Abstract
An R-plasmid-mediated metallo-β-lactamase was found in Klebsiella pneumoniae DK4 isolated in Japan in 1991. The nucleotide sequence of its structural gene revealed that the β-lactamase termed DK4 was identical to the IMP-1 metallo-β-lactamase which was mediated by a chromosomal gene of Serratia marcescens TN9106 isolated in Japan in 1991 (E. Osano et al., Antimicrob. Agents Chemother. 38:71–78, 1994). The dose effect of DK4 β-lactamase production on the resistance levels indicated a significant contribution of the enzyme to bacterial resistance to all the β-lactams except monobactams. The enzymatic characteristics of the DK4 β-lactamase and its kinetic parameters for nine β-lactams were examined. The DK4 β-lactamase was confirmed to contain 2 mol of zinc per mol of enzyme protein. The apoenzyme that lacked the two zincs was structurally unstable, and the activities of only 30% of the apoenzyme molecules could be restored by the addition of 1 mM zinc sulfate. The substitution of five conserved histidines (His28, His86, His88, His149, His210) and a cysteine (Cys168) for an alanine indicated that His86, His88, and His149 served as ligands to one of the zincs and that Cys168 played a role as a ligand to the second zinc. Both zinc molecules contribute to the enzymatic process. Mutant enzymes that lack only one of these retained some activity. Additionally, a conserved aspartic acid at position 90 was replaced by asparagine. This mutant enzyme showed an approximately 1,000 times lower kcat value for cephalothin than that of the wild-type enzyme but retained the two zincs even after dialysis against zinc-free buffer. The observed effect of pH on the activity suggested that Asp90 functions as a general base in the enzymatic process.
β-Lactamases (EC 3.5.2.6) are the bacterial enzymes that hydrolyze the β-lactam amide bond of β-lactam antibiotics, and their production is the most common mechanism of bacterial resistance to these antibiotics. β-Lactamases are divided into four classes, classes A to D, on the basis of their primary structures (1). The enzymes can also be classified into two groups on the basis of the differences in their catalytic mechanisms, i.e., serine β-lactamases (classes A, C, and D) and metallo-β-lactamases (class B). Metallo-β-lactamases have broad substrate specificity, including carbapenems, which are otherwise quite stable to serine β-lactamases. A limited number of bacterial species were known to produce metallo-β-lactamases; therefore, the enzymes were thought to be of little consequence clinically. However, we isolated an R-plasmid-mediated metallo-β-lactamase gene from Klebsiella pneumoniae DK4 in 1991 (GenBank accession number D29636). This was the first finding of an R-plasmid-mediated metallo-β-lactamase gene in a member of the family Enterobacteriaceae. On the basis of the nucleotide sequence of the DK4 β-lactamase gene, the DK4 enzyme was confirmed to be identical to a metallo-β-lactamase mediated by the blaIMP gene located in the chromosome of a Serratia marcescens strain isolated in 1991 (18).
Recently, the blaIMP gene has been found to be widely distributed in clinical isolates in Japan, including among members of the family Enterobacteriaceae and Pseudomonas aeruginosa isolates (2, 10, 24). Although kinetic parameters for common β-lactams were reported (13, 16, 18), little is known about other molecular characteristics of the metallo-β-lactamases. The study described here was carried out to understand the functional properties of the DK4 (IMP-1) β-lactamases by using site-directed mutagenesis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
K. pneumoniae DK4 is a clinical strain isolated in 1991 in Tokyo, Japan, and its β-lactam resistance is mediated by an R plasmid termed RDK4. Escherichia coli 1037 Rifr (11) was used as the recipient for R-plasmid transfer. E. coli AS226-51 (27) is an ampD mutant of strain C600 and a mutant with an ampC deletion and was used as the host for enzyme preparation. E. coli TG1 (5) was used as the host for the cloned β-lactamase gene. E. coli MV1184 (28) was used as a recipient for transformation in the site-directed mutagenesis study. Plasmid pHSG398 (25) was used as a cloning vector that carries a marker for resistance to chloramphenicol. Plasmid pKF18K (Takara Shuzo Co., Ltd.), which has a marker for resistance to kanamycin, was used as the vector for site-directed mutagenesis.
Media, chemicals, and enzymes.
Nutrient broth and Drigalski agar (Eiken Chemical Co., Tokyo, Japan) were used as the culture medium and the selective agar for R-plasmid transfer, respectively. For transformation, 2× yeast extract-tryptone (2×YT) broth and yeast extract-tryptone (YT) agar were used. For β-lactamase preparation, the bacteria were grown in heart infusion broth (Eiken Chemical Co.) or Terrific broth. Heart infusion agar was used to measure the susceptibilities of the bacteria to β-lactams.
The enzymes and enzyme kits used for DNA technology procedures were purchased from Takara Shuzo Co. (Shiga, Japan), Toyobo Co. (Osaka, Japan), and Wako Junyaku Co. (Tokyo, Japan). [α32-P]dCTP was purchased from Amersham (Buckinghamshire, United Kingdom). The following antibiotics and β-lactamase inhibitors used in this study were kindly provided by the indicated pharmaceutical companies: benzylpenicillin and kanamycin, Meiji Seika Kaisha, Ltd. (Tokyo, Japan); cephalothin, Shionogi & Co. (Osaka, Japan); cefuroxime, Nippon Glaxo Ltd. (Tokyo, Japan); imipenem, cefoxitin, and cilastatin, Banyu Pharmaceutical Co. (Tokyo, Japan); sulbactam, Pfizer Pharmaceuticals Inc. (Tokyo, Japan); clavulanic acid, SmithKline Beecham (Tokyo, Japan); chloramphenicol, Yamanouchi Pharmaceutical Co. (Tokyo, Japan); a chromogenic cephalosporin (FR18419), Fujisawa Pharmaceutical Co. (Osaka, Japan); rifampin, Daiichi Pharmaceutical Co. (Tokyo, Japan); and aztreonam, Eisai Co. (Tokyo, Japan).
