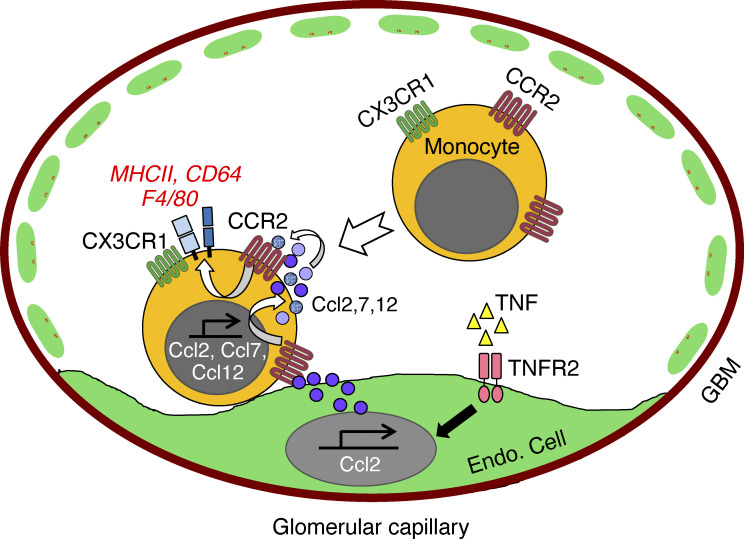
Changes in monocyte state occur during inflammation. We show that following glomerular inflammation, CCR2+CX3CR1+ double-positive monocytes, which are preferentially recruited, acquire macrophage features while still within the renal vasculature, in a process requiring CCR2 on monocytes and TNF-TNFR2–activated endothelial cells.
Abstract
Monocytes undergo phenotypic and functional changes in response to inflammatory cues, but the molecular signals that drive different monocyte states remain largely undefined. We show that monocytes acquire macrophage markers upon glomerulonephritis and may be derived from CCR2+CX3CR1+ double-positive monocytes, which are preferentially recruited, dwell within glomerular capillaries, and acquire proinflammatory characteristics in the nephritic kidney. Mechanistically, the transition to immature macrophages begins within the vasculature and relies on CCR2 in circulating cells and TNFR2 in parenchymal cells, findings that are recapitulated in vitro with monocytes cocultured with TNF-TNFR2–activated endothelial cells generating CCR2 ligands. Single-cell RNA sequencing of cocultures defines a CCR2-dependent monocyte differentiation path associated with the acquisition of immune effector functions and generation of CCR2 ligands. Immature macrophages are detected in the urine of lupus nephritis patients, and their frequency correlates with clinical disease. In conclusion, CCR2-dependent functional specialization of monocytes into macrophages begins within the TNF-TNFR2–activated vasculature and may establish a CCR2-based autocrine, feed-forward loop that amplifies renal inflammation.
Graphical Abstract
Introduction
Circulating monocytes are heterogeneous and play key roles in the induction and resolution of the inflammatory response (Mildner et al., 2013). Monocytes consist of two functionally distinct subsets distinguished by expression of chemokine receptors CCR2 and CX3CR1. Nonclassical, “patrolling” Ly6CloCCR2loCX3CR1hi monocytes are blood-resident cells that do not extravasate but survey the luminal side of endothelial cells, where they contribute to tissue repair and regulate vascular inflammation. Classical, “migratory” Ly6ChiCCR2hi monocytes remain in circulation for about a day before entering tissue under homeostasis or in response to inflammatory cues (Jakubzick et al., 2017). These cells can transition into CCR2+CX3CR1+ double-positive (DP) cells at a site of sterile injury (Dal-Secco et al., 2015) and/or be preferentially recruited from the blood, as was shown for skin infection (Romano et al., 2017). Classical monocytes exhibit considerable functional plasticity. Under steady state, classical monocytes traverse tissue and transport antigen to lymph nodes without differentiating, enter and establish a secondary reservoir in the spleen, or return to the bone marrow and convert into nonclassical monocytes (Jakubzick et al., 2017). During inflammation, they can maintain their monocytic state in tissues while differentiating into effector cells that polarize toward a proinflammatory or reparative phenotype depending on microenvironmental influences (Dal-Secco et al., 2015; Mildner et al., 2013). They can also transition to a classically activated (M1, proinflammatory) macrophage and, over time, to an alternatively activated (M2, reparative) macrophage and/or integrate into a longer-lasting resident macrophage pool, which in many organs is a separate lineage seeded in tissues before birth and maintained by self-renewal (Guilliams et al., 2018; Nelson et al., 2012).
Classical monocytes can progressively acquire macrophage-like markers (Guilliams et al., 2018; Jakubzick et al., 2017; Nelson et al., 2012). For example, Ly6C+ monocytes acquire MHCII and upregulate F4/80 and CD64 to attain the phenotype of mature macrophages in the colonic intestinal mucosa under steady state and during inflammation (Joeris et al., 2017; Tamoutounour et al., 2012). This single differentiation continuum from Ly6Chi monocytes to macrophages via a series of intermediate states is termed monocyte maturation. Ly6Chi monocytes and Ly6C− macrophages have also been reported to be independent lineages, with the latter being maintained by self-renewal in both the gut (De Schepper et al., 2018) and kidney (Lever et al., 2019). Thus, infiltrating monocyte-derived macrophages and tissue macrophages can coexist during inflammation.
Macrophages play a key role in glomerulonephritis (GN). In biopsies of patients with crescentic GN, macrophages associate with chronic renal damage and microvascular injury (Sean Eardley and Cockwell, 2005; Segerer et al., 2000). In a mouse model of GN, termed nephrotoxic nephritis (NTN; Artinger et al., 2017), disease is associated with the infiltration of bone marrow–derived monocytes and an expansion of renal macrophages (Chalmers et al., 2015; Munro and Hughes, 2017). CCR2 (Kurihara et al., 1997; Kuziel et al., 1997; Tsou et al., 2007) and CX3CR1 (Gautier et al., 2009; Prinz and Priller, 2010) are implicated in various monocyte functions including monocyte recruitment under some inflammatory conditions. A blockade or deficiency in CCR2 or its ligand, CCL2 (MCP-1), in most cases reduces renal macrophage accumulation and improves glomerular and interstitial injury in NTN (Lloyd et al., 1997; Panzer et al., 2001; Wada et al., 1996) and lupus nephritis (Pérez de Lema et al., 2005; Tesch et al., 1999), while the role of CX3CR1 is not as clear (Feng et al., 1999; Haskell et al., 2001; Hochheiser et al., 2013; Inoue et al., 2005). Generation of the cytokine TNF by the inflamed kidney drives GN, and a deficiency in parenchymal tissue of one of its receptors, TNFR2, prevents monocyte influx and associated renal injury (Al-Lamki and Mayadas, 2015).
Despite significant progress in understanding the role of monocytes and macrophages in homeostasis and disease pathogenesis (Ley et al., 2011; Moore et al., 2013; Nelson et al., 2012; Shan and Ju, 2020), the molecular cues that drive the maturation of classical monocytes into macrophages in inflammatory environments remain poorly understood (Jakubzick et al., 2017). Accordingly, it is unclear whether this is a stochastic process or is instructed by discrete stimuli generated by specific cell types: an important distinction, as the latter would provide important opportunities for therapeutic intervention. Here, we examine monocyte fate and phenotypes in the context of GN induced by nephrotoxic serum (NTS) and define the mechanisms that drive the observed monocyte accumulation and transition to immature macrophages using adoptive transfer approaches, multiphoton intravital microscopy (IVM), transcriptional profiling, and in vitro monocyte-endothelial cell cocultures.
Results
Renal accumulated monocytes express macrophage-like markers following GN
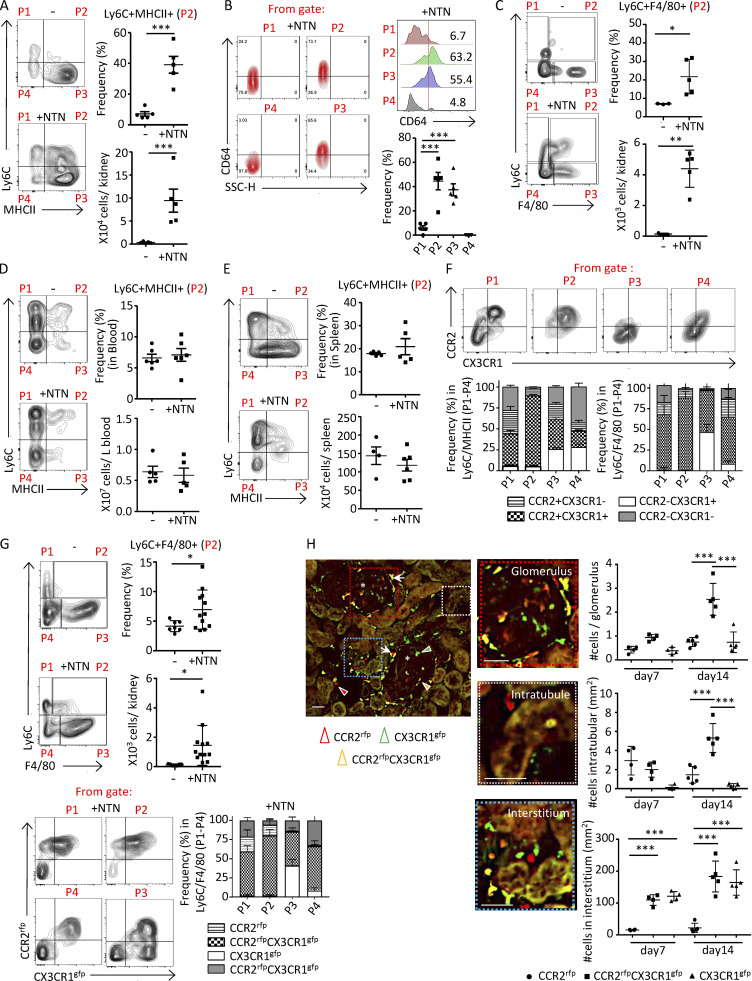
To characterize the nature of monocytes and macrophages that accumulate within the kidney during GN, wild-type (Wt) mice were left untreated or injected with heterologous rabbit NTS containing anti-glomerular basement membrane antibody after preimmunization with IgG in CFA to induce NTN. 14 d after NTS administration, cells from the kidney, blood, and spleen from mice with NTN and controls were compared by multiparameter flow cytometry. After excluding Ly6G+ neutrophils, CD11c+ dendritic cells, T cells, and NK cells within the CD11b+ gate (Fig. S1 A), cell surface expression of Ly6C and MHCII was evaluated (Fig. 1 A) on the remaining cells. In the control kidneys, we identified two major populations: Ly6C+MHC− cells (P1), which resemble the phenotype of circulating monocytes, and Ly6C−MHCII+ cells (P3), which are similar in phenotype to mature macrophages described in many other organs. Following NTN, we identified an abundant renal Ly6C+MHCII+ population (P2) in addition to the Ly6C+MHCII− and Ly6C−MHCII+ populations identified in the control kidneys. The majority of these Ly6C+MHCII+ cells expressed CD64 (Fig. 1 B), a widely accepted marker for cells of the macrophage lineage. A population of Ly6C+ cells that express the macrophage tissue marker F4/80 (Waddell et al., 2018) was also observed only within the kidneys of mice with NTN (Fig. 1 C). The frequency of Ly6C+MHCII+ cells within the blood (Fig. 1 D) and the spleen (Fig. 1 E) was similar before and after NTN, suggesting that the observed renal Ly6C+MHCII+ cells were generated from circulating Ly6C+ blood monocytes in response to signals generated within the inflamed kidney.
Figure S1.
Gating strategies, CCR2, and CX3CR1 antibody specificity and the analysis of CCR2 and CX3CR1 on blood monocytes. (A) Representative FACS plot of the gating strategy for the analysis of Ly6C and MHCII in monocyte/macrophage populations in the kidney 14 d after NTN induction. Live, CD45+ cells (not depicted) were analyzed under the Singlets+Ly6G−CD3−CD11c−NK−CD11b+ gate. Monocyte/macrophage populations were distinguished by Ly6C and MHCII. FSC, forward scatter; SSC, side scatter. (B) Specificity of monoclonal antibodies against CX3CR1 (1:100 dilution) and CCR2 (1:20 dilution) were confirmed by staining blood samples isolated from Wt, Cx3cr1 KO (Cx3cr1gfp/gfp), Ccr2 KO (Ccr2rfp/rfp), and DKO (Ccr2rfp/rfpCx3cr1gfp/gfp) mice. A representative FACS plot is shown. CX3CR1 and CCR2 expression assessed by antibodies and GFP and RFP were analyzed under a CD45+CD11b+ gate. (C) Blood was collected from Wt mice on day 14 after NTN induction and analyzed by flow cytometry. Blood leukocytes were divided based on CD45+CD11b+ and CD45+CD11b− gates and further stained for CX3CR1 and CCR2; monocyte markers Ly6C and CD115; and lineage-negative, CD11c, Ly6G, and CD3. Histogram plots (right) show that all CCR2+CX3CR1+ DP populations express Ly6C and CD115. None of the DP populations express CD11c, Ly6G, or CD3.
Figure 1.
Renal accumulated monocytes in nephritic mice express macrophage markers and are DP for CCR2 and CX3CR1. Mice were either untreated (−) or subjected to NTN (+NTN) and analyzed on day 14. CD11b+ lineage-negative cells were evaluated for Ly6C and MHCII or Ly6C and F4/80 as indicated. The defined P1–P4 populations were further analyzed for CD64 or CCR2 and CX3CR1. (A) Representative FACS plots of four subsets of monocytes in the kidney of Wt mice defined by Ly6C and MHCII distinguishes four populations (P1–P4; left). The frequency and absolute counts of the Ly6C+MHCII+ (P2) population (right) are shown. (B) Populations P1–P4 defined in A were further analyzed for CD64. Representative FACS profiles (left), histogram representation (right upper) and frequency graph (right lower) of CD64 in P1–P4 populations are shown. SSC, side scatter. (C) Representative FACS plots of Ly6C and F4/80 in Ccr2rfp+/−Cx3cr1gfp+/− reporter mice. The frequencies and absolute counts of the Ly6C+F4/80+ (P2) population are shown. (D and E) Representative FACS profiles, frequencies, and absolute counts of Ly6C+MHCII+ (P2) cells in blood (D) and spleen (E). (F) Representative FACS plots for P1–P4 populations defined for Ly6C and MHCII in nephritic mice in A and further analyzed for CCR2 and CX3CR1. The frequency of renal subpopulations defined by CCR2 and CX3CR1 in Ly6C and MHCII parent P1–P4 populations (A, +NTN) and Ly6C and F4/80 parent P1–P4 populations (C, +NTN), are shown in grouped bar graphs. (G) Analysis of radiation chimeras of Wt recipients reconstituted with bone marrow from Ccr2rfp+/−Cx3cr1gfp+/− reporter mice with or without NTN induction. Representative profiles of Ly6C monocytes evaluated for F4/80 expression (left) and the frequencies and absolute counts of the P2 (Ly6C+F4/80+) populations (right) are given. In bottom panels, frequency of populations P1–P4 further analyzed for CCR2 and CX3CR1 (as shown in the representative FACs plot) are shown in a grouped bar graph. (H) Histological analysis of three monocyte populations in kidney sections of radiation chimeric Wt recipients reconstituted with Ccr2rfp+/−Cx3cr1gfp+/− bone marrow identified by CCR2rfp and CX3CR1gfp in the glomerulus, intratubular and renal interstitial compartment on days 7 and 14 after NTN induction. Representative image of monocytes in glomeruli (asterisks) and juxtaglomerular regions (white arrows) are shown. Scale bar = 50 μm. Cells expressing CCR2 (red), CX3CR1 (green), or both fluorophores (yellow) are indicated by arrowheads. Enlarged areas of the glomerulus, interstitium, and intratubular space are shown. The number of cells in each of these compartments was determined. Two independent experiments were performed for A–H. Data are mean ± SEM. For A–E, G, and H, unpaired two-tailed t test (two data sets comparison) and one-way ANOVA (Tukey’s multiple comparison test) were performed. *, P < 0.05; **, P < 0.01; ***, P < 0.005 vs. normal basal values.
Notably, many Ly6C+MHCII− (P1) cells and virtually all Ly6C+MHCII+ (P2) cells within the nephritic kidney were positive for both CCR2 and CX3CR1 (using validated antibodies; Figs. S1 B and 1 F). The same was observed for Ly6C+F4/80+ (P2) cells (Fig. 1 F). This suggests that the majority of monocytes that acquire MHCII or F4/80 are CCR2 and CX3CR1 DP. Consistent with a potential key role for circulating monocytes in contributing to renal CCR2+CX3CR1+ cells, CD11b cells isolated from the kidneys of radiation chimeric Wt mice reconstituted with bone marrow from Ccr2rfp/+Cx3cr1gfp/+ reporter mice and subjected to NTN exhibited broad expression of both RFP and GFP on Ly6C+F4/80+ (P2) cells (Fig. 1 G). Cells with dual RFP and GFP were found within the glomerulus, tubules, and interstitium of the nephritic kidney (Fig. 1 H); the pronounced accumulation of these cells in the tubulointerstitium is significant, as inflammation in this area secondary to glomerular injury can result in the progressive loss of kidney function (Kriz and LeHir, 2005).
Together, these observations indicate that following NTN, circulating Ly6C+ monocytes that accumulate in the kidney acquire macrophage markers, MHCII, CD64, and F4/80 and are distinguished by the expression of both CCR2 and CX3CR1. These observations support the hypothesis that these cells, herein referred to as immature macrophages, accumulate in the nephritic kidney and contribute to renal injury.
