Abstract
The simple structure of phosphatidic acid (PA) belies its complex biological functions as both a key phospholipid biosynthetic intermediate and a potent signaling molecule. In the latter role, PA controls processes including vesicle trafficking, actin dynamics, cell growth, and migration. However, experimental methods to decode the pleiotropy of PA are sorely lacking. Because PA metabolism and trafficking are rapid, approaches to accurately visualize and manipulate its levels require high spatiotemporal precision. Here, we describe recent efforts to create a suite of chemical tools that enable imaging and perturbation of PA signaling. First, we describe techniques to visualize PA production by phospholipase D (PLD) enzymes, which are major producers of PA, called Imaging Phospholipase D Activity with Clickable Alcohols via Transphosphatidylation (IMPACT). IMPACT harnesses the ability of endogenous PLD enzymes to accept bioorthogonally tagged alcohols in transphosphatidylation reactions to generate functionalized reporter lipids that are subsequently fluorescently tagged via click chemistry. Second, we describe two light-controlled approaches for precisely manipulating PA signaling. Optogenetic PLDs use light-mediated heterodimerization to recruit a bacterial PLD to desired organelle membranes, and photoswitchable PA analogs contain azobenzene photoswitches in their acyl tails, enabling molecular shape and bioactivity to be controlled by light. We highlight select applications of these tools for studying GPCR–Gq signaling, discovering regulators of PLD signaling, tracking intracellular lipid transport pathways, and elucidating new oncogenic signaling roles for PA. We envision that these chemical tools hold promise for revealing many new insights into lipid signaling pathways.
Keywords: click chemistry, optogenetics, phosphatidic acid, phospholipase D, lipid signaling, chemical biology, bioorthogonal chemistry, induced proximity, photoswitchable lipids
Abbreviations: DAG, diacylglycerol; ER, endoplasmic reticulum; FACS, fluorescence-activated cell sorting; GPCR, G protein-coupled receptor; IEDDA, inverse electron-demand Diels–Alder; IMPACT, Imaging Phospholipase D Activity with Clickable Alcohols via Transphosphatidylation; LPA, lysophosphatidic acid; NAPE, N-acylphosphatidylethanolamine; optoPLD, optogenetic PLD; PA, phosphatidic acid; PC, phosphatidylcholine; PLC, phospholipase C; PLD, phospholipase D; RT-IMPACT, real-time IMPACT; RTK, receptor tyrosine kinase; TCO, trans-cyclooctene
Lipids have many important functions in cells, including as major components of membranes, compounds for energy storage, and messenger molecules for signal transduction (1). Lipids are typically classified by their head group and backbone, with each class containing multiple species with variable composition of their acyl tails. Signaling lipids often have pleiotropic effects, which can arise both from this acyl chain diversity and also from what we term locational diversity, that is, the diversity of subcellular localizations that most lipids can assume (2). In cell signaling pathways, protein localizations are carefully regulated by various factors, and proper lipid localization — both to and within the correct membrane — is crucial for enabling the intended lipid–protein interactions to occur. Both the hydrophilic head group and lipid acyl chains can be important in determining protein-binding affinities (3) and biophysical properties of lipids, such as lateral and flip-flop movements in the bilayer (4, 5), which affect their localization. As a result, it is critical to study lipids in native, cellular contexts containing their structural and locational diversity to decipher their pleiotropic roles in signaling. Consequently, tools that can visualize and manipulate lipid production with high specificity, in a physiological milieu, are essential for lipid research (6).
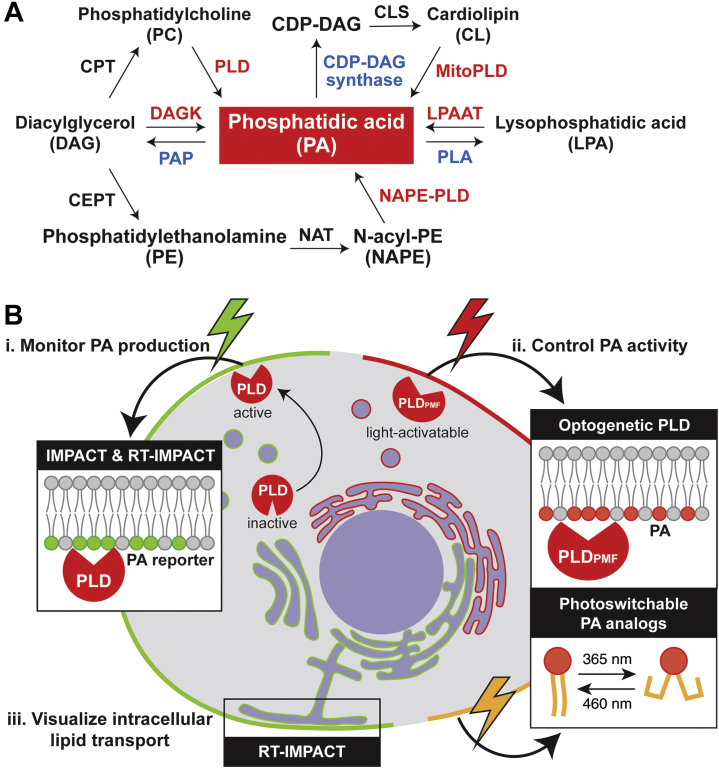
Phosphatidic acid (PA) is among the simplest glycerophospholipids. It mediates several functions via lipid–protein interactions and its unique cone-shaped geometry (7, 8, 9), and its metabolism is under a complex set of controls in mammalian cells (Fig. 1A) (10, 11, 12). PA is produced via three main pathways: (1) hydrolysis of phosphatidylcholine (PC) by phospholipase D (PLD), (2) phosphorylation of diacylglycerol (DAG) by diacylglycerol kinase, and (3) acylation of lysophosphatidic acid (LPA) by lysophosphatidic acid acyltransferase. A small amount of PA can also be formed in certain contexts via (4) hydrolysis of N-acylphosphatidylethanolamine (NAPE) by NAPE-specific PLD and (5) hydrolysis of cardiolipin by mitochondrial PLD (13, 14). Further metabolism of PA is catalyzed via three other pathways: (1) dephosphorylation to DAG by PA phosphatase/lipin, (2) hydrolysis to LPA by phospholipase A, and (3) conversion to CDP-DAG by CDP-DAG synthase. These enzymes, with their isoform-specific localizations and acyl chain preferences (10, 15), diversify the pools of PA in cells and thus contribute to its pleiotropy.
Figure 1.
Overview of chemical biology approaches to image and control PA signaling.A, PA metabolic pathways in mammalian cells. B, a summary of tools described in this article. IMPACT and RT-IMPACT visualize the active pools of PLD enzymes via fluorescent phospholipid reporters produced by PLD. Optogenetic PLD (optoPLD) and photoswitchable PA analogs enable spatiotemporal control of bioactive PA pools. RT-IMPACT can also be used for tracing intracellular phospholipid trafficking pathways. CEPT, choline/ethanolamine phosphotransferase; CL, cardiolipin; CLS, Cardiolipin synthase; CPT, choline phosphotransferase; DAGK, diacylglycerol kinase; IMPACT, Imaging Phospholipase D Activity with Clickable Alcohols via Transphosphatidylation; LPAAT, lysophosphatidic acid acyltransferase; MitoPLD, mitochondrial phospholipase D; NAPE-PLD, N-acylphosphatidylethanolamine specific phospholipase D; PAP, phosphatidic acid phosphatase; PE, phosphatidylethanolamine; PLA, phospholipase A; PLC, phospholipase C; PLD, phospholipase D.
Several recent findings have reinvigorated the field, helping us to better understand PA production and metabolism, notably the first high-resolution structures of mammalian PLDs (16, 17) and lipin (18). Still, many questions remain unanswered relating to how these enzymes affect different pools of PA and, in turn, how distinct PA pools are responsible for various cellular functions. As is the case for other pleiotropic lipids, tools for monitoring and perturbing specific pools of PA would be crucial for answering these questions. However, a major hurdle is the interconnectedness of the different PA metabolic pathways, as perturbations to individual steps are often compensated by changes to flux in other steps, much as a blocked road in a dense urban environment results in traffic being rerouted down other streets. Therefore, to minimize interference from such endogenous, homeostatic regulation, ideal tools should meet these criteria: (1) they should work rapidly, giving cells less time to accommodate the changes, and (2) they should be orthogonal to cellular systems, minimally influencing endogenous processes.