Antibiotic susceptibility testing.
Bacterial susceptibility to antibiotics was measured by the serial agar dilution method by a previously described procedure (23) and was expressed as the MIC of each drug.
Cloning of β-lactamase gene.
RDK4 DNA was prepared from E. coli AS226-51/RDK4 by the alkaline lysis method and was digested with a restriction endonuclease, PstI. The digested DNA fragments were ligated into the PstI site of a vector plasmid, pHSG398. The resultant recombinant plasmid DNA was confirmed to have incorporated a 9.7-kb DNA fragment and was designated pDK4-1. The pDK4-1 DNA was retransferred into E. coli AS226-51, which was confirmed to produce a β-lactamase with a pI of 8.3, identical to that of the DK4 β-lactamase. A 1.2-kb DNA fragment was finally prepared by digestion of pDK4-1 with StyI, and the fragment was confirmed to include the intact β-lactamase gene by blunt-ending analysis. The fragment was inserted antiparallel with respect to the lacZ direction into the HincII site of pHSG398. The cloned plasmid DNA was termed pDK4-5. The 1.2-kb DNA fragment was sequenced by the dideoxynucleotide chain-termination method (21) with a BcaBEST dideoxy sequencing kit (Takara Shuzo Co., Kyoto, Japan).
Site-directed mutagenesis.
Site-directed mutagenesis was carried out by use of the oligonucleotide-directed dual amber method (9) with a template plasmid clone, pKFDK4. The template plasmid was prepared by insertion of the 1.2-kb DNA fragment containing the β-lactamase gene into pKF18K antiparallel with respect to the lacZ direction. The entire mutant gene was sequenced by the dideoxy chain-termination method to confirm the desired exchange in the nucleotide sequence.
Isoelectric focusing.
Isoelectric focusing was carried out by use of a model 111 IEF cell (Bio-Rad Laboratories, Hercules, Calif.) and a gel plate containing 5% Ampholine (pH 3.5 to 9.5). The β-lactamase activity on the plate was detected by spraying with a chromogenic cephalosporin, FR18419. The β-lactamases from the following strains were used as isoelectric markers: Citrobacter freundii GN346, pI 8.9 (22); Proteus mirabilis N29/pCS229, pI 6.9 (20); and E. coli ML1410/RGN823, pI 5.4 (22).
β-Lactamase purification and β-lactamase assay.
E. coli AS226-51 cells carrying the wild-type or a mutant β-lactamase gene were grown overnight at 37°C in heart infusion broth or Terrific broth containing a sublethal concentration of kanamycin (50 μg/ml). The preculture was diluted with a 40-fold volume of fresh medium, followed by growth under aeration at 37 or 24°C. In the cases of the bacterial cells carrying the mutant gene with the H28A, H86A, H88A, H149A, and C168A mutations, bacterial growth was carried out at 24°C to achieve a higher yield of the active enzyme than that achieved at 37°C. At the mid-logarithmic phase the bacterial cells were disrupted with a French press in 50 mM MOPS [3-(N-morpholino)propanesulfonic acid] buffer (pH 7.0) containing 0.1 mM zinc sulfate. The disrupted cells were centrifuged for 1 h at 40,000 × g and 4°C, and the supernatant was used for further purification of the enzyme by ion-exchange chromatography on a CM-Sephadex C-50 column equilibrated in MOPS buffer with 0.1 mM zinc sulfate. The enzyme was eluted from the column by a linear NaCl gradient (0.1 to 0.4 M), followed by gel filtration on a Sephadex G-75 column equilibrated with MOPS buffer. The β-lactamase in the fractions was detected by cephalothin hydrolysis in 50 mM MOPS buffer (pH 7.0) with 1 mM zinc sulfate or by an immunoblotting technique with the polyclonal antiserum against the DK4 β-lactamase. The purity of the enzyme preparation was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The enzyme protein was determined from UV absorption at 280 nm with ɛ equal to 44,600 M−1 cm−1. Confirmation was obtained by the Bio-Rad version of Bradford's dye binding assay (4) with bovine serum albumin as a standard.
β-Lactamase activity, unless stated otherwise, was assayed by the UV spectrophotometric method (30) in 50 mM MOPS buffer (pH 7.0) at 30°C. For determination of the kcat and the Km values, the initial velocity at each substrate concentration was determined by using a spectrophotometer (U-3300; Hitachi Ltd., Tokyo, Japan), and the kinetic parameters were calculated by means of nonlinear regression.
Protein analysis.
The N-terminal sequence was determined by the use of a protein sequencer (model 477A; Applied Biosystems, Foster City, Calif.).
Atomic absorption spectroscopy.
The zinc content in the enzyme molecule was determined as follows. All the glassware was washed with 6 N HNO3 in order to eliminate contamination. Enzyme samples were subjected to 3 days of dialysis against zinc-free 50 mM MOPS buffer (pH 7.0) containing Chelex 100 resin at 4°C with multiple changes of the dialysis buffer, and then the enzyme samples were diluted with the same buffer to provide samples with the expected range of 0.05 to 1.00 ppm of zinc. The preparation was analyzed with a Z-8000 Polarized Zeeman Atomic Absorption Spectrophotometer (Hitachi Ltd.).
