Abstract
Entamoeba histolytica is the third leading cause of parasitic mortality globally. E. histolytica infection is generally asymptomatic but the parasite has potent pathogenic potential. The origins, benefits and triggers of amebic virulence are complex. Amebic pathogenesis entails depletion of the host mucosal barrier, adherence to the colonic lumen, cytotoxicity and invasion of the colonic epithelium. Parasite damage results in colitis and in some cases, disseminated disease. Both host and parasite genotypes influence the development of disease, as do the regulatory responses they govern at the host-pathogen interface. Host environmental factors determine parasite transmission and shape the colonic microenvironment E. histolytica infects. Here we highlight research that illuminates novel links in host, parasite and environmental factors in the regulation of E. histolytica virulence.
Keywords: Virulence, microbiota, pathobiota, mucous, carbohydrate utilization
INTRODUCTION
Evolution of E. histolytica virulence
Entamoeba histolytica is an ancient obligate parasite of humans. The first documented case of amebiasis may be a Sanskrit description of bloody mucoid diarrhea from 3000 BCE (67). Today E. histolytica is estimated cause ∼50 million cases of disease and 40,000–100,000 deaths annually (3, 56, 123). Evolutionary theories posit that obligate human parasites evolve toward commensalism but E. histolytica has maintained a potent capacity for virulence in comparison to the recently diverged avirulent Entamoeba dispar. The high frequency of chronic asymptomatic infection with E. histolytica and E. dispar indicates that virulence is dispensable for parasite survival, replication and transmission. In addition, virulence does not seem to confer a selective advantage in these processes. Similar levels of E. histolytica parasites were found in symptomatic and asymptomatic hosts, thus disease does not appear to enhance replicative capacity (58). Similarly, asymptomatic hosts passed high levels of infectious amebic cysts, apparently obviating virulence for transmission (84). E. histolytica virulence has been proposed to originate from coincidental selection by bacterial killing but coincidental selection does not explain why E. dispar which also kills and consumes bacteria is avirulent. Despite an uncertain evolutionary role, the mechanisms of E. histolytica virulence are increasingly understood. Virulence can also be a consequence of the immune responses in the host. E. histolytica has the capacity to cause serious infections in immunocompetent hosts but may be more severe in hosts with impaired immune function. E. histolytica virulence is not predictable and depends on multiple factors that determine the capacity of the parasite to damage its host.
Epidemiology of E. histolytica
E. histolytica is endemic to areas of Mexico, Central and South America, Asia, Africa, and the Pacific islands and where transmission occurs via fecally-contaminated food and water (reviewed in 8,9). E. histolytica is also common in parts of Asia and Australia in men who have sex with men (MSM) and can be transmitted sexually (reviewed in 10). A recent study in Japan detected antibodies to E. histolytica in 21% of 1303 HIV positive patients, 88% of E. histolytica infections were in MSM (138). HIV infection is associated with Entamoeba infection but HIV and AIDS do not appear to increase E. histolytica disease. E. histolytica infection extends beyond endemic regions as the third most common cause of chronic diarrhea in returning travelers (112).
Estimating the global burden of amebiasis is complicated by limited diagnosis capacities and surveillance in most endemic areas. In addition, there is extreme intra-host variability in incubation period and disease presentation. Only 10–20% of E. histolytica infections result in disease, which encompasses self-limiting colitis, invasive colitis, extra-intestinal infection and invasive organ abscess (56, 123). These clinical manifestations are not necessarily sequential and may appear after years of asymptomatic infection (56, 109, 123).
Multiple community based prospective studies in Bangladesh have revealed the consequences of E. histolytica infection in endemic regions, particularly for children (54, 55, 57, 90). In combination, these studies found that E. histolytica infection had a cumulative incidence of ∼30% by 1 year of age reaching 60–90% by four years of age (54, 55, 57, 90). Despite the high prevalence of infection, only 3–10% of diarrheal episodes in this population were attributable to E. histolytica (55, 57, 125) (Figure 1). The case-control Global Enteric Multicenter Study (GEMS) found a similar incidence of E. histolytica-diarrhea in hospitalized children (2.0% in Mali and 3.4% in Bangladesh) though E. histolytica was the pathogen with the highest hazard ratio for death in the second year of life (71). Likewise, E. histolytica was significantly increased in cases of severe diarrhea relative to all diarrhea in Bangladesh (6.6% versus 10.3%) (125). Thus while symptomatic E. histolytica disease is rare it is associated with severe outcomes likely due to specific host vulnerabilities in combination with parasite virulence attributes. It is also worth noting that some children had no evidence of E. histolytica infection (seroconversion and/or PCR-detection in monthly stool samples) in the first two years of life.
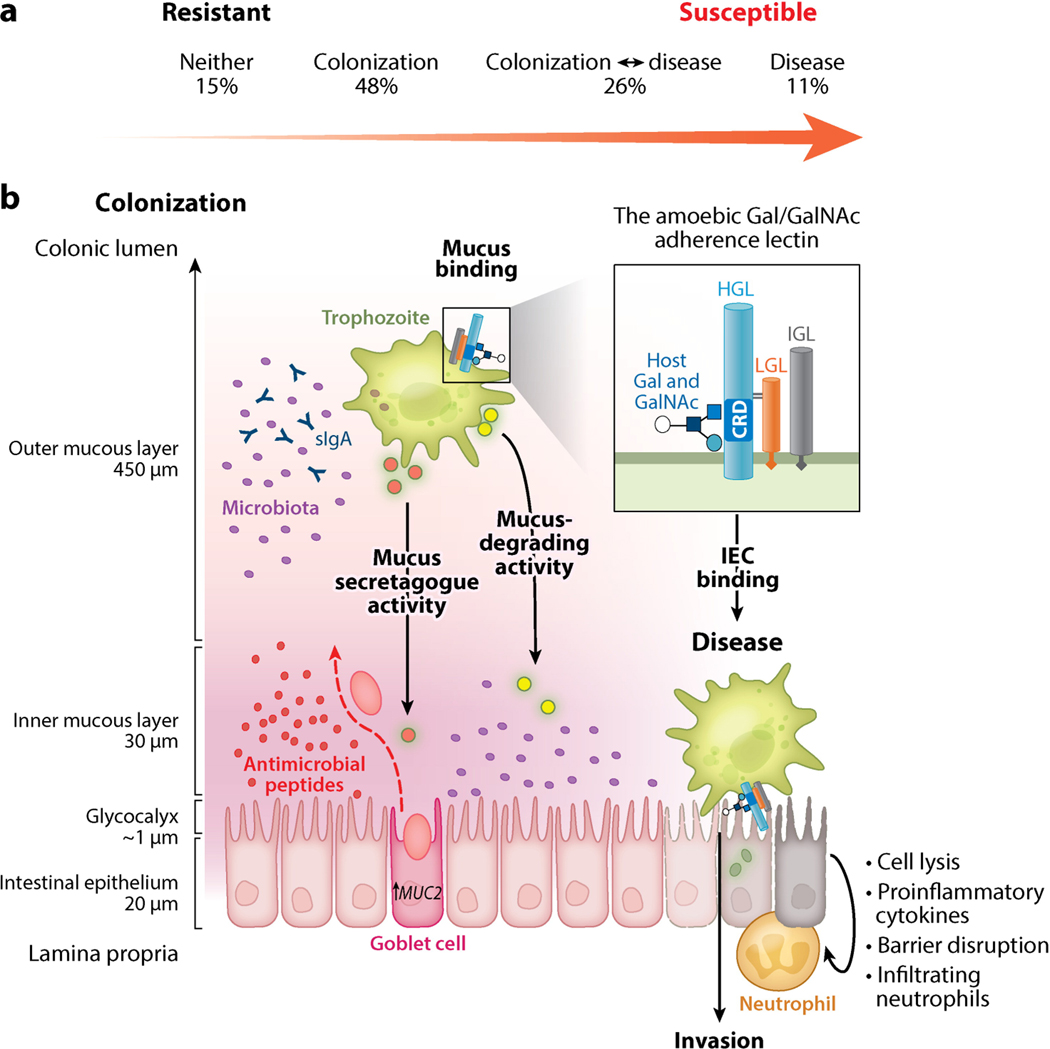
Figure 1.
E. histolytica virulence depends on a dynamic interaction in the infected host. (a) Continuum of E. histolytica disease in a natural population. Analysis of diarrheal and monthly surveillance stool samples for E. histolytica detected four possible outcomes in the first two years of life: (1) no evidence of infection, (2) colonization with no E. histolytica–associated diarrhea, (3) diarrhea with prior asymptomatic colonization and/or subsequent asymptomatic persistence, or (4) E. histolytica–associated diarrhea with no previous colonization. This pattern reinforces the importance of both parasite and host factors in the outcome of an E. histolytica infection. (b) To establish infection, E. histolytica must bind and adhere in the host colon. Adherence is mediated by an amoebic lectin with a carbohydrate-recognition domain (CRD) that binds galactose (Gal) and N-acetyl-d-galactosamine (GalNAc) on host glycoconjugates with high affinity. The Gal/GalNAc lectin is composed of heavy (HGL), intermediate (IGL), and light (LGL) subunits. The CRD is located on the HGL, which also contains a putative intracellular signaling domain. HGL forms a disulfide bond with LGL. The HGL-LGL heterodimer can associate with the glycosylphosphatidylinositol-anchored IGL, but this subunit does not have a well-defined function (inset). Colonic mucin forms a dense polymeric gel over the intestinal epithelium, which trophozoites bind to with high affinity. Trophozoites also induce mucin secretion by goblet cells. In colonization, mucin binding mediates attachment and provides a nutrient source for E. histolytica. Mucin polymers may be degraded by amoebic proteases and glycosidases for nutrients, and the mucosal microbiota provides a nutrient source via amoebic phagocytosis. The transition from colonization to disease is marked by destruction of the mucin barrier. Mucus depletion may result from enhanced amoebic degradation and/or depletion of mucin stores by continual secretion during chronic infection. Other factors including coinfections, host diet, and disruption of the microbiota can also mediate mucus depletion. Mucus depletion exposes the intestinal epithelium to E. histolytica trophozoites. The amoebic lectin CRD binds to Gal and GalNAc on exposed intestinal epithelial cells (IECs) and the cell-associated glycocalyx. Adherence to IECs results in amoebic cytotoxicity and the release of proinflammatory molecules. Abbreviation: sIgA, secretory immunoglobulin A.
Pathogenesis in the human host
The species name histolytica is derived from ancient Greek for tissue dissolving. Befitting its name, the parasite is capable of extensive tissue destruction. E. histolytica infection is established by parasite adherence to the colonic mucin layer. Trophozoites express a surface lectin with high affinity for galactose (Gal) and N-acetyl-D-galactosamine oligosaccharides (GalNAc) on host mucin and cells (reviewed in 22). Initial E. histolytica infection induces thickening of the mucosal layer, potentially due E. histolytica from contacting the intestinal epithelium. E. histolytica also produces a mucin secretagogue that stimulates goblet cell mucin secretion. Concerted glycosidase and protease activity can mediate degradation of the mucus polymer (Figure 1). In the absence of mucin, the amebic Gal/GalNAc lectin binds to Gal and GalNAc residues on the surface of exposed intestinal epithelial cells (IECs). Progressive disease is marked by mucin depletion, IEC flattening, and infiltrating neutrophils (104). In addition, E. histolytica secretory molecules disrupt tight junctions and intestinal ion transport provoking diarrhea. In accordance, E. histolytica pathology is significantly worse in the absence of mucin in the murine model of amebiasis. In MUC2 deficient mice, E. histolytica directly bound IECs causing greater pathology, barrier disruption, and secretory and pro-inflammatory responses (69). Amebic lesions in the intestinal epithelium can progress to necrotic flask-shaped ulcers containing trophozoites, bacteria and inflammatory cells. From these ulcers, trophozoites may invade the lamina propria and enter the bloodstream, often disseminating to the liver and causing amebic liver abscess (ALA) (37, 56). (Figure 1)
Differential virulence in Entamoeba
E. histolytica and E. dispar were classified as distinct species in 1993 based on the long-standing observation that E. dispar does not cause disease (31). This seperation redefined diagnostics, epidemiology and treatment of amebiasis (143). Since 1993, E. dispar has been infrequently detected in human colitis and ALA (142) and zenic E. dispar caused necrotic lesions in experimental ALA (32, 53). The free-living Entamoeba moshkovskii was initially believed to be avirulent (6) but was recently associated with human colitis and caused colitis in mice (119, 120). Entamoeba bangladeshi was discovered in cyst-containing diarrheal samples and may be a novel pathogenic species (114). In addition, differential virulence exists within E. histolytica- the Rahman strain was isolated from an asymptomatic individual and is considered avirulent. In vitro Rahman and E. dispar have reduced ability to ingest erythrocytes, damage colonic epithelia and cause ALA, relative to the virulent E. histolytica isolate HM1:IMSS. Infection with mixed Entamoeba species is also common (4, 143). Intra and interspecies competition for space and resources may increase parasite virulence. Co-infection could also be protective, however no cross-reactive immunity between E. histolytica and E. moshkovskii developed in mice (119).