Chemical tools have the potential to shine in this arena. Relative to genetic tools, chemical tools can have much higher temporal resolution and, in certain cases, specificity. Small-molecule inhibitors act more rapidly than genetic manipulations such as gene knockdown and knockout, and they can have better orthogonality by separating catalytic functions of target enzymes from noncatalytic roles (e.g., protein–protein interactions). In the realm of PA metabolism, isoform-selective PLD inhibitors (19) and protein-based fluorescent PA-binding probes (20, 21, 22) are very useful tools to understand loss-of-function effects of specific PA pools and to visualize the total cellular population of PA. However, these tools alone do not have enough specificity to answer several important questions: Where are PLD-derived pools of PA, as opposed to pools made by other routes, synthesized? How do cells regulate PLD activity? How do distinct, spatiotemporally defined pools of PA affect specific cellular processes?
To answer these questions, we have harnessed the power of synthetic chemistry and protein design and evolution to develop a suite of chemical biology tools for studying PA biology (Fig. 1B). The first set of methods enables visualization of the locations where PLD-derived pools of PA are produced. These approaches, termed IMPACT, for Imaging PLD Activity with Clickable Alcohols via Transphosphatidylation, combine chemoenzymatic labeling and click chemistry tagging to image and quantify the sites of PLD activity within live cells (23, 24, 25, 26). The second set of complementary enzyme- and small molecule–based tools enables spatiotemporal manipulation of PA signaling activity triggered by light (27, 28). The first of these uses an engineered, light-activated PLD to produce PA at specific subcellular locations, and the second uses photoswitchable PA analogs, whose shape and bioactivity is directly controlled by a light-mediated isomerization reaction. Throughout the review, we will highlight applications of these tools, showcasing their ability to elucidate biological functions of PA and, more generally, highlighting the power of bringing a chemical biology mindset to dissect dynamic lipid signaling pathways.
IMPACT: Tools to visualize PLD-mediated PA signaling
The functions of the two PLD isoforms primarily responsible for PA signaling, PLD1 and PLD2, are highly regulated in space and time (29). In addition to translocating to different subcellular locations in response to extracellular signals, PLD1 can also alter its catalytic activity through protein–protein interactions, protein–lipid interactions, and phosphorylation (29). Therefore, the localizations of the PLD enzymes themselves do not necessarily reflect the sites where they are actively producing PA. A method to selectively reveal the locations of active PLDs would allow determination of where these enzymes are initiating PA signaling events.
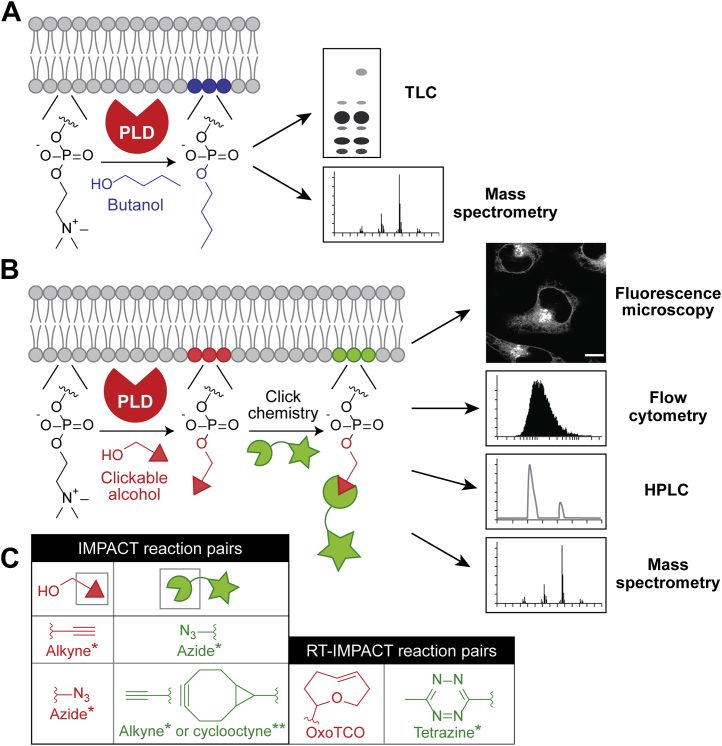
To achieve such an activity-based imaging method, we harnessed the unusually promiscuous reactivity of PLD enzymes. PLDs produce PA via hydrolysis of PC, releasing choline as a byproduct (30). When supplied with short, aliphatic primary alcohols, PLDs use these instead of water as a nucleophile to perform a transphosphatidylation of PC, where the alcohol replaces choline as the lipid head group. This reaction is traditionally used to label the PLD-derived pools of lipids using 1-butanol, 1-propanol, or ethanol (31). The resulting transphosphatidylation products, termed phosphatidyl alcohols, are typically analyzed by TLC or mass spectrometry, a bulk biochemical measurement (Fig. 2A). Yet, this lipid reporter of PLD activity is generated in situ on organelle membranes bearing active, endogenous PLD enzymes. We sought to develop a method capitalizing upon transphosphatidylation but which enabled detection of the phosphatidyl alcohol products within live cells. Such a method would reveal both the localization and amplitude of PLD activity at single-cell, and even subcellular, resolution. In our approach, termed IMPACT, primary alcohols with reactive, bioorthogonal handles are used as PLD substrates. The lipid products with these groups are subsequently tagged with appropriately labeled fluorescent probes, enabling detection by numerous methods.
Figure 2.
Schematic depictions of classical PLD activity assays and IMPACT.A, classical PLD activity assay using butanol followed by the detection of phosphatidyl butanol product by TLC or LC–MS, in combination with radiolabeling or stable isotope labeling, respectively. B, IMPACT using a bioorthogonally tagged alcohol for subsequent ligation with a detectable tag via click chemistry. Customizable tags enable single-cell readouts (i.e., fluorescence microscopy and flow cytometry) as well as bulk biochemical analyses. C, structures of alcohols and clickable detection tags used in IMPACT and RT-IMPACT. Asterisks denote commercial availability of IMPACT reagents. ∗, compounds that are commercially available, including various alkynols, azido alcohols, and alkynyl, azido, and tetrazine-conjugated fluorophores. ∗∗, compounds with similar functionalities to those used for IMPACT that are commercially available. (i.e., though, in our studies, we have used a bicyclononyne-functionalized BODIPY dye that is not commercially available (24), fluorescently conjugated dibenzocyclooctynes, which can also react with azide-tagged biomolecules via SPAAC, are commercially available.) The scale bar represents 10 μm. IMPACT, Imaging Phospholipase D Activity with Clickable Alcohols via Transphosphatidylation; PLD, phospholipase D.
We began by investigating primary alcohols bearing traditional click chemistry handles, alkynols and azido alcohols, and found that both are efficiently accepted by PLDs (23, 24). The resulting lipids can then be ligated to fluorescent molecules using the Cu(I)-catalyzed azide-alkyne cycloaddition or strain-promoted azide-alkyne cycloaddition reactions, which are performed in fixed or live cells, respectively (Fig. 2, B and C). The fluorescent signal in each cell can be detected by microscopy or flow cytometry, both of which enable single-cell analysis of PLD activity. Through this study, we found a striking heterogeneity in PLD activity across the cells in the same population (24), which could not have been previously observed using classic butanol transphosphatidylation assays. This finding suggests a complex regulation of PLD activity at the single-cell level within populations and highlights the importance of using reporters that enable single-cell analysis.
Instead of performing the click chemistry tagging reaction in cells, we also showed that the lipid product can be isolated and tagged in vitro for detection by HPLC, which enables a nonradioisotope-based, precise quantification of bulk PLD activity (23, 24). Alternatively, the extracted lipids can be tagged with a quaternary ammonium group, which improves detection by electrospray ionization-based mass spectrometry, to identify lipid species by LC–MS and determine their acyl chain compositions (24). Because of the versatility of click chemistry and the sensitivity of these detection methods, the concentration of alcohol used in IMPACT is typically 1 mM, which is low enough that the transphosphatidylation reaction does not outcompete PLD-mediated hydrolysis. Thus, IMPACT does not perturb endogenous PA production by PLDs.
