genetic differences between individuals. Over the course of the first month in this study, the number of participants shedding fecal viral RNA decreased to 11% (6%−20%), and the viral RNA concentration among those still shedding decreased from up to ∼3 log10 copies per microliter to <1 log10 copies per microliter. At the first time point, we found that a larger proportion of participants shed viral RNA in their OP swab compared with their feces; however, this trend reversed in the rest of the time points. This suggests that clearance of SARS-CoV-2 is more rapid in the respiratory tissue than it is in the GI tissue and that the GI tract may be a site of longer-term infection.
When considered in the context of previously documented evidence of a likely GI infection by SARS-CoV-2, our detection of SARS-CoV-2 sgRNA in fecal samples supports the model of an active infection in the GI tract. The presence of sgRNA, as opposed to gRNA, has been proposed as a marker of active infection and viral replication; however, subsequent work has now established that sgRNA outlives actively replicating virus in cell culture experiments and therefore may be an unreliable indicator of an ongoing, active infection.39 , 41 Therefore, although we detected sgRNA in stool up to 28 days after infection, whether or not this, on its own, is sufficient evidence of an ongoing infection remains unclear.
Beyond informing our understanding of SARS-CoV-2 pathobiology, the information we present on the frequency, amount, and duration of viral RNA shed in stool is valuable for inferring population-level prevalence of COVID-19 from wastewater studies. This may, in turn, help inform public health measures. For example, long-term fecal viral RNA shedders may contribute to prolonged elevated levels of SARS-CoV-2 RNA in wastewater. If transmission occurs largely or entirely through respiratory secretions, the continued presence of fecal viral RNA in wastewater from a prolonged GI infection may be mistakenly interpreted as evidence of the prevalence of infectious individuals in a community. Since wastewater viral RNA levels are being considered for use in guiding community level policies (e.g., shutdowns and re-openings),42, 43, 44, 45, 46 it is critical that we understand how respiratory viral shedding and transmissibility of SARS-CoV-2 RNA are temporally related to fecal viral RNA shedding.
Based on the available evidence, it is highly plausible that the presence of GI symptoms in patients with COVID-19 is due to infection of the GI tissues. With a comprehensive collection of clinical symptom data and fecal viral RNA concentrations, we find that over the course of the first month after enrollment, those who shed viral RNA in stool are more likely to also have GI symptoms including nausea, vomiting, and abdominal pain among other symptoms like runny nose, body aches, and headaches. It is notable that those who shed viral RNA in stool were not more likely to have diarrhea—this finding is contradicted by two prior studies (n = 59, 44), which found that patients with diarrhea were more likely to shed viral RNA in stool and, that too, at higher concentrations.2 Our finding of no association between diarrhea and fecal viral RNA shedding might be due to the relatively small number of participants who reported diarrhea in our study. When focusing on participants who had extended shedding of viral RNA in their stool even after their OP shedding had ceased, we found that fecal shedding of viral RNA is associated with a range of systemic and GI symptoms. On the other hand, for the duration that participants provided an OP swab positive for viral RNA, i.e., had an active respiratory infection, we did not find any association between fecal viral RNA shedding and symptomatology. We postulate that this is because participants who have an ongoing respiratory infection manifest classic COVID-19-related symptoms whether or not they have an infection in their GI tract. These observations support the hypothesis that there is likely a prolonged SARS-CoV-2 infection of the GI tract even after the upper respiratory infection is cleared. Since the GI tract is a highly immunoactive tissue,47 prolonged infections of the GI tissue may have consequences for patient health and may also be associated with the hitherto mysterious phenomenon of PASC or Long COVID. In fact, many studies following patients who have recovered from COVID-19 identify the prolonged presence of GI sequelae.48, 49, 50, 51, 52, 53, 54
In conclusion, we sought to address a key gap in our knowledge about the pathophysiology of a possible GI infection by SARS-CoV-2 by sampling stool over an extended period of time (10 months) and gathering paired symptomatology data. We have demonstrated the longest recorded shedding of fecal SARS-CoV-2 RNA in any COVID-19 patient: ∼210 days post-infection in two participants. Furthermore, we have found that extended shedding of SARS-CoV-2 RNA in participants who no longer have detectable viral RNA in OP swabs is closely associated with a host of systemic and GI symptoms, providing further evidence of a SARS-CoV-2 infection of the gut. Data presented here, when placed in the context of preliminary work that has suggested that the extended presence of SARS-CoV-2 viral antigen in gut biopsies from participants with COVID-19 may be associated with an improved immune response,21 urges follow-up immunological studies that investigate stool samples. Finally, initiatives such as Researching COVID to Enhance Recovery (RECOVER, NIH) that are poised to elucidate the hitherto elusive phenomenon of PASC should look closely at stool samples as an important factor of SARS-CoV-2 infection with potential long-term impact.
Limitations of the study
Despite its large size and longitudinal nature, this study has limitations. First, the study is limited in its resolution, having collected only six samples over a 10-month period. Follow-up studies with more frequent sampling, especially in the first 2 months after diagnosis, may help build a more nuanced model of decline of fecal viral RNA concentration over time. This will also allow a closer evaluation of the relative cessation of viral RNA in stool vis-a-vis other respiratory samples such as the OP swab. We were also unable to collect stool samples in a way that would enable recovery of live virus. As this was an outpatient study during the early part of the pandemic, we required participants to collect stool themselves at home and then mail the stool kits to us. For safety and practical purposes, we thus had to provide participants with kits that were rated for virus inactivation. Future studies, which facilitate the careful, consistent collection of stool samples from individuals with COVID-19 in a safe setting, might enhance the likelihood of more accurate measurement of live virus. This would be more direct evidence of SARS-CoV-2 being viable in the gut. Third, we did not obtain direct tissue evidence of infection; to do so would require intestinal biopsies. Of note, recent autopsy-4 and prior biopsy-based21 reports in limited numbers of patients have demonstrated evidence of direct intestinal infection and cytopathic changes. While intestinal biopsies from patients with mild to moderate COVID-19 would be highly informative, to date, these samples have been understandably difficult to obtain. In upcoming large studies, such as the RECOVER study, a subset of patients will be getting such biopsies, and the results of these large-scale studies will be illuminating.