Parasite factors that influence virulence
Differentiation
E. histolytica differentiates between an environmental cyst and replicative trophozoite. The chitin-rich cyst wall allows the parasite to survive outside the host and pass through the acidic stomach to establish infection. Excystation occurs in the small intestine and each quadrinucleate cyst releases eight motile trophozoites, which establish infection in the colon. During infection trophozoites encyst and are excreted in the stool. The cues for excystation and encystation are unknown, and in vitro differentiation of E. histolytica remains elusive. The related Entamoeba invadens is a model for stage conversion. Dramatic transcriptional remodeling occurred during encystation and excystation of E. invadens (28, 34). Excystation triggered upregulation of carbohydrate metabolism (glycoside, hydrolases, hexokinases), protein synthesis, lipid biosynthesis and virulence (proteases, lectin) genes while encystation was marked by downregulation of metabolic processes and upregulation of genes for meiosis, chitin biosynthesis and phospholipase D (PLD) (28, 34). PLD is a lipid second messenger and inhibition of PLD activity blocked encystation of E. invadens (39). Differentiation is essential for transmission of E. histolytica; as such the environmental cues and parasite genes required for differentiation are ideal targets for transmission-blocking interventions.
Genomic virulence
The E. histolytica genome is ∼24 MB with large differences in gene content, copy number, ploidy and intergenic regions between isolates (46, 139). Structural and sequence variation has been associated with clinical outcome of E. histolytica infection (5, 7, 8, 43, 63, 140). Paired colon and liver isolates from ALA patients had different genotypes suggesting either mixed infections and/or genetic alterations during an infection leading to a subpopulation of invasive trophozoites (8). Sequencing of clinical isolates found that sequence diversity varies by gene and single nucleotide polymorphisms (SNPs) in CYCLCIN2 and lectin genes were associated with disease (46, 139). The E. histolytica genome shows evidence of dynamic evolution marked by extensive recombination, rapid mutation, RNA silencing, and epigenetic silencing (129). E. histolytica retrotransposons (LINES/SINES) modify gene expression and structure. Retrotransposons were more highly expressed in virulent E. histolytica relative to non-virulent species and strains, indicating that rapid evolution occur during disease progression (80). Genomic evidence has indicated that that E. histolytica exchanges genetic information by homologous recombination. Recently, the first experimental demonstration of homologous recombination was reported and homologous recombination was induced by serum starvation (121). Increased characterization of circulating strains and sequential clinical isolates as patients progress from colonization to disease is needed to understand how genetic variation mediates E. histolytica virulence.
Metabolism and nutrient acquisition
E. histolytica has a unique reductive metabolism (77). E. histolytica lacks most amino acid biosynthetic pathways except those for serine and cysteine. Cysteine production may be conserved as a component of amebic oxidative stress resistance. E. histolytica also lacks purine, pyrimidine and thymidylate synthesis and utilizes salvage pathways. E. histolytica does not synthesize fatty acids but can synthesize a variety of phospholipids. E. histolytica imports galactose and glucose for fermentation in the anoxic colon and may also catabolize amino acids to produce ATP (9). E. histolytica possesses multiple amylases which can degrade starch (9). There is strong evidence for lateral gene transfer (LGT) from prokaryotes in the E. histolytica genome including genes for several glycosidases for the catabolism of fructose and galactose (77). Prokaryotic genes for carbohydrate transport and metabolism may allow E. histolytica to exploit glycan degradation by its microbial co-inhabitants in the colon. E. histolytica relies on host, dietary and bacterial for essential nutrients including amino acids, nucleic acids, carbohydrates, lipids and vitamins. Axenic culture of E. histolytica required adapting phagocytic trophozoites to uptake of nutrients by pinocytosis in culture. Interestingly E. dispar has been more difficult to establish in axenic culture (26).
Pathogenic mechanisms of E. histolytica
Adherence
Surface molecules control adherence, signaling, ingestion and immune modulation at the host-parasite interface. Many amebic surface proteins have functional diversity in these processes including the heterotrimeric Gal/GalNAc lectin. The lectin is composed of heavy (HGL), light (LGL) and intermediate (IGL) subunits. The HGL contains the carbohydrate recognition domain (CRD), which strongly binds Gal and GalNAc residues on mucus and host cells (Figure 1). Inhibition of HGL via genetic silencing, neutralizing antibodies and excess ligands (including Gal and mucins) blocked adherence and killing of host cells in vitro (100). Additionally, anti-CRD-IgA was protective against re-infection in humans (57). The HGL subunit also contains an intracellular domain with homology to β-integrins and may have a functional role in signaling after CRD engagement. These observations in combination with the finding that secreted products or amebic lysates were not sufficient for cytolysis (85, 86) lead to the model of contact-dependent amebic cytotoxicity (Figure 2).
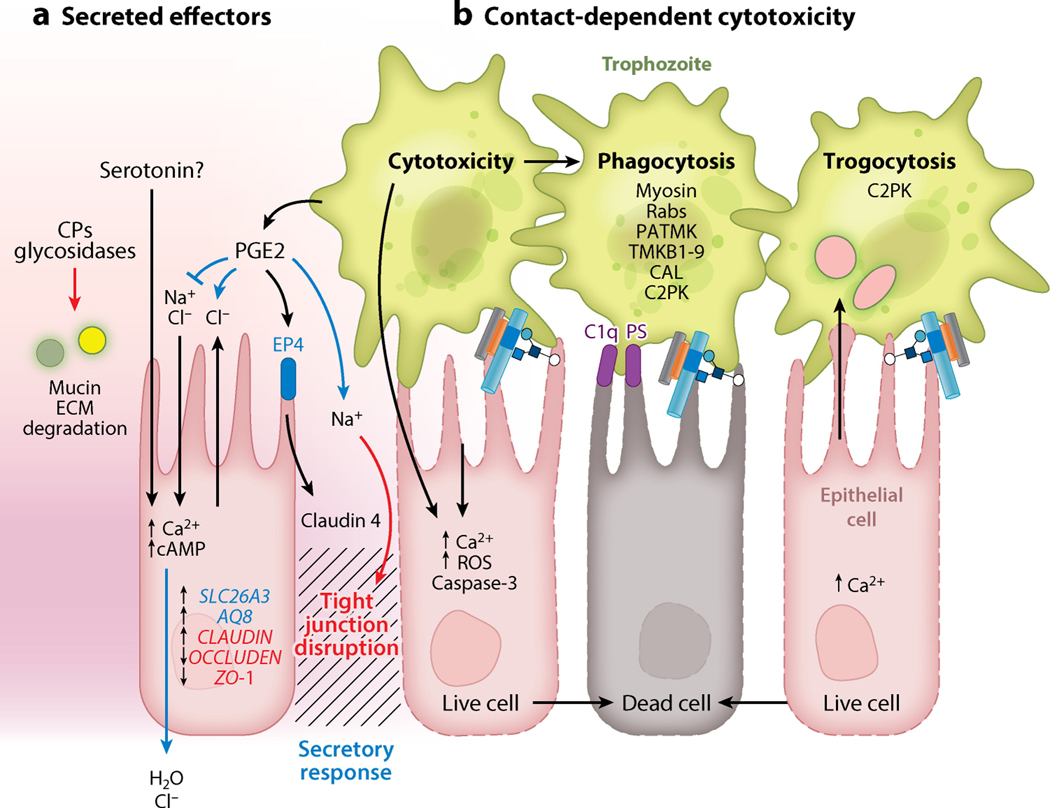
Figure 2.
Pathology at the intestinal epithelium. (a) Physiological mediators of E. histolytica diarrhea. Secreted amoebic effectors have contact-independent physiological effects on intestinal epithelial cells. (1) Proteases and glycosidases degrade mucus and extracellular matrix (ECM) proteins. (2) Amoebic PG2 disrupts barrier function by binding to host EP4, leading to altered expression and localization of tight junction proteins including zona occludens proteins and claudins. (3) PG2 also increased secretion and disrupted ion gradients, leading to decreased cellular Na2+ absorption and increased Na2+ and Cl− secretion at the apical surface. Amoebic serotonin is present in amoebic lysates, but it is not known if it is secreted. Serotonin elevates intracellular Ca2+ and cAMP, leading to increased H2O and Cl− secretion at the serosal surface. Disruption of barrier function further disrupts ion gradients at the intestinal epithelium, and these effects are likely the physiological mediators of amoebic diarrhea (secretory response, blue; tight junction disruption, red). (b) In vitro trophozoites must adhere to target cells to induce death. Contact-dependent killing can be mediated by amoebic activation of host caspase-3 through an undefined mechanism and a rapid apoptotic-like death, preceded by elevated intracellular Ca2+ and reactive oxygen species. E. histolytica phagocytosis is initiated by exposed C1q and phosphatidylserine (PS) on apoptotic cells, which are bound by amoebic calreticulin (CAL) and C2K, respectively. Amoebic kinases PATMK, TMKB1–9, and TMK39 are also involved in phagocytosis. Phagosome formation requires vesicular trafficking and cytoskeletal rearrangement controlled by G-proteins and amoebic myosin. Trogocytosis is a distinct contact-dependent mechanism of amoebic cytotoxicity. In trogocytosis, trophozoites actively ingest pieces of living cells, resulting in membrane disruption and rapid target cell death. Trogocytosis also requires amoebic C2PK and leads to increased intracellular Ca2+ prior to cell death; however, in trogocytosis E. histolytica does not ingest cells after killing. Other abbreviations: CP, cysteine protease; PGE2, prostaglandin 2; ROS, reactive oxygen species.
Contact-dependent cytotoxicity
Upon adherence E. histolytica has multiple cytotoxic effects including increased intracellular Ca2+, reactive oxygen species (ROS) production, loss of membrane integrity, DNA fragmentation, phosphatidylserine (PS) exposure and caspase-3 activation (Figure 2) (reviewed in 26). Early experiments established a link between amebic cytotoxicity and amebic phagocytosis (95, 111).
Subsequent work established a model of sequential adherence, contact-dependent cytotoxicity and phagocytosis. E. histolytica preferentially ingests apoptotic cells in vitro via recognition of PS and collectins-though the molecular interactions were not defined until recently. Amebic calreticulin (CAL) was found to be the surface receptor for host C1q. Calreticulin was required for phagocytosis of apoptotic cells but did not mediate killing (133). The amebic kinase C2PK was recently found to bind PS and was required for formation of the phagocytic cup by recruiting amebic actin (122). Several amebic transmembrane kinases are also important for phagocytosis of apoptotic cells and may have additional roles in cytotoxicity. TMKB1–9 and TMK96 (PATMK) mediated adherence and phagocytosis of apoptotic cells in vitro and were required in murine colitis but not ALA (2, 15).
Trogocytosis
The paradigm of sequential adherence, cytotoxicity and ingestion was overturned by the recent discovery that trophozoites mainly ingest pieces of intact living cell in a process named trogocytosis. Trogocytosis is an active process that resembles phagocytosis in some ways. Trogocytosis required adherence to the target cell, as well as C2PK. Trogocytosis also caused a rapid rise in intracellular Ca2+ and resulted in cell death but dead cells were not ingested (106). These similarities suggest that the relationship between amebic adherence, cytotoxicity and ingestion is much more dynamic than previously appreciated (Figure 2). Ingested erythrocytes are commonly observed in biopsies of amebiasis (104), thus contact-dependent trogocytosis and phagocytosis may both lead to tissue lysis at the intestinal epithelium.
Contact independent effects
Secreted amebic products can have contact independent effects on tight junction integrity and ion absorption. Amebic prostaglandin 2 (PGE2) bound IEC EP4 receptors and altered Claudin-4 localization diminishing tight junction integrity and increasing luminal Cl- secretion (72). E. histolytica lysates contain a serotonin analog which inhibited cellular Na+ and Cl- absorption while stimulating Cl- secretion in colonic tissue (85, 86). It is not known if E. histolytica serotonin is secreted. Accordingly intestinal expression of the Cl−/HCO3− exchanger SLC26A3, and the AQP8 aquaporin, were significantly upregulated during acute E. histolytica colitis in humans (98). (Figure 2).
Other mechanisms of damage appear to be contact-independent mediated by trophozoite dephosphorylated and degraded host tight junction zona occludens (ZO) (73), claudins and occluding (69, 72).
Proteases and hydrolases
Ingested material is degraded in the amebic phagolysosome. The bactericidal and digestive functions of the phagolysosome is mediated by proteases, lysozymes, glycosidases, cytolytic amoebapores and phospholipases (94, 118). Some degradative enzymes have been found to have dual functions in nutrient digestion and virulence. Notably, E. histolytica was recently found to have 6 surface-associated glycolytic enzymes (13). Surface glycolytic enzymes may degrade extracellular carbohydrates to forms that can be imported by the parasite. Surface glycosidases in other pathogenic parasites degrade host proteins and have roles in nutrient extraction, invasion and immune evasion (reviewed in 50). In addition, E. histolytica supernatants had glycosidase activity including β-N-acetyl-d-glucosaminidase, α-d-glucosidase β-d-galactosidase, β-l-fucosidase, and α-N-acetyl-d-galactosaminidase (88).
