Abstract
Exposure to environmental mutagens but also cell-endogenous processes can create DNA double-strand breaks (DSBs) in a cell’s genome. DSBs need to be repaired accurately and timely to ensure genomic integrity and cell survival. One major DSB repair mechanism, called homologous recombination, relies on the nucleolytic degradation of the 5’-terminated strands in a process termed end resection. Here, we review new insights into end resection with a focus on the mechanistic interplay of the nucleases, helicases, and accessory factors involved.
Keywords: homologous recombination, nuclease, MRX/N, Sae2
Introduction
DNA double-strand breaks (DSBs) can form in a cell’s genome due to exposure to irradiation or chemicals [1], but also as a consequence of cell-endogenous processes, such as DNA replication, transcription, and the generation of reactive oxygen species. Moreover, DSBs are necessary intermediates during certain cell developmental processes, such as meiosis [2]. DSBs are highly cytotoxic and need to be repaired timely and efficiently to ensure genomic integrity and cell survival.
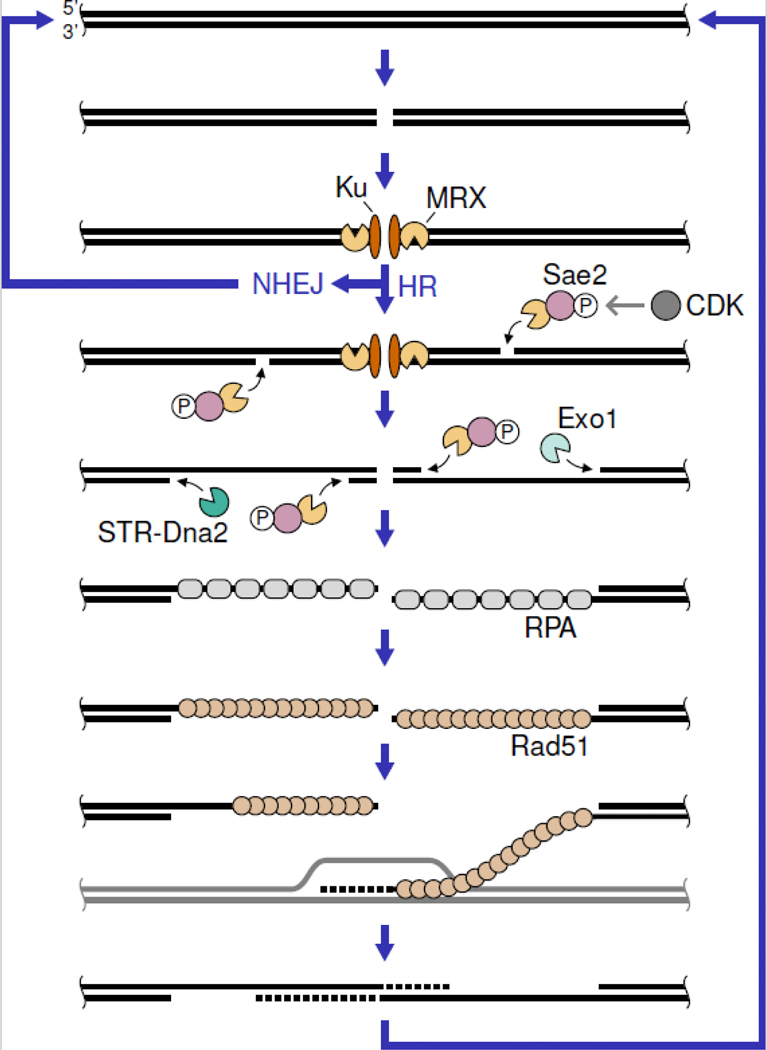
The two major classes of DSB repair pathways are non-homologous end-joining (NHEJ) and homologous recombination (HR) [3] (Figure 1). During NHEJ, DSB ends are bound by the Ku70-Ku80 (Ku) complex, which recruits DNA ligase 4 and accessory factors for mostly accurate re-ligation of the DNA ends. HR is more complex and initiates with the nucleolytic degradation of the DSB 5’ strands in a process called end resection [4]. The 3’ single-stranded DNA (ssDNA) overhangs generated are bound by the ssDNAbinding protein complex RPA, which is then replaced with the Rad51 recombinase in a process catalyzed by mediator proteins. The ssDNA-Rad51 nucleoprotein filament executes a homology search and invades into an intact copy of the broken sequence, which is usually the sister chromatid. The invading 3’ end is then extended by DNA synthesis. Reannealing with the other DSB end, fill-in synthesis, and ligation complete the repair process. End resection plays a crucial role in DSB repair pathway choice, as the ssDNA generated is a poor substrate for Ku binding and, thus, channels repair towards the HR pathway. Moreover, RPA-coated ssDNA plays an important role in a DSB signaling cascade that orchestrates the DNA damage response. In this review we discuss new insights into the mechanism of end resection with a focus on the interplay of the involved nucleases, helicases, and accessory factors. We will not cover resection at replication forks, and 53BP1-dependent resection suppression and fill-in synthesis in mammalian cells. We refer the interested reader to excellent reviews discussing these topics [5,6].
Figure 1: DSB are repaired by NHEJ or HR.
While NHEJ directly re-ligates DSBs, HR requires nucleolytic processing of the DSB ends by MRX-mediated initial and Exo1 or STR (Sgs1-Top3-Rmi1)-Dna2-mediated long-range resection. The ssDNA generated is coated by RPA and subsequently bound by the Rad51 recombinase for strand invasion and repair synthesis. The S. cerevisiae protein names are shown. See text for homologs in other organisms.
