Abstract
Horizontal gene transfer (HGT) in the microbiome has profound consequences for human health and disease. The spread of antibiotic resistance genes, virulence, and pathogenicity determinants predominantly occurs by way of HGT. Evidence exists of extensive horizontal transfer in the human gut microbiome. Phage transduction is a type of HGT event in which a bacteriophage transfers non-viral DNA from one bacterial host cell to another. The abundance of tailed bacteriophages in the human gut suggests that transduction could act as a significant mode of HGT in the gut microbiome. Here we review in detail the known mechanisms of phage-mediated HGT, namely specialized and generalized transduction, lateral transduction, gene-transfer agents, and molecular piracy, as well as methods used to detect phage-mediated HGT, and discuss its potential implications for the human gut microbiome.
Keywords: gut phageome, horizontal gene transfer, gene transduction, phage-mediated gene transfer
Introduction
The human gut microbiome is a complex community with a vast network of microbe–host interactions. The ability to change and adapt to acute events is essential to maintain long-term stability. The microbial community, with its short generation times and intense selective pressure, evolves at a speed unimaginable for multicellular organisms, and can therefore respond more quickly than its host to changing environmental conditions [1]. This is further enhanced by the ability of the microbiome to exchange genetic material that ensures swift access to an extensive bacterial pan-genome [2].
Genetic information is typically transferred vertically from parent to offspring and is subject to change through mutation. This is how a phylogenetic tree depicts evolution. Any other movement of genetic information is referred to as horizontal gene transfer (HGT). In recent years it has become increasingly clear that without taking HGT into account, it is impossible to describe the evolution of microbial communities [3], especially those as complex as the human gut microbiome. A phylogenetic web or network, rather than a tree, has been proposed to comprehensively depict these evolutionary relationships [1, 4]. Moreover, it has been shown that a dense network of HGT connects members of the human microbiome [2] and evidence of extensive horizontal transfer has been found in the human gut microbiome in particular [5].
HGT within the microbiome has potential consequences for human health and disease. The spread of antibiotic resistance genes happens mainly by way of HGT [6]. Virulence factors that determine a strain’s pathogenicity can also be transferred horizontally [7], as can genes involved in metabolic functions [8], including the catabolism of certain sugars [9].
Several types of HGT events are well known in bacteria; these include transformation, conjugation, and transduction, as well as nanotube contact and vesicle-mediated transfer. Transformation is the uptake of free extracellular DNA by a competent bacterial cell (any cell that possesses the ability to capture, “ingest,” and incorporate free DNA into its genome). Conjugation describes the transfer of genetic material (typically but not always plasmids) between two bacteria through cell-to-cell contact, using specialized pili or direct adhesion. Another mode of HGT that also involves cell-to-cell contact and nanotube structures has been shown to facilitate transfer of non-conjugative plasmids in Bacillus subtilis [10]. DNA transfer in membrane vesicles has been described in the marine environment [11, 12], including “serial transfer” that enables the recipient to produce identical DNA-transporting membrane vesicles [13, 14]. This review focuses on transduction, the process in which a bacteriophage particle transfers non-viral DNA from one bacterial host cell to another. This occurs when bacterial DNA gets packaged inside some of the viral particles either in place of or together with bacteriophage DNA.
A brief overview of phage replication and virion assembly
The number of viruses in the human gut virome is estimated to be >1012 [15] and the majority of these viruses are tailed bacteriophages (phages). This makes tailed phages the most abundant gene-transfer particles in the human gut microbiome. They are also perfectly suited for the task: all tailed bacteriophages (order Caudovirales) have double-stranded DNA (dsDNA) genomes with a virion structure optimized for carrying dsDNA. We can confidently predict that bacteriophages play a significant role in gene exchange in the human gut.
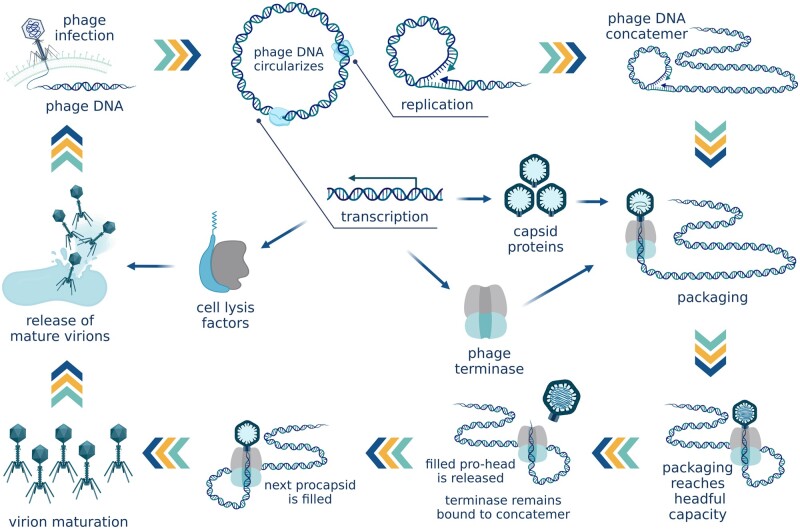
The normal propagation cycle of bacteriophages results in the production of viable virions that contain a single copy of the phage genome (Figure 1). Viable virions, or infectious particles, can infect and kill their host bacteria, producing phage progeny. The phage binds to the surface of the bacterial cell and injects its DNA into the cytoplasm. Lytic, or virulent, phages start propagating immediately and the progeny is released into the environment through cell lysis. Lysogenic (temperate) phages can also initiate the lytic cycle on entry into the cell. Alternatively, their genome can integrate into the host chromosome at a specific attachment (att) site. The integrated phage genome (prophage) replicates together with the host chromosome and is transferred vertically from the initial infected cell to its progeny through cell division. In an SOS response to DNA damage, the prophage excises from the host genome and enters the lytic cycle.
Figure 1.
Phage reproduction. Rolling-circle replication produces long concatemers of the phage genome. Phage small terminase subunit (TerS) recognizes a pac site and initiates packaging. After one virion is filled with DNA, it disengages from the terminase complex, which remains attached to the DNA concatemer and binds to the next virion. (Created with BioRender.com.)
In the process of phage propagation, the phage genome, either having entered the cytoplasm from the infecting virion or having been freshly excised from the host DNA in response to inducing factors, circularizes and starts replicating. The first copies are produced through theta-replication, which ensures the generation of high-fidelity genome copies. Each of these copies then becomes a matrix template for rolling-circle replication that produces long covalent end-to-end polymers of the phage genome called concatemers.
Meanwhile, the translation of phage genes produces all the proteins required for the formation of viable phage particles. These include structural “head” and “tail” proteins, scaffolding and chaperone proteins that assist the correct assembly of the capsids, regulatory proteins, and the phage terminase whose function is to govern the packaging of phage genomes inside the phage heads.
The phage terminase complex consists of two subunits. The large terminase subunit (TerL) performs most of the mechanical work. It possesses endonuclease activity to cut the DNA, a complex packaging motor that translocates the DNA into the procapsid, and an ATPase domain that generates the energy used by the packaging motor. The small terminase subunit (TerS) performs regulatory functions. TerS is responsible for the packaging specificity of the terminase complex; it contains a DNA-binding domain that recognizes a specific tag sequence in the phage genome that labels it for cleavage and packaging. Having translocated one copy of the genome into the procapsid, the large subunit makes a second cut. The terminase complex remains bound to the free end of the concatemer, docks on another empty prohead, and continues packaging further headfuls of phage DNA while the filled capsid disengages and becomes ready for tail maturation [16]. Several types of phage terminases determine different strategies to ensure packaging specificity.
