Abstract
Cytometric immunophenotyping is a powerful tool to discover and implement T-cell biomarkers of type 1 diabetes (T1D) progression and response to clinical therapy. Although many discovery-based T-cell biomarkers have been described, to date, no such markers have been widely adopted in standard practice. The heterogeneous nature of T1D and lack of standardized assays and experimental design across studies is a major barrier to the broader adoption of T-cell immunophenotyping assays. There is an unmet need to harmonize the design of immunophenotyping assays, including those that measure antigen-agnostic cell populations, such that data collected from different clinical trial sites and T1D cohorts are comparable, yet account for cohort-specific features and different drug mechanisms of action. In these Guidelines, we aim to provide expert advice on how to unify aspects of study design and practice. We provide recommendations for defining cohorts, method implementation, as well as tools for data analysis and reporting by highlighting and building on selected successes. Harmonization of cytometry-based T-cell assays will allow researchers to better integrate findings across trials, ultimately enabling the identification and validation of biomarkers of disease progression and treatment response in T1D.
Keywords: type 1 diabetes, flow cytometry, immune monitoring, biomarkers, T cells
Introduction
Immunophenotyping individuals with type 1 diabetes (T1D) is critical for understanding mechanistic links between immune cells and disease state, and for monitoring immune modulation in the growing number of interventional studies. T1D is a complex autoimmune disease with multiple disease endotypes related to stages of the disease process and variable ages of disease onset [1, 2]. Thus, the careful design of experiments with thoughtful sampling is critical for meaningful data collection and interpretation. In addition, technologies for immunophenotyping are rapidly advancing, and there is a need for data harmonization to effectively compare immunophenotyping data sets in multicenter trials and from trials testing different therapeutic agents. Here, we discuss factors and challenges related to study design, sample collection, and analysis that should be considered when conducting immunophenotyping in T1D for the purpose of robust biomarker measurement and discovery.
T-cell responses to self-antigens are widely thought to be pathogenic, conferring an autoimmune attack on the insulin producing β-cells in pancreatic islets, during the natural history of T1D [3-8]. Methods for effective enumeration and characterization of islet-specific T cells are addressed in detail elsewhere [9, 10]. While these measures constitute an important feature of tissue and beta-cell specificity, these measures still require specialized labs and reagents. Thus, we have chosen to discuss the measurement of antigen-agnostic T-cell features for broad adoption across multiple labs and countries. Many such features, including increased frequency of IL-17+CD4+ T cells [11-13], increased frequency of follicular helper T cells [14, 15], reduced function of Tregs [16, 17], “exhausted” CD8+ T cells [18-20], and functional CD4+ T-effector resistance to suppression [21-23], reviewed in [24] have been shown to distinguish T1D from healthy controls. However, many other studies linking T-cell phenotypes to disease state have not been replicated in independent cohorts. To validate these candidate biomarkers, standardized protocols must be developed to enable the future translation of results in the clinic.
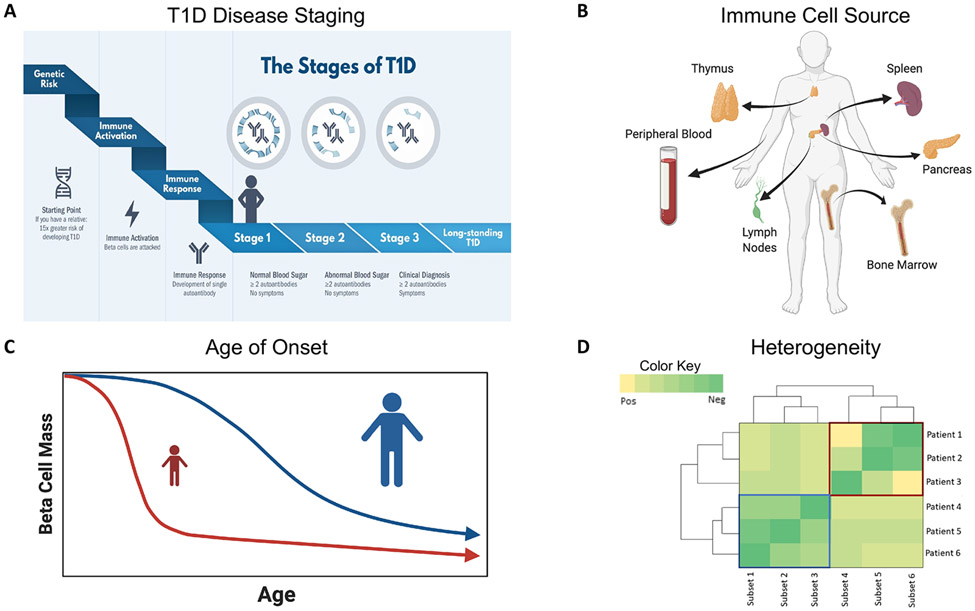
Biological heterogeneity and the small cohort sizes of most studies present a persistent challenge to linking immune phenotypes with T1D progression and treatment response. When utilizing samples collected in clinical trials, design of experimental and analytical approaches must incorporate key aspects of the autoimmune process including stage of disease progression, heterogeneity between individuals, environmental factors, age of diagnosis, and the source of immune cell sampling as depicted in Fig. 1. T1D is a disease that progresses in stages (Fig. 1A) demarcated by the appearance of multiple autoantibodies (Stage 1), abnormal blood sugar (Stage 2), and clinical diagnosis (Stage 3) [2]. It is increasingly evident that some immune profiles are disease stage specific, whereas others may mark activation or rate of progression regardless of the stage of disease. These include not only lineage-defining, costimulatory, inhibitory, and cytokine signaling markers related to general immune function, but also islet antigen specificity. Sampling and analysis of the pancreas and proximal tissues is rarely feasible, making the immune cell source a key consideration for data interpretation (Fig. 1B). Age at T1D onset, in particular, is a known driver of both clinical and immune heterogeneity (Fig. 1C). Furthermore, immune profiles can be profoundly influenced by genetics (e.g., HLA and other susceptibility loci) and environmental factors (e.g., viral infection), which can also drive individual heterogeneity (Fig. 1D). Collectively, these characteristics contribute to heterogeneity in disease phenotypes, such that observed differences in immune phenotypes can be driven by both disease-dependent and disease-independent factors. Strategies for dealing with this heterogeneity include holding defined parameters constant (e.g., age matching) and/or utilizing multivariate modeling approaches that include age at sampling, age at diagnosis, and T1D disease stage (Fig. 1) to separate out sources of variability [25, 26].
Figure 1. Considerations for assay and trial design in T1D.
When designing immunophenotyping assays, there are necessary considerations that depend on (A) the stages of T1D [those at-risk for T1D, those in the three progressive stages of T1D development, and those with long-standing T1D; https://www.trialnet.org/t1d-facts], (B) the sampling source, with peripheral blood being the most sampled in large trials, although other tissue sources may be sampled in other studies, (C) the impact of age of T1D onset on disease progression (with onset at young age associated with more rapid β-cell decline), and (D) individual heterogeneity influencing immunophenotypes. Red and blue boxes denote two sets of patients grouped together by subsets of an expression of a phenotype. Created with BioRender.com.
Use of biomarkers can broadly be divided into two categories: immune monitoring and discovery. The majority of these Guidelines focus on strategies to improve robust immune monitoring and validation of biomarkers of disease state, progression, and response to therapy. The importance of continued discovery in this rapidly evolving field is obvious, given the increasing number of new therapies being tested. Therefore, we also address considerations for biomarker discovery. Validating biomarkers in large complex T1D populations, with the goal of assay reproducibility across cohorts and trials, will require coordinated efforts. These may include collecting shared biological sample sets, establishing the technical and biological variability of each assay, and coordinating efforts to unify standard operating procedures and analytical approaches. Subsequently, the research community must then commit to data sharing between laboratories within the T1D research community. Through consortium efforts, the T1D community has already collaborated to define discrete disease stages preceding clinical diagnosis, marked by autoantibody production and metabolic changes (Fig. 1) [1, 2, 27, 28]. Collective efforts have also resulted in several workshops to advance the utility and multicenter reproducibility of immunoassays [29-37], and have driven groundbreaking studies to define the phenotype and function of T cells in the pancreas and LNs which can inform studies in the blood (Fig. 1) [7, 38-44).
The next step is to link these efforts, enabling consistent use of immunocytometry assays for use in clinical trials to measure and predict disease progression and/or response to therapy. In combination with biostatistical and unbiased discovery platforms, cytometric phenotyping makes biomarker discovery more tenable and serves as a feasible way to integrate biological mechanism with clinical response and disease progression. Here, we present practical considerations and examples for how to plan cytometry-based immune-cell profiling studies with recommendations on sample collection and appropriate controls, cytometry platform options, assay validation, data analysis tools, and subsequent interpretation including examples from the field of T1D. We then conclude with some additional considerations for biomarker discovery. The recommendations are in the context of T1D, but many are broadly applicable to any immune-mediated disease.
General approaches for cross-site and cross-study sample standardization
Clinical trials are now commonly conducted at more than one center to bolster the rate and feasibility of subject recruitment. Standardization of sample processing can be achieved in multicenter trials through the use of standard operating procedures and systematic training of each site by the coordinating center. Assay validation is a prerequisite for utilization of cytometry in the context of a clinical trial. This process assesses the precision, robustness, and stability of each technique, and allows the sensitivity of each assay to be defined, to ensure assessment of true biological differences, and not technical variability. Many well-defined guidelines for validation of cell-based assays have been published (https://members.aoac.org/AOAC_Docs/lptp/alacc_guide_2008.pdf, https://www.ich.org/page/quality-guidelines, and Refs. [45-47]). However, recommendations on how to standardize these validated assays across different sites and clinical trials are less common [48-50]. This is especially true in the context of unique considerations for T1D research, as noted in Fig. 1. Although many of these considerations are discussed in detail in the aforementioned guidelines, we address those that remain challenging to standardize in T1D clinical trial design including specifics on sample collection and handling as well as appropriate controls and detailed documentation.