Finally, it would be interesting to sequence fecal viral RNA from participants with extended shedding to evaluate the persistence of the original virus variant, evolution of the original variant, and/or potential re-infection by the same or a different SARS-CoV-2 variant. Unfortunately, one of the limitations of current technologies for sequencing variants from complex matrices such as stool is the requirement of an adequate concentration of virus to be able to either amplify or assemble the virus from direct or enriched sequencing. As future technologies are developed for sensitive determination of variant sequences from stool, this type of analysis should be feasible. Of note, this study was carried out prior to the emergence of the strains (Omicron, Delta) that are prevalent today. Different strains may have different relative tropisms to the respiratory versus GI tract and may exhibit differences in clearance rates. This may be the consequence of their inherent biology as well as the immune status of the host due to underlying disorders, prior COVID-19 disease, and natural immunization, or vaccination.
Of note, in this study we used samples that were collected as part of a previously published clinical trial.35 The original study reports the enrollment criteria applied to recruit participants. Briefly, the study actively sought to have equal male and female, racially and socioeconomically diverse participants between the ages of 18 and 75 years. The study did not collect information about self-reported gender in recruitment. Participants at risk of current or imminent hospitalization, with a respiratory rate >20 breaths per minute, room air oxygen saturation <94%, history of decompensated liver disease, recent use of interferons, antibiotics, anticoagulants, or other investigational and/or immunomodulatory agents for treatment of COVID-19, and prespecified laboratory abnormalities were excluded. Additionally, pregnant or breastfeeding participants were also excluded.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Stool from participants in Peginterferon Lambda-1a (IFN-λ) clinical trial (NCT04331899) | Stanford University | N/A |
| Oropharyngeal swabs from participants in Peginterferon Lambda-1a (IFN-λ) clinical trial (NCT04331899) | Stanford University35 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Phosphate buffered saline (PBS) | Fisher Scientific | BP399-500 |
| 0.8 mM Ethylenediaminetetraacetic Acid (EDTA) | Fisher Scientific | EC200-449-9 |
| Nuclease-free water | Ambion | AM9937 |
| Tris-HCl pH 8.0 | Invitrogen | 15567-027 |
| Critical commercial assays | ||
| QiaAMP Viral RNA Mini kit | Qiagen | 52906 |
| Custom ddPCR Assay Primer/Probe Mix | BioRad | 10031277 |
| One-Step RT-ddPCR Advanced Kit for Probes | BioRad | 1864021 |
| TaqPath 1-Step RT-qPCR Master Mix, CG | ThermoFisher | A15299 |
| Deposited data | ||
| A digital repository of all data supporting the findings of this study can be found at Zenodo | This study | https://zenodo.org/record/6374138 |
| Oligonucleotides | ||
| Primers for RT-qPCR and ddPCR used in this study, see Data S1 | This study | N/A |
| Probes for RT-qPCR and ddPCR used in this study, see Data S1 | This Study | N/A |
| Recombinant DNA | ||
| Synthetic SARS-CoV-2 RNA | ATCC | VR-3276SD |
| Zoetis Calf-Guard Bovine Rotavirus-Coronavirus Vaccine | Zoetis | VLN 190/PCN 1931.20 |
| Software and algorithms | ||
| Design and Analysis software | Thermo Fisher Scientific | Version 2.5.1 |
| REDCap Cloud | https://projectredcap.org/ | Version 1.5 |
| Python | https://www.python.org/ | Version 3.8.5 |
| Statsmodel package | https://www.statsmodels.org/stable/index.html | Version 0.12.0 |
| RStudio | https://www.rstudio.com/ | Version 1.3.959 |
| Other | ||
| Biomek-FX liquid handler | Biomek | N/A |
| 12k Flex Applied Biosystems qPCR machine | Applied Biosystems | N/A |
| QX200 AutoDG Droplet Digital PCR System | BioRad | N/A |
| BioRad C1000 thermocycler | BioRad | N/A |
| ddPCR reader | BioRad | QX200 |
| OMNIGene GUT collection tube | DNA Genotek | OM-200 |
| Toilet accessory | DNA Genotek | OM-AC1 |
| DNA/RNA shield fecal collection tube | Zymo | R1101-E |
| 96-well plates | BioRad | HSP9601 |
| Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines, see Data S1 | Bustin et al. (2009)55 | Quantitative Real-Time PCR Experiments (MIQE) guidelines |
| Digital Minimum Information for Publication of Quantitative Real-Time PCR Experiments (dMIQE) guidelines, see Data S1 | dMIQE Group & Huggett (2020)56 | Digital MIQE guidelines |
| Droplet Digital PCR Applications Guide on QX200 machines | BioRad | Droplet Digital PCR Applications Guide |
| MicroAmp Optical 384-well plates | FisherScientific | 43-098-49 |
| Optically clear seal | Applied biosystems | 4311971 |
Resource availability
Lead contact
Correspondence and requests for materials should be addressed to the lead contact, Ami S. Bhatt (269 Campus Dr, CCSR 1155b, Stanford University, Palo Alto, CA 94305. Tel: (650) 498-4438; e-mail: asbhatt@stanford.edu).
Materials availability
PCR primers sequences are reported in Data S1. Other resources are available upon request of the lead contact.
Experimental model and subject details
Study design and population
A total of 120 adults aged 18 - 71 years who had received a positive SARS-CoV-2 reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR) based respiratory swab test within the past 72 hours were recruited for enrollment in a single-blind, placebo controlled, phase 2 clinical trial of Peginterferon Lambda-1a (IFN-λ) as an intervention for uncomplicated coronavirus disease 2019 (COVID-19). Informed consent was obtained for all participants under Stanford University Institutional Review Board (IRB) approved protocol # 55619 (PIs: Upinder Singh, Prasanna Jagannathan).
The primary results of the null study, secondary outcomes, and the full details of study recruitment, inclusion and exclusion criteria were previously reported on and are only briefly summarized here.35 Individuals with study defined lab abnormalities, respiratory rate >20 breaths per minute, room air oxygen saturation levels <94%, pregnancy or breastfeeding, or recent history of hospitalization, uncontrolled liver disease, or use of COVID-19 interventional therapeutics, anticoagulants, antibiotics, and/or antivirals were excluded from the study. Subjects were randomized 1:1 to either the interventional or control study arm to receive a one-time subcutaneous injection of Peginterferon Lambda-1a or saline, respectively, on the first day of enrollment. Randomization was stratified by age (≥50 and <50 years old) and sex. The demographics of study participants are summarized in Table 1. Participant information on sex, age, race and ethnicity was self-reported and was reported in the original clinical manuscript describing this study.35 Information on gender and socioeconomic status was not collected.
In addition, healthy adults were recruited to provide stool samples for use as extraction controls under Stanford IRB protocol #42043 (PI: Ami Bhatt). All donors gave informed consent prior to donating stool samples. Information on sex, gender, age, socioeconomic status, race and ethnicity was not collected.