Moreover, in addition to simple, linear alkynols and azido alcohols, we also found that other functionalized alcohols, including an alkynyl choline analog and a bifunctional, photocrosslinkable diazirine alkyne alcohol, can be accepted by PLDs at decent efficiencies (32, 33). The alkynyl choline analog allowed either selective labeling of PLD activity or de novo PC biosynthesis via the Kennedy pathway by using different labeling protocols (32, 34), making it a versatile reporter for two different biosynthetic pathways. The diazirine alkyne alcohol enables the creation in situ of dual-functionalized photocrosslinkable and clickable lipid reporters of PLD activity. These lipids bear structural similarity to phosphatidyl ethanol, a PLD-derived lipid metabolite of ethanol that accumulates in human serum following alcohol consumption. By performing photocrosslinking, click chemistry tagging, and enrichment of lipid–protein complexes, we have identified several protein interactors of these lipids as a first step toward elucidating potential (patho)physiological functions of phosphatidyl ethanol and other phosphatidyl alcohols (33).
Whereas IMPACT is an effective tool to monitor PLD activity at the single-cell and bulk population levels, we made unexpected observations when imaging the subcellular localizations of PLD activity using fluorescence microscopy. We found that IMPACT labeling deriving from both basal and stimulated PLD activities appeared primarily at the endoplasmic reticulum (ER) and Golgi apparatus, as well as minor endosomal and lysosomal pools (24). Notably, no IMPACT labeling was observed at the plasma membrane (e.g., Fig. 2B), where PLD2 and, under certain circumstances, stimulated PLD1 are reported to localize (29, 35). We speculated that the lack of plasma membrane IMPACT labeling might arise from rapid trafficking of fluorescent lipids between organelle membranes occurring during the IMPACT procedure. The reaction kinetics of strain-promoted azide-alkyne cycloaddition tagging and the need for rinsing away excess fluorophore set a lower limit for the labeling time of 20 min, and we hypothesized that, during this time, lipids potentially generated at the plasma membrane were trafficking to intracellular organelles. To solve this issue, we designed a rapid version of IMPACT, which we have termed real-time IMPACT (RT-IMPACT), by taking advantage of an alternative click chemistry tagging reaction.
We found that a bulky, hydrophilic trans-cyclooctene (TCO)-containing primary alcohol could be accepted by PLDs, although with a lower efficiency than other simple alcohols (i.e., alkynols and azido alcohols) (25). Critically, the resultant TCO-containing lipids could be tagged with a fluorogenic tetrazine reagent via a no-rinse, inverse electron-demand Diels–Alder (IEDDA) reaction (36, 37), enabling their immediate visualization by confocal microscopy in real time, within seconds of administration of the tetrazine reagent. We screened various TCO compounds for high signal, low background, and negligible cell toxicity, and we found that trans-5-oxocene (38) performed appropriately (25). A short (3–5 min) incubation with the alcohol, followed by a real-time monitoring of the click chemistry tagging by time-lapse imaging, is sufficient to report on the PLD activity.
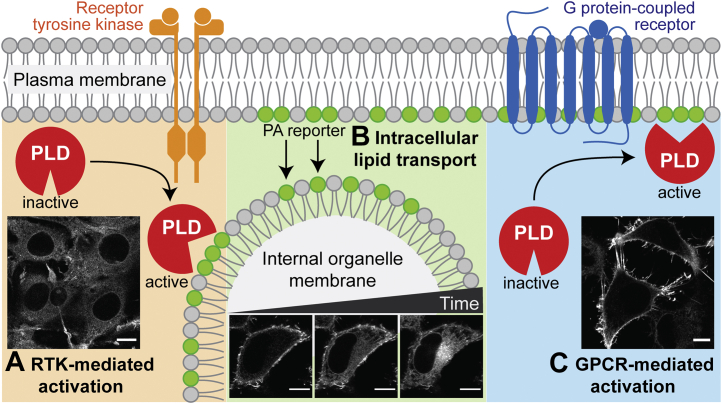
We then investigated if the temporal resolution of RT-IMPACT is high enough to capture the sites where reporter lipids are generated via transphosphatidylation before their trafficking to other organelle membranes (25). Excitingly, RT-IMPACT–derived reporter lipids were found predominantly at the plasma membrane when endogenous PLDs were stimulated with a phorbol ester, a strong stimulus of PKC, which activates PLDs at the plasma membrane (39). Strikingly, when we observed the fate of these fluorescent lipid reporters by time-lapse imaging over the next few minutes, we observed their rapid trafficking from the plasma membrane to the ER, and subsequently to the Golgi complex (Fig. 3B). Kinetics studies revealed that the trafficking of reporter lipids occurred more rapidly after the click chemistry tagging step, suggesting that the change in the head group upon IEDDA tagging to introduce the BODIPY moiety accelerated the phospholipid trafficking rate. Interestingly, the reporter lipids were internalized via apparent nonvesicular pathways rather than endocytosis, because the first intracellular destination of the IMPACT-derived fluorescent lipids was the ER, not endosomes. Thus, this finding not only highlights the importance of a rapid method for visualizing PLD-derived lipid probes but also suggests the application of RT-IMPACT for directly imaging and probing mechanisms of intracellular phospholipid transport.
Figure 3.
Applications of RT-IMPACT to visualize PLD signaling and intracellular phospholipid transport.A, RT-IMPACT reveals intracellular PLD activity downstream of PDGF receptor signaling, a prototypical receptor tyrosine kinase pathway. B, RT-IMPACT as a tool to visualize the rapid trafficking of BODIPY-tagged phosphatidyl alcohol reporters initially produced at the plasma membrane to internal organelle membranes. The time-lapse images are from 9, 30, and 75 s timepoints, respectively, of the fluorogenic click chemistry tagging reaction. C, RT-IMPACT reveals plasma membrane-associated PLD activity downstream of the muscarinic M1 receptor, a prototypical Gq-coupled GPCR. Confocal microscopy images are reproduced with permission from (25). The scale bars represent 10 μm. IMPACT, Imaging PLD Activity with Clickable Alcohols via Transphosphatidylation; GPCR, G protein-coupled receptor; PDGF, platelet-derived growth factor; PLD, phospholipase D.
Having validated RT-IMPACT, we set out to apply it to evaluate the subcellular localizations of PLD activity elicited by different physiological stimuli. PLD enzymes can receive input from multiple classes of cell-surface receptors, including G protein-coupled receptors (GPCRs) and receptor tyrosine kinases (RTKs) (35, 40). Both GPCRs and RTKs can stimulate PLDs via activation of several different intermediary, intracellular effectors, including those from the PKC, Rho GTPase, and Arf GTPase families (16, 41, 42, 43). As a result, they both lead to PLD-mediated PA biosynthesis but cause different physiological effects on cells. By performing RT-IMPACT with selective activators of GPCR and RTK signaling, we identified marked differences in the locations of PLD activity between two treatments (25). Stimulation of the M1 muscarinic receptor, a prototypical Gq-coupled GPCR, resulted in clear plasma membrane-localized IMPACT labeling, whereas stimulation of the platelet-derived growth factor receptor, a prototypical RTK, resulted in IMPACT labeling on predominantly intracellular membranes (Fig. 3, A and C). These results demonstrated that RT-IMPACT possesses the appropriate spatiotemporal resolution to pinpoint the diverse subcellular locations of endogenous PLD activity.