Sample collection, storage, and transportation
It is important to keep the type of anticoagulant in blood collection tubes consistent throughout trials and between sites, since the choice of anticoagulant tubes for blood collection can influence cell state and marker stability in downstream assays. For example, EDTA tubes are routinely used for general immunophenotyping, but as EDTA binds to divalent cations, they are not ideal for lymphocyte activation and cytokine measurements; sodium heparin tubes are better suited for functional assays. Accordingly, two ongoing, collaborative international T1D clinical trials [UST1D2 and USTEKID] to assess safety and efficacy of ustekinumab, a monoclonal antibody that targets the IL-12/IL-23 pathway in newly diagnosed subjects (https://clinicaltrials.gov/ct2/show/NCT03941132 and http://www.isrctn.com/ISRCTN14274380), utilize both EDTA and sodium heparin blood collection tubes. In these trials, EDTA tubes are used to measure general changes in frequencies and phenotypes of cell types of interest (e.g., subsets of B, T, and NK cells in peripheral blood), whereas sodium heparin tubes are being used to assess changes in cytokine production (e.g., IFN-γ and IL-17) and frequency of proinsulin-specific T cells using an activation-induced marker assay [51].
The temperature of sample storage and transportation can affect downstream assays and needs to be controlled to improve standardization. For example, transport of whole blood below ambient temperature (18–22°C) has been observed to compromise the PBMC function and yield, with an increase in granulocyte contamination (Supporting information Fig. S1A). While this granulocyte contamination can be eliminated with commercial kits (Supporting information Fig. S1B), phenotype and function of T cells, including the production of various T-cell cytokines upon short PMA and ionomycin stimulation, can still be affected when storage and transportation temperatures are below ambient temperature (Supporting information Fig. S1C). Indeed, a study by Westendorf et al. [52] showed a significant number of genes with altered expression after 2 h at ambient temperature as compared to 4°C. When cryopreservation is needed, the use of reagents, such as leucoSep (Greiner) or SepMate (STEMCELL Technologies) tubes and commercially available freezing medium, such as CryoStor, can simplify and harmonize cryopreservation at each center. Furthermore, shipment of cryopreserved samples to a single laboratory for processing can aid in standardization by decreasing variability in sample quality and viability.
It is well known that there are vast differences between the frequencies of many immune populations from different tissue sources [44] and that various factors, such as circadian rhythm [53], cryopreservation methods [54], and delays in processing time [33], can alter immune cell populations. It is also known that blood glucose levels may impact immune cell subsets and should be considered when taking samples from subjects with T1D [55]. Therefore, it is very important to keep these factors as consistent as possible throughout trials.
Appropriate controls and documentation
The inclusion of positive, negative, and quality controls (QC) is essential for standardizing assays across studies and sites. Using identical QC samples, such as cryopreserved PBMC aliquots from a single blood draw, across batches of experiments, and/or across sites, not only allows for the normalization of data across days or sites, but can also help control for batch effects, as further discussed in the “Analysis/reporting” section, and aid in the identification of batches of data that are affected by technical errors in procedure or reagent used.
It is important to define as many crucial parameters as possible before a trial and ensure that these are standardized throughout the trial. However, it is not always possible to control all variables that may affect experimental readouts; thus, it is essential to document them at every step of sample collection and processing so they can be controlled during data analysis. Recommendations of parameters that would be useful to be documented include (but are not limited to) anticoagulant used, time of blood collection and sample processing, minimum and maximum transport and storage temperature of samples, name of operator/analyst and machine used, reagent lot number, and comments to document instances where procedure deviates from protocol. Such documentation will not ensure a precise and accurate outcome, but simply reflect variabilities inherent in protocol. However, a robustness study can be performed after standardization of critical parameters to ensure that the assay behaves similarly within the allowed variation limits. Examples of this include storage temperature testing and time in transit, described in the sample collection, storage and transportation section.
Defining the current state of fluorescence flow cytometry assay standardization
When biomarkers are defined for immune phenotyping, consistent and robust measurement can be improved with cytometry standardization [56]. Although state-of-the-art standardization for all aspects of a clinical trial is ideal, this is unlikely to be achieved in reality. Here, we have suggested three levels of rigor for fluorescence cytometric assay standardization (fundamental, desirable, and ideal) and applied these three levels to four important categories to consider with their specific challenges when designing cytometric experiments: (A) equipment, (B) antibody panel design, (C) antibodies/staining reagents, and (D) analysis/reporting (Table 1). With the exception of those related to spectral spillover, these strategies are also relevant for mass cytometry and oligonucleotide-based antibody barcoding facilitated biomarker discovery. We follow this with a case study that demonstrates how standardization for immune monitoring can result in successful harmonization of assays in T1D clinical trials.
Table 1.
Staged approach to standardization of flow cytometric assays
| Three stan dardization stages |
Four flow cytometry standardization categories |
|||
|---|---|---|---|---|
| Equipment | Antibody panels | Antibodies/staining reagents | Analysis/reporting | |
| Fundamental | Different instruments Performance of instrument calibration using CS&T and application settings (BD), FSP (Beckman Coulter), or SpectroFlo (Cytek Aurora) beads Challenge: requires thorough data comparisons across different types of instruments |
Different antibody panel at each site, albeit same marker(s) to define cell types. Check full panel stain on samples relevant to the trial (e.g., subjects with T1Ds) Challenge: different clones can lead to differences in MFI and percentage of positive cells detected |
Perform antibody titrations and optimization of other key reagents. Ensure regents are used within expiry dates Challenge: time- and resource-consuming to check titration of different antibody lots |
Similar gating strategy and the use of appropriate controls. Report all deviations from protocol that may impact analysis Challenge: a rigid analysis may preclude unexpected results |
| Desirable | Similar instruments, despite different configurations (e.g., BD LSRFortessa 3 vs. 4 lasers) Back-up instruments at each site Challenges: requires thorough data comparisons across different instruments; some sites will limit and prioritize markers due to lack of number of parameters |
Similar panels across sites that use same markers and clones albeit a slight difference in marker/fluorochrome combination Challenge: different fluorochromes usually affect marker staining sensitivity; must be taken into account |
Marker evaluation across platforms to ensure that populations of interest can be detected by cytometers at each clinical trial site. Challenge: time- and resource-consuming |
Predefined gating strategy and data acceptance criteria along with normalization to controls Challenge: a rigid analysis may preclude unexpected results |
| Ideal | Identical instruments with the same configuration Levy-Jennings plots for cross-validating QC across sites & over time Challenge: same instrument can differ in daily performance and sensitivity; some comparisons of data is desirable |
Identical panels with consideration to minimize fluorochrome spillover and spread Challenge: there may be lot-to-lot variability between identical antibodies. Consider purchasing lots in bulk |
Use of lyophilized antibody tubes to reduce or eliminate lot-to-lot variation and pipetting error Perform lot-to-lot crossovers when using wet antibodies Challenge: when several panels are used for a study, lot-to-lot crossover testing can become tedious, expensive, and not feasible |
Use of automatic gating and independent analyses of same samples/data by two different sites. Internal controls to assess technical versus biological variation Challenge: a rigid analysis may preclude unexpected results |
| Case study | Desirable Site B: 5-laser BD LSRFortessa Site C: 3-laser BD LSRFortessa Ideal improvement: use of same instrument with identical configuration to allow for the use of identical flow panels |
Desirable Similar five flow panels using same antibody clones Ideal improvement: identical instrument used with identical flow panels |
Desirable Wet antibody titrated with documentation of reagent lot numbers Ideal improvement: validate the use of antibody master mixes and/or identify where possible the use of lyophilized antibody tubes to reduce pipetting |
Ideal Identical gating strategy, data analyzed by site each and all data reviewed by coordinating site Operator and time between sample collection and processing were documented to assess technical variation |
The application of three stages of standardization (fundamental, desirable, and ideal) to four categories (equipment, antibody panel design, antibodies/staining reagents, and analysis/reporting) when designing flow cytometric assays in a clinical trial and when applied to a case study with challenges noted.
Equipment
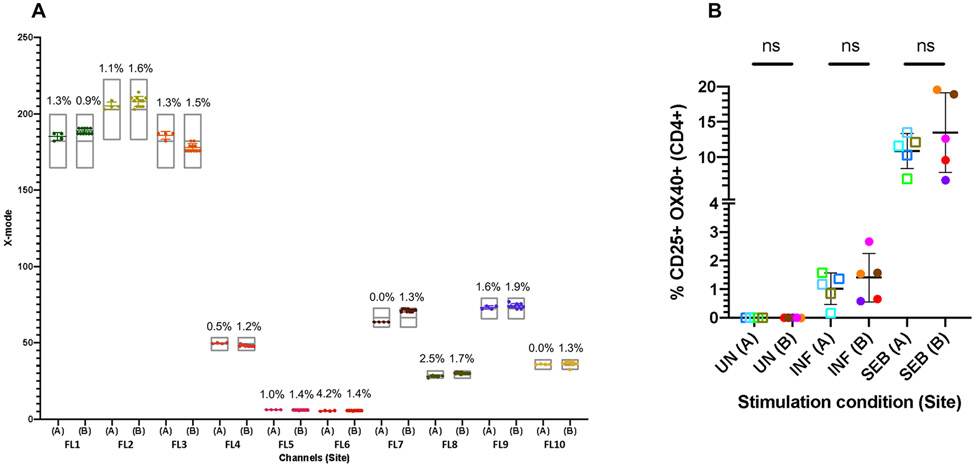
Cytometers resolve populations of interest differently due to instrument-specific filter configuration and instrument noise and sensitivity, even when acquiring the same samples. For this reason, it is ideal to use cytometers with identical configurations across all study sites. Furthermore, an identically configured back-up instrument on-site can prevent poor quality or missing data due to technical issues in primary cytometer performance. Cytometers should also be harmonized using target regions with fluorescence control beads, for example, Flow-Set Pro beads for Beckman Coulter machines, Cytometer Setup and tracking (CS&T) beads or Rainbow Calibration particles (eight-peak beads) for Becton Dickinson (BD) machines [57], and SpectroFlo QC beads for the Cytek Aurora (Fig. 2A).