Initiation of end resection
The Mre11-Rad50-Xrs2/Nbs1 (MRX in Saccharomyces cerevisiae/MRN in mammals) complex is one of the first repair factors to be recruited to DSBs. MRN can slide along DNA and bypass protein obstacles, such as nucleosomes, and this facilitated diffusion might support fast recruitment to DSBs [7]. Once localized at the DSB, MRX initiates end resection by nicking the 5’ strands internal to the ends and resecting in 3’−5’ direction back towards the DSB [8] (Figure 1). MRX nicking is particularly important if the DSB ends contain protein blocks, such as the meiotic DSB-forming Spo11 endonuclease (see section “Meiotic resection”), or hairpin-capped ends (see next section). In vitro data revealed that MRX nicking is in fact stimulated by artificial and physiological protein blocks, such as streptavidin, nuclease-dead restriction enzymes, Ku, and RPA [9–11]. A recent study implies that MRX bound to DSB ends can itself stimulate nicking by adjacent MRX complexes [12]. Upon MRX nicking, the long-range resection machineries further degrade the 5’ strand in a 5’−3’ direction (see next section and Figure 1).
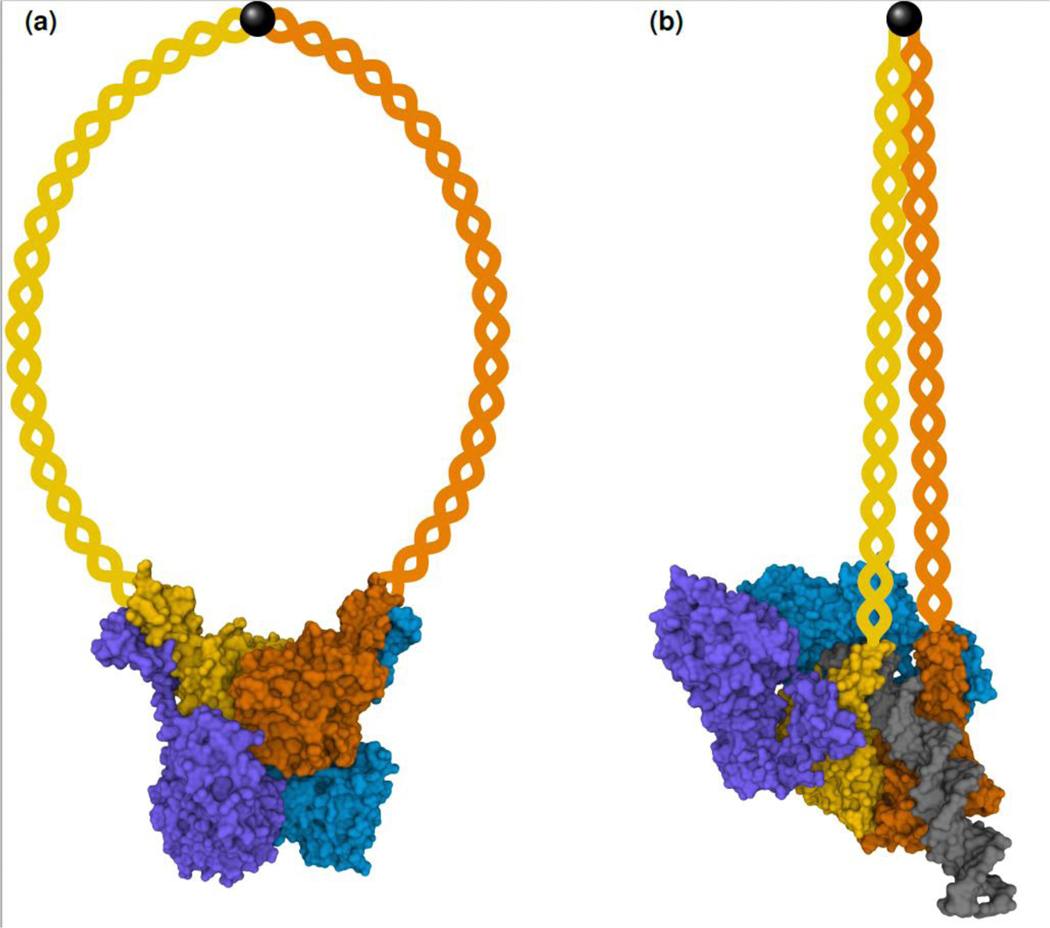
The MRX/N complex consists of two copies of each subunit [13] (Figure 2). The catalytic head region contains the Mre11 nuclease and the Rad50 ATPase domains. Two long Rad50 coiled coils protrude from the head region and interact at the apex via a zinc hook. The Xrs2/Nbs1 subunit is specific for eukaryotic organisms and supports nuclear localization, Mre11 dimerization, interaction with other proteins, and – in the case of higher eukaryotes – Mre11 nuclease activity [14,15]. MRX can adopt at least two conformations called “open” or “resting state” and “closed” or “cutting state”, and the transition between these involves ATP hydrolysis by Rad50 [16]. Previous structural studies used complexes with truncated Rad50 coiled coils to investigate these states. A recent cryo-electron microscopy study presented the two conformations for the full-length Escherichia coli MR homolog SbcCD [17]. In the E. coli MR resting state, the Mre11 nuclease domain is positioned below the Rad50 ATPase domain, which contains two ATP molecules, and the Rad50 coiled coils adopt a ring-shaped conformation (Figure 2a). Upon addition of DNA and ATP hydrolysis, the complex adopted the cutting state, where the Mre11 nuclease domains are repositioned to the site of the Rad50 ATPase domains (Figure 2b). DNA is bound by the Rad50 subunits and one of the Mre11 nuclease domains and positioned for nucleolytic attack. Interestingly, the Rad50 coiled coils are zipped up, adopt a rod-like conformation, and participate in DNA binding at their base. Only a single DNA duplex end can fit into the cutting state complex and this might explain how Mre11 nuclease activity is restricted to DSB ends and does not attack internal DNA.
Figure 2: “Resting state” (a) and “cutting state” (b) of the E. coli MR (SbcCD) complex.