In addition to glycosidase activity, E. histolytica possesses an armamentarium of proteases. The cysteine proteases (CPs) have been of particular interest due to their in vitro activity against a variety of host molecules. Whether these activities are relevant at in vivo may depend on concentration, localization and pH in the colonic environment and remains to be determined. Inhibition of certain CPs either chemically or by gene silencing blocked E. histolytica adherence, cytotoxicity, and motility in vitro and in vivo assays (118). These experiments have particularly implicated CP5 in virulence. CP5 degraded mucin in vitro but CP-5 silenced parasites were still able to degrade mucin on colonic explants but were unable to invade the epithelium (11). The absence of a CP5 homolog in E. dispar further supports a role in invasion. E. dispar also lacks a CP1 homolog, the absence of CP1 and CP5 may partially account for the dramatically lower overall protease activity relative to HM1:IMSS of E. dispar (140).
E. histolytica cysteine protease-binding proteins (CPBFs) were recently identified as regulators of protease activity and trafficking to phagosomes (40, 41, 92). CPBF1 bound the virulence-associated CP5 and was required for its activity (92). CPBF6 bound and trafficked α-amylase and γ-amylase to the amebic phagosomes (92). CPBF8 was required for localization of β-hexosaminidase and lysozyme to phagosomes. In addition silencing of CPBF8 reduced cytotoxicity (78). The emerging dual roles of CPBFs in digestion in the phagosome and potentially virulence may impart functional flexibility of degradative enzymes depending on the available nutrient sources,
The recent characterization of the surface metalloprotease MSP-1 and the rhomboid intramembrane protease ROM1 has highlighted the importance of proteolysis in regulating adherence, phagocytosis and motility. Silencing of MSP1 increased adherence to live and apoptotic cells but reduced motility, phagocytosis and cytotoxicity (126). Interestingly, ROM1 was only required for adherence to live cells-but was necessary for phagocytosis of live and apoptotic cells suggesting ROM1 has independent roles in these processes. ROM1 co-localized with the Gal/NAc lectin was reported to control shedding of host antibodies and complement (12). In both these studies however the phenotypes were apparently independent of alterations in Gal/GalNAc surface expression (57). The substrates of both MSP-1 and ROM1 are unknown. Potential candidates might be the serine, threonine, isoleucine-rich proteins (STIRPs) which were required for adherence and cytotoxicity in E. histolytica and are notably absent in E. dispar (81). Proteolytic mechanisms to rapidly regulate adherence may allow parasites to rapidly shift from an adherent phenotype during colonization to a motile invasive form in response to changing conditions. The activity, expression, disease phenotype and localization of amebic glycosidases and proteases and their chaperones are summarized in Table 1.
Table 1:
Activity, expression and localization of proteases
| Name | Substrate | Virulence phenotypes and expression | Localization | Ref |
|---|---|---|---|---|
| CP1 | Collagen pro-IL-1β IL-1β(active) villin C3C3b (active) |
Upregulated in murine colitis, ALA, HM1:IMSS v Rahman. Upregulated by mucin. KD did not prevent monolayer destruction. Absent in Ed. | Surface Phagosome | (30, 48, 118, 145) |
| CP2 | Proteoglycan | Upregulated by mucin. Overexpression increased monolayer destruction but KD did not prevent monolayer destruction. | Membrane-associated Phagosome | (30, 118) |
| CP3 | Nutrients in phagosome | Upregulated in Rahman v. HM1:IMSS. present in Ed | Cytoplasmic Phagosome |
(94, 127, 140) |
| CP4 | C3 IgA Lamanin Pro-IL-18 (degrades) |
Upregulated in murine colitis and experimental ALA. KD blocked ALA, Chemical inhibition blocked murine colitis upregulated in HM1:IMSS v Rahman. Induced by cell contact, mucin. | Secreted Nuclear Phagosome | (30, 48, 118) |
| CP5 | IgG Mucin BSA Integrin binding |
KD prevents lamina propria invasion in colonic explants and monolayer destruction. Overexpression increased monolayer destruction but KD did not prevent monolayer destruction. Upregulated in HM1:IMSS and ALA isolates. Upregulated by mucin. Absent in Ed | Surface Phagosome |
(30, 48, 118) |
| CPADH (CP112 +ADH112) |
Collagen Fibronectin, Hemoglobin Integrin binding |
Antibodies block adherence, phagocytosis, monolayer destruction and ALA. | Cytoplasmic vesicles Plasma- membrane Secreted |
(118) |
| CPBF1 | Binds CP5 | Required for CP5 activity | ER Phagosomes |
(92) |
| CPBF6 | Binds α-amylase and γ-amylase | Transports α-amylase and γ-amylase to phagosome | Lysosomes Phagosomes |
(41) |
| CPBF8 | Binds β-hexosaminidase and lysozymes | KD decreased digestion of bacteria in phagosomes and cytotoxicity | Lysosomes Phagosomes |
(40) |
| MSP-1 | Metalloprotease Homology to leishmanolysin |
KD increased adherence / reduced motility, phagocytosis and cytotoxicity. Upregulated in HM1:IMSS v. Rahman. Absent in Ed | Surface | (126, 127) |
| ROM1 | Intramembrane protease, binds HGL | KD reduced adherence/cytotoxicity |
Surface Cap Vesicles |
(12) |
| β-amylase | Starch | Upregulated in HM1:IMSS v. Rahman. Induced by explant contact. KD reduced mucin degradation. | Surface, Cytoplasmic vesicles | (127) |
| Lysozyme | Nutrient degradation | Upregulated in murine colitis. Upregulated in Rahman v. HM1:IMSS. | Surface, Phagosome | (48, 127) |
KD-knockdown, Ed-E. dispar
Virulence regulation at the host-pathogen interface
Many E. histolytica virulence factors are genomically encoded and expressed in avirulent Entamoeba strains and species. Differences in gene content and expression that might mediate virulence have been extensively analyzed (49, 140). Overall, virulent HM1:IMSS has increased expression of adherence genes including LGL1/5, STIRPs, SREHP, and KERP1 (127). LGL3 is an exception with much higher expression in Rahman (140) but the significance of multiple copies of lectin genes is unclear, and complicated by the finding that LGL2 and LGL3 were downregulated during intestinal colonization of mice (48). Cysteine protease genes are differentially regulated in HM1:IMSS and Rahman. Regulation of specific CPs via expression and trafficking by CPBFs may control their degradative abilities. CP5 was more highly expressed in HM1:IMSS relative to Rahman in a recent analysis (127). Previous work reported increased CP4, CP6 and CP1 and decreased CP3, CP7 and CP9 expression in HM1:IMSS relative to Rahman (140). Adaptation to the murine colon increased expression of CP4, CP6, and lysozyme (48). In interpreting these studies it is important to note that HM1:IMSS and Rahman were isolated over 30 years ago from different continents, therefore distinct expression profiles likely also reflect mutations and epigenetic alterations from prolonged axenic in vitro growth that are unrelated to virulence capacity (49).
Metabolic flexibility
Analysis of E. histolytica adaptation from in vitro culture to murine colitis and human colon explants has shown that trophozoites rapidly adapt to massive changes in nutrient availability and oxidative stress (48, 127, 136). This adaptation does not accurately reflect adaptation to the colon during excystation; nonetheless adaptive capacity is critical for survival in the chaotic host environment. During murine colitis trophozoites initially downregulated glycolysis genes and induced lipase genes genes-likely in response to the scarcity of monosaccharaides in the colon relative to culture media (48). Adaptation to the mucosal surface of colonic explants induced genes involved in metabolism of complex carbohydrates and in glycolysis in HM1:ISS while Rahman expressed higher levels of lipases (127). HM1:IMSS rapidly bound and dissolved mucus on colonic explants, while Rahman and E. dispar bound but did not degrade mucus (10, 127). HM1:IMSS induced β-amylase expression during contact with colonic explants and silencing of β-amylase reduced mucin degradation (127). It has been proposed that the ability to catabolize host mucin as a carbon source may mediate invasive capacity (127).
The pathways of amebic mucin degradation have not been elucidated. The secreted glycosidase activity of E. histolytica is thought to mediate mucin degradation by exposing mucin peptides to amebic proteolysis (13, 76, 88). Trophozoites in vitro internalized and released host mucins without proteolytic degradation (23) though transcriptome studies have demonstrated that the regulation of metabolism genes is associated with mucin degradation (127)-thus the ability to sense nutrient sources and induce appropriate pathways may determine metabolic flexibility in the host environment.
Several lines of experimental evidence indicate that the ability to sense and respond to nutrient levels may mediate E. histolytica invasion. E. histolytica motility was highly increased by nutrient depletion and trophozoites were repelled by the byproducts of their own glycolysis-suggesting motility may be a competitive response (144). Increased motility was not observed upon starvation in E. dispar, suggesting this response may be associated with invasion (144). E. histolytica displays chemotactic movement toward serum, fibronectin and TNF-α, potentially directing trophozoites to the intestinal epithelium upon starvation (14). Further, glucose starvation dramatically increased E. histolytica motility, adherence and cytolysis in vitro. Low glucose activated the transcription factor URE3-BP and increased expression of LGL1 (130). URE3-BP has previously been shown to regulate HGL5 and ferredoxin during murine colitis and has been suggested to coordinate motility, adherence and oxidative stress resistance during infection (47, 48).
Stress resistance
G-protein signaling
In addition to surviving nutrient stress, adaptation is key for surviving other stresses in the mutable host environment. G-proteins signaling controls many processes including adherence, protease activity and phagocytosis and may mediate rapid adaptation in E. histolytica (reviewed in 95). E. histolytica encodes 8 putative ligand-activated G-protein-coupled receptors (GPCRs) but only GPCR1 was expressed in vitro (103). GPCR-1 bound LPS in phagocytic cups (21) and was reported to bind RabB in vitro, which regulates amebic phagocytosis (61). E. histolytica also encodes the G-protein subunits Gα1 and Gβγ which interacted with RGS-RhoGEF. Overexpression of Gα1 increased trophozoite motility, adherence and cytotoxicity (17) while overexpression of RGS-RhoGEF had the opposite effect (16). The multiple effects of Gα1 signaling in virulence was attributed to their global regulation of parasite protease activity, but the mechanism remains to be defined (17). The finding that G-protein signaling was upregulated in HM1:IMSS relative to Rahman upon introduction to colonic explants further supports their role in mediating adaptation to host stress (127).
Oxidative stress
Trophozoites must detoxify reactive oxygen and nitrogen species (ROS/NOS) produced by infiltrating immune cells and during invasive disease beyond the anoxic colon. E. histolytica produces high levels of cysteine and numerous enzymes to combat oxidative stress which include: peroxiredoxin (PRX), superoxide dismutase (SOD), flavoprotein A, ferredoxin (FRX), thioredoxin(TRX) and TRX reductase (108, 136). TRX was crucial for buffering sensitive proteins during oxidative assault (117) and the amebicidal activities of metronidazole and auroanofin are mediated by disruption of TRX (29). The ability to survive oxidative stress is also associated with increased virulence. Oxidative stress induced upregulation of a stress-induced adhesion factor (SIAF) and a phospholipid transporting P-type ATPase/flippase (PTPA) which both have roles in adhesion and phagocytosis (108). Oxidative stress also induced metabolic alterations including glycerol and chitin biosynthesis potentially triggering encystation (64). Overall, HM1:IMSS responded more strongly to oxidative stress than either E. dispar or Rahman and surface localization of PRX in HM1:IMSS was associated with virulence (136).
Regulation of E. histolytica virulence by the colonic microbiota
Influence of enteric microbiota on E. histolytica virulence
The enteric microbiota is a nutrient source for E. histolytica and bacteria have long been recognized as a crucial determinant of the pathophysiology of E. histolytica infection (74). The microbiota is generally protective for enteric infection, however E. histolytica virulence seems to require the presence of other enteric organisms. In 1946 it was discovered that ameba caused similar colonic ulcers in symptomatic and asymptomatic individuals but symptomatic individuals had increased ulcer-associated bacteria suggesting that ulcerations in the absence of bacteria were not sufficient to cause disease (36). While provocative this result is likely confounded by infection with non-invasive E. dispar, which could not be distinguished from E. histolytica. Subsequent experiments found that germ-free animals were resistant to E. histolytica infection but the introduction of a single bacterial species restored amebic pathogenesis (101, 102). Axenization decreased parasite virulence (141) and incubation of axenic trophozoites with live bacteria increased virulence depending on the bacterial species (87). Incubation of E histolytica with enteropathogenic E. coli or Shigella increased adherence and cytotoxicity of E. histolytica, but had no effect on E. dispar (42). Conversely, incubation with specific E. coli strains decreased parasite virulence, and these effects have been attributed to regulation of the amebic lectin in response to distinct bacterial surface lipopolysaccharides (87). These observations indicate the enteric microbiota likely regulate E. histolytica virulence during infection though this awaits confirmation in vivo.