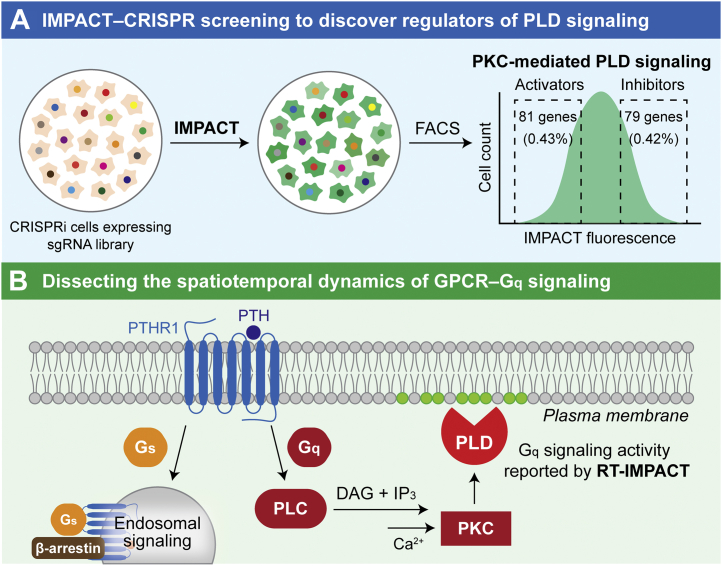
We have recently carried out additional studies that capitalize upon the single-cell and subcellular-level resolution of IMPACT and RT-IMPACT. The combination of IMPACT-based fluorescent labeling with fluorescence-activated cell sorting (FACS) is particularly powerful, as it enables enrichment of cells from a mixed population with high or low PLD activity. We envision applications to combine this tagging-and-enrichment strategy with single-cell technologies or other perturbations to understand the nature of cell-to-cell variance of PLD signaling that we have observed in a cancer cell line (24). Separately, we have recently used IMPACT-enabled FACS enrichment as a phenotypic selection step in a genome-wide CRISPR interference screen to elucidate new regulators of PLD signaling (Fig. 4A) (44). At a mechanistic level, these studies revealed glycogen synthase kinase 3 as a positive regulator of PKC-mediated PLD signaling via effects on de novo gene expression. Going forward, these and other IMPACT-enabled CRISPR screens have the potential to reveal new regulatory circuits controlling PLD signaling. Further, this work illustrates how our bioorthogonal, activity-based fluorescent tagging tool can expand the power of genome-wide CRISPR screening to elucidate mechanisms regulating a specific enzyme-driven signaling pathway in mammalian cells.
Figure 4.
Applications of IMPACT and RT-IMPACT to understand PLD-mediated cell signaling.A, IMPACT-enabled CRISPR screens have the potential to reveal new regulatory circuits controlling PLD signaling. CRISPRi cells expressing a dead Cas9–KRAB fusion and one member of a genome-wide sgRNA library are labeled with IMPACT, followed by FACS-based enrichment of the IMPACT-high and IMPACT-low cell populations. Cells with a higher extent of IMPACT labeling contain sgRNAs targeting putative PLD-inhibitory genes, and those with lower levels of IMPACT labeling contain sgRNAs targeting putative PLD-activating genes. Identities of such putative PLD inhibitors and activators are determined by next-generation sequencing of sgRNAs in enriched populations relative to each other or unsorted cells. B, RT-IMPACT enables visualization and quantification of the subcellular locations and duration of PLD signaling elicited by different stimuli. PTH-induced activation of PTHR1 leads to two separate downstream signaling pathways, Gs and Gq, that are spatiotemporally controlled. RT-IMPACT can be used to dissect the two signaling events by selectively revealing PLD activation that occurs downstream of Gq signaling. FACS, fluorescence-activated cell sorting; GPCR, G protein-coupled receptor; IMPACT, Imaging PLD Activity with Clickable Alcohols via Transphosphatidylation; PLC, phospholipase C; PLD, phospholipase D; PTHR1, parathyroid hormone receptor.
At the subcellular level, RT-IMPACT is poised to reveal long-sought answers to questions about the subcellular locations where PLD signaling takes place downstream of different stimuli. Initially, we examined these localizations for only a handful of stimuli, including the pleiotropic phorbol 12-myristate 13-acetate and well-established, canonical pathways such as a Gq-coupled GPCR (muscarinic M1 receptor) and an RTK (the platelet-derived growth factor receptor) (25). Many other agonists have been reported to activate PLDs (29), and an understanding of the localizations of particular PLD isoforms downstream of these stimuli would reveal important mechanistic information in these pathways. Toward this end, we recently used RT-IMPACT to carry out a mechanistic examination of the spatiotemporal dynamics of Gq signaling downstream of the parathyroid hormone receptor (Fig. 4B) (45). This GPCR, important in development and multiple diseases, signals via both Gs and Gq, and whereas the former can signal noncanonically from endosomes following internalization (46), the localization and kinetic behavior of the Gq pathway remained unknown. We validated RT-IMPACT as a faithful and selective reporter of Gq signaling and determined that such signaling occurs transiently and exclusively at the plasma membrane, upstream of sustained intracellular Gs signaling. We anticipate future applications of RT-IMPACT as a complementary tool to Ca2+ imaging for visualizing and quantifying GPCR–Gq signaling pathways.
Controlling PA signaling with light using optogenetic PLDs and photoswitchable PA analogs
A key complement to tools for visualizing spatiotemporally distinct pools of PA are approaches for perturbing these pools, with the goal of ascribing specific biological functions to them. Useful tools for loss-of-function studies include PLD knockout and a panel of potent pharmacological inhibitors of PLDs, both pan and isoform-selective (19, 47). Such tools allow identification of scenarios where PLD-mediated PA is necessary for downstream events. Gain of function would be optimal to enable tests for sufficiency, but it is harder to achieve using conventional approaches. To enable gain-of-function studies with high spatiotemporal control, we took two parallel approaches, one based on engineered proteins and the other small-molecule lipid mimetics.
First, to develop a tool to enable spatiotemporally defined production of PA, we focused on engineering a PLD whose localization could be controlled on demand. We desired an orthogonal system for PA production in cells, so we avoided using mammalian PLDs, whose activities are under complex regulation and whose noncatalytic functions are notable (29, 35). Therefore, we turned to a bacterial PLD from Streptomyces sp. PMF (PLDPMF), which shares catalytic activity with mammalian PLDs in generating PA via PC hydrolysis. Yet, PLDPMF has low overall homology with mammalian PLDs and lacks all reported regulatory domains, including PI(4,5)P2-binding pocket, N-terminal PKC interaction site, and C-terminal RhoA interaction site, and sites for eukaryotic posttranslational modifications such as palmitoylation (16). Thus, PLDPMF could potentially be an orthogonal catalyst in mammalian cells, one whose activity could be manipulated in an independent manner. Whereas the activation of mammalian endogenous PLDs by upstream effectors (e.g., PKC and Rho) is typically accompanied by additional pleiotropic effects, the activation of PLDPMF is bioorthogonal, enabling the dissection of the outcome of PA production from other effects of such pharmacological stimulation of endogenous PLD signaling.
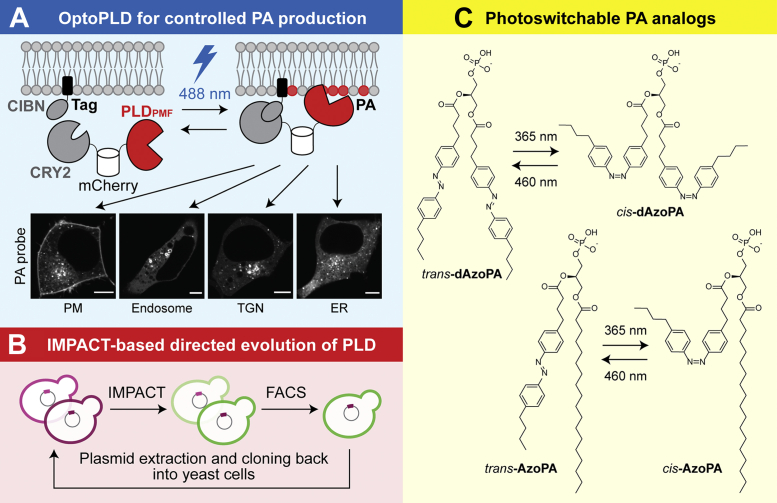
We found that untagged PLDPMF was localized in the cytosol — that is, not on any membrane-bound compartments — when expressed in mammalian cells. To enable recruitment of PLDPMF from the cytosol to a target membrane for activation, we applied an optogenetic dimerization system that uses CRY2 and CIBN (48). We fused CRY2 to PLDPMF together with the mCherry fluorescent protein. CIBN was fused, separately, to various genetically encoded, organelle-targeting tags to target it constitutively to a desired organelle membrane. When kept in the dark, CRY2 and CIBN remain dissociated. Upon activation by blue light, CRY2–CIBN dimerization brings PLDPMF to the desired organelle membrane (Fig. 5A). We termed this tool optogenetic PLD (optoPLD) and confirmed its ability to produce spatially and temporally regulated pools of PA by using a genetically encoded PA-binding probe (27).