Headful packaging, the mechanism used by pac-type terminases (also called headful terminases), recognizes a single site in the phage genome called the pac site. When the small subunit of the terminase complex recognizes the pac sequence, the large subunit proceeds to package the DNA into the procapsid until the capacity of the phage head is reached, determining the site of the second cleavage by procapsid volume rather than a specific DNA sequence. Having packaged a full “headful,” which typically amounts to 102%–110% of the full phage genome size, the large subunit severs the packaged DNA from the rest of the concatemer [17]. This terminal redundancy created by packaging slightly more than a full-length bacteriophage genome becomes useful when the phage injects its DNA into the target cell where, once in the cytoplasm, the DNA will use its matching ends to circularize through recombination [18]. Among the phages, Salmonella phage P22 and Escherichia coli phages P1 and T4 are use this mechanism [19].
cos-Type terminases recognize two cos sites on the phage genome. The first cos site is required to initiate packaging and the second identical sequence acts as a signal for the terminase complex to introduce a second cut that separates the packaged DNA from the rest of the concatemer. As the DNA is not merely carried within the capsid but is itself an important part of the capsid structure, packaging of exactly the right length of DNA is crucial for virion assembly and maturation [16]. For this reason the terminase complex only becomes capable of recognizing the second cos sequence and making the cut once the procapsid is filled nearly to its capacity [16]. When cutting the DNA, cos-type terminases introduce two staggered cuts, first on one DNA strand and then the other, generating complementary, sticky ends used for circularization (cohesive ends). Bacteriophage lambda is the best studied model for cos-type phages [20]. A similar mechanism is used by phages with direct terminal repeats. After the terminase introduces staggered nicks in the phage DNA, the 3' end is extended to form identical blunt ends, as seen in E. coli phage T3 and B. subtilis phage SPO1 [21, 22].
Some bacteriophages, such as B. subtilis phage Φ29, differ not only in their packaging strategy but also in their mode of replication. They use protein-primed replication and do not produce concatemers. The terminase complex, which lacks the small subunit and endonuclease activity in the large subunit, binds to the terminal protein attached to each viral genome monomer and packages the genome copy [16].
Another notable strategy in the context of HGT belongs to E. coli bacteriophage Mu that replicates by transposition. Similarly to Φ29, its packaging substrates are genome monomers. On entry into the cell, the phage integrates randomly into the host chromosome and, when induced, produces multiple copies of its genome that re-integrate into random locations in the host genome. As a result the genome of phage Mu is always flanked by random fragments of host DNA [23]. Notably, the induction of phage Mu is low-frequency and spontaneous; no physical or chemical treatment is known to trigger lytic development of wild-type Mu [24].
As the human gut microbiome is a rich and diverse microbial community, it is likely to contain examples of all of the above strategies to ensure packaging specificity. Yet none of the mechanisms can completely exclude mispackaging and encapsidation of host DNA, and mechanisms of HGT by phage are even more diverse than the systems working against erroneous packaging.
Mechanisms of phage HGT
Specialized transduction
Specialized transduction can only be carried out by temperate phages and is normally restricted to those genes adjacent to the att site of the prophage. The induced prophage will occasionally excise imprecisely, cutting out adjacent host genes together with the phage genome. This host DNA becomes part of the circularized phage genome and is replicated. As a result of this aberrant excision event, all the virions produced by the cell will carry a fragment of host DNA (Figure 2). However, imprecise prophage excision is a rare event. In phage lambda for example, a transducing particle is produced by ∼104 virions [25], while the reported rate of successful transduction is ∼1 in 106 [26], which indicates that only one in ∼100 transducing particles leads to a completed transduction event.
Figure 2.
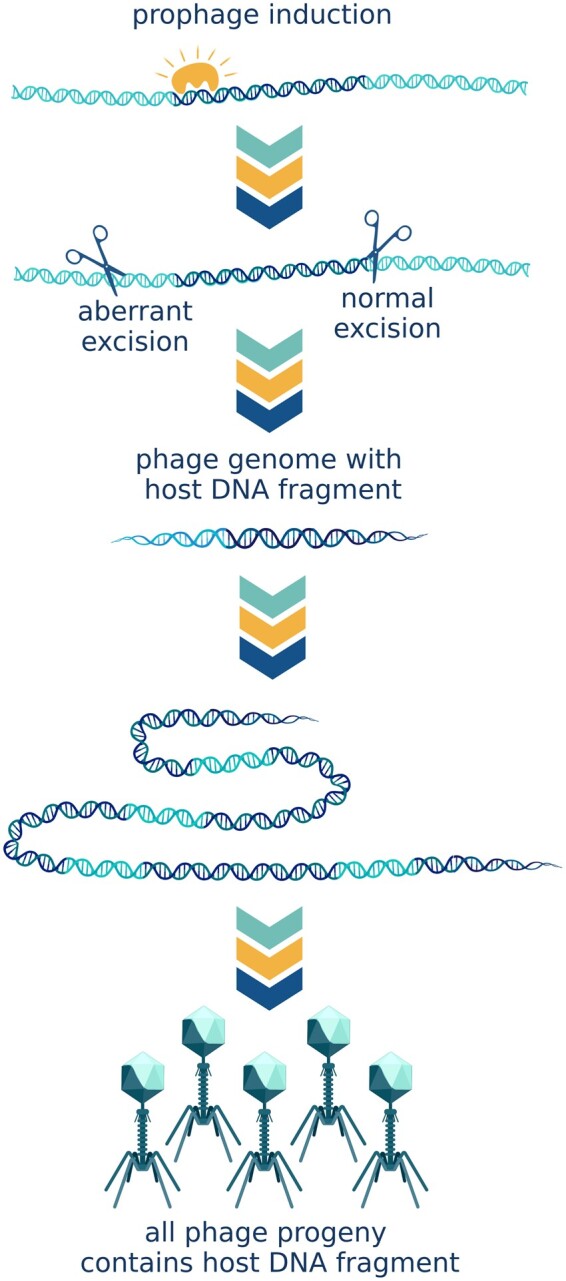
Specialized transduction. As a result of aberrant excision, adjacent host genes are cut out of the chromosome together with the phage genome. Host DNA becomes part of the circularized phage genome and is replicated. As a result, all the virions produced by the cell will carry a fragment of host DNA. (Created with BioRender.com.)
Bacteriophage lambda, a classic example of specialized transducing phage, integrates into the genome of its host, E. coli, between the galactose metabolism and biotin biosynthesis operons. The phage can consequently transfer these genes, conferring a fitness advantage in certain environments. Likewise, in the gut microbiome, horizontal transfer of such valuable metabolic functions could facilitate adaptive changes in response to dietary alterations. Temperate bacteriophages constitute, from various estimates, 20%–50% of human gut phages [27]. Such a high prevalence of temperate phage supports the likelihood of significant levels of specialized transduction in the human gut.