The Mre11 subunits are in blue and purple and the Rad50 subunits are in yellow and orange. The DNA duplex in the “cutting state” is in gray. The protruding Rad50 coiled coils and the zinc hook, whose structures could not be determined due to their flexibility, are schematically depicted. Molecular surface structures are from [17] (PDB IDs 6S6V and 6S85 [59]) and were rendered with Mol* [60].
A crucial Mre11-Rad50 interaction in the cutting state of the E. coli MR complex relies on a specific Mre11 peptide loop. Eukaryotic Mre11 lacks this loop, but the corresponding Rad50 region interacts with the regulatory cofactor Sae2 (in S. cerevisiae, Ctp1 in Schizosaccharomyces pombe, CtIP in mammals). Sae2 is the target of multiple kinases and its phosphorylation status is a crucial determinant for MRX nuclease activity. Sae2 phosphorylation by cyclin-dependent kinase (CDK) is necessary to stimulate MRX nicking and restricts end resection initiation to S and G2 cell cycle stages, when a sister chromatid is available to template repair. Additional Sae2 phosphorylation at multiple sites by DNA damage signaling kinases (see section “Resection and the DNA damage response”) is necessary for full MRX nicking stimulation, likely by regulating the transition from an inactive multimeric to an active tetrameric state [18]. Tetramer formation depends on the N-terminal region of Ctp1 [19]. Besides nicking, the 3’−5’ exonuclease activity of MRX has also been shown to be stimulated by phosphorylated Sae2 [12]. Sae2 phosphorylation is important for binding to the MRX complex [14,18,20]. Once bound to the MRX complex, an evolutionarily conserved C-terminal fifteen-amino-acid peptide is necessary and sufficient for nuclease stimulation [20]. Altogether, these studies suggest that phosphorylated Sae2 stimulates MRX nuclease activities by stabilizing its cutting state conformation. Recent work has shown that the telomeric protein Rif2 has the opposite effect on MRX activity [21–23]. A Rif2 N-terminal motif destabilizes the MRX cutting state at telomeres to prevent detrimental telomere processing and DNA damage signaling. Interestingly, both, Rif2 and Sae2 bind to the same Rad50 surface. Further studies are needed to elucidate the molecular and structural details of MRX nicking regulation and to clarify how nicking is restricted to the 5’ strand of DSB ends.
Long-range end resection
After MRX nicking, long-range resection machineries extend the resection tracts [4] (Figure 1). One of the two partially redundant machineries is the 5’−3’ exonuclease Exo1. The other machinery consists of the endonuclease Dna2, which works in concert with Sgs1 (in S. cerevisiae, Rqh1 in S. pombe, and BLM or WRN in mammals) and accessory factors, such as Top3, Rmi1 (and RMI2 in mammals), and RPA. The entry site for long-range resection can be 5’ resected or gapped DNA substrates. Exo1 has also been shown to resect from nicks and recent work established that the same is true for the Sgs1-Top3-Rmi1-Dna2 (STR-Dna2) complex [24].
RPA plays multiple important roles during DNA end resection. In vitro experiments have shown that interaction with RPA stimulates Dna2 degradation of the 5’-terminated strands, while protecting 3’-terminated strands [25,26], and the Dna2 crystal structure has elucidated the underlying molecular mechanism [27]. RPA supports recruitment of Dna2 and Sgs1 [27–29] and in vitro studies have demonstrated that RPA stimulates the Sgs1 helicase activity [24,29] as well as resection by BLM in combination with EXO1 or DNA2 [30]. These data confirm previous in vivo findings showing that RPA depletion inhibits resection by both Exo1 and STR-Dna2 [28]. The regulatory role of RPA in end resection suggests a mechanism ensuring that the generated ssDNA is immediately coated with RPA. In fact, RPA coating of resected DNA has been shown to play important safeguarding roles. It prevents formation of DNA secondary structures, which are subject to nucleolytic attack, for example by the MRX complex [28], and prevents promiscuous microhomology annealing, which can give rise to genomic rearrangements [31,32]. Moreover, RPA-coated ssDNA plays important functions downstream of resection during DNA damage signaling (see next section) as well as Rad51 filament formation. Interestingly, another downstream HR factor has recently been shown to regulate long-range resection. Rad52 is known to help RPA replacement with Rad51, but also restricts loading and activity of Sgs1 in S. cerevisiae and Rqh1 in S. pombe [33].
The MRX complex not only initiates resection, but also stimulates long-range resection. As discussed above, MRX nuclease activities generate ssDNA, which is a suitable substrate for both long-range resection machineries and stimulates STR-Dna2 recruitment and activity via RPA binding. Moreover, MRX physically interacts with Sgs1 and MRN has recently been shown to physically interact with EXO1 [7]. Correspondingly, MRX directly recruits the long-range resection machineries to DSBs and stimulates resection [24–26]. Recent single-molecule microscopy experiments suggest that MRN stays in close proximity to resecting EXO1 [7]. Although MRN did not influence resection kinetics, it prevented an inhibitory effect of RPA previously seen under these conditions [34]. More work is needed to clarify if MRX stays in direct contact with resection factors during long-range resection. A potential role of MRX in this context could be to nick behind resection-stalling lesions or protein-adducts to restart long-range resection.
Another example for crosstalk between resection initiation and long-range resection has recently been discovered. Besides its long-known role in activating MRN nicking, CtIP also stimulates long-range resection via DNA2. CtIP helps to recruit DNA2 to DSBs [35] and recent work shows that it also activates DNA2-mediated resection [36]. DNA2 possesses a helicase domain in addition to its nuclease activity, and the former is stimulated by phosphorylated CtIP. Moreover, phosphorylated CtIP stimulates BLM helicase activity [36,37]. Interestingly, the S. cerevisiae CtIP homolog Sae2 does not stimulate yeast Dna2 but supports long-range resection by regulating DNA damage signaling (see next section).