Figure 2. Flow cytometer standardization using Flow-Set pro (FSP) beads at two study sites.
(A) FSP beads were used to set mean fluorescence intensity (MFI) target values (indicated by grey floating bars [target values±10%]) for all 10 fluorescence channels (FL1 to FL10) at Site A. The identical lot of FSP beads was used at Site B to match the target values. After machine standardization, the same lot of FSP beads was acquired prior to study sample acquisition at both sites (n = 4 for Site A; n = 11 for Site B), with the resulting MFI of values shown in (A). Data from each channel are in a different color; error bars indicate mean ± SD; percentages indicate CV. (B) Blood from 10 healthy subjects (n = 5 per site) was collected in sodium heparin tubes then stimulated with INFANRIX (pentavalent vaccine) vaccine or 1 μg/mL Staphylococcal enterotoxin B (SEB) or left unstimulated for 48 h. Samples were stained using the same lot of custom-made Duraclone tubes to assess the frequency of antigen-induced CD25+OX40+ cells within gated CD4+ T cells. Samples were acquired on Navios machines at each site after machine standardization. Each individual is depicted by a color. Student’s unpaired t-tests were performed (ns = not significant).
In reality, it is rare that all study sites involved in a given trial have the same cytometer and configuration, and rarer still that this cytometer is present in duplicate at each study site. Assay qualification, however, can include more than one center, including the use of different instruments as exemplified by Ivison and colleagues who successfully compared data acquired on BD LSRFortessa and Beckman Coulter Navios machines [58].
Antibody panel design
For each immune monitoring biomarker study, drug effects and marker detection should be considered when designing antibody panels (examples of published panels used in T1D studies and trials are shown in Supporting information Table S1). When studying subjects receiving biologics, the antibody clone used for staining should not block or interact with the therapeutic antibody clone. Monitoring immune populations that are directly related to the drug mechanism of action is crucial. For example, aldesleukin (recombinant human IL-2) enhanced CD25+FOXP3+ Treg frequency [59, 60]; ustekinumab (antagonist for IL-12 and IL-23 binding to their respective receptors) reduced marker expression of IFN-γ and IL-17-producing T cells (UST1D: Clinical Trials.gov ID NCT02117765) [61, 62]; alefacept (LFA3-Ig) depleted CD4+ and CD8+ memory T cells in at-risk or T1D subjects [63]; and teplizumab (anti-CD3) augmented exhausted CD8+ T cells [20, 64, 65]. It is also important to include markers for immune populations that can help the interpretation of other clinical and mechanistic measures such as partnering B-cell phenotyping with autoantibody clinical readouts or islet antigen-specific T-cell analyses. Monitoring off-target populations may also be valuable, as shown with NK expansion in an IL-2/rapamycin trial [66]. Last, given the heterogeneity of T1D, it is advisable to include any populations previously associated with age, activation, rate of progression, or stage of disease.
It can be beneficial to include many of these parameters in a standardized base panel that can be used across different sites or between trials and leaving channels open for drop-ins. This allows the inclusion of study-specific markers of interest, while retaining the ability to compare basic immune parameters. One key example is the use of a T-cell panel across many Immune Tolerance Network T1D trials, along with others [20, 36, 63-65, 67], that reproducibly identifies T-cell differentiation, activation, and hyporesponsiveness (Supporting information Table S1).
One of the major limitations of conventional flow cytometry is the spectral overlap and spread of fluorochromes. Pairing bright fluorochromes with low-density target antigens and dim fluorochromes with high-density target antigens will limit unnecessary fluorescent spillover. Spectral compensation and spreading matrices are valuable tools that should be used to carefully and effectively design panels to ensure that all cell populations of interest can be resolved beyond the background signal [68]. Clones and fluorochromes are known to affect the level of antigen expression detected, so it is ideal to keep these consistent when designing panels for a clinical trial across different sites. At the very least, cell populations of interest should be identified using the same cell markers/antigens.
Antibodies/staining reagents
Titrations should be performed to ensure that the appropriate amount of antibody is used in each assay to improve the resolution of all of the parameters in the panel [69]. Since target antigen varies across samples, it is recommended to use an antibody titration just above target saturation levels. In instances where combinations of high antigen density paired with bright fluorochromes cannot be avoided, tittering using ratios of fluorochrome-labeled antibody with unlabeled antibody of the same clone may help to control fluorescence spillover. Antibody titration is particularly important as immune composition can vary dramatically in T1D studies due to treatment and age. Wet antibodies often have lot-to-lot variation and can be subject to degradation over time, an attribute strongly influenced by the stability of the fluorochrome. Nevertheless, some of these effects can be controlled by performing lot-to-lot crossovers and performing stability testing. In addition, off-the-shelf master mixes, which have been tested for antibody titration and stability, can aid in reproducibility, albeit at an increased cost. A more expensive alternative is the use of lyophilized antibody tubes that are much more stable than wet antibodies and can eliminate pipetting error. Upon standardization of the same machines at two trial sites (Fig. 2A) using same lot of custom-made lyophilized tubes, we observed similar levels of antigen-specific CD4+ T cells after stimulation of blood overnight (n = 5 age-matched samples; Fig. 2B). In comparison to fluorochrome-conjugated reagents, metal-tagged antibodies used for mass cytometry (CyTOF) studies can be frozen as master mixes that are stable and represent a more economical solution [70]. It is of utmost importance to perform full panel testing between sites to ensure similar staining quality. The choice of using samples from healthy controls versus individuals with T1D for panel testing should also be considered as some panels, such as those that detect frequencies or phenotypes of islet-specific cells, will be most effective if samples from individuals with T1D are used.
Analysis/reporting
For standardized immune monitoring in subjects with T1D, subtle changes in frequency of rare cells or incremental changes in the level of expression of markers are often observed. Use of standard staining controls, such as fluorescence minus one, to improve the accuracy of gating populations is a key. A predefined gating strategy and template is an effective way to streamline and standardize analysis for manual (Boolean) gating within and between study sites. Automatic gating also streamlines analysis and can minimize subjectivity and maximize reproducibility; however, it is less effective at identifying rare and “shoulder” populations. The importance of accurately capturing these rare/shoulder populations should be balanced with improved workflow and consistency when considering automated gating.
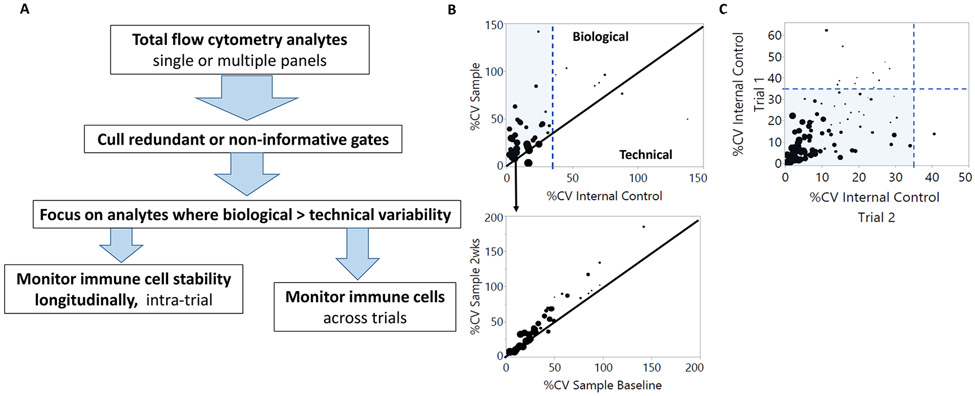
Analysis of large flow cytometry datasets can benefit from a standardized workflow that focuses on biologically relevant immune analytes within and between clinical trials. Critical to this process is using an internal control sample to parse technical from biological variability, the former generally arises when samples are divided into batches to manage sample processing. A standardized analysis workflow begins by compiling batch data, and limiting noise by removing redundant gates that can include negative populations or Boolean gate parents (Fig. 3A). The percent coefficient of variation (%CV) is determined per immune cell analyte within the compiled internal control (technical variability), and among donor samples (biological variability; Fig. 3B). The provided graphs represent a means of parsing technical from biological variation where high technical variation is defined here as more than 35% CV from the internal control. High technical variation often results from analytes with poor resolution between positive and negative populations or low frequency of detection (Fig. 3B, C) [47, 71, 72]. Biologically relevant immune subsets of interest are identified where biological variability exceeds that of technical variability. These subsets can be further explored for patient response via intratrial analyte comparisons (Fig. 3B). This same workflow can be used to explore autoimmune signatures across trials where trial design incorporates the same antibody panel and immune cell readouts. In this example, we focus on analytes with CVs less than 35%; however, other CV cut-offs may be determined/used on a per trial basis. A review by Hedley and Keeney discusses the number of flow events required for a given level of precision, a factor which is particularly relevant for rare subset analysis [72] and should be used to determine the lower limit of acquired events. Predefined criteria for how to exclude inaccurate data due to insufficient events are essential.
Figure 3. Using thresholds of technical variability to identify biologically informative cytometry data.