Figure 5.
Light-controlled protein- and small-molecule-based gain-of-function tools to manipulate PA signaling.A, schematic depiction of optoPLD. CRY2–CIBN dimerization, triggered by blue light, brings PLDPMF to the desired organelle membrane. Organelle-specific PA production was confirmed using a genetically encoded PA probe. B, directed evolution platform for engineering PLD mutants with altered catalytic activities. A PLD library with random mutations was expressed in yeast cells, followed by FACS-based selection using IMPACT. C, chemical structures of azobenzene-containing photoswitchable PA analogs, AzoPA, and dAzoPA, whose acyl tail structures can be controlled by light. Confocal microscopy images are reproduced from Tei and Baskin (2020), originally published in Journal of Cell Biology (27). The scale bars represent 5 μm. ER, endoplasmic reticulum; FACS, fluorescence-activated cell sorting; IMPACT, Imaging PLD Activity with Clickable Alcohols via Transphosphatidylation; optoPLD, optogenetic phospholipase D; PA, phosphatidic acid; PM, plasma membrane; TGN, trans-Golgi network.
Similar to mammalian PLDs, PLDPMF catalyzes both hydrolysis and transphosphatidylation (49, 50); thus, IMPACT labeling is also useful for monitoring and quantifying optoPLD activity. We generated a PLD1/2 double knockout HEK293T (PLD1/2KO) cell line to eliminate any undesired signal from endogenous PLD activities. Using IMPACT coupled with flow cytometry in PLD1/2KO cells expressing optoPLD, we found that optoPLD exhibits light-dependent PA production, with a signal-to-noise ratio of greater than 10:1 for optoPLDs targeted to four different organelle locations (27). Moreover, via IMPACT coupled with LC–MS, we confirmed that optoPLD produces lipids with similar acyl chain compositions as mammalian PLDs, indicating that it can function as a mimic of mammalian PLDs and produce biologically relevant pools of PA in cells.
IMPACT enabled not only the characterization of our panel of optoPLDs but also the further engineering of the tool. By combining IMPACT with yeast-based directed evolution, we developed a selection system for engineering PLDPMF mutants with different catalytic efficiencies (Fig. 5B) (27). We expressed a PLDPMF library with randomly generated mutations in yeast cells, fluorescently labeled them using IMPACT, and subjected the cells to FACS to isolate cells with higher IMPACT labeling. The optoPLD mutants obtained from this selection exhibited wide-ranging activities both higher and lower than WT PLDPMF, which adds finer tunability to the system and would be potentially useful to reproduce different activation states of mammalian PLDs in cells. We are currently adapting this directed evolution strategy to a mammalian cell-based engineering platform to develop super-active optoPLD mutants with optimal properties for use in mammalian cells.
Although our collection of optoPLDs enables rapid and highly tunable control of the amount and subcellular localization of PA, it requires genetic manipulation of cells for its expression, which limits its application in certain systems such as hard-to-transfect cell lines. Thus, as a complementary approach to optoPLD, we developed synthetic, small-molecule PA analogs that can be reversibly activated and deactivated by light. We focused on photoswitchable lipids, which exhibit a hydrophobic azobenzene photoswitch in their lipid tail (51). Photoswitchable lipids have emerged as powerful tools to optically control functions of bioactive lipids while keeping their native head group structures. The light-controlled cis–trans isomerization of azobenzene units allows optical switching of lipid tail structure between its straight and bent forms, which are often associated with different bioactivities in various biological systems and where the bent, cis forms have usually been found to be more active (52, 53, 54, 55).
Based on this concept, we developed PA analogs containing azobenzene moieties in one or both of their lipid tails (Fig. 5C) (28). These PA analogs, termed AzoPA and dAzoPA, respectively, were efficiently incorporated into both HEK293T and NIH3T3 cells, as confirmed by HPLC quantification of whole-cell lipid extracts. We observed no noticeable differences in the uptake efficiency between the light-activated cis forms and dark-adopted trans forms. Moreover, we found that the PA analogs could be subjected to cellular lipid metabolism and become converted to other azobenzene-containing species that corresponded to photolipid analogs of DAG, LPA, free fatty acids, and PC. For example, in NIH3T3 cells, 40 to 60% of AzoPA and dAzoPA were left unmetabolized after a 1-h treatment of cells with each compound, with the cis and trans forms exhibiting different conversion rates (28). These analyses confirm that the PA analogs can be effectively incorporated into cells and recognized by endogenous lipid-modifying enzymes as well as highlight the rapid and complex metabolism of PA in cells (10, 11).
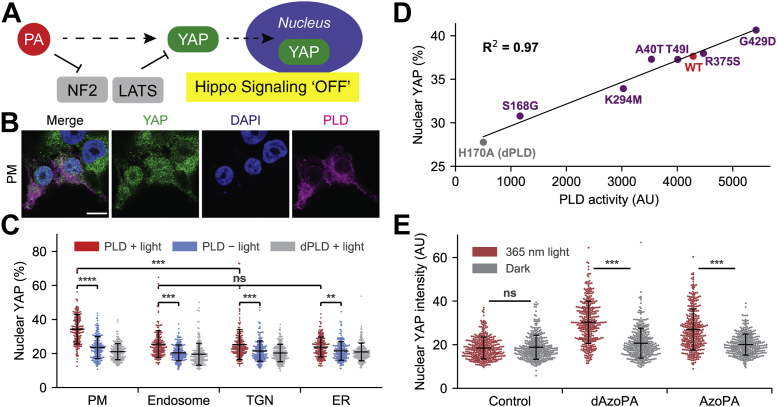
Finally, having established both genetically encoded and small-molecule tools for light-controlled production of PA within cells, we sought to explore their utility to direct endogenous, PLD-dependent PA signaling events. To highlight one example, we applied them to study the effects of PA on the Hippo signaling pathway (Fig. 6A). Hippo signaling is a mechanism by which cells can control growth and proliferation (56). When it is in the “on” state, it reduces cell growth and proliferation by activating the large tumor suppressor 1/2 kinases, which phosphorylate a key progrowth transcription factor, Yes-associated protein (YAP), and prevent its translocation to the nucleus (57). In the Hippo “off” state, nuclear translocation of YAP is permitted, leading to the activation of cell growth and proliferative gene expression programs. A recent study demonstrated that PLD-derived PA attenuates Hippo signaling, leading to the decreased phosphorylation and correspondingly increased nuclear translocation of YAP (58). The subcellular localization of the relevant PA pool, as well as a definitive identification of the relevant PA effector protein(s), however, remained unknown.
Figure 6.
Application of optoPLD and photoswitchable PA analogs to reveal localization and dose-dependent functions of PA in Hippo signaling.A, schematic depiction of PA as a regulator of Hippo signaling. PA binds and inhibits the effectors of Hippo signaling including NF2 and LATS, promoting the nuclear localization of YAP. B, OptoPLD targeting to the plasma membrane significantly increased the level of YAP localized in nuclei in optoPLD-transfected cells compared to nontransfected cells. The scale bar represents 10 μm. C, quantification of YAP localization changes induced by active optoPLD (red) at four different membranes compared to negative controls (blue and gray). D, OptoPLD mutants obtained from PLD engineering (Fig. 5B) reveal that the extent of YAP translocation depends on the catalytic activity of the PM-optoPLD construct and thus the amount of PA produced in the plasma membrane. E, light-activated cis form of photoswitchable PA analogs (AzoPA and dAzoPA) significantly increased the level of nuclear YAP in NIH3T3 cells. B–D are reproduced from ©Tei and Baskin (2020), originally published in Journal of Cell Biology (27). ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. AU, arbitrary units; dPLD, catalytically dead PLD; ER, endoplasmic reticulum; ns, not significant; optoPLD, optogenetic PLD; PA, phosphatidic acid; PLD, phospholipase D; PM, plasma membrane; TGN, trans-Golgi network.