CCR2+CX3CR1+ DP monocytes are preferentially recruited and exhibit a TNFR2-dependent increase in intravascular retention time in the glomerulus
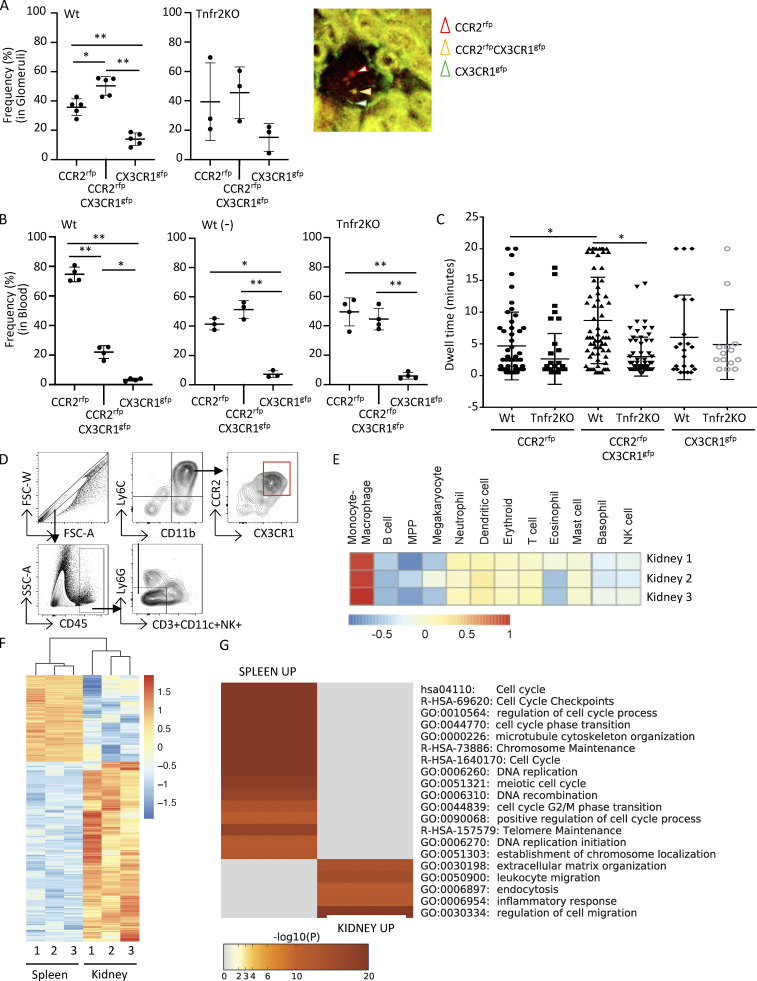
The peripheral blood of nephritic mice contains CCR2 and CX3CR1 DP cells, with almost all expressing Ly6C+ and CD115+, which confirms their monocyte lineage (Fig. S1 C). As renal immature macrophages are DP for CCR2 and CX3CR1, we determined whether monocytes expressing both these receptors are preferentially recruited from blood and/or exhibit a longer retention time within glomerular capillaries following NTN, both of which would lead to their enhanced accumulation in the tissue. Given that endothelial TNFR2 promotes monocyte accumulation in NTN and signals the generation of monocyte chemokines (Al-Lamki and Mayadas, 2015; Venkatesh et al., 2013), we also determined whether parenchymal TNFR2 is required for monocyte recruitment and/or retention. To examine monocyte interactions within glomerular capillaries, we conducted multiphoton IVM in the kidney of radiation chimeric Wt and Tnfr2 KO mice reconstituted with bone marrow from Ccr2rfp/+Cx3cr1gfp/+ reporter mice and subjected to NTN 10 d before IVM. In parallel, flow cytometric analysis of peripheral blood was undertaken to calculate the frequency of blood monocyte populations, as it was not possible to accurately count freely flowing leukocytes by IVM. We observed that the frequency of CCR2rfpCX3CR1gfp cells was significantly higher in glomeruli (Fig. 2 A) and lower in the blood (Fig. 2 B), compared with CCR2rfp cells in chimeric Wt mice. This suggests that the DP blood monocytes are preferentially recruited to the glomerulus during NTN, a result further supported by the overall reduction of this population in the blood of nephritic vs. untreated mice (Fig. 2 B). In contrast, chimeric mice deficient in parenchymal TNFR2 showed comparable, rather than enhanced, recruitment of CCR2rfpCX3CR1gfp vs. CCR2rfp monocytes in glomeruli (Fig. 2 A). In the blood, these two monocyte populations were equally represented and mirrored those observed in untreated Wt animals (Fig. 2 B). These data are consistent with the reported requirement of parenchymal TNFR2 to develop NTN (Venkatesh et al., 2013; Vielhauer et al., 2005) and, expectedly, a reactive change in the peripheral blood inflammatory cell composition. Thus, TNFR2 deficiency has no effect on the recruitment of DP monocytes per se. Next, we evaluated the dwell time of recruited monocytes. We found that recruited CCR2rfpCX3CR1gfp cells exhibited a longer retention time within glomerular capillaries of Wt mice compared with those expressing CCR2rfp or CX3CR1gfp alone, with many of them dwelling for >20 min (Fig. 2 C and Video 1). Importantly, this was not observed in chimeric mice lacking parenchymal TNFR2 (Fig. 2 C and Video 2).
Figure 2.
Renal accumulated Ly6C+CCR2+CX3CR1+ monocytes exhibit an increase in intravascular dwell time within glomerular capillaries and have a proinflammatory transcriptional profile. (A–C) Radiation chimeric Wt and Tnfr2 KO recipient mice reconstituted with Ccr2rfp/+Cx3cr1gfp/+ (red signal, CCR2-rfp; green signal, CX3CR1-gfp) reporter bone marrow were analyzed on day 10 after NTN induction. (A) Two-photon IVM of glomeruli was undertaken, and images were acquired every 30 s for 20 min. The frequency of monocyte subpopulations among recruited cells, defined as a cell adherent for two frames (60 s) and a representative image of a glomerulus with cells positive for RFP (CCR2rfp), GFP (CX3CR1gfp), or both fluorophores (CCR2rfpCX3CR1gfp), is shown. Three to five mice per group and three to four glomeruli were imaged per mouse across six independent experiments. (B) Blood taken before (−) and after induction of NTN in Wt chimeras and after NTN in Tnfr2 KO chimeras were analyzed by flow cytometry. (C) The dwell time of individual monocytes within glomeruli analyzed in A is given. One-way ANOVA (Tukey’s multiple comparison test) was performed. *, P < 0.005; **, P < 0.0001. (D) FACS sorting strategy for isolating Ly6C+ monocytes expressing both CCR2 and CX3CR1 using lineage-negative markers to exclude T cells, dendritic cells, and NK cells and gating remaining cells for CD11b and Ly6C and then CCR2 and CX3CR1. CCR2 and CX3CR1 DP cells were collected. FSC, forward scatter; SSC, side scatter. (E) Haemopedia cell type fingerprint heatmap. Shown are the mean row-normalized log2 CPM values for the indicated gene sets, clustered by gene set for the renal DP population. (F) Heatmap of DEGs in renal DP cells vs. spleen DP cells. Shown are row-normalized log2 CPM values. (G) Top 20 GO terms enriched in the DEGs of E. Data are from three biological replicates, with each replicate containing sorted kidney and spleen/bone marrow cells from 10 mice.
Video 1.
IVM imaging showing dwell time of CCR2rfp (red), CX3CR1gfp (green), and CCR2rfpCX3CR1gfp (yellow) cells within capillaries of glomeruli in a radiation chimeric Wt recipient reconstituted with Ccr2rfp/+Cx3cr1gfp/+ reporter bone marrow subjected to NTN and imaged 14 d later. Frame rate is 8 frames/s.
Video 2.
IVM imaging showing dwell time of CCR2rfp (red), CX3CR1gfp (green), and CCR2rfpCX3CR1gfp (yellow) cells within capillaries of glomeruli in a radiation chimeric Tnfr2 KO recipient reconstituted with Ccr2rfp/+Cx3cr1gfp/+ reporter bone marrow subjected to NTN and imaged 14 d later. Frame rate is 8 frames/s.
Together, our data show that CCR2 and CX3CR1 DP monocytes are preferentially recruited and have a longer dwell time within glomerular capillaries following NTN, with the enhanced retention being dependent on parenchymal TNFR2. An NTS-induced increase in the dwell time of neutrophils and monocytes within the glomerular capillaries has been shown to correlate with their activation (Devi et al., 2013; Nishi et al., 2017; Turner-Stokes et al., 2020). Thus, TNFR2 within inflamed glomerular capillaries may facilitate the activation and subsequent transition of recruited CCR2+CX3CR1+ monocytes to immature macrophages.
Renal-derived Ly6C+CCR2+CX3CR1+ DP monocytes exhibit a proinflammatory and migratory transcriptional profile
To determine whether the renal accumulated CCR2+CX3CR1+ monocytes exhibit proinflammatory features, we sorted Ly6C+CCR2+CX3CR1+ cells from both the kidneys and spleen/bone marrow of Wt mice with NTN (Fig. 2 D) and analyzed gene expression using bulk RNA sequencing (RNA-seq). Both spleen and bone marrow were needed to retrieve sufficient cell numbers for analysis due to the low frequency of this specific monocyte population in these two organs. The monocyte/macrophage lineage of the sorted population from three independent pooled nephritic kidney samples was assessed by a fingerprint of distinctive Haemopedia gene sets (de Graaf et al., 2016; Fig. 2 E). Differentially expressed genes (DEGs) between spleen/bone marrow and kidney CCR2+CX3CR1+ cells were identified (Fig. 2 F and Table S1). Interestingly, although cells isolated from the kidney were clearly of the monocyte-macrophage lineage, inflammatory processes were higher in cells isolated from the kidney than the spleen (Fig. 2 G and Table S1). Gene Ontology (GO) enrichment analysis of these DEGs revealed a downregulation of cell cycle genes and an increase in the expression of genes associated with leukocyte migration and the inflammatory response in the kidney vs. the spleen/bone marrow populations (Fig. 2 G). These experiments strongly support the hypothesis that CCR2+CX3CR1+ DP monocytes that accumulate within the nephritic kidney acquire a proinflammatory phenotype.
Ly6C+CCR2+CX3CR1+ monocytes are pathogenic
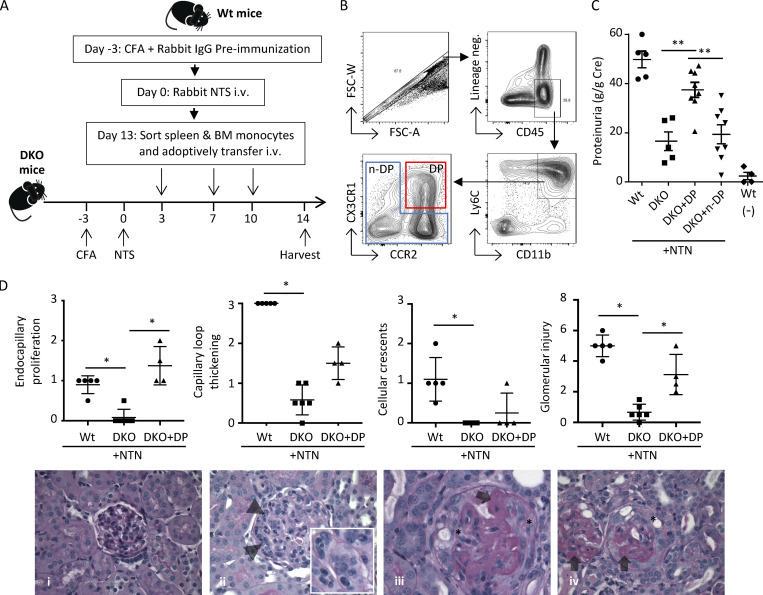
To examine whether Ly6C+CCR2+CX3CR1+ cells that accumulate in the kidney are pathogenic, we pursued an adoptive transfer approach in Ccr2 and Cx3cr1 double-knockout (Ccr2rfp/rfpCx3cr1gfp/gfp DKO) mice, which are largely resistant to developing disease (see Fig. S2 C). As depicted in Fig. 3 A, Ly6C+ monocytes were isolated from the spleen and bone marrow of Wt mice subjected to NTN and sorted for CCR2+CX3CR1+ DP monocytes. The remaining non-DP (n-DP) cells were collected as our comparator. The sorting strategy is shown in Fig. 3 B. Each population was injected i.v. into cohorts of DKO recipient mice that had been subjected to NTN. Controls included unmanipulated DKO and Wt mice that were subjected to NTN and sham-injected Wt mice. Urine was collected and kidneys were harvested from recipient mice on day 14 after NTN induction. DKO mice that were subjected to NTN and sham injected or given Ly6C+ n-DP monocytes had low proteinuria, whereas those given DP monocytes exhibited proteinuria that was comparable to that observed in Wt mice subjected to NTN (Fig. 3 C). Histologically, there was an increase in endocapillary proliferation and glomerular injury score in kidneys of DKO recipient mice that received DP monocytes vs. those that were sham-injected (Fig. 3 D).
Figure S2.
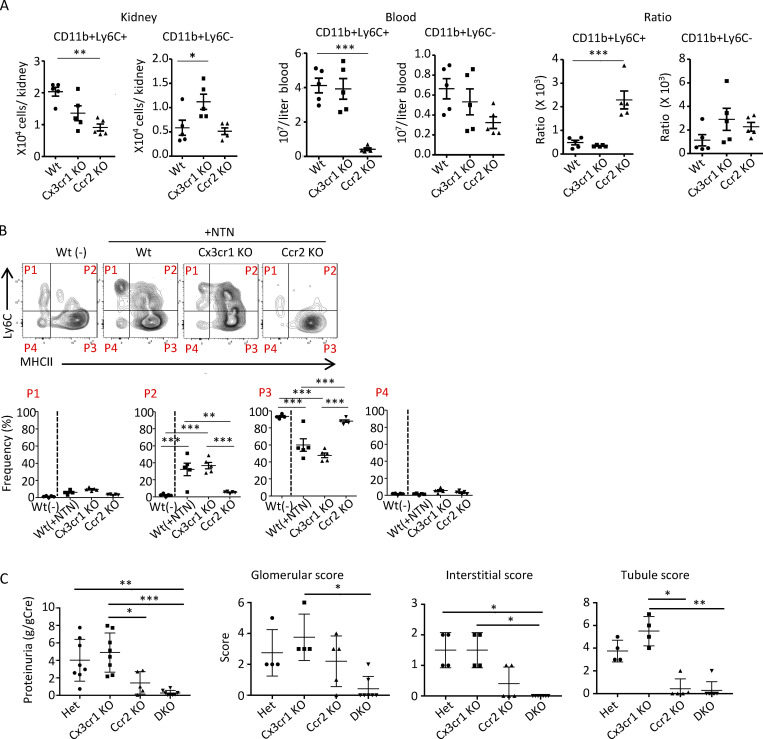
NTN-induced monocyte recruitment per se is not affected, but proteinuria and histological evidence of renal injury are reduced in CCR2-deficient mice. (A) The number of CD11b+Ly6C+ and CD11b+Ly6C− cells in the kidney and blood of Wt, Cx3cr1 KO, and Ccr2 KO mice 60 min after NTS injection (in the absence of priming with CFA). As CCR2 KO mice had a reduction in circulating blood monocytes, a ratio of monocytes in kidney vs. blood was calculated as an index of monocyte recruitment. (B) NTN was induced in Wt, Cx3cr1 KO, and Ccr2 KO mice and analyzed along with untreated Wt (−) animals. Representative FACS plots using Ly6C and MHCII to distinguish mononuclear populations P1–P4 and frequencies in each population are shown. (C) Proteinuria in radiation chimeras of Wt recipients reconstituted with Het (Ccr2rfp/+Cx3cr1gfp/+, n = 8), Cx3cr1 KO (Cx3cr1gfp/gfp, n = 8), Ccr2 KO (Ccr2rfp/rfp, n = 5), or DKO (Ccr2rfp/rfpCx3cr1gfp/gfp, n = 8) bone marrow on day 14 after NTN induction. Histological scores for glomerular, interstitial, and tubular injury in Het (n = 4), Cx3cr1 KO (n = 4), Ccr2 KO (n = 5), and DKO (n = 7) on day 14 after NTN induction. Two independent experiments were performed. *, P < 0.05, **, P < 0.01, ***, P < 0.005 (Tukey’s multiple comparison test).
Figure 3.
Adoptively transferred Ly6C+CCR2+CX3CR1+ monocytes promote glomerular injury. (A) Schematic for adoptive transfer of cells and induction of NTN. Spleen/bone marrow monocytes were sorted from cohorts of Wt donor mice on day 13 after induction of NTN and injected at indicated days into Ccr2 and Cx3cr1 DKO recipient mice that had been immunized with CFA and NTS to induce NTN. Urine samples and both kidneys of DKO recipient mice were harvested on day 14 for analysis of proteinuria and histological scoring of GN. (B) Flow cytometry gating strategy for sorting CD45+, lineage-negative (CD3−NK−Ly6G−CD11c−), CD11b+Ly6C+CX3CR1+CCR2+ DP (red box), and the remaining non-DP fractions (n-DP, blue box). FSC, forward scatter. (C) Proteinuria in Wt mice with (n = 5) and without (n = 4) NTN, DKO mice (n = 5), and recipient DKO mice transferred with DP (n = 8) or n-DP (n = 8) cells. Two independent experiments were performed. (D) Histological scores for endocapillary proliferation, capillary loop thickening, cellular crescents, and glomerular injury in Wt, DKO, and DKO + DP cells on day 14 after NTN induction. Representative histological images of periodic acid–Schiff-stained slides are shown. (i) A normal glomerulus with normal glomerular capillary loops (thickness) and normocellular without evidence of an active glomerulitis (i.e., endocapillary proliferation or cellular crescents). (ii) A glomerulus with global endocapillary proliferation, with inflammatory cells within the glomerular capillary loops indicated by arrowheads. (iii and iv) Glomeruli with severe glomerular capillary loop thickening (indicated with arrows) and extracapillary proliferation of cellular crescents (indicated with asterisks). Original magnifications for i, ii, and iv, 600×; for iii, 400×. Data are mean ± SEM. *, P < 0.05; **, P < 0.01.
Monocyte CCR2 and parenchymal TNFR2 are required for the generation of immature macrophages in the nephritic kidney
To begin to understand the molecular underpinnings of monocyte conversion to immature macrophages, we assessed whether CCR2 and/or CX3CR1 were required for acquisition of F4/80 and MHCII. First, we determined whether these two chemokine receptors were essential for monocyte recruitment in GN. We examined monocyte accumulation for only 2 h vs. longer time periods after NTS injection to exclude indirect effects of receptor deficiency on monocyte recruitment. Cx3cr1 KO and Wt mice had comparable monocyte accumulation in the kidney, while a reduction was observed in Ccr2 KO mice (Fig. S2 A). However, Ccr2 KO mice also exhibited a marked reduction in monocytes in the blood, consistent with a role for this receptor in their emergency release during inflammation (Tsou et al., 2007). Nonetheless, a ratio of renal vs. blood monocytes as an index of recruitment showed no effect of CCR2 deficiency on renal monocyte accumulation per se (Fig. S2 A). Thus, CX3CR1 and CCR2 are not required for early NTS-induced renal monocyte recruitment.
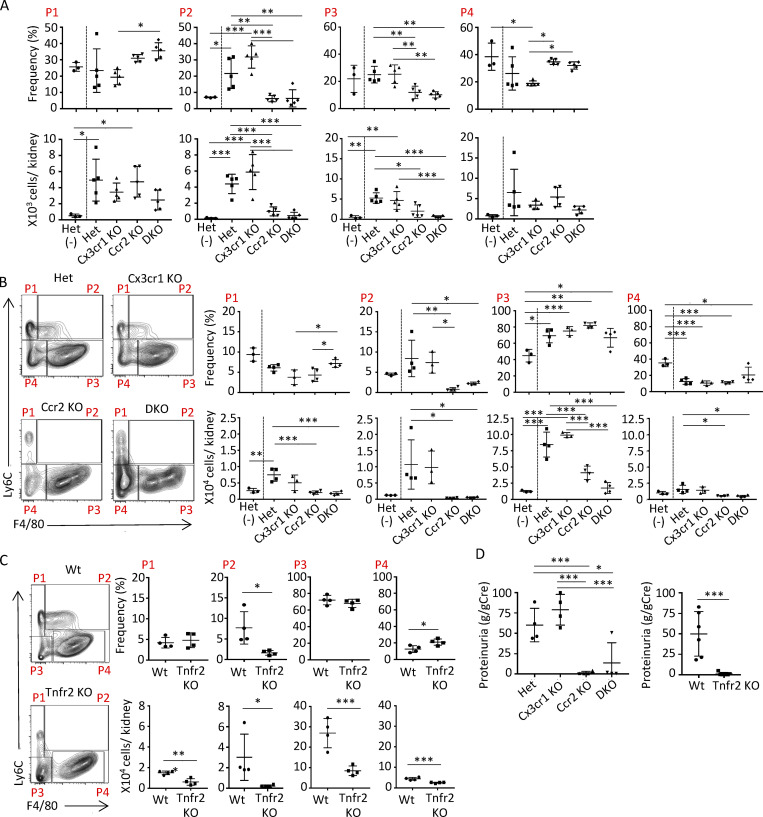
Next, we examined the acquisition of F4/80 and MHCII on Ly6C+ monocytes on day 10 after the induction of NTN in heterozygous controls (Ccr2rfp/+Cx3cr1gfp/+, heterozygous-reporter mice) and mice with a KO of one or both chemokine receptors: Ccr2 KO (Ccr2rfp/rfpCx3cr1gfp/+), Cx3cr1 KO (Ccr2rfp/+Cx3cr1gfp/gfp), and DKO (Ccr2rfp/rfpCx3cr1gfp/gfp) mice. We found that the frequency of Ly6C+F4/80+ (P2) monocytes was significantly reduced in CCR2 and DKO mice compared with Cx3cr1 KO and heterozygous controls (Fig. 4 A). Similar results were obtained using MHCII to define immature macrophages (Fig. S2 B). Ccr2 KO mice exhibited a significant reduction in proteinuria, a hallmark of glomerular injury, and histological indices of renal injury while disease parameters in Cx3cr1 KO mice were comparable to those in heterozygous controls and largely absent in mice lacking both chemokine receptors (Fig. S2 C). CCR2 is present on both hematopoietic and renal cells (Lee et al., 2009). Thus, we examined the role of CCR2 in the hematopoietic compartment in generating immature macrophages by generating radiation chimeras of Wt recipient mice reconstituted with bone marrow from heterozygous, Ccr2 KO, Cx3cr1 KO, or DKO mice. The absence of hematopoietic CCR2 in chimeric mice significantly reduced the accumulation of Ly6C+F4/80+ immature macrophages (Fig. 4 B).