E. histolytica perturbs the composition of the enteric microbiota
There is some evidence that E. histolytica significantly alters the principal phyla of the host microbiota during disease. E. histolytica-induced dysbiosis was characterized by significantly less Bacteroides, Clostridia, Lactobacillus, Campylobacter and Eubacterium and significantly increased Bifdobacterium species (135). It will be interesting to determine if dysbiosis results from disruption of intestinal physiology by amebic pathology and peristalsis. E. histolytica could also directly induce dysbiosis through specific predation, lysis, or modulation of the host immune response. In vitro experiments have found that:
E. histolytica displayed preferential ingestion of some bacterial species (19, 87).
Cytolytic amoebapores had differential activity against certain bacterial species and eukaryotic cells in vitro (22).
E. histolytica induced and degraded colonic antimicrobial peptides but is resistant to their activity (27).
It remains to be seen if amebic dysbiosis is similar to dysbiosis induced by other enteric infections, as well as if asymptomatic E. histolytica infection results in comparable dysbiosis.
Regulation of E. histolytica virulence by the microbiota
The composition of the intestinal microbiota is mechanistically linked to the nutritional and immune status of the host. In E. histolytica endemic areas infants are also chronically infected with multiple enteric pathogens, termed the pathobiota (115, 125). Experimentally, pretreatment of intestinal epithelial cells with enteropathogenic bacteria prior to E. histolytica infection increased inflammatory cytokine production, decreased epithelial barrier integrity and resulted in enhanced trophozoite adherence and subsequent cytotoxicity (42, 43). During human infection the enteric pathobiota may also lead to greater inflammation and amebic tissue damage by enhancement of the inflammatory response, decreased barrier function and specific modulation of the amebic lectin. The composition of the enteric microbiota controlled susceptibility to infectious colitis in mice via modulation of intestinal ion channel genes including SLC26A3 (45) and aquaporin activity (128). Both SLC26A3 and AQP8 aquaporin were significantly upregulated during acute E. histolytica colitis in humans (98) further implicating specific microbiota in mediating host susceptibility.
E. histolytica depends on host and microbial nutrients to survive. The microbiota produce glycosidases that degrade complex polysaccharides into forms available for host absorption (96). Microbial glycosidase activity determines levels of free colonic carbohydrates (the glycobiome). The microbiota-dependent glycobiome has an emerging role in regulating the virulence of enteric pathogens (39). The FusKR signaling pathway is a novel fucose-responsive regulator of virulence genes in Enterohemorrhagic Escherichia coli (EHEC) (97). Clostridium difficile disease was mediated by sialic acid levels in vivo, while Salmonella typimurium virulence depended on both fucose and sialic acid in vivo (93). The finding that glucose starvation enhanced E. histolytica virulence, motility and lectin expression by URE-3BP(130), a transcription factor previously linked to virulence capacity (47) suggests similar mechanisms could exist in E. histolytica (Figure 3).
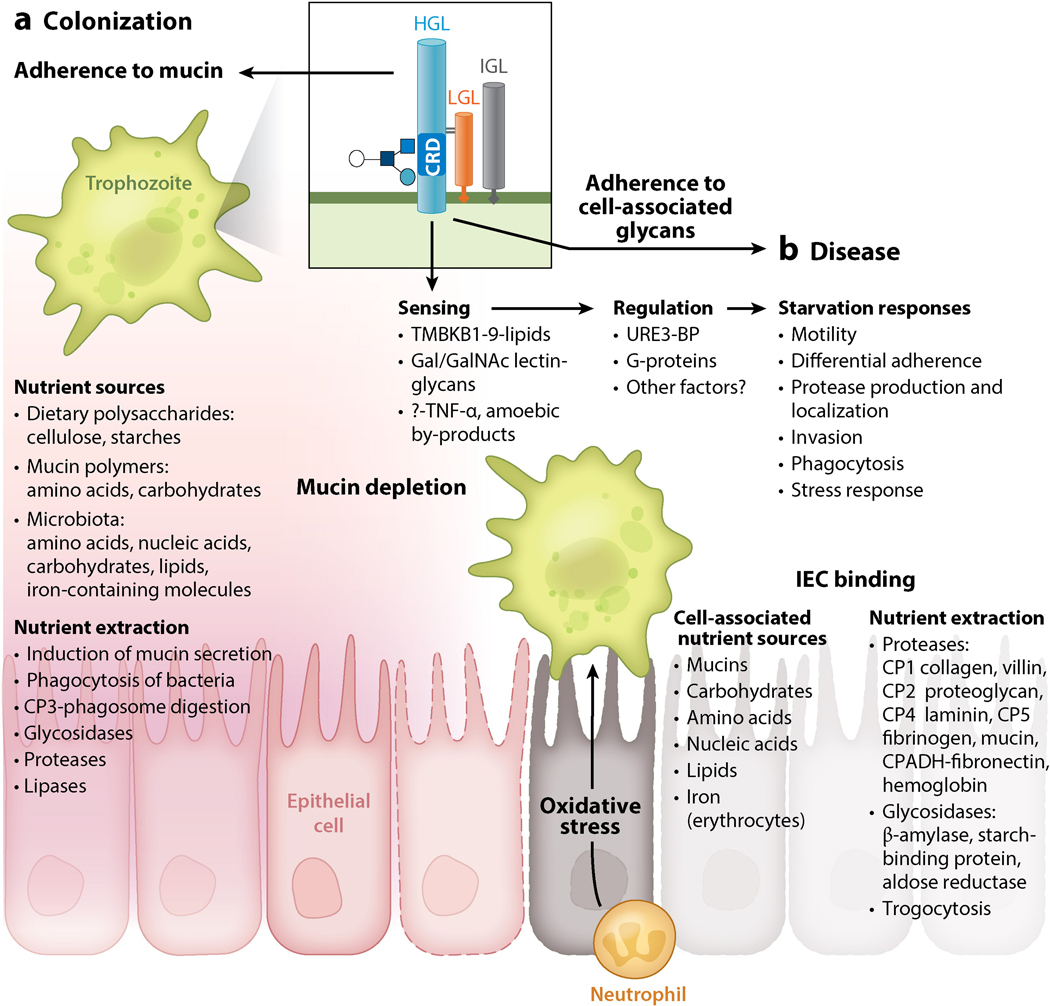
Figure 3.
E. histolytica adaptive ability mediates nutrient extraction and survival in the fluctuating colonic environment. (a) Colonic nutrient sources include dietary polysaccharides, microbiota, and human cellular molecules including mucin. Microbial glycosidases degrade complex polysaccharides into fatty acids absorbed by intestinal epithelial cells (IECs) and sugars for their own metabolism. An intact mucous layer provides plentiful nutrients from mucin and microbiota, inducing a parasite program for colonization. (b) Upon mucus depletion, nutrient starvation induces virulence and activates the transcriptional regulator URE3-BP. Starvation responses include decreased adherence and enhanced motility and oxidative stress resistance. Upon adherence to host cells, virulence factors are induced to enable extraction of cell-associated nutrients. Abbreviations: CP, cysteine protease; CRD, carbohydrate-recognition domain; Gal, galactose; GalNAc, N-acetyl-d-galactosamine; HGL, heavy lectin subunit; IGL, intermediate lectin subunit; LGL, light lectin subunit.
Host factors that influence E. histolytica virulence regulation
Immune response and immune evasion
Cell Mediated
The immune response is a critical mediator of amebic virulence, however E. histolytica infects immunocompetent hosts. Disease seems to be enhanced by the immunodeficiency of malnutrition while defects in T-cell immunity in HIV/AIDS does not seem to mediate increased disease. Another mystery is the particular disposition to ALA in men, despite equivalent susceptibility to infection. The mucin barrier is the first layer of defense blocking trophozoite adherence and cytotoxicity to the intestinal epithelium (24). In the absence of mucin, trophozoites contact the intestinal epithelium (Figure 1). IECs recognized the CRD of the Gal/GalNAc lectin via TLR-2/4 and activated NF-κB leading to the production of inflammatory cytokines including IL-8, IL-6, IL-12, IL-1β, IFN-γ and TNF-α (10, 43). In vivo neutrophils predominated in amebic lesions while macrophages were infrequent (37). In vitro activated neutrophils and macrophages have amebicidal activity however E. histolytica displayed reciprocal killing (44). Clearance of infection was associated with IFN-γ, while IL-4 and TNF-α are correlated with disease (60, 78, 99, 116). IFN-γ production by peripheral mononuclear cells (PMNs) significantly correlated with protection from future E. histolytica disease in children (59). In vaccinated mice protection required IFN-γ-producing CD4+ T-cells and IL-17-producing CD8+ T-cells (51).
The predominance of ALA in men may also be due to IFN-γ. In experimental ALA, protection was mediated by IFN-γ from natural killer T-cells (NKT) while TNF-α producing macrophages increased tissue damage (60, 78). Female mouse NKTs produced more IFN-γ, in a testosterone dependent fashion, mediating ALA protection (79). Experimental ALA introduces trophozoites directly into the liver and does not model upstream immune responses prior to invasive disease in humans. Nonetheless, human and experimental studies have indicated that impaired cell-mediated immunity can worsen host damage by E. histolytica. In vitro evidence indicates trophozoites are capable suppressing cell mediated immunity by: killing immune cells (107) proteolytic cleavage of pro-IL-1 β (causing activation) and IL-18 (causing degradation) (118). In addition, E. histolytica secrets PGE2 which downregulated induced IL-8, decreased macrophage MHC II expression and may inhibit T-cell activation and oxidative capacity (137).
Adaptive Immunity
Adaptive immunity is also protective against E. histolytica as previous infection and vaccination reduced susceptibility to subsequent infections in mice (51, 120). In humans, protection from reinfection is associated with the sIgA response (1, 54, 55, 57). Conversely serum antibodies were associated with increased frequency and severity of amebic disease, though the reason for this is unclear (20,12). In vitro antibodies bound E. histolytica and blocked attachment to host cells and molecules (75). Surface-bound antibodies also activated the complement membrane attack complex (MAC) (20). E. histolytica evaded antibody-mediated defenses by rapidly shedding bound antibodies in a Gal/GalNAc lectin cap (25). The Gal/GalNAc lectin also inhibited MAC formation on trophozoites (20) and E. histolytica CPs degraded the complement factors C3a and C5a (110) and host IgA and IgG (68, 131).
Nutrition
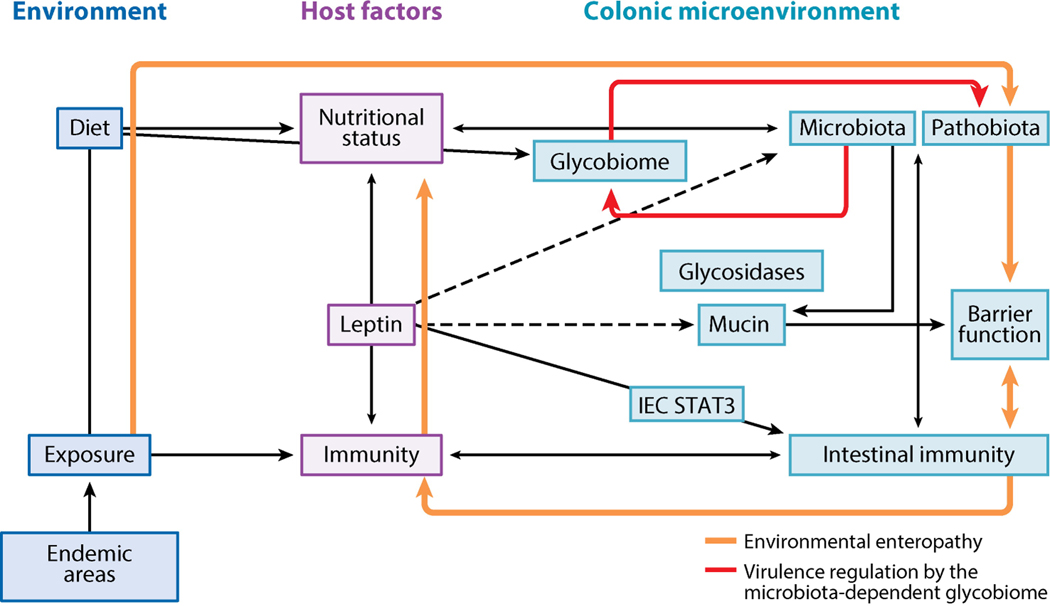
Malnutrition causes immunodeficiency and increased susceptibility to E. histolytica (reviewed in 92). Children with E. histolytica diarrhea are significantly more likely to be malnourished or stunted (91). In addition, malnourished children had three times more E. histolytica-associated diarrheal episodes than children without malnutrition (89). Malnutrition was specifically and significantly associated with E. histolytica compared to all enteric infections (89). The nutritional cytokine leptin is a critical link between nutritional status and immunity (38). Reduced leptin signaling due to a leptin receptor (LEPR) polymorphism (Q223R) is associated with increased susceptibility to E. histolytica diarrhea in children and ALA in adults (33). Leptin-deficient mice (52) and LEPR Q223R (82) mice are more susceptible to E. histolytica infection. Infection of intestinal epithelial LEPR-knockout mice and in vitro studies demonstrated that protection was dependent on leptin activation of STAT3 in intestinal epithelial cells (52, 83). Mice lacking LEPR at the intestinal epithelium had similar body weight, food intake, fecal microbiota and antimicrobial peptide expression (105) indicating that protection is mediated by specific leptin-regulated immune mechanisms which are known to include prevention of apoptosis, increased mucin secretion and enhanced intestinal cell repair and proliferation (35, 113, 124). There is emerging evidence that E. histolytica infection may cause nutritional deficits. E. histolytica infection is associated with intestinal inflammation, mucosal disruption, diminished barrier integrity, ion secretion and dysbiosis, with potentially compounding effects on the nutritional status of the infected host (62). E. histolytica intestinal damage, in particular chronic inflammation and mucus depletion may be triggers for environmental enteropathy further impairing nutrient absorption (70) (Figure 4). Breast-fed infants are at lower risk of E. histolytica infections. It was recently shown that human milk oligosaccharides as well as synthetic galacto-oligosaccharides protected human IECs from amebic cytotoxicity in vitro. Galacto-oligosaccharides are stable, inexpensive and commonly added to infant formula thus these results have implications for nutritional interventions for E. histolytica (66).