Resection and the DNA damage response
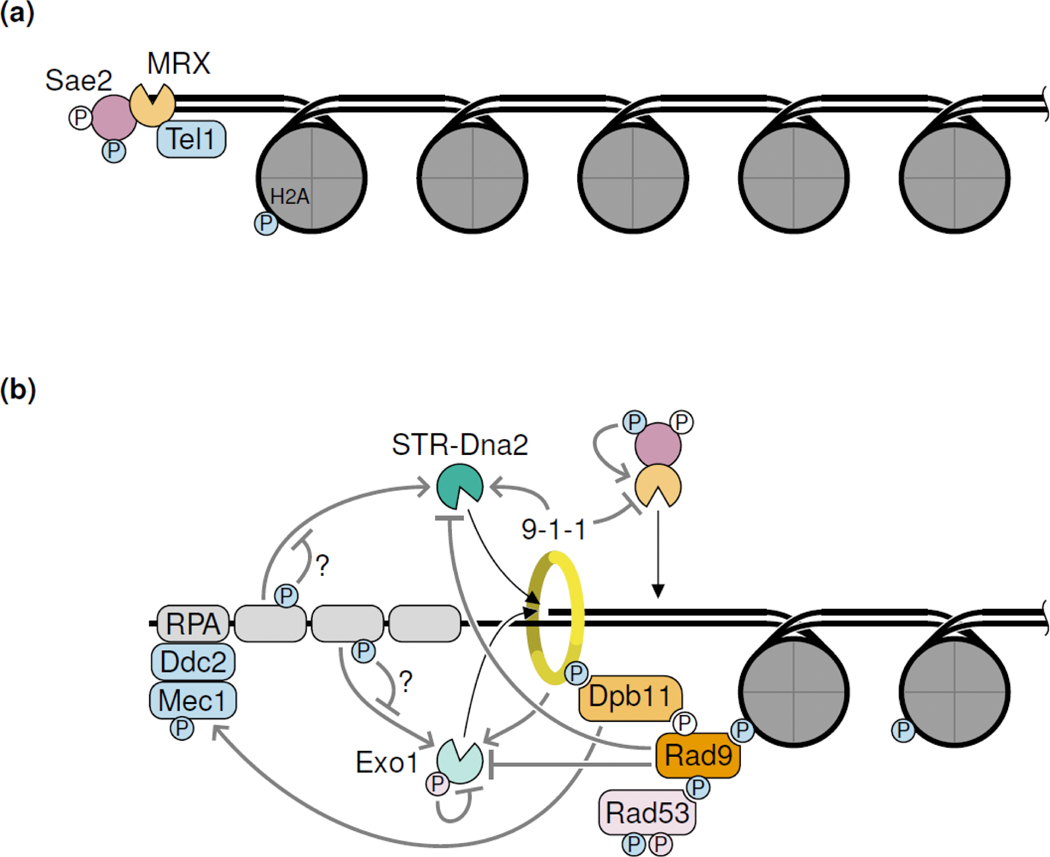
DSBs trigger a complicated signaling response [38] (Figure 3). The apical kinases Tel1 (in S. cerevisiae, ATM in mammals) and Mec1 (in S. cerevisiae, ATR in mammals) initiate the DNA damage response by phosphorylating common target proteins. Tel1 is recruited to DSBs via interaction with the MRX complex, while Mec1 is recruited via Ddc2 (in S. cerevisiae, ATRIP in mammals) to RPA-coated ssDNA resulting from end resection. Thus, DNA damage signaling transitions from Tel1 to Mec1 when end resection initiates. DNA damage signaling has both positive and negative effects on resection. As mentioned above, Tel1 and Mec1 phosphorylation of Sae2 stimulates resection initiation by MRX [18]. Tel1 and Mec1 also phosphorylate the histone subunit H2A and the 9–1-1 complex, which support recruitment and Tel1 or Mec1-mediated phosphorylation of the adapter protein Rad9 (53BP1 in mammals) [38]. Chromatin-bound Rad9 next to DSBs inhibits long-range resection via both Exo1 and STR-Dna2 [39,40]. Rad9 also helps recruitment and activation of the effector kinase Rad53 (CHK2 in mammals), which phosphorylates and inhibits Exo1 [41].
Figure 3: Resection and the DNA damage response (DDR).
(a) Tel1 is recruited to DSBs by MRX. (b) Upon resection and RPA binding, Mec1 is recruited. Multiple DDR proteins as well as DDR-induced phosphorylation events regulate the resection nucleases both in a stimulatory and inhibitory fashion, giving rise to a complex regulatory network. Phosphorylation events (P) are colored according to the responsible kinase. White Ps denote CDK-mediated phosphorylation. Question marks denote regulations that were described for mammals but have not yet been established in other organisms. The S. cerevisiae protein names are shown. See text for homologs in other organisms.
Recent work has elucidated new functional interactions between components of the DNA damage response and resection factors. Sae2 phosphorylation by Tel1 and Mec1 not only activates MRX nicking, but also dampens phosphorylation of other Tel1 and Mec1 targets, such as Rad9, to counteract resection suppression [42]. Besides recruiting Rad9, the 9–1-1 complex also recruits Exo1 and STR-Dna2 and stimulates long-range resection in regions of low Rad9 abundance, such as at uncapped telomeres [40]. In contrast, initial resection by MRX is suppressed by the 9–1-1 complex independently of its role in DNA damage signaling [43]. Finally, the stimulatory role of RPA on BLM-EXO1 and BLM-DNA2-mediated resection has recently been found to be suppressed upon RPA phosphorylation by DNA damage kinases [30]. It will be interesting to see if these functional interactions are conserved between different organisms.