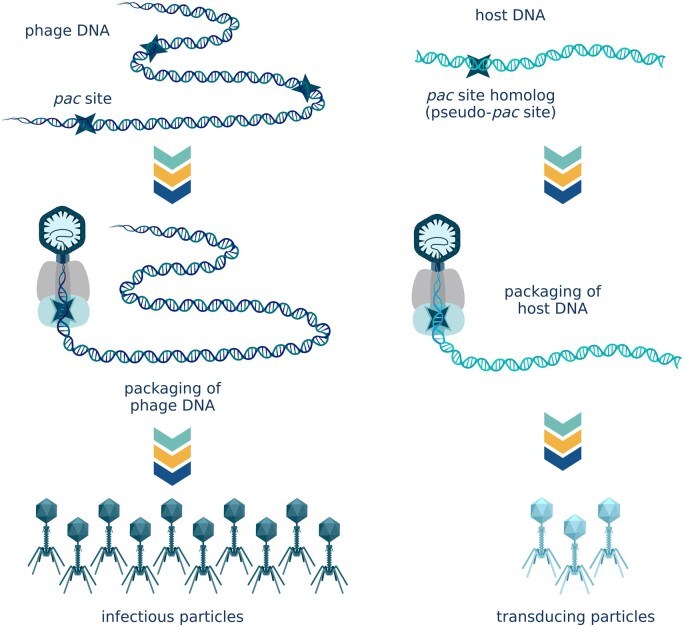
Generalized transduction
In generalized transduction, instead of recognizing a bona fide pac site, the terminase complex recognizes a pac site homolog present in the host chromosome and introduces a double-stranded break in its vicinity. Once the terminase machinery has encapsidated a “headful” of host DNA, a second break is introduced in the DNA. The filled virion is ready for tail maturation and detaches from the terminase complex. The terminase complex attaches to the free end of the bacterial chromosome and proceeds to encapsidate phage-sized fragments of host genome until it disengages from the chromosome [16] (Figure 3). The farther away from the initial pseudo-pac site, the higher the likelihood of the terminase complex detaching from the chromosome, and so transduction frequencies steadily decrease with increasing distance [25].
Figure 3.
Generalized transduction. Instead of recognizing a bona fide pac site, the terminase complex recognizes a pac site homolog present in the host chromosome and proceeds to encapsidate phage-sized fragments of host genome until it disengages from the chromosome. (Created with BioRender.com.)
Generalized transduction is almost exclusively the domain of pac phages because headful (pac-type) terminases only require one pac site homolog in the host chromosome to package host DNA fragments into phage particles. For cos phages to engage in generalized transduction, the host genome would have to contain not one, but two cos site homologs located at an appropriate distance from one another [16]. Even though constrained by the location of host pseudo-pac sites, generalized transduction is unpredictable and thus inevitably adds yet another layer of complexity to HGT in the human gut microbiome. Generalized transduction is a low-frequency event. Only 1%–6% of virions contain host DNA [25, 28–30]. Of these transducing particles, 10%–15% are shown to inject their DNA into the host cells [31]. The number of complete transductions will be one to three orders of magnitude lower, depending on whether the recipient cells are recombinase-positive [31, 32]. Rare as it is, generalized transduction can facilitate significant adaptational changes in the human gut. If a transduction event involves genetic traits that dramatically increase fitness, it can initiate rapid spread of these traits in the population.
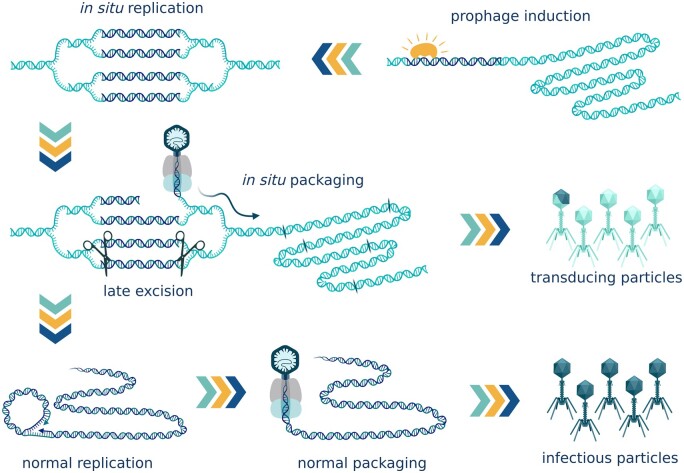
Lateral transduction
Unlike generalized transduction, lateral transduction, such as observed in the prophages of Staphylococcus aureus [33] and Salmonella phage P22 [34], allows high-frequency host DNA transfer. Laterally transducing phages do not follow the typical excision–replication–packaging life cycle. Instead, they start replicating while still integrated in the host chromosome. In situ replication creates multiple integrated phage genomes. Some of these genome copies later get excised and follow the typical cycle forming infectious particles. In those genomes that stay integrated, the terminase complex recognizes the pac site located near the middle of the phage genome. It cuts the DNA at the pac site and proceeds to package without stopping at the junction between phage and host genomes. The resulting virion contains a chimeric DNA molecule that is half phage and half bacterial DNA. The terminase then continues through the adjacent host chromosome efficiently packing up to seven headfuls before the frequencies decrease to the rates of generalized transduction. In this manner, in situ packaging and the typical excision–replication–packaging cycle happen in parallel, producing both the transducing particles and the actual infectious virions [33] (Figure 4).
Figure 4.
Lateral transduction. Lateral transducing phages start replicating while still integrated in the host chromosome. In situ replication creates multiple integrated phage genomes. Some of these genome copies later get excised and follow the typical cycle forming infectious particles. In those that stay integrated, the terminase complex initiates in situ packaging from the pac site near the middle of the phage genome and proceeds to package towards the host genome. The first virion contains a chimeric DNA molecule that is half phage and half bacterial. The terminase continues through the adjacent host chromosome efficiently packaging up to seven headfuls before the frequencies decrease. In situ packaging and the typical excision–replication–packaging cycle happen in parallel, producing both the transducing particles and the actual infectious virions. (Created with BioRender.com.)
It has been observed that lateral transduction is a factor driving genome organization and the location of genes carrying useful adaptive traits. Virulence factors, pathogenicity determinants, resistance genes, and other genes responsible for fast adaptation to the change in conditions can be clustered in the regions of the chromosome adjacent to the phage integration sites in the direction of in situ packaging [33]. Lateral transduction was first described in the prophages of S. aureus, in which it creates a high-throughput gene-transfer channel promoting genome mobility and thus facilitating the swift adaptation of S. aureus to changing environments. The same need to constantly change and adjust to change is characteristic of the gut microbiome. Even though lateral transduction has so far only been observed in Staphylococcus and Salmonella, it is likely to occur in members of the gut microbiome, as well as in other environments.