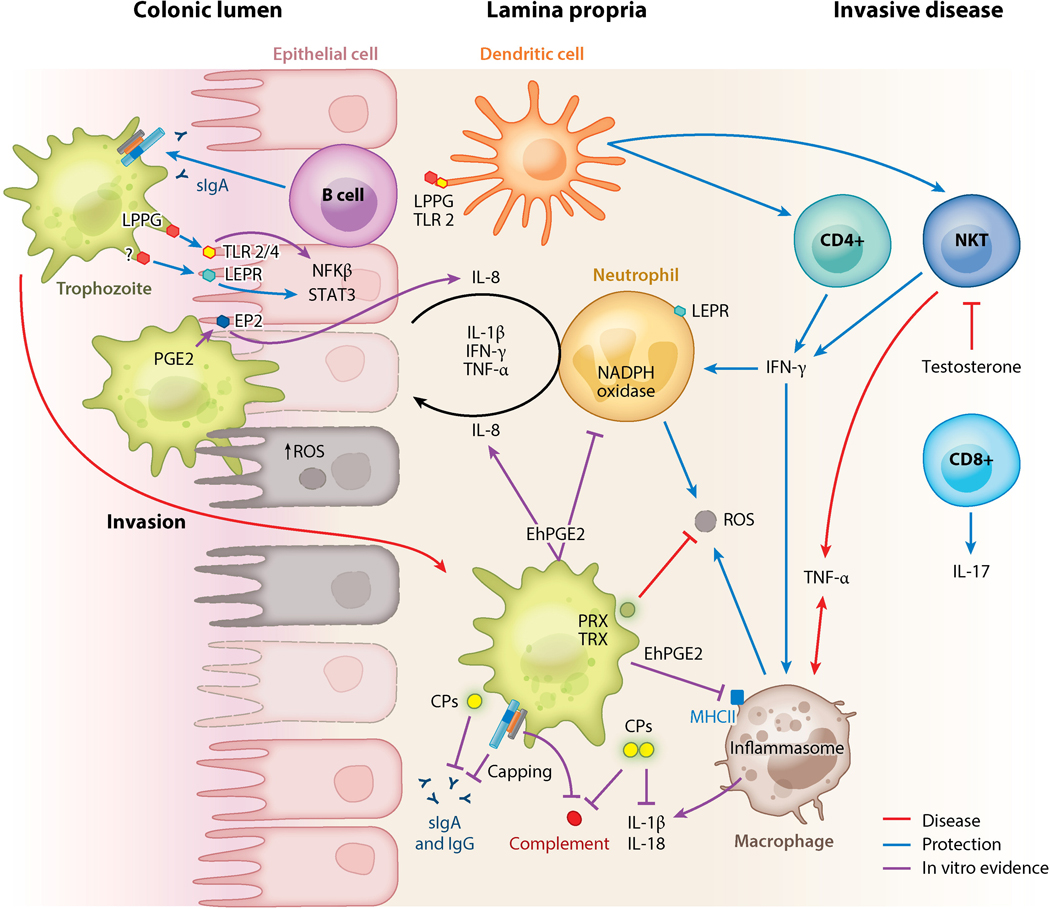
Figure 4.
Immune regulators of E. histolytica virulence. The mucin barrier, mucosal sIgA to the carbohydrate-recognition domain of the galactose (Gal)/N-acetyl-d-galactosamine (GalNAc) lectin, and leptin signaling via a leptin receptor (LEPR) at the epithelium are critical determinates of protection in the colonic lumen. The Gal/GalNAc lectin activates Toll-like receptors (TLRs) on intestinal epithelial cells (IECs), leading to IL-8 secretion and neutrophil recruitment. IEC damage from amoebic cytotoxicity further induces secretion of proinflammatory mediators and disrupts tight junctions, enhancing neutrophil infiltration. Neutrophil reactive oxygen species (ROS) can kill trophozoites; however, trophozoites also kill immune cells. ROS can also exacerbate host tissue damage. Trophozoite peroxiredoxin (PRX) and thioredoxin (TRX) detoxify ROS. Amoebic PG2 can suppress ROS production and impair major histocompatibility complex II (MHCII) expression on macrophages, inhibiting their antigen-presenting ability. Dendritic cells (DCs) in the lamina propria also act as antigen-presenting cells and recognize amoebic LPPG via TLR-2. DCs can activate natural killer and CD+ T cells. In invasive disease IFN-γ-producing natural killer T cells (NKTs) are associated with production. TNF-α production by NKTs and macrophages is associated with increased disease severity. The Gal/GalNAc lectin activates the NLRP3 inflammasome and secretion of IL-1β and IL-18 in macrophages in vitro, though it is not known if this is protective or deleterious. Amoebic proteases cleave complement, IgA, IgG, pro-IL-1β, and IL-18. The Gal/GalNAc lectin inhibits formation of complement membrane attack complex (MAC) and can mediate the capping and shedding of bound antibodies. Processes associated with protection in vivo (animals and/or humans) are indicated with blue arrows; processes associated with disease in vivo are shown in red. Purple lines indicate in vitro evidence. Other abbreviations: CP, cysteine protease; PGE2, prostaglandin 2; sIgA, secretory immunoglobulin A.
Conclusion
E. histolytica virulence depends on a complex interaction of parasite, host and environmental factors, When E. histolytica progresses to virulence the destruction of colonic environment can lead to degradation of protective mucus, disrupted epithelial barrier function, deregulated ion transport, local and systemic inflammation, impaired nutrient absorption, and disruption of the microbiota. The host processes altered by E. histolytica virulence are inherently and reciprocally linked and infection has severe impacts in vulnerable hosts. This capacity is unique to E. histolytica and while many of the mechanisms for virulence are defined the benefits for E. histolytica survival relative to closely related avirulent strains and species are not understood. A salient feature of E. histolytica is the capacity to sense and adapt to diverse host nutrient sources and stresses. The multifunctional Gal/GalNAc lectin is critical for adherence in the host colon and may also mediate downstream signaling upon ligand binding. Engagement of lectin by distinct host glycans may prime parasite gene expression for survival in a particular host niche. Mucin binding may induce a phagocytic, mucus dwelling lifestyle where host mucin and microbiota are the primary nutrient sources. Upon mucin depletion, parasites may sense carbohydrate scarcity and display enhanced motility and directional migration to the epithelium. Engagement of cell-associated glycans may lead to priming of an invasive phenotype that prepares the parasite for oxidative stress and nutrient extraction from host cells. The recent discovery of trogocytosis suggests that E. histolytica does not ingest intact living cells-but ingests cellular pieces leading to cell death, thus cytotoxicity may be collateral damage from nutrient extraction from host IECs. It is known that an appropriate cell mediated immune response can lead to clearance of E. histolytica, while an inappropriate response can increase tissue damage, allowing parasites to invade the lamina propria. Colonization may be specifically regulated to allow parasites to compete in dense colonic microbiota. Thus far, research has focused defining the events that are associated with pathogenesis. Further attention will be required to understand parasite persistence and colonization—which have developed through co-evolution within the human host. The multilayered effects of diet, nutrition, microbiota, immunity and glycobiome may create a colonic environment that induces E. histolytica virulence due to mucus, nutrient and immune depletion. Host nutrition, microbiota and immunity exist in a delicate network that is critical for overall host health. Disruption of these intrinsically linked processes by E. histolytica may have compounding effects for host susceptibility to disease and for parasite virulence regulation.
Figure 5.
The vicious cycle of enteric infection and malnutrition. E. histolytica cysts enter the host via fecally contaminated food and water. In areas where E. histolytica infection is endemic, amoebic infection is pervasive and accompanied by multiple other enteric pathogens. The linked immune and nutritional status of the host determines whether infections will be resolved or established in the host intestine. Leptin levels regulate the immunodeficiency of malnutrition, and reduced leptin increases susceptibility to E. histolytica and other pathogens. Leptin signaling is critical for protection from E. histolytica at the intestinal epithelium. In the absence of immune clearance or treatment, E. histolytica and other enteric pathogens establish chronic infection as part of the pathobiota. The pathobiota causes chronic intestinal inflammation and mucus depletion. Chronic inflammatory responses disrupt the absorptive and barrier functions of the intestine, worsening malnutrition and leading to environmental enteropathy. E. histolytica causes dysbiosis with potential consequences for host nutrition and immunity, as the microbiota mediates intestinal immune homeostasis and nutrient extraction. The microbiota stimulates antimicrobial peptide and mucin production by intestinal epithelial cells (IECs), leading to exclusion of pathogens. Microbial metabolism of dietary and host-derived carbohydrates is essential for host nutrient absorption and for microbial metabolism and leads to competitive exclusion of some enteric pathogens. Microbial metabolism of complex polysaccharides in the colon produces short-chain fatty acids and oligosaccharides, which are critical for host nutrition. E. histolytica encodes sugar transporters and glycosidase genes from prokaryotes, indicating that E. histolytica is capable of exploiting free sugars produced by microbial metabolism. The microbially derived glycobiome is also an emerging regulator of enteric pathogen virulence. Dashed lines are hypothetical.
Summary points: Virulence regulation of E. histolytica by the enteric microbiota
Direct effects
The microbiota and associated metabolome provide essential nutrient sources for E. histolytica that are required for survival in the colon.
The levels of specific microbial products or species regulate virulence of E. histolytica parasites via regulation of virulence factors involved in binding, ingestion and killing of bacterial species.
Indirect effects
Dysbiosis alters host nutritional status and immune function that correlate with susceptibility to E. histolytica including lower IFN-γ production
Pathobiota induce an inflammatory cascade, barrier dysfunction and nutrient malabsorption, which increase susceptibility to E. histolytica infection.
E. histolytica depletion of host mucous eliminates the spatial separation between the microbiota/pathobiota and the intestinal epithelium aggravating inflammation and associated amebic tissue damage during E. histolytica infection.
E. histolytica degradation of host antimicrobial peptides compromises intestinal immune homeostatic interactions
- E. histolytica alteration of the host microbiota. Thee microbiota controls:
- Host nutrient availability and absorption
- Mucins level and integrity
- Leptin production
- Competitive exclusion of pathogens
- Intestinal barrier function and ion transport
Future issues: Which unanswered questions are most important for improving human health?
Parasite factors
-
1
What is the mechanism of carbohydrate sensing in E. histolytica? What are the roles of specific E. histolytica glycosidases in virulence? What is the function of secreted and surface-associated glycosidases?
Host factors
-
2
Do genetic (Q223R), immune (IgG, IFN-γ), specific co-infections, or other biomarkers predict which E. histolytica infected individuals will develop disease? Why are serum antibodies to E. histolytica correlated with disease?
-
3
Why is E. histolytica specifically associated with human malnutrition? Does E. histolytica infection cause malnutrition and environmental enteropathy?
Parasite Environment
-
4
How does E. histolytica impact the composition of the enteric microbiota and regulation of the microbiome? Are there differences between colonization and disease? What are the implications of E. histolytica infection in infants on the maturing microbiota?
-
5
Do specific species of the host microbiota and/or pathobiota alter E. histolytica virulence?
-
6
Do differences in the glycobiome due to dietary intake, mucin level, and microbial glycosidases regulate virulence in E. histolytica?
-
7Which interventions will be most effective to limit E. histolytica mortality and morbidity? Can E. histolytica be eradicated?
- Improved sanitation
- Increased diagnosis and treatment of asymptomatic infection
- Screening for individuals likely to develop invasive disease (IgG?)
- Development and implementation of a vaccine
- Nutritional therapy. Specific carbohydrates?