(A) Workflow for separating biological data and technical noise. Data are filtered to remove redundant or noninformative gates. Variability (%CV) is detected in immune populations within samples at baseline and internal batch controls. (B) Representative immune cell analytes (n = 39) from a published trial data set are simplified here as a workflow example. Each dot represents one immune parameter ranging in detection frequency from 0.8 to 93%, where dot size corresponds continuously with magnitude of detection from low to high and the minimum number of events acquired was 150 000 [98]. Samples to the right of the blue dotted line at 35% have high technical variability and often tend to be immune subsets or phenotypes with low frequency (smaller dots). Black diagonal lines indicate that variation in analyte detection is equivalent between technical reproducibility (internal control) and sample biology (top panel) or between longitudinal trial time points (bottom panel). The shaded area represents analytes with greater biological than technical variation. (C) Analytes between multiple trials utilizing the same antibody panel and immune cell readouts have the potential to identify common and/or unique immune phenotypes. Shown are representative immune analytes (n = 127) published for two trials with a common flow cytometry panel. Analytes were detected with frequencies between 0.04 and 97%, reflected by dot size. The first step in analyzing these data sets is removal of analytes with high technical variability (≥35% CV) using the respective internal controls. Analytes with low technical variability (shaded) can then be explored for biological relationships between trials. Unique technical variability that arises between trials (≥35% CV) can be used to identify areas for protocol improvement between trial sites. Graphs were generated using JMP software (SAS).
When comparing MFI of samples that are acquired across multiple days, it is worth noting that although machines may be calibrated prior to sample acquisition, day-to-day variation of machines is still observed. It may be useful to consider normalization of MFI to molecules of equivalent soluble fluorochrome by acquiring beads such as Rainbow Calibration particles (eight-peak beads), Bang beads, or other appropriate equivalents on each experimental day. This approach was successfully used to study the differences in CD25 expression on CD4+ conventional T cells and Treg cells in individuals with T1D [59, 73]. In cases where different cytometers and fluorochrome combinations are used across centers, direct comparability of changes in MFI is a challenge.
To reiterate the importance of documentation, there are many variables that may affect the quality of flow cytometric data such as who processed the samples, time between collection and staining of samples, time between poststained and acquisition of samples, the rate and number of events that are recorded during sample acquisition, and importantly, the quality and viability of the cells.
Crucial aspects of data reporting are described in the Minimum Information about a Flow Cytometry Experiment criteria [74] and include location of data deposition, publication of full gating strategies used, labelling of flow plots, inclusion of all antibody clones, fluorochromes, antibody titrations, and associated reagents used. Many journals now require this information for publication including Research Resource Identifiers.
Case study: Standardization of a fresh whole blood flow cytometry assay across multiple sites
As part of a multicenter longitudinal sample collection initiative called INNODIA, blood samples from newly diagnosed T1D subjects (ND-T1D) were collected from several clinical sites in Europe and shipped to sites for sample processing. Samples were collected, mainly in the morning, in EDTA vacutainers from subjects recruited within 6 weeks since diagnosis, and shipped at ambient temperature (18–22°C) [75]. Upon use, viability and recovery of each sample was evaluated, in a similar manner as in other studies [65, 76].
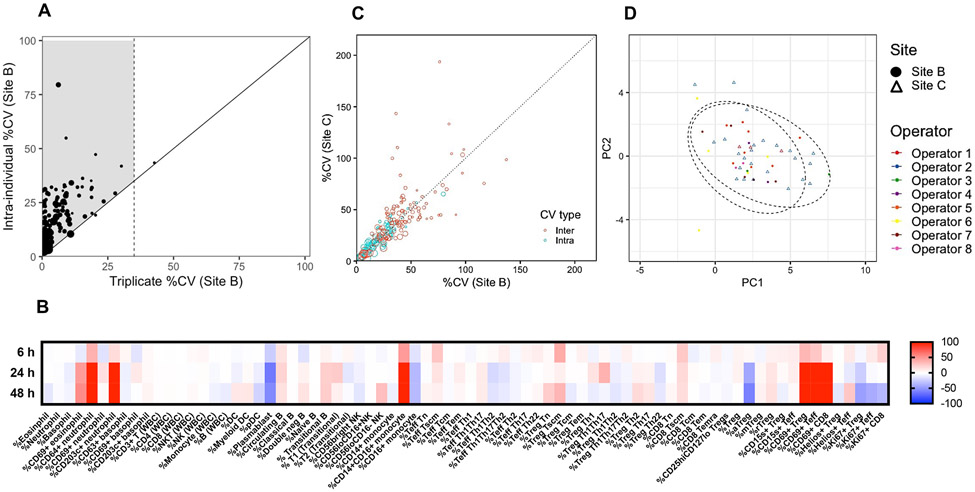
Prior to the initiation of this study, three representative samples were stained using five flow panels in triplicate to assess technical reproducibility for a broad range of cell subsets, including T and B lymphocytes, NK cells, monocytes, DC, and granulocytes. Only 1 out of 118 cell subsets measured, namely CD45RA+CCR7− Tregs, was excluded from further analyses due to technical variation over 35% CV (Fig. 4A). To assess stability of blood collected in EDTA tubes, samples were stained at different time points after sample collection. Samples stained within 6 h from blood draw were stable (within 20% difference compared to fresh) while post-6 h, deviations from fresh were observed (Fig. 4B). The activation state of granulocytes and lymphocytes and the frequency of monocytes were increased and selected features of T-cell subsets and plasmablasts were decreased due to prolonged storage. A maximum of 6 h from blood drawn then defined the time frame criteria for performing flow staining at each site, which was similar to the criteria set for the ONE study [50].
Figure 4. Flow cytometry standardization of five antibody panels using whole blood in the INNODIA multicenter study.
Stained blood samples from newly diagnosed (ND)-T1D donors collected in EDTA tubes were acquired on BD LSRFortessa flow cytometers with either five or three lasers at Sites B and C, respectively. Averaged intraindividual CV per parameter was determined using data at three time points0 (baseline, 3 and 6 months) from 13 and 21 ND-T1D subjects at Sites B and C, respectively. (A) Interindividual CV per parameter was determined using baseline data from the same cohorts. Technical variability (triplicate staining of three samples for all five flow panels) compared to biological intraindividual variability was assessed using averaged data from three samples. A total of 118 cell subset parameters were measured as depicted by each data point. Size of data point = mean frequency of each cell population (ranging in frequency of parent population from 0.03 to 93%). For samples to the right of the black diagonal line, technical and biological variability cannot be discriminated (i.e., >35% CV). Shaded area indicates biologically relevant immune subsets of interest with less than 35% technical CV and biological variability exceeding that of technical. (B) Two samples were stained at 0 (fresh), 6, 24, and 48 h after blood collection. Differences in frequencies of selected cell subsets (columns) were calculated for each time point compared to fresh values and data for the two samples were averaged, where red and blue indicate increased or decreased frequency, respectively. (C) Intra- and interindividual percent CV are compared between Sites B and C for N = 117 cell subsets. Size of data point = mean frequency of each cell population (ranging in frequency of parent population from 0.03 to 94%). (D) Complete baseline flow data from N = 24 and N = 27 samples from ND-T1D donors at Sites B and C, respectively, were used to perform PCA using base R functions and ggplot2 R packages in an unsupervised approach. Ellipse drawn assumed multivariate t-distribution at a level of 0.95 for each site (depicted by shape). Each operator is depicted by a color.
All INNODIA sites used the same antibody clones, but there was variation in fluorophore combinations due to different BD LSRFortessa configurations. The protocol included the use of CS&T beads and application settings prior to samples acquisition. Unlike the use of batched, cryopreserved PBMCs, when using whole blood, it is more challenging to include a consistent sample for QC. Thus, for this study using machines with different configurations and antibody panels, the study opted to not use QC samples, and to only assess the frequency of cell subsets (determined using an identical gating strategy), and not MFIs. All gating and datasets were quality controlled by one operator to maintain data consistency. To investigate whether data obtained by different sites could be combined, both intra- (averaged variation across three time points within an individual) and interindividual CV (variation between individuals at baseline time point) were compared for each cell subset measured and observed both types of CVs were similar between sites (Fig. 4C). Principal component analysis (PCA) further confirmed the lack of site and operator-specific effects (Fig. 4D). Ideally the use of identical cytometer and flow panels across centers would improve this study further, but by focusing on the four categories listed above a desirable level of standardization was obtained despite differences in panels, machines, sites, and operators.
Integrating modern immunophenotyping platforms in T1D studies
Biomarker discovery shares many of the considerations discussed above for standardized immune monitoring, but also presents some additional concerns due to the exploratory nature of these studies that are often performed in specialized labs with continually evolving techniques and analysis platforms. There is an ever-increasing range of tools for measuring single-cell protein expression, many of which have specific strengths and weaknesses that make them well suited to tackle specific scientific challenges including those in the realm of T1D research and T1D trials (Table 2). These tools generally require trade-offs between the number of parameters that can be simultaneously analyzed on a single cell, throughput in terms of both the number of cells analyzed from a given biological sample and the total number of samples analyzed, and the cost of any given study. In this section, recent innovations in cytometry are summarized for three fundamental technologies (fluorescence-based cytometry, mass cytometry, and oligonucleotide-based profiling), with the objective of informing decisions on platform selection and experimental design.
Table 2.
Immunophenotyping platforms
| Platform | Number of Parameters |
Throughput (events/second) |
Scalability |
|---|---|---|---|
| Standard Flow Cytometry | Typically: 1–20 Maximum published: 30 [99] |
Typically: 1000—10 000 Maximum: 100 000 |
|
| Spectral Flow Cytometry | Typically: 20–30 Maximum published: 43 [78] |
Typically: 1000–3000 Maximum: 30 000 |
|
| CyTOF | Typical: 30–40 Maximum published: 47 [100] |
Typically: 200–500 Maximum: 1000 |
|
| CITE-Seq & REAP-Seq | Typical: 30–40 Maximum published: 228 [101] |
Not acquired in real time; typical experiments will only evaluate <10 000/sample |
|
Costs will be affected by number of antibodies. Isotope conjugation for mass cytometry increases costs/antibody.
