Abstract
Egg production is an important economic trait in laying chickens as higher yields bring higher profits. Small yellow follicle (SYFL) development is a key determinant of chicken reproductive performance; however, the majority of SYFLs are not selected during the process of chicken reproduction and thus, atresia occurs. Although there have been numerous omic studies focused on egg production, the molecular mechanisms involved are still not well-understood. In this study, we used high-throughput technology to analyze the differences between the SYFL mRNA transcriptomes of high– (H) and low–egg-yielding (L) Taihang layer hens, with the aim of identifying the potential candidate genes involved in controlling the rate of egg production. We constructed six cDNA libraries, three from H and three from L Taihang hens and then performed high-throughput sequencing. Comparison of the H and L groups showed 415 differentially expressed genes (DEGs). In the high-yield group, 226 were upregulated and 189 were downregulated. Differentially enriched biological functions and processes were identified using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) database analysis. Ten of the candidate DEGs we identified (DRD1, MC5R, PCK1, CTSA, TGFBR3, AGO4, SLIT2, RGS1, SCNN1B, and ZP3) have been identified in previous studies as being involved in the development of small yellow follicles. DRD1 was significantly enriched in the gap junction pathway, which is an important pathway in chicken granulosa cells (GCs) to pass nutrition to an oocyte. Homology analysis showed that DRD1 was highly conserved in numerous species, indicating that it may be a productive target for improving egg production. Evidence from bioinformatics analysis revealed that gga-miR-302a-3p putatively targets the 3′UTR region of DRD1. We then identified the functions of gga-miR-302a-3p in follicular granulosa cell proliferation by targeting DRD1. RT-qPCR analysis showed that DRD1 and miR-302a-3p expression were inversely related in the SYLs of high and low egg-yielding chickens. Luciferase assays showed that miR-302a-3p targets the 3′UTR of DRD1, and overexpression of miR-302a-3p significantly inhibits the expression of DRD1 in chicken GCs (p < 0.01). Functional experiments revealed that by targeting DRD1, miR-302a-3p acts as an inhibitor of GC proliferation. Taken together, we concluded that miR-302a-3p affects chicken GC proliferation by targeting DRD1. Our data expanded the knowledge base of genes whose functions are important in egg production and the molecular mechanisms of high-yield egg production in chicken small yellow follicles.
Keywords: DRD1, GC proliferation, Taihang chicken, small yellow follicles, miR-302a-3p
Introduction
Follicle development, a key factor in determining reproductive performance, is influenced by physiology and the environment. The characteristics of follicle maturation in hens are like those in mammals, making them a useful model for studying follicle development in general (Bahr, 1991). The process of ovulation occurs through the recruitment of primordial follicles followed by primary follicles, small white follicles, large white follicles, small yellow follicles (SYFLs), and hierarchal follicles that progress toward maturity (Johnson, 2012). During the peak laying period of hens, ovarian hierarchal follicles develop from a pool of SYFLs each day (Hocking, 2009). In the process of hen egg production, the molecular and cellular mechanisms by which the pre-hierarchical follicles are selected to enter the preovulatory stage are well-known (Fortune et al., 2004; Johnson and Lee, 2016). It is estimated that there are approximately 12,000 oocytes in a sexually mature hen; however, only a few hundred of them are selected to mature and reach the ovulation stage (Onagbesan et al., 2009). Most SYFLs are not selected and undergo atresia (Tilly et al., 1991; Johnson et al., 1996). The SYFs continue to develop into the eight largest orderly arranged pre-ovulatory follicles, F8, F7, F6, F5, F4, F3, F2, F1 and proceed to ovulation (Huang et al., 2008). The development of SYFLs is crucial for ovulation in poultry (Dong et al., 2014).
Follicle development is a complex process, with numerous genes and pathways involved in their proliferation and differentiation (Woods and Johnson, 2005; Johnson et al., 2008). Dopamine receptor D1 (DRD1) is a type of D1-like receptor. In avians, dopamine participates in stimulating and inhibiting the secretion of prolactin (PRL) and plays a key role in the onset and maintenance of incubation behavior (Sharp et al., 1988; March et al., 1994; Youngren et al., 1995). Tempfli et al. found that DRD1 promotes ovarian development by PRL and induces SYF differentiation in hens. Xu et al. reported that polymorphisms in DRD1 affect the egg- laying performance of hens and that its haplotypes are significantly associated with some egg production traits in chickens (Xu et al., 2010; Tempfli et al., 2015). Wang et al. reported that DRD1 may be a target gene for improving the characteristics of duck reproduction (Wang et al., 2012). Schnell et al. and Chaiseha et al. found that DRD1 is expressed broadly in the hypothalamus and pituitary of turkeys and that its expression is related to reproductive function (Schnell and You, 1999; Chaiseha et al., 2003). In mammals, DRD1 is expressed in many tissues, including the hypothalamus and thalamus (Fremeau et al., 1991; Jackson and Westlind-Danielsson, 1994), though not in the cerebellum, hippocampus, mesencephalon, or pituitary tissues (Monsma et al., 1990; Niznik and Van Tol, 1992; Vallone et al., 2000). The distribution of DRD1 in the avian forebrain is largely the same as in mammals (Schnabel et al., 1997). Put together, these studies have revealed that DRD1 is likely involved in regulating reproduction in birds.
The Taihang chicken, which produces high-quality eggs, is a breed native to Hebei Province in China. In this study, we used RNA-seq to investigate the molecular mechanisms of ovary follicle growth in high– and low–egg-yielding Taihang chickens. Differentially expressed genes were identified and then further examined to elucidate the potential functions and mode of regulation. We identified the DRD1 as an important player in ovary follicle growth and miR-302a-3p as being potentially associated with follicle growth. These results provided a better understanding of chicken SYFL development and offered a target for subsequent investigation into improving the SYFL selected ratios.