Figure 4.
CCR2 on hematopoietic lineages and TNFR2 on parenchymal cells are required for the generation of immature macrophages and kidney injury. (A) NTN was induced in mice heterozygous for CCR2 and CX3CR1 (Het), Cx3cr1 KO, Ccr2 KO, or lacking both receptors (DKO) and analyzed along with Het untreated animals (−). On day 14, Ly6C and F4/80 were examined on kidney samples to distinguish P1–P4 monocyte-macrophage populations. (B) NTN was induced in radiation chimeric Wt recipients reconstituted with heterozygous, Cx3cr1 KO, Ccr2 KO, or DKO bone marrow. Representative FACS plots for indicated chimeric mice subjected to NTN and the frequency and absolute counts of P1–P4 populations are shown. (C) NTN was induced in radiation chimeras of Wt or Tnfr2 KO recipients reconstituted with bone marrow heterozygous for CCR2 and CX3CR1 and analyzed as in A. FACS plots and frequencies and absolute counts of P1–P4 populations are shown. (D) Proteinuria evaluated in radiation chimeras examined in B (left) and C (right). Two independent experiments were performed. *, P < 0.05; **, P < 0.01; ***, P < 0.005 (Tukey’s multiple comparison test).
TNF promotes the generation of CCR2 ligands by the endothelium (Rollins et al., 1990; Strieter et al., 1989) and podocytes (Chung et al., 2015; Ohta et al., 2000), and its receptor TNFR2 in parenchymal cells contributes to monocyte recruitment and associated renal injury in NTN (Al-Lamki and Mayadas, 2015). Thus, we hypothesized that renal TNFR2 contributes to the CCR2-dependent accumulation of immature macrophages following NTN. As TNFR2 is present on both hematopoietic and renal cells (Al-Lamki and Mayadas, 2015), we examined radiation chimeras of Tnfr2 KO recipients reconstituted with bone marrow from Ccr2rfp/+Cx3cr1gfp/+ reporter mice. The absence of TNFR2 on parenchymal cells significantly decreased the accumulation of Ly6C+F4/80+ immature macrophages following NTN (Fig. 4 C).
Proteinuria was significantly attenuated in chimeric Ccr2 KO and DKO mice compared with Cx3cr1 KO and heterozygous, reporter animals (Fig. 4 D) and mice with Tnfr2 KO in parenchymal cells (Fig. 4 D). The observed decrease in immature macrophages in the CCR2 and TNFR2 chimeras likely contributes to the reduced proteinuria, but it is expected that the primary reason is the reduction in the absolute number of renal monocytes in these animals.
Ly6C+ monocytes acquire MHCII and F4/80 within the vasculature of the nephritic kidney
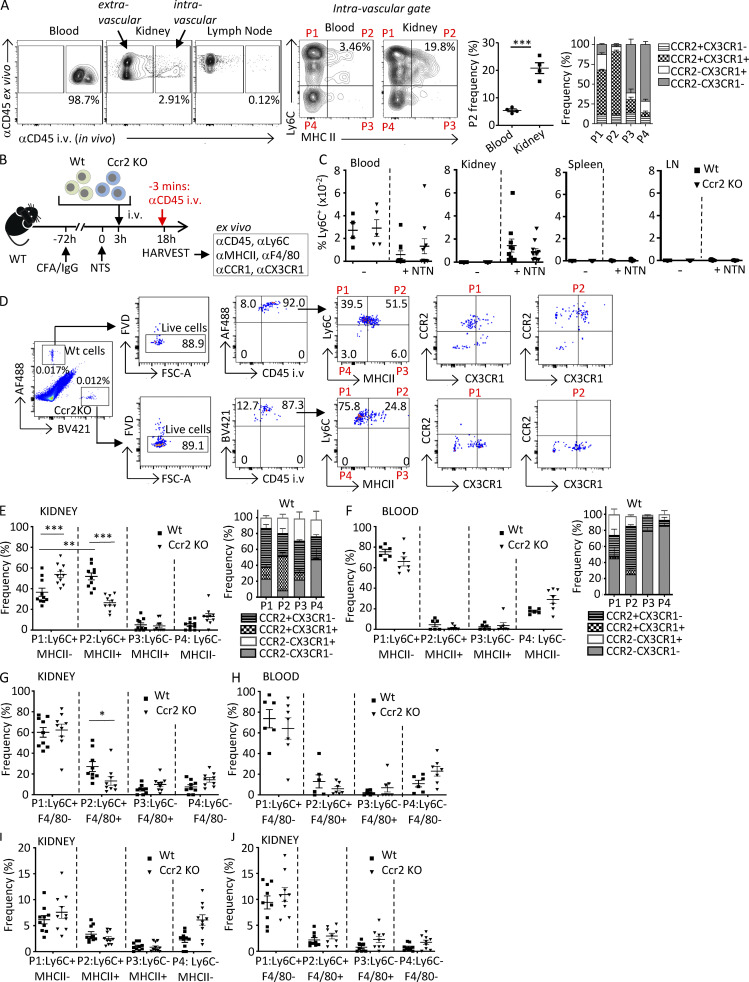
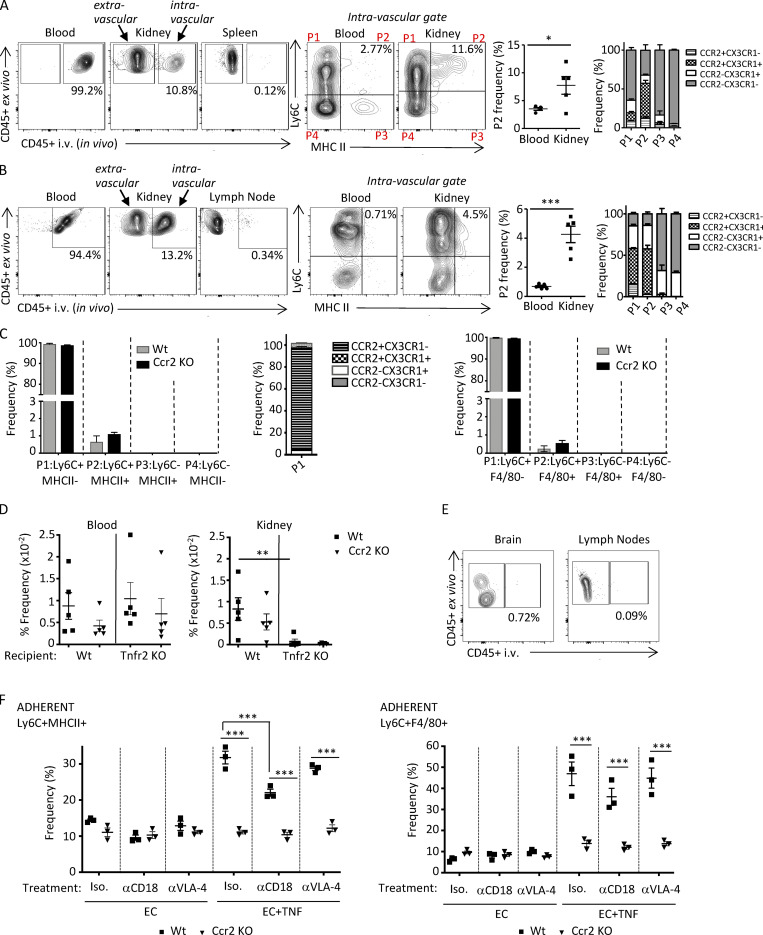
Our radiation chimeric studies suggest that CCR2 monocyte intrinsic functions drive monocyte transition to immature macrophages within the inflamed kidney. However, an outstanding question with important mechanistic implications is whether the initial steps of monocyte maturation occur after their extravasation into tissue or whether maturation begins upon interaction of monocytes with the inflamed endothelium before extravasation. To distinguish between these possibilities, 14 d after induction of NTN, mice were injected i.v. with fluorophore-labeled CD45 antibody for in vivo labeling and euthanized 3 min later. This short time period of in vivo CD45 labeling restricts staining to circulating leukocytes and those adherent to the vessel wall and therefore discriminates between vascular and tissue leukocytes (Anderson et al., 2014). Single-cell suspensions were prepared from blood, lymph node, spleen, and kidney and then stained ex vivo with Ly6C, MHCII, CCR2, and CX3CR1 as well as a CD45 antibody labeled with a different fluorophore than the antibody used for in vivo labeling. While virtually all cells in the blood that were positive for the ex vivo CD45 antibody were also labeled with the in vivo CD45 antibody, minimal in vivo CD45 labeling was detected in the lymph node, confirming that the in vivo CD45 antibody was exclusively labeling leukocytes located within the intravascular compartment. Importantly, we identified in vivo CD45 antibody–labeled Ly6C+MHCII+ macrophages in the kidney that were additionally CCR2 and CX3CR1 positive, a population that was significantly higher in the kidney vs. the blood (Fig. 5 A). This differential distribution in the kidney vs. the blood suggests that the Ly6C+ monocytes acquire macrophage markers while still within the inflamed renal vasculature. A similar analysis in nephritic kidneys of lupus-prone mice analyzed 42 d after IFNγ-induced accelerated nephritis also showed an intravascular Ly6C+MHCII+ population that was CCR2+CX3CR1+ and was significantly more frequent in the kidney than in the blood (Fig. S3 A).
Figure 5.
Monocytes acquire MHCII and F4/80 within the vasculature following NTN in a CCR2-dependent process. (A) On day 14 after induction of NTN in Wt mice, anti-CD45 was injected i.v. via the tail vein 3 min before harvesting blood, kidney, and lymph nodes to label intravascular cells. Single-cell suspensions of samples were further labeled ex vivo with antibodies to CD45 and monocyte and macrophage markers. Representative images of intravascular (i.e., αCD45 in vivo and ex vivo positive) and extravascular (i.e., αCD45 in vivo negative and ex vivo positive) cells are shown. In vivo–labeled CD45+, lineage-negative (CD3−CD11c−NK1.1−Ly6G−) cells further analyzed for Ly6C and MHCII defined four populations (P1–P4). The frequency of Ly6C+MHCII+ (P2) is shown in a scatter plot, and CX3CR1 and CCR2 frequency in P1–P4 populations is shown in a grouped bar graph. n = 4 mice. Significance was determined by two-tailed unpaired t test. (B) Schematic of timeline of adoptive transfer and induction of NTN for C–J. Ly6C+ monocytes purified from bone marrow of Wt and Ccr2 KO mice were labeled with CellTrace CFSE dye (AF488-green) and CellTrace violet dye (BV421-blue), respectively, and adoptively transferred into Wt mice, which were preimmunized with CFA plus rabbit IgG at −72 h and NTS at 0 h to induce NTN. Blood, kidney, spleen, and lymph nodes (LN) were harvested after 18 h. 3 min before harvest, mice were i.v. injected with anti-CD45. Harvested samples were stained ex vivo with indicated antibodies. (C) The frequencies of labeled Wt and Ccr2 KO monocytes in indicated organs of recipient Wt mice that were untreated (−) or subjected to NTN (+NTN) are shown. (D) Gating strategy for Wt and Ccr2 KO monocytes detected by CellTrace tracker dyes in whole-kidney homogenates from Wt recipients subjected to NTN. Cells assessed for in vivo CD45 labeling and then Ly6C and MHCII distinguished four populations (P1–P4) that were further analyzed for CCR2 and CX3CR1, for which representative profiles are only shown for P1 and P2, as they were the predominant populations. FSC, forward scatter. (E and F) Frequency of Ly6C+MHCII− (P1), Ly6C+MHCII+ (P2), Ly6C−MHCII+ (P3), and Ly6C−MHCII− (P4) for kidney (E) and blood (F) are shown. Frequency of CX3CR1 and CCR2 in P1–P4 populations are shown in grouped bar graphs. (G and H) Kidney (G) and blood (H) cells were also stained for Ly6C and F4/80 and analyzed as in E and F. (I and J) Frequency of Ly6C and MHCII (I) or F4/80 (J) extravascular populations, which are negative for the in vivo and positive for the ex vivo CD45 antibody, are shown. Two to three independent experiments were performed for all experiments. Data are expressed as mean ± SEM. Statistical significance was determined by one-way ANOVA with Dunnett’s t test correction. *, P < 0.05; **, P < 0.01; ***, P < 0.005.
Figure S3.
Intravascular monocyte populations following lupus nephritis and acute NTN, analysis of isolated monocytes for adoptive transfer, analysis of renal intravascular monocyte accumulation in Tnfr KO, in vivo CD45 antibody controls, and effect of blocking adhesion on monocyte acquisition of MHCII and F4/80. (A and B) Intravascular monocyte populations were determined in NZB/W lupus-prone mice with accelerated nephritis (A) and Wt mice subjected to acute NTN (B) using approaches described in Fig. 5. (C) Left: Monocytes isolated from the bone marrow of Wt and Ccr2 KO mice were examined for Ly6C and MHCII. All cells were Ly6C+, and >95% were Ly6C+MHCII− (P1) and <1.5% were Ly6C+MHCII+ (P2). Middle: The two Ly6C+ populations were further analyzed for CX3CR1 and CCR2. The majority of Ly6C+MHCII− (P1) cells were CCR2+ and CX3CR1−. Right: Monocytes from isolated bone marrow of Wt and Ccr2 KO were also examined for Ly6C and F4/80. All cells were Ly6C+: >95% were Ly6C+F4/80− and ≤0.5% were Ly6C+F4/80+. (D) Wt and TNFR2-deficient (Tnfr2 KO) mice subjected to NTN received CellTrace CFSE dye (AF488)–labeled Wt and CellTrace violet dye (BV421)–labeled Ccr2 KO monocytes i.v., and the frequencies of the labeled monocytes in blood and kidney were calculated. (E) Intravascular accumulation of leukocytes in lymph nodes and brain of mice subjected to acute NTN. Data are expressed as mean ± SEM. Statistical significance was determined by one-way ANOVA with Dunnett’s t test correction. (F) Monocytes treated with isotype (Iso.) or functional blocking antibody to CD18 (aCD18) or VLA-4 (aVLA-4) were incubated with endothelial cells or endothelial cells treated with TNF (EC + TNF). Adherent monocytes were evaluated for acquisition of MHCII (left) or F4/80 (right). *, P < 0.05; **, P < 0.01; ***, P < 0.005.
To confirm that the observed in vivo labeling of Ly6C+MHCII+ cells in the nephritic kidney did not result from extravasation of the i.v. administered CD45 due to NTN-induced increase in leakiness of the vascular endothelium on day 14 after NTS treatment (as evident by proteinuria; Figs. 4 D and S2 C), we decided to also conduct experiments 18 h after administering NTS, a time point at which no proteinuria is detected. Again, we identified in vivo CD45-labeled immature macrophages in the nephritic kidney that were present at a much higher frequency than in blood (Fig. S3 B). These results strongly suggest that monocytes acquire MHCII in the inflamed kidney while still within the renal vasculature and before they extravasate into the tissue.
Renal intravascular acquisition of MHCII and F4/80 by Ly6C+ monocytes is CCR2 dependent
To assess whether the intravascular monocyte acquisition of macrophage markers is dependent on the presence of CCR2 on monocytes, monocytes isolated from the bone marrow of Wt and Ccr2 KO mice (>99% MHCII and F4/80 negative; Fig. S3 C) were labeled with different fluorescent cell-staining dyes and injected i.v. into Wt mice that had been subjected to NTN 3 h previously. The blood, kidney, spleen, and lymph nodes were harvested 15 h later and 3 min after the i.v. injection of anti-CD45 antibody (Fig. 5 B). Cell suspensions of the organs were analyzed. Adoptively transferred Wt and Ccr2 KO Ly6C+ monocytes similarly accumulated in the inflamed kidney and were not detected in the spleen and lymph nodes (Fig. 5 C). Next, MHCII and CCR2 and CX3CR1 on intravascular Wt and Ccr2 KO monocytes was examined in the kidney (gating strategy, Fig. 5 D) and blood. A high frequency of in vivo CD45 antibody–labeled, Wt Ly6C+ monocytes had MHCII and were positive for both CCR2 and CX3CR1 (Fig. 5 E), while the frequency of this Ly6C+MHCII+ population was very low in the blood (Fig. 5 F). This result aligns with the observed enrichment of endogenous Ly6C+MHCII+ monocytes in the kidney of Wt mice 14 d after induction of NTN (Fig. 1 A). Importantly, in the kidney, the frequency of Ly6C+MHCII+ among in vivo labeled Ccr2 KO monocytes was significantly lower compared with Wt, and this was accompanied by an increase in the Ly6C+ population lacking MHCII (Fig. 5 E), consistent with the idea that Ly6C+ monocytes acquire MHCII within the vasculature of the inflamed kidney and that this is CCR2 dependent. Similar results were obtained in the kidney (Fig. 5 G) and blood (Fig. 5 H), using F4/80 to define the monocyte-macrophage populations. To determine whether the immature macrophages migrate into tissues, we examined MHCII (Fig. 5 I) and F4/80 (Fig. 5 J) on fluorophore-labeled adoptively transferred Wt and Ccr2 KO monocytes that were negative for in vivo CD45 antibody and were thus extravascular. The percentage of extravascular monocytes averaged <15%. This could mean that monocytes remain primarily intravascular within the first 18 h of NTN and/or that the adoptively transferred monocytes are compromised in their ability to migrate. Among the extravascular cells, the relative distribution of monocyte-macrophage populations defined by MHCII (Fig. 5 I) and F4/80 (Fig. 5 J) was similar to that found in the renal intravascular space (Fig. 5, E and G). This suggests that the identified intravascular populations are equally capable of extravasating, although we cannot rule out that differentially transmigrated populations acquire or lose Ly6C and/or MHCII once they are in the tissue.
To assess the role of renal TNFR2 in intravascular monocyte conversion, Wt and Ccr2 KO monocytes were injected into Wt and Tnfr2 KO mice. Very little accumulation of adoptively transferred Wt and Ccr2 KO monocytes was observed in Tnfr2 KO mice (Fig. S3 D). This is consistent with a role for parenchymal TNFR2 in monocyte accumulation in crescentic GN (Venkatesh et al., 2013; Vielhauer et al., 2005), potentially by increasing monocyte retention within the glomerular capillaries (Fig. 2 C). However, the large overall reduction in accumulation of adoptively transferred monocytes precluded the assessment of TNFR2’s role in monocyte conversion to immature macrophages. Together, these data suggest that the intravascular compartment of the inflamed kidney plays an active role in monocyte transition to immature macrophages and that this is instructed by CCR2 on monocytes.
