Abstract
Molecules and combinations of molecules are the natural communication currency of microbes; microbes have evolved and been engineered to sense a variety of compounds, often with exquisite sensitivity. The availability of microbial biosensors, combined with the ability to genetically engineer biological circuits to process information, make microbes attractive bionanomachines for propagating information through molecular communication (MC) networks. However, MC networks built entirely of biological components suffer a number of limitations. They are extremely slow due to processing and propagation delays and must employ simple algorithms due to the still limited computational capabilities of biological circuits. In this work, we propose a hybrid bio-electronic framework which utilizes biological components for sensing but offloads processing and computation to traditional electronic systems and communication infrastructure. This is achieved by using tools from the burgeoning field of optogenetics to trigger biosensing through an optoelectronic interface, alleviating the need for computation and communication in the biological domain.
Keywords: Molecular communication, Optogenetics, Biological nanomachines, Bio-electronic framework, Hardware → Bio-embedded electronics;, Networks → Network protocol design
1. INTRODUCTION
Conventional electromagnetic- (EM) based communication systems embed information in the properties of EM waves. In contrast, molecular communication (MC) networks use molecules to encode, transmit, and receive information. At the highest level of abstraction, MC networks propagate information from a source to a destination using MC links composed of a transmitter, channel, and receiver [1, 2]. MC networks are suitable for communication at small scale and in environments where EM-based communication is inefficient or impossible. Microorganisms are attractive components in MC networks as they naturally use molecules and combinations of molecules (such as peptides and proteins) to communicate; they are molecular transceivers [3].
Microbes have evolved to rapidly sense their environmental conditions, and therefore they contain a vast array of biosensing proteins. These biosensors are often very specific, as most biosensors involve binding of the target molecule to an active site with a complementary shape that has been carefully tuned through natural selection. Typically, natural biosensors contain both sensing and reporter domains, with the reporting domain generating an intracellular signal that drives the cellular response. The natural repertoire of microbial biosensors has been further expanded by successes in protein engineering and synthetic biology [4]. Various underlying natural biomolecular scaffolds, including RNA and protein, have been engineered to detect a broad array of inputs including small molecules and proteins with increased specificity, sensitivity, and dynamic range [5].
Due to the innate capabilities of cells to sense important biological compounds, particularly compounds relevant for disease diagnosis and environmental monitoring, they are attractive as sensing components for interfacing molecular communication networks with the natural world. However, attempting to engineer all aspects of an MC network with cells or cellular components comes with particular challenges. While cells and organisms are amazing biological computers in their natural milieu, the capabilities of engineered biological circuits (despite much progress in the field of synthetic biology) are still more limited [4]. To circumvent this challenge, in this work we propose Bio-electronic network, a new framework that leverages the innate sensing capabilities of biological compounds with the power of electrical and electronic systems.
The bio-electronic framework takes advantage of the strength of biological sensors while offloading the processing and computation to electronic systems and EM networks. Specifically, we propose a bio-electronic network with biosensors controlled by an external light stimulus. We utilize tools from the burgeoning field of optogenetics to engineer light-control of the desired sensor. The light stimulus acts as an external controller that turns the biosensor on or off. The ability to light-gate sensing reduces the burden on the bio-transmitter to implement complex communication algorithms.
While optogenetics has been a steadily developing field in the last several decades [6], to the best of our knowledge, no optogenetic methodologies have been used to resolve outstanding problems in developing MC networks. In this work, we present an introduction to the power of this technology and its potential for alleviating some of the bottlenecks in MC networks.
The remainder of this paper is organized as follows. In Section 2, we provide an overview of the systemic and algorithmic challenges in building a molecular communication network. We introduce Bio-electronic network in Section 3, followed by an overview of optogenetics, and a proposed experimental setup of our bio-electronic network. We also discuss how our framework can simplify some of the open challenges in MC networks. In Section 4, we identify the opportunities for future research in this new framework that can further improve and make it practical for real-time biosensing, and conclude the paper in Section 5.
2. BACKGROUND ON CHALLENGES IN MOLECULAR COMMUNICATION SYSTEMS
Communication between sensors, and to the external world is essential to build an autonomous network; it enables the sensors to convey information in real-time. Existing research on MC system identifies three building blocks, in line with digital communication systems viz., a transmitter, a receiver, and a channel. The transmitter consists of a Sensor, Processing unit with or without Storage, Modulator, followed by the Channel that allows information to be carried through it to a Receiver that consists of a Demodulator, Decoder, Processor. Figure 1 summarizes the broad categories of MC research [1, 2]. In this section we provide an overview of progress in each of the modules, both in system design and communication algorithms, and identify the open research problems.
Figure 1:
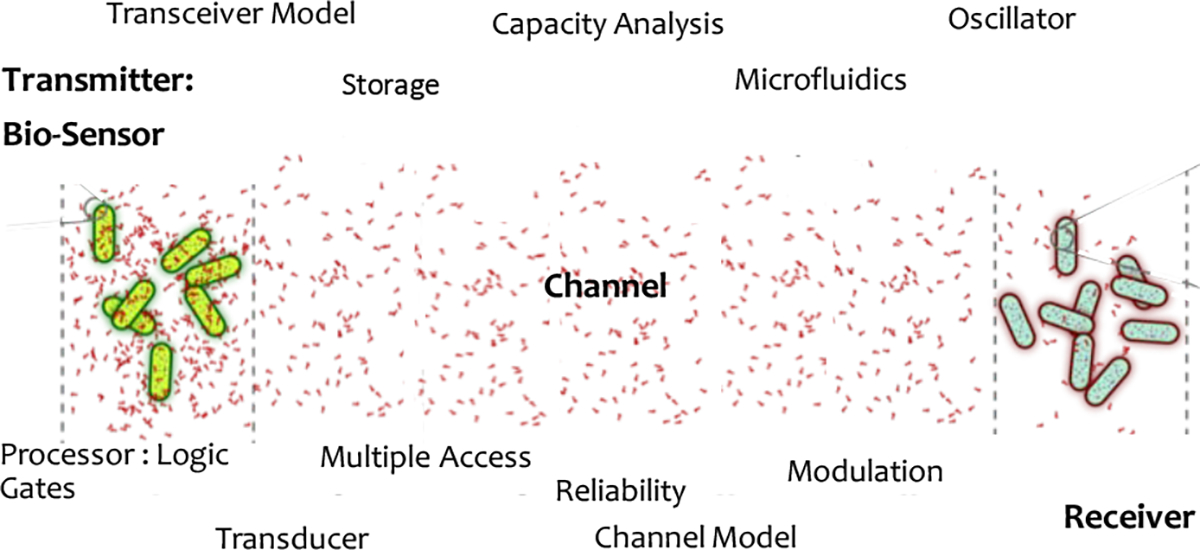
Molecular Communications : An overview
2.1. Building blocks of MC system
Bio-Sensor.