Materials and Methods
Animals and Tissue Preparation
Four hundred Taihang hens raised at the Taihang Chicken Industry Co., Ltd. poultry breeding farm in Hebei Province were used in this study. At 33 weeks of age, three high-yield (high egg production, H) and three low-yield (low egg production, L) hens were selected from the same batch of laying hens. The laying rate in the H group was 68.33 ± 0.40%, and in the L group, it was 48.12 ± 0.40% (p < 0.05). The chickens had free access to food and water throughout the experiment and 12 h of natural light with temperatures between 17 and 25°C. The egg number was recorded daily at 16:00. The chickens were slaughtered by exsanguination, and the SYFLs were immediately harvested from the ovaries and stored in liquid nitrogen.
RNA Isolation, Library Construction, and RNA-Seq
In accordance with the manufacturer’s instructions, total RNA was isolated from the six samples using TRIzol reagent (Life Technologies, Carlsbad, CA). The integrity of the isolated RNA (degradation and contamination) was determined by 1% agarose gels. RNA purity and concentration were determined by spectrophotometry using a NanoDrop ND-2000 (Implen, Westlake Village, CA). The RNA integrity number (RIN) of the samples ranged from 8.0 to 9.2; an RIN greater than 8.0 was considered acceptable for RNA-seq.
3 μg of the total RNA from each sample was used as the input material for library construction. Six libraries were generated using the NEB Next Ultra RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA) according to the manufacturer’s instruction. We added index codes to each sample for later identification. An Agilent Bioanalyzer 2100 system was used to determine the insert size of the libraries. The index-coded samples were clustered using the cBot Cluster Generation System and the TruSeq PE Cluster Kit v3-cBot-HS (Illumina, San Diego, CA), following the manufacturer’s instructions. The libraries were sequenced using an Illumina Hiseq X Ten platform to generate paired-end reads 150 bp in length (BGI Genomics Co., Ltd.).
Raw Data Processing and Sequence Alignment
Raw reads were filtered using Trimmomatic (v0.36) (Bolger et al., 2014). In this step, the reads were removed from the data set if they included adapters, poly-N, or were of low quality. The clean reads were mapped to the Gallus gallus (v6.0) genome using HISAT2 (ver.2.2.1) with default parameters (Kim et al., 2015). Cufflinks were used to predict novel transcripts by comparing reconstructed transcripts with known transcripts (ver. 2.1.1) (Trapnell et al., 2012).
Analysis of Differentially Expressed Genes
The reads mapped to each gene were counted by HiSeq (ver. 0.6.1). Gene expression levels were normalized by the Fragments Per Kilobase of transcript per Million map reads (FPKM) method (Trapnell et al., 2010). DESeq2 was used to analyze the differentially expressed genes from high– and low–egg-yielding chicken SYFLs (Anders and Huber, 2010). Genes with adjusted p ≤ 0.05 and |log2 (fold change) | ≥ 1 were classified as differentially expressed genes (DEGs).
Confirmation of Differentially Expressed Genes
To validate the RNA-seq data, the expression levels of eight randomly selected DEGs were determined by RT-qPCR. Using a Fast First-Strand cDNA Synthesis kit (Tiangen, Beijing, China), RNA was reverse-transcribed onto cDNA. The primers used in the RT-qPCR were designed by Primer Premier 5 software (Premier Biosoft, Palo Alto, CA) and are shown in Supplementary Table S1. Each PCR reaction consisted of 1 μL of the cDNA template, 0.5 μL of forward and reverse primer (10 μmol), 10 μL of SYBR Green Master Mix (Tiangen, Beijing, China), and 8 μL of RNAase-free water. The amplification conditions were as follows: denaturation at 95°C for 5 min and then 40 cycles of amplification (95°C for 10 s and 60°C for 30 s). After amplification, melting curve analysis was performed by heating the samples to 95°C for 15 s, then cooling to 60°C for 1 min, followed by heating to 95°C at a rate of 0.3°C/s. Each sample was tested in triplicate. The expression level of the genes was calculated by the 2 −ΔΔCt method using GAPDH as a reference control (Livak and Schmittgen, 2001).
DEG Functional Annotation Using GO and KEGG Enrichment Analyses
Gene Ontology (GO) enrichment analysis of the DEGs was carried out using the GOseq R package (ver. 2.12), with correction for gene length bias. Wallenius’ noncentral hypergeometric distribution was used to calculate p-values. GO terms with a corrected p value <0.05 were considered significantly enriched (Young et al., 2010). KOBAS (2.0) (Mao et al., 2005) was used to test for statistically significant enrichment of DEGs in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database.
Prediction of miRNAs Targeting DRD1
The miRNA-targeting DRD1 gene was predicted using TargetScan (http://www.targetscan.org/) and miRanda (http://www.miranda.org). miRNA–target relationships were considered significant at p < 0.05. To predict the conserved function of the DRD1 gene, the sequencing results were subjected to a BLAST search (NCBI, http://blast.ncbi.nlm.nih.gov/Blast.cgi) to retrieve gene sequences homologous to DRD1. Multiple amino acid sequences were compared using DNAMAN software, and a phylogenetic tree was constructed using the Neighbor-Joining method in MEGA7 software (Kumar et al., 2016).
Functional Assays
Vector Construction
Two recombinant psiCHECK2 vectors containing the predicted miR-302a-3p target site in the 3′- UTR sequence of DRD1 were constructed using XhoI and NotI (Takara, Dalian, China). Wild and mutant-type recombinants are referred to as DRD1-3′UTR-WT and DRD1-3′UTR-MUT, respectively. The sequences of the wild and mutant- types were synthesized, and the sequences are shown in Supplementary Table S2. The miR-302a-3p sequence (CCACCACUUAAACGUGGAUGUACUUGCUUUGAAACUAAAGAAGUAAGUGCUUCCAUGUUUUGGUGAUGG), was synthesized, digested with BamHI and EcoRI, and then inserted into pHBLV-U6-MCS-CMV-ZsGreen-PGK-PURO to obtain the LV021-miR-302a-3p. An empty vector served as a negative control (NC).
Cell Culture and Transfection
Following the methods described by Sharma et al., chicken primary granulosa cells (GCs) were isolated from SYFLs of ovarian tissues (Sharma and Sharma, 2020). The cells were seeded in 6-mm plates at a density of 105 cells and maintained with a complete medium (DMEM/F12 (1:1), 10% FBS, and 1% penicillin/streptomycin) as described by Yang et al. (2017). HEK293T cells, obtained from our laboratory stock were seeded in 6-mm plates at a density of 105 cells and maintained with a complete medium (DMEM/F12 (1:1), 10% FBS, and 1% penicillin/streptomycin).