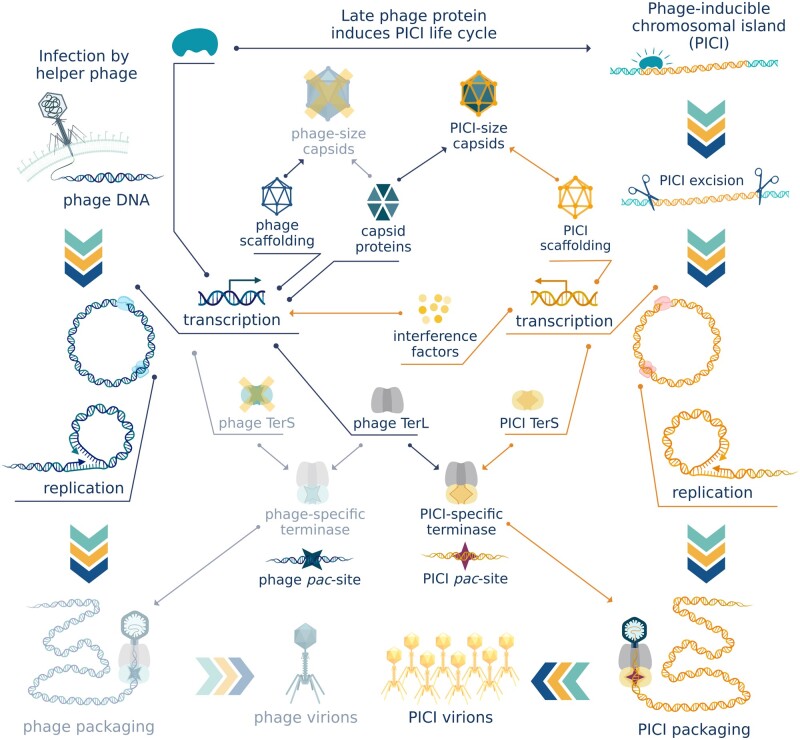
Molecular piracy
Another extraordinary mode of transduction was also first described in S. aureus [35]. Phage-inducible chromosomal islands (PICIs) are a family of phage satellites that exploit actively replicating phages for their dissemination. PICIs have a conserved genetic organization [36], but do not possess genes encoding phage structural proteins. Like prophages, PICIs are latent in the host genome until they are derepressed. However, unlike prophages, phage-inducible islands are not directly SOS-induced; instead, each responds only to a single-phage-encoded protein that serves as a PICI-specific derepressor [37]. The phage antirepressor protein disrupts the complex between the PICI master repressor and its binding site on the PICI genome [37], enabling the transcription of most of the PICI genes. Transcription of early genes is followed by PICI genome excision, circularization, and replication. The derepression mechanism ensures that PICIs only get induced in the presence of an actively propagating helper phage, whose capsids they hijack for transfer to a new host cell (Figure 5).
Figure 5.
Phage-inducible chromosomal islands. Phage-inducible chromosomal islands (PICIs) are latent in the host genome until they are derepressed in the presence of an actively growing helper phage. PICIs hijack structural proteins and packaging machinery of the helper phage for transfer of PICI genes. PICI-specific small terminase subunit (TerS) forms a functional complex with phage large terminase subunit (TerL) while PICI interference inhibits the expression of phage TerS ensuring preferential packaging of PICI genes. PICI scaffolding proteins redirect capsid size to fit the smaller size of PICI genomes. (Created with BioRender.com.)
Some PICIs replace phage-encoded scaffolding proteins during the making of procapsids and redirect the capsid size to fit their considerably smaller genomes [38, 39], while other PICIs simply use normal-sized helper phage capsids, packaging more than one genome copy inside each virion [40]. Most known PICIs encode their own TerS but no TerL [36]. Two key features are characteristics of PICI-encoded TerS. First is the formation of a functional complex with the TerL encoded by the helper phage. The second is the recognition of the pac sequence of the PICI instead of that of the helper phage. TerS will guide the large subunit to package the chromosomal island genome while ignoring the helper phage genome entirely. To increase the probability of phage TerL forming a complex with PICI TerS, PICI-encoded repressor proteins target the transcription of phage-encoded TerS [41]. However, PICIs do not encode a TerS sequence. Instead, they contain a pac site recognizable by phage-encoded terminase-like PICI elements of Enterococcus faecalis, or a pair of cos sites located at appropriate distance from each other as observed in the chromosomal islands of E. coli [42]. Both features can also be present, as seen in the S. aureus PICI SaPIbov5 [43].
PICIs severely impair helper phage reproduction, ensuring that only functions that benefit the satellite are preserved [41, 44]. As a result, most of the produced virus-like particles (VLPs) contain a PICI genome. For instance, in the E. faecalis EfCIV583-Φ1 pirate-helper system, almost 10 PICI-carrying particles are produced for each bona fide helper phage virion [25, 45]. An even more extreme case is the Vibrio cholerae PICI-like element PLE1. When cells carrying this element are infected with its helper phage ICP1, no phage progeny are produced [46, 47].
Helper phages and their satellites undergo rapid co-evolution with phages developing resistance to the chromosomal islands and the PICIs in turn evolving to evade that resistance. Mutations give rise to allelic polymorphism in the genes encoding derepressor proteins for PICIs. It has been observed that E. faecalis phages that had interacted with PICI elements accumulated mutations in the gene acting as the inducer for the chromosomal island [48]. PICIs, in turn, mutate the master repressor binding site in a way that allows its complex with the master repressor to be disrupted by the new allelic variant of the derepressor protein [48]. Vibriocholerae bacteriophage ICP1 encodes its own CRISPR/Cas system against PICI-like elements that parasitize ICP1 and prevent it from producing progeny [46].
While PICIs are detrimental to helper phage, they can benefit their bacterial hosts, not only by reducing the number of infectious particles produced by their helper phage, but also by carrying genes that increase the host’s fitness. The discovery of the first-phage-inducible islands was prompted by studies of toxic shock syndrome toxin encoded by a PICI element [35]. Many more accessory genes that encode virulence and pathogenicity factors, resistance pathways [49], and other instruments of swift adaptation have been identified in PICIs. For instance, in S. aureus, PICIs determine animal host specificity [50]. Since their discovery in S. aureus, phage-inducible islands have proven to be ubiquitous among both Gram-positive and Gram-negative bacteria. Evidence points to known PICIs not sharing a common ancestor. Known PICI elements seem to belong to at least two distinct lineages, likely evolved on separate occasions from different prophage ancestors [42].
PICI elements have been described in human gut commensals such as E. faecalis. The presence of phage-inducible mobile elements in the genomes of human gut residents can impact the data we obtain from sequencing fecal viromes. For instance, an actively reproducing PICI element will produce high-coverage contigs that lack structural proteins typically used to identify a sequence as viral, yet contain other common markers of viral origin. This situation may further complicate our attempts to shed light on what we know as “viral dark matter.”
PICIs are known to use multiple phages as helpers [36]. Even a non-helper phage can be used by a PICI element for transfer by means of generalized transduction if the PICI genome is located in the proximity of a non-helper phage pseudo-pac site in the direction of packaging. Due to their ability to integrate into the host genome, a higher frequency of successful transduction events is observed as compared to the transfer of a random genome fragment [36].
As shown in S. aureus [36], chromosomal islands that encode their own TerS can mediate low-frequency generalized transduction. The PICI-encoded TerSwill recognizes the PICI pac site homologs and packages adjacent fragments of host DNA. Interestingly, PICI-mediated generalized transduction is not restricted to cases of active PICI reproduction. PICI TerS gene is part of operon I and is activated by the SOS response independently from PICI induction, even in the absence of an actively growing helper phage. This allows the chromosomal island to hijack any non-helper phage induced by the same SOS response for PICI-driven generalized transduction, so long as that phage uses pac-type packaging and possesses a TerL that can form a functional complex with PICI-encoded TerS. Predictably, this phenomenon drives genome organization, with pathogenicity and resistance genes clustering near to PICI pseudo-pac sites [51].
It has been shown that PICIs of S. aureus can be transferred to other Staphylococcus species and even to Listeria monocytogenes [52, 53]. The helper phage does not need to be able to reproduce in the recipient strain; it need only attach to the cell surface and inject its DNA into the cytoplasm. In this case, intergeneric transfer is possible because the helper phage uses non-specific teichoic acids for binding to its host cell and many Gram-positive genera share the structure of these polymers [54, 55]. Once in the cytoplasm, the PICI element will be able to integrate into the host genome if there is a serviceable att site present in the genome [52].