Sidebar: Disentangling the virulence networks of Entamoeba
Entamoeba must dynamically sense, respond and exploit host and microbiota compounds to acquire nutrients, evade immunity and survive in the host. The integration of existing E. histolytica genomic, transcriptional, biochemical and proteomic datasets into interacting pathways may illuminate novel parasite responses associated with virulence. These pathways can also be compared to datasets from E. dispar and E. histolytica Rahman to highlight global responses that are specific to invasion. As analytical and experimental tools improve it will be possible to incorporate ‘omics’ data from the infecting parasite, infected host and colonic environment (including microbiota, microbiome, pathobiota, pathbiome and colonic metabolome). Response-related networks of parasite and host data will be insightful into the dynamic regulation of E. histolytica virulence in susceptible and resistant hosts. The potential applications of large datasets include: the identification of host susceptibility biomarkers, understanding the relationship of enteric co-infections and the discovery of novel parasite virulence pathways induced in vivo. Integrating and incorporating diverse host/pathogen data could direct targeted host and pathogen therapeutics interventions and permit extrapolative prediction of the functions of hypothetical and unknown Entamoeba genes that may be important in virulence.
Sidebar: Measuring E. histolytica virulence
A brief overview of common methods for assaying virulence phenotypes of E. histolytica.
In vitro
In vitro assays using cultured cells can be useful for measuring virulence traits including adherence, cytotoxicity, protease activity, monolayer destruction, motility, phagocytosis and trogocytosis. It is important to note that some cell types may not reflect the cells encountered by E. histolytica during natural infection. In addition, the linked nature of these phenotypes can make it difficult to ascribe a specific functionality in vitro.
Ex vivo
In human colon explant E. histolytica degrades colonic mucus, migrates along collagen networks, degrades ECM, kills host cells and invades tissue. In addition, colonic explants produced a potent inflammatory response. This technique, in combination with imagining and transcriptome studies has provided key insights into E. histolytica’s invasion and virulence potential of human tissue. However, the explant model is limited to understanding early stages of infection.
In vivo
E. histolytica naturally infects humans and some other primates and the development of animal models that reproduce natural infection has been problematic. A variety of animals have been used including dogs, kittens, primates, rabbits, guinea pigs, hamsters and gerbils. Mouse strains display differential susceptibility to amebic colitis. Natural resistance in animals has been informative to understanding protection in humans and pointed to immune cells (mainly neutrophils), pro-inflammatory cytokine production and mucin content as mediators of innate protection. Experimental studies of invasion beyond the intestine have focused on ALA in susceptible hamsters and gerbils, which develop hepatic lesions. Encystation of E. histolytica in vitro has not been successful, thus both colitis and ALA models rely on direct introduction of trophozoites into the colon or liver. To date an animal model that captures the complete cycle of natural E. histolytica infection does not exist. Reviewed in (132).
Acknowledgements
Work cited from our lab and the preparation of this review was supported by NIH AI-26649. C.M. is the recipient of an NIH iNRSA grant F32AI09304 and The Hartwell Foundation biomedical research fellowship.
Acronyms and Definitions
- Zenic
co-culture with one or more unidentified organisms
- Axenic
a pure culture of single species
- Pathogenic potential
the ability to cause disease in a given environment
- Trogocytosis
contact-dependent amebic ingestion of pieces of living cells (from the Greek, trogo-nibble)
- Microbiota
the collection of microbes colonizing a host
- Microbiome
The collection of genes of the microbiota
- Pathobiota
The collection of pathogens in a host
- Glycobiome
the glycan composition of a host niche
- ALA
Amebic liver abscess
- IEC
intestinal epithelial cell
- Gal/GalNAc
galactose/N-acetyl-D-galactosamine
- PRX
peroredoxin
- TRX
thioredoxin
- ROS
reactive oxygen species
- CAL
amebic calreticulin
- CP
cysteine protease
- PS
phosphatidylserine
Literature cited
- 1.Abd-Alla MD, Jackson TFGH, Rogers T, Reddy S, Ravdin JI. 2006. Mucosal immunity to asymptomatic entamoeba histolytica and entamoeba dispar infection is associated with a peak intestinal anti-lectin immunoglobulin a antibody response. Infect. Immun. 74(7):3897–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abhyankar MM, Shrimal S, Gilchrist CA, Bhattacharya A, Petri WA Jr. 2012. The entamoeba histolytica serum-inducible transmembrane kinase ehtmkb1–9 is involved in intestinal amebiasis. Int. J. Parasitol. Drugs Drug Resist. 2:243–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ackers J, Clark G, Diamond L, Duchene M, Cantellano M, et al. 1997. Who/paho/unesco report of a consultation of experts on amoebiasis
- 4.Ali IKM, Clark CG, Petri WA Jr. 2008. Molecular epidemiology of amebiasis. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 8(5):698–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ali IKM, Haque R, Alam F, Kabir M, Siddique A, Petri WA Jr. 2012. Evidence for a link between locus r-r sequence type and outcome of infection with entamoeba histolytica. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 18(7):E235–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ali IKM, Hossain MB, Roy S, Ayeh-Kumi PF, Petri WA Jr, et al. 2003. Entamoeba moshkovskii infections in children, bangladesh. Emerg. Infect. Dis. 9(5):580–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ali IKM, Mondal U, Roy S, Haque R, Petri WA Jr, Clark CG. 2007. Evidence for a link between parasite genotype and outcome of infection with entamoeba histolytica. J. Clin. Microbiol. 45(2):285–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ali IKM, Solaymani-Mohammadi S, Akhter J, Roy S, Gorrini C, et al. 2008. Tissue invasion by entamoeba histolytica: evidence of genetic selection and/or dna reorganization events in organ tropism. PLoS Negl. Trop. Dis. 2(4):e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson IJ, Loftus BJ. 2005. Entamoeba histolytica: observations on metabolism based on the genome sequence. Exp. Parasitol. 110(3):173–77 [DOI] [PubMed] [Google Scholar]
- 10.Bansal D, Ave P, Kerneis S, Frileux P, Boché O, et al. 2009. An ex-vivo human intestinal model to study entamoeba histolytica pathogenesis. PLoS Negl. Trop. Dis. 3(11):e551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bansal D, Sehgal R, Chawla Y, Malla N, Mahajan RC. 2005. Cytokine mrna expressions in symptomatic vs. asymptomatic amoebiasis patients. Parasite Immunol. 27(1–2):37–43 [DOI] [PubMed] [Google Scholar]
- 12.Baxt LA, Rastew E, Bracha R, Mirelman D, Singh U. 2010. Downregulation of an entamoeba histolytica rhomboid protease reveals roles in regulating parasite adhesion and phagocytosis. Eukaryot. Cell. 9(8):1283–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biller L, Matthiesen J, Kühne V, Lotter H, Handal G, et al. 2014. The cell surface proteome of entamoeba histolytica. Mol. Cell. Proteomics MCP. 13(1):132–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blazquez S, Zimmer C, Guigon G, Olivo-Marin J-C, Guillen N, Labruyere E. 2006. Human tumor necrosis factor is a chemoattractant for the parasite entamoeba histolytica. Infect. Immun. 74(2):1407–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boettner DR, Huston CD, Linford AS, Buss SN, Houpt E, et al. 2008. Entamoeba histolytica phagocytosis of human erythrocytes involves patmk, a member of the transmembrane kinase family. PLoS Pathog. 4(1):e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosch DE, Kimple AJ, Manning AJ, Muller RE, Willard FS, et al. 2013. Structural determinants of rgs-rhogef signaling critical to entamoeba histolytica pathogenesis. Struct. Lond. Engl. 1993. 21(1):65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bosch DE, Kimple AJ, Muller RE, Giguère PM, Machius M, et al. 2012. Heterotrimeric g-protein signaling is critical to pathogenic processes in entamoeba histolytica. PLoS Pathog. 8(11):e1003040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bosch DE, Siderovski DP. 2013. G protein signaling in the parasite entamoeba histolytica. Exp. Mol. Med. 45:e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bracha R, Kobiler D, Mirelman D. 1982. Attachment and ingestion of bacteria by trophozoites of entamoeba histolytica. Infect. Immun. 36(1):396–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braga LL, Ninomiya H, McCoy JJ, Eacker S, Wiedmer T, et al. 1992. Inhibition of the complement membrane attack complex by the galactose-specific adhesion of entamoeba histolytica. J. Clin. Invest. 90(3):1131–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brewer MT, Agbedanu PN, Zamanian M, Day TA, Carlson SA. 2013. Evidence for a bacterial lipopolysaccharide-recognizing g-protein-coupled receptor in the bacterial engulfment by entamoeba histolytica. Eukaryot. Cell. 12(11):1433–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruhn H, Riekens B, Berninghausen O, Leippe M. 2003. Amoebapores and nk-lysin, members of a class of structurally distinct antimicrobial and cytolytic peptides from protozoa and mammals: a comparative functional analysis. Biochem. J. 375(Pt 3):737–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chadee K, Johnson ML, Orozco E, Petri WA Jr, Ravdin JI. 1988. Binding and internalization of rat colonic mucins by the galactose/n-acetyl-d-galactosamine adherence lectin of entamoeba histolytica. J. Infect. Dis. 158(2):398–406 [DOI] [PubMed] [Google Scholar]
- 24.Chadee K, Petri WA Jr, Innes DJ, Ravdin JI. 1987. Rat and human colonic mucins bind to and inhibit adherence lectin of entamoeba histolytica. J. Clin. Invest. 80(5):1245–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chávez-Munguía B, Talamás-Rohana P, Castañón G, Salazar-Villatoro L, Hernández-Ramírez V, Martínez-Palomo A. 2012. Differences in cap formation between invasive entamoeba histolytica and non-invasive entamoeba dispar. Parasitol. Res. 111(1):215–21 [DOI] [PubMed] [Google Scholar]
- 26.Clark CG, Diamond LS. 2002. Methods for cultivation of luminal parasitic protists of clinical importance. Clin. Microbiol. Rev. 15(3):329–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cobo ER, He C, Hirata K, Hwang G, Tran U, et al. 2012. Entamoeba histolytica induces intestinal cathelicidins but is resistant to cathelicidin-mediated killing. Infect. Immun. 80(1):143–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Cádiz AE, Jeelani G, Nakada-Tsukui K, Caler E, Nozaki T. 2013. Transcriptome analysis of encystation in entamoeba invadens. PloS One. 8(9):e74840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Debnath A, Parsonage D, Andrade RM, He C, Cobo ER, et al. 2012. A high-throughput drug screen for entamoeba histolytica identifies a new lead and target. Nat. Med. 18(6):956–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Debnath A, Tashker JS, Sajid M, McKerrow JH. 2007. Transcriptional and secretory responses of entamoeba histolytica to mucins, epithelial cells and bacteria. Int. J. Parasitol. 37(8–9):897–906 [DOI] [PubMed] [Google Scholar]
- 31.Diamond LS, Clark CG. 1993. A redescription of entamoeba histolytica schaudinn, 1903 (emended walker, 1911) separating it from entamoeba dispar brumpt, 1925. J. Eukaryot. Microbiol. 40(3):340–44 [DOI] [PubMed] [Google Scholar]
- 32.Dolabella SS, Serrano-Luna J, Navarro-García F, Cerritos R, Ximénez C, et al. 2012. Amoebic liver abscess production by entamoeba dispar. Ann. Hepatol. 11(1):107–17 [PubMed] [Google Scholar]
- 33.Duggal P, Guo X, Haque R, Peterson KM, Ricklefs S, et al. 2011. A mutation in the leptin receptor is associated with entamoeba histolytica infection in children. J. Clin. Invest. 121(3):1191–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehrenkaufer GM, Weedall GD, Williams D, Lorenzi HA, Caler E, et al. 2013. The genome and transcriptome of the enteric parasite entamoeba invadens, a model for encystation. Genome Biol. 14(7):R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El Homsi M, Ducroc R, Claustre J, Jourdan G, Gertler A, et al. 2007. Leptin modulates the expression of secreted and membrane-associated mucins in colonic epithelial cells by targeting pkc, pi3k, and mapk pathways. Am. J. Physiol. Gastrointest. Liver Physiol. 293(1):G365–373 [DOI] [PubMed] [Google Scholar]