File size will be dependent upon number of events acquired.
The number of parameters required will vary based on experimental needs and reagent availability, cost, and in the case of flow cytometry platforms, marker coexpression.
Innovations in fluorescence-based flow cytometry
Fluorescence-based flow cytometry continues to represent one of the most accessible, affordable, and high-throughput approaches for multiparameter single-cell analysis. It is also a rapidly growing technology, with modern spectral cytometers that may now resolve over 40 markers in a panel [77, 78]. This is accomplished through improved resolution of fluorescence emission patterns, utilization of spectral unmixing algorithms, and fluorophore innovations. However, designing high-parameter fluorescence-based panels requires considerable investment in careful panel design, optimization, and an extensive number of controls relative to lower parameter flow cytometry. Further, panels designed for a single lineage are subject to increased spectral spill from coexpressed markers. Consequently, high-dimensional flow cytometry exploratory studies more typically leverage panels in the 20–30 marker range.
Mass cytometry
Mass cytometry overcomes the limitations of fluorescence spectral overlap by substituting fluorophore-conjugated antibodies with purified-isotope-conjugated antibodies, which can be detected using time-of-flight mass spectrometry (CyTOF). The technology has enabled detection of up to 47 antibodies with additional channels used for viability detection and sample multiplexing reagents [79]. This increased dimensionality comes at the cost of reduced throughput (<500 events/s vs. >10, 000 events/s for typical flow cytometers), lower recovery of quantifiable events as compared to flow cytometry, and reduced sensitivity for low abundance antigens. However, due to use of premixed antibody cocktails and internal bead and bar-coding controls [80], mass cytometry data often has increased reproducibility over time. A growing list of T1D-associated cellular features has been defined using mass cytometry including two recently identified exhausted-like subsets associated with response to therapy [81] and spatially distinct populations identified by imaging mass cytometry [82, 83]. To date, the use of mass cytometry in T1D research has been somewhat circumscribed [84], but more wide-spread application (including pursuit of the TrialNet Key Question 1, https://www.trialnet.org/researchers; Rahman and Homann, unpublished) of this mature technology platform is expected in the near future.
Oligonucleotide-based antibody profiling
The recent advent of oligonucleotide-based antibody profiling using approaches, such as Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-Seq), presents a new frontier in high dimensional single-cell analysis [85, 86]. Antibodies are conjugated to oligonucleotides and detected using massively parallel single-cell sequencing approaches. Given that the potential range of oligonucleotide barcodes exceeds the number of existing commercial antibodies, the number of targets that can be simultaneously detected is technically limitless. While the development and optimization of large CITE-Seq panels can present significant practical challenges, the technology has enabled simultaneous profiling of 228 antibodies, coupled with concurrent detection of thousands of gene expression transcripts from the same cells [87]. Currently, the cost and complexity of the approach limits most studies to analyze a few thousand cells per sample in relatively small sample cohorts and, thus far, no T1D studies employing this technology have been published. That said, applications of this technology are underway, including the use of CITEseq labels to define donor or time point, as well as considerations of donor variants for sample demultiplexing [88].
Choosing a platform
After discovery, most biomarkers transition to more traditional flow cytometers for validation and standard immune monitoring, as only limited flow cytometry panels have been validated in Clinical Laboratory Improvement Amendments certified diagnostic laboratories. However, multiple platforms should be considered for discovery. While no single platform stands out as optimally suited for T1D phenotyping per se, scalability, cost associated with custom conjugation, target cell frequency, and sample availability should be considered when selecting a platform (Table 2).
High parameter data analysis tools
The considerable increase of discrete single-cell parameters quantified by polychromatic and spectral flow cytometry, CyTOF, and CITE-Seq necessitates a structured parsing of the high-dimensional data space to permit systems-level analysis of immune cell subsets, their phenotypic properties, and functional capacities [89]. These advances have driven the field of computational flow cytometry, which is now focused on data processing and high-parameter data analysis in a way that accounts for both technical and biological variability [90]. With regard to T1D, analyses must also incorporate clinical stage, subject age, T1D duration or age at onset, and other study-related metadata. In this section, we highlight commonly used packages created to handle high-throughput sample handling, batch processing, and downstream data analysis, and discuss the various capabilities and ongoing needs while noting examples of usage in T1D.
Expanding horizons
The advancements in technology noted above have moved the 24 possible combinations in a simple four-color flow cytometry assay into what today could represent an astounding 9157 possible combinations in a 100-parameter antibody panel. The net result is that data analysis, which was once a niche skill for a single individual, has now transformed into a complex and interactive discipline that often requires an integrated team including bench and data scientists with programming and biostatistical knowledge. This transition has also now mandated two dichotomous approaches to flow analyses: hypothesis-driven with manually supervised gating strategies and hypothesis generating with unsupervised or semisupervised sample analysis. The former is used in standardized immune monitoring and biomarker validation, where consistency and robustness are keys. In the latter approach, the goal is to distill complex high-parameter datasets in an unbiased manner to identify informative subsets and phenotypes. The data matrix output is refined using tools and packages to reduce dimensionality and display trends in a way that highlights reportable observations in an informative and user-friendly manner. Some examples of collaborative systems and cytometry discovery in T1D include phenotypes of rare cells identified using DISCOV-R (an extension of Phenograph) [19], unique subsets of exhausted-like cells associated with clinical response to treatment using FlowSOM cluster correlations with gene sets [81], and tempospatial relationships using SCORPIUS [82].
Big data challenges
As the flow cytometry parameter space grows, so too does data file size. In the context of large trials and meta-analyses, data compilation becomes cumbersome due to lengthy transfer times, limitations of storage space, as well as potential problems with file name and feature name misalignment. Therefore, the handling of large datasets requires careful planning before the trial begins to negate the risk of data loss and compilation errors. A major challenge of big data analysis is maintaining standardized, reproducible data formats, and annotations between collaborating institutions, across individual analysts, and over the course of multiyear studies. Naming and formatting standards should be defined as part of the initial experiment design to ensure reproducibility. The challenges related to storage and direct transfer of large data files can overcome through cloud-based platforms like FlowRepository [91], ImmPort [92], or Cytobank. Cloud-based databases may be preferable to direct file transfer, as they generally require some degree of quality checks and annotation standards. For example, FlowRepository requires that FCS files and their associated annotation adhere to the Minimum Information about a Flow Cytometry standard [93].
Distilling meaning from the noise
Ultimately, the power of computational cytometry comes from the application of the knowledge gained in an unbiased manner, leading to biomarker discovery that can be further validated and applied to immune monitoring in interventional trials. This fundamental need for actionable insight includes the need to apply machine-learning algorithms to flow cytometric data. Examples of these approaches, as well as various advantages and disadvantages are summarized (Table 3). The main objective of these tools is to identify cell populations in exponentially growing parameter space using unsupervised clustering methods. This is followed by dimensionality reduction methods to allow two-dimensional visualization of population structure, usually as a tree or scatter plot. A number of these tools have been used to identify immune signatures, pathways, and therapeutic responses from high-parameter flow cytometry data sets in T1D studies [19, 81-83, 94-96].
Table 3.
High throughput data analysis tools
| Method | Description | Years (Citations) |
|---|---|---|
| Cytobank1 |
|
2010 [102] |
| SPADE2 |
|
2011 [103] |
| CITRUS2 |
|
2014 [104] |
| FlowSOM1 |
|
2015 [105] |
| PhenoGraph2 |
|
2015 [106] |
| CellCnn |
|
2017 [107] |
| MetaCyto |
|
2018 [108] |
| Astrolabe1,2 |
|
2019 [109] |
Fee for use.
Available as a plug-in for some commercial flow cytometry analysis software.
Features of available data analysis tools are presented with launch years and citations.
Expanding data parameter space presents major challenges for biomarker identification and validation to avoid type I and II error. Thus, as the field of computational flow cytometry continues to evolve, it is increasingly important to implement data harmonization standards beginning with sample collection and processing, batch processing, and metadata standards, and robust systems for downstream data analytics and reporting. Equally as important, the field must plan for and follow through with data sharing as recommended by the Findable, Accessible, Interoperable, and Reusable data standards [97]. Ultimately, we expect the field to advance more rapidly when multiple groups can replicate observations. Together, these approaches will help move discovery of cell populations to validated T1D biomarkers [24].
Conclusions
Identification and validation of robust biomarkers is a critical next step to link immune mechanisms with T1D clinical measures that will guide development of therapeutic strategies and cohort selection criteria. Generation of relevant and potentially actionable data requires an accurate biological knowledge of T1D to better anchor and leverage the interpretation of highly technical approaches for data acquisition and analysis, as well as careful study design and implementation. Our aim has been to provide a cogent overview that will serve as an effective starting point for further dialogue within the field, with the overarching goal of adopting “universal best practices” for immunophenotyping within the field of T1D. To that end, we have provided recommendations about study design, cytometry platform options, assay validation, data analysis tools, and subsequent data interpretation in the context of immune monitoring and biomarker discovery. Standardization of these elements of practice will be essential to allow data sharing between laboratories, effective multicenter data acquisition for multisite clinical trials, and integrated meta-analyses of findings across multiple studies to draw the high-level insights that will be crucial to drive the field forward.
Supplementary Material
Acknowledgments:
We appreciate the Immunology of Diabetes Society T-Cell Workshop and the Immunology of Diabetes Society T-Cell Cytometry Group for fostering discussions that launched this manuscript. We thank Thinzar Myint (University of Florida) for aid with graphics.