Biological sensors can reach and in turn provide access to domains that are otherwise a mystery to the conventional electromagnetic world. Efforts in synthetic biology and protein engineering [7, 8] have developed sensors to detect toxic chemicals [9], water quality [10, 11], pathogen contamination [12], human hormones [7, 13] and cancerous cell growth [14] among many others.
In an MC network, the transmitter, which is typically co-located with the sensor, is event-triggered i.e., it initiates communication when the sensed signal is above a threshold. Therefore, the sensor remains “on” all the time. An always-on biological sensor must incorporate mechanisms to maintain the properties of the sensor. For whole-cell sensors, this includes the need for mechanisms to maintain cell viability while controlling population size, which in turn require the hardware such as the microfluidic chip to be capable of adapting to these dynamic changes [15].
Processing unit.
A sensor is followed by a processing unit, with optional storage units, that maps sensed signal to information units, and a modulator to embed information to chemical signal. Biological circuits to emulate the function of NAND and NOR gates (the building blocks of a digital processor) have been developed in-lab and are promising to build a bio-processing unit [16, 17]. Not only do the gates with increasing inputs display low fidelity, interconnecting gates with fewer inputs in series or in parallel remains an open research problem due to the high latency and stochasticity of biological circuits [18–20] (though see [21] for a summary of recent progress). Preliminary studies on integrating multiple gates have been shown to be unstable in the presence of multiple layers and changing environments [22–24]. Similarly, access to storage or memory remains an open problem. DNA as a biological memory or storage unit has been studied and implemented in laboratories; but read and write operations on the storage unit using another bio-circuit remains an unsolved and a challenging problem. Despite the feasibility of constructing biological gates and memory, realizing complex biological circuits in practice requires significant advancements in synthetic biology.
Channel.
A variety of channel models have been proposed and validated for MC channels [1]. MC channels can be broadly classified into diffusion-based, flow-based, and active transport based paths. A number of works focusing on the modeling of the above identified type of channels have been developed and validated experimentally. Research on microfludic chips/arrays to house and carry information molecules have resulted in a range of channel geometries and architecture [25, 26]. In case of a living biosensor, the channel must also allow nutrients to pass through, but not the transceivers themselves. In a flow-based propagation, channel geometry and length also determine the maximum propagation speed, in turn limiting the distance [15].
Despite promising advancements in the individual modules to envision a biological communication system, interconnecting the individual blocks remains an open problem. Therefore, communication algorithms for molecular systems must be designed to overcome the challenges of the underlying system.
2.2. Algorithms for MC networks
While MC has similarities to other communication systems in the architecture and communication paradigms, it differs significantly in channel conditions, channel geometry, complexity, stability, availability, and feasibility of system hardware. Most importantly, MC utilizes chemical molecules as carrier signals to communicate. These differences have driven the need for novel algorithms and protocols for molecular communication. Here, we summarize the open problems in molecular communication and briefly discuss the approaches that have been proposed to address the above problems as well as the challenges in implementing them in practice.
Information theoretic analysis.
Early research on MC focused on the theoretical analysis of the new communication paradigm [27, 28]. Following the path of EM communication, researchers derived maximum achievable rates/capacity for various channel models [29, 30]. Based on experimental results, channel models were developed [31] to represent and simulate a MC channel for varying transmitter, receiver, and channel conditions. Despite the varieties of works on the theoretical analysis, development of a unified channel model that is consistent and reproducible remains an open area of research.
Physical Layer.
A practical deployment of a communication link requires transmission of data from one end to the other. Studies of data particles [32], modulation schemes to encode information [33, 34], encoders and decoders [35, 36], and models for simulating the behavior of biological transceivers [37, 38] broadly fall in the category of physical layer approaches to MC. Majority of the transceiver models and the algorithms are specific to an experimental setup and are difficult to generalize and recreate.
Link Layer.
When more than one transmitter communicates to the receiver and/or multiple senders communicate to multiple receivers, the need for addressing and medium access control (MAC) arises. [39] provides an overview of addressing and MAC algorithms designed for MC networks. MAC protocols that utilize the type of signal [40], spatial diversity [41], or characteristics of the signal [39] have been proposed and studied. Systems requirements to implement these solutions is a bottleneck for practical deployment. Scalability and the associated challenges remain unexplored.
Network Layer.
An increase in the distance between the sender and the receiver in traditional communication systems result in multiple hops between transceivers, in need of routing algorithms [42]. Engineering existing routing algorithms on biological circuits will be challenging due to the complexity of the algorithms and need for feedback, among other issues [43]. Reliability mechanisms in traditional systems to ensure accurate delivery of information are also challenging to adapt to MC systems as the reliability of the engineered bio-circuits decreases with increasing complexity [44]. Error correction codes [45] that are simple and practical have been proposed for MC. Although these reliability mechanisms provide codes targeting MC channel noise, errors introduced by the underlying system and its dynamic changes make them impractical.
It can be noted that a majority of the innovations in communication algorithms requires and relies on the ability of biological circuits to perform computation as well as exchange information using chemical molecules. Despite the advancements in bio-engineering and synthetic biology, biological circuitry to consistently perform designed tasks remains the bottleneck. The modular approach of traditional communication systems is therefore non-ideal for molecular systems and MC algorithms. An integrated approach that incorporates the constraints and optimizes the systemic and algorithmic design would be beneficial for practical biological communication.
3. BIO-ELECTRONIC NETWORKS
To overcome these drawbacks, we propose Bio-Electronic network, a framework that integrates the strength of biosensors with the power of electronics and optogenetics. A bio-electronic network consists of a biosensor, biological transmitter, an electronic receiver, and an opto-electronic stimulus/trigger as illustrated in Figure 2.