- 36.ELLENBERG M 1946. Amoebiasis; the role of bacteria in symptomatology; sigmoidoscopic findings in symptomatic and asymptomatic cases; the effect of sulfadiazine on symptoms and sigmoidoscopic findings. Am. J. Dig. Dis. 13(11):356–60 [DOI] [PubMed] [Google Scholar]
- 37.Espinosa-Cantellano M, Martinez-Palomo A. 2000. Pathogenesis of intestinal amebiasis: from molecules to disease. Clin. Microbiol. Rev. 13(2):318–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faggioni R, Feingold KR, Grunfeld C. 2001. Leptin regulation of the immune response and the immunodeficiency of malnutrition. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 15(14):2565–71 [DOI] [PubMed] [Google Scholar]
- 39.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. 2012. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 3(4):289–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Furukawa A, Nakada-Tsukui K, Nozaki T. 2012. Novel transmembrane receptor involved in phagosome transport of lysozymes and β-hexosaminidase in the enteric protozoan entamoeba histolytica. PLoS Pathog. 8(2):e1002539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Furukawa A, Nakada-Tsukui K, Nozaki T. 2013. Cysteine protease-binding protein family 6 mediates the trafficking of amylases to phagosomes in the enteric protozoan entamoeba histolytica. Infect. Immun. 81(5):1820–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galván-Moroyoqui JM, Del Carmen Domínguez-Robles M, Franco E, Meza I. 2008. The interplay between entamoeba and enteropathogenic bacteria modulates epithelial cell damage. PLoS Negl. Trop. Dis. 2(7):e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galván-Moroyoqui JM, Del Carmen Domínguez-Robles M, Meza I. 2011. Pathogenic bacteria prime the induction of toll-like receptor signalling in human colonic cells by the gal/galnac lectin carbohydrate recognition domain of entamoeba histolytica. Int. J. Parasitol. 41(10):1101–12 [DOI] [PubMed] [Google Scholar]
- 44.Ghadirian E, Denis M. 1992. In vivo activation of macrophages by ifn-gamma to kill entamoeba histolytica trophozoites in vitro. Parasite Immunol. 14(4):397–404 [DOI] [PubMed] [Google Scholar]
- 45.Ghosh S, Dai C, Brown K, Rajendiran E, Makarenko S, et al. 2011. Colonic microbiota alters host susceptibility to infectious colitis by modulating inflammation, redox status, and ion transporter gene expression. Am. J. Physiol. Gastrointest. Liver Physiol. 301(1):G39–49 [DOI] [PubMed] [Google Scholar]
- 46.Gilchrist CA, Ali IKM, Kabir M, Alam F, Scherbakova S, et al. 2012. A multilocus sequence typing system (mlst) reveals a high level of diversity and a genetic component to entamoeba histolytica virulence. BMC Microbiol. 12:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilchrist CA, Baba DJ, Zhang Y, Crasta O, Evans C, et al. 2008. Targets of the entamoeba histolytica transcription factor ure3-bp. PLoS Negl Trop Dis. 2:e282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilchrist CA, Houpt E, Trapaidze N, Fei Z, Crasta O, et al. 2006. Impact of intestinal colonization and invasion on the entamoeba histolytica transcriptome. Mol. Biochem. Parasitol. 147(2):163–76 [DOI] [PubMed] [Google Scholar]
- 49.Gilchrist CA, Petri WA Jr. 2009. Using differential gene expression to study entamoeba histolytica pathogenesis. Trends Parasitol. 25(3):124–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gómez-Arreaza A, Acosta H, Quiñones W, Concepción JL, Michels PAM, Avilán L. 2014. Extracellular functions of glycolytic enzymes of parasites: unpredicted use of ancient proteins. Mol. Biochem. Parasitol. 193(2):75–81 [DOI] [PubMed] [Google Scholar]
- 51.Guo X, Barroso L, Becker SM, Lyerly DM, Vedvick TS, et al. 2009. Protection against intestinal amebiasis by a recombinant vaccine is transferable by t cells and mediated by gamma interferon. Infect. Immun. 77(9):3909–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo X, Roberts MR, Becker SM, Podd B, Zhang Y, et al. 2011. Leptin signaling in intestinal epithelium mediates resistance to enteric infection by entamoeba histolytica. Mucosal Immunol. 4(3):294–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guzmán-Silva MA, Santos HLC, Peralta RS, Peralta JM, de Macedo HW. 2013. Experimental amoebic liver abscess in hamsters caused by trophozoites of a brazilian strain of entamoeba dispar. Exp. Parasitol. 134(1):39–47 [DOI] [PubMed] [Google Scholar]
- 54.Haque R, Ali IM, Sack RB, Farr BM, Ramakrishnan G, Petri WA. 2001. Amebiasis and mucosal iga antibody against the entamoeba histolytica adherence lectin in bangladeshi children. J. Infect. Dis. 183(12):1787–93 [DOI] [PubMed] [Google Scholar]
- 55.Haque R, Duggal P, Ali IM, Hossain MB, Mondal D, et al. 2002. Innate and acquired resistance to amebiasis in bangladeshi children. J. Infect. Dis. 186(4):547–52 [DOI] [PubMed] [Google Scholar]
- 56.Haque R, Huston CD, Hughes M, Houpt E, Petri WA Jr. 2003. Amebiasis. N. Engl. J. Med. 348(16):1565–73 [DOI] [PubMed] [Google Scholar]
- 57.Haque R, Mondal D, Duggal P, Kabir M, Roy S, et al. 2006. Entamoeba histolytica infection in children and protection from subsequent amebiasis. Infect. Immun. 74(2):904–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haque R, Mondal D, Karim A, Molla IH, Rahim A, et al. 2009. Prospective case-control study of the association between common enteric protozoal parasites and diarrhea in bangladesh. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 48(9):1191–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haque R, Mondal D, Shu J, Roy S, Kabir M, et al. 2007. Correlation of interferon-gamma production by peripheral blood mononuclear cells with childhood malnutrition and susceptibility to amebiasis. Am. J. Trop. Med. Hyg. 76(2):340–44 [PubMed] [Google Scholar]
- 60.Helk E, Bernin H, Ernst T, Ittrich H, Jacobs T, et al. 2013. Tnfα-mediated liver destruction by kupffer cells and ly6chi monocytes during entamoeba histolytica infection. PLoS Pathog. 9(1):e1003096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hernandes-Alejandro M, Calixto-Gálvez M, López-Reyes I, Salas-Casas A, Cázares-Ápatiga J, et al. 2013. The small gtpase ehrabb of entamoeba histolytica is differentially expressed during phagocytosis. Parasitol. Res. 112(4):1631–40 [DOI] [PubMed] [Google Scholar]
- 62.Hoque KM, Chakraborty S, Sheikh IA, Woodward OM. 2012. New advances in the pathophysiology of intestinal ion transport and barrier function in diarrhea and the impact on therapy. Expert Rev. Anti Infect. Ther. 10(6):687–99 [DOI] [PubMed] [Google Scholar]
- 63.Hung C-C, Chang S-Y, Ji D-D. 2012. Entamoeba histolytica infection in men who have sex with men. Lancet Infect. Dis. 12(9):729–36 [DOI] [PubMed] [Google Scholar]
- 64.Husain A, Sato D, Jeelani G, Soga T, Nozaki T. 2012. Dramatic increase in glycerol biosynthesis upon oxidative stress in the anaerobic protozoan parasite entamoeba histolytica. PLoS Negl. Trop. Dis. 6(9):e1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jaiswal V, Ghoshal U, Mittal B, Dhole TN, Ghoshal UC. 2014. Association between allelic variation due to short tandem repeats in trna gene of entamoeba histolytica and clinical phenotypes of amoebiasis. Acta Trop. [DOI] [PubMed]
- 66.Jantscher-Krenn E, Lauwaet T, Bliss LA, Reed SL, Gillin FD, Bode L. 2012. Human milk oligosaccharides reduce entamoeba histolytica attachment and cytotoxicity in vitro. Br. J. Nutr. 108(10):1839–46 [DOI] [PubMed] [Google Scholar]
- 67.Jarnagin WR. 2012. Blumgart’s Surgery of the Liver, Pancreas and Biliary Tract: Expert Consult - Online. Elsevier Health Sciences. 8086 pp. [Google Scholar]
- 68.Kelsall BL, Ravdin JI. 1993. Degradation of human iga by entamoeba histolytica. J. Infect. Dis. 168(5):1319–22 [DOI] [PubMed] [Google Scholar]
- 69.Kissoon-Singh V, Moreau F, Trusevych E, Chadee K. 2013. Entamoeba histolytica exacerbates epithelial tight junction permeability and proinflammatory responses in muc2(−/−) mice. Am. J. Pathol. [DOI] [PubMed]
- 70.Korpe PS, Petri WA. 2012. Environmental enteropathy: critical implications of a poorly understood condition. Trends Mol. Med. 18(6):328–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, et al. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the global enteric multicenter study, gems): a prospective, case-control study. Lancet. 382(9888):209–22 [DOI] [PubMed] [Google Scholar]
- 72.Lejeune M, Moreau F, Chadee K. 2011. Prostaglandin e2 produced by entamoeba histolytica signals via ep4 receptor and alters claudin-4 to increase ion permeability of tight junctions. Am. J. Pathol. 179(2):807–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leroy A, Lauwaet T, De Bruyne G, Cornelissen M, Mareel M. 2000. Entamoeba histolytica disturbs the tight junction complex in human enteric t84 cell layers. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 14(9):1139–46 [DOI] [PubMed] [Google Scholar]
- 74.Lesh FA. 1975. Massive development of amebas in the large intestine. Am. J. Trop. Med. Hyg. 24(3):383–92 [DOI] [PubMed] [Google Scholar]
- 75.Leyva O, Rico G, Ramos F, Morán P, Melendro EI, Ximénez C. 1992. Inhibition of adhesion process mediated by anti-entamoeba histolytica specific monoclonal iga antibodies. Arch. Med. Res. 23(2):227–29 [PubMed] [Google Scholar]
- 76.Lidell ME, Moncada DM, Chadee K, Hansson GC. 2006. Entamoeba histolytica cysteine proteases cleave the muc2 mucin in its c-terminal domain and dissolve the protective colonic mucus gel. Proc. Natl. Acad. Sci. 103(24):9298–9303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Loftus B, Anderson I, Davies R, Alsmark UCM, Samuelson J, et al. 2005. The genome of the protist parasite entamoeba histolytica. Nature. 433(7028):865–68 [DOI] [PubMed] [Google Scholar]
- 78.Lotter H, González-Roldán N, Lindner B, Winau F, Isibasi A, et al. 2009. Natural killer t cells activated by a lipopeptidophosphoglycan from entamoeba histolytica are critically important to control amebic liver abscess. PLoS Pathog. 5(5):e1000434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lotter H, Helk E, Bernin H, Jacobs T, Prehn C, et al. 2013. Testosterone increases susceptibility to amebic liver abscess in mice and mediates inhibition of ifnγ secretion in natural killer t cells. PloS One. 8(2):e55694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.MacFarlane RC, Singh U. 2006. Identification of differentially expressed genes in virulent and nonvirulent entamoeba species: potential implications for amebic pathogenesis. Infect. Immun. 74(1):340–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.MacFarlane RC, Singh U. 2007. Identification of an entamoeba histolytica serine-, threonine-, and isoleucine-rich protein with roles in adhesion and cytotoxicity. Eukaryot. Cell. 6(11):2139–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mackey-Lawrence NM, Guo X, Sturdevant DE, Virtaneva K, Hernandez MM, et al. 2013. Effect of the leptin receptor q223r polymorphism on the host transcriptome following infection with entamoeba histolytica. Infect. Immun. 81(5):1460–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marie CS, Verkerke HP, Paul SN, Mackey AJ, Petri WA Jr. 2012. Leptin protects host cells from entamoeba histolytica cytotoxicity by a stat3-dependent mechanism. Infect. Immun. 80(5):1934–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McAuley JB, Herwaldt BL, Stokes SL, Becher JA, Roberts JM, et al. 1992. Diloxanide furoate for treating asymptomatic entamoeba histolytica cyst passers: 14 years’ experience in the united states. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 15(3):464–68 [DOI] [PubMed] [Google Scholar]
- 85.McGowan K, Kane A, Asarkof N, Wicks J, Guerina V, et al. 1983. Entamoeba histolytica causes intestinal secretion: role of serotonin. Science. 221(4612):762–64 [DOI] [PubMed] [Google Scholar]
- 86.McGowan K, Piver G, Stoff JS, Donowitz M. 1990. Role of prostaglandins and calcium in the effects of entamoeba histolytica on colonic electrolyte transport. Gastroenterology. 98(4):873–80 [DOI] [PubMed] [Google Scholar]
- 87.Mirelman D, Feingold C, Wexler A, Bracha R. 1983. Interactions between entamoeba histolytica, bacteria and intestinal cells. Ciba Found. Symp. 99:2–30 [DOI] [PubMed] [Google Scholar]
- 88.Moncada D, Keller K, Chadee K. 2005. Entamoeba histolytica-secreted products degrade colonic mucin oligosaccharides. Infect. Immun. 73(6):3790–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mondal D, Haque R, Sack RB, Kirkpatrick BD, Petri WA. 2009. Attribution of malnutrition to cause-specific diarrheal illness: evidence from a prospective study of preschool children in mirpur, dhaka, bangladesh. Am. J. Trop. Med. Hyg. 80(5):824–26 [PMC free article] [PubMed] [Google Scholar]
