Abstract
Swarming motility is one of three distinct modes of motility observed in the gram-negative bacterium Pseudomonas aeruginosa. Swarming motility is defined as the movement across a semisolid surface, and in P.aeruginosa requires flagellar motility and the production of biosurfactants. Swarming motility is thought to occur on gelatinous/viscous surfaces inside a host, such as on epithelial cells. There is currently no standardized in vitro assay to visualize and study swarming motility, and the assays used can vary greatly between laboratory groups. Here, we describe a detailed, reproducible in vitro swarming motility assay for P.aeruginosa. While different protocols have previously been reported in the literature, we hope that adopting this method will improve the reproducibility of these swarming motility assays and allow comparisons of swarming motility findings between and among groups.
Keywords: Swarming motility, Flagellum, Biosurfactants, Surface motility
1. Introduction
In addition to flagellum-dependent swimming and type IV pili-dependent twitching motility, Pseudomonas aeruginosa is also capable of swarming motility, which occurs on semisolid surfaces. Unlike swimming motility, swarming motility has been shown to require the production of two biosurfactants — rhamnolipids (RL) and 3-hydroxyalkanoic acids (HAA) — along with a functional flagellum [1-4].
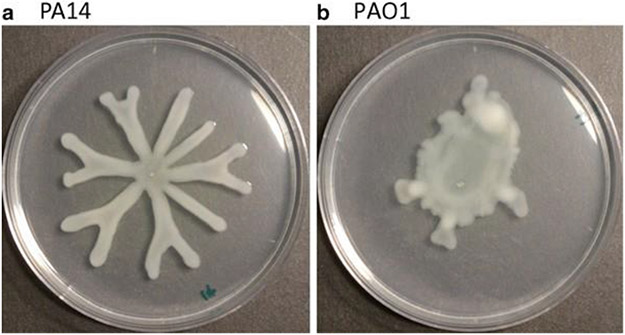
Different strains of P. aeruginosa show distinct swarming motility phenotypes, for example, P.aeruginosa PAO1 is associated with a circular pattern, whereas P.aeruginosa PA14 forms a unique flower-like pattern, with noticeable tendril projections from the point of inoculation (see Fig. 1).
Fig. 1.
Swarming motility of Pseudomonas aeruginosa (a) PA14 vs. (b) PAO1
At present, one of the major drawbacks with swarming motility assays is the lack of consistency and reproducibility. That is, each swarming assay, even from a single batch of plates, can yield drastically different outcomes based on multiple factors, including (but not limited to): freshness of the bacterial culture, agar concentration (%), thickness of the plate, dryness of the plate, pH, incubation temperature [5], and location within the incubator. Compounding the issue is the variability in medium composition used among the laboratories investigating swarming motility. To date, however, investigators have yet to systemically investigate the relative merits of each medium, leaving the novice investigators somewhat bewildered when attempting this assay for the first time.
Here, we provide a useful method that has been optimized by our laboratory group through extensive experimentation over the past decade, and yields reproducible swarming motility phenotypes. While our laboratory has adopted the M8-supplemented swarming medium [1], we believe our protocol will also improve the reproducibility and consistency in other swarming media as well [5,6].
2. Materials
Prepare all solutions using ultrapure, distilled deionized water. All solutions used in this assay are sterilized (autoclaved at least 45 min at 121 °C, 15 psi per 1 L solution) prior to use and are kept at room temperature (unless indicated otherwise). Follow all waste disposal procedures when disposing of contaminated solutions at the completion of the experiments.
Tube(s) of P.aeruginosa strain(s) grown overnight at 37 °C in lysogeny broth (LB: 0.5 g yeast extract, 1 g tryptone, 0.5 g NaCl in 100 mL water) from a fresh streak plate, <7 days old.
Petri plates: sterile, 100 mm × 15 mm.
Autoclavable bottle: 1 L.
Autoclavable flask: 2 L.
Automatic pipettor.
Disposable, sterile serological pipettes: 10, 25, 50 mL.
20 % Casamino acids solution in water (see Note 1).
20 % Glucose solution in water.
1 M MgSO4 solution in water.
Pipette: P10, P1000.
Agar.