T.M.B. is supported by the National Institutes of Health (P01 AI042288, R01 DK106191, HIRN UC4 DK104194), and The Leona M. and Harry B. Helmsley Charitable Trust. D.H. and A.H.R are supported by Juvenile Diabetes Research Foundation Strategic Research Agreement JDRF 2-SRA-2018-643-M-B and D.H. is supported by National Institutes of Health (NIH) grants U01DK123716, UC4DK116284, and P30DK02054141. E.A.J. is supported by grants from the JDRF (1-SRA-2017-344-S-B and 2-SRA-2018-551-S-B). S.C.K. is supported by the National Institute of Diabetes and Digestive and Kidney Diseases (UC4 DK116284) and S.C.K. is the George F. and Sybil H. Fuller Term Chair in Diabetes. M.K.L and K.A.W.-H are supported by JDRF (3-SRA-2018-629-S-B). K.A.W.-H is supported by the JDRF (1-PDF-2019-709-A-N), the Canadian Institutes of Health Research Canadian (CIHR), and the Association of Gastroenterology (CAG) (201711FGA-398491-294541). S.A.L is supported by a grant from the JDRF (3-SRA-2019-792-S-B) and the National Institutes of Health (R01 AI141952). C.S. is supported by a grant from the JDRF (3-SRA-2019-791-S-B). T.I.M.T. and J.H.M.Y. are supported by the T1DUK Immunotherapy Consortium funded by grants from Diabetes UK (Ref: 15/0005232 and 15/0005233) and from the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Center award to Guy’s & St Thomas’ NHS Foundation Trust in partnership with King’s College London and King’s College Hospital NHS Foundation Trust. This project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement No. 115797 (INNODIA), which receives support from the European Union’s Horizon 2020 research and innovation programme and “EFPIA,” ‘JDRF,” and “The Leona M. and Harry B. Helmsley Charitable Trust.” All authors are the guarantors of this work.
Abbreviations.
- BD
Becton Dickinson
- CITE-Seq
Cellular Indexing of Transcriptomes and Epitopes by Sequencing
- CS&T
cytometer setup and tracking
- %CV
coefficient of variation
- ND-T1D
newly diagnosed T1D subjects
- PCA
principal component analysis
- QC
quality controls
- T1D
type 1 diabetes
Footnotes
Conflict of Interest: The authors declare that no commercial or financial conflict of interest.
Permission to Reproduce Material from Other Sources: We gratefully thank Carla Greenbaum, M.D. for use of The Stages of T1D description (Fig. 1A). Figure 1 panels B-D were created with BioRender.com.
Additional supporting information may be found online in the Supporting Information section at the end of the article.
Immunology of Diabetes Society T-Cell Cytometry GroupArja Vuorela, Linnea Reinert-Hartwall, Juho Hämäläinen, Jarno Honkanen, and Mikael KnipResearch Program for Clinical and Molecular Metabolism, University of Helsinki, Helsinki, FloridaEvangelia Williams, Leena Khatri, Eleni Christakou, Yasaman Shahrabi-Taylor, Emily Pollock, Clara Domingo-VilaDepartment of Immunobiology, Faculty of Life Sciences & Medicine, King’s College London, London, UKNational Institute of Health Research Biomedical Research Centre at Guy’s and St. Thomas’ National Health Service Foundation Trust and King’s College London, London, UK
Data availability statement:
The data that support the findings of this study are available on request from the corresponding author.
References
- 1.Battaglia M, Ahmed S, Anderson MS, Atkinson MA, Becker D, Bingley PJ et al. , Introducing the endotype concept to address the challenge of disease heterogeneity in type 1 diabetes. Diabetes Care. 2020. 43, 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Insel RA, Dunne JL, Atkinson MA, Chiang JL, Dabelea D, Gottlieb PA et al. , Staging presymptomatic type 1 diabetes: A scientific statement of JDRF, the Endocrine Society, and the American Diabetes Association. Diabetes Care. 2015. 38,1964–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kent SC, Mannering SI, Michels AW and Babon JAB, Deciphering the pathogenesis of human type 1 diabetes (T1D) by interrogating T cells from the “scene of the crime”. Curr Diab Rep. 2017. 17 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mannering S, Pathiraja V and Kay T, The case for an autoimmune aetiology of type 1 diabetes. Clin. Exp. Immunol 2016. 183 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landry L, Anderson A, Russ H, Yu L, Kent S, Atkinson M et al. , Proinsulin-reactive CD4 T cells in the islets of type 1 diabetes organ donors. Frontiers in Endocrinology. 2021. 12, 622647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michels AW, Landry LG, McDaniel KA, Yu L, Campbell-Thompson M, Kwok WW et al. , Islet-derived CD4 T cells targeting proinsulin in human autoimmune diabetes. Diabetes. 2017. 66 722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babon JAB, DeNicola ME, Blodgett DM, Crèvecoeur I, Buttrick TS, Maehr R et al. , Analysis of self-antigen specificity of islet-infiltrating T cells from human donors with type 1 diabetes. Nat. Med 2016. 22, 1482–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coppieters KT, Dotta F, Amirian N, Campbell PD, Kay TW, Atkinson MA et al. , Demonstration of islet-autoreactive CD8 T cells in insulitic lesions from recent onset and long-term type 1 diabetes patients. J. Exp. Med 2012. 209, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carré A, Richardson SJ, Larger E and Mallone R, Presumption of guilt for T cells in type 1 diabetes: Lead culprits or partners in crime depending on age of onset? Diabetologia. 2021, 64, 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mallone R and Eizirik DI, Presumption of innocence for beta cells: why are they vulnerable autoimmune targets in type 1 diabetes? Diabetologia. 2020, 63, 1999–2006. [DOI] [PubMed] [Google Scholar]
- 11.Ferraro A, Socci C, Stabilini A, Valle A, Monti P, Piemonti L et al. , Expansion of Th17 cells and functional defects in T regulatory cells are key features of the pancreatic lymph nodes in patients with type 1 diabetes. Diabetes. 2011. 60, 2903–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marwaha AK, Crome SQ, Panagiotopoulos C, Berg KB, Qin H, Ouyang Q et al. , Cutting edge: Increased IL-17-secreting T cells in children with new-onset type 1 diabetes. J. Immunol 2010. 185, 3814–3818. [DOI] [PubMed] [Google Scholar]
- 13.Honkanen J, NJ K, Gao R, Luopajarvi K, Luopajarvi K, Salo HM et al. , IL-17 immunity in human type 1 diabetes. J. Immunol 2010. 185, 1959–1967. [DOI] [PubMed] [Google Scholar]
- 14.Kenefeck R, Wang CJ, Kapadi T, Wardzinski L, Attridge K, Clough LE et al. , Follicular helper T cell signature in type 1 diabetes. J. Clin. Invest 2015. 125, 292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viisanen T, Ihantola EL, Näntö-Salonen K, Hyöty H, Nurminen N, Selvenius J et al. , Circulating CXCR5+PD-1+ICOS+ follicular T helper cells are increased close to the diagnosis of type 1 diabetes in children with multiple autoantibodies. Diabetes. 2017. 66, 437–447. [DOI] [PubMed] [Google Scholar]
- 16.McClymont SA, Putnam AL, Lee MR, Esensten JH, Liu W, Hulme MA et al. , Plasticity of human regulatory T cells in healthy subjects and patients with type 1 diabetes. J. Immunol 2011. 186, 3918–3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long SA, Cerosaletti K, Pl B, Tatum M, Shilling H, Zhang S et al. , Defects in IL-2R signaling contribute to diminished maintenance of FOXP3 expression in CD4(+)CD25(+) regulatory T-cells of type 1 diabetic subjects. Diabetes. 2010, 59. 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linsley PS and Long SA, Enforcing the checkpoints: Harnessing T-cell exhaustion for therapy of T1D. Curr. Opin. Endocrinol. Diabetes Obes 2019, 26. 231–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiedeman AE, Muir VS, Rosasco MG, DeBerg HA, Presnell S, Haas B et al. , Autoreactive CD8+ T cell exhaustion distinguishes subjects with slow type 1 diabetes progression. J. Clin. Invest 2020, 130. 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long SA, Thorpe J, DeBerg HA, Gersuk V, Eddy J, Harris KM et al. , Partial exhaustion of CD8 T cells and clinical response to teplizumab in new-onset type 1 diabetes. Science Immunology. 2016. 1, eaai7793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ihantola EL, Viisanen T, Gazali AM, Näntö-Salonen K, Juutilainen A, Moilanen L et al. , Effector T cell resistance to suppression and STAT3 signaling during the development of human type 1 diabetes. J. Immunol 2018. 201, 1144–1153. [DOI] [PubMed] [Google Scholar]
- 22.Lawson JM, Tremble J, Dayan CF, Beyan H, Leslie RD, Peakman M et al. , Increased resistance to CD4+CD25hi regulatory T cell-mediated suppression in patients with type 1 diabetes. Clin. Exp. Immunol 2008. 154, 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider A, Rieck M, Sanda S, Pihoker C, Greenbaum C and Buckner JH, The effector T cells of diabetic subjects are resistant to regulation via CD4+ FOXP3+ regulatory T cells. J. Immunol 2008. 181, 7350–7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmed S, Cerosaletti K, James EA, Long SA, Mannering S, Speake C et al. , Standardizing T-cell biomarkers in type 1 diabetes: Challenges and recent advances. Diabetes. 2019. 68, 1366–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dufort MJ, Greenbaum CJ, Speake C and Linsley PS, Cell type-specific immune phenotypes predict loss of insulin secretion in new-onset type 1 diabetes. JCI Insight. 2019. 4, e125556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Speake C, Bahnson HT, Wesley JD, Perdue N, Friedrich D, Pham MN et al. , Systematic assessment of immune marker variation in type 1 diabetes: A prospective longitudinal study. Front Immunol. 2019. 10, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bingley PJ, Wherrett DK, Shultz A, Rafkin LE, Atkinson MA and Greenbaum CJ, Type 1 diabetes TrialNet: A multifaceted approach to bringing disease-modifying therapy to clinical use in type 1 diabetes. Diabetes Care. 2018. 41, 653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Couper JJ, Haller MJ, Greenbaum CJ, Ziegler AG, Wherrett DK, Knip M et al. , ISPAD Clinical Practice Consensus Guidelines 2018: Stages of type 1 diabetes in children and adolescents. Pediatric Diabetes. 2018. 27, 20–27. [DOI] [PubMed] [Google Scholar]