Nucleotide sequence accession number.
The nucleotide sequence of the DK4 β-lactamase gene was entered into the GSDB, DDBJ, EMBL, and NCBI nucleotide sequence databases under the accession number D29636.
RESULTS AND DISCUSSION
The DK4 β-lactamase gene and its product.
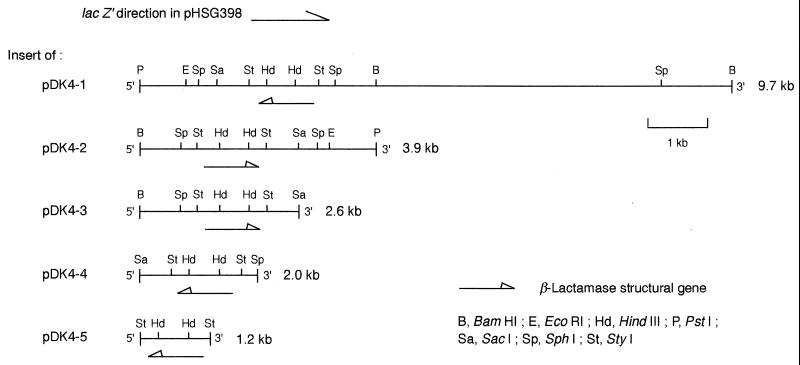
K. pneumoniae DK4 exhibited a high level of resistance to extended-spectrum cephalosporins and a moderate level of resistance to carbapenens, i.e., imipenen. The resistance was conjugally transferred to E. coli 1037 Rifr. The β-lactam resistance was due to a β-lactamase with a pI of 8.6 and was mediated by an R plasmid termed RDK4. The activity of the DK4 β-lactamase was completely inhibited by 10 mM EDTA. The DK4 β-lactamase gene in a 9.7-kb DNA fragment was cloned into a plasmid vector, pHSG398, and the recombinant plasmid was termed pDK4-1. From the 9.7-kb DNA fragment, four shorter DNA fragments (of 3.9, 2.6, 2.0, and 1.2 kb) containing the β-lactamase gene were prepared, and each fragment was cloned into the plasmid vector (Fig. 1). The recombinant plasmids obtained were termed pDK4-2, pDK4-3, pDK4-4, and pDK4-5, respectively. The complete nucleotide sequence of the 1.2-kb fragment inserted into pDK4-5 was determined. The sequenced region contained an open reading frame, and an amino acid sequence comprising 246 amino acids was deduced. Through determination of the N-terminal amino acid sequence of the purified enzyme protein, a signal peptide with 18 amino acids and a mature enzyme with 228 amino acids were detected.
FIG. 1.
Restriction maps of inserts of the clone and subclones carrying the target β-lactamase gene.
Alignment of the mature enzyme with known metallo-β-lactamase amino acid sequences indicated that the DK4 β-lactamase has 38.4% homology to the enzymes of Bacillus cereus 5/B/6 (15), 35.4% homology to the enzyme of Bacteroides fragilis TAL2480 (26), 32.3% homology to the enzyme of Aeromonas hydrophila AE036 (17), and 19.5% homology to the enzyme of Stenotrophomonas maltophilia IID1275 (29). On the other hand, the amino acid sequence of the DK4 β-lactamase is identical to that of a metallo β-lactamase termed IMP-1 and produced by S. marcescens TN9106 (18). The IMP-1 β-lactamase gene (blaIMP) was previously reported to be located on the chromosome of S. marcescens. Our detection of an identical β-lactamase gene incorporated in an R plasmid suggests the insertion of this gene into the chromosome of S. marcescens.
Dose effect of DK4 β-lactamase on resistance to β-lactam antibiotics in E. coli cells.
During the preparation of the subcloned β-lactamase gene, we observed that the β-lactamase activity in the E. coli AS226-51 cells was inversely proportional to the size of the DNA fragment inserted into the vector plasmid (Table 1). This phenomenon may be due to the difference in copy number of the plasmid in the host cells because all the β-lactamase genes were expressed by their own promoters, and the pDK4 series can be used to provide an understanding of the dose effect of metallo-β-lactamase production with respect to the level of resistance of the bacterial cells to β-lactams. The β-lactamase activity of K. pneumoniae DK4 was 0.12 U per mg of bacterial protein, as determined with cephalothin as a substrate. When RDK4 was transferred to E. coli AS226-51 cells, the enzyme activity was 0.098 U per mg of bacterial protein, about 80% of that for the original strain. For the series of E. coli subclones, the MICs for the cells increased up to 8 times and the level of enzyme production increased up to 13 times. The results in Table 1 show quantitatively that the metallo-β-lactamase contributed to the resistance of the bacteria to all β-lactams except the monobactam aztreonam.
TABLE 1.