Figure 2:
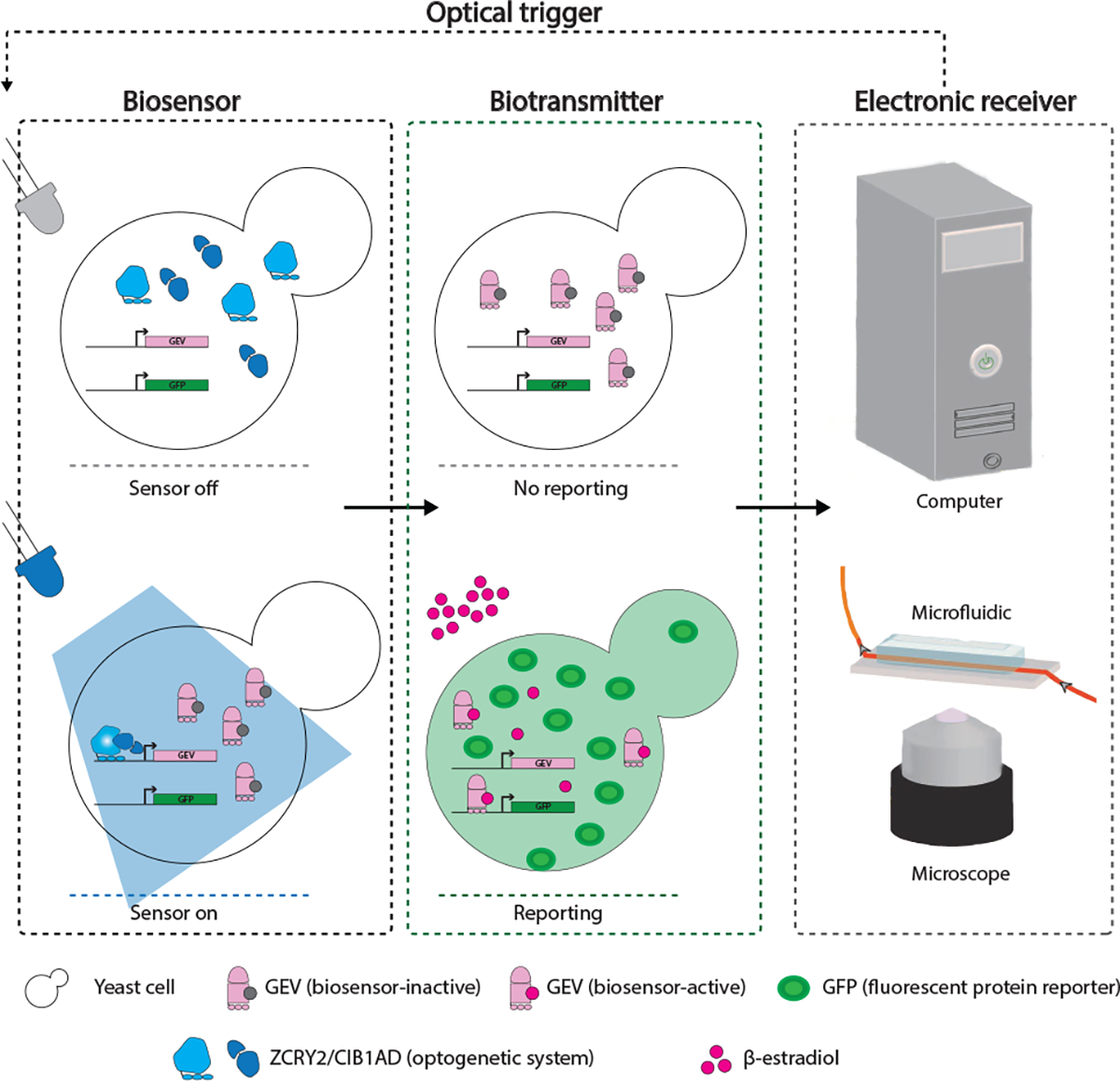
Illustration of optogenetic MC scheme
Biosensors are the driving force to the inception of biological communication networks. Sensors give networks access to new domains, and communication allows this information to be relayed. Bio-electronic network retains the benefits of a biosensor, without overwhelming the biological circuitry with computation and communication needs. The majority of the challenges identified in Section 2 arise from the need for synthetically engineered biological computation circuits. A bio-electronic network eliminates this need by using an external trigger that drives the biosensor.
We propose an optical stimulus to a light sensitive biosensor. In its default operation, the sensor remains in sleep or idle mode, and is woken up by an external light trigger which guides the sensor to sense. On trigger, the sensed information is encoded, modulated, and transmitted, which is then received by an electronic receiver. A bio-electronic network therefore offloads computational and communication complexity to electronic circuitry while biological circuitry is dedicated for sensing. We believe that bio-electronic networks not only make biosensors accessible and practical, but are also a step towards fully autonomous biological networks. In the rest of the section, we provide an overview of optogenetics that enables us to envision a practical bio-electronic network. We also discuss our in-house experimental setup as a proof-of-concept for optical trigger, biosensing, transmission, and reception.
3.1. Introduction to Optogenetics
The ability to turn a biosensor on or off at specific times or in specific populations of cells would alleviate some of the challenges of a purely biological MC network. Stimuli such as pH, temperature, and chemical inducers are frequently used to control biological processes and could be used to turn a biosensor on or off. Such control could happen either directly, through activation or inhibition of the sensor, or indirectly through control of gene expression or protein circuits capable of regulating the sensor. Chemically induced systems, in particular, are routinely used in basic research and synthetic biology to control microbial cells [8]. As discussed in Section 2, chemical stimuli suffer several disadvantages. Chemical inducers are difficult to add and remove from cultures of cells, and the precision of spatial control is limited by fluid handling, compound lifetime, and diffusion. Additionally, pH, temperature, and certain chemical inducers often have pleiotropic effects on cells beyond regulating the desired process. This is particularly severe in microbes, which, as single-celled organisms have evolved to be exquisitely sensitive to changes in the environment.
Light provides an attractive alternative to most commonly employed stimuli. Light can be precisely controlled in space and time, with tunable wavelength and intensity. Compared to chemical inducers, light is inexpensive and can be controlled with readily available electronic and optical components [46–48]. The control of cellular processes with genetically encoded light-sensitive proteins is called ‘optogenetics’ [6, 49]. Light-sensitive proteins have evolved naturally in many organisms, such as plants and light-dependent microbes, to sense and respond to light [50]. These naturally occurring proteins have different sensitivities to both the wavelength and intensity of light, with many proteins sensitive to blue (~ 450nm), red (~ 650nm) and far-red (~ 750nm) wavelengths [51]. Optogenetics takes advantage of these naturally occurring light-sensitive proteins to actuate processes inside of a cell.