- 29.Brooks-Worrell B, Tree T, Mannering SI, Durinovic-Bello I, James E, Gottlieb P et al. , Comparison of cryopreservation methods on T-cell responses to islet and control antigens from type 1 diabetic patients and controls. Diabetes Metabolism Research Review. 2011. 27, 737–745. [DOI] [PubMed] [Google Scholar]
- 30.Greenbaum CJ and Harrison LC, IoD Society. Guidelines for intervention trials in subjects with newly diagnosed type 1 diabetes. Diabetes. 2003. 52, 1059–1065. [DOI] [PubMed] [Google Scholar]
- 31.James EA, Mallone R, Kent SC and DiLorenzo TP, T-cell epitopes and neo-epitopes in type 1 diabetes: A comprehensive update and reappraisal. Diabetes. 2020. 69, 1311–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.James EA, Mallone R, Schloot NC, Gagnerault MC, Thorpe J, Fitzgerald-Miller L et al. , Immunology of Diabetes Society T-Cell Workshop: HLA class II tetramer-directed epitope validation initiative. Diabetes and Metabolism Research Review. 2011. 27, 727–736. [DOI] [PubMed] [Google Scholar]
- 33.Mallone R, Mannering SI, Brooks-Worrell BM, Durinovic-Belló I, Cilio CM, Wong FS et al. , Isolation and preservation of peripheral blood mononuclear cells for analysis of islet antigen-reactive T cell responses: Position statement of the T-Cell Workshop Committee of the Immunology of Diabetes Society. Clin. Exp. Immunol 2011. 163, 33–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mannering SI, Wong FS, Durinovic-Belló I, Brooks-Worrell B, Tree TIM, Cilio CM et al. , Current approaches to measuring human islet-antigen specific T cell function in type 1 diabetes. Clin. Exp. Immunol 2010. 163, 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinkse GG, Boitard C, Tree TI, Peakman M and Roep BO, HLA class I epitope discovery in type 1 diabetes: Independent and reproducible identification of proinsulin epitopes of CD8 T cells–report of the IDS T Cell Workshop Committee. Ann. N. Y. Acad. Sci 2006. 1079, 19–23. [DOI] [PubMed] [Google Scholar]
- 36.Speake C, Skinner SO, Berel D, Whalen E, Dufort MJ, Young WC et al. , A composite immune signature parallels disease progression across T1D subjects. JCI Insight. 2019. 4, e126917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tree TI, RB O and Peakman M, A mini meta-analysis of studies on CD4+CD25+ T cells in human type 1 diabetes: Report of the Immunology of Diabetes Society T Cell Workshop. Ann. N. Y. Acad. Sci 2006. 1079, 9–18. [DOI] [PubMed] [Google Scholar]
- 38.Delong T, Wiles TA, Baker RL, Bradley B, Barbour G, Reisdorph R et al. , Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science. 2016. 351, 711–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kent SC, Chen Y, Bregoli L, Clemmings SM, Kenyon NS, Ricordi C et al. , Expanded T cells from pancreatic lymph nodes of type 1 diabetic subjects recognize an insulin epitope. Nature. 2006. 435, 224–248. [DOI] [PubMed] [Google Scholar]
- 40.Marre ML, McGinty JW, Chow IT, DeNicola ME, Beck NW, Kent SC et al. , Modifying enzymes are elicited by ER stress, generating epitopes that are selectively recognized by CD4(+) T cells in patients with type 1 diabetes. Diabetes. 2018. 67, 1356–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michels AW, Landry LG, McDaniel KA, Yu L, Campbell-Thompson M, Kwok WW et al. Islet-derived CD4 T cells targeting proinsulin in human autoimmune diabetes. Diabetes. 2017. 66, 722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pathiraja V, Kuehlich JP, Campbell PD, Krishnamurthy B, Loudovaris T, Coates PT et al. , Proinsulin-specific, HLA-DQ8, and HLA-DQ8-transdimer-restricted CD4+ T cells infiltrate islets in type 1 diabetes. Diabetes. 2016. 64, 172–182. [DOI] [PubMed] [Google Scholar]
- 43.Seay HR, Yusko E, Rothweiler SJ, Zhang L, Posgai AL, Campbell-Thompson M et al. , Tissue distribution and clonal diversity of the T and B cell repertoire in type 1 diabetes. JCI Insight. 2016. 1, e88242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang JHM, Khatri L, Mickunas M, Williams E, Tatovic D, Alhadj Ali M et al. , Phenotypic analysis of human lymph nodes in subjects with new-onset type 1 diabetes and healthy individuals by flow cytometry. Frontiers in Immunology. 2019. 10, 2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis BH, Dasgupta A, Kussick S, Han JY, Estrellado A,Group. IIW. Validation of cell-based fluorescence assays: practice guidelines from the ICSH and ICCS- part I-preanalytical issues. Cytometry B Clinical Cytometry. 2013. 84, 286–290. [DOI] [PubMed] [Google Scholar]
- 46.der Strate BV, Longdin R, Geerlings M, Bachmayer N, Cavallin M, Litwin V et al. , Best practices in performing flow cytometry in a regulated environment: feedback from experience within the European Bioanalysis Forum. Bioanalysis. 2017. 9, 1253–1264. [DOI] [PubMed] [Google Scholar]
- 47.Selliah N, Eck S, Green C, Oldaker T, Stewart J, Vitaliti A et al. , Flow cytometry method validation protocols. Current Protocols in Cytometry. 2019. 87, e53. [DOI] [PubMed] [Google Scholar]
- 48.Finak G, Langweiler M, Jaimes M, Malek M, Taghiyar J, Korin Y et al. , Standardizing flow cytometry immunophenotyping analysis from the human immunophenotyping consortium. Scienctific Reports. 2016. 6, 20686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jimenez Vera E, Chew YV, Nicholson L, Burns H, Anderson P, Chen HT et al. , Standardisation of flow cytometry for whole blood immunophenotyping of islet transplant and transplant clinical trial recipients. PLoS One. 2019. 14, e0217163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Streitz M, Miloud T, Kapinsky M, Reed MR, Magari R, Geissler EK et al. , Standardization of whole blood immune phenotype monitoring for clinical trials: panels and methods from the ONE study. Transplant Research. 2013. 2, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zaunders JJ, Munier MI, Seddiki N, Pett S, Ip S, Bailey M et al. , High levels of human antigen-specific CD4+ T cells in peripheral blood revealed by stimulated coexpression of CD25 and CD134 (OX40). J. Immunol 2009. 183, 2827–2836. [DOI] [PubMed] [Google Scholar]
- 52.Westendorf K, Okhrimenko A, Grün J, Schliemann H, Chang H, Dong J et al. , Unbiased transcriptomes of resting human CD4+ CD45RO+ T lymphocytes. Eur. J. Immunol 2014. 44 1866–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beam CA, Wasserfall C, Woodwyk A, Akers M, Rauch H, Blok T et al. , Synchronization of the normal human peripheral immune system: A comprehensive circadian systems immunology analysis. Sci. Rep 2020. 10, 672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tree TI, Roep BO and Peakman M, Enhancing the sensitivity of assays to detect T cell reactivity: The effect of cell separation and cryopreservation media. Ann. N. Y. Acad. Sci 2004. 1037, 26–32. [DOI] [PubMed] [Google Scholar]
- 55.Wijsman CA, Mooijaart SP, Westendorp RG and Maier AB, Responsiveness of the innate immune system and glucose concentrations in the oldest old. Age (Dordr). 2012. 34, 983–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cossarizza A, Chang HD, Radbruch A, Abrignani S, Addo R, Akdis M et al. , Guidelines for the use of flow cytometry and cell sorting in immunological studies (third edition). Eur. J. Immunol 2021. 51 2708–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Degheidy H, Bauer S, Marti G and Wang L, Flow cytometer performance characterization, standardization and calibration against CD4 on T lymphocytes enables quantification of biomarker expressions for immunological applications. Scientific Research. 2014. 7, 756–768. [Google Scholar]
- 58.Ivison S, Malek M, Garcia RV, Broady R, Halpin A, Richaud M et al. , A standardized immune phenotyping and automated data analysis platform for multicenter biomarker studies. JCI Insight. 2018. 3, e121867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Long SA, Rieck M, Sanda S, Bollyky JO, Samuels PL, Goland R et al. , Rapamycin/IL-2 combination therapy in patients with type 1 diabetes augments Tregs yet transiently impairs β-cell function. Diabetes. 2012. 61, 2340–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seelig E, Howlett J, Porter L, Truman L, Heywood J, Kennet J et al. , The DIL frequency study is an adaptive trial to identify optimal IL-2 dosing in patients with type 1 diabetes. JCI Insight. 2018. 3, e99306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marwaha A, Chow S, Pesenacker A, Cook L, Sun A, Long S et al. , A phase 1b open-label dose-finding study of ustekinumab in young adults with type 1 diabetes. Immunotherapy Advances. 2021. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marwaha AK, Tan S and Dutz JP, Targeting the IL-17/IFN-γ axis as a potential new clinical therapy for type 1 diabetes. Clin. Immunol 2014. 154, 84–89. [DOI] [PubMed] [Google Scholar]