Dose effect of DK4 β-lactamase on resistance of bacteria to typical β-lactams
| E. coli strain | β-Lactamase activity (U/mg of bacterial protein) | MIC (μg/ml)a
|
|||||
|---|---|---|---|---|---|---|---|
| AMP | CET | CXM | CFX | ATM | IMP | ||
| AS226-51 (host cell) | <10−5 | 0.8 | 0.8 | 1.6 | 1.6 | <0.2 | <0.2 |
| AS226-51/RDK4 | 0.098 | 25 | 100 | 100 | 200 | <0.2 | 1.6 |
| AS226-51/RDK4-1 | 0.14 | 100 | 400 | 400 | 800 | <0.2 | 3.1 |
| AS226-51/RDK4-2 | 0.62 | 200 | 400 | 800 | 800 | <0.2 | 6.3 |
| AS226-51/RDK4-3 | 0.67 | 200 | 400 | 800 | 1,600 | <0.2 | 12.5 |
| AS226-51/RDK4-4 | 0.90 | 200 | 400 | 800 | 1,600 | <0.2 | 12.5 |
| AS226-51/RDK4-5 | 1.30 | 200 | 400 | 800 | 1,600 | <0.2 | 12.5 |
The following β-lactams were used: ampicillin (AMP), cephalothin (CET), cefuroxime (CXM), cefoxitin (CFX), aztreonam (ATM), and imipenen (IMP).
Characteristics of DK4 β-lactamase and its kinetic parameters.
The DK4 β-lactamase was extracted from the E. coli cells carrying pDK4-5 and was completely purified. From 18 liters of the bacterial culture, 18 mg of the purified enzyme protein was obtained, and its purity was confirmed by SDS-PAGE. The presence or absence of 0.1 mM zinc sulfate throughout the purification processes did not affect the recovery of active enzyme, indicating a tight binding of zinc to the enzyme. The purified enzyme showed a high degree of thermostability, as the enzyme retained its full activity even after 30 min of incubation at 60°C in 50 mM MOPS buffer (pH 7.0) containing 0.1 mM zinc sulfate. The activity was assayed within 30 s of the enzyme reaction at 30°C. The effect of pH on the activity of the enzyme against cephalothin at between pH 5.0 and 8.0 was measured in the following buffers: 50 mM MES buffer with 0.5 M NaCl and 1 mM zinc sulfate (pH 5.0 to 6.5) and 50 mM MOPS buffer with 0.5 M NaCl (pH 7.0 to 80). The β-lactamase activity increased concomitantly with pH, such that at pHs 7.0, 6.0, and 5.0 the activities were 75, 49, and 32%, respectively, of that at pH 8.0. The enzyme was found to be unstable at a pH lower than 6.0, and this phenomenon may be attributable to the presence of 2-(N-morpholino)ethanesulfonic acid (MES) (8). Stability at lower pH could be protected by the presence of 1 mM zinc sulfate. We failed to measure activity at a pH higher than 8.0 because of alkaline hydrolysis of the substrate in the reaction mixture.
The zinc content in the purified enzyme molecule was determined by means of atomic absorption spectrophotometry. The results indicated that the DK4 β-lactamase contains 2.0 zinc atoms per mature enzyme protein. Further testing indicated that enzyme activity was not influenced by the addition of 5 mM zinc sulfate to the reaction mixture, and for complete inactivation of the enzyme, an EDTA concentration greater than 1 mM was required. The 50% inhibitory dose of EDTA was 0.62 mM.
During the UV spectrophotometric assay for enzyme activity, we observed that the molar extinction coefficient decreased upon the addition of zinc sulfate. In the case of cephalothin as the substrate, the coefficient was changed from 7,200 to 6,300 M−1 cm−1 by the addition of 1 mM zinc sulfate. This effect of zinc could be negated in the presence of 0.5 M NaCl. This phenomenon may be attributable to an ionic interaction between the zinc cation and the cleaved β-lactam ring, and it may lead to an erroneous conclusion that the activity of the metallo-β-lactamase is inhibited by a high concentration of zinc.
The kinetic parameters of the purified DK4 β-lactamase for nine β-lactams were determined in MOPS buffer (pH 7.0) at 30°C, and the results are summarized in Table 2. The DK4 β-lactamase has a broad substrate specificity, including penicillins, cephalosporins, cephamycin, and carbapenen, similar to those of known metallo-β-lactamases. When the kinetic parameters of the DK4 β-lactamase for typical β-lactams were compared with those for IMP-1 from a more recent report (13), some differences in the parameters, especially in the kcat value for ampicillin and Km values for cephalosporins including cefoxitin, were observed. The kcat value of the DK4 enzyme for ampicillin was about 1/16 of that of the IMP-1 enzyme. The Km values of the IMP-1 enzyme for the cephalosporins were 3 to 46 times greater than those of the DK4 enzyme. We confirmed that the kinetic data in this paper were reproducible under the conditions used in the study.
TABLE 2.
Kinetic parameters of DK4 β-lactamase for β-lactams
| β-Lactam group (β-lactam) | kcat (s−1) | Km (μM) | kcat/Km (μM s−1) |
|---|---|---|---|
| Penicillins | |||
| Benzylpenicillin | 212 ± 36 | 280 ± 61 | 0.76 ± 0.07 |
| Ampicillin | 59.0 ± 9.8 | 176 ± 3.2 | 0.34 ± 0.06 |
| Carbenicillin | >330 | >1,000 | |
| Cephalosporins | |||
| Cephalothin | 26.4 ± 2.5 | 1.6 ± 0.1 | 16.3 ± 1.7 |
| Cephaloridine | 32.6 ± 1.4 | 7.6 ± 0.3 | 4.3 ± 0.1 |
| Cefuroxime | 58.2 ± 3.25 | 0.8 ± 0.2 | 70.1 ± 11.8 |
| Cefotaxime | 11.9 ± 0.7 | 1.4 ± 0.2 | 8.8 ± 1.0 |
| Cefamycin (cefoxitin) | 12.7 ± 1.4 | 1.6 ± 0.36 | 8.0 ± 1.0 |
| Carbapenem (imipenen) | 40.7 ± 1.4 | 18.8 ± 1.0 | 2.2 ± 0.1 |
The effects of the serine β-lactamase inhibitors and a renal membrane dipeptidase inhibitor (cilastatin) on the DK4 β-lactamase were examined. The activity of the DK4 enzyme was not affected by 10 mM sulbactam, 10 mM clavulanic acid, or 1 mM aztreonam; and it had undetectable hydrolytic activity against the three β-lactams. Cilastatin, which is known to be an inhibitor of dipeptidase, showed weak inhibitory activity against the DK4 β-lactamase. The 50% inhibitory concentration of cilastatin was about 3 mM, and this value is about 104 times the 50% inhibitory concentration of cilastatin for dipeptidase (12).