Neutrophil depletion reduces the intravascular acquisition of MHCII and F4/80 by Ly6C+ monocytes
Neutrophils play a key role in the induction of NTN (Flores-Mendoza et al., 2018). Thus, we examined whether neutrophils were required for the intravascular monocyte acquisition of MHCII and F4/80 18 h after the induction of NTN using neutrophil depletion approaches and in vivo labeling methods described in Fig. 5. Neutrophil anti-Ly6G depleting antibody markedly depleted neutrophils in blood without significantly affecting monocyte counts (Fig. 6 A) or the frequency and numbers of in vivo anti-CD45–labeled monocyte-macrophage populations defined by MHCII, F4/80, or both (Fig. 6 B). Anti-Ly6G antibody partially depleted neutrophils in the kidney and resulted in a nonsignificant trend of reduction in the accumulation of monocytes following NTN (Fig. 6 C). Importantly, neutrophil depletion significantly decreased the frequency and number of renal intravascular monocytes acquiring MHCII and/or F4/80 (Fig. 6 D). Minimal in vivo CD45 antibody labeling was observed in the lymph node and brain, which shows vascular retention of the antibody (Fig. S3 E). Thus, neutrophils contribute to the transition of monocytes to immature macrophages. Although the mechanism for this was not further pursued, it is possible that neutrophil, membrane-bound TNF engages TNFR2 to induce TNFR2-dependent autocrine signaling (Venkatesh et al., 2013) and subsequent monocyte maturation.
Figure 6.
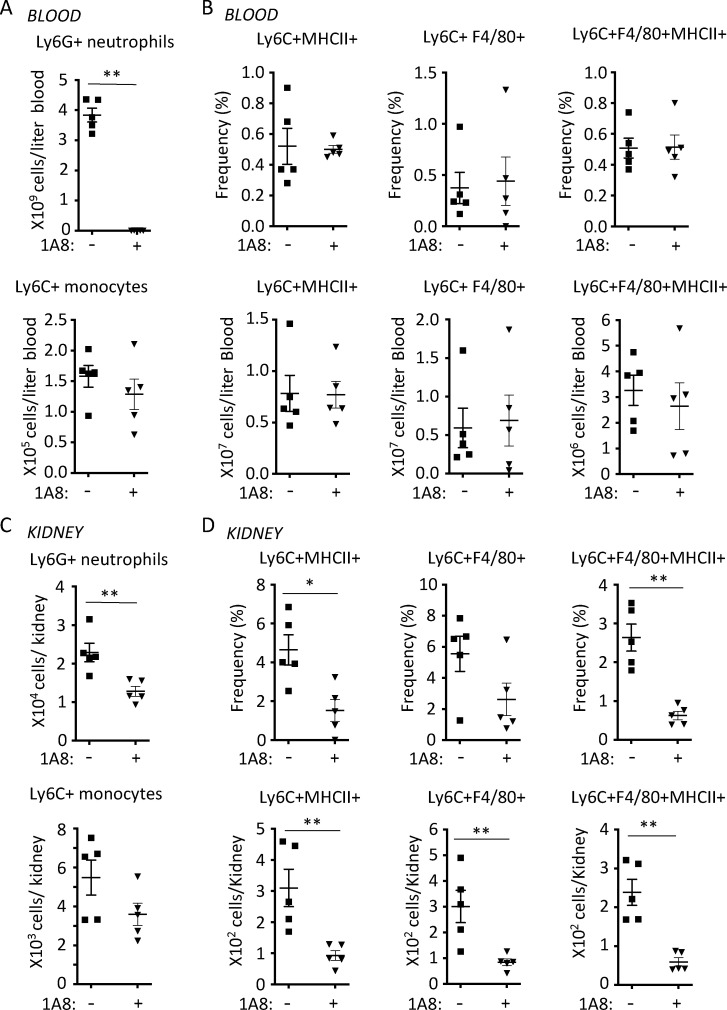
Neutrophil depletion attenuates the intravascular conversion of monocytes to immature macrophages. Mice were left untreated (−) or given the neutrophil-depleting antibody 1A8 before inducing NTN. 18 h after induction of NTN, anti-CD45 mAb was injected i.v. 3 min before harvesting the blood and kidney. Total neutrophil and monocyte counts (A and C) and the frequency and number of Ly6C+ monocytes expressing MHCII and/or F4/80 (B and D) in the blood (A and B) and kidney (C and D) are given. Data are from a single experiment with n = 5 individual mice per group. Statistical significance was determined by two-tailed Mann–Whitney U test. *, P < 0.05; **, P < 0.01; ***, P < 0.005.
CCR2-dependent monocyte conversion to immature macrophages is recapitulated in cocultures of monocytes and TNF-TNFR2–activated endothelial cells
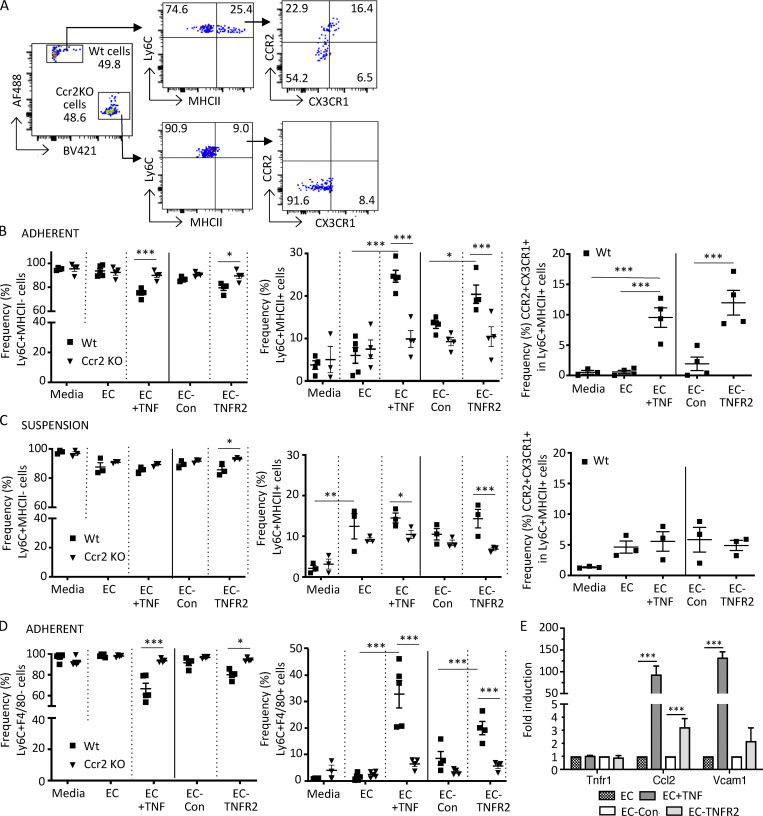
To determine whether the endothelium is a key driver of the CCR2-dependent intravascular acquisition of MHCII and F4/80 by Ly6C+ monocytes, we pursued approaches in vitro. We cultured rat fat pad endothelial cells (RFPECs) that were left untreated or treated with TNF for 6 h (and washed) with isolated Wt and Ccr2 KO monocytes. RFPECs rather than murine microvascular endothelial cells were used because the latter had significant levels of activation markers (e.g., VCAM-1) in the absence of TNF treatment (Lim and Luscinskas, 2006). As a control, monocytes were also incubated in medium in the absence of RFPECs. After 18 h, monocytes in medium alone, monocytes that were adherent to the endothelium (including transmigrated cells), and nonadherent cells in suspension were collected separately and analyzed. Ly6C+ monocytes that acquired MHCII and were additionally CCR2 and CX3CR1 DP (Fig. 7 A, gating strategy) were observed, and their frequency was significantly higher among those adhering to the TNF-stimulated endothelial cells than among those cultured with unstimulated endothelium or in medium alone (Fig. 7 B). In contrast, the percentage of Ly6C+MHCII+ cells among Ccr2 KO monocytes adherent to TNF-activated and unactivated endothelial cells was similar (Fig. 7 B).
Figure 7.
Monocytes adherent to TNF-activated or TNFR2-transduced endothelial cells acquire macrophage markers. CellTrace CFSE dye (AF488)–labeled Wt and CellTrace violet dye (BV421)–labeled Ccr2 KO monocytes harvested from mice were incubated for 18 h in medium alone (media); with endothelial cells that were untreated (EC), stimulated with rat TNFα for 6 h and washed (EC + TNF), or transduced with control lentivirus (EC-Con); or with TNFR2 lentivirus (EC-TNFR2). Endothelial adherent and nonadherent (suspension) monocytes were harvested separately and processed for FACS analysis. (A) Flow cytometry gating strategy for detection of differentially labeled Wt and Ccr2 KO monocytes adherent to TNF-activated endothelial cells. Cells were gated for viability and stained for Ly6C and MHCII and the Ly6C+MHCII+ population was further analyzed for CCR2 and CX3CR1. (B and C) Analysis of adherent (B) and nonadherent (C) monocytes. Frequency of Ly6C+MHCII− (left) and Ly6C+MHCII+ (middle), and CCR2 and CX3CR1 in the Ly6C+MHCII+ populations (right, only for CellTrace CFSE dye [AF488]–labeled Wt cells), are shown. (D) Adherent cells were analyzed for Ly6C and F4/80 and the frequency of Ly6C+F4/80− (left) and Ly6C+F4/80+ (right) in Wt and Ccr2 KO cells were determined as in B. (E) Analysis of Tnfr1, Ccl2, and Vcam1 message in endothelial cells treated as indicated by quantitative real-time PCR using the indicated gene-specific primers. The normalized fold-change vs. untreated cells (EC) or control virus (EC-Con) was calculated using the ΔΔCt method, with GAPDH as a control gene. Data are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.005. Four independent experiments were performed. Statistical significance was determined by multiple t test and one-way ANOVA with Dunnett’s t test correction.
Next, the contribution of endothelial TNFR2 to monocyte differentiation was examined. Cultured endothelial cells express very low levels of TNFR2 before and after TNF stimulation, while experimentally induced TNFR2 overexpression leads to spontaneous receptor clustering and ligand-independent signaling (Gaeta et al., 2000; Venkatesh et al., 2013). We found that a higher percentage of Wt monocytes expressed MHCII when incubated with TNFR2- vs. control-transduced endothelial cells, while this was not observed with Ccr2 KO monocytes (Fig. 7 B). Interestingly, in cocultures with TNF-activated endothelial cells, nonadherent monocytes (in suspension) had a profile that was similar to monocytes in contact with unstimulated endothelial cells (Fig. 7 C), which suggests that monocyte adhesion to the activated endothelium contributes to the acquisition of macrophage markers. The frequency of Ly6C+MHCII+ cells in suspension retrieved from unactivated endothelial cells did increase compared with those in medium alone (Fig. 7 C), which aligns with a previous study (Jakubzick et al., 2013). However, the frequency was lower than for monocytes adherent to TNF-activated endothelial cells (Fig. 7 B), and it was not CCR2 dependent (Fig. 7 C). As observed with MHCII, F4/80 was induced on Ly6C+ monocytes adherent to TNF-stimulated or TNFR2-transduced endothelial cells (Fig. 7 D). Unlike our analysis with MHCII, monocytes adherent to unactivated endothelial cells or in medium alone did not express F4/80 (Fig. 7 D), which suggests that this macrophage marker is specific for monocytes in contact with activated endothelial cells. Next, we assessed whether adhesion to the activated endothelium, per se, contributes to monocyte conversion. Monocyte firm adhesion to the activated endothelium in vitro is mediated by CCL2, IL-8, and partially, integrin VLA-4 (Gerhardt and Ley, 2015; Schenkel et al., 2004), while the CD18 integrins are required for monocyte locomotion to junctions and transmigration (Schenkel et al., 2004). We found that functional blocking antibody to CD18 but not VLA-4 partially reduced monocyte acquisition of MHCII. Neither treatment had any effect on the acquisition of F4/80 (Fig. S3 F). Thus, CD18-dependent adhesion and/or transmigration contributes to some aspects of monocyte conversion to immature macrophages.
Activation of human endothelial cells with TNFα or IL-1β induces the release of the CCR2 ligand, CCL2 (Rollins et al., 1990; Strieter et al., 1989). Consistent with this, TNF stimulation of RFPECs (used in our in vitro cocultures) resulted in a large increase in CCL2 (MCP-1) mRNA (Fig. 7 E). TNFR1 expression did not increase (Fig. 7 E), and TNFR2 and another CCR2 ligand, CCL7, were undetectable (not depicted). The monocyte adhesion molecule VCAM-1 was also highly upregulated (Fig. 7 E) after exposure to TNF, as expected. Importantly, higher levels of CCL2 mRNA were observed in TNFR2 compared with control-transduced cells (Fig. 7 E), indicating that TNFR2 signaling alone can induce endothelial expression of CCL2. In summary, a CCR2-dependent acquisition of macrophage markers is observed on monocytes adherent to TNF-activated or TNFR2-transduced endothelial cells, which express the CCR2 ligand, CCL2.
Single-cell transcriptional profiling reveals monocyte states and gene programs induced by TNF-activated endothelial cells and monocyte CCR2
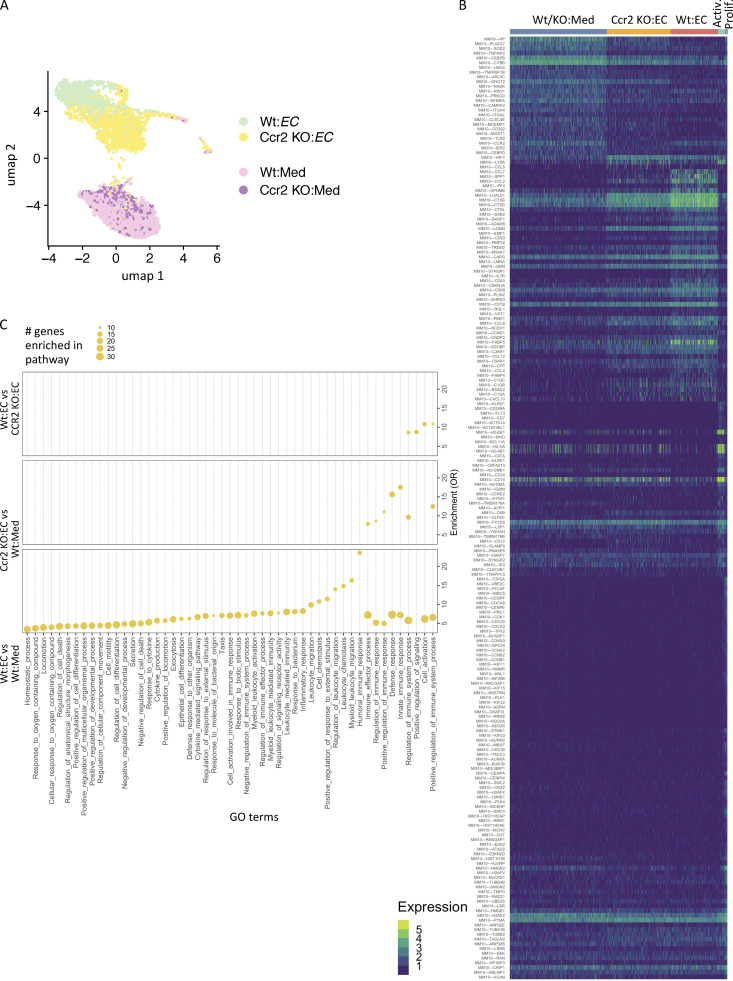
Next, we used single-cell RNA-seq (scRNA-seq) to define monocyte states induced by the TNF-activated endothelium, identify CCR2-dependent states, and infer relationships between them. For this, Wt and Ccr2 KO monocytes cocultured with TNF-activated endothelial cells or in medium alone for 24 h were collected. We chose medium alone vs. untreated endothelial cells as the comparator because the latter can influence monocyte phenotypes (Jakubzick et al., 2013; Fig. 7 C) and be inadvertently activated in vitro. Samples of Wt and Ccr2 KO monocytes separately cultured in medium alone were combined for the scRNA-seq analysis (Wt/KO:Med). We observed five transcriptionally distinct clusters (Fig. 8 A, Fig. S4 A, Table S2, and Table S3) that were comprised of Wt or Ccr2 KO monocytes cultured with TNF-activated endothelium and in medium alone and a smaller cluster of proliferating monocytes as well as activated monocytes in which cells from all the different conditions were represented. The relative localization of clusters suggests that Ccr2 KO monocytes cultured on the TNF-activated endothelium represent a transitional state between the monocytes cultured in medium alone and the Wt monocytes cultured on TNF-activated endothelial cells (Fig. 8 A). Supporting this conclusion, 90% of monocytes in this cluster were Ccr2 KO and 9.8% were Wt monocytes. The remaining 0.2% were Wt and Ccr2 KO monocytes cultured in medium alone. The distinct clustering with minimal overlap of the three conditions (Wt or Ccr2 KO monocytes on activated endothelial cells, and Wt and Ccr2 KO monocytes in medium) suggests little heterogeneity in the system.
Figure 8.
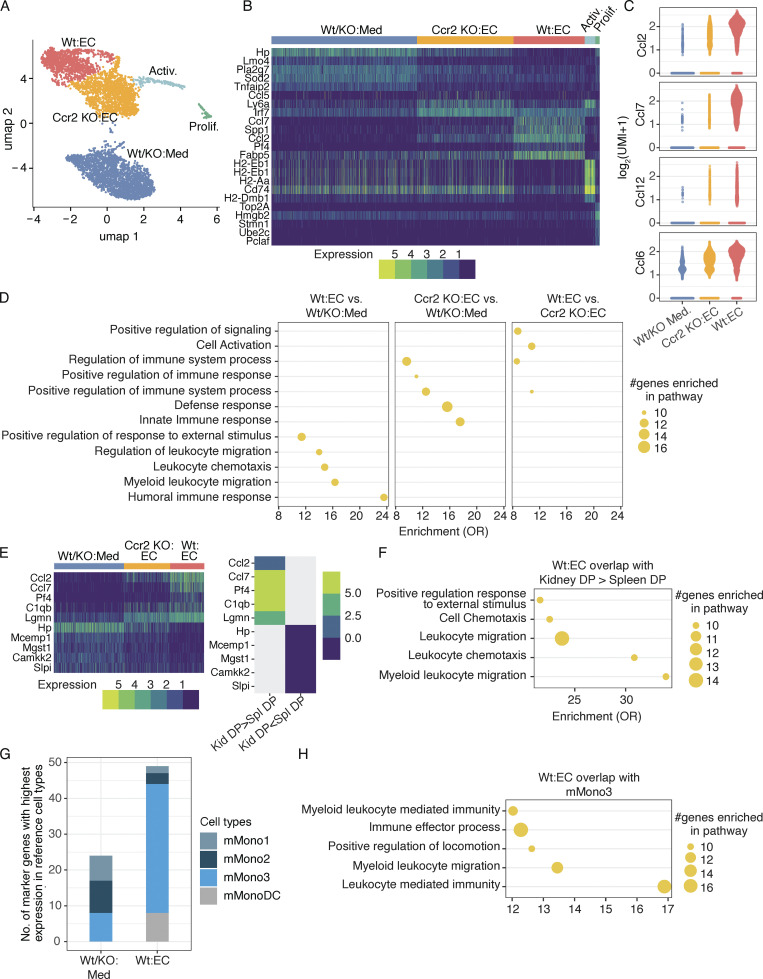
scRNA-seq characterizes monocytes in contact with activated endothelial cells. (A) UMAP comprising five clusters of Wt and Ccr2 KO monocytes cultured on TNF-activated endothelial cells (EC) and medium alone containing Wt and Ccr2 KO monocytes (Wt/KO:Med) represented in our data set (n = 5,608 cells). (B) Heatmap of the top markers of identified clusters. The number of UMIs as the expression of markers is shown. Top markers are shown here, and all markers are shown in Fig. S4 B. (C) The expression of chemokines Ccl2, Ccl7, Ccl12, and Ccl6 across monocytes cultured on TNF-activated endothelial cells or medium alone. (D) Gene set enrichment in GO terms related to biological processes (GSEA-GO-Bp) for the genes overexpressed in Wt monocytes cultured with activated endothelial cells in comparison to monocytes cultured in medium, Ccr2 KO monocytes cultured with activated endothelial cells in comparison to monocytes cultured in medium alone, and Wt monocytes in comparison to Ccr2 KO monocytes cultured with activated endothelial cells. Top enriched terms are shown here, and all terms are shown in Fig. S4 C. OR, odds ratio. (E) Heatmap of cluster markers (left) which are overexpressed and underexpressed in nephritic kidney DP (Kid-DP) in comparison to spleen DP cells (Spl-DP; right). Top shared markers are shown here, and all shared markers are shown in Fig. S5 A. (F) GSEA-GO-Bp for those markers of Wt monocytes cultured with activated endothelial cells, which were overexpressed in Kid-DP in comparison to Spl-DP. Only top enriched terms are shown here, and all terms are shown in Fig. S5 B. (G) Number of the markers of Wt monocytes cultured on activated endothelial cells and monocytes cultured in medium alone having highest expression in three monocyte populations (mMono1-3) and monocyte-derived dendritic cell population (mMonoDC) from Zilionis et al. (2019). (H) GSEA-GO-Bp for those markers of Wt monocytes cultured with endothelial cells that had higher expression in mMono3 than mMono1, mMono2, and mDC populations in Zilionis et al. (2019). Only top enriched terms are shown here; all terms are shown in Fig. S5 C.