- 63.Rigby MR, Harris KM, Pinckney A, DiMeglio LA, Rendell MS, Felner EI et al. , Alefacept provides sustained clinical and immunological effects in new-onset type 1 diabetes patients. J. Clin. Invest 2015. 125, 3285–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Herold KC, Bundy BN, Long SA, Bluestone JA, DiMeglio LA, Dufort MJ et al. , An Anti-CD3 antibody, teplizumab, in relatives at risk for type 1 diabetes. N. Engl. J. Med 2019. 381, 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Long SA, Thorpe J, Herold KC, Ehlers M, Sanda S, Lim N et al. , Remodeling T cell compartments during anti-CD3 immunotherapy of type 1 diabetes. Cell. Immunol 2017. 319, 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Long SA, Rieck M, Sanda S, Bollyky JB, Samuels PL, Goland R et al. , Rapamycin/IL-2 combination therapy in patients with type 1 diabetes augments Tregs yet transiently impairs beta-cell function. Diabetes. 2012. 61, 2340–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haller MJ, Long SA, Blanchfield JL, Schatz DA, Skyler JS, Krischer JP et al. , Low-dose anti-thymocyte globulin preserves C-peptide, reduces HbA(1c), and increases regulatory to conventional T-cell ratios in new-onset type 1 diabetes: Two-year clinical trial data. Diabetes. 2019. 68, 1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen R, Perfetto S, Mahnke YD, Chattopadhyay P and Roederer M, Quantifying spillover spreading for comparing instrument performance and aiding in multicolor panel design. Cytometry A. 2013. 83, 306–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ashhurst TM, Smith AL and King NJC, High-dimensional fluorescence cytometry. Curr. Protoc. Immunol 2017. 119, 5.8.1–5.8.38. [DOI] [PubMed] [Google Scholar]
- 70.Schulz A, Baumgart S, Schulze J, Urbicht M, Grützkau A and Mei H, Stabilizing antibody cocktails for mass cytometry. Cytometry A. 2019.95 910–916. [DOI] [PubMed] [Google Scholar]
- 71.Burel JG, Qian Y, Lindestam Arlehamn C, Weiskopf D, Zapardiel-Gonzalo J, Taplitz R et al. , An integrated workflow to assess technical and biological variability of cell population frequencies in human peripheral blood by flow cytometry. J. Immunol 2017. 198, 1748–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hedley BD and Keeney M, Technical issues: flow cytometry and rare event analysis. International Journal of Laboratory Hematology. 2013. 35, 344–350. [DOI] [PubMed] [Google Scholar]
- 73.Dendrou CA, Plagnol V, Fung E, Yang JH, Downes K, Cooper JD et al. , Cell-specific protein phenotypes for the autoimmune locus IL2RA using a genotype-selectable human bioresource. Nat. Genet 2009. 41, 1011–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee J, Spidlen J, Boyce K, Cai J, Crosbie N, Dalphin M et al. , MIFlowCyt: The minimum information about a Flow Cytometry Experiment. Cytometry A. 2008. 73 926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mathieu C, Lahesmaa R, Bonifacio E, Achenbach P and Tree T, Immunological biomarkers for the development and progression of type 1 diabetes. Diabetologia. 2018. 61, 2252–2258. [DOI] [PubMed] [Google Scholar]
- 76.Habib T, Long SA, Samuels PL, Brahmandam A, Tatum M, Funk A et al. , Dynamic immune phenotypes of B and T helper cells mark distinct stages of T1D progression. Diabetes. 2019. 68, 1240–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Park LM, Lannigan J and Jaimes MC, OMIP-069: Forty-color full spectrum flow cytometry panel for deep immunophenotyping of major cell subsets in human peripheral blood. Cytometry A. 2020. 97, 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sahir F, Mateo J, Steinhoff M & Siveen K Development of a 43 color panel for the characterization of conventional and unconventional T-cell subsets,B cells, NK cells,monocytes, dendritic cells, and innate lymphoid cells using spectral flow cytometry. Cytometry A. 2020. 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rodriguez L, Pekkarinen PT, Lakshmikanth T, Tan Z, Consiglio CR, Pou C et al. Systems-level immunomonitoring from acute to recovery phase of severe COVID-19. Cell Reports Medicine. 2020. 1, 100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Geanon D, Lee B, Kelly G, Handler D, Upadhyaya B, Leech J et al. , A streamlined CyTOF workflow to facilitate standardized multi-site immune profiling of COVID-19 patients. medRxiv. 2020. 99, 446–461. [Google Scholar]
- 81.Diggins KE, Serti E, Muir VS, Rosasco MG, Lu T, Balmas E et al. , Exhausted-like CD8 T cell phenotypes linked to C-peptide preservation in alefacept-treated T1D subjects. JCI Insight. 2020. 6, e142680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Damond N, Engler S, Zanotelli VRT, Schapiro D, Wasserfall CH, Kusmartseva I et al. , A map of human type 1 diabetes progression by imaging mass cytometry. Cell Metab. 2019. 29, 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang YJ, Traum D, Schug J, Gao L, Liu C, Atkinson MA et al. , Multiplexed in situ imaging mass cytometry analysis of the human endocrine pancreas and immune system in type 1 diabetes. Cell Metabolism. 2019. 29, 769–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rahman AH and Homann D, Mass cytometry and type 1 diabetes research in the age of single-cell data science. Current Opinion in Endocrinology, Diabetes, and Obesity. 2020. 27, 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peterson VM, Zhang KX, Kumar N, Wong J, Li L, Wilson DC et al. , Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol 2017. 35, 936–939. [DOI] [PubMed] [Google Scholar]
- 86.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H et al. , Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 2017. 14, 865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A et al. , Integrated analysis of multimodal single-cell data. bioRxiv. 2020. 184, 3573–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kang H, Subramaniam M, Targ S, Nguyen M, Maliskova L, McCarthy E et al. , Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol 2018. 36 89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eberhardt M, Lai X, Tomar N, Gupta S, Schmeck B, Steinkasserer A et al. , Third-kind encounters in biomedicine: immunology meets mathematics and informatics to become quantitative and predictive. Methods in Molecular Biology. 2016. 1386, 135–179. [DOI] [PubMed] [Google Scholar]
- 90.Saeys Y, Van Gassen S and Lambrecht BN, Computational flow cytometry: Helping to make sense of high-dimensional immunology data. Nat. Rev. Immunol 2016. 16, 449–462. [DOI] [PubMed] [Google Scholar]
- 91.Spidlen J and Brinkman RR, Use FlowRepository to share your clinical data upon study publication. Cytometry Part B, Clinical Cytometry. 2018. 94, 196–198. [DOI] [PubMed] [Google Scholar]
- 92.Bhattacharya S, Dunn P, Thomas C, Smith B, Schaefer H, Chen J et al. , ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Scientific data. 2018. 5, 180015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee JA, Spidlen J, Boyce K, Cai J, Crosbie N, Dalphin M et al. , MIFlowCyt: The minimum information about a Flow Cytometry Experiment. Cytometry A. 2008. 73, 926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alhadj AM, Liu YF, Arif S, Tatovic D, Shariff HA, Gibson VB et al. , Metabolic and immune effects of immunotherapy with proinsulin peptide in human new-onset type 1 diabetes. Sci. Transl. Med 2017. 9, eaaf7779. [DOI] [PubMed] [Google Scholar]
- 95.Edner NM, Heuts F, Thomas N, Wang CJ, Petersone L, Kenefeck R et al. , Follicular helper T cell profiles predict response to costimulation blockade in type 1 diabetes. Nat. Immunol 2020. 21, 1244–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD et al. , Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 2015. 162, 184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Clarke DJB, Wang L, Jones A, Wojciechowicz ML, Torre D, Jagodnik KM et al. , FAIRshake: Toolkit to evaluate the FAIRness of research digital resources. Cell Systems. 2019. 9, 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roederer M, How many events is enough? Are you positive? Cytometry A. 2008. 73 384–385. [DOI] [PubMed] [Google Scholar]
- 99.Liechti T and Roederer M, OMIP-060: 30-Parameter flow cytometry panel to assess T cell effector functions and regulatory T cells. Cytometry A. 2019. 95 1129–1134. [DOI] [PubMed] [Google Scholar]
- 100.Rodriguez L, Pekkarinen P, Lakshmikanth T, Tan Z, Consiglio C, Pou C et al. , Systems-level immunomonitoring from acute to recovery phase of severe COVID-19. Cell Reports Medicine. 2020. 1 100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hao Y, Hao S, Andersen-Nissen E, Mauck W, Zheng S, Butler A et al. , Integrated analysis of multimodal single-cell data. Cell. 2021. S0092-8674 00583–00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kotecha N, Krutzik P and Irish J, Web-based analysis and publication of flow cytometry experiments. Current Protocols in Cytometry. 2010. Chapter 10:Unit10.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bendall S, Simonds E, Qiu P, e-A A, Krutzik P, Finck R et al. , Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011. 332 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bruggner R, Bodenmiller B, Dill D, Tibshirani R and Nolan G, Automated identification of stratifying signatures in cellular subpopulations. Proc Natl Acad Sci U S A. 2014. 111 E2770–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Van Gassen S, Callebaut B, Van Helden M, Lambrecht B, Demeester P, Dhaene T et al. , FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 2015. 87 636–645. [DOI] [PubMed] [Google Scholar]
- 106.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD et al. , Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015, 162. 184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Arvaniti E and Claassen M, Sensitive detection of rare disease-associated cell subsets via representation learning. Nat. Commun 2017. 8, 14825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hu Z, Jujjavarapu C, Hughey J, Andorf S, Lee H, Gherardini P et al. , MetaCyto: A tool for automated meta-analysis of mass and flow cytometry data. Cell Rep. 2018. 24 1377–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hartmann F, Babdor J, Gherardini P, Amir E, Jones K, Sahaf B et al. , Comprehensive immune monitoring of clinical trials to advance human immunotherapy. Cell Rep. 2019. 28 819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.