- 90.Mondal D, Minak J, Alam M, Liu Y, Dai J, et al. 2012. Contribution of enteric infection, altered intestinal barrier function, and maternal malnutrition to infant malnutrition in bangladesh. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 54(2):185–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mondal D, Petri WA Jr, Sack RB, Kirkpatrick BD, Haque R. 2006. Entamoeba histolytica-associated diarrheal illness is negatively associated with the growth of preschool children: evidence from a prospective study. Trans. R. Soc. Trop. Med. Hyg. 100(11):1032–38 [DOI] [PubMed] [Google Scholar]
- 92.Nakada-Tsukui K, Tsuboi K, Furukawa A, Yamada Y, Nozaki T. 2012. A novel class of cysteine protease receptors that mediate lysosomal transport. Cell. Microbiol. 14(8):1299–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, et al. 2013. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 502(7469):96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Okada M, Huston CD, Oue M, Mann BJ, Petri WA Jr, et al. 2006. Kinetics and strain variation of phagosome proteins of entamoeba histolytica by proteomic analysis. Mol. Biochem. Parasitol. 145(2):171–83 [DOI] [PubMed] [Google Scholar]
- 95.Orozco E, Guarneros G, Martinez-Palomo A, Sánchez T. 1983. Entamoeba histolytica. phagocytosis as a virulence factor. J. Exp. Med. 158(5):1511–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ouwerkerk JP, de Vos WM, Belzer C. 2013. Glycobiome: bacteria and mucus at the epithelial interface. Best Pract. Res. Clin. Gastroenterol. 27(1):25–38 [DOI] [PubMed] [Google Scholar]
- 97.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, et al. 2012. Fucose sensing regulates bacterial intestinal colonization. Nature. 492(7427):113–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Peterson KM, Guo X, Elkahloun AG, Mondal D, Bardhan PK, et al. 2011. The expression of reg 1a and reg 1b is increased during acute amebic colitis. Parasitol. Int. 60(3):296–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Peterson KM, Shu J, Duggal P, Haque R, Mondal D, Petri WA Jr. 2010. Association between tnf-alpha and entamoeba histolytica diarrhea. Am. J. Trop. Med. Hyg. 82(4):620–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Petri WA Jr, Haque R, Mann BJ. 2002. The bittersweet interface of parasite and host: lectin-carbohydrate interactions during human invasion by the parasite entamoeba histolytica. Annu. Rev. Microbiol. 56:39–64 [DOI] [PubMed] [Google Scholar]
- 101.Phillips BP, Gorstein F. 1966. Effects of different species of bacteria on the pathology of enteric amebiasis in monocontaminated guinea pigs. Am. J. Trop. Med. Hyg. 15(6):863–68 [DOI] [PubMed] [Google Scholar]
- 102.PHILLIPS BP, WOLFE PA, REES CW, GORDON HA, WRIGHT WH, REYNIERS JA. 1955. Studies on the ameba-bacteria relationship in amebiasis; comparative results of the intracecal inoculation of germfree, monocontaminated, and conventional guinea pigs with entamoeba histolytica. Am. J. Trop. Med. Hyg. 4(4):675–92 [PubMed] [Google Scholar]
- 103.Picazarri K, Luna-Arias JP, Carrillo E, Orozco E, Rodriguez MA. 2005. Entamoeba histolytica: identification of ehgpcr-1, a novel putative g protein-coupled receptor that binds to ehrabb. Exp. Parasitol. 110(3):253–58 [DOI] [PubMed] [Google Scholar]
- 104.Prathap K, Gilman R. 1970. The histopathology of acute intestinal amebiasis. a rectal biopsy study. Am. J. Pathol. 60(2):229–46 [PMC free article] [PubMed] [Google Scholar]
- 105.Rajala MW, Patterson CM, Opp JS, Foltin SK, Young VB, Myers MG. 2014. Leptin acts independently of food intake to modulate gut microbial composition in male mice. Endocrinology. 155(3):748–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ralston KS, Michael Solga, Mackey-Lawrence Somlata, Bhattacharya A, Petri WA Jr. Trogocytosis-like ingestion by entamoeba histolytica contributes to human cell killing and tissue invasion. Nature. In Press: [DOI] [PMC free article] [PubMed]
- 107.Ralston KS, Petri WA Jr. 2011. Tissue destruction and invasion by entamoeba histolytica. Trends Parasitol. [DOI] [PMC free article] [PubMed]
- 108.Rastew E, Vicente JB, Singh U. 2012. Oxidative stress resistance genes contribute to the pathogenic potential of the anaerobic protozoan parasite, entamoeba histolytica. Int. J. Parasitol. 42(11):1007–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Redaèlli M, Mahmoohdzad J, Lang R, Schencking M. 2013. Globalised world, globalised diseases: a case report on an amoebiasis-associated colon perforation. World J. Clin. Cases. 1(2):79–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Reed SL, Ember JA, Herdman DS, DiScipio RG, Hugli TE, Gigli I. 1995. The extracellular neutral cysteine proteinase of entamoeba histolytica degrades anaphylatoxins c3a and c5a. J. Immunol. Baltim. Md 1950. 155(1):266–74 [PubMed] [Google Scholar]
- 111.Rodríguez MA, Orozco E. 1986. Isolation and characterization of phagocytosis- and virulence-deficient mutants of entamoeba histolytica. J. Infect. Dis. 154(1):27–32 [DOI] [PubMed] [Google Scholar]
- 112.Ross AGP, Olds GR, Cripps AW, Farrar JJ, McManus DP. 2013. Enteropathogens and chronic illness in returning travelers. N. Engl. J. Med. 368(19):1817–25 [DOI] [PubMed] [Google Scholar]
- 113.Rouet-Benzineb P, Aparicio T, Guilmeau S, Pouzet C, Descatoire V, et al. 2004. Leptin counteracts sodium butyrate-induced apoptosis in human colon cancer ht-29 cells via nf-kappab signaling. J. Biol. Chem. 279(16):16495–502 [DOI] [PubMed] [Google Scholar]
- 114.Royer TL, Gilchrist C, Kabir M, Arju T, Ralston KS, et al. 2012. entamoeba bangladeshi nov. sp., bangladesh. Emerg. Infect. Dis. 18(9):1543–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ryan ET. 2013. The intestinal pathobiome: its reality and consequences among infants and young children in resource-limited settings. J. Infect. Dis, p. jit509 [DOI] [PubMed]
- 116.Sánchez-Guillén MDC, Pérez-Fuentes R, Salgado-Rosas H, Ruiz-Argüelles A, Ackers J, et al. 2002. Differentiation of entamoeba histolytica/entamoeba dispar by pcr and their correlation with humoral and cellular immunity in individuals with clinical variants of amoebiasis. Am. J. Trop. Med. Hyg. 66(6):731–37 [DOI] [PubMed] [Google Scholar]
- 117.Schlosser S, Leitsch D, Duchêne M. 2013. Entamoeba histolytica: identification of thioredoxin-targeted proteins and analysis of serine acetyltransferase-1 as a prototype example. Biochem. J. 451(2):277–88 [DOI] [PubMed] [Google Scholar]
- 118.Serrano-Luna J, Piña-Vázquez C, Reyes-López M, Ortiz-Estrada G, de la Garza M. 2013. Proteases from entamoeba spp. and pathogenic free-living amoebae as virulence factors. J. Trop. Med. 2013:890603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shimokawa C, Culleton R, Imai T, Suzue K, Hirai M, et al. 2013. Species-specific immunity induced by infection with entamoeba histolytica and entamoeba moshkovskii in mice. PloS One. 8(11):e82025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shimokawa C, Kabir M, Taniuchi M, Mondal D, Kobayashi S, et al. 2012. Entamoeba moshkovskii is associated with diarrhea in infants and causes diarrhea and colitis in mice. J. Infect. Dis. 206(5):744–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Singh N, Bhattacharya A, Bhattacharya S. 2013. Homologous recombination occurs in entamoeba and is enhanced during growth stress and stage conversion. PLoS ONE. 8(9):e74465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Somlata Bhattacharya S, Bhattacharya A 2011. A c2 domain protein kinase initiates phagocytosis in the protozoan parasite entamoeba histolytica. Nat. Commun. 2:230. [DOI] [PubMed] [Google Scholar]
- 123.Stanley SL Jr. 2003. Amoebiasis. Lancet. 361(9362):1025–34 [DOI] [PubMed] [Google Scholar]
- 124.Sukhotnik I, Vadasz Z, Coran AG, Lurie M, Shiloni E, et al. 2006. Effect of leptin on intestinal re-growth following massive small bowel resection in rat. Pediatr. Surg. Int. 22(1):9–15 [DOI] [PubMed] [Google Scholar]
- 125.Taniuchi M, Sobuz SU, Begum S, Platts-Mills JA, Liu J, et al. 2013. Etiology of diarrhea in bangladeshi infants in the first year of life analyzed using molecular methods. J. Infect. Dis. 208(11):1794–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Teixeira JE, Sateriale A, Bessoff KE, Huston CD. 2012. Control of entamoeba histolytica adherence involves metallosurface protease 1, an m8 family surface metalloprotease with homology to leishmanolysin. Infect. Immun. 80(6):2165–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Thibeaux R, Weber C, Hon C-C, Dillies M-A, Avé P, et al. 2013. Identification of the virulence landscape essential for entamoeba histolytica invasion of the human colon. PLoS Pathog. 9(12):e1003824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tomas J, Wrzosek L, Bouznad N, Bouet S, Mayeur C, et al. 2013. Primocolonization is associated with colonic epithelial maturation during conventionalization. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 27(2):645–55 [DOI] [PubMed] [Google Scholar]
- 129.Tovy A, Ankri S. 2010. Epigenetics in the unicellular parasite entamoeba histolytica. Future Microbiol. 5(12):1875–84 [DOI] [PubMed] [Google Scholar]
- 130.Tovy A, Hertz R, Siman-Tov R, Syan S, Faust DM, Guillén N. 2011. Glucose starvation boosts entamoeba histolytica virulence. PLoS Negl Trop Dis. 5:e1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tran VQ, Herdman DS, Torian BE, Reed SL. 1998. The neutral cysteine proteinase of entamoeba histolytica degrades igg and prevents its binding. J. Infect. Dis. 177(2):508–11 [DOI] [PubMed] [Google Scholar]
- 132.Tsutsumi V, Shibayama M. 2006. Experimental amebiasis: a selected review of some in vivo models. Arch. Med. Res. 37(2):210–20 [DOI] [PubMed] [Google Scholar]
- 133.Vaithilingam A, Teixeira JE, Miller PJ, Heron BT, Huston CD. 2012. Entamoeba histolytica cell surface calreticulin binds human c1q and functions in amebic phagocytosis of host cells. Infect. Immun. 80(6):2008–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Verkerke HP, Petri WA Jr, Marie CS. 2012. The dynamic interdependence of amebiasis, innate immunity, and undernutrition. Semin. Immunopathol. 34(6):771–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Verma AK, Verma R, Ahuja V, Paul J. 2012. Real-time analysis of gut flora in entamoeba histolytica infected patients of northern india. BMC Microbiol. 12:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vicente JB, Ehrenkaufer GM, Saraiva LM, Teixeira M, Singh U. 2009. Entamoeba histolytica modulates a complex repertoire of novel genes in response to oxidative and nitrosative stresses: implications for amebic pathogenesis. Cell. Microbiol. 11(1):51–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang W, Chadee K. 1995. Entamoeba histolytica suppresses gamma interferon-induced macrophage class ii major histocompatibility complex ia molecule and i-a beta mrna expression by a prostaglandin e2-dependent mechanism. Infect. Immun. 63(3):1089–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Watanabe K, Aoki T, Nagata N, Tanuma J, Kikuchi Y, et al. 2013. Clinical significance of high anti-entamoeba histolytica antibody titer in asymptomatic hiv-1-infected individuals. J. Infect. Dis. [DOI] [PubMed]
- 139.Weedall GD, Clark CG, Koldkjaer P, Kay S, Bruchhaus I, et al. 2012. Genomic diversity of the human intestinal parasite entamoeba histolytica. Genome Biol. 13(5):R38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wilson IW, Weedall GD, Hall N. 2012. Host-parasite interactions in entamoeba histolytica and entamoeba dispar: what have we learned from their genomes? Parasite Immunol. 34(2–3):90–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wittner M, Rosenbaum RM. 1970. Role of bacteria in modifying virulence of entamoeba histolytica. studies of amebae from axenic cultures. Am. J. Trop. Med. Hyg. 19(5):755–61 [DOI] [PubMed] [Google Scholar]
- 142.Ximénez C, Cerritos R, Rojas L, Dolabella S, Morán P, et al. 2010. Human amebiasis: breaking the paradigm? Int. J. Environ. Res. Public. Health. 7(3):1105–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ximénez C, Morán P, Rojas L, Valadez A, Gómez A. 2009. Reassessment of the epidemiology of amebiasis: state of the art. Infect. Genet. Evol. 9(6):1023–32 [DOI] [PubMed] [Google Scholar]
- 144.Zaki M, Andrew N, Insall RH. 2006. Entamoeba histolytica cell movement: a central role for self-generated chemokines and chemorepellents. Proc. Natl. Acad. Sci. U. S. A. 103(49):18751–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang Z, Wang L, Seydel KB, Li E, Ankri S, et al. 2000. Entamoeba histolytica cysteine proteinases with interleukin-1 beta converting enzyme (ice) activity cause intestinal inflammation and tissue damage in amoebiasis. Mol. Microbiol. 37(3):542–48 [DOI] [PubMed] [Google Scholar]