A somewhat similar yet unique case of molecular piracy is the E. coli integrative plasmid P4 (also known as satellite phage P4) [56, 57]. Similarly to PICIs, this element uses structural proteins of its helper phage P2 for transfer, redirecting the capsid size to fit its smaller genome [58, 59]. P4 is induced in the presence of its helper phage and, interestingly, can induce the helper [60]. In the absence of its helper, phage P4 can reside quiescently integrated in the host genome. Alternatively, P4 can excise and persist as a multiple copy plasmid [61]. P4 converts to its plasmid state in only ∼1% of cases [61]. Sequences similar to P4 are widespread among E. coli strains [57]. A similar but unrelated integrative plasmid has also been described in archaea [62].
Gene-transfer agents
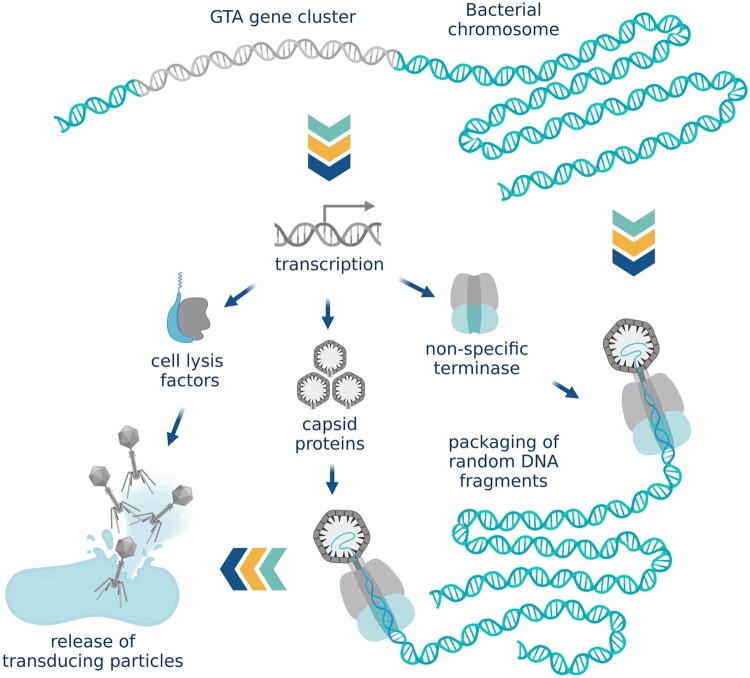
Whereas PICIs are “selfish” mobile elements with no structural genes that hijack helper phage capsids to spread in the population, gene-transfer agents (GTAs) are phage-like particles [63] that encapsidate random fragments of their “host” DNA and are released into the environment by cell lysis to facilitate adaptation and survival of the surrounding cells, but without transferring the GTA-encoding genes (Figure 6). With their typical headful capacity of 4–14 kb, GTAs are too small to carry their own full set of genes and GTAs also lack specificity in DNA packaging [64]. They are vehicles of genetic mobility rather than mobile genetic elements. Gene transfer by GTAs does not technically involve bacteriophages. However, practically speaking, GTAs are morphologically similar to tailed phages and are subsequently found in viral metagenomes [25].
Figure 6.
Gene-transfer agents. Gene-transfer agents (GTAs) are phage-like particles that encapsidate random fragments of bacterial DNA and are released into the environment by cell lysis. The size of GTA particles is not sufficient to transfer the full cluster of genes that encodes them. (Created with BioRender.com.)
GTA genes typically contain complete bacteriophage morphogenesis and lysis modules but lack the genes for replication, integration, and excision [28]. The expression of the GTA gene cluster is regulated by cellular systems and modulated by quorum sensing pathways [64, 65] rather than a phage-typical repressor mechanism. GTAs most likely use headful packaging strategy for encapsidating the genomic DNA but no specific pac sites have been identified [66].
Interestingly, it has been shown in Rhodobacter capsulatus that DNA carried by GTAs is only injected as far as the cell’s periplasm (the space between outer and inner membrane in Gram-negative bacteria) [63]. Further uptake of the DNA into the cell’s cytoplasm requires homologs of competence pathway proteins that are involved in DNA uptake via transformation [67]. The process is regulated by the same protein that controls GTA production [63].
Several unrelated families of GTAs have been identified in proteobacteria, spirochetes [64], and archaea [68]. In B. subtilis, GTA-like elements are produced by the defective resident phage PBSX [69]. Similarly to the GTAs, PBSX contains the full morphogenesis module but lacks the ability to replicate [70]. When induced, PBSX randomly packages 13-kb fragments of the host chromosome into VLPs that are released through cell lysis. The important distinction between PBSX GTA-like particles and bona fide GTAs is that PBSX particles do not seem to produce any detectable transductants, instead acting similarly to bacteriocins by inhibiting the growth of PBSX-negative cells and thus creating selective pressure for the PBSX element [64].
Similar to lateral transduction, GTAs promote genome mobility and can potentially increase the adaptability of the gut microbial community.
Fate of the transducing DNA
Unlike transformation and conjugation that can transfer DNA across large phylogenetic distances, transduction is constrained by the host range of the carrier phage particle. The transducing particles carrying cellular DNA are morphologically identical to the parent phage, including tail receptors. It will subsequently be able to attach to the cell surface of any strains or species that the original phage can attach to and can then inject the packaged DNA into the cytoplasm. However, the bacteriophage does not need to be able to successfully reproduce in the recipient cell for the transduction event to take place. In consequence, the bacteriophage’s potential taxonomic range for transduction may differ significantly from its host range as certain bacteriophages are known to adsorb on bacterial cells in which they are unable to reproduce [53]. Still, most transduction events appear to take place within a narrow taxonomic range restricted to closely related strains and species with highly similar genomes [71] and are further constrained by ecological barriers.
Once the transducing DNA enters the cell, it is either degraded by intracellular nucleases, lost in subsequent cell divisions, or gets incorporated into the host chromosome. In the case of the E. coli generalized transducing phage P1, <3% of the DNA from transducing particles is integrated into the recipient chromosome [31]. Only a fragment of the transducing DNA is typically integrated, usually significantly smaller than the entire phage genome-sized transducing DNA [31]. Between 10% and 15% of the DNA is degraded and recycled. The rest persists without replications or integration for at least several generations, becoming gradually diluted in the population.
Transducing DNA is integrated into the chromosome by homologous recombination, performed by RecA, a highly conserved DNA-repair protein. The process can only integrate DNA that has some degree of sequence identity to the recipient’s DNA, thus restricting transduction events to homologous regions. After the transducing DNA has integrated into the chromosome, there are fitness consequences to be considered. The integration itself appears to come at a cost. While the typical generation time of an E. coli culture is 30 min after a 30-min lag phase, the transductants take 180 min to double in number, meaning a lag of about three generation times. The same has been observed for Salmonella phage P22 transductants [72]. In consequence, the transductants can be outcompeted by faster-growing cells.
Genetic traits carried by the transducing DNA may also impact cell fitness. The acquired traits can be beneficial, neutral, or detrimental to the recipient. In case of the latter, the recipient cell’s progeny will be outcompeted and the acquired genes will be quickly lost in the population. Conversely, if the acquired trait dramatically increases fitness, it has the potential to rapidly spread throughout the microbial community. It has been shown that most horizontally acquired genes are neutral or nearly neutral [1] and that most completed transduction events introduce genes that are highly similar to those of the recipient [71]. Such duplicating transductions are believed to happen more often than internal gene duplication and provide the material for later adaptation, rather than directly introducing genetic innovations into the genome [71].