Figure S4.
scRNA-seq characterizes monocytes in contact with TNF-activated endothelial cells. (A) UMAP showing the distribution of monocytes which are Wt and Ccr2 KO cultured with TNF-activated endothelial cells (EC) or medium (Med) alone (n = 5,608 cells in total). (B) Heatmap of all markers of identified monocyte clusters. The number of UMIs as the expression of markers is shown. (C) Gene set enrichment in GO terms related to biological processes (GSEA-GO-Bp) for the genes overexpressed in Wt monocytes cultured with activated endothelial cells in comparison to monocytes cultured in medium alone (Wt:EC vs. Wt:Med), Ccr2 KO monocytes cultured with endothelial cells in comparison to monocytes cultured in medium alone (Ccr2 KO:EC vs. Wt:Med), and Wt monocytes in comparison to Ccr2 KO monocytes cultured with activated endothelial cells (Wt:EC vs. Ccr2 KO:EC). OR, odds ratio.
The markers of individual clusters, defined as genes overexpressed in a given cluster compared with all other clusters, shed light on their functional characteristics. Ccl7, Ccl2, Pf4, and Spp1, identified as markers of Wt monocytes cultured on TNF-activated endothelium, were reduced in Ccr2 KO monocytes (Fig. 8 B; all markers in Fig. S4 B). CCL7 and CCL2 are well-described CCR2 ligands. A comparison of expression level of chemokines across these clusters reiterated the CCR2-dependent enhancement of Ccl2 and Ccl7 and added additional monocyte chemoattractants, Ccl12 (closely related to Ccl2) and Ccl6 (Fig. 8 C) to the list. Surprisingly, MHCII genes (H2-EB1, H2-AB1, H2-AA, and H2-DMB1) and MHCII-related CD74 were reduced in Wt monocytes in contact with the activated endothelium compared with other clusters and were highly upregulated in the activated monocyte cluster, which had cells from all conditions represented (Fig. 8 B). This is contrary to the increase in surface expression of MHCII evaluated by flow cytometry in monocytes in contact with the TNF/TNFR2-activated endothelium (Fig. 7 B). An early increase in translation and/or reduced endocytosis of MHC molecules and their mobilization to the surface membrane, as observed in mature dendritic cells (Hennies et al., 2015), might explain this discordance. The observed small proliferating cluster was enriched in cell cycle genes; this population may represent contaminating monocyte progenitors or macrophages.
The DEGs (genes overexpressed in a given cluster compared with another cluster) and their enrichment in GO terms identified among the different monocyte clusters allowed a comparison of biological processes that are influenced by exposure to TNF-activated endothelial cells, and those that are altered by the presence of CCR2 (top GO terms in Fig. 8 D and all in Fig. S4 C). We identified an increase in myeloid leukocyte activation and positive regulation of signaling, and upregulation of immune responses in Wt compared with Ccr2 KO monocytes exposed to TNF-activated endothelial cells. On the other hand, Ccr2 KO monocytes on endothelial cells vs. medium had a separate set of associated GO terms from the other two comparisons, suggesting a distinct phenotype for these monocytes that are impaired in their transition. Thus, CCR2-dependent factors in monocytes may function as regulators of leukocyte recruitment and effector function.
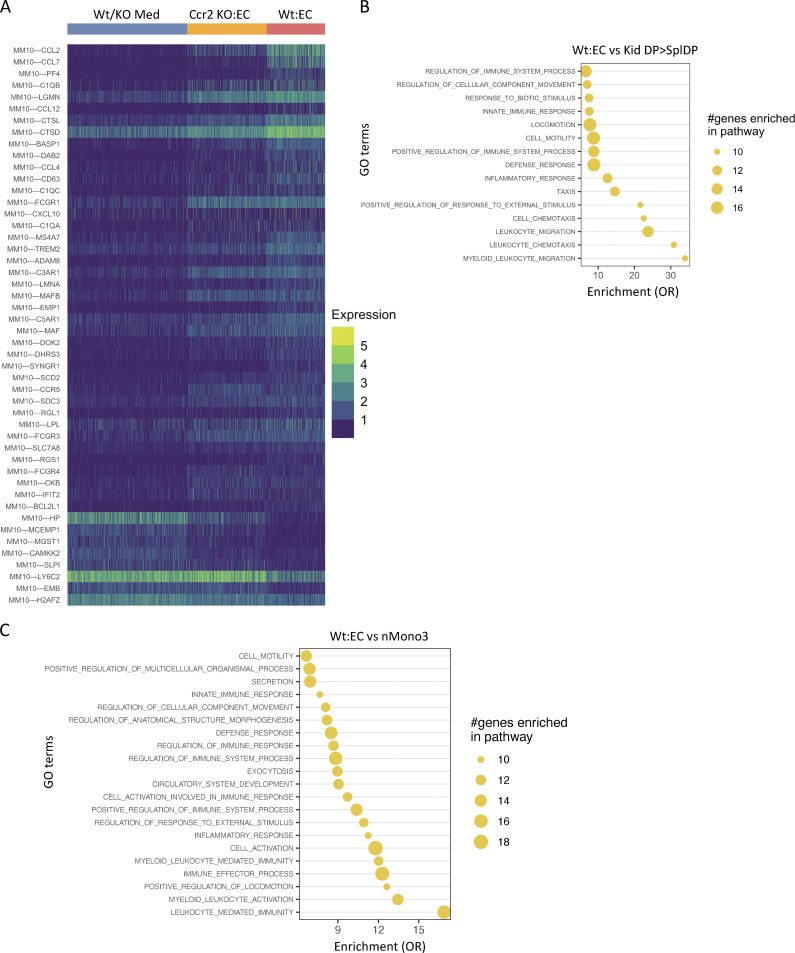
Having defined DEGs enriched in monocytes exposed to TNF-activated endothelial cells, we asked whether they overlap with DEGs in the inflammatory CCR2+CX3CR1+ monocyte population of the nephritic kidney. Several DEGs that were overexpressed in this population in the inflamed, nephritic kidney vs. the spleen overlapped with the markers of Wt monocytes in contact with activated endothelial cells (top shared genes in Fig. 8 E; all in Fig. S5 A). This suggests that the activated endothelium induces a monocyte state that parallels the renal inflammatory CCR2+CX3CR1+ monocytes that accumulate in the kidney following GN. These shared genes were enriched in GO terms related to leukocyte activation and migration and the regulation of immune responses (top GO terms in Fig. 8 F; all in Fig. S5 B). For reference, we compared identified monocyte clusters with a previous analysis of murine and human lung cancer tissues that reported four monocyte subsets that were analogous in both species (mMono1-3 and mMonoDC; Zilionis et al., 2019). This dataset was selected due to the extensive analysis of each of the monocyte species in both mouse and humans: mMono1 and mMono2 express canonical classical and nonclassical monocyte-associated genes, respectively, in murine and human tumors (Zilionis et al., 2019) and human blood (Villani et al., 2017), mMono3 is an unappreciated subtype of classical monocytes (Villani et al., 2017; Zilionis et al., 2019), and mMonoDCs are enriched in dendritic cell–related genes in lung tumor tissue. We found that markers of monocytes in medium alone overlapped with mMono1-3, distributing equivalently across all three clusters, while Wt monocytes in contact with activated endothelial cells primarily overlapped with mMono3 and mMonoDC (Fig. 8 G). GO term enrichment of the genes shared between mMono3 and Wt monocytes in contact with activated endothelial cells revealed overrepresentation of functional networks related to immune cell activation and effector functions (top GO terms in Fig. 8 H; all in Fig. S5 C). Thus, the TNF-activated endothelium induces monocyte phenotypes that resemble not only inflammatory monocytes in the nephritic kidney but potentially in other inflammatory environments such as in lung cancer.
Figure S5.
Overlap and GO term enrichment of genes with CCR2+CX3CR1+ DP monocytes of the nephritic kidney and genes reported in tumor-associated monocytes. (A) Heatmap of cluster markers overexpressed in kidney DP cells (Kid DP) in comparison to spleen DP (Spl DP) cells plotted across clusters of Wt/KO Med, Ccr2 KO:EC, and Wt:EC. (B) GSEA-GO-Bp for those markers of Wt monocytes cultured on endothelium which were overexpressed in kidney in comparison to spleen DPs (Kid DP > Spl DP). OR, odds ratio. (C) GSEA-GO-Bp for those markers of Wt monocytes cultured with activated endothelial cells, which had higher expression in mMono3 than mMono1, mMono2, and mMonoDC populations identified in Zilionis et al. (2019).
Urinary CD14+CD163+ monocytes expressing MHCII accumulate in patients with lupus nephritis and correlate with disease scores
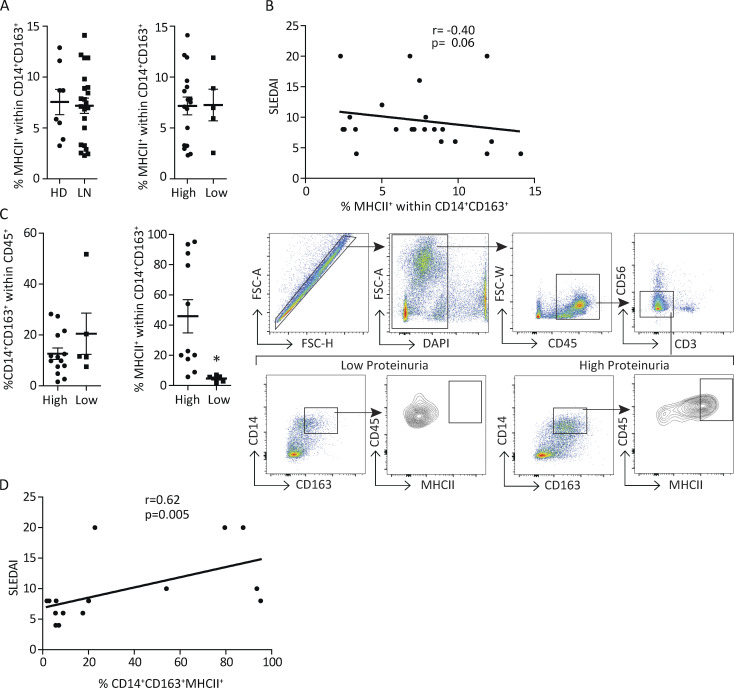
GN caused by systemic lupus erythematosus is the major cause of morbidity and mortality in lupus patients (Yu et al., 2017). In humans, CD14 and CD163 are expressed on CD16− and not CD16+ nonclassical monocytes (Ingersoll et al., 2010). CD163 is restricted to the monocytic–macrophage lineage (Etzerodt and Moestrup, 2013) and is associated with polarization conditions that are anti-inflammatory as well as proinflammatory (Barros et al., 2013; Etzerodt and Moestrup, 2013; Van Gorp et al., 2010). Here, we examined whether immature macrophages can be detected in human GN and are associated with clinical disease by evaluating MHCII on CD14+CD163+ monocytes in blood and urine of patients with lupus nephritis, stratifying them as having high or low proteinuria, and correlating the results with Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) scores, which is a global assessment of disease activity. We found a similar frequency of CD14+CD163+MHCII+ monocytes in the peripheral blood of healthy donors, patients with active lupus nephritis, and lupus patients with high or low proteinuria (Fig. 9 A). Furthermore, the frequency of these cells in blood of lupus patients did not correlate with SLEDAI scores (Fig. 9 B). Urinary cellular composition is known to reflect the inflammatory status of the kidney. In urine samples, the abundance of urinary CD14+CD163+ cells was similar in patients with high and low proteinuria. However, the frequency of CD14+CD163+ cells expressing MHCII was significantly higher in the high- vs. low-proteinuria SLE patient group (≥532 mg in 24 h analysis; Fig. 9 C) and, importantly, correlated with SLEDAI scores (Fig. 9 D). Together, these data demonstrate an association between the frequency of renal but not blood immature macrophages and glomerular damage and therefore predict that their accumulation in the kidney correlates with active GN.
Figure 9.
MHCII+ monocytes are present in the urine of patients with active lupus nephritis, and their frequency correlates with disease activity. (A) Cumulative percentage of MHCIIhi cells within the CD14+CD163+ population in blood from healthy donors (HD) and patients with active lupus nephritis (LN; left) and lupus patients with high and low proteinuria (right). (B) Correlation of percentage of blood MHCII+ monocytes and disease index activity (SLEDAI). (C) Cumulative data and representative gating strategy showing the percentages of CD14+CD163+ cells within the live CD45+ cells (left panel) and MHCIIhi cells within the CD14+CD163+ population in urine from patients with active LN (right panel), separated based on the level of proteinuria (≥532 mg/24 h = high proteinuria). *, P < 0.05. FSC, forward scatter. (D) The correlation of percentage urine MHCIIhi monocytes and SLEDAI is shown.
Discussion
Monocytes play essential roles in homeostasis, infection and sterile inflammation and acquire macrophage phenotypes by transitioning through intermediate states in several settings. However, the mechanisms leading to the accumulation of these monocyte intermediates within tissues were not well understood; in the intestine, it has been attributed to a disruption in differentiation along the monocyte–macrophage continuum under steady-state conditions and following inflammation (Bain et al., 2013; Platt et al., 2010; Tamoutounour et al., 2012; Zigmond et al., 2012). We demonstrate that monocyte-to-macrophage conversion is an active process that begins within the vasculature before their extravasation and is instructed by CCR2 on monocytes and a TNF-TNFR2–activated endothelium. The TNF-activated endothelium induces the generation of proinflammatory immature macrophages that are enriched in genes associated with cell activation and immune responses, as well as CCR2 ligands. The latter could potentially establish a monocyte CCR2 intrinsic feed-forward loop for monocyte differentiation within the vasculature that in turn fuels inflammation. Several studies demonstrate that the CCR2-CCL2 axis contributes to monocyte recruitment and tissue injury (Wada et al., 1996; Lloyd et al., 1997; Tesch et al., 1999; Bird et al., 2000; Segerer et al., 2000; Panzer et al., 2001; Bianconi et al., 2018). Our results indicate additional roles for monocyte CCR2 and parenchymal TNFR2 in the generation of immature proinflammatory macrophages and further demonstrate that this specialization occurs within the vasculature.
We demonstrate that CCR2 and CX3CR1 DP cells are preferentially recruited to the glomerulus and hypothesize that these recruited cells are the precursors of renal immature macrophages. The increased dwell time of CCR2+CX3CR1+ monocytes within the glomerular capillaries may allow them to better detect inflammatory modulators on the surface of the activated endothelium, such as CCL2 and other chemokines known to be retained by glycosaminoglycans on the endothelial surface (Celie et al., 2009). The TNF-stimulated endothelium has been previously reported to generate CCL2 (Rollins et al., 1990; Strieter et al., 1989), and we confirm and extend these findings to show that TNFR2 can signal the same. We propose that CCL2 retained on the surface of the activated endothelium locally engages CCR2 on dwelling monocytes to begin their conversion to immature macrophages and acquisition of proinflammatory characteristics. Transcriptional profiles of monocytes cocultured with TNF-activated endothelial cells revealed a CCR2-dependent enrichment in genes associated with leukocyte activation and immune responses, as well as CCR2 ligands, including Ccl2, Ccl7, and Ccl12. The other genes that increased in a CCR2-dependent manner are also potential regulators of monocyte function (Fig. 8 B); PF4 is a chemokine for monocytes and neutrophils, SPP1 (secreted phosphoprotein 1, osteopontin) may contribute to monocyte differentiation (Zhang et al., 2014), and FABP5 is an intracellular lipid chaperone that suppresses nuclear hormone peroxisome proliferator-activated receptor γ, which in turn may increase CCR2 on monocytes (Babaev et al., 2011). Together, our findings coalesce around the concept that CCR2 engagement on monocytes signals the expression of proteins that establish a CCR2-mediated feed-forward loop that promotes monocyte maturation and function and thereby fuels renal inflammation. In vivo, it is possible that CCL2 generated by TNF-stimulated podocytes and mesangial cells (Chung et al., 2015; Ohta et al., 2000) diffuses into the vasculature to also contribute to monocyte conversion.
Monocyte adhesion within the renal vasculature may also contribute to their conversion to immature macrophages, as monocytes adherent to TNF-activated endothelial cells and not nonadherent cells recovered in the supernatant acquire significant MHCII and F4/80. A role for direct contact with the endothelium is further suggested by the partial inhibition of conversion by a functional blocking antibody to the leukocyte adhesion receptor, CD18. Neutrophil depletion approaches indicate that these cells, recruited soon after the in situ formation of immune complexes induced by NTS (Flores-Mendoza et al., 2018), contribute to intravascular monocyte conversion to immature macrophages. As is the case for neutrophils (Mayadas et al., 2009), FcγRs on monocytes can support monocyte recruitment to intravascular immune complexes (Olaru et al., 2018). Although our in vitro studies demonstrate that the TNF-activated endothelium alone promotes conversion, FcγRs engaging immune complexes within the vasculature could further enhance monocyte transition to immature macrophages by directly inducing monocyte recruitment and/or dwell time or indirectly recruiting neutrophils.
Monocytes that accumulate in other organ-specific inflammatory diseases often display augmented expression of gene sets associated with inflammation (Guilliams and Svedberg, 2021; Robinson et al., 2021). In comparing the transcriptome of the small number of CCR2+CX3CR1+ DP cells in the spleen/bone marrow with those in the nephritic kidney, we found that the nephritic kidney imprints a proinflammatory phenotype on these cells. This adds to the mounting evidence that local cues dictate monocyte fate. For example, under steady state, tissue-derived TGFβ is the principal determinant of local monocyte differentiation into resident macrophages (Schridde et al., 2017), and endothelial Notch ligand δ-like protein 1 promotes Ly6C monocyte conversion to patrolling cells (Jakubzick et al., 2017). Consistent with these findings, we find that the spleen/bone marrow pool of these cells was able to reconstitute disease when adoptively transferred into largely disease-resistant Ccr2 and Cx3cr1 DKO recipient mice, indicating that they contribute to glomerular damage when recruited to the inflamed kidney and exposed to the local renal inflammatory environment. This result also demonstrates their sufficiency to induce disease and thus uncovers a critical role for this mononuclear population in the progression of GN. However, an additional role for other described tissue-resident mononuclear phagocytes with potential inflammatory capabilities during disease (Huen and Cantley, 2017; Kawakami et al., 2013; Lee et al., 2018; Li et al., 2008; Stamatiades et al., 2016; Yang et al., 2019) cannot be excluded.