To address these questions, we expressed optoPLDs targeted to different organelle membranes in HEK293T cells. Upon light-mediated recruitment and activation of optoPLD to trigger PA production at different membrane locations, we found that this effect was strongly regulated by a plasma membrane pool of PA (Fig. 6, B and C) (27). Moreover, with plasma membrane-targeting optoPLD mutants bearing a range of catalytic efficiencies, derived from our directed evolution studies, we demonstrated that this effect increases proportionally to the amount of PA produced, indicating that inhibition of Hippo signaling is exquisitely sensitive to the amount of PA produced in the plasma membrane (Fig. 6D). To confirm these findings using small-molecule, photoswitchable PA analogs in a second and hard-to-transfect cell line (NIH3T3 cells), we similarly evaluated YAP localization. Excitingly, we found that light-activated, cis forms of both AzoPA and dAzoPA efficiently increased the nuclear translocation of YAP, verifying the bioactivity of the PA analogs, as well as their ability to be controlled by light-mediated azobenzene isomerization (Fig. 6E) (28). Collectively, optoPLD and photoswitchable PA analogs are tools capable of producing functional pools of PA with spatiotemporal precision.
Conclusion and outlook
Decoding the pleiotropy of lipids that act as signaling molecules has been a major motivating challenge for those who study the cell biology of lipid signaling. PA is a class of molecules with simple structures yet complex functions. Historically, PA has been difficult to study because of its interconnected metabolic network, which is subject to strong homeostatic regulation. To study PA functions with minimal perturbations to endogenous signaling, we have applied the concepts of molecular design to develop small-molecule and protein-based tools that enable rapid monitoring and control of PA production in cells with high spatiotemporal resolution.
For visualizing and quantifying PA production, we have developed IMPACT. This approach uses a small-molecule alcohol bearing a bioorthogonal chemical handle for labeling PLD-produced pools of lipids by click chemistry, enabling both single-cell and bulk analyses. To precisely visualize the subcellular locations of PLD signaling, we have developed a variant, RT-IMPACT, which uses the rapid, fluorogenic IEDDA reaction to achieve real-time imaging of the click chemistry tagging reaction and enable visualization before rapid inter-organelle trafficking of the IMPACT-derived fluorescent lipids.
The development of IMPACT and RT-IMPACT paved the way to build a novel gain-of-function tool to perturb PA production. OptoPLD, which uses light-triggered protein dimerization to recruit PLDPMF to desired membranes, enables the production of bioactive PA pools with spatiotemporal control. IMPACT was a critical tool not only to evaluate and characterize the activity of optoPLD but also to engineer optoPLD mutants that exhibit various catalytic efficiencies and thus can mimic different activation states of mammalian PLDs. Moreover, as a complementary tool to optoPLD, we synthesized photoswitchable PA analogs bearing azobenzene photoswitches in their acyl tails. These PA analogs dispense with the need for genetic manipulation and could thus enable applications in a broader set of biological systems.
Collectively, IMPACT, RT-IMPACT, optoPLD, and photoswitchable PA analogs provide complementary approaches to understand PLD-mediated PA signaling, either by monitoring or perturbing PA activity in cells. Potential applications of these tools abound. For the IMPACT-based tools, future avenues should capitalize upon the unique strengths of IMPACT and RT-IMPACT at visualizing and quantifying PLD activity at the single-cell and subcellular levels, respectively. In parallel to studies that harness the ability of IMPACT to precisely track PLD signaling dynamics, the application of optoPLD to other PA-dependent signaling pathways beyond Hippo signaling could be a powerful approach to determine the localization of functionally relevant pools of PA. For instance, the involvement of PA in mTOR signaling has long been debated (59, 60). PA binds to and allosterically regulates mTOR and its components for their activation (61, 62, 63), but the precise location(s) where this activation takes place and which of the biological functions of mTOR are controlled by PA (i.e., using stringent tests for necessity and sufficiency) remain open questions in many contexts (63, 64). We envision that the panel of optoPLDs with varying catalytic efficiencies will serve as useful tools to address these questions, as well as questions regarding the role of PA at other biological sites, including mitochondrial dynamics (65, 66) and nuclear envelope maintenance (67).
In closing, we believe that lipid signaling represents fertile ground for the development and application of chemical biology-based approaches to fuel the discovery of new biological findings. As molecules that are not directly genetically encoded but instead are the products of complex and intertwined metabolic pathways, lipids represent sometimes devilishly elusive targets for tool development. We have devoted our efforts thus far to produce a collection of tools for the precise visualization and manipulation of one such signaling lipid, PA. These approaches are inspired by foundational work across multiple disciplines including lipid biochemistry, cell biology, genetics, optogenetics, protein directed evolution, and of course chemical biology. We hope that they will in turn spur the parallel development of tools for probing other types of lipids and widen the possibilities for what types of longstanding questions about the biological functions of lipids can be answered.
Conflict of interest
The authors declare that they no conflicts of interest with the contents of this article.
Acknowledgments
Work in the Baskin lab discussed here is supported by the National Science Foundation (CAREER CHE-1749919), the Arnold and Mabel Beckman Foundation (Beckman Young Investigator), and the Alfred P. Sloan Research Foundation (Sloan Research Fellowship).
Author contributions
R. T. and J. M. B. conceptualization; R. T. and J. M. B. writing–original draft; R. T. and J. M. B. writing–review and editing; J. M. B. supervision.
Funding and additional information
R. T. acknowledges support from by Honjo International, Funai Overseas, and Cornell Fellowships.
Edited by Phyllis Hanson
References
- 1.Eyster K.M. The membrane and lipids as integral participants in signal transduction: Lipid signal transduction for the non-lipid biochemist. Adv. Physiol. Educ. 2007;31:5–16. doi: 10.1152/advan.00088.2006. [DOI] [PubMed] [Google Scholar]
- 2.Harayama T., Riezman H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 2018;19:281–296. doi: 10.1038/nrm.2017.138. [DOI] [PubMed] [Google Scholar]
- 3.Corradi V., Sejdiu B.I., Mesa-Galloso H., Abdizadeh H., Noskov S. Yu., Marrink S.J., Tieleman D.P. Emerging diversity in lipid–protein interactions. Chem. Rev. 2019;119:5775–5848. doi: 10.1021/acs.chemrev.8b00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coreta-Gomes F.M., Vaz W.L.C., Moreno M.J. Effect of acyl chain length on the rate of phospholipid flip-flop and intermembrane transfer. J. Membr. Biol. 2018;251:431–442. doi: 10.1007/s00232-017-0009-4. [DOI] [PubMed] [Google Scholar]