Resection in the chromatin context
Resection of naked DNA in vitro is much faster than resection of chromatinized DNA in vivo [44]. In a reconstituted system, nucleosomes were shown to impede STR-Dna2 and especially Exo1-mediated resection [45]. Recent in vitro studies showed that the long-range resection machineries can mobilize, but not evict nucleosomes, and are stalled upon encountering a dense nucleosome array [30,46]. Accordingly, several chromatin remodelers have been implicated in supporting resection in the chromatin context. Both the RSC and SWI/SNF complexes support MRX recruitment to DSBs and early resection with redundant roles of the INO80 complex [47–50]. Further, a recent study implicated the RSC and SWI/SNF remodeler complexes in long-range resection [51]. Another chromatin remodeler supporting long-range resection is Fun30, which counteracts the inhibitory effect of Rad9 [49,52,53]. Interestingly, Fun30 recruitment to DSBs is regulated by CDK phosphorylation-dependent binding to the 9–1-1 interactor Dbp11, establishing another layer of cell cycle-regulated resection control [54]. While an in vitro study suggested that histones are not evicted upon resection [55], a recent in vivo study showed that nucleosomes are not present on resected DNA [51]. More work is needed to fully understand the redundancy and synergism of chromatin remodelers in resection and to clarify if nucleosome eviction precedes or is coupled to end resection.
Meiotic Resection
Meiosis generates haploid gametes from diploid precursor cells and relies on HR for proper homologous chromosome pairing and segregation. Meiotic HR initiates with Spo11-mediated DSB formation and resection of these breaks [2]. A deep sequencing-based approach termed S1-seq was developed to monitor meiotic resection genome-wide in S. cerevisiae [56]. Meiotic resection initiates by MRX nicking and 3’−5’ resection to remove the covalently linked topoisomerase-like protein Spo11 from DSB ends. Limited long-range resection in meiosis depends on Exo1, while STR-Dna2 is dispensable in yeast. The S1-seq method and an alternative deep sequencing-based method (END-seq) have recently been applied to study resection during mouse meiosis [57,58]. Surprisingly, mouse meiotic resection depends only mildly on EXO1. It will be interesting to see if DNA2 or another nuclease mediates long-range resection in mouse meiosis.
Interestingly, meiotic resection tracts seem to reach their final length quickly (ca. 0.8 kb and 1.1 kb in yeast and mouse, respectively), after which resection ceases [56–58]. Meiotic resection is not considerably increased upon depletion of strand invasion activities. In contrast, resection of a single DSB in mitotically dividing, recombination-deficient yeast cells continues without cessation at a constant speed of ca. 4 kb/h, generating tracts of ssDNA extending to tens of kb [44]. These differences might be due to the large number of DSBs, specialized chromatin context, and the activities and regulation of resection factors and chromatin remodelers during meiosis. It is possible that limited meiotic resection could prevent exhaustion of recombination factors. More work is needed to clarify the common and unique features of mitotic and meiotic resection.
Conclusion
End resection serves important functions in HR, repair pathway choice, and DNA damage signaling. However, unscheduled or extensive resection can pose risks to genomic integrity, as ssDNA is more vulnerable to mutagenesis than dsDNA and long ssDNA tracts can trigger aberrant recombination. Thus, end resection is tightly controlled, and recent studies have added to our understanding of the complex interaction networks that integrate end resection, DNA damage signaling, and downstream HR steps. Many mechanistic details of these interactions are still unknown, but we anticipate new and exciting discoveries in the coming years.
Acknowledgements
We apologize to investigators whose research could not be appropriately cited due to space limitations. Research in our laboratory is supported by grants from the National Institutes for Health (GM126997 and CA174653). The authors declare no conflict of interest.
2. Funding
Funding was received for this work.
All of the sources of funding for the work described in this publication are acknowledged below:
[List funding sources and their role in study design, data analysis, and result interpretation]
NIH GM126997, NIH CA174653
No funding was received for this work.
Footnotes
Author declaration
[Instructions: Please check all applicable boxes and provide additional information as requested.]
Conflict of Interest
Potential conflict of interest exists:
We wish to draw the attention of the Editor to the following facts, which may be considered as potential conflicts of interest, and to significant financial contributions to this work:
The nature of potential conflict of interest is described below:
No conflict of interest exists.
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Intellectual Property
We confirm that we have given due consideration to the protection of intellectual property associated with this work and that there are no impediments to publication, including the timing of public ation, with respect to intellectual property. In so doing we confirm that we have followed the regulations of our institutions concerning intellectual property.
Research Ethics
We further confirm that any aspect of the work covered in this manuscript that has involved huma patients has been conducted with the ethical approval of all relevant bodies and that such approvals are acknowledged within the manuscript.
IRB approval was obtained (required for studies and series of 3 or more cases)
Written consent to publish potentially identifying information, such as details or the case and photographs, was obtained from the patient(s) or their legal guardian(s).
Authorship
The International Committee of Medical Journal Editors (ICMJE) recommends that authorship be based on the following four criteria:
1. Substantial contributions to the conception or design of the work; or the acquisition, analysis, or interpretation of data for the work; AND
2. Drafting the work or revising it critically for important intellectual content; AND
3. Final approval of the version to be published; AND
4. Agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
All those designated as authors should meet all four criteria for authorship, and all who meet the four criteria should be identified as authors. For more information on authorship, please see http://www.icmje.org/recommendations/browse/roles-and-responsibilities/defining-the-role-of-authors-and-contributors.html#two.
All listed authors meet the ICMJE criteria. We attest that all authors contributed significantly to the creation of this manuscript, each having fulfilled criteria as established by the ICMJE.
One or more listed authors do(es) not meet the ICMJE criteria.
We believe these individuals should be listed as authors because:
[Please elaborate below]
We confirm that the manuscript has been read and approved by all named authors.
We confirm that the order of authors listed in the manuscript has been approved by all named authors.
Contact with the Editorial Office
The Corresponding Author declared on the title page of the manuscript is:
Lorraine Symington
This author submitted this manuscript using his/her account in EVISE.
We understand that this Corresponding Author is the sole contact for the Editorial process (including EVISE and direct communications with the office). He/she is responsible for communicating with the other authors about progress, submissions of revisions and final approval of proofs.