Optogenetic tools were first developed for control of ion-flux in neuroscience [6]. Light-sensitive transmembrane ion-channel proteins called opsins, primarily from microbes, are used in neuroscience to control neural activity by changing the action potential across the cell membrane. The development of hardware and algorithms for optogenetically controlling neuronal activity has been extensively discussed elsewhere [52], and is not the focus of this work. Non-neural optogenetics, which is generally not focused on controlling ion flux, primarily uses nonopsin photoactivatable proteins or gene switches to develop light-sensitive tools to control cellular processes such as gene expression and signaling [51]. Optogenetic tools have been shown to be effective in controlling a variety of cellular processes, from gene regulation to protein localization, in microbes such as Escherichia coli and Saccharomyces cerevisiae [53].
3.2. Adapting Optogenetics to Bio-electronic networks
We propose to utilize optogenetic tools to turn specific biosensors on (or off). This requires the light stimulus to change the activity of a sensor protein either by directly modifying it or by changing its expression or localization within the cell. There are several routes to implementing optogenetic control of a biosensor. Optogenetic tools [49, 53] that have been developed to drive gene expression could be used to change the expression level of a biosensor protein. Tools that control molecular targeting signals within a cell can be used to change the localization of a biosensor protein. Since a biosensor’s location in the cell (e.g. at the plasma membrane) often determines its activity, this light-based targeting also serves to control activity. Light-induced clustering of biosensors could also be used to control their activity. More advanced protein engineering can be used to control the activity of a biosensor itself by incorporating a light sensitive protein into the structure of the biosensor protein. Finally, optogenetic approaches for targeting specific proteins for degradation could be used to reduce the level of a biosensor, only allowing sensing under illumination conditions that allow biosensor expression levels to recover.
The range of optogenetic tools available for manipulating protein concentration and protein activity in microbes [54, 55] make it feasible to practically employ this technology in molecular communication networks to control the activity of biosensors and the sensing state of the system. This opens up exciting possibilities for using an optoelectronic interface to control sensing status, thus moving most computation and processing into conventional electronic systems and requiring only sensing from the biological components.
3.3. Experimental setup and methods
Integrating optogenetics into MC networks requires that an optical trigger be able to access cells to modify their sensing properties. These cells must then be able to sense a signal and transmit it to a receiver. The receiver must further be capable of coordinating the optical trigger to determine the sensing properties desired at a given moment based on the underlying communication algorithm.
The explosion of optogenetic technologies in the last several decades, both biological technologies for generating light-dependent cellular behavior [54–56] and instrumentation technologies for stimulating cells with light [46–48], have made an optogenetically-controlled sensor scheme decidedly possible. However, to date none of these methodologies have been put into practice to address the outstanding problems in developing MC networks.
We present here a scheme for light-based gating of sensing capabilities that requires feasible modifications to an existing scheme for computer-controlled light-triggering of yeast cells in culture [57, 58]. The updated scheme is outlined in Figure 2. Yeast, specifically Saccharomyces cerevisiae are an excellent sensing organism as they have been engineered to sense a wide variety of compounds, are a standard chassis organism in synthetic biology amenable to further engineering, are generally non-pathogenic, and can survive a variety of environments, including in vivo niches in the human body and harsh environmental conditions [7].
Sensor:
One of the most prolific classes of yeast biosensors are based on heterologous expression of human steroid receptors. The human estrogen receptor (hER) has been known to function in yeast since 1988 [59] and further efforts have developed additional human receptors into yeast biosensors of human hormones and hormone disruptors [7, 60, 61]. These sensors have been applied to test compounds or environmental samples for endocrine disruptive compounds (EDCs) that pose a significant risk to human health [62].
Optical Stimulus:
Yeast cells as estrogen sensors is controlled by exposure to blue light. This would be achieved by putting protein expression of a yeast biosensor for estrogen, for example the GEV sensor [13], under the control of the ZCRY2/CIB1AD optogenetic system. In this optogenetic system, dimerization of a split transcription factor (ZCRY2/CIB1AD) is controlled by blue light (through CRY2/CIB1 binding) and genes put under the control of a responsive promoter (e.g. pZF-GEV) are expressed in response to blue light [56, 63]. In this way, yeast would only be capable of sensing estrogen after exposure to blue light.
Receiver/Readout Mechanism:
When GEV is expressed, it is sequestered in the cytoplasm of the yeast cell by a chaperone complex (Hsp90). Estrogen is sensed by displacing Hsp90, allowing GEV to localize to the nucleus of the yeast cells, where it binds a GEV-responsive promoter to drive expression of a fluorescent protein. Thus, estrogen levels are converted to a fluorescent signal.
In keeping with a previously published scheme [57, 58], yeast carrying the GEV biosensor under the control of the ZCRY2/CIB1AD optogenetic system would be cultured in a bioreactor that is illuminated by a blue (450nm) LED. Estrogen or EDCs present in the media would be sensed by the yeast cells in culture when the GEV biosensor is present. The corresponding fluorescence signal could be read out by several methods, including by automatically sampling yeast into a microfluidic device and using fluorescence microscopy, as was previously reported [57].
Our experimental setup with a biosensor that is triggered using blue light and read-out using fluorescence is a promising step towards the feasibility of a bio-electronic network. It is not only a new framework for biosensing and data collection, it is a stepping stone towards an fully autonomous biological network. This framework simplifies many of the open communication problems in MC by simplifying the hardware requirements on the biological domain. In the rest of the section, we describe in detail two of the challenges in MC identified in Section 2 and discuss how they are simplified in a bio-electronic network.
3.4. Simplifying MC using Optogenetics
Addressing and Medium Access Control.
A bio-electronic network simplifies addressing and MAC by outsourcing it to the external trigger. The optical trigger specifically turns on and off the biosensor and hence does not require additional addressing. Consider a group of biosensors deployed spatially. The optical trigger will focus on one sensor at a time and hence the sensor does not have to convey address with the information. The external trigger can thus implement other MAC protocols without requiring any coordination between the senders. The external control therefore simplifies the system design as well as the communication algorithm design.