- 5.Schuhmacher M., Grasskamp A.T., Barahtjan P., Wagner N., Lombardot B., Schuhmacher J.S., Sala P., Lohmann A., Henry I., Shevchenko A., Coskun Ü., Walter A.M., Nadler A. Live-cell lipid biochemistry reveals a role of diacylglycerol side-chain composition for cellular lipid dynamics and protein affinities. Proc. Natl. Acad. Sci. U. S. A. 2020;117:7729–7738. doi: 10.1073/pnas.1912684117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tei R., Baskin J.M. Induced proximity tools for precise manipulation of lipid signaling. Curr. Opin. Chem. Biol. 2021;65:93–100. doi: 10.1016/j.cbpa.2021.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Tanguy E., Wang Q., Moine H., Vitale N. Phosphatidic acid: From pleiotropic functions to neuronal pathology. Front. Cell. Neurosci. 2019;13:2. doi: 10.3389/fncel.2019.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhukovsky M.A., Filograna A., Luini A., Corda D., Valente C. Phosphatidic acid in membrane rearrangements. FEBS Lett. 2019;593:2428–2451. doi: 10.1002/1873-3468.13563. [DOI] [PubMed] [Google Scholar]
- 9.Tanguy E., Kassas N., Vitale N. Protein–phospholipid interaction motifs: A focus on phosphatidic acid. Biomolecules. 2018;8:20. doi: 10.3390/biom8020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zegarlinska J., Piaścik M., Sikorski A.F., Czogalla A. Phosphatidic acid – a simple phospholipid with multiple faces. Acta Biochim. Pol. 2018;65:163–171. doi: 10.18388/abp.2018_2592. [DOI] [PubMed] [Google Scholar]
- 11.Thakur R., Naik A., Panda A., Raghu P. Regulation of membrane turnover by phosphatidic acid: Cellular functions and disease implications. Front. Cell Dev. Biol. 2019;7:83. doi: 10.3389/fcell.2019.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lutkewitte A.J., Finck B.N. Regulation of signaling and metabolism by lipin-mediated phosphatidic acid phosphohydrolase activity. Biomolecules. 2020;10:1386. doi: 10.3390/biom10101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmid H.H.O., Schmid P.C., Natarajan V. The N-acylation-phosphodiesterase pathway and cell signalling. Chem. Phys. Lipids. 1996;80:133–142. doi: 10.1016/0009-3084(96)02554-6. [DOI] [PubMed] [Google Scholar]
- 14.Choi S.-Y., Huang P., Jenkins G.M., Chan D.C., Schiller J., Frohman M.A. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat. Cell Biol. 2006;8:1255–1262. doi: 10.1038/ncb1487. [DOI] [PubMed] [Google Scholar]
- 15.Massart J., Zierath J.R. Role of diacylglycerol kinases in glucose and energy homeostasis. Trends Endocrinol. Metab. 2019;30:603–617. doi: 10.1016/j.tem.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Bowling F.Z., Salazar C.M., Bell J.A., Huq T.S., Frohman M.A., Airola M.V. Crystal structure of human PLD1 provides insight into activation by PI(4,5)P2 and RhoA. Nat. Chem. Biol. 2020;16:400–407. doi: 10.1038/s41589-020-0499-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metrick C.M., Peterson E.A., Santoro J.C., Enyedy I.J., Murugan P., Chen T., Michelsen K., Cullivan M., Spilker K.A., Kumar P.R., May-Dracka T.L., Chodaparambil J.V. Human PLD structures enable drug design and characterization of isoenzyme selectivity. Nat. Chem. Biol. 2020;16:391–399. doi: 10.1038/s41589-019-0458-4. [DOI] [PubMed] [Google Scholar]
- 18.Khayyo V.I., Hoffmann R.M., Wang H., Bell J.A., Burke J.E., Reue K., Airola M.V. Crystal structure of a lipin/Pah phosphatidic acid phosphatase. Nat. Commun. 2020;11:1309. doi: 10.1038/s41467-020-15124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez-Cambronero J., Shah K.N. In: Gomez-Cambronero J., Frohman M.A., editors. vol. 259. Springer International Publishing; Cham: 2019. Phospholipase D and the mitogen phosphatidic acid in human disease: Inhibitors of PLD at the crossroads of phospholipid biology and cancer; pp. 89–113. (Lipid Signaling in Human Diseases). Handbook of Experimental Pharmacology. [DOI] [PubMed] [Google Scholar]
- 20.Kassas N., Tryoen-Tóth P., Corrotte M., Thahouly T., Bader M.-F., Grant N.J., Vitale N. In: Di Paolo G., Wenk M.R., editors. vol. 108. Academic Press; Cambridge, MA: 2012. Chapter 20 - genetically encoded probes for phosphatidic acid; pp. 445–459. (Methods in Cell Biology). Lipids. [DOI] [PubMed] [Google Scholar]
- 21.Zhang F., Wang Z., Lu M., Yonekubo Y., Liang X., Zhang Y., Wu P., Zhou Y., Grinstein S., Hancock J.F., Du G. Temporal production of the signaling lipid phosphatidic acid by phospholipase D2 determines the output of extracellular signal-regulated kinase signaling in cancer cells. Mol. Cell. Biol. 2014;34:84–95. doi: 10.1128/MCB.00987-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kassas N., Tanguy E., Thahouly T., Fouillen L., Heintz D., Chasserot-Golaz S., Bader M.-F., Grant N.J., Vitale N. Comparative characterization of phosphatidic acid sensors and their localization during frustrated phagocytosis. J. Biol. Chem. 2017;292:4266–4279. doi: 10.1074/jbc.M116.742346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bumpus T.W., Baskin J.M. A chemoenzymatic strategy for imaging cellular phosphatidic acid synthesis. Angew. Chem. Int. Ed. Engl. 2016;55:13155–13158. doi: 10.1002/anie.201607443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bumpus T.W., Baskin J.M. Clickable substrate mimics enable imaging of phospholipase D activity. ACS Cent. Sci. 2017;3:1070–1077. doi: 10.1021/acscentsci.7b00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang D., Wu K., Tei R., Bumpus T.W., Ye J., Baskin J.M. A real-time, click chemistry imaging approach reveals stimulus-specific subcellular locations of phospholipase D activity. Proc. Natl. Acad. Sci. U. S. A. 2019;116:15453–15462. doi: 10.1073/pnas.1903949116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bumpus T.W., Liang D., Baskin J.M. In: Chenoweth D.M., editor. vol. 641. Academic Press; Cambridge, MA: 2020. Chapter four - IMPACT: Imaging phospholipase D activity with clickable alcohols via transphosphatidylation; pp. 75–94. (Methods in Enzymology). Chemical Tools for Imaging, Manipulating, and Tracking Biological Systems: Diverse Chemical, Optical and Bioorthogonal Methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tei R., Baskin J.M. Spatiotemporal control of phosphatidic acid signaling with optogenetic, engineered phospholipase Ds. J. Cell Biol. 2020;219 doi: 10.1083/jcb.201907013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tei R., Morstein J., Shemet A., Trauner D., Baskin J.M. Optical control of phosphatidic acid signaling. ACS Cent. Sci. 2021;7:1205–1215. doi: 10.1021/acscentsci.1c00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowling F.Z., Frohman M.A., Airola M.V. Structure and regulation of human phospholipase D. Adv. Biol. Regul. 2021;79:100783. doi: 10.1016/j.jbior.2020.100783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brailoiu E., Chakraborty S., Brailoiu G.C., Zhao P., Barr J.L., Ilies M.A., Unterwald E.M., Abood M.E., Taylor C.W. Choline is an intracellular messenger linking extracellular stimuli to IP3-evoked Ca2+ signals through sigma-1 receptors. Cell Rep. 2019;26:330–337.e4. doi: 10.1016/j.celrep.2018.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang S.F., Freer S., Benson A.A. Transphosphatidylation by phospholipase D. J. Biol. Chem. 1967;242:477–484. [PubMed] [Google Scholar]
- 32.Bumpus T.W., Liang F.J., Baskin J.M. Ex uno plura: Differential labeling of phospholipid biosynthetic pathways with a single bioorthogonal alcohol. Biochemistry. 2018;57:226–230. doi: 10.1021/acs.biochem.7b01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu W., Lin Z., Woo C.M., Baskin J.M. A chemoproteomics approach to profile phospholipase D-derived phosphatidyl alcohol interactions. ACS Chem. Biol. 2021 doi: 10.1021/acschembio.1c00584. [DOI] [PubMed] [Google Scholar]