Another factor working against transductants is the concurring presence of intact infectious particles. Experimental studies of transduction are usually carried out under conditions that prevent the production of infectious particles and subsequent superinfection, which is not the case for naturally occurring transduction events. The cell “infected” with the transducing particle will have no bacteriophage-encoded immunity against superinfection. As the cells around it become infected with infectious particles and phage progeny is produced, the likelihood of the recipient cell getting infected and lysed becomes higher. Overall, the number of transducing particles is typically 10- to 1,000-fold higher than the number of completed transduction events [25, 31, 32].
Finding evidence of phage HGT in gut metagenomes
Reconstructing historical HGT events
Historical HGT events can be identified by observing phylogenetic incongruence; the genes that were acquired by horizontal transfer may display an evolutionary descent pattern different from that of the rest of the genome [3]. Where transduction events take place between closely related species or strains, close relation of donor and recipient will mask this phylogenetic evidence. The spatial distribution of the horizontally transferred genes in closely related lineages will be inconsistent with common ancestor patterns, and the order and composition of transferred gene clusters will often be indicative of HGT [3]. Distinguishing historical transduction events from other types of HGT poses a challenge. In the case of specialized transduction or phage morons (genes of cellular function encoded by a bacteriophage), these events can be detected by association of the acquired genes with prophage-related elements [71]. However, transduction events mediated by non-temperate phages or transduction events in which transducing DNA arrives unassociated with bacteriophage genome, as is the case for generalized transduction and GTAs, are essentially indistinguishable from other types of horizontal transfer.
Recognizing ongoing phage-mediated HGT in the gut microbiome
It is possible to observe nascent transduction events in the gut by purifying VLPs from fecal samples, sequencing the packaged DNA, and looking for bacterial sequences in the resulting viral metagenome. In theory, any bacterial DNA found in the purified VLP metagenome would constitute evidence of ongoing transduction. However, this approach is constrained by both imperfections of purification methods [73, 74] and a lack of comprehensive knowledge of bacteriophage genomes that would allow precise identification of a sequence as being of viral or bacterial origin [15, 75]. Even with the most rigorous purification techniques (e.g. using CsCl gradient centrifugation), a certain level of “noise” can produce non-specific coverage signals [25]. Genuine transduction signals can be distinguished from non-specific noise when coverage patterns of bacterial contigs in the viral metagenome are compared to bacterial genome coverage patterns from single-phage preparations of transducing bacteriophages. This approach, termed “transductomics,” makes it possible to detect coverage patterns characteristic of a specific transduction mechanism in the overwhelming amount of sequencing data from a gut virome [25]. Unlike the historical approach, this method can be used to observe all ongoing potential gene transfer through phages, including incomplete HGT events that will not lead to retention of the acquired DNA, but does not make it possible to determine the transduction efficacy. For example, GTA-like particles produced by defective B. subtilis phage PBSX do not produce transductants, but they carry host DNA and will be identified as transducing particles using this approach [25].
Discussion
Transduction frequencies are known to vary between habitats [71]. No reliable data exist on the frequency of transduction in human gut microbiome, but some findings point to the likelihood of high transduction frequencies. As observed by reconstruction of historical HGT events, both overall horizontal transfer [2] and transduction events [71] are enriched between human-associated bacteria as compared with bacteria from other environments. Moreover, it has been shown that prophages are induced during gastrointestinal transit [76], which would be expected to result in increased frequency of transduction.
Transductomics analysis suggests that 8.6% of bacterial contigs in murine gut metagenome display transduction signals, of which one-quarter have patterns inconsistent with any known mode of transduction but that are distinct from non-specific contamination or erroneous read mapping [25]. Such a high occurrence of unidentified transduction patterns illustrates that our understanding of phage-mediated gene transfer remains incomplete. An experimental gut-colonization study in mice suggests that evolution by phage HGT precedes and overshadows evolution by mutation in E. coli [77]. The same study demonstrates a multifaceted context for phage HGT events in gut microbiomes involving the interplay between environmental conditions, phage–host and phage–phage interactions, prophage immunity, and metabolic adaptations.
It is highly likely that phage-mediated horizontal transfer in the gut microbiome is involved in health and disease outcomes in humans, but no reliable data exist to confirm or refute this hypothesis. Gut microbiome and virome alterations have been linked to the immune system [78, 79], mental health [80–83], obesity [84–86], type 2 diabetes [84, 85], and potentially coronary heart disease [87]. Specific mechanisms of gut microbiome involvement have not yet been determined. It is possible that a balance of certain metabolic functions or virulence factors underlies some of the above correlations, and the genes determining such functions might be mobilized through phage-mediated transfer. Transfer of antimicrobial resistance genes by phages remains a subject of debate [75, 88–90], showcasing the need for further research and better understanding of phage-mediated gene transfer, as well as the need to develop more reliable methodology to analyse viral metagenomes.
We believe that understanding phage-mediated gene transfer is crucial in interpreting complex ecological interactions within the human gut microbiome and is therefore worthy of further investigation.
Authors’ Contributions
Conceptualization: A.N.S. and T.B.; Writing—original draft: T.B.; Writing—revision and editing: T.B., A.N.S., R.P.R., and C.H.; funding acquisition: A.N.S., R.P.R., and C.H. The authors read and approved the final manuscript.
Funding
This publication has emanated from financial support from Science Foundation Ireland [grant number SFI/12/RC/2273_P2] and Wellcome Trust under a Wellcome Trust Research Career Development Fellowship [220646/Z/20/Z] (A.N.S.). This research was funded in whole, or in part, by the Wellcome Trust [220646/Z/20/Z]. For the purpose of open access, the authors have applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Acknowledgements
We thank Bianca Govi (APC Microbiome Ireland, UCC) for discussion and comments that greatly improved the manuscript.
Conflict of Interest
None declared.