Luciferase Assays
To validate the miRNA targets, 293T cells were cultured into 24-well plates and cotransfected with 200 ng DRD1-3′UTR-wt or DRD1-3′UTR-mut and 10 µL of miR-302a-3p, miR-302a-3p mimic, or negative control (NC) using Lipofectamine 2000 (Invitrogen). After 48 h, the cells were collected to measure the luciferase activity using a Dual-Luciferase Reporter Assay System Kit (Promega, WI, United States). All the experiments were performed in triplicates.
Cell Counting Kit-8 Assay
GCs were aliquoted into 96-well plates with approximately 100 μL of the cell suspension (about 103−104 cells) per well in three replicates. The cells were incubated for 2–4 h at 37°C after cell apposition for the CCK-8 assay. A Cell Counting Kit-8 (Beyotime, Beijing, China) was used to detect the GC proliferation following the manufacturer’s protocol. Absorbance at 450 nm was read at 0, 6, 12, 24, 48, and 72 h after the addition of CCK-8 solution.
Statistical Analysis
Relative gene expression obtained from the RT-qPCR data was calculated using the 2 −∆∆Ct method (Stocks et al., 2012). All data were expressed as the mean ± SE (standard error) with at least three independent replicates and visualized using the “ggplot2” package in R (version 3.2.2, the University of Auckland, Auckland, New Zealand) and GraphPad Prism 7 software (San Diego, CA, United States). Student’s t-test was used to evaluate significant differences between the H and L groups. All data are shown as means ± standard error. P values <0.05 were defined as significant, and p values <0.01 were highly significant.
Results
RNA-Sequencing Data
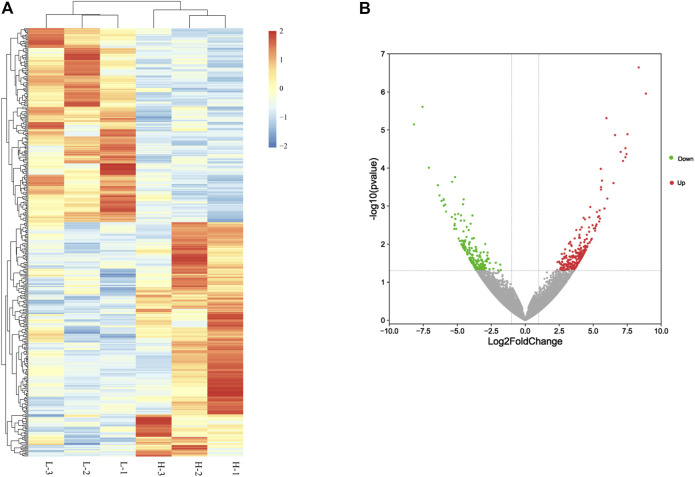
A summary of the sequencing data (Supplementary Table S3) shows that these libraries have highly consistent bulk statistics. More than 7 Gb of data from each library were obtained after filtering to remove low-quality reads and adapter sequences. The libraries yielded an average of 93.66% clean reads, and more than 93% of the total reads were mapped to the chicken reference genome in every library. The Q30 value was over 91% for every sample. These results demonstrated that the sequencing data from each library were of high quality. We used DESeq2 to screen for key candidate genes involved in ovary development and identified 415 differentially expressed genes between the H and L egg-laying groups (Figure 1A; Supplementary Table S4), of which 226 were upregulated and 189 were downregulated in the H group (Figure 1B). The log2 fold change values ranged from −7.08 to 8.87.
FIGURE 1.
Differentially expressed genes between high– and low–egg-yielding groups of Taihang chicken. (A) Hierarchical clusters of differentially expressed genes. H represents the high–egg-producing group and L represents the low–egg-producing group. (B) Volcano plot of differentially expressed genes in chicken small yellow follicles. Red dots represent upregulated genes and green dots represent downregulated genes.
Characterization of DEGs Using GO and KEGG Pathway Analyses
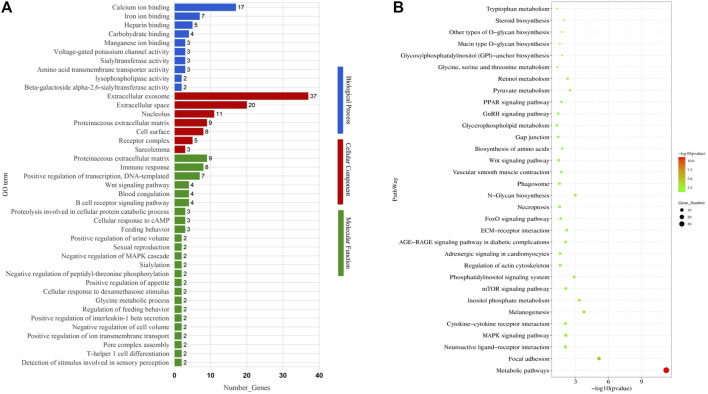
To identify the potential functional roles for the DEGs, we used GOseq to perform GO enrichment analysis. Within the category “biological process”, the most abundant GO terms were “extracellular exosome,” “positive regulation of protein kinase B signaling,” and “proteolysis involved in the cellular protein catabolic process.” For the “cellular component” and “molecular function” categories, the GO terms included “integral component of plasma membrane,” “proteinaceous extracellular matrix,” and “extracellular exosome” (Figure 2A; Supplementary Table S5).
FIGURE 2.
Gene Ontology (GO) terms and KEGG pathways enriched of differentially expressed genes. (A) GO terms. (B) KEGG pathways.
To further identify gene functions and interactions, we also subjected the DEGs to KEGG pathway analysis, resulting in the identification of 105 enriched pathways (p < 0.05). Out of these, 32 enriched pathways were related to follicular development, including the GnRH signaling pathway, MAPK signaling pathway, steroid biosynthesis, gap junction, and mTOR signaling pathway (Figure 2B; Table 1).
TABLE 1.