We confirm that the email address shown below is accessible by the Corresponding Author, is the address to which Corresponding Author’s EVISE account is linked, and has been configured to accept email from the editorial office of Current Opinion in Structural Biology: lss5@cumc.columbia.edu
Someone other than the Corresponding Author declared above submitted this manuscript from his/her account in EVISE:
[Insert name below]
We understand that this author is the sole contact for the Editorial process (including EVISE and direct communications with the office). He/she is responsible for communicating with the other authors, including the Corresponding Author, about progress, submissions of revisions and final approval of proofs.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mehta A, Haber JE: Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol 2014, 6:a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lam I, Keeney S: Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb Perspect Biol 2014, 7:a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ranjha L, Howard SM, Cejka P: Main steps in DNA double-strand break repair: an introduction to homologous recombination and related processes. Chromosoma 2018, 127:187–214. [DOI] [PubMed] [Google Scholar]
- 4.Symington LS: Mechanism and regulation of DNA end resection in eukaryotes. Crit Rev Biochem Mol Biol 2016, 51:195–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rickman K, Smogorzewska A: Advances in understanding DNA processing and protection at stalled replication forks. J Cell Biol 2019, 218:1096–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirman Z, de Lange T: 53BP1: a DSB escort. Genes Dev 2020, 34:7–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Myler LR, Gallardo IF, Soniat MM, Deshpande RA, Gonzalez XB, Kim Y, Paull TT, Finkelstein IJ: SingleMolecule Imaging Reveals How Mre11-Rad50-Nbs1 Initiates DNA Break Repair. Molecular cell 2017, 67:891–898.e894. * Using purified proteins, single molecule microscopy, and DNA curtain technology the authors reveal that the MRN complex can bind to DSBs, but also slide along DNA and bypass internal protein obstacles, such as nucleosomes. The authors also find that MRN physically interacts with EXO1 and supports processive EXO1-mediated resection in the presence of RPA.
- 8.Garcia V, Phelps SE, Gray S, Neale MJ: Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 2011, 479:241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cannavo E, Cejka P: Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature 2014, 514:122–125. [DOI] [PubMed] [Google Scholar]
- 10.Reginato G, Cannavo E, Cejka P: Physiological protein blocks direct the Mre11-Rad50-Xrs2 and Sae2 nuclease complex to initiate DNA end resection. Genes & development 2017, 31:2325–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W, Daley JM, Kwon Y, Krasner DS, Sung P: Plasticity of the Mre11-Rad50-Xrs2-Sae2 nuclease ensemble in the processing of DNA-bound obstacles. Genes & development 2017, 31:2331–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cannavo E, Reginato G, Cejka P: Stepwise 5’ DNA end-specific resection of DNA breaks by the Mre11-Rad50-Xrs2 and Sae2 nuclease ensemble. Proceedings of the National Academy of Sciences of the United States of America 2019, 116:5505–5513. * Using reconstituted in vitro systems the authors detect repetitive MRX nicking and 3’−5’ resection at DSBs. ATP-bound Rad50 restricts and phosphorylated Sae2 releases the Mre11 exonuclease activity leading to preferential resection of the 5’ strand. Interestingly, MRX resection occurs independently of a protein block at the DSB, indicating that DSB-bound MRX itself can stimulate nicking by an adjacent MRX complex.
- 13.Oh J, Symington LS: Role of the Mre11 Complex in Preserving Genome Integrity. Genes (Basel) 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anand R, Jasrotia A, Bundschuh D, Howard SM, Ranjha L, Stucki M, Cejka P: NBS1 promotes the endonuclease activity of the MRE11-RAD50 complex by sensing CtIP phosphorylation. The EMBO journal 2019, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anand R, Ranjha L, Cannavo E, Cejka P: Phosphorylated CtIP Functions as a Co-factor of the MRE11-RAD50-NBS1 Endonuclease in DNA End Resection. Mol Cell 2016, 64:940–950. [DOI] [PubMed] [Google Scholar]
- 16.Deshpande RA, Williams GJ, Limbo O, Williams RS, Kuhnlein J, Lee JH, Classen S, Guenther G, Russell P, Tainer JA, et al. : ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling. EMBO J 2014, 33:482–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Käshammer L, Saathoff J-H, Lammens K, Gut F, Bartho J, Alt A, Kessler B, Hopfner K-P: Mechanism of DNA End Sensing and Processing by the Mre11-Rad50 Complex. Molecular cell 2019, 76:382–394.e386. ** The authors present cryo-electron microscopy structures of the full-length E. coli MR (SbcCD) complex in the DNA-free “resting state” and DNA-bound “cutting state”. Interestingly, in the “cutting state” the Rad50 coiled coils zip up into a rod-like confirmation and participate in DNA binding. Crucial Mre11-Rad50 interactions are identified that might be functionally replaced by Rad50-Sae2 interactions in eukaryotes.
- 18. Cannavo E, Johnson D, Andres SN, Kissling VM, Reinert JK, Garcia V, Erie DA, Hess D, Thomä NH, Enchev RI, et al. : Regulatory control of DNA end resection by Sae2 phosphorylation. Nature communications 2018, 9:4016. ** The authors delineate how Cyclin-dependent kinase (CDK) and DNA damage response (DDR) kinases phosphorylate multiple sites on Sae2 to stimulate MRX nicking. Interestingly, Sae2 phosphorylation supports an interaction with Rad50 and the long-known Rad50S mutants are defective for this interaction, suggesting an explanation for the nicking defect of MRX complexes contain Rad50S.
- 19.Andres SN, Appel CD, Westmoreland JW, Williams JS, Nguyen Y, Robertson PD, Resnick MA, Williams RS: Tetrameric Ctp1 coordinates DNA binding and DNA bridging in DNA double-strand-break repair. Nat Struct Mol Biol 2015, 22:158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zdravković A, Daley JM, Dutta A, Niwa T, Murayama Y, Kanamaru S, Ito K, Maki T, Argunhan B, Takahashi M, et al. : A conserved Ctp1/CtIP C-terminal peptide stimulates Mre11 endonuclease activity. Proceedings of the National Academy of Sciences of the United States of America 2021, 118. * The authors show that the evolutionary conserved C-terminal 15/19 amino acids of S. pombe Ctp1/human CtIP are necessary and sufficient to activate the MRN endonuclease activity.