Lupus nephritis is a major driver of renal dysfunction leading to morbidity and mortality in SLE patients despite potent antiinflammatory and immunosuppressive therapies (Almaani et al., 2017). Our data in lupus patients suggest that the frequency of immature macrophages in the urine, a sampling of the kidney infiltrate, but not the blood, is associated with clinical disease scores. These findings, together with the reported role of the CCR2-CCL2 axis and TNFR2 in the pathogenesis of lupus nephritis (Mok et al., 2016; Rovin et al., 1996; Segerer et al., 2000; Tucci et al., 2004; Wu et al., 2016), suggest the possibility that CCR2-mediated monocyte transition to immature macrophages at the surface of the activated endothelium plays a critical role in the pathogenesis of GN in humans. The immature macrophages generated in the inflamed kidney, or by the TNF-activated endothelium in vitro, resemble mMono3, a monocyte subset classified as an unappreciated subtype of classical monocytes in inflamed tumor tissues of mice and humans (Zilionis et al., 2019). Thus, the immature macrophage population and the CCR2-dependent mechanisms responsible for its generation in GN described herein may be relevant in other inflammatory settings.
In summary, we show that CCR2+CX3CR1+ monocytes are preferentially recruited to the glomerular capillaries of the nephritic kidney, acquire proinflammatory characteristics within the kidney, and may be the precursor to immature macrophages. We demonstrate that CCR2 promotes monocyte conversion to immature macrophages within the vasculature and identify a molecular pathway initiated by the TNF-TNFR2–activated endothelium that drives this process. In doing so, our studies indicate that the transition of monocytes to macrophages is an instructive process, and we assign a role for a canonical monocyte chemokine receptor, previously shown to primarily regulate monocyte trafficking and the endothelium, in regulating monocyte fate during inflammation.
Materials and methods
Mice
8–10-wk-old female or male mice were used for all the experiments. WT C57BL/6J (Wt) mice and mice deficient in TNFR2 (Tnfrsf1b−/−), CX3CR1 (Cx3cr1gfp/gfp), and CCR2 (Ccr2rfp/rfp) were purchased from The Jackson Laboratory. All KO and transgenic mice were backcrossed to C57Bl/6 for a minimum of nine generations and maintained as colonies alongside C57Bl/6 Wt mice (originally purchased from the Jackson Laboratory). Age- and sex-matched Wt animals were used as controls. Ccr2rfp/rfpCx3cr1gfp/gfp DKO mice were a generous gift from Dr. Paul Kubes (University of Calgary, Calgary, Canada). They were mated with Wt mice to generate reporter mice (Cx3cr1gfp/+Ccr2rfp/+). For some experiments, they were also mated with Cx3cr1 (Cx3cr1gfp/gfp) or Ccr2 (Ccr2rfp/rfp) KO mice to generate CCR2 or CX3CR1 deficiency on the dual reporter background. All transgenic mice used in this study were cohoused in specific pathogen–free conditions.
Induction and analysis of NTN
To induce chronic NTN, mice were preimmunized with a subcutaneous injection of 0.05 mg rabbit IgG (Jackson ImmunoResearch Laboratories) in 1:1 emulsion with Freund’s incomplete adjuvant (Sigma-Aldrich) and nonviable desiccated Mycobacterium tuberculosis H37Ra (Difco) on day −3. On day 0, 100 μl of heat-inactivated, filter-sterilized rabbit NTS containing anti-glomerular basement membrane antibody (NTS) was given i.v. Urine, blood, kidney, and other organ samples were harvested on day 13 or 14 as indicated except for IVM, which was done on day 10, as mice did not tolerate anesthesia well at later times of disease. Urine albumin was determined and normalized to creatinine as described (Tsuboi et al., 2008). For neutrophil depletion experiments, mice were i.p. administered 350 μg anti-mouse Ly6G antibody (clone 1A8, #BE0061; BioXCell) on days −2, −1, and 1 and analyzed 18 h after NTS as described above. Acute glomerular inflammation was induced to evaluate leukocyte recruitment by injecting 200 μl of NTS i.v. without any preimmunization, and samples were collected at 2 h. Accelerated nephritis was induced in NZB/W lupus prone mice (Bethunaickan et al., 2012; Liu et al., 2011; Mathian et al., 2005) after injection of adenovirus expressing IFNγ.
Generation of bone marrow chimeras
Bone marrow obtained from reporter (Ccr2rfp/+Cx3cr1gfp/+), Ccr2 KO (Ccr2rfp/rfpCx3cr1gfp/+), Cx3cr1 KO (Ccr2rfp/+Cx3cr2gfp/gfp), or Ccr2 and Cx3cr1 DKO (Cx3cr1gfp/gfpCcr2rfp/rfp) mice were transplanted into 9.5-Gy-irradiated Wt or Tnfr2 KO mice. 8 wk after transplantation, mice were subjected to NTN and evaluated 14 d later.
Flow cytometry analysis
Mice were anesthetized and perfused with 60 ml of cold 10-mM EDTA solution in DPBS. Kidney and spleen were cut into small pieces and digested in HBSS (Ca+) containing 0.5 mg/ml Liberase TL (Roche) and 0.1 mg/ml DNAse I (Sigma-Aldrich) or 2 mg/ml Collagenase type 1 at 37°C. Single-cell suspensions were made by mechanical disruption and passage through a 70-µm cell strainer. Cells were blocked with anti-CD16/CD32 mAbs in FACS buffer, PBS (Ca−, Mg−) + 2% FCS + 1 mM EDTA for 10 min at 4°C, followed by staining with fluorophore-conjugated antibodies for 30 min at 4°C. Cells were washed twice with FACS buffer and fixed in 1% formaldehyde. CountBright Absolute Counting Beads (Invitrogen) were used to assess cell number. Samples were run on a BD LSRII or BD FACS Symphony A5 and analyzed by BD FACS Diva or FlowJo software.
Antibodies and dyes
The following antibodies and stains were used for all murine studies. Antibodies were purchased from BioLegend (Fc block, 2.4G2, 553142; CD45, 30-F11, 103134; CD3, 145-2C11, 100351; B220, RA3-6B2, 103244; NK1.1, PK136, 108739, CD11c, N418, 117333; Ly6G, 1A8, 127639; CD11b, M1/70, 101216; CD64, X54-5/7.1, 139311; F4/80, BM8, 123135; Ly6C, HK1.4, 128037; MHCII, AF6-120.1, 116415; CX3CR1, SA011F11, and 149008), Bio-Rad (F4/80, Cl:A3-1, and MCA-497), and R&D Systems (CCR2, 475301, and FAB5538P). Cell tracker dyes were purchased from Thermo Fisher Scientific (CellTracker Blue CMF2HC Dye, C12881; CellTracker Green CMFDA Dye, C2925; CellTrace violet [BV421], C34557; CellTrace CFSE [AF488], and C34554).
IVM of the kidney
Mice were anesthetized and placed on a heating pad to maintain constant body temperature. The left kidney was exteriorized and immobilized in a custom-made holder. The kidney was imaged with a multiphoton microscope (Prairie Technologies) equipped with a 20× water-immersion objective (Olympus), driven by a MaiTai Ti:sapphire laser (Spectra-Physics) tuned at 780–1,000 nm for multiphoton excitation and second-harmonic generation. For dynamic analysis of recruitment and the dwell time of cells in the glomeruli, images were acquired every 30 s for 20 min. Cells present in two consecutive frames (60 s) were considered recruited. After acquisition image analysis, volume rendering and 4D time-lapse videos were performed using Imaris software (Bitplane). For lower-time-resolution imaging experiments, cells captured in only a single frame were excluded from the analysis, as two frames are required to calculate the duration of an adhesion event.
Histology
Kidneys were collected, fixed, embedded in paraffin, and stained with H&E or periodic acid–Schiff using standard methods. For immunofluorescence, kidneys were fixed in 1% paraformaldehyde, 0.1 M L-lysine (Sigma-Aldrich), and PBS (pH 7.4), embedded in optimal cutting temperature compound (Sakura Finetek), and snap frozen. 5-μm kidney sections were permeabilized in PBS containing 0.1% Triton X-100 (Sigma-Aldrich) and incubated with 20% Blocking One solution (Nacalai Tesque). Sections were analyzed for RFP and GFP fluorescence on an Olympus Fluo View FV 1000 high-speed, confocal microscope. RFP-only positive red cells were counted as positive for CCR2, GFP-only positive green cells were counted as CX3CR1 positive, and RFP and GFP orange to yellow cells were counted as CCR2 and CX3CR1 DP cells. At least five images were taken at 200× magnification for each mouse, and then cells in the glomerulus, interstitium, and tubules were counted. Data are presented as the number of cells per glomerulus and the number of cells per mm2 in the tubules and interstitium.
Monocyte enrichment, cell sorting, and adoptive transfer into Ccr2rfp/rfpCx3cr1gfp/gfp DKO mice
After NTN induction, bone marrow and spleen were removed from 10-wk-old donor Wt-C57BL/6J mice (n = 8). Single-cell suspensions from bone marrow and spleens were pooled together, followed by RBC lysis for 5 min at room temperature with ACK lysis buffer. Monocytes from bone marrow and spleen were enriched by negative selection using EasySep Mouse Monocyte Isolation Kit (StemCell Technologies) as per the manufacturer instruction. Untouched, isolated monocytes were blocked with anti-CD16/CD32 mAbs, followed by staining with conjugated antibodies for 30 min at 4°C. Enriched monocytes were resuspended in RPMI 1640 supplemented with 2% FCS and sorted by FACSAria cytometer (Beckton Dickinson). Two cell populations were obtained after sorting: Ly6C+CD11b+CX3CR1+CCR2+ (DP) and the remaining population (negative; n-DP). NTN was induced in 10-wk-old female Ccr2rfp/rfpCx3cr1gfp/gfp recipient mice. 5 × 106 sorted monocytes harvested from Wt donor mice on day 13 after NTN, in 200 μl of DPBS, were injected i.v. in the tail immediately after FACS sorting on days 3, 7, and 10 after induction of NTN in recipient mice. Urine samples and both kidneys were harvested on day 14 for analysis of albuminuria and histology, respectively. The histopathologic parameters of endocapillary proliferation, capillary loop thickening, and cellular crescents, features of active immune complex–mediated GN, were scored. Capillary loop thickening reflects the deposits of immune complexes within the glomerulus, while endocapillary proliferation and cellular crescents are inflammatory responses to the immune complex deposition. These parameters were scored in a semiquantitative manner on a scale from 0 to 3 (0, no evidence of features; 3, severe and diffuse endocapillary proliferation, capillary loop thickening, or cellular crescents).
Monocyte enrichment and cell adoptive transfer in WT and Tnfr2 KO mice
Monocytes from Wt and Ccr2 KO donor mice were isolated from bone marrow by negative selection using EasySep Mouse Monocyte isolation kit (Stem Cell Technologies). Briefly, femurs and tibias were harvested, and marrow was flushed out using cold RPMI 1640. Bone marrow cell suspensions were passed through a 70-µm cell strainer to obtain uniform single-cell suspension. This cell suspension was treated with EasySep reagents according to the manufacturer’s instructions, and cell purity was assessed by flow cytometry. CellTrace violet (BV421) and CellTrace CFSE (488) proliferation kit (ThermoFischer Scientific) fluorescent dyes were dissolved in DMSO as 5-mM stock solutions. Monocytes were resuspended in RPMI 1640 medium (Invitrogen) supplemented with 2% FCS, and dyes were added at a final concentration of 5 µM. Wt-C57BL/6J cells were labeled with CellTrace CFSE (488) and Ccr2 KO cells were labeled with CellTrace violet (BV421; Quah and Parish, 2010). After labeling, cells were washed three times with PBS. Cell viability, which was routinely 99%, was determined by trypan blue exclusion and flow cytometry. 1 × 106 labeled Wt and Ccr2 KO monocytes were mixed 1:1 and adoptively transferred retroorbitally.
Intravascular staining in mice with nephritis
On day 14 after NTN or day 42 after induction of accelerated lupus nephritis in NZB/NZW mice, 3 µg BV421-conjugated CD45 antibody (diluted in DPBS) was injected via the retroorbital plexus. 3 min after antibody injection, blood was collected. Mice were immediately sacrificed, and kidney, inguinal lymph nodes, and spleen were collected for the ex vivo staining with the fluorophore-conjugated antibodies, followed by FACS analysis. To assess phenotypic changes in adoptively transferred monocytes, Wt or Tnfr2 KO mice were immunized with CFA on day −3 and 100 μl NTS on day 0. 5 h later, each recipient mouse received an i.v. injection of 1 × 106 CellTrace CFSE (488)–labeled Wt and 1 × 106 CellTrace violet (BV421)–labeled Ccr2 KO monocytes prepared as detailed below. 18 h after NTS injection, mice were injected retroorbitally with 4 µg of Pe-Cy7–conjugated anti-CD45 antibody and processed 3 min later as described above. Single-cell suspensions of organs and blood samples after RBC lysis were stained with LIVE/DEAD dye to assess viability and antibodies to Ly6C, MHCII, CX3CR1, CCR2, and F4/80. Samples were also stained with antibodies to lineage markers B220, Ly6G, CD11c, NK1.1, and CD3 and analyzed using flow cytometry.
Endothelial cell culture, DNA constructs, and lentiviral transduction
RFPECs (Ebong et al., 2011), generously provided by Dr. John M. Tarbell (The City College of New York, New York, NY) were cultured in DMEM containing 10% (vol/vol) FBS supplemented with 1% penicillin-streptomycin. RFPECs were cultured on gelatin-coated dishes. For lentiviral transduction of RFPECs, HEK293T packaging cells were transfected with control or TNFR2 lentiviral vectors as previously published (Venkatesh et al., 2013). Supernatants from packaging cells were collected 48 h after transfection, filtered through 0.45-μm low-protein binding filters (Millex-HV; Merck Millipore), and used to transduce RFPECs on day 0. The cells were washed 12 h later, rested overnight, and reinfected on day 1. 12 h later, the monolayer was washed, and cells were used for assays 36 h later.
In vitro endothelial and monocyte cultures
RFPECs seeded at 2 × 105 cells/well in gelatin-coated 6-well plates were cultured overnight, stimulated with 10 ng/ml rat TNFα or left untreated for 6 h, and washed with PBS before coculture with monocytes. Wt and Ccr2 KO monocytes were isolated from bone marrow using EasySep Mouse Monocyte isolation kit and labeled with CellTrace CFSE (488) or CellTrace violet (BV421). 6 × 105 CellTrace CFSE (488)–labeled Wt monocytes and 6 × 105 CellTrace violet (BV421)–labeled Ccr2 KO monocytes were resuspended in DMEM with 10% FCS and 1% penicillin-streptomycin and cocultured with RFPECs or medium alone (control) in tissue culture plates for 20 h. Nonadherent monocytes present in the supernatant (in suspension) and monocytes adherent to the endothelial cells were separately harvested, stained with Ly6C, MHCII, CX3CR1, CCR2, and F4/80 antibodies, and processed for flow cytometric analysis. For blocking experiments, monocytes were pretreated with 10 μg/ml of anti-VLA-4 (BE0071; BioXCell) antibody and/or 5 μg/ml of anti-CD18 (101417; BioLegend) antibody or isotype control for 20 min. The antibodies were also added to the coculture.
Quantitative RT-PCR (qPCR)
RFPECs stimulated with rat TNF (10 ng/ml) for 6 h or TNFR2-transduced cells were washed twice with PBS (pH 7.4) and lysed with RLT buffer using Qiagen RNeasy mini kit following the manufacturer’s protocol. RNA was quantified using nanodrop and reverse transcribed (1–2 µg) into cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems). qPCR was carried out on StepOnePlus thermal cycler (Applied Biosystems) using gene-specific primers and Sybr Green master mix (Applied Biosystems). Primers were MCP1: forward, 5′-GTGCTGACCCCAATAAGGAA-3′, and reverse, 5′-TGAGGTGGTTGTGGAAAAGA-3′; TNFR1: forward, 5′-GGGATTCAGCTCCTGTCAAA-3′, and reverse, 5′-ATGAACTCCTCCCAGCGTGT-3′; VCAM1: forward, 5′-TTGCAAGAAAAGCCAACATGGGGG-3′, and reverse, 5′-TCTCCAACAGTTCAGACGTTAGC-3′; and GAPDH: forward, 5′-CGACCCCTTCATTGACCTCAACTACATG-3′, and reverse, 5′-CCCCGGCCTTCTCCATGGTGGTGAAGAC-3′.
RNA isolation and sequencing and library preparation of tissue monocytes
NTN was induced in 10 8–10-wk-old normal Wt mice. On day 13, mice were sacrificed to collect the kidneys, spleen, and bone marrow. Renal monocytes were enriched using the EasySep Mouse Monocyte isolation kit (Stem Cell Technologies); stained with BV421-anti-CD45, BV605-anti-CD3, anti-Ly6G anti-CD11c, anti-NK1.1, PE-Cy7-anti-CD11b, APC-Cy7-anti-Ly6C, PE-anti-CCR2, and FITC-CX3CR1 for 30 min on ice; and washed twice with FACS buffer. After washing, cells were FACS sorted and collected in RPMI 1640 supplemented with 10% FBS. Similarly, monocytes from pooled single-cell suspensions of spleen and bone marrow were enriched and flow sorted. Approximately 0.5–1.0 × 104 cells were FACS sorted and washed with PBS, and cell pellets were immediately resuspended in RLT buffer (RNeasy mini RNA isolation kit; Qiagen) and frozen at −70°C. Total RNA was purified using the RNeasy Mini Kit according to the manufacturer’s instructions (Qiagen), and RNA concentration was estimated by an Agilent Bio analyzer. RNA samples were processed for low-input RNA-seq to Molecular Biology Core Services, Dana Farber Cancer Institute (Boston, MA). For library preparation and sequencing, cDNA was synthesized from 2 ng of RNA using Clontech SmartSeq v4 reagents. Full-length cDNA was fragmented to a mean size of 150 bp with a Covaris M220 ultrasonicator, and Illumina libraries were prepared from 2 ng of sheared cDNA using Rubicon Genomics Thruplex DNAseq reagents according to the manufacturer’s protocol. The finished dsDNA libraries were quantified by Qubit fluorometer, Agilent TapeStation 2200, and RT-qPCR using the Kapa Biosystems library quantification kit. Uniquely indexed libraries were pooled in equimolar ratios and sequenced with single-end 75-bp reads on an Illumina NextSeq500 run by the Dana-Farber Cancer Institute Molecular Biology Core Facilities. The Gene Expression Omnibus data accession no. is GSE196730.
RNA-seq analysis of tissue monocytes
Sequenced reads were aligned to the UCSC mm9 reference genome assembly, and gene counts were quantified using STAR (v2.5.1b; Dobin et al., 2013). Differential gene expression testing was performed with DESeq2 (v1.10.1; Love et al., 2014), and normalized read counts (FPKM) were calculated using cufflinks (v2.2.1; Trapnell et al., 2010). RNA-seq analysis was performed using the VIPER snakemake pipeline (Cornwell et al., 2018). Raw counts were loaded into R (R Core Team, 2017) and analyzed with DESeq2 (Love et al., 2014). Genes with an adjusted P value <0.05 and fold-change >2 were considered differentially expressed. Regularized log2 counts per million mapped reads (rlog CPM) were used for figures. The Metascape (Clarke, 1986) web server (http://metascape.org) was used for pathway analysis using the 14,403 genes with >1 CPM in at least one sample as the background. Mean column-normalized log2 CPM values are shown for genes from the indicated Haemopedia (de Graaf et al., 2016) signatures that were present in our RNA-seq data and had a minimum rlog CPM >5 in at least one sample.