Significantly enriched pathways with functions in follicle development.
| Pathway ID | Pathway term | Target gene list |
|---|---|---|
| gga01100 | Metabolic pathways | NADK, MSMO1, ASL2, PCK1, ST6GAL2, NSDHL, ST6GAL1, DHTKD1, SMPD1, PGAM1, PLA2G4A, CYP27A1, GCNT2, ACAT2, MGAT5B, BCO1, NDUFA12, GLO1, PLCB2, ISYNA1, CRYL1, CYP21A1, PIGK, MAN2A2, PIGS, PHGDH, GGT5, BLVRA, GALNT7, HPSE, NME6, INPP5D, GALNT10, PIP5K1B, CDS2, INPP5K, ELOVL6, CYP26A1, ATP6V1C2 |
| gga04510 | Focal adhesion | PGF, LAMA1, LAMA2, PDGFC, VAV3, PARVG, CAPN2, COL6A2, EGF, THBS4 |
| gga04916 | Melanogenesis | PLCB2, FZD4, GNAO1, WNT6, EDN1, WNT16 |
| gga00562 | Inositol phosphate metabolism | PIP5K1B, PLCB2, INPP5K, INPP5D, ISYNA1 |
| gga00510 | N-glycan biosynthesis | MGAT5B, ST6GAL2, MAN2A2, ST6GAL1 |
| gga04070 | Phosphatidylinositol signaling system | PIP5K1B, PLCB2, INPP5K, INPP5D, CDS2 |
| gga00620 | Pyruvate metabolism | GLO1, ACAT2, PCK1 |
| gga00830 | Retinol metabolism | PNPLA4, BCO1, CYP26A1 |
| gga04512 | ECM–receptor interaction | LAMA1, LAMA2, COL6A2, THBS4 |
| gga04010 | MAPK signaling pathway | PGF, TGFA, PDGFC, CSF1R, MAPK12, PLA2G4A, EGF |
| gga04150 | mTOR signaling pathway | ATP6V1C2, SLC7A5, FZD4, WNT6, WNT16 |
| gga04933 | AGE-RAGE signaling pathway in diabetic complications | EDN1, PLCB2, PRKCZ, MAPK12 |
| gga04080 | Neuroactive ligand–receptor interaction | OPRM1, CCKAR, LEPR, GRIN2C, NPY2R, EDN1, MC5R, P2RX7 |
| gga04060 | Cytokine–cytokine receptor interaction | CCL4, CSF1R, IL18R1, LEPR, TNFRSF11B, CXCL14 |
| gga00100 | Steroid biosynthesis | MSMO1, NSDHL |
| gga01230 | Biosynthesis of amino acids | ASL2, PHGDH, PGAM1 |
| gga00514 | Other types of O-glycan biosynthesis | ST6GAL2, ST6GAL1 |
| gga00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | PIGS, PIGK |
| gga03320 | PPAR signaling pathway | CYP27A1, PCK1, FABP3 |
| gga04270 | Vascular smooth muscle contraction | EDN1, PLCB2, PLA2G4A, RAMP2 |
| gga04068 | FoxO signaling pathway | MAPK12, PCK1, EGF, FBXO32 |
| gga04810 | Regulation of actin cytoskeleton | BRK1, PIP5K1B, VAV3, PDGFC, EGF |
| gga04261 | Adrenergic signaling in cardiomyocytes | PLCB2, MAPK12, PPP2R2C, ATP1B1 |
| gga00512 | Mucin-type O-glycan biosynthesis | GALNT7, GALNT10 |
| gga04217 | Necroptosis | PLA2G4A, HMGB1, SMPD1, CAPN2 |
| gga04145 | Phagosome | ATP6V1C2, THBS4, MARCO, DMB2 |
| gga04310 | Wnt signaling pathway | WNT16, PLCB2, FZD4, WNT6 |
| gga04540 | Gap junction | PLCB2, PDGFC, EGF, DRD1 |
| gga04912 | GnRH signaling pathway | PLCB2, PLA2G4A, MAPK12 |
| gga00260 | Glycine, serine, and threonine metabolism | PHGDH, PGAM1 |
| gga00380 | Tryptophan metabolism | DHTKD1, ACAT2 |
| gga00564 | Glycerophospholipid metabolism | CDS2, LPGAT1, PLA2G4A |
DEGs Validation by RT-qPCR
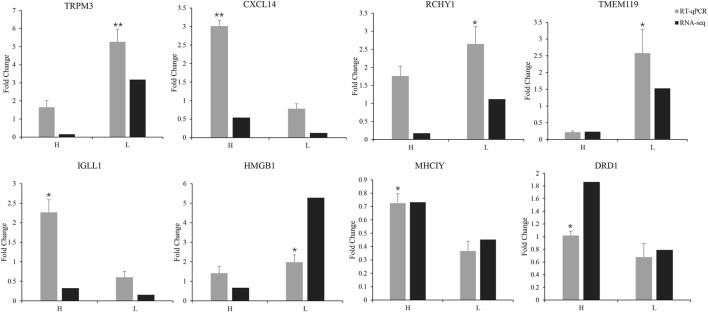
To validate the expression levels of the DEGs, we performed RT-qPCR analysis on four upregulated and four downregulated genes. The upregulated genes were D (1) dopamine receptor protein (DRD1), MHC-like class I Y protein (MHCIY), C-X-C motif chemokine 14 (CXCL14), and immunoglobulin lambda-like polypeptide 1 (IGLL1). The downregulated genes were RING finger and CHY zinc finger domain-containing protein 1 (RCHY1), high- mobility group protein B1 (HMGB1), transmembrane protein 119 (TMEM119), and transient receptor potential cation channel subfamily M member 3 protein (TRPM3). The expression levels of these genes were significantly different between the H and L groups (shown in Figure 3, p < 0.05 or p < 0.01). The RT-qPCR results were consistent with our RNA-seq data and demonstrated that the DEGs identified by RNA-seq were reliable and accurate.
FIGURE 3.
RT-qPCR validation of the expression of eight differentially expressed genes compared with mRNA transcriptome sequencing results (*p < 0.05, **p < 0.01).