The apo-DK4 β-lactamase and its restoration.
A DK4 β-lactamase that was missing the two zincs was prepared by 3 days of dialysis of the EDTA-treated enzyme against zinc-free MOPS buffer. The enzyme, termed apo-DK4, was completely lacking enzyme activity, and its activity was estimated to be less than 0.004% of that of the holoenzyme. The activity of the apo-DK4 enzyme was restored to about 30% of its original activity by the addition of 1 mM zinc sulfate to the enzyme solution. This restoration was hindered in the presence of a sulfhydryl reagent, methylmethane thiosulfonate, suggesting the contribution of a cysteine to zinc binding. The reactivated enzyme had about the same Km value for cephalothin as that of the native enzyme, and it was therefore thought that about 70% of the enzyme molecules were irreversibly denatured during production of the apoenzyme state.
Functions of His28, His86, His88, His149, His210, and Cys168 as zinc ligands.
The alignment of the amino acid sequences of the metallo-β-lactamases indicated that histidines at positions 86, 88, 149, and 210 were conserved in all the enzymes, and a histidine at position 28 was found in some metallo-β-lactamases. All of these histidine residues except His28 were estimated to be located at positions close to the zincs by reference to a three-dimensional structure of a metallo-β-lactamase from B. fragilis (6). Cys168 was also presumed to be situated in the vicinity of the zinc. To establish the functions of these residues in the DK4 β-lactamase, these five histidines and the cysteine were individually replaced by an alanine by site-directed mutagenesis, and the mutant genes on the vector plasmid were transformed into E. coli AS226-51.
The mutant β-lactamases were purified in the same way as the wild-type enzyme. All the mutant enzymes except the enzyme with the H28A mutation showed decreased activity following dialysis against zinc-free MOPS buffer. Therefore, purification of these mutant enzymes was carried out in the presence of 0.1 mM zinc sulfate.
The wild-type and His mutant β-lactamases purified were dialyzed in zinc-free MOPS buffer (pH 7.0) containing Chelex 100 resin. The wild-type and the mutant with the H28A mutation retained their activities even after the dialysis, and the 2 mol of zinc per mol of enzyme was detected in the dialyzed enzymes (Table 3). This result agreed with the metal/enzyme ratio for the wild-type enzyme reported by Laraki et al. (13).
TABLE 3.
Zinc contents of DK4 metallo-β-lactamase and its mutantsa
| β-Lactamase mutation | Zinc content (mol/mol of enzyme) with dialysis at:
|
|
|---|---|---|
| pH 7.0 | pH 9.5 | |
| Wild type | 2.00 ± 0.02 | |
| H28A | 1.90 ± 0.06 | |
| H86A | 1.20 ± 0.02 | |
| H88A | 0.90 ± 0.09 | |
| H149A | 1.00 ± 0.02 | |
| H210A | 0.5 | |
| C168A | 0.82 ± 0.01 | 1.01 ± 0.01 |
| C168S | 1.07 ± 0.02 | 1.85 ± 0.06 |
| C168D | 2.04 ± 0.03 | |
The enzymes were dialyzed against zinc-free 50 mM MOPS buffer (pH 7.0) or 100 mM Tris-HCl buffer (pH 9.5) for 3 days at 4°C. The zinc content was determined by atomic absorption spectrophotometry.
On the other hand, the enzymes with the H86A, H88A, H149A, and C168A mutations showed significantly decreased activity, and this residual activity remained constant 24 h after the dialysis. The zinc content after exhaustive dialysis of the enzymes with H86A, H88A, H149A and C168A mutation was about half that of the wild-type enzyme, suggesting that all the enzymes except that with the H28A mutation had one zinc atom per one enzyme molecule (Table 3). In the case of the mutant with the H210A mutation, activity was completely absent following dialysis, and the zinc content was estimated to be 0.5 mol per enzyme mol. These data suggest a significant distortion of the active site in the mutant with the H210A mutation.
Cys168 is the only cysteine residue in the DK4 β-lactamase. Its replacement by alanine resulted in a significant lowering of activity, as measured after enzyme purification, and the missing activity was only slightly restored even by 1 mM zinc (Table 4). This result is consistent with that for the mutant of the B. cereus Zn2+ β-lactamase with the C168A mutation (19). On the basis of the fact that serine is situated at position 168 of a metallo-β-lactamase from S. maltophilla (29), a mutant with the C168S mutation was prepared from the DK4 β-lactamase. The zinc content of the enzyme with the C168S mutation was 1.07 mol after dialysis at pH 7.0, but the zinc content was increased to 1.85 mol by dialysis at pH 9.5. This observation suggested that a negative charge at position 168 is necessary for retention of the second zinc atom. This assumption was confirmed by the fact that a mutant with a C168D mutation that we prepared had a zinc content of 2.04 mol after dialysis at pH 7.0 (Table 3).