- 34.Jao C.Y., Roth M., Welti R., Salic A. Metabolic labeling and direct imaging of choline phospholipids in vivo. Proc. Natl. Acad. Sci. U. S. A. 2009;106:15332–15337. doi: 10.1073/pnas.0907864106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selvy P.E., Lavieri R.R., Lindsley C.W., Brown H.A. Phospholipase D: Enzymology, functionality, and chemical modulation. Chem. Rev. 2011;111:6064–6119. doi: 10.1021/cr200296t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blackman M.L., Royzen M., Fox J.M. Tetrazine ligation: Fast bioconjugation based on inverse-electron-demand Diels−Alder reactivity. J. Am. Chem. Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Devaraj N.K., Weissleder R., Hilderbrand S.A. Tetrazine-based cycloadditions: Application to pretargeted live cell imaging. Bioconjug. Chem. 2008;19:2297–2299. doi: 10.1021/bc8004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang M., Vannam R., Lambert W.D., Xie Y., Wang H., Giglio B., Ma X., Wu Z., Fox J., Li Z. Hydrophilic 18F-labeled trans-5-oxocene (oxoTCO) for efficient construction of PET agents with improved tumor-to-background ratios in neurotensin receptor (NTR) imaging. Chem. Commun. 2019;55:2485–2488. doi: 10.1039/c8cc09747j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du G., Altshuller Y.M., Vitale N., Huang P., Chasserot-Golaz S., Morris A.J., Bader M.-F., Frohman M.A. Regulation of phospholipase D1 subcellular cycling through coordination of multiple membrane association motifs. J. Cell Biol. 2003;162:305–315. doi: 10.1083/jcb.200302033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du G., Altshuller Y.M., Kim Y., Han J.M., Ryu S.H., Morris A.J., Frohman M.A. Dual requirement for Rho and protein kinase C in direct activation of phospholipase D1 through G protein-coupled receptor signaling. Mol. Biol. Cell. 2000;11:4359–4368. doi: 10.1091/mbc.11.12.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mozzicato S., Joshi B.V., Jacobson K.A., Liang B.T. Role of direct RhoA-phospholipase D interaction in mediating adenosine-induced protection from cardiac ischemia. FASEB J. 2004;18:1–13. doi: 10.1096/fj.03-0592fje. [DOI] [PubMed] [Google Scholar]
- 42.Powner D.J., Wakelam M.J.O. The regulation of phospholipase D by inositol phospholipids and small GTPases. FEBS Lett. 2002;531:62–64. doi: 10.1016/s0014-5793(02)03410-5. [DOI] [PubMed] [Google Scholar]
- 43.Everett P.B., Senogles S.E. D3 dopamine receptor signals to activation of phospholipase D through a complex with Rho. J. Neurochem. 2010;112:963–971. doi: 10.1111/j.1471-4159.2009.06508.x. [DOI] [PubMed] [Google Scholar]
- 44.Bumpus T.W., Huang S., Tei R., Baskin J.M. Click chemistry–enabled CRISPR screening reveals GSK3 as a regulator of PLD signaling. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2025265118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang D., Cheloha R.W., Watanabe T., Gardella T.J., Baskin J.M. Activity-based, bioorthogonal imaging of phospholipase D reveals spatiotemporal dynamics of GPCR–Gq signaling. Cell Chem. Biol. 2022;29:67–73.e3. doi: 10.1016/j.chembiol.2021.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferrandon S., Feinstein T.N., Castro M., Wang B., Bouley R., Potts J.T., Gardella T.J., Vilardaga J.-P. Sustained cyclic AMP production by parathyroid hormone receptor endocytosis. Nat. Chem. Biol. 2009;5:734–742. doi: 10.1038/nchembio.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salazar C., Frohman M.A. In: Gomez-Cambronero J., Frohman M.A., editors. vol. 259. Springer International Publishing; Cham: 2019. Prospects for PLD inhibition in cancer and thrombotic disease; pp. 79–88. (Lipid Signaling in Human Diseases). Handbook of Experimental Pharmacology. [Google Scholar]
- 48.Kennedy M.J., Hughes R.M., Peteya L.A., Schwartz J.W., Ehlers M.D., Tucker C.L. Rapid blue light induction of protein interactions in living cells. Nat. Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leiros I., McSweeney S., Hough E. The reaction mechanism of phospholipase D from Streptomyces sp. strain PMF. Snapshots along the reaction pathway reveal a pentacoordinate reaction intermediate and an unexpected final product. J. Mol. Biol. 2004;339:805–820. doi: 10.1016/j.jmb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 50.Leiros I., Secundo F., Zambonelli C., Servi S., Hough E. The first crystal structure of a phospholipase D. Structure. 2000;8:655–667. doi: 10.1016/s0969-2126(00)00150-7. [DOI] [PubMed] [Google Scholar]
- 51.Morstein J., Impastato A.C., Trauner D. Photoswitchable lipids. ChemBioChem. 2021;22:73–83. doi: 10.1002/cbic.202000449. [DOI] [PubMed] [Google Scholar]
- 52.Frank J.A., Yushchenko D.A., Hodson D.J., Lipstein N., Nagpal J., Rutter G.A., Rhee J.-S., Gottschalk A., Brose N., Schultz C., Trauner D. Photoswitchable diacylglycerols enable optical control of protein translocation, PKC activity, and vesicle release. Nat. Chem. Biol. 2016;12:755–762. doi: 10.1038/nchembio.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kol M., Williams B., Toombs-Ruane H., Franquelim H.G., Korneev S., Schroeer C., Schwille P., Trauner D., Holthuis J.C., Frank J.A. Optical manipulation of sphingolipid biosynthesis using photoswitchable ceramides. Elife. 2019;8 doi: 10.7554/eLife.43230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morstein J., Hill R.Z., Novak A.J.E., Feng S., Norman D.D., Donthamsetti P.C., Frank J.A., Harayama T., Williams B.M., Parrill A.L., Tigyi G.J., Riezman H., Isacoff E.Y., Bautista D.M., Trauner D. Optical control of sphingosine-1-phosphate formation and function. Nat. Chem. Biol. 2019;15:623–631. doi: 10.1038/s41589-019-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morstein J., Dacheux M.A., Norman D.D., Shemet A., Donthamsetti P.C., Citir M., Frank J.A., Schultz C., Isacoff E.Y., Parrill A.L., Tigyi G.J., Trauner D. Optical control of lysophosphatidic acid signaling. J. Am. Chem. Soc. 2020;142:10612–10616. doi: 10.1021/jacs.0c02154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu F.-X., Zhao B., Guan K.-L. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu F.-X., Zhao B., Panupinthu N., Jewell J.L., Lian I., Wang L.H., Zhao J., Yuan H., Tumaneng K., Li H., Fu X.-D., Mills G.B., Guan K.-L. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han H., Qi R., Zhou J.J., Ta A.P., Yang B., Nakaoka H.J., Seo G., Guan K.-L., Luo R., Wang W. Regulation of the Hippo pathway by phosphatidic acid-mediated lipid-protein interaction. Mol. Cell. 2018;72:328–340.e8. doi: 10.1016/j.molcel.2018.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fang Y., Vilella-Bach M., Bachmann R., Flanigan A., Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- 60.Foster D.A. Regulation of mTOR by phosphatidic acid? Cancer Res. 2007;67:1–4. doi: 10.1158/0008-5472.CAN-06-3016. [DOI] [PubMed] [Google Scholar]
- 61.Lehman N., Ledford B., Fulvio M.D., Frondorf K., McPhail L.C., Gomez-Cambronero J. Phospholipase D2-derived phosphatidic acid binds to and activates ribosomal p70 S6 kinase independently of mTOR. FASEB J. 2007;21:1075–1087. doi: 10.1096/fj.06-6652com. [DOI] [PubMed] [Google Scholar]
- 62.Toschi A., Lee E., Xu L., Garcia A., Gadir N., Foster D.A. Regulation of mTORC1 and mTORC2 complex assembly by phosphatidic acid: Competition with rapamycin. Mol. Cell. Biol. 2009;29:1411–1420. doi: 10.1128/MCB.00782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weng Z., Shen X., Zheng J., Liang H., Liu Y. Structural basis of DEPTOR to recognize phosphatidic acid using its tandem DEP domains. J. Mol. Biol. 2021;433:166989. doi: 10.1016/j.jmb.2021.166989. [DOI] [PubMed] [Google Scholar]
- 64.Frias M.A., Mukhopadhyay S., Lehman E., Walasek A., Utter M., Menon D., Foster D.A. Phosphatidic acid drives mTORC1 lysosomal translocation in the absence of amino acids. J. Biol. Chem. 2020;295:263–274. doi: 10.1074/jbc.RA119.010892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ha E.E.-J., Frohman M.A. Regulation of mitochondrial morphology by lipids. Biofactors. 2014;40:419–424. doi: 10.1002/biof.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kameoka S., Adachi Y., Okamoto K., Iijima M., Sesaki H. Phosphatidic acid and cardiolipin coordinate mitochondrial dynamics. Trends Cell Biol. 2018;28:67–76. doi: 10.1016/j.tcb.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thaller D.J., Tong D., Marklew C.J., Ader N.R., Mannino P.J., Borah S., King M.C., Ciani B., Lusk C.P. Direct binding of ESCRT protein Chm7 to phosphatidic acid–rich membranes at nuclear envelope herniations. J. Cell Biol. 2021;220 doi: 10.1083/jcb.202004222. [DOI] [PMC free article] [PubMed] [Google Scholar]