Routing and Reliability.
In a bio-electronic network, routing is performed on the receiver end to steer the trigger to the corresponding sensor. The sensor is thus unaware of the network and eliminates overheads to determine the best route. Reliability mechanisms, similarly, can be offloaded to the electronic domain, greatly simplifying the bioengineering requirements. The receiver, aware of the sensor that is triggered is responsible for resolving data from the sensor. Feedback requirements of traditional reliability protocols are replaced by follow up stimulus from the external controller.
Distributed Sensing.
The power of light controlled sensing comes in allowing parallel sensing without additional overheads. Consider the case of wanting to sense multiple EDCs. Yeast biosensors for a variety of human EDCs have been developed [64, 65]. A mixed culture of yeast cells, each containing a specific biosensor could be grown in the bioreactor. By serially deciding which sensor should be on (using different colored LEDs as stimuli ), the output for each sensor (e.g. fluorescence) could be the same. Light can also be exquisitely controlled in space. If individual sensors are spatially separated, this would allow all of the yeast colonies to report their information using the same signal and channel, as the receiver could deconvolve this by knowing which biosensors had been turned on.
4. OPPORTUNITIES IN BIO-ELECTRONIC NETWORK
As we have discussed so far, a bio-electronic network offloads computation and communication to the receiver/controller and the biosensor is only responsible for sensing and encoding information. This framework opens up a number of new research opportunities including developing communication techniques to enhance the throughput and delay performance, diversifying the sensors and environments, improving the scale of the network, and simplifying hardware for practical deployments. We discuss some of the opportunities in system design and algorithmic design in detail in this section.
4.1. Communication Algorithms for Bio-Electronic Networks
Modulation.
Modulation techniques developed for MC can be applied here to encode information in response to a light trigger. The proposed framework offers a larger set of parameters to modulate the data such as properties of the colony, characteristics of the output signal, and optical properties of the colony, among others.
Scalability.
The proposed framework allows us to collect data from a large number of sensors without any coordination on the transmitter end. Thus, the receiver must be capable of coordinating the trigger and resolving the responses. Research in the areas of queuing theory, cooperative communication, and parallel processing can be adapted to maximize the scale and hence the throughput. Optimizing the number of sensors that can be triggered and processed in parallel in turn will increase the throughput of the network.
Cross-talk and interference.
While techniques from synthetic biology and optogenetics can allow biosensor activity to be controlled by a specific wavelength of light, difficulties start to arise when we want to control multiple sensors with different wavelengths. There is crosstalk between the light-responsive proteins used to construct optogenetic systems, in that they may respond to a range of light wavelengths [51]. This could result in crosstalk or interference. Error correction mechanisms on the receiver side to overcome these effects can significantly improve the reliability of a bio-electronic network. Communication techniques to leverage spatial diversity and co-operative sensing that are aware of interference can be developed to overcome this challenge.
Optical Network Architecture.
We envision a bio-electronic network to consist of an array of optical triggers interacting with a large set of biosensors. Network architecture design to optimize the number of optical trigger systems for a given array of sensors and vice versa will be needed for practical deployments. Compressed sensing, and graph coloring techniques adapted to incorporate the latency due to switching and response time can pave the way for a large-scale, practical bio-electronic network.
4.2. Future research in Optogenetics and Biological Implementation
Management of live cell biosensors.
The external stimulus in a bio-electronic network lets the sensor remain in idle mode except while sensing. In order to allow sensors to rapidly wake up in response to a trigger, dormant and senescent cells must be avoided. Recent developments in synthetic biology technologies that provide this capability can reduce the latency of the sensing system [66, 67]. Microfluidic chip design to handle cell cultures and ensure constant colony size will improve the usability of this framework. Future research on optogenetic or chemical protocols to set sensors back to sleep will also improve the sensing system.
Spectrum expansion.
Optogenetic tools are built around naturally light-sensitive proteins. There are a limited number of wavelengths to which these proteins respond, due to chromophore properties [51], which reduces the number of distinct wavelengths that can be combined independently. Recent successes in hybrid optogenetic circuitry that responds to the rate and intensity of light, in addition to the wavelength [68] open up the possibility of controlling multiple sensors with the same color of light. Further development of these approaches will allow a limited number of light wavelengths to control a much larger number of sensors
Biological sensor stability.
Bio-electronic networks simplify MC by reducing the engineering burden on the sensor. There is still room for improvement in the stability of the biosensor. Expressing heterologous protein biosensors can be expensive for cells, incentivizing them to lose the sensor through mutation, resulting in inaccuracies. Research into approaches that more carefully account for resource allocation to reduce the deleterious effect of heterologous protein expression [69] and engineering stem-cell like regenerative properties into microbial communities are some of the possible future research directions to ensure sensor stability [66, 67].
Expanding biological readouts.
Fluorescence is a well received readout owing to its ease-of-use in laboratories. The latest low cost measurement technologies show promise for practical use [70]. Expanding the repertoire of readouts beyond fluorescence would allow the output of biosensors to be measured with more rapid, inexpensive, and miniaturizable technology. Potential advantageous readouts include colony growth, luminescence, and production of hydrogen ions that can be measured using a pH electrode [71].
Harnessing microbial diversity.
Currently, a limited number of chassis organisms that can be easily engineered are used for sensing. However, non-domesticated microbes provide advantages over the commonly used organisms; they contain unique natural sensors and are adapted for a range of environments and environmental stressors [72]. Approaches that are expanding the synthetic biology toolkit into non-model organisms [73] will allow these cells to be plugged in as sensors, potentially expanding the environmental niches into which bio-electronic networks can be deployed.
5. CONCLUSION
In this work, we proposed bio-electronic network, a new framework to realize biological communication networks. Our framework consists of an external controller/trigger, a biosensor, and an electronic receiver. Specifically, we propose an optical controller that uses light stimulus to turn on (or off) a light sensitive protein which in turn initiates sensing. Such a network offloads computation and communication complexity to the receiver, which has access to electrical and electronic circuitry to process. This framework leverages advancements in optogenetics that have proven light-stimulus based biosensors to be a viable and efficient option. We identified the challenges in using chemical stimulus for sensing, computation, and communication. We also present a proof-of-concept experimental setup for optical stimulus and a light-sensitive biosensor that can be generalized to other sensors and wavelengths. We also identify potential research opportunities to make a bio-electronic network easy-to-use and practical. We strongly believe that a bio-electronic network is a step towards a fully autonomous biological network.