TABLE 4.
Kinetic parameters of mutant metallo-β-lactamases for cephalothin in the presence or absence of 1 mM zinc sulfatea
| Zinc content and β-lactamase mutation | kcat (s−1) | Km (μM) | kcat/Km (μM s−1) |
|---|---|---|---|
| Zinc free | |||
| Wild type | 26.4 ± 2.5 | 1.6 ± 0.1 | 16.3 ± 1.7 |
| H28A | 33 ± 2 | 2.3 ± 0.3 | 14.3 ± 1.0 |
| H86A | 0.29 ± 0.02 | 33 ± 2 | 0.0100 ± 0.0001 |
| H88A | <0.001 | NDb | |
| H149A | 0.002 | ND | |
| C168A | 0.040 ± 0.009 | 91 ± 26 | 0.00040 ± 0.00003 |
| C168S | 0.72 ± 0.08 | 17 ± 3 | 0.038 ± 0.003 |
| C168D | <0.001 | ND | |
| 1 mM Zinc | |||
| Wild type | 35 ± 2 | 2.7 ± 0.2 | 13.5 ± 0.4 |
| H28A | 43 ± 1 | 4.3 ± 0.1 | 10.6 ± 0.3 |
| H86A | 89 ± 4 | 8.7 ± 0.9 | 8.1 ± 0.5 |
| H88A | 230 ± 18 | 48 ± 7 | 5.0 ± 0.4 |
| H149A | 191 ± 27 | 131 ± 26 | 1.4 ± 0.1 |
| C168A | 0.34 ± 0.01 | 0.82 ± 0.13 | 0.54 ± 0.08 |
| C168S | 0.76 ± 0.07 | 2.2 ± 1.0 | 0.47 ± 0.17 |
| C168D | 660 ± 44 | 70 ± 7 | 9.5 ± 0.4 |
Enzyme preparations dialyzed against zinc-free MOPS buffer (pH 7.0) were used.
ND, not determined.
The mutant enzymes with the H86A and C168A mutations had detectable activity even after dialysis, and their Michaelis-Menten constants for cephalothin were significantly greater than that of the wild-type enzyme (Table 4). Residual activity in the mutant enzymes was essentially missing following treatment with 1 mM EDTA. This observation indicated that a little activity may still be retained in the enzyme lacking one of the two zincs.
The enzymes with the alanine substitutions showed increased specific activity with an increase in the zinc concentration in the reaction medium. The maximum activity was achieved with 1 mM zinc sulfate, in the presence of which the enzyme probably retains two zincs at the active site. The kinetic parameters of the dialyzed enzymes for cephalothin in the presence or absence of 1 mM zinc sulfate are summarized in Table 4.
Three histidines at positions 86, 88, and 149 and the cysteine at position 168 are thought to be the residues that function as zinc ligands. Alteration of the kinetic parameters of the His mutant enzymes by the addition of 1 mM zinc sulfate was most remarkable in the cases of the enzymes with the H86A, H88A and H149A mutations; however, the activity of the enzyme with the C168A mutation was only slightly increased in the presence of 1 mM zinc sulfate. The difference in the effect of zinc between the mutants with the histidine mutations and the mutant with the cysteine mutation may indicate a difference in the zinc atom associated with ligands. The mutant enzyme with the C168D mutation retained two zinc atoms, but its activity could not be detected in the absence of 1 mM zinc sulfate, suggesting that the zincs combine irregularly or weakly to the active site. On the other hand, the mutant enzyme with the C168D mutation exhibited significantly higher kcat and Km values than the wild-type enzyme in the reaction medium with 1 mM zinc sulfate, and its kcat/Km value was restored up to 70% of that for the wild-type enzyme. It can be presumed that the kinetic properties of a metallo-β-lactamase are dependent on the situation of the zincs in the active site.
In order to compare the structural stabilities of the wild type and the mutants with His mutations, their thermal stabilities were examined. After various incubation times in 50 mM MOPS buffer containing 0.1 mM zinc sulfate at 50°C, aliquots of the enzyme solution were withdrawn and the residual activity was determined. The β-lactamases with the H86A, H88A, and H149A mutations lost nearly all of their activities within 20 min of incubation. On the other hand, the wild-type β-lactamase retained its activity, and the β-lactamase with the H28A mutation had about 60% of its original activity even after 60 min (data not shown). These observations suggest a close relationship between structural stability and appropriate binding of the zinc atoms in the active site.
Lim et al. (14) claimed that His28 of the B. cereus metallo-β-lactamase is essential for enzyme activity on the basis of the observation that E. coli cells with the H28Y mutant gene showed high levels of susceptibility to ampicillin and cephalosporin C (14). In the case of the DK4 β-lactamase, we could not observe significant differences in the enzymatic properties and zinc contents between the wild type and the mutant with the H28A mutation. It may be concluded that His28 of the DK4 enzyme is not a functional residue.
Function of Asp90 as a general base in the enzyme reaction.
Aspartic acid at position 90 is one of the conserved residues in known metallo-β-lactamases. Concha et al. (6) claimed that Asp90 of a metallo-β-lactamase from B. fragilis is one of the zinc ligands. The B. fragilis β-lactamase with the D90V mutation, in fact, had a lower zinc content than the wild-type enzyme (7). We observed that the kcat value of the DK4 β-lactamase increased with an increase in the pH from 5.0 to 8.0, suggesting the existence of a general base in the reaction. A candidate for the general base was Asp90, which was assumed to be localized to the active-site area. In order to confirm this assumption, Asp90 was replaced by asparagine.