Trial registration
ClinicalTrials.gov Identifier: NCT04331899
Method details
Study samples and data
Stool and other data and samples were collected from each set of study participants as outlined below.
For the first 28 days following enrollment, participants in the clinical trial completed daily symptom questionnaires administered via REDCap Cloud (version 1.5)36 and self-performed daily measurements of temperature and oxygen saturation using study provided at-home devices. Participants returned to the study site on 1, 3, 5, 7, 10, 14, 21, 28 days (all +/− 1 day) and 120, 210, and 300 days (all +/− 3 weeks) post-enrollment for follow-up visits during which oropharyngeal (OP) swabs were collected, symptoms were queried, and vital signs were recorded. All clinical trial participants were provided a fecal sample collection kit on 0, 5, 21, 28, 120, and 210 days after enrollment and were asked to collect a stool sample in the provided kit, store at room temperature, and drop off for processing at their subsequent study visit or mail back to the study site at the long term follow up time points. We define the following six time points based on when participants returned the stool samples: days 3 (range 0 - 7 days), 14 (8 - 21), 28 (22 - 35), 120 (75 - 165), 210 (166 - 255) and 300 (>255) (Figure 1A).
At the start of study enrollment on 25 April 2020, the collection kit consisted of the OMNIGene GUT collection tube (OG), toilet accessory, gloves and Spanish and English translations of manufacturer instructions. Later, starting 14 May 2020, the Zymo DNA/RNA shield fecal collection tube (ZY) was included in the fecal sample collection kit in addition to the OG collection tube. Spanish and English translations of manufacturer instructions specific to the ZY collection tube were also added. Subsequently, all participants were asked to collect a portion of the same stool sample in both of the two kits for each time point.
The OG and ZY collection tubes are both marketed to preserve stool samples at ambient temperatures for up to 30 days. This eliminated the burden of sample refrigeration requirements for study participants. Fecal samples were processed within 24 hours of receipt by the lab. Samples collected in the OG and ZY collection tubes were processed similarly, by first vortexing the collection tube for 30 seconds to thoroughly homogenize the sample. Each sample was then aliquoted into 1.8 mL cryovials, labeled with the patient study ID and study time point, and then frozen at −80°C.
Healthy control stool samples for use in every batch of RNA extractions were obtained from a healthy individual without prior history of COVID-19 exposure or positive SARS-CoV-2 respiratory test. Healthy stool samples for the limit of blank (LoB) determination were collected in 2018 well prior to the onset of the pandemic. All healthy donors self-collected fecal samples fresh and stored them at 4°C until processing. Within 24 hours of sample collection, samples were aliquoted into cryovials without preservative and frozen immediately at −80°C.
Extraction of RNA
Stool samples were randomly assigned a sample ID and processed for RNA extraction in batches of 18 following a previously optimized method,10 which is summarized here and in Figure S1.
Two positive controls (OG and ZY) were included in each extraction batch for a total of 20 extractions per batch. Positive controls were prepared by adding biopsy punches of stool collected from a healthy individual to OG (4 biopsy punches) and ZY (8 biopsy punches) tubes. Each tube was then spiked with 10 μL of synthetic SARS-CoV-2 RNA at 104 copies/μL, vortexed for 30 seconds for homogenization, transferred in 500 μL aliquots to eppendorf tubes and frozen −80°C.
Samples were gradually thawed on ice and vortexed for five seconds to ensure thorough homogenization. 500 μL of the stool-buffer slurry was transferred to an eppendorf tube, spun at 10,000 x g for 2 minutes at room temperature, and 140 μL of the supernatant was transferred to a fresh eppendorf tube for RNA extraction using the QiaAMP Viral RNA Mini kit. RNA extraction was performed as per manufacturer’s protocol and eluted in 100 μL of the elution buffer EB from the kit. Extracted RNA was then transferred to 96 well plates, briefly spun down, sealed and stored at −80 °C until further analysis.
Samples collected at the 4, 7 and 10 month timepoints and associated batch controls were additionally spiked with 10 μL of attenuated BCoV vaccine as recommended.10 BCoV was prepared by resuspending one vial of lyophilized Zoetis Calf-Guard Bovine Rotavirus-Coronavirus Vaccine in 3 mL of phosphate buffered saline as per the manufacturer’s instructions.
Quantification and statistical analysis
RT-qPCR quantification of RNA
An RT-qPCR assay to detect and quantify SARS-CoV-2 genomic RNA (gRNA)57, 58 was developed using primer probe sets recommended by the United States Centers for Disease Control and prevention (CDC)59 targeting the Envelope protein (E), Nucleocapsid proteins (N1, N2), and RNA-dependent RNA polymerase protein (RdRP) of the viral genome. To quantify SARS-CoV-2 subgenomic RNA (sgRNA) from stool samples as previously described39 an additional primer probe set targeting the N1 gene with the forward primer annealing to the canonical leader sequence at the 5′ end was included in the assay. All RNA extracts were assayed for all four gRNA targets and the single sgRNA target. Primer and probe sequences are listed in Data S1.
Each 20 μL RT-qPCR reaction was composed of 5 μL TaqPath 1-Step RT-qPCR Master Mix, CG, 1.5 μL of primer/probe mix, 8.5 μL of nuclease-free water. The primer/probe mix was prepared with a final concentration of 400 nM of each of the forward and reverse primers and 200 nM of the corresponding probe in 8.5 mM Tris-HCl pH 8.0 and 0.8 mM EDTA. Reactions were prepared in Micro-Amp Optical 384-well plates with 5 μL of stool RNA samples, synthetic RNA standards, or nuclease free water using a Biomek-FX liquid handler. Every assay plate also included standard curves. Standard curves were prepared by serially diluting quantitative synthetic SARS-CoV-2 RNA from 105-10−1 copies per μL. For standard curves in the sgRNA assays, a purified PCR product corresponding to the target gene39 was diluted from 106-10−1 copies per reaction. Nuclease-free water was used as a negative control.
RNA extracted from each stool sample was assayed in two technical replicates for each target. Standard curves were run in technical duplicates for all targets on every RT-qPCR assay plate. Eight negative controls were included in each assay plate. Prior to the assay, plates were sealed with an optically clear seal and spun down at room temperature. The samples were assayed in a 12k Flex Applied Biosystems qPCR machine in standard mode using the following cycling conditions: 25°C for 2 minutes, 50°C for 15 minutes, and 95°C for 2 minutes, followed by 45 cycles of 95°C, 3 seconds, and 55°C, 30 seconds.