References
- 1. Soucy SM, Huang J, Gogarten JP.. Horizontal gene transfer: building the web of life. Nat Rev Genet 2015;16:472–82. [DOI] [PubMed] [Google Scholar]
- 2. Smillie CS, Smith MB, Friedman J. et al. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 2011;480:241–4. [DOI] [PubMed] [Google Scholar]
- 3. Zhaxybayeva O, Doolittle WF.. Lateral gene transfer. Curr Biol 2011;21:R242–6. [DOI] [PubMed] [Google Scholar]
- 4. Puigbò P, Wolf YI, Koonin EV.. The tree and net components of prokaryote evolution. Genome Biol Evol 2010;2:745–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yaffe E, Relman DA.. Tracking microbial evolution in the human gut using Hi-C reveals extensive horizontal gene transfer, persistence, and adaptation. Nat Microbiol 2020;5:343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baquero F, Martínez JL, F Lanza V Evolutionary pathways and trajectories in antibiotic resistance. Clin Microbiol Rev 2021;34. doi:10.1128/CMR.00050-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Penadés JR, Chen J, Quiles-Puchalt N. et al. Bacteriophage-mediated spread of bacterial virulence genes. Curr Opin Microbiol 2015;23:171–8. [DOI] [PubMed] [Google Scholar]
- 8. Hurwitz BL, U'Ren JM.. Viral metabolic reprogramming in marine ecosystems. Curr Opin Microbiol 2016;31:161–8. [DOI] [PubMed] [Google Scholar]
- 9. Hehemann J-H, Correc G, Barbeyron T. et al. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 2010;464:908–12. [DOI] [PubMed] [Google Scholar]
- 10. Dubey GP, Ben-Yehuda S.. Intercellular nanotubes mediate bacterial communication. Cell 2011;144:590–600. [DOI] [PubMed] [Google Scholar]
- 11. Mashburn-Warren LM, Whiteley M. Special delivery: vesicle trafficking in prokaryotes. Mol Microbiol 2006;61:839–46. [DOI] [PubMed] [Google Scholar]
- 12. Bitto NJ, Chapman R, Pidot S.. Bacterial membrane vesicles transport their DNA cargo into host cells. Sci Reports 2017;7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Velimirov B, Hagemann S.. Mobilizable bacterial DNA packaged into membrane vesicles induces serial transduction. Mob Genet Elements 2011;1:80–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiura HX, Kogure K, Hagemann S et al. Evidence for particle-induced horizontal gene transfer and serial transduction between bacteria. FEMS Microbiol Ecol 2011;76:576–91. [DOI] [PubMed] [Google Scholar]
- 15.Shkoporov AN, Hill C. Bacteriophages of the human gut: the “known unknown” of the microbiome. Cell Host Microbe 2019;25:195–209. [DOI] [PubMed] [Google Scholar]
- 16. Rao VB, Feiss M.. Mechanisms of DNA packaging by large double-stranded DNA viruses. Annu Rev Virol 2015;2:351–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Casjens SR, Gilcrease EB.. Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Methods Mol Biol 2009;502:91–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet 2008;42:647–81. [DOI] [PubMed] [Google Scholar]
- 19.Lin H, Black LW. DNA requirements in vivo for phage T4 packaging. Virology 1998;242:118–27. [DOI] [PubMed] [Google Scholar]
- 20.Feiss M, Widner W, Miller G et al. Structure of the bacteriophage lambda cohesive end site: location of the sites of terminase binding (cosB) and nicking (cosN). Gene 1983;24:207–18. [DOI] [PubMed] [Google Scholar]
- 21. Stewart CR, Casjens SR, Cresawn SG. et al. The genome of Bacillus subtilis bacteriophage SPO1. J Mol Biol 2009;388:48–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pajunen MI, Elizondo MR, Skurnik M. et al. Complete nucleotide sequence and likely recombinatorial origin of bacteriophage T3. J Mol Biol 2002;319:1115–32. [DOI] [PubMed] [Google Scholar]
- 23. Golais F, Hollý J, Vítkovská J.. Coevolution of bacteria and their viruses. Folia Microbiol 2012;58:177–86. [DOI] [PubMed] [Google Scholar]
- 24. Ranquet C, Toussaint A, De Jong H. et al. Control of bacteriophage mu lysogenic repression. J Mol Biol 2005;353:186–95. [DOI] [PubMed] [Google Scholar]
- 25. Kleiner M, Bushnell B, Sanderson KE. et al. Transductomics: sequencing-based detection and analysis of transduced DNA in pure cultures and microbial communities. Microbiome 2020;8:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morse ML, Lederberg EM, Lederberg J.. Transduction in Escherichia coli K-12. Genetics 1956;41:142–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sausset R, Petit MA, Gaboriau-Routhiau V. et al. New insights into intestinal phages. Mucosal Immunol 2020;13:205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Novick RP, Ram G.. The floating (pathogenicity) island: a genomic dessert. Trends Genet 2016;32:114–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hanks MC, Newman B, Oliver IR. et al. Packaging of transducing DNA by bacteriophage P1. Mol Gen Genet MGG 1988;214:523–32. [DOI] [PubMed] [Google Scholar]
- 30.Ikeda H, Tomizawa JI. Transducing fragments in generalized transduction by phage P1. I. Molecular origin of the fragments. J Mol Biol 1965;14:85–109. [DOI] [PubMed] [Google Scholar]
- 31. Sandri RM, Berger H.. Bacteriophage P1-mediated generalized transduction in Escherichia coli: Fate of transduced DNA in Rec+ and RecA− recipients. Virology 1980;106:14–29. [DOI] [PubMed] [Google Scholar]
- 32. Hertman I, Luria SE.. Transduction studies on the role of a rec+ gene in the ultraviolet induction of prophage lambda. J Mol Biol 1967;23:117–33. [DOI] [PubMed] [Google Scholar]
- 33. Chen J, Quiles-Puchalt N, Chiang YN. et al. Genome hypermobility by lateral transduction. Science 2018;362:207–12. [DOI] [PubMed] [Google Scholar]
- 34. Fillol-Salom A, Bacigalupe R, Humphrey S. et al. Lateral transduction is inherent to the life cycle of the archetypical Salmonella phage P22. Nat Commun 2021;12:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lindsay JA, Ruzin A, Ross HF. et al. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol Microbiol 1998;29:527–43. [DOI] [PubMed] [Google Scholar]
- 36. Penadés JR, Christie GE.. The phage-inducible chromosomal islands: a family of highly evolved molecular parasites. Annu Rev Virol 2015;2:181–201. [DOI] [PubMed] [Google Scholar]
- 37.Tormo-Más MA, Mir I, Shrestha A et al. Moonlighting bacteriophage proteins derepress staphylococcal pathogenicity islands. Nature 2010;465:779–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dearborn AD, Wall EA, Kizziah JL. et al. Competing scaffolding proteins determine capsid size during mobilization of Staphylococcus aureus pathogenicity islands. Elife 2017;6. doi: 10.7554/ELIFE.30822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Poliakov A, Chang JR, Spilman MS. et al. Capsid size determination by Staphylococcus aureus pathogenicity island SaPI1 involves specific incorporation of SaPI1 proteins into procapsids. J Mol Biol 2008;380:465–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maiques E, Ubeda C, Tormo MA. et al. Role of staphylococcal phage and SaPI integrase in intra- and interspecies SaPI transfer. J Bacteriol 2007;189:5608–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ram G, Chen J, Kumar K et al. Staphylococcal pathogenicity island interference with helper phage reproduction is a paradigm of molecular parasitism. Proc Natl Acad Sci USA 2012;109:16300–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fillol-Salom A, Martínez-Rubio R, Abdulrahman RF. et al. Phage-inducible chromosomal islands are ubiquitous within the bacterial universe. ISME J 2018;12:2114–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Quiles-Puchalt N, Carpena N, Alonso JC. et al. Staphylococcal pathogenicity island DNA packaging system involving cos-site packaging and phage-encoded HNH endonucleases. Proc Natl Acad Sci USA 2014;111:6016–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ram G, Chen J, Ross HF. et al. Precisely modulated pathogenicity island interference with late phage gene transcription. Proc Natl Acad Sci USA 2014;111:14536–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Matos R. Enterococcus faecalis prophage dynamics and contributions to pathogenic traits. PLoS Genet 2013;9. doi: 10.1371/journal.pgen.1003539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Seed KD, Lazinski DW, Calderwood SB. et al. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature 2013;494:489–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McKitterick AC, Seed KD.. Anti-phage islands force their target phage to directly mediate island excision and spread. Nat Commun 2018;9. doi: 10.1038/S41467-018-04786-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Frígols B, Quiles-Puchalt N, Mir-Sanchis I. et al. Virus satellites drive viral evolution and ecology. PLoS Genet 2015;11. doi: 10.1371/JOURNAL.PGEN.1005609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuroda M, Yamashita A, Hirakawa H et al. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc Natl Acad Sci USA 2005;102:13272–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viana D, Blanco J, Tormo-Más MA et al. Adaptation of Staphylococcus aureus to ruminant and equine hosts involves SaPI-carried variants of von Willebrand factor-binding protein. Mol Microbiol 2010;77:1583–94. [DOI] [PubMed] [Google Scholar]