Contributor Information
Bhuvana Krishnaswamy, University of Wisconsin-Madison.
Megan N. McClean, University of Wisconsin-Madison
REFERENCES
- [1].Nariman Farsad, Yilmaz Birkan H, Eckford Andrew, Chae Chan-Byoung, and Guo Weisi. A comprehensive survey of recent advancements in molecular communication. IEEE Communications Surveys & Tutorials, 2016. [Google Scholar]
- [2].Krishnaswamy Bhuvana. Algorithms for molecular communication networks. PhD thesis, Georgia Institute of Technology, 2018. [Google Scholar]
- [3].Akyildiz Ian F, Pierobon Massimiliano, and Balasubramaniam Sasitharan. Moving forward with molecular communication: from theory to human health applications. Proceedings of the IEEE, 107(5):858–865, 2019. [Google Scholar]
- [4].Slomovic Shimyn, Pardee Keith, and Collins James J. Synthetic biology devices for in vitro and in vivo diagnostics. Proceedings of the National Academy of Sciences of the United States of America, 112:14429–14435, November 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Snoek Tim, Chaberski Evan K, Ambri Francesca, Kol Stefan, Bj?rn Sara P, Pang Bo, Barajas Jesus F, Welner Ditte H, Jensen Michael K, and Keasling Jay D. Evolution-guided engineering of small-molecule biosensors. Nucleic acids research, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Boyden Edward S. A history of optogenetics: the development of tools for controlling brain circuits with light. F1000 biology reports, 3, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Adeniran Adebola, Sherer Michael, and Tyo Keith E J. Yeast-based biosensors: design and applications. FEMS yeast research, 15, February 2015. [DOI] [PubMed] [Google Scholar]
- [8].Ford Tyler J and Silver Pamela A. Synthetic biology expands chemical control of microorganisms. Current opinion in chemical biology, 28:20–28, October 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Moser Felix, Horwitz Andrew, Chen Jacinto, Lim Wendell, and Voigt Christopher A. Genetic sensor for strong methylating compounds. ACS synthetic biology, 2:614–624, October 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Trang Pham Thi Kim, Berg Michael, Viet Pham Hung, Van Mui Nguyen, and van der Meer Jan Roelof. Bacterial bioassay for rapid and accurate analysis of arsenic in highly variable groundwater samples. Environmental science & technology, 2005. [DOI] [PubMed] [Google Scholar]
- [11].van der Meer Jan Roelof and Belkin Shimshon. Where microbiology meets microengineering: design and applications of reporter bacteria. Nature reviews. Microbiology, 8:511–522, July 2010. [DOI] [PubMed] [Google Scholar]
- [12].Nguyen C, Makkar R, Sharp NJ, Page MA, Molineux IJ, and Schofield DA. Detection of bacillus anthracis spores from environmental water using bioluminescent reporter phage. Journal of applied microbiology, 123:1184–1193, November 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Scott McIsaac R, Silverman Sanford J, McClean Megan N, Gibney Patrick A, Macinskas Joanna, Hickman Mark J, Petti Allegra A, and Botstein David. Fast-acting and nearly gratuitous induction of gene expression and protein depletion in saccharomyces cerevisiae. Mol Biol Cell, 22(22):4447–4459, Nov 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chien Tiffany, Doshi Anjali, and Danino Tal. Advances in bacterial cancer therapies using synthetic biology. Current opinion in systems biology, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Austin Caitlin Marie. Dynamics of molecular communication in bacteria within microfluidic environments. PhD thesis, Georgia Institute of Technology, 2016. [Google Scholar]
- [16].Tamsir Alvin, Tabor Jeffrey J, and Voigt Christopher A. Robust multicellular computing using genetically encoded nor gates and chemical ‘wires’. Nature, 469(7329):212, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Siuti Piro, Yazbek John, and Lu Timothy K. Synthetic circuits integrating logic and memory in living cells. Nature biotechnology, 31(5):448, 2013. [DOI] [PubMed] [Google Scholar]
- [18].Khalil Ahmad S and Collins James J. Synthetic biology: applications come of age. Nature Reviews Genetics, 11(5):367–379, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Marchisio Mario A and Stelling Jörg. Automatic design of digital synthetic gene circuits. PLoS computational biology, 7(2), 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Xiang Yiyu, Dalchau Neil, and Wang Baojun. Scaling up genetic circuit design for cellular computing: advances and prospects. Natural computing, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].McBride Cameron, Shah Rushina, and Vecchio Domitilla Del. The effect of loads in molecular communications. Proceedings of the IEEE, 107(7):1369–1386, 2019. [Google Scholar]
- [22].Lin Xiaodong, Liu Yaqing, Deng Jiankang, Lyu Yanlong, Qian Pengcheng, Li Yunfei, and Wang Shuo. Multiple advanced logic gates made of dna-ag nanocluster and the application for intelligent detection of pathogenic bacterial genes. Chemical science, 9(7):1774–1781, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bradley Robert W, Buck Martin, and Wang Baojun. Recognizing and engineering digital-like logic gates and switches in gene regulatory networks. Current opinion in microbiology, 33:74–82, 2016. [DOI] [PubMed] [Google Scholar]
- [24].Miyamoto Takafumi, Razavi Shiva, DeRose Robert, and Inoue Takanari. Synthesizing biomolecule-based boolean logic gates. ACS synthetic biology, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Luo Xiaolong, Wu Hsuan-Chen, Tsao Chen-Yu, Cheng Yi, Betz Jordan, Payne Gregory F, Rubloff Gary W, and Bentley William E. Biofabrication of stratified biofilm mimics for observation and control of bacterial signaling. Biomaterials, 33(20):5136–5143, 2012. [DOI] [PubMed] [Google Scholar]
- [26].Luo Xiaolong, Tsao Chen-Yu, Wu Hsuan-Chen, Quan David N, Payne Gregory F, Rubloff Gary W, and Bentley William E. Distal modulation of bacterial cell–cell signalling in a synthetic ecosystem using partitioned microfluidics. Lab on a Chip, 15(8):1842–1851, 2015. [DOI] [PubMed] [Google Scholar]