- 21.Khayat F, Cannavo E, Alshmery M, Foster WR, Chahwan C, Maddalena M, Smith C, Oliver AW, Watson AT, Carr AM, et al. : Inhibition of MRN activity by a telomere protein motif. Nat Commun 2021, 12:3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roisne-Hamelin F, Pobiega S, Jezequel K, Miron S, Depagne J, Veaute X, Busso D, Du ML, Callebaut I, Charbonnier JB, et al. : Mechanism of MRX inhibition by Rif2 at telomeres. Nat Commun 2021, 12:2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marsella A, Gobbini E, Cassani C, Tisi R, Cannavo E, Reginato G, Cejka P, Longhese MP: Sae2 and Rif2 regulate MRX endonuclease activity at DNA double-strand breaks in opposite manners. Cell reports 2021, 34:108906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W, Daley JM, Kwon Y, Xue X, Krasner DS, Miller AS, Nguyen KA, Williamson EA, Shim EY, Lee SE, et al.: A DNA nick at Ku-blocked double-strand break ends serves as an entry site for exonuclease 1 (Exo1) or Sgs1-Dna2 in long-range DNA end resection. The Journal of biological chemistry 2018, 293:17061–17069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC: DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 2010, 467:112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, et al. : Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 2010, 467:108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou C, Pourmal S, Pavletich NP: Dna2 nuclease-helicase structure, mechanism and regulation by Rpa. eLife 2015, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, Lisby M, Symington LS: RPA coordinates DNA end resection and prevents formation of DNA hairpins. Molecular cell 2013, 50:589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasaciunaite K, Fettes F, Levikova M, Daldrop P, Anand R, Cejka P, Seidel R: Competing interaction partners modulate the activity of Sgs1 helicase during DNA end resection. The EMBO journal 2019, 38:e101516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Soniat MM, Myler LR, Kuo H-C, Paull TT, Finkelstein IJ: RPA Phosphorylation Inhibits DNA Resection. Molecular cell 2019, 75:145–153.e145. * Using purified proteins, single molecule microscopy, and DNA curtain technology the authors observe how RPA stimulates resection. Besides the known stimulation of resection by BLM-DNA2, RPA also stimulates BLM-EXO1-mediated resection. Interestingly the stimulatory RPA effect is abrogated upon its DNA damage-induced phosphorylation.
- 31.Deng SK, Gibb B, de Almeida MJ, Greene EC, Symington LS: RPA antagonizes microhomologymediated repair of DNA double-strand breaks. Nature structural & molecular biology 2014, 21:405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng SK, Yin Y, Petes TD, Symington LS: Mre11-Sae2 and RPA Collaborate to Prevent Palindromic Gene Amplification. Molecular cell 2015, 60:500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yan Z, Xue C, Kumar S, Crickard JB, Yu Y, Wang W, Pham N, Li Y, Niu H, Sung P, et al. : Rad52 Restrains Resection at DNA Double-Strand Break Ends in Yeast. Molecular cell 2019, 76:699–711.e696. * The authors identify an unexpected role of the recombination mediator Rad52 in controlling longrange resection. In both fission and budding yeast, Rad52 suppresses the Dna2-dependent longrange resection pathway. Single molecule experiments with purified S. cerevisiae proteins show that Rad52 prevents Sgs1 from binding and translocating along DNA.
- 34.Myler LR, Gallardo IF, Zhou Y, Gong F, Yang S-H, Wold MS, Miller KM, Paull TT, Finkelstein IJ: Singlemolecule imaging reveals the mechanism of Exo1 regulation by single-stranded DNA binding proteins. Proceedings of the National Academy of Sciences of the United States of America 2016, 113:E1170–E1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoa NN, Kobayashi J, Omura M, Hirakawa M, Yang SH, Komatsu K, Paull TT, Takeda S, Sasanuma H: BRCA1 and CtIP Are Both Required to Recruit Dna2 at Double-Strand Breaks in Homologous Recombination. PLoS One 2015, 10:e0124495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ceppi I, Howard SM, Kasaciunaite K, Pinto C, Anand R, Seidel R, Cejka P: CtIP promotes the motor activity of DNA2 to accelerate long-range DNA end resection. Proceedings of the National Academy of Sciences of the United States of America 2020, 117:8859–8869. ** The authors show that CtIP stimulates the DNA2 helicase activity to promote long-range resection. The CtIP region responsible for DNA2 helicase stimulation is internal and separate from the N- and C-terminal regions necessary for MRN stimulation. This work indicates a role of CtIP in coupling MRN-mediated initial and DNA2-mediated long-range resection and suggests an explanation for the severe resection defect observed upon CtIP inactivation in vivo.
- 37.Daley JM, Jimenez-Sainz J, Wang W, Miller AS, Xue X, Nguyen KA, Jensen RB, Sung P: Enhancement of BLM-DNA2-Mediated Long-Range DNA End Resection by CtIP. Cell Rep 2017, 21:324–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lanz MC, Dibitetto D, Smolka MB: DNA damage kinase signaling: checkpoint and repair at 30 years. EMBO J 2019, 38:e101801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clerici M, Trovesi C, Galbiati A, Lucchini G, Longhese MP: Mec1/ATR regulates the generation of single-stranded DNA that attenuates Tel1/ATM signaling at DNA ends. EMBO J 2014, 33:198216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ngo GHP, Lydall D: The 9–1-1 checkpoint clamp coordinates resection at DNA double strand breaks. Nucleic acids research 2015, 43:5017–5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morin I, Ngo HP, Greenall A, Zubko MK, Morrice N, Lydall D: Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J 2008, 27:2400–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yu T-Y, Kimble MT, Symington LS: Sae2 antagonizes Rad9 accumulation at DNA double-strand breaks to attenuate checkpoint signaling and facilitate end resection. Proceedings of the National Academy of Sciences of the United States of America 2018, 115:E11961–E11969. * The authors show that Sae2 not only activates MRX nicking, but also dampens the DNA damage checkpoint to alleviate long-range resection suppression. These observations suggest an explanation for the more severe phenotype of sae2 mutants compared to mre11 nuclease-dead mutants.