Expression of miR-302a-3p and DRD1 in SYFLs
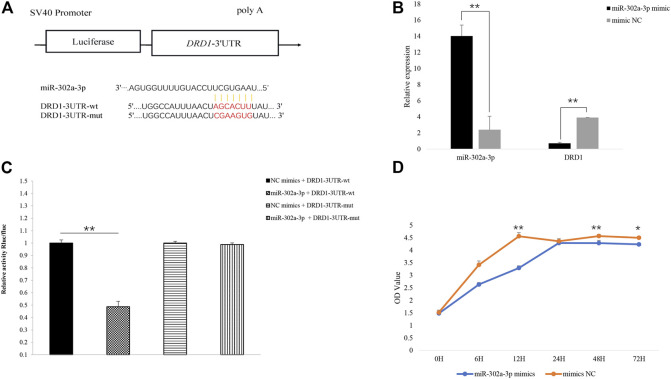
The gap junction channels are an important way for the GCs to pass nutrition to an oocyte (Starich et al., 2014). In the gap junction pathway, DRD1 was significantly enriched. Cluster analysis showed that chicken DRD1 is highly homologous with that of ducks and second with that of mice (Figure 4A). As can be seen from the relative expression of miR-302a-3p and DRD1 in the H and L groups (Figure 4B), there is a negative correlation between the expression of miR-302a-3p and DRD1. This analysis indicated that DRD1 may be a promising target for the genetic improvement of chicken egg production. A predicted binding site for miR-302a-3p was found in the DRD1 3′-UTR region (Figure 5A), and dual-luciferase reporter assays demonstrated that miR-302a-3p binds to the 3′UTR of DRD1 (Figure 5C).
FIGURE 4.
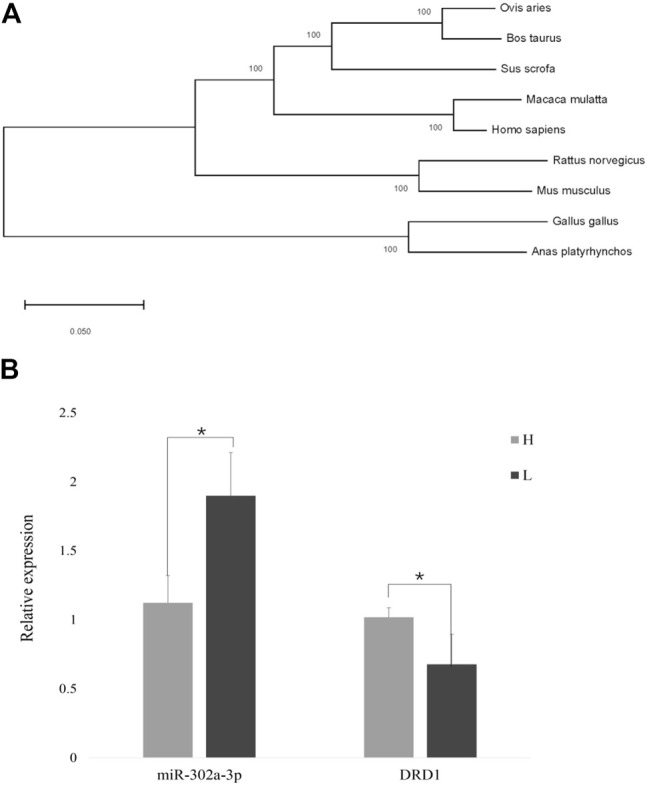
(A) Cluster plot showing DRD1 conservation in various species. (B) Expression of DRD1 and miR-302a-3p in small yellow follicles of high– and low–egg-yielding Taihang chickens (*p < 0.05).
FIGURE 5.
(A) Predicted binding site of miR-302a-3p on the 3′UTR of DRD1 and vector construction. (B) Overexpression of miR-302a-3p inhibited the expression of DRD1 in chicken granulosa cells. (C) Verification of miR-302a-3p binding to the 3′UTR of DRD1 using dual-luciferase assay. (D) Overexpression of miR-302a-3p promoted chicken GC proliferation. *p < 0.05, **p < 0.01.
Overexpression of miR-302a-3p Inhibits GC Proliferation by Targeting DRD1
To validate the function of miR-302a-3p, miR-302a-3p was overexpressed in chicken GCs by transfection. We found that DRD1 expression was significantly inhibited in the miR-302a-3p overexpressing GCs (p < 0.01) (Figure 5B). Overexpression of miR-302a-3p inhibited cell proliferation, which was measured by CCK-8 (Figure 5D). These data indicated that miR-302a-3p inhibits the proliferation of GCs.
Discussion
Egg production is a complex trait determined by genetics and numerous other factors (Zhao et al., 2019). In recent years, the understanding of egg production mechanisms has been greatly expanded as RNA-seq has revealed numerous genes that appear to be related to egg production, for example, DRD1, MC5R, PCK1, CTSA, and TGFBR3 (Mishra et al., 2020). As we report here, a proportion of the DEGs we identified between high-yielding and low-yielding egg layers also play an important role in follicle development.
Several pathways involved in egg production were significantly enriched, including the GnRH signaling pathway (gga04912), oocyte meiosis (gga04114), progesterone-mediated oocyte maturation (gga04914), and the calcium signaling pathway (gga04020). The GnRH signaling pathway is an important signal transduction pathway for fertility in animals. GnRH stimulates the synthesis and release of gonadotropins and induces estrogen production and ovulation (Park et al., 2018). The calcium signaling pathway drives key events surrounding fertilization and activation of development in all species studied to date. Elevated intracellular Ca2+ concentrations are associated with processes that release unfertilized eggs from meiosis and trigger embryonic development (Miao and Williams, 2012). In egg production and reproduction pathways, the epidermal growth factor (EGF) is also involved in the regulation of the actin cytoskeleton, while PGF functions in corpus luteum proliferation and cell death (Ding et al., 2020).