- [27].Pierobon Massimiliano and Akyildiz Ian F. A physical end-to-end model for molecular communication in nanonetworks. IEEE Journal on Selected Areas in Communications, 28(4):602–611, 2010. [Google Scholar]
- [28].Pierobon Massimiliano and Akyildiz Ian F. Information capacity of diffusion-based molecular communication in nanonetworks. In 2011 Proceedings IEEE INFOCOM, pages 506–510. IEEE, 2011. [Google Scholar]
- [29].Farsad Nariman, Murin Yonathan, Eckford Andrew W, and Goldsmith Andrea. Capacity limits of diffusion-based molecular timing channels with finite particle lifetime. IEEE Transactions on Molecular, Biological and Multi-Scale Communications, 4(2):88–106, 2018. [Google Scholar]
- [30].Kim Na-Rae, Farsad Nariman, Lee Changmin, Eckford Andrew W, and Chan-Byoung Chae. An experimentally validated channel model for molecular communication systems. IEEE Access, 7:81849–81858, 2019. [Google Scholar]
- [31].Guo Weisi, Asyhari Taufiq, Farsad Nariman, Yilmaz H Birkan, Li Bin, Eckford Andrew, and Chae Chan-Byoung. Molecular communications: Channel model and physical layer techniques. IEEE Wireless Communications, 2016. [Google Scholar]
- [32].Hiyama Satoshi and Moritani Yuki. Molecular communication: Harnessing biochemical materials to engineer biomimetic communication systems. Nano Communication Networks, 1(1):20–30, 2010. [Google Scholar]
- [33].Kuran Mehmet S, Yilmaz Huseyin Birkan, Tugcu Tuna, and Akyildiz Ian F. Modulation techniques for communication via diffusion in nanonetworks. In 2011 IEEE international conference on communications (ICC), pages 1–5. IEEE, 2011. [Google Scholar]
- [34].Krishnaswamy Bhuvana, Austin Caitlin M, Bardill J Patrick, Russakow Daniel, Holst Gregory L, Hammer Brian K, Forest Craig R, and Sivakumar Raghupathy. Time-elapse communication: Bacterial communication on a microfluidic chip. IEEE Transactions on Communications, 61(12):5139–5151, 2013. [Google Scholar]
- [35].Fang Yuting, Noel Adam, Yang Nan, Eckford Andrew W, and Kennedy Rodney A. Symbol-by-symbol maximum likelihood detection for cooperative molecular communication. IEEE Transactions on Communications, 2019. [Google Scholar]
- [36].Mai Trang C, Egan Malcolm, Duong Trung Q, and Renzo Marco Di. Event detection in molecular communication networks with anomalous diffusion. IEEE Communications Letters, 21(6):1249–1252, 2017. [Google Scholar]
- [37].Vasconcelos Marcos M, Mitra Urbashi, Câmara Odilon, Gangan Manasi S, and Boedicker James. A continuous-time decision-making model for bacterial growth via quorum sensing: theory and evidence. In Proceedings of the Sixth Annual ACM International Conference on Nanoscale Computing and Communication, 2019. [Google Scholar]
- [38].Pierobon Massimiliano. A systems-theoretic model of a biological circuit for molecular communication in nanonetworks. Nano Comm. Networks, 2014. [Google Scholar]
- [39].Krishnaswamy Bhuvana, Jian Yubing, Austin Caitlin M, Perdomo Jorge E, Patel Sagar C, Hammer Brian K, Forest Craig R, and Sivakumar Raghupathy. Adma: Amplitude-division multiple access for bacterial communication networks. IEEE Transactions on Molecular, Biological and Multi-Scale Communications, 2017. [Google Scholar]
- [40].Atakan Baris and Akan Ozgur B. Single and multiple-access channel capacity in molecular nanonetworks. In International Conference on Nano-Networks, 2009. [Google Scholar]
- [41].Moore Michael J and Nakano Tadashi. Addressing by beacon distances using molecular communication. Nano Communication Networks, 2011. [Google Scholar]
- [42].Kurose James F and Ross Keith W. Computer networking, 1991. [Google Scholar]
- [43].Balasubramaniam Sasitharan et al. Opportunistic routing through conjugation in bacteria communication nanonetwork. Nano Communication Networks, 2012. [Google Scholar]
- [44].Katz Evgeny. Biomolecular information processing: from logic systems to smart sensors and actuators. John Wiley & Sons, 2013. [Google Scholar]
- [45].He Peng, Mao Yuming, Liu Qiang, and Yang Kun. Improving reliability performance of diffusion-based molecular communication with adaptive threshold variation algorithm. International Journal of Communication Systems, 2016. [Google Scholar]
- [46].Gerhardt Karl P, Olson Evan J, Castillo-Hair Sebastian M, Hartsough Lucas A, Landry Brian P, Ekness Felix, Yokoo Rayka, Gomez Eric J, Ramakrishnan Prabha, Suh Junghae, et al. An open-hardware platform for optogenetics and photobiology. Scientific Reports, 6(35363), 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Bugaj Lukasz J and Lim Wendell A. High-throughput multicolor optogenetics in microwell plates. Nature protocols, 14:2205–2228, July 2019. [DOI] [PubMed] [Google Scholar]
- [48].Sweeney Kieran, Morales Neydis Moreno, Burmeister Zachary, Nimunkar Amit J, and McClean Megan N. Easy calibration of the light plate apparatus for optogenetic experiments. MethodsX, 6:1480–1488, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mansouri Maysam, Strittmatter Tobias, and Fussenegger Martin. Light-controlled mammalian cells and their therapeutic applications in synthetic biology. Advanced science (Weinheim, Baden-Wurttemberg, Germany), 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Repina Nicole A, Rosenbloom Alyssa, Mukherjee Abhirup, Schaffer David V, and Kane Ravi S. At light speed: Advances in optogenetic systems for regulating cell signaling and behavior. Annual review of chemical and biomolecular engineering, 8:13–39, June 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kolar Katja, Knobloch Christian, Stork Hendrik, Žnidarič Matej, and Weber Wilfried. Optobase: A web platform for molecular optogenetics. ACS synthetic biology, 2018. [DOI] [PubMed] [Google Scholar]