The purified β-lactamase with the D90N mutation was extensively dialyzed against MOPS buffer without zinc, and its zinc content was determined to be 2.29 mol of zinc per mol of enzyme. Kinetic parameters of the β-lactamase with the D90N mutation for cephalothin were determined at pH 7.0 and compared with those of the wild-type enzyme (Table 5). It was also noted that varying the zinc concentration in the reaction mixture did not affect the parameters of either the wild-type or the mutant β-lactamase. The β-lactamase with the D90N mutation showed an approximately 1,000 times lower kcat value for cephalothin than that of the wild-type β-lactamase; however, no difference in Km values was detected.
TABLE 5.
Kinetic parameters of the β-lactamase with the D90N mutation for cephalothin and its zinc content
| β-Lactamase | kcat (s−1) | Km (μM) | kcat/Km (μM s−1) | Zinc content (mol/mol of enzyme) |
|---|---|---|---|---|
| Wild type | 35 ± 2 | 2.7 ± 0.2 | 13.5 ± 0.4 | 2.00 ± 0.02 |
| D90N | 0.04 ± 0.001 | 2.81 ± 0.50 | 0.017 ± 0.003 | 2.29 ± 0.18 |
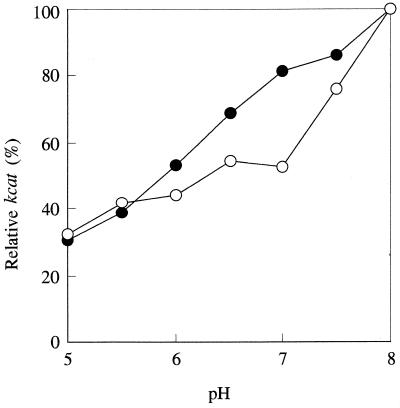
The kcat value was determined from pH 5.0 to 8.0 by using cephalothin as the substrate. Figure 2 shows the effect of pH on the kcat value, which is expressed as the percentage of the kcat determined at pH 8.0. The β-lactamase with the D90N mutation exhibited a lower value than the wild-type enzyme from pH 5.5 to 8.0. This result suggests that Asp90 acts as a general base in the enzyme reaction and is consistent with the results obtained with the B. cereus 569/H/9 β-lactamase (3).
FIG. 2.
Effect of pH on the relative kcat values of the wild-type and the D90N β-lactamases. The following buffers were used for the assay; 50 mM MES buffers containing 0.5 M NaCl and 1 mM zinc sulfate (pH 5.0 to 6.5) and 50 mM MOPS buffer containing 0.5 M NaCl (pH 7.0 to 8.0). The activity was measured with cephalothin as the substrate. ●, wild-type enzyme; ○, enzyme with D90N mutation.
ACKNOWLEDGMENT
This study was supported in part by a grant for the study on drug-resistant bacteria funded by the Ministry of Health and Welfare of Japan.
REFERENCES
- 1.Ambler R P. The structure of β-lactamases. Philos Trans R Soc London (Biol) 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 2.Arakawa Y, Murakami M, Suzuki K, Ito H, Wacharotayankun R, Ohsuka S, Kato N, Ohta M. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob Agents Chemother. 1995;39:1612–1615. doi: 10.1128/aac.39.7.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bounaga S, Laws A P, Galleni M, Page M I. The mechanism of catalysis and the inhibition of the Bacillus cereus zinc-dependent β-lactamase. Biochem J. 1998;331:703–711. doi: 10.1042/bj3310703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Carter P, Bedouelle H, Waye M M Y, Winter G. Oligonucleotide-directed mutagenesis in M13. Experiment manual. London, United Kingdom: Medical Research Council; 1984. [Google Scholar]
- 6.Concha M W, Rasmussen B A, Bush K, Herzberg O. Crystal structure of the wide-spectrum binuclear zinc β-lactamase from Bacteroides fragilis. Strucure. 1996;4:823–836. doi: 10.1016/s0969-2126(96)00089-5. [DOI] [PubMed] [Google Scholar]
- 7.Crowder M W, Wang Z G, Franklin S L, Zovinka E P, Benkovic S J. Characterization of the metal-binding sites of the β-lactamase from Bacteroides fragilis. Biochemistry. 1996;35:12126–12132. doi: 10.1021/bi960976h. [DOI] [PubMed] [Google Scholar]
- 8.Fitzgerald P M D, Wu J K, Tony J H. Unanticipated inhibition of the metallo-lactamase from Bacteroides fragilis by 4-morpholineethane-sulfonic acid (MES): a crystallographic study at 1.85-Å resolution. Biochemistry. 1998;37:6791–6800. doi: 10.1021/bi9730339. [DOI] [PubMed] [Google Scholar]
- 9.Hashomoto-Gotoh T, Mizuno T, Ogasahara Y, Nakagawa M. An oligodeoxyribonucleotide-directed dual amber method for site-directed mutagenesis. Gene. 1995;152:271–275. doi: 10.1016/0378-1119(94)00750-m. [DOI] [PubMed] [Google Scholar]