DRD1 is one of seven highly homologous cross-domain members of the G-protein–coupled receptor family (Wang et al., 2012). Dopamine promotes prolactin (PRL) secretion via DRD1 at the hypothalamic level, while DRD2 inhibits the secretion of PRL (Youngren et al., 1998; Al et al., 2003). In a study of the production traits of the Hungarian yellow chicken, DRD1 was associated with egg strength and body weight (Tempfli et al., 2015). The expression of DRD1 undoubtedly increases PRL secretion, resulting in an increase in brooding behavior frequency. In a transcriptomic analysis of atrophic ovaries, brooding hens had significantly higher PRL secretion levels than normal laying hens (Liu et al., 2018). Hens stop laying during brooding (Jiang et al., 2010), resulting in ovarian atrophy and an increase in white follicles, which slows development making it difficult for SYFLs to develop (Liu et al., 2009). The laying performance depends to some extent on ovary development, and DRD1 may be related to brooding behavior (Xu et al., 2010). In this study, a homology analysis showed that DRD1 was highly conserved in various species, which indicates that the function of DRD1 is similar in these species. Our RNA-seq data and RT-qPCR analysis showed that the expression of DRD1 was significantly higher in SYFs of high–egg-yielding chickens than in low–egg-yielding chickens. That DRD1 plays a role in egg production and follicle development was consistent with previous studies.
Our results showed that DRD1 is a target of gga-miR-302a-3p. miRNAs regulate gene expression by translational inhibition or by binding to the 3′ UTR of targeted genes. They regulate a variety of processes involved in development, organ formation, cell proliferation, and apoptosis (Kang et al., 2013). miRNAs participate in the production of steroids and can regulate human ovary GC proliferation and apoptosis (Sirotkin et la., 2009; Sirotkin et al., 2010). Eleven miRNAs have been identified in the chicken ovary that play regulatory roles in ovary and follicle development (Wu et al., 2017). miR-503, miR-672, and miR-465 are expressed in the mouse ovary and play an important role in the regulation of follicle development (Ahn et al., 2010). A reduction of miR-302 expression may be involved in reduced development in early embryogenesis (Parvin et al., 2019). Therefore, the role of miRNAs in follicle development cannot be ignored.
Conclusion
In summary, RNA-seq analysis indicates that DRD1 is involved in egg production. Functional assays showed that miR-302a-3p affects follicle differentiation by targeting DRD1. These results provide a foundation for improving chicken egg production, and further, our understanding of the development of chicken follicles.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI, PRJNA796736, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA796736/.
Ethics Statement
All experiments involving animals were approved by the Institutional Animal Care and Use Committee (IACUC) of Henan Agricultural University (Permit Number: 17-0118).
Author Contributions
YL and KW designed this study, and YL, ZZ, and HZ conducted animal experiments. YL, HH, and WL performed the transcriptome data analysis. YL conducted sample analysis and wrote the manuscript. ZZ, HZ, JY, and HH assisted with data analysis. All authors approved the final version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China Youth Program (31902144), Hebei Provincial Education Department Project Fund (QN2020105), and the Science and Technology Research and Development Plan Project of Handan (19422011008-54). These funding bodies had no roles in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.832762/full#supplementary-material
Abbreviations
CCK-8, Cell Counting Kit-8; DEGs, differentially expressed genes; DRD1, dopamine receptor; FPKM, Fragments per Kilobase Millions; GCs, granulosa cells; GO, Gene Ontology; H, high egg production group; KEGG, Kyoto Encyclopedia of Genes and Genomes; L, low egg production group; MUT, mutant type; NC, negative control; PRL, prolactin; SYFLs, small yellow follicles; WT, wild type.
References
- Ahn H. W., Morin R. D., Zhao H., Harris R. A., Coarfa C., Chen Z.-J., et al. (2010). MicroRNA Transcriptome in the Newborn Mouse Ovaries Determined by Massive Parallel Sequencing. Mol. Hum. Reprod. 16, 463–471. 10.1093/molehr/gaq017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Kahtane A., Chaiseha Y., El Halawani M. (2003). Dopaminergic Regulation of Avian Prolactin Gene Transcription. J. Mol. Endocrinol. 31 (1), 185–196. 10.1677/jme.0.0310185 [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential Expression Analysis for Sequence Count Data. Genome Biol. 11, R106. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahr J. (1991). The Chicken Ovary as a Model of Follicular Development. Semin. Reprod. Med. 9 (04), 352–359. 10.1055/s-2007-1019427 [DOI] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a Flexible Trimmer for Illumina Sequence Data. Bioinformatics 30 (15), 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiseha Y., Youngren O., Al-Zailaie K., El Halawani M. (2003). Expression of D1 and D2 Dopamine Receptors in the Hypothalamus and Pituitary during the turkey Reproductive Cycle: Colocalization with Vasoactive Intestinal Peptide. Neuroendocrinology 77 (2), 105–118. 10.1159/000068649 [DOI] [PubMed] [Google Scholar]
- Ding C., Zou Q., Wu Y., Lu J., Qian C., Li H., et al. (2020). EGF Released from Human Placental Mesenchymal Stem Cells Improves Premature Ovarian Insufficiency via NRF2/HO-1 Activation. Aging 12 (3), 2992–3009. 10.18632/aging.102794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Liu H. H., Wang J. W., Xiao Q. H., Yuan X., Li L., et al. (2014). Histological and Developmental Study of Prehierarchical Follicles in Geese. Folia Biol. (Krakow) 62, 171–177. 10.3409/fb62_3.171 [DOI] [PubMed] [Google Scholar]