In the RT-qPCR assays, quantification cycle (Cq) value was calculated using the Design and Analysis software. On a plate-by-plate basis, assays with a Cq value greater than the Cq value of the synthetic RNA standard at 1 copy per μL were called undetermined. Cq values for each sample were converted to viral RNA concentration in copies/μL using the linear regression model fit to the standard curve for each plate. We used a statistical model to average over the results of all the technical replicates, and more details about the model are available in the Statistical analysis section.
Finally, we calculated the LoB of the assay (additional details are available in the in the following sections of the STAR Methods) and converted all viral RNA concentrations equal to or lower than the LoB to be undetermined, because these were beyond the reliable specificity of the assay. All viral RNA concentrations were expressed on a logarithmic scale by applying the transformation log10(viral RNA concentration+1).
SARS-CoV-2 viral RNA concentrations from oropharyngeal swabs were derived from a previously published companion study.35 This study measured the E gene in the SARS-CoV-2 genomic RNA and RNaseP in the human genome in a multiplexed assay. RNaseP was used as an internal control for the extraction of RNA and to monitor the effect of RT-qPCR inhibitors in these samples. Only samples where RNaseP was detected were evaluated. As a requirement for the Stanford FDA Emergency Use Authorization for the SARS-CoV-2 RNA diagnostic test, the sensitivity of the assay for nasopharyngeal swab testing was determined to be 1000 copies/mL. While the FDA did not require the assessment of assay sensitivity for different respiratory tissues, we believe that the assay sensitivity for nasopharyngeal vs. oropharyngeal swabs to be comparable. Similarly, based on previously reported benchmarking and Limit of Detection (LoD) assays, the sensitivity of fecal sample testing for SARS-CoV-2 RNA is 1000 copies/mL.10 Moreover, the assay sensitivity of fecal testing was highly concordant between the tested genes, particularly for the N1, N2, and E genes; RdRP has a slightly lower sensitivity by comparison.10 Therefore, we are confident that the sensitivity of SARS-CoV-2 RNA testing is highly comparable in stool and respiratory biospecimen of the study subjects (1000 copies/mL).
ddPCR quantification of RNA
Droplet digital PCR (ddPCR) is resilient to PCR inhibitors prevalent in stool, enables absolute quantification without the need for an exhaustive standard curve, and is also more sensitive than traditional qPCR.10 , 37 Therefore, we quantified viral RNA using this orthogonal method as previously described.10 The ddPCR reactions were prepared with the One-Step RT-ddPCR Advanced Kit for Probes. Using a Biomek FX liquid handler, each reaction well was loaded with 5.5 μL of extracted RNA to 5.5 μL Supermix, 2.2 μL reverse transcriptase, 1.1 μL of 300 nM dithiothreitol (DTT), 1.1 μL of 20× Custom ddPCR Assay Primer/Probe Mix and 6.6 μL of nuclease-free water per the manufacturer instructions. For multiplexed reactions, we added 1.1 μL of each of the primer/probe mixes and reduced the amount of nuclease free water to 5.5 μL.
We then used a QX200 AutoDG Droplet Digital PCR System to partition reaction samples into droplets of 1 nL using default settings. PCR amplification of the templates was performed on a BioRad T100 thermocycler using the following thermocycling program: 50 °C for 60 min, 95 °C for 10 min, 40 cycles of 94 °C for 30 s and 55 °C for 1 min, followed by 1 cycle of 98 °C for 10 min and 4 °C for 30 min with ramp speed of 1.6 °C/s at each step. Finally, amplified reactions were quantified using a ddPCR reader.
The ddPCR analysis was guided by the Droplet Digital PCR Applications Guide on QX200 machines (BioRad)60 and the digital MIQE guidelines.56 We have included the recommended associated checklist in Data S1. We applied a rigorous strategy to threshold the assays and identify true positive reactions as previously described10 and summarized below. Briefly, we analyzed the standards and negative controls in a plate-by-plate fashion and applied a suitable threshold to these samples. This threshold was applied such that the number of positive droplets in the negative control was minimal and the concentration of RNA in the standard matched the theoretical expectation most closely. We then calculated the difference in amplitude between the negative droplets and the threshold in the reactions with the negative control, and applied a threshold to all the other wells such that this same difference in amplitude was maintained. Finally, as with the RT-qPCR reactions, we established an LoB for this assay (additional details are available in the following sections of the STAR Methods), and any sample with viral RNA concentration less than or equal to the LoB was considered to be undetermined. All viral RNA concentrations were expressed on a logarithmic scale by applying the transformation log10(viral RNA concentration+1).
Ensuring high specificity in RT-qPCR and ddPCR assays of fecal SARS-CoV-2 RNA
In assays to quantify viral RNA, we took a conservative approach at every step to ensure high specificity. First, we adopted a method to determine the limit of blank (LoB) that is based on guidelines set out by the Clinical and Laboratory Standards Institute (CLSI),61 as summarized in the next section. We systematically identified the LoB for stool collected in the OG and ZY kits against each of the four target genes in independent combinations. All samples with an RNA concentration equal to or lower than the corresponding LoB are considered to have an undetermined amount of viral RNA, since this is below a reliable specificity threshold for that assay (example in Data S1). Second, we identified the linear detection range of our assays. A six-point 10-fold dilution series of synthetic SARS-CoV-2 RNA from the American Type Culture Collection (ATCC) starting at 104 log10 copies per μL was used here as previously described.10 Resulting standard curves generated for each of the genes in the genomic RNA measured using RT-qPCR and those measured by ddPCR are shown in Data S1. In assays that detected sgRNA, we used a six-point 10-fold dilution series with pre-quantified sgRNA starting at 106 log10 copies per μL from a previously reported study39 and provide standard curves in Data S1. All samples that yield a viral RNA concentration below the lowest detectable concentration in the linear range of standards are considered to have an undetermined amount of viral RNA. Third, anticipating that few if any stool samples collected beyond the 28 day time point were going to be positive for SARS-CoV-2 RNA, we incorporated a control to guard against false negatives that could result from incomplete or inefficient extraction of RNA, as previously described.10 Briefly, all long-term stool samples were spiked with 10 μL of attenuated Bovine coronavirus (BCoV) prior to RNA extraction. The extracted RNA was then tested for the M gene from BCoV in addition to the regular SARS-CoV-2 based assays. This served to determine if RNA extractions were successful, ensuring we did not falsely report negative SARS-CoV-2 assays as a consequence of ineffective RNA extraction. Out of 239 samples, 237 yielded BCoV RNA, and those that did not were left out of further analysis. Together, these experimental checkpoints increase confident that our reported fecal viral RNA concentrations are accurate.