RNA-seq analysis and quantification of gene expression and cell clustering for in vitro monocyte cultures
Cultured RFPECs were activated with rat TNF for 6 h, and the TNF was then washed off. Monocytes were isolated from Wt mice or Ccr2 KO mice and incubated with plastic or TNF-activated endothelial cells for 18 h. Monocytes and endothelial cell cocultures along with monocytes on plastic were harvested. Sample populations were stained with Total-Seq A hashtag antibodies #1–#10. The populations were pooled together and loaded onto the 10x Chromium controller.
We mapped raw sequencing reads to a customized genome, the combination of Mus musculus (GRCm38 assembly), Rattus (Rnor6 assembly), and red fluorescent protein gene (GenBank accession AF506027) protein, using CellRanger v3.1.0. We quantified gene expression and the number of unique molecular identifiers (UMIs), in all cells that yielded a gene by cell expression matrix. We then performed all downstream analysis using custom scripts and Seurat v3.1.4 package (Hafemeister and Satija, 2019) in R v3.5.1. For quality control, we filtered out cells in which >20% of total UMIs were derived from mitochondrial genes. To remove potential doublets, we retained those cells that expressed 1,250–3,000 unique genes. Because of sequence homology between mouse and rat genomes, we retained those cells having ≥2,000 UMIs mapped to mouse genome and ≤100 UMIs mapped to rat genome. We then normalized each cell to 10,000 reads and log-transformed them. We identified the top 2,000 highly variable genes based on dispersion. Based on the highly variable genes, we scaled the data matrix to have mean 0 and variance 1. We normalized the expression matrix using L2 norm and calculated cosine distance. For dimensionality reduction, we performed principal component analysis (PCA) to obtain 50 principal components using Seurat’s RunPCA function and used them to project cells onto Uniform Manifold Approximation and Projection (UMAP) space using the umap package (McInnes et al., 2018). We clustered the cells using graph-based clustering at resolution 0.05 in Seurat’s FindClusters function (Hafemeister and Satija, 2019).
Identification of cluster markers and DEGs for in vitro monocyte cultures
The markers of individual clusters, i.e., genes overexpressed in a cluster compared with all others, were identified using Wilcoxon test in Seurat’s FindAllMarkers function (Hafemeister and Satija, 2019). We retained those genes that were overexpressed in a given cluster by ≥2 log-folds between compared with the rest and had a Bonferroni-adjusted P value <0.05. The genes that were differentially expressed in a cluster compared with another cluster were identified by comparing their expression using Wilcox test in Seurat’s FindMarkers function (Hafemeister and Satija, 2019). We considered those genes that had a difference of ≥2 log-folds between two clusters and had a Bonferroni adjusted P value <0.05.
Gene set enrichment analysis (GSEA) for in vitro monocyte cultures
For given gene sets, e.g., DEGs between clusters, we performed GSEA using for GO terms related to biological processes taken from the Molecular Signatures Database v7.1 (Liberzon et al., 2015; Liberzon et al., 2011). We considered those terms that had ≥10 genes in common with given gene sets. We calculated enrichment (odds ratio) using Fisher’s exact test against the background of all expressed genes minus DEGs. Subsequently, we performed 10,000 permutations to derive the statistical significance P value of observed enrichment. The P values were Bonferroni corrected for total number of tested pathways (n = 7,350 GO terms related to biological processes). The Gene Expression Omnibus data accession no. is GSE196930.
Analysis of samples from lupus patients
Peripheral blood samples were obtained from healthy controls (HC, n = 8) and patients with active lupus nephritis (n = 22). Patients with SLE fulfilled the 1997 ACR classification criteria and presented with active sediment. Healthy volunteers without a family history of autoimmune disease served as controls. To evaluate the cell phenotype in blood, 100 μl of blood was incubated with an antibody cocktail for 30 min on ice. RBCs were lysed using a lysis buffer (BD Pharm Lyse; 555899). For urine analysis, 15 ml of freshly collected urine was buffered with RPMI 10% serum and washed with TBS + 1% BSA. The cell pellet was stained with Ghost Dye (Tonbo Bioscience) to eliminate cell debris and further stained with an antibody cocktail for 30 min in TBS + 1% BSA on ice. The samples were analyzed by flow cytometry in an LSR Fortessa cytometer (BD Bioscience). Analysis of the samples was performed using FlowJo (TreeStar).
The following antibodies (and dilutions) and related reagents for flow cytometric analysis were used: purchased from Tonbo Bioscience: Ghost Dye, Fluorophore UV 450, 130868, 1:1,000, and anti-human CD45, FITC, 350459, 1:100; from BioLegend: human TruStain FcX, 422302, 1:100; anti-human HLA-DR, PE-Cy7, 307616, 1:100; anti-human CD3, AF700, 300424, 1:100; anti-human CD56, APC-Cy7, 318332, 1:100; anti-human CD14, Pacific Blue, 301828, 1:100; and anti-human CD163, BV605, 333616, 1:100).
Statistical analysis
Statistical analyses were performed using GraphPad Prism. The statistical significance of the differences was determined by a nonparametric test using Tukey’s multiple comparisons analysis. Data are expressed as mean ± SEM. Statistical significance was determined by multiple t test. For group analysis, one-way ANOVA with Dunnett’s t test correction was used. The nonparametric test using Mann–Whitney U analysis was used to evaluate statistics in the human samples. Correlation was analyzed using Spearman test. P < 0.05 was considered significant.
Study approval
All studies in mice were done in accordance with the ethical regulations and animal care guidelines preapproved by the Harvard Medical School and Brigham and Women’s Hospital standing committee on animals. The study assessing the blood and urine of lupus patients was approved by the institutional review board of the Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán (IRE-2297), and all participants signed informed consent forms.
Online supplemental material
Fig. S1 shows gating strategies and CCR2 and CX3CR1 antibody specificity, and demonstrates that the majority of blood CCR2+CX3CR1+ cells are monocytes. Fig. S2 shows that NTN-induced monocyte recruitment per se is not affected, but proteinuria and histological evidence of renal injury is reduced in Ccr2 KO mice. Fig. S3 shows intravascular monocyte populations following lupus nephritis and acute NTN, the analysis of isolated Wt and Ccr2 KO monocytes for adoptive transfer, results in Tnfr2 KO and in vivo CD45 antibody controls and effects of blocking adhesion of monocytes to endothelial cells in vitro on acquisition of MHCII and F4/80. Fig. S4 shows the scRNA-seq characterization of monocytes in contact with the activated endothelium. Fig. S5 shows overlap and GO-term enrichment of genes with CCR2+CX3CR1+ monocytes of the nephritic kidney and genes reported in tumor associated monocytes. Video 1 shows IVM imaging showing dwell time of CCR2rfp (red), CX3CR1gfp (green), and CCR2rfpCX3CR1gfp (yellow) cells within capillaries of glomeruli in a radiation chimeric Wt recipient reconstituted with Ccr2rfp/+Cx3cr1gfp/+ reporter bone marrow subjected to NTN and imaged 14 d later. Video 2 shows IVM imaging showing dwell time of CCR2rfp (red), CX3CR1gfp (green), and CCR2rfpCX3CR1gfp (yellow) cells within capillaries of glomeruli in a radiation chimeric Tnfr2 KO recipient reconstituted with Ccr2rfp/+Cx3cr1gfp/+ reporter bone marrow subjected to NTN and imaged 14 d later. Table S1 shows the bulk RNA-seq nonredundant list of DEGs in kidney DP vs. spleen DP monocyte/macrophage populations. Table S2 shows the scRNA-seq list of cluster marker genes for Wt monocytes on TNF-activated endothelial cells, Ccr2 KO monocytes on activated endothelial cells, and Wt and Ccr2 KO monocytes in medium alone. Table S3 shows the scRNA-seq nonredundant list of DEGs in Wt monocytes on TNF-activated endothelial cells, Ccr2 KO monocytes on activated endothelial cells, and Wt and Ccr2 KO monocytes in medium alone.
Supplementary Material
shows the bulk RNA-seq nonredundant list of DEGs in kidney DP vs. spleen DP monocyte/macrophage populations.
shows the scRNA-seq list of cluster marker genes for WT monocytes on TNF-activated endothelial cells, CCR2 KO monocytes on activated endothelial cells, and WT and CCR2 KO monocytes in medium alone.
shows the scRNA-seq nonredundant list of DEGs in WT monocytes on TNF-activated endothelial cells, CCR2 KO monocytes on activated endothelial cells, and WT and CCR2 KO monocytes in medium alone.
Acknowledgments
RFPEC and the adenovirus expressing IFNγ were kindly provided by Dr. John M. Tarbell (The City College of New York, New York, NY) and Dr. Anne Davidson (Feinstein Institute for Medical Research, Manhasset, NY), respectively. The Ccr2rfp/rfpCx3cr1gfp/gfp mice were generously provided by Dr. Paul Kubes (University of Calgary, Calgary, Alberta, Canada).
This work was supported by National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases grant DK099507 (T.N. Mayadas) and National Heart, Lung, and Blood Institute grant HL065095 (T.N. Mayadas).
Author contributions: V. Mysore, S. Tahir, K. Furuhashi, X. Cullere, and P. Yazbeck conducted and analyzed experiments. The order among the co-first authors (V. Mysore, S. Tahir, and K. Furuhashi) was assigned based on the individuals who brought the project to completion. J. Arora and M.E. Lemieux conducted the transcriptomic analysis of in vitro cultured monocytes and kidney/spleen, respectively. F. Rosetti conducted the studies on lupus patient samples. M. Sekulic completed the pathological analysis of mouse kidney specimens. S. Raychaudhuri, B.H. Horwitz, and T.N. Mayadas contributed to the study design and data interpretation. S. Raychaudhuri supervised the transcriptomic analysis, and T.N. Mayadas conceived and supervised the study.
References
- Al-Lamki, R.S., and Mayadas T.N.. 2015. TNF receptors: Signaling pathways and contribution to renal dysfunction. Kidney Int. 87:281–296. 10.1038/ki.2014.285 [DOI] [PubMed] [Google Scholar]
- Almaani, S., Meara A., and Rovin B.H.. 2017. Update on lupus nephritis. Clin. J. Am. Soc. Nephrol. 12:825–835. 10.2215/CJN.05780616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, K.G., Mayer-Barber K., Sung H., Beura L., James B.R., Taylor J.J., Qunaj L., Griffith T.S., Vezys V., Barber D.L., and Masopust D.. 2014. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc. 9:209–222. 10.1038/nprot.2014.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artinger, K., Kirsch A.H., Aringer I., Moschovaki-Filippidou F., Eller P., Rosenkranz A.R., and Eller K.. 2017. Innate and adaptive immunity in experimental glomerulonephritis: A pathfinder tale. Pediatr. Nephrol. 32:943–947. 10.1007/s00467-016s0043404-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babaev, V.R., Runner R.P., Fan D., Ding L., Zhang Y., Tao H., Erbay E., Gorgun C.Z., Fazio S., Hotamisligil G.S., and Linton M.F.. 2011. Macrophage Mal1 deficiency suppresses atherosclerosis in low-density lipoprotein receptor-null mice by activating peroxisome proliferator-activated receptor-gamma-regulated genes. Arterioscler Thromb. Vasc. Biol. 31:1283–1290. 10.1161/ATVBAHA.111.225839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain, C.C., Scott C.L., Uronen-Hansson H., Gudjonsson S., Jansson O., Grip O., Guilliams M., Malissen B., Agace W.W., and Mowat A.M.. 2013. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 6:498–510. 10.1038/mi.2012.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros, M.H.M., Hauck F., Dreyer J.H., Kempkes B., and Niedobitek G.. 2013. Macrophage polarisation: An immunohistochemical approach for identifying M1 and M2 macrophages. PLoS One. 8:e80908. 10.1371/journal.pone.0080908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethunaickan, R., Sahu R., Liu Z., Tang Y.T., Huang W., Edegbe O., Tao H., Ramanujam M., Madaio M.P., and Davidson A.. 2012. Anti-tumor necrosis factor alpha treatment of interferon-alpha-induced murine lupus nephritis reduces the renal macrophage response but does not alter glomerular immune complex formation. Arthritis Rheum. 64:3399–3408. 10.1002/art.34553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianconi, V., Sahebkar A., Atkin S.L., and Pirro M.. 2018. The regulation and importance of monocyte chemoattractant protein-1. Curr. Opin. Hematol. 25:44–51. 10.1097/moh.0000000000000389 [DOI] [PubMed] [Google Scholar]
- Bird, J.E., Giancarli M.R., Kurihara T., Kowala M.C., Valentine M.T., Gitlitz P.H., Pandya D.G., French M.H., and Durham S.K.. 2000. Increased severity of glomerulonephritis in C-C chemokine receptor 2 knockout mice. Kidney Int. 57:129–136. 10.1046/j.1523-1755.2000.00848.x [DOI] [PubMed] [Google Scholar]
- Celie, J.W.A.M., Beelen R.H.J., and van den Born J.. 2009. Heparan sulfate proteoglycans in extravasation: Assisting leukocyte guidance. Front. Biosci. (Landmark Ed). 14:4932–4949. 10.2741/3578 [DOI] [PubMed] [Google Scholar]
- Chalmers, S.A., Chitu V., Ramanujam M., and Putterman C.. 2015. Therapeutic targeting of macrophages in lupus nephritis. Discov. Med. 20:43–49. [PubMed] [Google Scholar]
- Chung, C.H., Fan J., Lee E.Y., Kang J.S., Lee S.J., Pyagay P.E., Khoury C.C., Yeo T.K., Khayat M.F., Wang A., and Chen S.. 2015. Effects of tumor necrosis factor-α on podocyte expression of monocyte chemoattractant protein-1 and in diabetic nephropathy. Nephron Extra. 5:1–18. 10.1159/000369576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, P.N. 1986. Theoretical and measurement issues in the study of field phenomena. ANS Adv. Nurs. Sci. 9:29–39. 10.1097/00012272-198610000-00006 [DOI] [PubMed] [Google Scholar]
- Cornwell, M., Vangala M., Taing L., Herbert Z., Koster J., Li B., Sun H., Li T., Zhang J., Qiu X., et al. 2018. VIPER: Visualization Pipeline for RNA-seq, a Snakemake workflow for efficient and complete RNA-seq analysis. BMC Bioinformatics. 19:135. 10.1186/s12859-018-2139-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal-Secco, D., Wang J., Zeng Z., Kolaczkowska E., Wong C.H., Petri B., Ransohoff R.M., Charo I.F., Jenne C.N., and Kubes P.. 2015. A dynamic spectrum of monocytes arising from the in situ reprogramming of CCR2+ monocytes at a site of sterile injury. J. Exp. Med. 212:447–456. 10.1084/jem.20141539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf, C.A., Choi J., Baldwin T.M., Bolden J.E., Fairfax K.A., Robinson A.J., Biben C., Morgan C., Ramsay K., Ng A.P., et al. 2016. Haemopedia: An expression atlas of murine hematopoietic cells. Stem Cell Rep. 7:571–582. 10.1016/j.stemcr.2016.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schepper, S., Verheijden S., Aguilera-Lizarraga J., Viola M.F., Boesmans W., Stakenborg N., Voytyuk I., Schmidt I., Boeckx B., Dierckx de Casterle I., et al. 2018. Self-maintaining gut macrophages are essential for intestinal homeostasis. Cell. 175:400–415.e13. 10.1016/j.cell.2018.07.048 [DOI] [PubMed] [Google Scholar]
- Devi, S., Li A., Westhorpe C.L.V., Lo C.Y., Abeynaike L.D., Snelgrove S.L., Hall P., Ooi J.D., Sobey C.G., Kitching A.R., and Hickey M.J.. 2013. Multiphoton imaging reveals a new leukocyte recruitment paradigm in the glomerulus. Nat. Med. 19:107–112. 10.1038/nm.3024 [DOI] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebong, E.E., Macaluso F.P., Spray D.C., and Tarbell J.M.. 2011. Imaging the endothelial glycocalyx in vitro by rapid freezing/freeze substitution transmission electron microscopy. Arterioscler Thromb. Vasc. Biol. 31:1908–1915. 10.1161/ATVBAHA.111.225268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzerodt, A., and Moestrup S.K.. 2013. CD163 and inflammation: Biological, diagnostic, and therapeutic aspects. Antioxid. Redox Signal. 18:2352–2363. 10.1089/ars.2012.4834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, L., Chen S., Garcia G.E., Xia Y., Siani M.A., Botti P., Wilson C.B., Harrison J.K., and Bacon K.B.. 1999. Prevention of crescentic glomerulonephritis by immunoneutralization of the fractalkine receptor CX3CR1 rapid communication. Kidney Int. 56:612–620. 10.1046/j.1523-1755.1999.00604.x [DOI] [PubMed] [Google Scholar]
- Flores-Mendoza, G., Sanson S.P., Rodriguez-Castro S., Crispin J.C., and Rosetti F.. 2018. Mechanisms of tissue injury in lupus nephritis. Trends Mol. Med. 24:364–378. 10.1016/j.molmed.2018.02.003 [DOI] [PubMed] [Google Scholar]
- Gaeta, M.L., Johnson D.R., Kluger M.S., and Pober J.S.. 2000. The death domain of tumor necrosis factor receptor 1 is necessary but not sufficient for Golgi retention of the receptor and mediates receptor desensitization. Lab. Invest. 80:1185–1194. 10.1038/labinvest.3780126 [DOI] [PubMed] [Google Scholar]
- Gautier, E.L., Jakubzick C., and Randolph G.J.. 2009. Regulation of the migration and survival of monocyte subsets by chemokine receptors and its relevance to atherosclerosis. Arterioscler Thromb. Vasc. Biol. 29:1412–1418. 10.1161/ATVBAHA.108.180505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt, T., and Ley K.. 2015. Monocyte trafficking across the vessel wall. Cardiovasc. Res. 107:321–330. 10.1093/cvr/cvv147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams, M., Mildner A., and Yona S.. 2018. Developmental and functional heterogeneity of monocytes. Immunity. 49:595–613. 10.1016/j.immuni.2018.10.005 [DOI] [PubMed] [Google Scholar]
- Guilliams, M., and Svedberg F.R.. 2021. Does tissue imprinting restrict macrophage plasticity? Nat. Immunol. 22:118–127. 10.1038/s41590-020s4100849-2 [DOI] [PubMed] [Google Scholar]
- Hafemeister, C., and Satija R.. 2019. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20:296. 10.1186/s13059-019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskell, C.A., Hancock W.W., Salant D.J., Gao W., Csizmadia V., Peters W., Faia K., Fituri O., Rottman J.B., and Charo I.F.. 2001. Targeted deletion of CX(3)CR1 reveals a role for fractalkine in cardiac allograft rejection. J. Clin. Invest. 108:679–688. 10.1172/JCI12976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennies, C.M., Lehn M.A., and Janssen E.M.. 2015. Quantitating MHC class II trafficking in primary dendritic cells using imaging flow cytometry. J. Immunol. Methods. 423:18–28. 10.1016/j.jim.2015.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheiser, K., Heuser C., Krause T.A., Teteris S., Ilias A., Weisheit C., Hoss F., Tittel A.P., Knolle P.A., Panzer U., et al. 2013. Exclusive CX3CR1 dependence of kidney DCs impacts glomerulonephritis progression. J. Clin. Invest. 123:4242–4254. 10.1172/JCI70143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen, S.C., and Cantley L.G.. 2017. Macrophages in renal injury and repair. Annu. Rev. Physiol. 79:449–469. 10.1146/annurev-physiol-022516-034219 [DOI] [PubMed] [Google Scholar]