- Fortune J. E., Rivera G. M., Yang M. Y. (2004). Follicular Development: the Role of the Follicular Microenvironment in Selection of the Dominant Follicle. Anim. Reprod. Sci. 82-83, 109–126. 10.1016/j.anireprosci.2004.04.031 [DOI] [PubMed] [Google Scholar]
- Fremeau R. T., Duncan G. E., Fornaretto M. G., Dearry A., Gingrich J. A., Breese G. R., et al. (1991). Localization of D1 Dopamine Receptor mRNA in Brain Supports a Role in Cognitive, Affective, and Neuroendocrine Aspects of Dopaminergic Neurotransmission. Proc. Natl. Acad. Sci. U.S.A. 88, 3772–3776. 10.1073/pnas.88.9.3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocking P. M. (2009). Biology of Breeding Poultry. CABI, 45. [Google Scholar]
- Huang Y. M., Shi Z. D., Liu Z., Liu Y., Li X. W. (2008). Endocrine Regulations of Reproductive Seasonality, Follicular Development and Incubation in Magang Geese. Anim. Reprod. Sci. 104 (2-4), 344–358. 10.1016/j.anireprosci.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Jackson D. M., Westlind-Danielsson A. (1994). Dopamine Receptors: Molecular Biology, Biochemistry and Behavioural Aspects. Pharmacol. Ther. 64, 291–370. 10.1016/0163-7258(94)90041-8 [DOI] [PubMed] [Google Scholar]
- Jiang R. S., Chen X. Y., Geng Z. Y. (2010). Broodiness, Egg Production, and Correlations between Broody Traits in an Indigenous Chicken Breed. Poult. Sci. 89 (6), 1094–1096. 10.3382/ps.2009-00621 [DOI] [PubMed] [Google Scholar]
- Johnson A. L., Bridgham J. T., Witty J. P., Tilly J. L. (1996). Susceptibility of Avian Ovarian Granulosa Cells to Apoptosis Is Dependent upon Stage of Follicle Development and Is Related to Endogenous Levels of Bcl-Xlong Gene Expression. Endocrinology 137 (5), 2059–2066. 10.1210/endo.137.5.8612548 [DOI] [PubMed] [Google Scholar]
- Johnson A. L., Lee J. (2016). Granulosa Cell Responsiveness to Follicle Stimulating Hormone during Early Growth of Hen Ovarian Follicles. Poult. Sci. 95 (1), 108–114. 10.3382/ps/pev318 [DOI] [PubMed] [Google Scholar]
- Johnson P. A., Kent T. R., Urick M. E., Giles J. R. (2008). Expression and Regulation of Anti-mullerian Hormone in an Oviparous Species, the Hen1. Biol. Reprod. 78, 13–19. 10.1095/biolreprod.107.061879 [DOI] [PubMed] [Google Scholar]
- Johnson P. (2012). Follicle Selection in the Avian Ovary. Reprod. Domest. Anim. 47 (4), 283–287. 10.1111/j.1439-0531.2012.02087.x [DOI] [PubMed] [Google Scholar]
- Kang L., Cui X., Zhang Y., Yang C., Jiang Y. (2013). Identification of miRNAs Associated with Sexual Maturity in Chicken Ovary by Illumina Small RNA Deep Sequencing. BMC genomics 14, 352. 10.1186/1471-2164-14-352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 12, 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Xiao Q., Gilbert E. R., Cui Z., Zhao X., Wang Y., et al. (2018). Whole-transcriptome Analysis of Atrophic Ovaries in Broody Chickens Reveals Regulatory Pathways Associated with Proliferation and Apoptosis. Sci. Rep. 8 (1), 7231. 10.1038/s41598-018-25103-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R. Z., Huang Y. M., Li W. L., Tian Y. B., Shi Z. D. (2009). Endocrine Regulation of Follicular Development in the Lay-Incubation Cycle of Magang Geese. Chin. J. Anim. Vet. Sci. 40 (5), 652–657. [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mao X., Cai T., Olyarchuk J. G., Wei L. (2005). Automated Genome Annotation and Pathway Identification Using the KEGG Orthology (KO) as a Controlled Vocabulary. Bioinformatics 21, 3787–3793. 10.1093/bioinformatics/bti430 [DOI] [PubMed] [Google Scholar]
- March J. B., Sharp P. J., Wilson P. W., Sang H. M. (1994). Effect of Active Immunization against Recombinant-Derived Chicken Prolactin Fusion Protein on the Onset of Broodiness and Photoinduced Egg Laying in bantam Hens. Reproduction 101 (1), 227–233. 10.1530/jrf.0.1010227 [DOI] [PubMed] [Google Scholar]
- Miao Y.-L., Williams C. J. (2012). Calcium Signaling in Mammalian Egg Activation and Embryo Development: The Influence of Subcellular Localization. Mol. Reprod. Dev. 79 (11), 742–756. 10.1002/mrd.22078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S. K., Chen B., Zhu Q., Xu Z., Ning C., Yin H., et al. (2020). Transcriptome Analysis Reveals Differentially Expressed Genes Associated with High Rates of Egg Production in Chicken Hypothalamic-Pituitary-Ovarian axis. Sci. Rep. 10, 5976. 10.1038/s41598-020-62886-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsma F. J., Mahan L. C., McVittie L. D., Gerfen C. R., Sibley D. R. (1990). Molecular Cloning and Expression of a D1 Dopamine Receptor Linked to Adenylyl Cyclase Activation. Proc. Natl. Acad. Sci. U.S.A. 87, 6723–6727. 10.1073/pnas.87.17.6723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niznik H. B., Van Tol H. H. (1992). Dopamine Receptor Genes: New Tools for Molecular Psychiatry. J. Psychiatry Neurosci. 17, 158–180. [PMC free article] [PubMed] [Google Scholar]
- Onagbesan O., Bruggeman V., Decuypere E. (2009). Intra-ovarian Growth Factors Regulating Ovarian Function in Avian Species: a Review. Anim. Reprod. Sci. 111 (2-4), 121–140. 10.1016/j.anireprosci.2008.09.017 [DOI] [PubMed] [Google Scholar]
- Park S.-R., Cho A., Park S. T., Park C. H., Lim S., Jin M., et al. (2018). Double-edged Sword of Gonadotropin-Releasing Hormone (GnRH): A Novel Role of GnRH in the Multiple Beneficial Functions of Endometrial Stem Cells. Cell Death Dis 9 (8), 828. 10.1038/s41419-018-0892-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvin D., Marefat Ghaffari N., Mohammad S., Fatane F. (2019). Expression of miR-302 in Human Embryo Derived from In-Vitro Matured Oocyte. Int. J. Reprod. Biomed. 17 (6), 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel R., Metzger M., Jiang S., Hemmings H. C., Greengard P., Braun K. (1997). Localization of Dopamine D1 Receptors and Dopaminoceptive Neurons in the Chick Forebrain. J. Comp. Neurol., 388146. [DOI] [PubMed] [Google Scholar]