Estimating limits of blanks
Understanding the specificity of the assays used in this study to quantify viral RNA is critical to evaluate confidence in results derived thereof. Therefore, we used a strategy based on guidelines set out by the Clinical and Laboratory Standards Institute (CLSI)61 to quantify the limit of blank (LoB) of our stool preservation and detection protocol.
To this end, we used stool samples collected from four healthy donors in the Fall of 2018. Since this was from before the emergence of SARS-CoV-2, these samples are confidently negative for SARS-CoV-2 RNA. One stool sample from each of the four donors was aliquoted into separate OG and ZY tubes as per manufacturer instructions. This was performed in independent duplicates by two different operators yielding 16 stool samples. Next, RNA was extracted from each of these samples in duplicate by the two operators resulting in 64 total RNA extracts. The sample preparation protocol is summarized in Data S1.
The 64 RNA extracts were assayed for the E, N1, N2 and RdRP genes in the gRNA in duplicate reactions identical to how clinical samples were assayed in this study. Next, these samples were also assayed for the N1 genes in ddPCR assays. Taken together, we calculated the LoB for relevant combinations of stool preservation (OG, ZY), target gene (E, N1, N2 and RdRP), and detection method (RT-qPCR, ddPCR).
It was notable that across all targeted genes in both RT-qPCR and ddPCR assays, the LoB measured in the OG kit was higher than that measured in the ZY kit. Specifically, RT-qPCR assays targeting the N1 gene yielded 0.487 log10 copies per μL of viral RNA in samples preserved in OG and 0.237 log10 copies per μL of RNA in those preserved in ZY. These corresponded to 0.429 copies per μL and 0.164 copies per μL of RNA in ddPCR assays targeting the N1 gene. Finally, while targeting the N2 gene via RT-qPCR also yielded low RNA concentrations in these negative controls, E and RdRP were highly specific and yielded no detectable RNA for these targets in the negative controls (Data S1). The RNA concentration derived here is used as the LoB in all further data analysis. Thus, all samples that bear an RNA concentration equal to or lower than the corresponding LoB are considered to have an undetermined amount of viral RNA, since this is below a reliable specificity threshold for that assay (example in Data S1).
Guarding against PCR inhibitors for the reliable detection of viral RNA
PCR inhibitors are often present in stool. Thus, we wanted to estimate the degree to which our RT-qPCR assays were impacted by PCR inhibition. We posited that diluting the stool RNA extracts prior to assaying for SARS-CoV-2 RNA would dilute any potential PCR inhibitors derived from the stool matrix. Thus, we would expect a higher positivity rate from assaying the diluted extracts. To this end, we assayed 72 clinical samples by RT-qPCR at the concentration they were extracted at (1X), and at a ten-fold dilution of the same samples (0.1X). In aggregate across the 4 RT-qPCR target genes, assaying the samples at 0.1X resulted in a gain of 4 positive samples but a loss of 15 positive samples, likely due to viral RNA concentration falling below the detection limit of the RT-qPCR assay with dilution (Data S1). Thus, the RT-qPCR analysis of the stool RNA extracts likely does not exhibit a high degree of PCR inhibition.
Statistical analysis
Absolute standardized differences (ASD),62 expressed in units of standard deviations, are displayed in Table 1 to compare the distribution of characteristics in participants reporting GI symptoms at enrollment or not. We interpreted ASDs using Cohen’s guidelines (d: 0.2 = small difference; 0.5 = medium difference; 0.8 = large difference; d < 0.2 = trivial difference).63
Our primary statistical analyses examined associations between participant characteristics and whether the RT-qPCR based detection of SARS-CoV-2 gRNA was positive, focusing only on the stool samples collected during the main study at the first three time points, and including fixed effects to account for the different positivity rates of the four target genes (E, N1, N2 and RdRP) and the two collection kits (OG and ZY). We augmented this with two sensitivity analyses. First, we conducted a subgroup analysis that included samples from all six time-points but that focused on the subset of participants who returned at least one sample during the long-term follow-up; we made decision to focus our primary analysis on the first three time points and to supplement it with this sensitivity analysis to avoid the concern that the decision to join the extended study might correlate with certain patient risk. Second, we conducted subset analyses that focused on individual genes separately. In all cases, we used logistic regression models fit with generalized estimating equations (GEE)64 to account for the correlation between samples and replicates within a participant.
To examine whether Peginterferon Lambda-1a (IFN-λ) had an effect on fecal viral RNA shedding, we fit a logistic regression to estimate the odds ratio of fecal shedding in participants receiving the IFN-λ intervention versus those that received a saline placebo. We adjusted the odds ratio by collection kit type (OG and ZY) and gene (E, N1, N2 and RdRP), to account for systematic differences between measurements, and as well as by the patient’s age and sex, because randomization had been stratified by those features.65 We included statistical interaction terms between study arms and indicators for time of collection in the model to estimate the difference between study arms at each time of collection. In addition to the two sensitivity analyses described above, we also used a negative binomial model to assess the association between the IFN-λ intervention and the total viral RNA concentration, whereas before we used GEE to account for correlation within individual patients.
In analyses to estimate association between fecal SARS-CoV-2 RNA and symptoms, we regressed the presence of symptoms reported at the time of sample collection on an indicator of the presence of fecal SARS-CoV-2 RNA, adjusted for age, sex, log of the number of days since symptom onset, collection kit type (OG and ZY), and gene (E, N1, N2 and RdRP). We fit a separate logistic regression for each of the symptoms. We additionally fit models including an interaction between fecal SARS-CoV-2 RNA shedding and an indicator of OP shedding to estimate associations among participants with or without an ongoing presence of viral RNA in their OP swabs.
All tests were two-sided and conducted at the 0.05 level of significance. Analyses were performed in Python version 3.8.5, using the Statsmodel package, version 0.12.0.
IFN-λ does not impact fecal SARS-CoV-2 RNA shedding
Exposure to IFN-λ appears to present lower odds of fecal viral RNA shedding at the first time point, around 3 days after receiving the intervention (Figure 3B). However, this association failed to replicate upon closer examination using several sensitivity analyses, as follows.