5 × M8 solution: dissolve 64 g Na2HPO4·7H2O, or alternatively, 30 g Na2HPO4; 15 g KH2PO4; 2.5 g NaCl in water, and bring the final volume to 1 L (see Note 2).
3. Methods
All procedures are performed at room temperature (RT) unless specified otherwise. The following protocol is scaled to make 1 L batch of agar. In our experience, 1 L batch should yield 30–36 swarm motility plates. For a different volume, scale accordingly to maintain the appropriate concentration for each component.
Add 5–8 g (for final concentration of 0.5–0.8 %) of agar to 800 mL of water. Autoclave sufficiently in 2 L flask to yield a sterile, homogeneous agar suspension; 45 min cycle is sufficient for a 1 L batch (see Note 3).
Autoclave 5× M8 solution in 1 L bottles. Add 200 mL of 5×M8 solution to the melted agar (see Note 4).
Add 10 mL of 20 % glucose to the melted agar; final concentration: 0.2 % glucose.
Add 25 mL of 20 % casamino acids to the melted agar; final concentration: 0.5 % casamino acids.
Add 1 mL of 1 M MgSO4 to the melted agar; final concentration: 1 mM MgSO4.
Mix the agar medium and cool prior to pouring onto petri plates (see Note 5).
Pour thick plates (~25 mL/plate) and let plates solidify at RT for a few hours (see Note 6).
Inoculate the center of the plate(s) using the overnight bacterial culture, 2.5 μL per inoculum. Pipette tip should be close to the agar surface during inoculation (see Note 7).
Incubate plate(s) upright at 37 °C for 16–24 h (see Notes 8 and 9) and observe for phenotype (see Note 10).
4. Notes
Since 5× M8 solution lacks any source of nitrogen, supplementation with a nitrogen source is key in a swarm motility assay. Therefore, casamino acids can be substituted with a different source of nitrogen depending on the investigator’s experimental question. As for 20 % stock solution of casamino acids, precipitates do accrue in older solutions at RT. In our experience, while these precipitates do not redissolve with increased temperature (~50 °C), they also do not impact the final outcome of swarm motility phenotypes.
Completely dissolving Na2HPO4 (both for heptahydrate- and anhydrous forms) into water takes some time; however, warming the solution is unnecessary. Add reagents to 800 mL of water before bringing up the final volume to 1 L.
We find it is best to prepare a fresh batch of molten agar on the day of the assay. Addition of a magnetic stir bar to the agar is recommended, as it will assist in mixing other components.
Depending on the temperature of the molten agar, addition of the 5× M8 solution could either prematurely solidify the agar, or speedup the cooling process. We find that the best approach is to pre-warm the 5× M8 solution in a warm water bath (~55 °C) prior to adding into the molten agar. If working on a shorter time frame, however, 5× M8 solution at RT can be added to a hot molten agar to decrease the time needed to cool the molten agar prior to pouring plates.
-
Since swarm motility occurs on the surface of the agar, it is best to eliminate any irregularities on the surface. Aggressive mixing of the components tends to cause foaming, which can later result in unwanted air bubbles on the plate surface. Magnetic stir bars, at low-to-medium rotational speed will achieve both homogeneous mixing and bubble-free medium. Alternatively, remaining air bubbles can be “popped” by gently running a flame over a poured plate (not recommended on batch volumes >1 L, as the plates begin solidifying rather quickly once poured).
Agar suspension that is cool to the touch (5 s to the touch without burning one’s hand) is when we generally start pouring plates. We feel this temperature is warm enough to prevent any premature solidifying of the agar, but also cool enough to pour manually without feeling too hot, nor melting the plastic petri plates.
Plate thickness does not have to be exact, but the general rule of thumb is that thicker plates will produce better swarm results. Thicker agar layer retains moisture longer during the incubation step, which prevents premature drying of the plate. When plates have been poured, we find it best to solidify them either as single plates or in small stacks of plates (<4 plates/stack) for approximately 3–4 h, as plates on the upper level of the stack tend to solidify and dry more slowly than those at the bottom. A previous report demonstrated increased reproducibility of swarming motility phenotypes by drying under laminar flow [5]; however, the uneven distribution of laminar flow resulted in different dryness of the agar based on the plate’s location within the flow, which can impact swarming phenotypes. For this reason, and the lack of laminar flow equipment in certain labs, we prefer drying our plates at RT as was described here. When plates are dry, make sure to remove any condensation that has accumulated on the underside of the petri plate cap. Incubation at 37 °C will further increase condensation, which may fall back onto the agar and alter the development of swarming motility.