- Schnell S. A., You S. (1999). El Halawani ME. D1 and D2 Dopamine Receptor Messenger Ribonucleic Acid in Brain and Pituitary during the Reproductive Cycle of the turkey Hen. Biol. Reprod. 388, 1378–1468. [DOI] [PubMed] [Google Scholar]
- Sharma A. K., Sharma R. K. (2020). Effect of Prostaglandins E2 and F2α on Granulosa Cell Apoptosis in Goat Ovarian Follicles. Iran J. Vet. Res. 21 (2), 97–102. [PMC free article] [PubMed] [Google Scholar]
- Sharp P. J., Macnamee M. C., Sterling R. J., Lea R. W., Pedersen H. C. (1988). Relationships between Prolactin, LH and Broody Behaviour in bantam Hens. J. Endocrinol. 118 (2), 279–286. 10.1677/joe.0.1180279 [DOI] [PubMed] [Google Scholar]
- Sirotkin A. V., Lauková M., Ovcharenko D., Brenaut P., Mlyncek M. (2010). Identification of microRNAs Controlling Human Ovarian Cell Proliferation and Apoptosis. J. Cel Physiol 223, 49–56. 10.1002/jcp.21999 [DOI] [PubMed] [Google Scholar]
- Sirotkin A. V., Ovcharenko D., Grossmann R., Lauková M., Mlynček M. (2009). Identification of MicroRNAs Controlling Human Ovarian Cell Steroidogenesis via a Genome-Scale Screen. J. Cel. Physiol. 219, 415–420. 10.1002/jcp.21689 [DOI] [PubMed] [Google Scholar]
- Starich T. A., Hall D. H., Greenstein D. (2014). Two Classes of gap junction Channels Mediate Soma-Germline Interactions Essential for Germline Proliferation and Gametogenesis in Caenorhabditis elegans . Genetics 198 (3), 1127–1153. 10.1534/genetics.114.168815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocks M. B., Moxon S., Mapleson D., Woolfenden H. C., Mohorianu I., Folkes L., et al. (2012). The UEA sRNA Workbench: A Suite of Tools for Analysing and Visualizing Next Generation Sequencing microRNA and Small RNA Datasets. Bioinformatics 28, 2059–2061. 10.1093/bioinformatics/bts311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempfli K., Konrád S., Kovácsné Gaál K., Pongrácz L., Bali Papp Á. (2015). Prolactin, Dopamine Receptor D1 and Spot14α Polymorphisms Affect Production Traits of Hungarian Yellow Hens. Livestock Sci. 174, 26–30. 10.1016/j.livsci.2015.01.012 [DOI] [Google Scholar]
- Tilly J. L., Kowalski K. I., Johnson A. L. (1991). Stage of Ovarian Follicular Development Associated with the Initiation of Steroidogenic Competence in Avian Granulosa Cells1. Biol. Reprod. 44 (2), 305–314. 10.1095/biolreprod44.2.305 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., et al. (2012). Differential Gene and Transcript Expression Analysis of RNA-Seq Experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., et al. (2010). Transcript Assembly and Quantification by RNA-Seq Reveals Unannotated Transcripts and Isoform Switching during Cell Differentiation. Nat. Biotechnol. 28, 511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallone D., Picetti R., Borrelli E. (2000). Structure and Function of Dopamine Receptors. Neurosci. Biobehavioral Rev. 24, 125–132. 10.1016/s0149-7634(99)00063-9 [DOI] [PubMed] [Google Scholar]
- Wang C., Li S., Li C., Feng Y., Peng X., Gong Y. (2012). Molecular Cloning, Expression Profile, Polymorphism and the Genetic Effects of the Dopamine D1 Receptor Gene on Duck Reproductive Traits. Mol. Biol. Rep. 39 (9), 9239–9246. 10.1007/s11033-012-1797-3 [DOI] [PubMed] [Google Scholar]
- Woods D. C., Johnson A. L. (2005). Regulation of Follicle-Stimulating Hormone-Receptor Messenger RNA in Hen Granulosa Cells Relative to Follicle Selection1. Biol. Reprod. 72, 643–650. 10.1095/biolreprod.104.033902 [DOI] [PubMed] [Google Scholar]
- Wu N., Gaur U., Zhu Q., Chen B., Xu Z., Zhao X., et al. (2017). Expressed microRNA Associated with High Rate of Egg Production in Chicken Ovarian Follicles. Anim. Genet. 48 (2), 205–216. 10.1111/age.12516 [DOI] [PubMed] [Google Scholar]
- Xu H., Shen X., Zhou M., Fang M., Zeng H., Nie Q., et al. (2010). The Genetic Effects of the Dopamine D1 Receptor Gene on Chicken Egg Production and Broodiness Traits. BMC Genet. 11 (1), 17. 10.1186/1471-2156-11-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Wang L., Lin P., Jiang T., Wang N., Zhao F., et al. (2017). An Immortalized Steroidogenic Goat Granulosa Cell Line as a Model System to Study the Effect of the Endoplasmic Reticulum (ER)-stress Response on Steroidogenesis. J. Reprod. Dev. 63 (1), 27–36. 10.1262/jrd.2016-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. D., Wakefield M. J., Smyth G. K., Oshlack A. (2010). Gene Ontology Analysis for RNA-Seq: Accounting for Selection Bias. Genome Biol. 11, R14. 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngren O. M., Chaiseha Y., El Halawani M. E. (1998). Regulation of Prolactin Secretion by Dopamine and Vasoactive Intestinal Peptide at the Level of the Pituitary in the turkey. Neuroendocrinology 68 (5), 319–325. 10.1159/000054380 [DOI] [PubMed] [Google Scholar]
- Youngren O. M., Pitts G. R., Phillips R. E., El Halawani M. E. (1995). The Stimulatory and Inhibitor Effects of Dopamine on Prolactin Secretion in the turkey. Gen. Comp. Endocrinol. 98, 111–117. 10.1006/gcen.1995.1049 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Chen J., Zhang X., Xu Z., Lin Z., Li H., et al. (2019). Genome-wide Association Analysis Reveals Key Genes Responsible for Egg Production of Lion Head Goose. Front. Genet. 10, 1391. 10.3389/fgene.2019.01391 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI, PRJNA796736, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA796736/.