-
1)
We calculated the adjusted odds ratio (aOR) that a person who received the IFN-λ intervention would also be shedding viral RNA in stool at the first three time points, limiting our attention to the subset of individuals who elected to participate in the extended study. Amongst these participants there was no association between the intervention and fecal shedding during any of the six time points (Figures S7A and S7B).
-
2)
We looked at an analysis that was restricted to just individual genes and kits. In this analysis, we find that the association at the first time point is being driven entirely by samples collected in the OG kit, which has previously been shown to have lower sensitivity for fecal SARS-CoV-2 RNA detection10 (Data S1).
-
3)
An analysis that looked at viral RNA concentrations instead of binary test results (positive vs. negative) found no association at any of the three time points (Figure S7C).
Acknowledgments
We thank Alexandria Boehm, Marlene Wolfe, and Nasa Sinnott-Armstrong for guidance on processing stool samples and detection of RNA; Angela Rogers for providing stool samples from participants admitted at Stanford Hospital; Rebecca Osbourne, Tiffany Nguyen, and the members of the Stanford Clinical and Translational Research Unit for assistance with stool sample receipt and processing; Elizabeth Ponder for coordinating initial stool sample collection kit distribution to study participants and providing information about funding from Chem-H; Catherine Blish and members of the Blish Lab for receiving and temporary storage of stool samples prior to biobanking; Dean Felsher for access to the QuantStudio 12K Flex qPCR machine; Yvonne Maldonado and Jonathan Altamirano for helping acquire funding to support this work; Said Attiya and Dhananjay Wagh for guidance on applying ddPCR assays; David Solow-Cordero for assistance setting up the Biomek FX and providing access; Luisa Jiminez and Sopheak Sim for assistance in using the Stanford Functional Genomics Facility and High-Throughput Bioscience Center; and Frida Salcedo for help acquiring reagents from Bio-Rad. We are grateful to the Peg-interferon-λ1a clinical trial team for coordinating procurement of stool samples from outpatients enrolled in this trial. Biorender has been a valuable resource for creating schematic illustrations. This work was supported by a ChemH-IMA grant (to A.S.B. and P.J.), the Stanford Dean’s Postdoctoral Fellowship (to A.N.), an AACR Fellowship (to S.Z.), and a NSF Graduate Research Fellowship Program grant (to A.H. and D.T.S.). The laboratory of A.S.B. is supported by NIH R01 AI148623 and R01 AI143757, and H.H. and the research reported in this publication are supported by the National Center for Advancing Translational Sciences of NIH award UL1TR003142. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
A.N., S.Z., E.F.B., and S.E.V. contributed equally to this work. A.N. designed experiments, extracted RNA from stool samples, assayed viral RNA using RT-qPCR and ddPCR, analyzed data, and wrote the manuscript. S.Z. designed experiments, assayed viral RNA using RT-qPCR and ddPCR, analyzed data, generated plots in R, and wrote the manuscript. E.F.B. and S.E.V. designed experiments, biobanked stool samples, extracted RNA from stool samples, and wrote the manuscript. A.D. and H.H. designed experiments, performed statistical analyses, generated plots in Python, and wrote the manuscript. R.M.P. analyzed data. A.H., D.T.S., and R.V. helped design experiments. K.B.J., J.P., H.F.B., U.S., B.A.P., J.A., and P.J. helped collect samples through the Lambda clinical trial and guided data analysis. A.S.B. helped design experiments, analyze data, and write the manuscript. S.Z., A.D., and H.H. performed and replicated the statistical analysis. A.N., E.F.B., S.E.V., and A.S.B. oversaw the statistical analysis. A.N., S.Z., E.F.B., S.E.V., A.D., H.H., and A.S.B. have unrestricted access to all data. A.N., S.Z., E.F.B., S.E.V., A.D., H.H., and A.S.B. prepared the first draft and reviewed and edited the manuscript. All authors read and approved the final manuscript and take responsibility for its content.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity statement
We worked to ensure gender balance in the study arms, recruited participants from diverse ethnic and socioeconomic backgrounds, and provided the study questionnaire and stool collection protocol in Spanish and English.
Published: April 13, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.medj.2022.04.001.
Supplemental information
Data and code availability
-
•
All data supporting the findings of this study have been deposited at Zenodo (https://zenodo.org/record/6374138) and are publicly available as of the date of publication.
-
•
All custom code and mathematical models have been deposited at Zenodo (https://zenodo.org/record/6374138) and are publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Merola E., Armelao F., de Pretis G. Prevalence of gastrointestinal symptoms in coronavirus disease 2019: a meta-analysis. Acta Gastroenterol. Belg. 2020;83:603–615. [PubMed] [Google Scholar]
- 2.Cheung K.S., Hung I.F.N., Chan P.P.Y., Lung K.C., Tso E., Liu R., Ng Y.Y., Chu M.Y., Chung T.W.H., Tam A.R., et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159:81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parasa S., Desai M., Thoguluva Chandrasekar V., Patel H.K., Kennedy K.F., Roesch T., Spadaccini M., Colombo M., Gabbiadini R., Artifon E.L.A., et al. Prevalence of gastrointestinal symptoms and fecal viral shedding in patients with coronavirus disease 2019: a systematic review and meta-analysis. JAMA Netw. Open. 2020;3:e2011335. doi: 10.1001/jamanetworkopen.2020.11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chertow D., Stein S., Ramelli S., Grazioli A., Chung J.-Y., Singh M., et al. SARS-CoV-2 infection and persistence throughout the human body and brain. Res. Square. 2021 doi: 10.21203/rs.3.rs-1139035/v1. [DOI] [Google Scholar]
- 5.Mao R., Qiu Y., He J.-S., Tan J.-Y., Li X.-H., Liang J., Shen J., Zhu L.-R., Chen Y., Iacucci M., et al. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: a systematic review and meta-analysis. The Lancet. Gastroenterol. Hepatol. 2020;5:667–678. doi: 10.1016/S2468-1253(20)30126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sultan S., Altayar O., Siddique S.M., Davitkov P., Feuerstein J.D., Lim J.K., Falck-Ytter Y., El-Serag H.B., AGA Institute AGA Institute rapid review of the gastrointestinal and liver manifestations of COVID-19, meta-analysis of International data, and recommendations for the consultative management of patients with COVID-19. Gastroenterology. 2020;159:320–334.e27. doi: 10.1053/j.gastro.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu C.L.H., Raval M., Schnall J.A., Kwong J.C., Holmes N.E. Duration of respiratory and gastrointestinal viral shedding in children with SARS-CoV-2: a systematic review and synthesis of data. Pediatr. Infect. Dis. J. 2020;39:e249–e256. doi: 10.1097/INF.0000000000002814. [DOI] [PubMed] [Google Scholar]