- 43. Gobbini E, Casari E, Colombo CV, Bonetti D, Longhese MP: The 9–1-1 Complex Controls Mre11 Nuclease and Checkpoint Activation during Short-Range Resection of DNA Double-Strand Breaks. Cell reports 2020, 33:108287. * The authors show that long-range resection suppression induces a 9–1-1-dependent DNA damage checkpoint response. Interestingly, besides checkpoint activation, 9–1-1 also limits short-range resection by the MRX complex.
- 44.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G: Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 2008, 134:981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adkins NL, Niu H, Sung P, Peterson CL: Nucleosome dynamics regulates DNA processing. Nat Struct Mol Biol 2013, 20:836–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xue C, Wang W, Crickard JB, Moevus CJ, Kwon Y, Sung P, Greene EC: Regulatory control of Sgs1 and Dna2 during eukaryotic DNA end resection. Proceedings of the National Academy of Sciences of the United States of America 2019, 116:6091–6100. * Using purified proteins, single molecule microscopy, and DNA curtain technology the authors monitor STR-Dna2-mediated resection. They find that the complete complex and RPA are necessary for efficient DNA binding and resection. Interestingly, Sgs1 can mobilize but not evict nucleosomes in vitro.
- 47.Shim EY, Hong SJ, Oum JH, Yanez Y, Zhang Y, Lee SE: RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol Cell Biol 2007, 27:1602–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiest NE, Houghtaling S, Sanchez JC, Tomkinson AE, Osley MA: The SWI/SNF ATP-dependent nucleosome remodeler promotes resection initiation at a DNA double-strand break in yeast. Nucleic acids research 2017, 45:5887–5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen X, Cui D, Papusha A, Zhang X, Chu C-D, Tang J, Chen K, Pan X, Ira G: The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature 2012, 489:576–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kent NA, Chambers AL, Downs JA: Dual chromatin remodeling roles for RSC during DNA double strand break induction and repair at the yeast MAT locus. J Biol Chem 2007, 282:27693–27701. [DOI] [PubMed] [Google Scholar]
- 51. Peritore M, Reusswig K-U, Bantele SCS, Straub T, Pfander B: Strand-specific ChIP-seq at DNA breaks distinguishes ssDNA versus dsDNA binding and refutes single-stranded nucleosomes. Molecular cell 2021. ** Using strand-specific ChIP-seq, the authors show that histones are depleted from ssDNA, suggesting that nucleosomes are not present on resected DNA. The authors also show that the RSC and SWI/SNF chromatin remodeler complexes support long-range resection in addition to their previously known role in short-range resection.
- 52.Costelloe T, Louge R, Tomimatsu N, Mukherjee B, Martini E, Khadaroo B, Dubois K, Wiegant WW, Thierry A, Burma S, et al. : The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature 2012, 489:581–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eapen VV, Sugawara N, Tsabar M, Wu WH, Haber JE: The Saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end resection and checkpoint deactivation. Mol Cell Biol 2012, 32:4727–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bantele SC, Ferreira P, Gritenaite D, Boos D, Pfander B: Targeting of the Fun30 nucleosome remodeller by the Dpb11 scaffold facilitates cell cycle-regulated DNA end resection. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adkins NL, Swygert SG, Kaur P, Niu H, Grigoryev SA, Sung P, Wang H, Peterson CL: Nucleosome-like, Single-stranded DNA (ssDNA)-Histone Octamer Complexes and the Implication for DNA Double Strand Break Repair. J Biol Chem 2017, 292:5271–5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mimitou EP, Yamada S, Keeney S: A global view of meiotic double-strand break end resection. Science (New York, N.Y.) 2017, 355:40–45. ** The authors present a new deep sequencing-based method (S1-seq) and apply it to monitor meiotic resection in S. cerevisiae. After MRX-mediated short-range resection, Exo1 extends the resection-tracts with unexpectedly fast kinetics. However, resection is rather limited even in recombination-suppressed backgrounds, indicating differences between mitotic and meiotic resection.
- 57. Yamada S, Hinch AG, Kamido H, Zhang Y, Edelmann W, Keeney S: Molecular structures and mechanisms of DNA break processing in mouse meiosis. Genes & development 2020, 34:806–818. * Using the S1-seq method the authors monitor resection during mouse meiosis. Surprisingly and in contrast to S. cerevisiae, EXO1 is largely dispensible for meiotic resection in mice. Interestingly, PRDM9, which is know to direct meiotic DSB formation in mice an humans, mediates increased chromatin accessibility at meiotic recombination sites.
- 58. Paiano J, Wu W, Yamada S, Sciascia N, Callen E, Paola Cotrim A, Deshpande RA, Maman Y, Day A, Paull TT, et al. : ATM and PRDM9 regulate SPO11-bound recombination intermediates during meiosis. Nature communications 2020, 11:857. * Using a deep-sequencing based method (End-seq) the authors detect resection tracts in mouse spermatocytes and find evidence for coupled SPO11 cleavage and limited resection. Surprisingly and in contrast to S. cerevisiae, EXO1 is largely dispensible for meiotic resection in mice. Interestingly, the authors find that SPO11 remains attached to considerable fractions of DSBs and recombination intermediates.
- 59.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE: The Protein Data Bank. Nucleic Acids Res 2000, 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sehnal D, Bittrich S, Deshpande M, Svobodova R, Berka K, Bazgier V, Velankar S, Burley SK, Koca J, Rose AS: Mol* Viewer: modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]