- 51. Chen J, Ram G, Penadés JR. et al. Pathogenicity island-directed transfer of unlinked chromosomal virulence genes. Mol Cell 2015;57:138–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen J, Novick RP.. Phage-mediated intergeneric transfer of toxin genes. Science 2009;323:139–41. [DOI] [PubMed] [Google Scholar]
- 53. Chen J, Carpena N, Quiles-Puchalt N. et al. Intra- and inter-generic transfer of pathogenicity island-encoded virulence genes by cos phages. ISME J2015;9:1260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Winstel V, Liang C, Sanchez-Carballo P. et al. Wall teichoic acid structure governs horizontal gene transfer between major bacterial pathogens. Nat Commun 2013;4. doi: 10.1038/NCOMMS3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Novick RP, Ram G.. Staphylococcal pathogenicity islands—movers and shakers inthe genomic firmament. Curr Opin Microbiol 2017;38:197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Christie GE, Dokland T.. Pirates of the caudovirales. Virology 2012;434:210–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lindqvist BH, Dehò G, Calendar R. Mechanisms of genome propagation and helper exploitation by satellite phage P4. Microbiol Rev 1993;57:683–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shore D, Dehò G, Tsipis J. et al. Determination of capsid size by satellite bacteriophage P4. Proc Natl Acad Sci USA 1978;75:400–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kizziah JL, Rodenburg CM, Dokland T.. Structure of the capsid size-determining scaffold of “satellite” bacteriophage P4. Viruses 2020;12. doi: 10.3390/V12090953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Six EW, Lindqvist BH.. Mutual derepression in the P2–P4 bacteriophage system. Virology 1978;87:217–30. [DOI] [PubMed] [Google Scholar]
- 61. Briani F, Dehò G, Forti F. et al. The plasmid status of satellite bacteriophage P4. Plasmid 2001;45:1–17. [DOI] [PubMed] [Google Scholar]
- 62.Arnold HP, She Q, Phan H et al. The genetic element pSSVx of the extremely thermophilic crenarchaeon Sulfolobus is a hybrid between a plasmid and a virus. Mol Microbiol 1999;34:217–26. [DOI] [PubMed] [Google Scholar]
- 63. Bárdy P, Füzik T, Hrebík D. et al. Structure and mechanism of DNA delivery of a gene transfer agent. Nat Commun 2020;11:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lang AS, Zhaxybayeva O, Beatty JT.. Gene transfer agents: phage-like elements of genetic exchange. Nat Rev Microbiol 2012;10:472–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schaefer AL, Taylor TA, Beatty JT. et al. Long-chain acyl-homoserine lactone quorum-sensing regulation of rhodobacter capsulatus gene transfer agent production. J Bacteriol 2002;184:6515–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tomasch J, Wang H, Hall ATK. et al. Packaging of Dinoroseobacter shibae DNA into gene transfer agent particles is not random. Genome Biol Evol 2018;10:359–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Brimacombe CA, Ding H, Johnson JA. et al. Homologues of genetic transformation DNA import genes are required for rhodobacter capsulatus gene transfer agent recipient capability regulated by the response regulator CtrA. J Bacteriol 2015;197:2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bertani G. Transduction-like gene transfer in the methanogen Methanococcus voltae. J Bacteriol 1999;181:2992–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Anderson LM, Bott KF.. DNA packaging by the Bacillus subtilis defective bacteriophage PBSX. J Virol 1985;54:773–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wood HE, Dawson MT, Devine KM. et al. Characterization of PBSX, a defective prophage of Bacillus subtilis. J Bacteriol 1990;172:2667–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Popa O, Landan G, Dagan T.. Phylogenomic networks reveal limited phylogenetic range of lateral gene transfer by transduction. 2017;11:543–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ebel-Tsipis J, Fox MS, Botstein D.. Generalized transduction by bacteriophage P22 in Salmonella typhimurium. II. Mechanism of integration of transducing DNA. J Mol Biol 1972;71:449–69. [DOI] [PubMed] [Google Scholar]
- 73. Kleiner M, Hooper LV, Duerkop BA.. Evaluation of methods to purify virus-like particles for metagenomic sequencing of intestinal viromes. BMC Genomics 2015;16:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Shkoporov AN, Ryan FJ, Draper LA. et al. Reproducible protocols for metagenomic analysis of human faecal phageomes. Microbiome 2018;6:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Enault F, Briet A, Bouteille L. et al. Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J2017;11:237–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Oh J-H, Alexander LM, Pan M. et al. Dietary fructose and microbiota-derived short-chain fatty acids promote bacteriophage production in the gut symbiont lactobacillus reuteri. Cell Host Microbe 2019;25:273–84.e6. [DOI] [PubMed] [Google Scholar]
- 77. Frazão N, Sousa A, Lässig M. et al. Horizontal gene transfer overrides mutation in Escherichia coli colonizing the mammalian gut. Proc Natl Acad Sci USA 2019;116:17906–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mukhopadhya I, Segal JP, Carding SR. et al. The gut virome: the “missing link” between gut bacteria and host immunity? Therap Adv Gastroenterol 2019;12:1756284819836620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Neil JA, Cadwell K.. The intestinal virome and immunity. J Immunol 2018;201:1615–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Clapp M, Aurora N, Herrera L. et al. Gut microbiota’s effect on mental health: the gut-brain axis. Clin Pract 2017;7:131–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Järbrink-Sehgal E, Andreasson A.. The gut microbiota and mental health in adults. Curr Opin Neurobiol 2020;62:102–14. [DOI] [PubMed] [Google Scholar]
- 82. Lai J, Jiang J, Zhang P. et al. Gut microbial clues to bipolar disorder: state‐of‐the‐art review of current findings and future directions. Clin Transl Med 2020;10. doi: 10.1002/CTM2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Malan-Muller S, Valles-Colomer M, Raes J. et al. The gut microbiome and mental health: Implications for anxiety- and trauma-related disorders. 2018;22:90–107. [DOI] [PubMed] [Google Scholar]
- 84. Yang K, Niu J, Zuo T. et al. Alterations in the gut virome in obesity and type 2 diabetes mellitus. Gastroenterology 2021;161:1257–69.e13. [DOI] [PubMed] [Google Scholar]
- 85. Rasmussen TS, Mentzel CMJ, Kot W. et al. Faecal virome transplantation decreases symptoms of type 2 diabetes and obesity in a murine model. Gut 2020;69:2122–30. [DOI] [PubMed] [Google Scholar]
- 86. Bikel S, López-Leal G, Cornejo-Granados F. et al. Gut dsDNA virome shows diversity and richness alterations associated with childhood obesity and metabolic syndrome. iScience 2021;24:102900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Guo L, Hua X, Zhang W. et al. Viral metagenomics analysis of feces from coronary heart disease patients reveals the genetic diversity of the Microviridae. Virol Sin 2017;32:130–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Modi SR, Lee HH, Spina CS. et al. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 2013;499:219–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Calero-Cáceres W, Ye M, Balcázar JL.. Bacteriophages as environmental reservoirs of antibiotic resistance. Trends Microbiol 2019;27:570–7. [DOI] [PubMed] [Google Scholar]
- 90. Colavecchio A, Cadieux B, Lo A. et al. Bacteriophages contribute to the spread of antibiotic resistance genes among foodborne pathogens of the Enterobacteriaceae family—a review. Front Microbiol 2017;8:1108. [DOI] [PMC free article] [PubMed] [Google Scholar]