- 10.Ito H, Arakawa Y, Ohsuka S, Wacharotayankun R, Kato N, Ohta M. Plasmid-mediated dissemination of the metallo-β-lactamase gene blaIMP among clinically isolated strains of Serratia marcescens. Antimicrob Agents Chemother. 1995;39:824–829. doi: 10.1128/aac.39.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iyobe S, Sagai H, Mitsuhashi S. TN2001, a transposon encoding chloramphenicol resistance in Pseudomonas aeruginosa. J Bacteriol. 1981;146:141–148. doi: 10.1128/jb.146.1.141-148.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keynan S, Hooper N M, Felici A, Amicosante G, Turner A J. The renal membrane dipeptidase (dehydropeptidase I) inhibitor, cilastatin, inhibits the bacterial metallo-β-lactamase enzyme CphA. Antimicrob Agents Chemother. 1995;39:1629–1631. doi: 10.1128/aac.39.7.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laraki N, Franceschini N, Rossolini G M, Santucci P, Meunier C, De Pauw E, Amicosante G, Frere J-M, Galleni M. Biochemical characterization of the Pseudomonas aeruginosa 101/1477 metallo-β-lactamase IMP-1 produced by Escherichia coli. Antimicrob Agents Chemother. 1999;43:902–906. doi: 10.1128/aac.43.4.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim H M, Pene J J. Mutations affecting the catalytic activity of Bacillus cereus 5/B/6 β-lactamase II. J Biol Chem. 1989;264:11682–11687. [PubMed] [Google Scholar]
- 15.Lim H M, Pene J J, Shaw R W. Cloning, nucleotide sequence, and expression of the Bacillus cereus 5/B/6 β-lactamase II structural gene. J Bacteriol. 1988;170:2873–2878. doi: 10.1128/jb.170.6.2873-2878.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marumo K, Takeda A, Nakamura Y, Nakaya K. Purification and characterization of metallo-β-lactamase from Serratia marcescens. Microbiol Immunol. 1995;39:27–33. doi: 10.1111/j.1348-0421.1995.tb02164.x. [DOI] [PubMed] [Google Scholar]
- 17.Massidda O, Rossolini G M, Satta G. The Aeromonas hydrophilia cphA gene: molecular heterogeneity among class B metallo-β-lactamases. J Bacteriol. 1991;173:4611–4617. doi: 10.1128/jb.173.15.4611-4617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osano E, Arakawa Y, Wacharotayankun R, Ohta M, Horii T, Ito H, Yoshomura F, Kato N. Molecular characterization of an entrobacterial metallo β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenen resistance. Antimicrob Agents Chemother. 1994;38:71–78. doi: 10.1128/aac.38.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paul-Soto R, Bauer R, Frere J-M, Galleni M, Meyer-Klaucke W, Nolting H, Rossolin G M, De Seny D, Hernandez-Valladares M, Zeppezauer M, Adolph H-W. Mono- abd binuclear Zn2+-β-lactamase; role of the conserved cysteine in the catalytic mechanism. J Biol Chem. 1999;274:13242–13249. doi: 10.1074/jbc.274.19.13242. [DOI] [PubMed] [Google Scholar]
- 20.Sakurai Y, Tsukamoto K, Sawai T. Nucleotide sequence and characterization of a carbenicillin-hydrolyzing penicillinase gene from Proteus mirabilis. J Bacteriol. 1991;173:7038–7041. doi: 10.1128/jb.173.21.7038-7041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawai T, Kanno M, Tsukamoto K. Characterization of eight β-lactamases of gram-negative bacteria. J Bacteriol. 1982;152:567–571. doi: 10.1128/jb.152.2.567-571.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sawai T, Yoshida T, Tsukamoto K, Yamagishi S. A set of bacterial strains for evaluation of β-lactamase-stability of β-lactam antibiotics. J Antibiot. 1981;34:1318–1326. doi: 10.7164/antibiotics.34.1318. [DOI] [PubMed] [Google Scholar]
- 24.Senda K, Arakawa Y, Ichiyama S, Nakashima K, Ito H, Ohsuka S, Shimokawa K, Kato N, Ohta M. PCR detection of metallo-β-lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum β-lactams. J Clin Microbiol. 1996;34:2909–2913. doi: 10.1128/jcm.34.12.2909-2913.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeshita S, Sato M, Toda M, Masahashi W, Hashimoto-Gotoh T. High-copy-number vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61:63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- 26.Thompson J S, Malamy M H. Sequencing the gene for an imipenen-cefoxitin-hydrolyzing enzyme (CfiA) from Bacteroides fragilis TAL2480 reveals strong similarity between CfiA and Bacillus cereus β-lactamase II. J Bacteriol. 1990;172:2584–2593. doi: 10.1128/jb.172.5.2584-2593.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsukamoto K, Tachibana K, Yamazaki N, Ishii Y, Ujiie K, Nishida N, Sawai T. Role of lysine-67 in the active site of class C β-lactamase from Citrobacter freundii GN346. Eur J Biochem. 1990;188:15–22. doi: 10.1111/j.1432-1033.1990.tb15365.x. [DOI] [PubMed] [Google Scholar]
- 28.Viera J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 29.Walsh T R, Hall L, Assinder S J, Nichols W W, Cartwright S J, Macgowan A P, Bennett P M. Sequence analysis of the L1 metallo-β-lactamase from Xanthomonas maltophilia. Biochim Biophys Acta. 1994;1218:199–201. doi: 10.1016/0167-4781(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 30.Yamaguchi A, Hirata T, Sawai T. Kinetic studies on inactivation of Citrobacter freundii cephalosporinase by sulbactam. Antimicrob Agents Chemother. 1983;24:23–30. doi: 10.1128/aac.24.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]