- 8.Wang J.-G., Cui H.-R., Tang H.-B., Deng X.-L. Gastrointestinal symptoms and fecal nucleic acid testing of children with 2019 coronavirus disease: a systematic review and meta-analysis. Sci. Rep. 2020;10:17846. doi: 10.1038/s41598-020-74913-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooks E.F., Bhatt A.S. The gut microbiome: a missing link in understanding the gastrointestinal manifestations of COVID-19? Cold Spring Harb. Mol. Case Stud. 2021;7:a006031. doi: 10.1101/mcs.a006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Natarajan A., Han A., Zlitni S., Brooks E.F., Vance S.E., Wolfe M., Singh U., Jagannathan P., Pinsky B.A., Boehm A., et al. Standardized preservation, extraction and quantification techniques for detection of fecal SARS-CoV-2 RNA. Nat. Commun. 2021;12:5753. doi: 10.1038/s41467-021-25576-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 12.Oran D.P., Topol E.J. The proportion of SARS-CoV-2 infections that are asymptomatic : a systematic review. Ann. Intern. Med. 2021;174:655–662. doi: 10.7326/M20-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo M., Tao W., Flavell R.A., Zhu S. Potential intestinal infection and faecal-oral transmission of SARS-CoV-2. Nat. Rev. Gastroenterol. Hepatol. 2021;18:269–283. doi: 10.1038/s41575-021-00416-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y., Chen C., Zhu S., Shu C., Wang D., Song J., Song Y., Zhen W., Feng Z., Wu G., et al. Isolation of 2019-nCoV from a stool specimen of a laboratory-confirmed case of the coronavirus disease 2019 (COVID-19) China CDC Wkly. 2020;2:123–124. [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao F., Sun J., Xu Y., Li F., Huang X., Li H., Zhao J., Huang J., Zhao J. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg. Infect. Dis. 2020;26:1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong H.W., Kim S.-M., Kim H.-S., Kim Y.-I., Kim J.H., Cho J.Y., Kim S.-H., Kang H., Kim S.-G., Park S.-J., et al. Viable SARS-CoV-2 in various specimens from COVID-19 patients. Clin. Microbiol. Infect. 2020;26:1520–1524. doi: 10.1016/j.cmi.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Chen C., Song Y., Zhu S., Wang D., Zhang H., Han G., Weng Y., Xu J., Xu J., et al. Excretion of SARS-CoV-2 through faecal specimens. Emerg. Microbes Infect. 2020;9:2501–2508. doi: 10.1080/22221751.2020.1844551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 20.Albert S., Ruíz A., Pemán J., Salavert M., Domingo-Calap P. Lack of evidence for infectious SARS-CoV-2 in feces and sewage. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40:2665–2667. doi: 10.1007/s10096-021-04304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin L., Jiang X., Zhang Z., Huang S., Zhang Z., Fang Z., Gu Z., Gao L., Shi H., Mai L., et al. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69:997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- 23.Bradley B.T., Maioli H., Johnston R., Chaudhry I., Fink S.L., Xu H., Najafian B., Deutsch G., Lacy J.M., Williams T., et al. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet. 2020;396:320–332. doi: 10.1016/S0140-6736(20)31305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qian Q., Fan L., Liu W., Li J., Yue J., Wang M., Ke X., Yin Y., Chen Q., Jiang C. Direct evidence of active SARS-CoV-2 replication in the intestine. Clin. Infect. Dis. 2021;73:361–366. doi: 10.1093/cid/ciaa925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Britton G.J., Chen-Liaw A., Cossarini F., Livanos A.E., Spindler M.P., Plitt T., Eggers J., Mogno I., Gonzalez-Reiche A.S., Siu S., et al. Limited intestinal inflammation despite diarrhea, fecal viral RNA and SARS-CoV-2-specific IgA in patients with acute COVID-19. Sci. Rep. 2021;11:13308. doi: 10.1038/s41598-021-92740-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cholankeril G., Podboy A., Aivaliotis V.I., Tarlow B., Pham E.A., Spencer S.P., Kim D., Hsing A., Ahmed A. High prevalence of concurrent gastrointestinal manifestations in patients with severe acute respiratory syndrome coronavirus 2: early experience from California. Gastroenterology. 2020;159:775–777. doi: 10.1053/j.gastro.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Effenberger M., Grabherr F., Mayr L., Schwaerzler J., Nairz M., Seifert M., Hilbe R., Seiwald S., Scholl-Buergi S., Fritsche G., et al. Faecal calprotectin indicates intestinal inflammation in COVID-19. Gut. 2020;69:1543–1544. doi: 10.1136/gutjnl-2020-321388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jang K.K., Kaczmarek M.E., Dallari S., Chen Y.-H., Tada T., Axelrad J., Landau N.R., Stapleford K.A., Cadwell K. Variable susceptibility of intestinal organoid-derived monolayers to SARS-CoV-2 infection. bioRxiv. 2022 doi: 10.1101/2021.07.16.452680. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saif L.J., Jung K. Comparative pathogenesis of bovine and porcine respiratory coronaviruses in the animal host species and SARS-CoV-2 in humans. J. Clin. Microbiol. 2020;58:e01355-20. doi: 10.1128/JCM.01355-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nalbandian A., Sehgal K., Gupta A., Madhavan M.V., McGroder C., Stevens J.S., Cook J.R., Nordvig A.S., Shalev D., Sehrawat T.S., et al. Post-acute COVID-19 syndrome. Nat. Med. 2021;27:601–615. doi: 10.1038/s41591-021-01283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jagannathan P., Andrews J.R., Bonilla H., Hedlin H., Jacobson K.B., Balasubramanian V., Purington N., Kamble S., de Vries C.R., Quintero O., et al. Peginterferon Lambda-1a for treatment of outpatients with uncomplicated COVID-19: a randomized placebo-controlled trial. Nat. Commun. 2021;12:1967. doi: 10.1038/s41467-021-22177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim M.Y., Hong S., Kim B.-M., Ahn Y., Kim H.-J., Nam Y.-D. Changes in microbiome and metabolomic profiles of fecal samples stored with stabilizing solution at room temperature: a pilot study. Sci. Rep. 2020;10:1789. doi: 10.1038/s41598-020-58719-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coryell M.P., Iakiviak M., Pereira N., Murugkar P.P., Rippe J., Williams D.B., Heald-Sargent T., Sanchez-Pinto L.N., Chavez J., Hastie J.L., et al. A method for detection of SARS-CoV-2 RNA in healthy human stool: a validation study. Lancet Microbe. 