- [52].Wirdatmadja Stefanus Arinno, Barros Michael Taynnan, Koucheryavy Yevgeni, Jornet Josep Miquel, and Balasubramaniam Sasitharan. Wireless optogenetic nanonetworks for brain stimulation: Device model and charging protocols. IEEE transactions on nanobioscience, 16(8):859–872, 2017. [DOI] [PubMed] [Google Scholar]
- [53].Zhang Kai, Duan Liting, Ong Qunxiang, Lin Ziliang, Varman Pooja Mahendra, Sung Kijung, and Cui Bianxiao. Light-mediated kinetic control reveals the temporal effect of the raf/mek/erk pathway in pc12 cell neurite outgrowth. PloS one, 9(3):e92917, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Liu Zedao, Zhang Jizhong, Jin Jiao, Geng Zilong, Qi Qingsheng, and Liang Quanfeng. Programming bacteria with light-sensors and applications in synthetic biology. Frontiers in microbiology, 9:2692, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Salinas Francisco, Rojas Vicente, Delgado Verónica, Agosin Eduardo, and Larrondo Luis F. Optogenetic switches for light-controlled gene expression in yeast. Applied microbiology and biotechnology, 101(7):2629–2640, 2017. [DOI] [PubMed] [Google Scholar]
- [56].An-Adirekkun Jidapas My, Stewart Cameron J, Geller Stephanie H, Patel Michael T, Melendez Justin, Oakes Benjamin L, Noyes Marcus B, and McClean Megan N. A yeast optogenetic toolkit (yotk) for gene expression control in saccharomyces cerevisiae. Biotechnology and bioengineering, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Melendez Justin, Patel Michael, Oakes Benjamin L., Xu Ping, Morton Patrick, and McClean Megan N.. Real-time optogenetic control of intracellular protein concentration in microbial cell cultures. Integr Biol (Camb), 6(3), Mar 2014. [DOI] [PubMed] [Google Scholar]
- [58].Stewart Cameron J and McClean Megan N. Design and implementation of an automated illuminating, culturing, and sampling system for microbial optogenetic applications. JoVE (Journal of Visualized Experiments), (120), 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Metzger D, White JH, and Chambon P. The human oestrogen receptor functions in yeast. Nature, 334, July 1988. [DOI] [PubMed] [Google Scholar]
- [60].Sanseverino John, Eldridge Melanie L, Layton Alice C, Easter James P, Yarbrough Jason, Schultz Terry Wayne, and Sayler Gary S. Screening of potentially hormonally active chemicals using bioluminescent yeast bioreporters. Toxicological sciences: an official journal of the Society of Toxicology, 2009. [DOI] [PubMed] [Google Scholar]
- [61].Rajasärkkä Johanna, Hakkila Kaisa, and Virta Marko. Developing a compound-specific receptor for bisphenol a by directed evolution of human estrogen receptor α. Biotechnology and bioengineering, 108(11):2526–2534, 2011. [DOI] [PubMed] [Google Scholar]
- [62].Gore AC, Chappell VA, Fenton SE, Flaws JA, Nadal A, Prins GS, Toppari J, and Zoeller RT. Edc-2: The endocrine society’s second scientific statement on endocrine-disrupting chemicals. Endocrine reviews, 36:E1–E150, December 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, and Tucker CL. Rapid blue-light-mediated induction of protein interactions in living cells. Nature Methods, 7:973–975, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Xu Tingting, Young Anna, Narula Jasleen, Sayler Gary, and Ripp Steven. High-throughput analysis of endocrine-disrupting compounds using blyes and blyas bioluminescent yeast bioassays. In Bioluminescent Imaging. Springer, 2020. [DOI] [PubMed] [Google Scholar]
- [65].Shiizaki Kazuhiro, Asai Shota, Ebata Shingo, Kawanishi Masanobu, and Yagi Takashi. Establishment of yeast reporter assay systems to detect ligands of thyroid hormone receptors α and β. Toxicology in Vitro, 24(2):638–644, 2010. [DOI] [PubMed] [Google Scholar]
- [66].Mushnikov Nikolai V, Fomicheva Anastasia, Gomelsky Mark, and Bowman Grant R. Inducible asymmetric cell division and cell differentiation in a bacterium. Nature chemical biology, 15:925–931, September 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Molinari Sara, Shis David L, Bhakta Shyam P, Chappell James, Igoshin Oleg A, and Bennett Matthew R. A synthetic system for asymmetric cell division in escherichia coli. Nature chemical biology, 15:917–924, September 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Benzinger Dirk and Khammash Mustafa. Pulsatile inputs achieve tunable attenuation of gene expression variability and graded multi-gene regulation. Nature communications, 9(1):3521, 2018. Printed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Darlington Alexander P S, Kim Juhyun, Jim?nez Jos? I, and Bates Declan G. Dynamic allocation of orthogonal ribosomes facilitates uncoupling of co-expressed genes. Nature communications, 9:695, February 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Dai Bo, Jiao Ziao, Zheng Lulu, Bachman Hunter, Fu Yongfeng, Wan Xinjun, Zhang Yule, Huang Yu, Han Xiaodian, Zhao Chenglong, Huang Tony Jun, Zhuang Songlin, and Zhang Dawei. Colour compound lenses for a portable fluorescence microscope. Light, science & applications, 8:75, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Grebenstein L, Kirchner J, Peixoto RS, Zimmermann W, Irnstorfer F, Wicke W, Ahmadzadeh A, Jamali V, Fischer G, Weigel R, Burkovski A, and Schober R. Biological optical-to-chemical signal conversion interface: A small-scale modulator for molecular communications. IEEE Transactions on NanoBioscience, 2019. [DOI] [PubMed] [Google Scholar]
- [72].Russell James J, Theriot Julie A, Sood Pranidhi, Marshall Wallace F, Landweber Laura F, Fritz-Laylin Lillian, Polka Jessica K, Oliferenko Snezhana, Gerbich Therese, Gladfelter Amy, Umen James, Bezanilla Magdalena, Lancaster Madeline A, He Shuonan, Gibson Matthew C, Goldstein Bob, Tanaka Elly M, Hu Chi-Kuo, and Brunet Anne. Non-model model organisms. BMC biology, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Yan Qiang and Fong Stephen S. Challenges and advances for genetic engineering of non-model bacteria and uses in consolidated bioprocessing. Frontiers in microbiology, 8:2060, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]