- Ingersoll, M.A., Spanbroek R., Lottaz C., Gautier E.L., Frankenberger M., Hoffmann R., Lang R., Haniffa M., Collin M., Tacke F., et al. 2010. Comparison of gene expression profiles between human and mouse monocyte subsets. Blood. 115:e10–e119. 10.1182/blood-2009b07-235028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, A., Hasegawa H., Kohno M., Ito M.R., Terada M., Imai T., Yoshie O., Nose M., and Fujita S.. 2005. Antagonist of fractalkine (CX3CL1) delays the initiation and ameliorates the progression of lupus nephritis in MRL/lpr mice. Arthritis Rheum. 52:1522–1533. 10.1002/art.21007 [DOI] [PubMed] [Google Scholar]
- Jakubzick, C., Gautier E.L., Gibbings S.L., Sojka D.K., Schlitzer A., Johnson T.E., Ivanov S., Duan Q., Bala S., Condon T., et al. 2013. Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity. 39:599–610. 10.1016/j.immuni.2013.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubzick, C.V., Randolph G.J., and Henson P.M.. 2017. Monocyte differentiation and antigen-presenting functions. Nat. Rev. Immunol. 17:349–362. 10.1038/nri.2017.28 [DOI] [PubMed] [Google Scholar]
- Joeris, T., Muller-Luda K., Agace W.W., and Mowat A.M.. 2017. Diversity and functions of intestinal mononuclear phagocytes. Mucosal Immunol. 10:845–864. 10.1038/mi.2017.22 [DOI] [PubMed] [Google Scholar]
- Kawakami, T., Lichtnekert J., Thompson L.J., Karna P., Bouabe H., Hohl T.M., Heinecke J.W., Ziegler S.F., Nelson P.J., and Duffield J.S.. 2013. Resident renal mononuclear phagocytes comprise five discrete populations with distinct phenotypes and functions. J. Immunol. 191:3358–3372. 10.4049/jimmunol.1300342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriz, W., and LeHir M.. 2005. Pathways to nephron loss starting from glomerular diseases-insights from animal models. Kidney Int. 67:404–419. 10.1111/j.1523-1755.2005.67097.x [DOI] [PubMed] [Google Scholar]
- Kurihara, T., Warr G., Loy J., and Bravo R.. 1997. Defects in macrophage recruitment and host defense in mice lacking the CCR2 chemokine receptor. J. Exp. Med. 186:1757–1762. 10.1084/jem.186.10.1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuziel, W.A., Morgan S.J., Dawson T.C., Griffin S., Smithies O., Ley K., and Maeda N.. 1997. Severe reduction in leukocyte adhesion and monocyte extravasation in mice deficient in CC chemokine receptor 2. Proc. Natl. Acad. Sci. USA. 94:12053–12058. 10.1073/pnas.94.22.12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E.Y., Chung C.H., Khoury C.C., Yeo T.K., Pyagay P.E., Wang A., and Chen S.. 2009. The monocyte chemoattractant protein-1/CCR2 loop, inducible by TGF-beta, increases podocyte motility and albumin permeability. Am. J. Physiol. Renal Physiol. 297:F85–F94. 10.1152/ajprenal.90642.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.A., Noel S., Sadasivam M., Allaf M.E., Pierorazio P.M., Hamad A.R.A., and Rabb H.. 2018. Characterization of kidney CD45intCD11bintF4/80+MHCII+CX3CR1+Ly6C- “intermediate mononuclear phagocytic cells”. PLoS One. 13:e0198608. 10.1371/journal.pone.0198608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever, J.M., Hull T.D., Boddu R., Pepin M.E., Black L.M., Adedoyin O.O., Yang Z., Traylor A.M., Jiang Y., Li Z., et al. 2019. Resident macrophages reprogram toward a developmental state after acute kidney injury. JCI Insight. 4:e125503. 10.1172/jci.insight.125503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley, K., Miller Y.I., and Hedrick C.C.. 2011. Monocyte and macrophage dynamics during atherogenesis. Arterioscler Thromb. Vasc. Biol. 31:1506–1516. 10.1161/atvbaha.110.221127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Huang L., Sung S.-S.J., Vergis A.L., Rosin D.L., Rose C.E. Jr., Lobo P.I., and Okusa M.D.. 2008. The chemokine receptors CCR2 and CX3CR1 mediate monocyte/macrophage trafficking in kidney ischemia-reperfusion injury. Kidney Int. 74:1526–1537. 10.1038/ki.2008.500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon, A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., and Tamayo P.. 2015. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1:417–425. 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon, A., Subramanian A., Pinchback R., Thorvaldsdottir H., Tamayo P., and Mesirov J.P.. 2011. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 27:1739–1740. 10.1093/bioinformatics/btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, Y.C., and Luscinskas F.W.. 2006. Isolation and culture of murine heart and lung endothelial cells for in vitro model systems. Methods Mol. Biol. 341:141–154. 10.1385/1-59745113-4:141 [DOI] [PubMed] [Google Scholar]
- Liu, Z., Bethunaickan R., Huang W., Lodhi U., Solano I., Madaio M.P., and Davidson A.. 2011. Interferon-alpha accelerates murine systemic lupus erythematosus in a T cell-dependent manner. Arthritis Rheum. 63:219–229. 10.1002/art.30087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, C.M., Dorf M.E., Proudfoot A., Salant D.J., and Gutierrez-Ramos J.C.. 1997. Role of MCP-1 and RANTES in inflammation and progression to fibrosis during murine crescentic nephritis. J. Leukoc. Biol. 62:676–680. 10.1002/jlb.62.5.676 [DOI] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathian, A., Weinberg A., Gallegos M., Banchereau J., and Koutouzov S.. 2005. IFN-alpha induces early lethal lupus in preautoimmune (New Zealand Black x New Zealand White) F1 but not in BALB/c mice. J. Immunol. 174:2499–2506. 10.4049/jimmunol.174.5.2499 [DOI] [PubMed] [Google Scholar]
- Mayadas, T.N., Tsokos G.C., and Tsuboi N.. 2009. Mechanisms of immune complex-mediated neutrophil recruitment and tissue injury. Circulation. 120:2012–2024. 10.1161/CIRCULATIONAHA.108.771170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes, L., Healy J., and Melville J.. 2018. UMAP: Uniform manifold approximation and projection for dimension reduction. arXiv: 1802.03426. [Google Scholar]
- Mildner, A., Yona S., and Jung S.. 2013. A close encounter of the third kind: Monocyte-derived cells. Adv. Immunol. 120:69–103. 10.1016/B978-012-417028-5.00003-X [DOI] [PubMed] [Google Scholar]
- Mok, C.C., Ding H.H., Kharboutli M., and Mohan C.. 2016. Axl, ferritin, insulin-like growth factor binding protein 2, and tumor necrosis factor receptor type II as biomarkers in systemic lupus erythematosus. Arthritis Care Res. (Hoboken). 68:1303–1309. 10.1002/acr.22835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, K.J., Sheedy F.J., and Fisher E.A.. 2013. Macrophages in atherosclerosis: A dynamic balance. Nat. Rev. Immunol. 13:709–721. 10.1038/nri3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro, D.A.D., and Hughes J.. 2017. The origins and functions of tissue-resident macrophages in kidney development. Front. Physiol. 8:837. 10.3389/fphys.2017.00837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, P.J., Rees A.J., Griffin M.D., Hughes J., Kurts C., and Duffield J.. 2012. The renal mononuclear phagocytic system. J. Am. Soc. Nephrol. 23:194–203. 10.1681/ASN.2011070680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi, H., Furuhashi K., Cullere X., Saggu G., Miller M.J., Chen Y., Rosetti F., Hamilton S.L., Yang L., Pittman S.P., et al. 2017. Neutrophil FcγRIIA promotes IgG-mediated glomerular neutrophil capture via Abl/Src kinases. J. Clin. Invest. 127:3810–3826. 10.1172/JCI94039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, M.Y., Nagai Y., Takamura T., Nohara E., and Kobayashi K.. 2000. Inhibitory effect of troglitazone on tumor necrosis factor alpha-induced expression of monocyte chemoattractant protein-1 in human mesangial cells. Metabolism. 49:163–166. 10.1016/s0026-0495(00)91143-0 [DOI] [PubMed] [Google Scholar]
- Olaru, F., Dobel T., Lonsdorf A.S., Oehrl S., Maas M., Enk A.H., Schmitz M., Grone E.F., Grone H.J., and Schakel K.. 2018. Intracapillary immune complexes recruit and activate slan-expressing CD16+ monocytes in human lupus nephritis. JCI Insight. 3:e96492. 10.1172/jci.insight.96492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzer, U., Thaiss F., Zahner G., Barth P., Reszka M., Reinking R.R., Wolf G., Helmchen U., and Stahl R.A.. 2001. Monocyte chemoattractant protein-1 and osteopontin differentially regulate monocytes recruitment in experimental glomerulonephritis. Kidney Int. 59:1762–1769. 10.1046/j.1523-1755.2001.0590051762.x [DOI] [PubMed] [Google Scholar]
- Pérez de Lema, G., Maier H., Franz T.J., Escribese M., Chilla S., Segerer S., Camarasa N., Schmid H., Banas B., Kalaydjiev S., et al. 2005. Chemokine receptor Ccr2 deficiency reduces renal disease and prolongs survival in MRL/lpr lupus-prone mice. J. Am. Soc. Nephrol. 16:3592–3601. 10.1681/asn.2005040426 [DOI] [PubMed] [Google Scholar]
- Platt, A.M., Bain C.C., Bordon Y., Sester D.P., and Mowat A.M.. 2010. An independent subset of TLR expressing CCR2-dependent macrophages promotes colonic inflammation. J. Immunol. 184:6843–6854. 10.4049/jimmunol.0903987 [DOI] [PubMed] [Google Scholar]
- Prinz, M., and Priller J.. 2010. Tickets to the brain: Role of CCR2 and CX3CR1 in myeloid cell entry in the CNS. J. Neuroimmunol. 224:80–84. 10.1016/j.jneuroim.2010.05.015 [DOI] [PubMed] [Google Scholar]
- Quah, B.J., and Parish C.R.. 2010. The use of carboxyfluorescein diacetate succinimidyl ester (CFSE) to monitor lymphocyte proliferation. J. Vis. Exp. 44:2259. 10.3791/2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2017. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. [Google Scholar]
- Robinson, A., Han C.Z., Glass C.K., and Pollard J.W.. 2021. Monocyte regulation in homeostasis and malignancy. Trends Immunol. 42:104–119. 10.1016/j.it.2020.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins, B.J., Yoshimura T., Leonard E.J., and Pober J.S.. 1990. Cytokine-activated human endothelial cells synthesize and secrete a monocyte chemoattractant, MCP-1/JE. Am. J. Pathol. 136:1229–1233. [PMC free article] [PubMed] [Google Scholar]
- Romano, A., Carneiro M.B.H., Doria N.A., Roma E.H., Ribeiro-Gomes F.L., Inbar E., Lee S.H., Mendez J., Paun A., Sacks D.L., and Peters N.C.. 2017. Divergent roles for Ly6C+CCR2+CX3CR1+ inflammatory monocytes during primary or secondary infection of the skin with the intra-phagosomal pathogen Leishmania major. PLoS Pathog. 13:e1006479. 10.1371/journal.ppat.1006479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovin, B.H., Doe N., and Tan L.C.. 1996. Monocyte chemoattractant protein-1 levels in patients with glomerular disease. Am. J. Kidney Dis. 27:640–646. 10.1016/s0272-6386(96)90097-9 [DOI] [PubMed] [Google Scholar]
- Schenkel, A.R., Mamdouh Z., and Muller W.A.. 2004. Locomotion of monocytes on endothelium is a critical step during extravasation. Nat. Immunol. 5:393–400. 10.1038/ni1051 [DOI] [PubMed] [Google Scholar]
- Schridde, A., Bain C.C., Mayer J.U., Montgomery J., Pollet E., Denecke B., Milling S.W.F., Jenkins S.J., Dalod M., Henri S., et al. 2017. Tissue-specific differentiation of colonic macrophages requires TGFβ receptor-mediated signaling. Mucosal Immunol. 10:1387–1399. 10.1038/mi.2016.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sean Eardley, K., and Cockwell P.. 2005. Macrophages and progressive tubulointerstitial disease. Kidney Int. 68:437–455. 10.1111/j.1523-1755.2005.00422.x [DOI] [PubMed] [Google Scholar]
- Segerer, S., Cui Y., Hudkins K.L., Goodpaster T., Eitner F., Mack M., Schlondorff D., and Alpers C.E.. 2000. Expression of the chemokine monocyte chemoattractant protein-1 and its receptor chemokine receptor 2 in human crescentic glomerulonephritis. J. Am. Soc. Nephrol. 11:2231–2242. 10.1681/ASN.V11122231 [DOI] [PubMed] [Google Scholar]
- Shan, Z., and Ju C.. 2020. Hepatic macrophages in liver injury. Front. Immunol. 11:322. 10.3389/fimmu.2020.00322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatiades, E.G., Tremblay M.E., Bohm M., Crozet L., Bisht K., Kao D., Coelho C., Fan X., Yewdell W.T., Davidson A., et al. 2016. Immune monitoring of trans-endothelial transport by kidney-resident macrophages. Cell. 166:991–1003. 10.1016/j.cell.2016.06.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strieter, R.M., Wiggins R., Phan S.H., Wharram B.L., Showell H.J., Remick D.G., Chensue S.W., and Kunkel S.L.. 1989. Monocyte chemotactic protein gene expression by cytokine-treated human fibroblasts and endothelial cells. Biochem. Biophys. Res. Commun. 162:694–700. 10.1016/0006-291x(89)92366-8 [DOI] [PubMed] [Google Scholar]
- Tamoutounour, S., Henri S., Lelouard H., de Bovis B., de Haar C., van der Woude C.J., Woltman A.M., Reyal Y., Bonnet D., Sichien D., et al. 2012. CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. Eur. J. Immunol. 42:3150–3166. 10.1002/eji.201242847 [DOI] [PubMed] [Google Scholar]
- Tesch, G.H., Maifert S., Schwarting A., Rollins B.J., and Kelley V.R.. 1999. Monocyte chemoattractant protein 1-dependent leukocytic infiltrates are responsible for autoimmune disease in MRL-Fas(lpr) mice. J. Exp. Med. 190:1813–1824. 10.1084/jem.190.12.1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., and Pachter L.. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28:511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou, C.L., Peters W., Si Y., Slaymaker S., Aslanian A.M., Weisberg S.P., Mack M., and Charo I.F.. 2007. Critical roles for CCR2 and MCP-3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. J. Clin. Invest. 117:902–909. 10.1172/JCI29919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi, N., Asano K., Lauterbach M., and Mayadas T.N.. 2008. Human neutrophil Fcgamma receptors initiate and play specialized nonredundant roles in antibody-mediated inflammatory diseases. Immunity. 28:833–846. 10.1016/j.immuni.2008.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucci, M., Barnes E.V., Sobel E.S., Croker B.P., Segal M.S., Reeves W.H., and Richards H.B.. 2004. Strong association of a functional polymorphism in the monocyte chemoattractant protein 1 promoter gene with lupus nephritis. Arthritis Rheum. 50:1842–1849. 10.1002/art.20266 [DOI] [PubMed] [Google Scholar]
- Turner-Stokes, T., Garcia Diaz A., Pinheiro D., Prendecki M., McAdoo S.P., Roufosse C., Cook H.T., Pusey C.D., and Woollard K.J.. 2020. Live imaging of monocyte subsets in immune complex-mediated glomerulonephritis reveals distinct phenotypes and effector functions. J. Am. Soc. Nephrol. 31:2523–2542. 10.1681/ASN.2019121326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gorp, H., Delputte P.L., and Nauwynck H.J.. 2010. Scavenger receptor CD163, a Jack-of-all-trades and potential target for cell-directed therapy. Mol. Immunol. 47:1650–1660. 10.1016/j.molimm.2010.02.008 [DOI] [PubMed] [Google Scholar]
- Venkatesh, D., Ernandez T., Rosetti F., Batal I., Cullere X., Luscinskas F.W., Zhang Y., Stavrakis G., Garcia-Cardena G., Horwitz B.H., and Mayadas T.N.. 2013. Endothelial TNF receptor 2 induces IRF1 transcription factor-dependent interferon-beta autocrine signaling to promote monocyte recruitment. Immunity. 38:1025–1037. 10.1016/j.immuni.2013.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielhauer, V., Stavrakis G., and Mayadas T.N.. 2005. Renal cell-expressed TNF receptor 2, not receptor 1, is essential for the development of glomerulonephritis. J. Clin. Invest. 115:1199–1209. 10.1172/jci200523348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani, A.C., Satija R., Reynolds G., Sarkizova S., Shekhar K., Fletcher J., Griesbeck M., Butler A., Zheng S., Lazo S., et al. 2017. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 356:eaah4573. 10.1126/science.aah4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, T., Yokoyama H., Furuichi K., Kobayashi K.I., Harada K., Naruto M., Su S.B., Akiyama M., Mukaida N., and Matsushima K.. 1996. Intervention of crescentic glomerulonephritis by antibodies to monocyte chemotactic and activating factor (MCAF/MCP-1). FASEB J. 10:1418–1425. 10.1096/fasebj.10.12.8903512 [DOI] [PubMed] [Google Scholar]
- Waddell, L.A., Lefevre L., Bush S.J., Raper A., Young R., Lisowski Z.M., McCulloch M.E.B., Muriuki C., Sauter K.A., Clark E.L., et al. 2018. ADGRE1 (EMR1, F4/80) is a rapidly-evolving gene expressed in mammalian monocyte-macrophages. Front. Immunol. 9:2246. 10.3389/fimmu.2018.02246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, T., Ding H., Han J., Arriens C., Wei C., Han W., Pedroza C., Jiang S., Anolik J., Petri M., et al. 2016. Antibody-array-based proteomic screening of serum markers in systemic lupus erythematosus: A discovery study. J. Proteome Res. 15:2102–2114. 10.1021/acs.jproteome.5b00905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q., Wang Y., Pei G., Deng X., Jiang H., Wu J., Zhou C., Guo Y., Yao Y., Zeng R., and Xu G.. 2019. Bone marrow-derived Ly6C(-) macrophages promote ischemia-induced chronic kidney disease. Cell Death Dis. 10:291. 10.1038/s41419-019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F., Haas M., Glassock R., and Zhao M.H.. 2017. Redefining lupus nephritis: Clinical implications of pathophysiologic subtypes. Nat. Rev. Nephrol. 13:483–495. 10.1038/nrneph.2017.85 [DOI] [PubMed] [Google Scholar]
- Zhang, G., Zhang H., Liu Y., He Y., Wang W., Du Y., Yang C., and Gao F.. 2014. CD44 clustering is involved in monocyte differentiation. Acta Biochim. Biophys. Sin (Shanghai). 46:540–547. 10.1093/abbs/gmu042 [DOI] [PubMed] [Google Scholar]
- Zigmond, E., Varol C., Farache J., Elmaliah E., Satpathy A.T., Friedlander G., Mack M., Shpigel N., Boneca I.G., Murphy K.M., et al. 2012. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity. 37:1076–1090. 10.1016/j.immuni.2012.08.026 [DOI] [PubMed] [Google Scholar]
- Zilionis, R., Engblom C., Pfirschke C., Savova V., Zemmour D., Saatcioglu H.D., Krishnan I., Maroni G., Meyerovitz C.V., Kerwin C.M., et al. 2019. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity. 50:1317–1334.e10. 10.1016/j.immuni.2019.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the bulk RNA-seq nonredundant list of DEGs in kidney DP vs. spleen DP monocyte/macrophage populations.
shows the scRNA-seq list of cluster marker genes for WT monocytes on TNF-activated endothelial cells, CCR2 KO monocytes on activated endothelial cells, and WT and CCR2 KO monocytes in medium alone.
shows the scRNA-seq nonredundant list of DEGs in WT monocytes on TNF-activated endothelial cells, CCR2 KO monocytes on activated endothelial cells, and WT and CCR2 KO monocytes in medium alone.