-
Unless the investigator is performing high-throughput screens of swarm motility mutants, we generally recommend one point of inoculation per plate in the center of the plate. As was seen in the example figure, 2.5 μL of P.aeruginosa spotted on the center of the plate, then incubated overnight at 37 °C tends to create a robust swarm phenotype spanning almost the entire plate. Two inoculations can be performed when investigating tendril-tendril interactions between two strains/mutants, but this is the maximum we recommend. As for screening purposes, inoculum size should be ≤1 μL with sufficient space between each inoculum to distinguish the phenotypes of each mutant/strain. We recommend 96 well replicators or the like for this purpose.
Regarding inoculation, it is recommended that pipet tips be as close to the agar surface prior to inoculating. Direct contact with the agar could result in a squirt of bacterial culture over the agar surface, rather than a uniform point of inoculum. Conversely, if the pipet tip is distant from the agar surface while inoculating, multiple inoculation points can inadvertently result in points of inoculation due to splashing.
Numerous factors can contribute to variability in swarm phenotypes. To mitigate the effects of these variables, consistency is extremely important. We generally stack the plates of a given strain, with each stack being of same height as other strains being tested within the same batch. Each stack is composed of <8 plates. These stacks of plates are placed near each other within the incubator at the same time, and also removed at the same time. At the end of the 16–24 h of incubation, place them at RT for 1–2 h for coloration of the swarm tendrils. This additional incubation is only for aesthetic purposes as green pigments of P.aeruginosa are accentuated during this RT treatment.
Controlled humidity in the incubation chamber is key in producing reproducible swarming phenotypes. Ideally, a humidity-controlled incubation chamber will yield the best set of results. Other non-humidity controlled chambers will require investigators to identify region(s) with (relatively) consistent humidity. For example, with incubation chambers utilizing circulating airflow, preliminary experiments will be necessary to identify regions within the chamber that do not over-dry swarming motility plates during the 16–24 h incubation period. Another solution we have tried and have found success with is covering the stack of swarming motility plates with an inverted tray. It is also important to incubate swarm plates on an even surface as even a slight slant could trigger the flow of the inoculum from its initial point of inoculation. The result will resemble a streak, rather than tendril formations and flower-like pattern, typically associated with positive swarming motility in P.aeruginosa. Should this happen, the investigator should not score the streak phenotype as a “positive” swarming phenotype.
Swarming motility continues at room temperature post-incubation at 37 °C. We have noticed that prolonged incubation thickens tendrils and accentuates the coloration of green pigments. It has also been observed that continued incubation at room temperature can cause negative-swarming mutants to start to produce tiny tendrils. In our hands, <24 h at room temperature following 37 °C incubation accentuates swarm tendrils for documenting purposes.
Acknowledgement
This work was supported by the NIH grant R01A1003256 to G.A.O., and the Rosaline Borison predoctoral fellowship awarded to D.G.H.
References
- 1.Caiazza NC, Shanks RM, O'Toole GA (2005) Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J Bacteriol 187:7351–7361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deziel E, Lepine F, Milot S, Villemur R (2003) rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149:2005–2013 [DOI] [PubMed] [Google Scholar]
- 3.Kohler T, Curty LK, Barja F, van Delden C, Pechere JC (2000) Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol 182:5990–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tremblay J, Richardson AP, Lepine F, Deziel E (2007) Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ Microbiol 9:2622–2630 [DOI] [PubMed] [Google Scholar]
- 5.Tremblay J, Deziel E (2008) Improving the reproducibility of Pseudomonas aeruginosa swarming motility assays. J Basic Microbiol 48:509–515 [DOI] [PubMed] [Google Scholar]
- 6.Overhage J, Lewenza S, Marr AK, Hancock RE (2007) Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 mini-Tn5-lux mutant library. J Bacteriol 189:2164–216 [DOI] [PMC free article] [PubMed] [Google Scholar]