2021;2:e259–e266. doi: 10.1016/S2666-5247(21)00059-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuypers J., Jerome K.R. Applications of digital PCR for clinical microbiology. J. Clin. Microbiol. 2017;55:1621–1628. doi: 10.1128/JCM.00211-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verma R., Kim E., Martínez-Colón G.J., Jagannathan P., Rustagi A., Parsonnet J., Bonilla H., Khosla C., Holubar M., Subramanian A., et al. SARS-CoV-2 subgenomic RNA kinetics in longitudinal clinical samples. Open Forum Infect. Dis. 2021;8:ofab310. doi: 10.1093/ofid/ofab310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saif L.J. Bovine respiratory coronavirus. Vet. Clin. North Am. Food Anim. Pract. 2010;26:349–364. doi: 10.1016/j.cvfa.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alexandersen, S., Chamings, A., and Bhatta, T.R. SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat. Commun. 11 6059 [DOI] [PMC free article] [PubMed]
- 42.McClary-Gutierrez J.S., Mattioli M.C., Marcenac P., Silverman A.I., Boehm A.B., Bibby K., Balliet M., de Los Reyes F.L., 3rd, Gerrity D., Griffith J.F., et al. SARS-CoV-2 wastewater surveillance for public health action. Emerg. Infect. Dis. 2021;27:1–8. doi: 10.3201/eid2709.210753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polo D., Quintela-Baluja M., Corbishley A., Jones D.L., Singer A.C., Graham D.W., Romalde J.L. Making waves: wastewater-based epidemiology for COVID-19 - approaches and challenges for surveillance and prediction. Water Res. 2020;186:116404. doi: 10.1016/j.watres.2020.116404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Medema G., Been F., Heijnen L., Petterson S. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Curr. Opin. Environ. Sci. Health. 2020;17:49–71. doi: 10.1016/j.coesh.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mallapaty S. How sewage could reveal true scale of coronavirus outbreak. Nature. 2020;580:176–177. doi: 10.1038/d41586-020-00973-x. [DOI] [PubMed] [Google Scholar]
- 46.CDC . Centers for Disease Control and Prevention; 2022. National Wastewater Surveillance System (NWSS)https://www.cdc.gov/healthywater/surveillance/wastewater-surveillance/wastewater-surveillance.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019-ncov%2Fcases-updates%2Fwastewater-surveillance.html [Google Scholar]
- 47.Mowat A.M., Agace W.W. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 2014;14:667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 48.Osikomaiya B., Erinoso O., Wright K.O., Odusola A.O., Thomas B., Adeyemi O., Bowale A., Adejumo O., Falana A., Abdus-Salam I., et al. Long COVID”: persistent COVID-19 symptoms in survivors managed in Lagos State, Nigeria. BMC Infect. Dis. 2021;21:304. doi: 10.1186/s12879-020-05716-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taquet M., Dercon Q., Luciano S., Geddes J.R., Husain M., Harrison P.J. Incidence, co-occurrence, and evolution of long-COVID features: a 6-month retrospective cohort study of 273,618 survivors of COVID-19. PLoS Med. 2021;18:e1003773. doi: 10.1371/journal.pmed.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramakrishnan R.K., Kashour T., Hamid Q., Halwani R., Tleyjeh I.M. Unraveling the mystery surrounding post-acute sequelae of COVID-19. Front. Immunol. 2021;12:686029. doi: 10.3389/fimmu.2021.686029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weng J., Li Y., Li J., Shen L., Zhu L., Liang Y., Lin X., Jiao N., Cheng S., Huang Y., et al. Gastrointestinal sequelae 90 days after discharge for COVID-19. Lancet Gastroenterol. Hepatol. 2021;6:344–346. doi: 10.1016/S2468-1253(21)00076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carfì A., Bernabei R., Landi F., Gemelli, Against COVID-19 post-acute care study group persistent symptoms in patients after acute COVID-19. JAMA. 2020;324:603–605. doi: 10.1001/jama.2020.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kayaaslan B., Eser F., Kalem A.K., Kaya G., Kaplan B., Kacar D., Hasanoglu I., Coskun B., Guner R. Post-COVID syndrome: a single-center questionnaire study on 1007 participants recovered from COVID-19. J. Med. Virol. 2021;93:6566–6574. doi: 10.1002/jmv.27198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Al-Aly Z., Xie Y., Bowe B. High-dimensional characterization of post-acute sequelae of COVID-19. Nature. 2021;594:259–264. doi: 10.1038/s41586-021-03553-9. [DOI] [PubMed] [Google Scholar]
- 55.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 56.dMIQE Group. Huggett J.F. The digital MIQE guidelines update: minimum information for publication of quantitative digital PCR experiments for 2020. Clin. Chem. 2020;66:1012–1029. doi: 10.1093/clinchem/hvaa125. [DOI] [PubMed] [Google Scholar]
- 57.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel (2020). (Division of Viral Diseases, Centers for Disease Control and Prevention).
- 59.Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S.A., Harcourt J., et al. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26:1654–1665. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Droplet Digital PCR Applications Guide https://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_6407.pdf.
- 61.Pierson-Perry J.F., Vaks J.E., Vore T.E.K., Durham A.P., Fischer C., Gutenbrunner C., Hillyard D., Kondratovich M.V., Ladwig P., Middleberg R.A. Clinical Laboratory Standards Institute; 2012. Evaluation of Detection Capability for Clinical Laboratory Measurement Procedures; Approved Guideline. [Google Scholar]
- 62.Austin P.C. Using the standardized difference to compare the prevalence of a binary variable between two groups in observational research. Commun. Stat. - Simulation Comput. 2009;38:1228–1234. [Google Scholar]
- 63.Cohen J. Routledge Academic; 1988. The Effect Size Index: D. Statistical Power Analysis for the Behavioral Sciences. [Google Scholar]
- 64.Liang K.-Y., Zeger S.L. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 65.Kernan W.N., Viscoli C.M., Makuch R.W., Brass L.M., Horwitz R.I. Stratified randomization for clinical trials. J. Clin. Epidemiol. 1999;52:19–26. doi: 10.1016/s0895-4356(98)00138-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data supporting the findings of this study have been deposited at Zenodo (https://zenodo.org/record/6374138) and are publicly available as of the date of publication.
-
•
All custom code and mathematical models have been deposited at Zenodo (https://zenodo.org/record/6374138) and are publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.