Abstract
Current serological antibody tests for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) require enzyme or fluorescent labels, and the titer well plates cannot be reused. By immobilizing histidine (His)-tagged SARS-CoV-2 spike (S1) protein onto tris‒nitrilotriacetic acid (tris-NTA) sensor and using the early association phase for mass-transfer-controlled concentration determination, we developed a rapid and regenerable surface plasmon resonance (SPR) method for quantifying anti-SARS-CoV-2 antibody. On a five-channel SPR instrument and with optimized S1 protein immobilization density, each of the four analytical channels is sequentially used for multiple measurements, and all four channels can be simultaneously regenerated once they have reached a threshold value. Coupled with a programmable autosampler, each sensor can be regenerated at least 20 times, enabling uninterrupted assays of more than 800 serum samples. The accuracy and speed of our method compare well with those of the enzyme-linked immunosorbent assay (ELISA), and the detection limit (0.057 μg mL−1) can easily meet the requirement for screening low antibody levels such as those in convalescent patients. In addition, our method exhibits excellent channel-to-channel (RSD = 1.9%) and sensor-to-sensor (RSD = 2.1%) reproducibility. Obviation of an enzyme label drastically reduced the assay cost, rending our method (<60 cents) much more cost effective than those of commercial ELISA kits ($4.4–11.4). Therefore, our method offers a cost-effective and high-throughput alternative to the existing methods for serological measurements of anti-SARS-CoV-2 antibody levels, holding great promise for rapid screening of clinical samples without elaborate sample pretreatments and special reagents.
Keywords: Surface plasmon resonance, tris-NTA chip, Anti-SARS-CoV-2 antibody, High-throughput
Graphical abstract
1. Introduction
The novel coronavirus or severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has created the COVID-19 pandemic that is still ravaging the world [[1], [2], [3]]. As of the end of March 2022, over 480 million confirmed cases of COVID-19 have been reported with more than 6 million deaths worldwide [4]. SARS-CoV-2 is a large single-strand positive-sense RNA virus with a spike protein (S) that facilitates its entry into different cells by binding the angiotensin-converting enzyme 2 (ACE2) receptor at the cell membrane. The S protein comprises two subunits (S1 and S2), with the S1 subunit encompassing the receptor binding domain [5,6]. Compared to the severe acute respiratory syndrome (SARS-CoV) and Middle East respiratory syndrome coronaviruses (MERS-CoV), SARS-CoV-2 exhibits a longer viral incubation period and much higher transmissibility even before the onset of symptoms, the latter of which is responsible to the infection of nearly 6% of the total world population thus far [7,8].
In addition to developing effective therapeutic cures and vaccines, the availability of and accessibility to facile and high-throughput screening methods for SARS-CoV-2 is key to contain this pandemic. Many methods have been reported [[9], [10], [11], [12], [13], [14], [15], [16], [17], [18]] and some have been widely implemented for mass testing. The methods in use can be categorized into two types: molecular tests for viral RNA detection and serological tests for antibodies. Real-time reverse transcription-polymerase chain reaction (RT-PCR) is the most widely accepted method for diagnosing SARS-CoV-2 [11,12]. However, as viruses are present in the respiratory tract and RNA molecules are unstable, accurate diagnoses are highly dependent on careful nasal and/or oropharyngeal sampling and proper implementation of the RT-PCR, which are often difficult to carry out in developing countries [19]. The relatively long analysis time of RT-PCR also causes a delay in quarantine notification, which is critical to counter an outbreak in populated areas [20,21]. In addition to the high false negative/positive rates (up to 38% [22,23]), there are at least three scenarios where serological antibody tests are indispensable or at least complementary to the molecular tests. First, antibody levels in serum samples of convalescent patients must be determined to be higher than 1 μg mL−1 for the convalescent serum therapy to be effective [[24], [25], [26]]. However, the antibody level in convalescent patients spans a wide range of antibody concentrations (0.7–45000 μg mL−1) and dwindles with time of recovery [[13], [14], [15],27]. Second, serological tests can identify individuals who were infected and recovered without symptoms, providing a better estimate of the infectiousness of the disease [28,29]. Third, despite the increasing number of people being vaccinated, the immunities given rise by the various vaccines over a period of time are still unknown [[30], [31], [32]]. The third scenario is highly consequential and a better understanding or assessment of the duration of immunity conferred by a vaccine requires time-lapse serological antibody assays among different people [33,34].
Three major types of serological tests have been commercialized, which are enzyme-linked immunosorbent assay (ELISA), chemiluminescence immunoassay (CLIA), and lateral flow assay (LFA) [12,28,35]. While LFA is a qualitative method that only determines the presence of antibodies, ELISA and CLIA can quantify amounts of antibodies in serum samples. However, ELISA requires enzyme-linked antibodies and CLIA relies on the use of fluorescent labels [28,35,36]. Degradations of enzymatic activities and fluorescent intensities compromise their analytical performances. Besides the relatively high cost in manufacturing and storing the test kits, a major limitation of these two methods is the long time required for the incubation/washing steps [[37], [38], [39]]. Without these long steps and delicate reagents, the fully automated analysis and high sensitivity inherent in ELISA and CLIA would have been ideal for serological tests of antibodies in mass-screening efforts.
Perhaps owing to the abovementioned problems, many reports have compared and/or evaluated the efficacies of the three commercial methods [40,41], while others have focused on developing alternative methods. An attractive candidate is surface plasmon resonance (SPR), which is a sensitive and label-free technique for studying biomolecular interactions [14,15,[42], [43], [44], [45], [46], [47]]. However, the SPR methods developed thus far resort to measuring signals from the equilibrium phases of each antibody-protein interaction. The calibration curves are also constructed using signals at the equilibrium phases using antibody standards. Attaining the equilibrium phase is time- and sample-consuming, and is frequently impractical for low antibody concentrations (>hours or even longer). To our surprise, the use of the early stage of the association phase of a biomolecular interaction under mass-transfer control for analyte quantification [48], has not been widely used for facile SPR concentration determinations. The quantitative aspect of this approach is based on the following relation:
| R = Rmaxkat C |
where R is the SPR signal at a short time (t) of the association phase, R max is the maximum binding signal for a sensor covered with a fixed amount of ligand, k a is the association rate constant, and C is the analyte concentration. Although signals at an earlier stage of the association phase are smaller than those at the equilibrium phases, each analysis can be shorter than 1 min and requires a much smaller sample solution. Furthermore, the analytical signals can be augmented by using a relatively high density of ligand immobilized on the sensor so that R max in the above equation is greater. Measuring R at a short t is certainly a more viable approach for the purpose of serological antibody assays of samples from a huge populace.
By immobilizing histidine (His)-tagged protein G onto a dextran-based tris‒nitrilotriacetic acid (tris-NTA) sensor and using a dual-channel SPR instrument, we recently demonstrated the feasibility of using the early association phase for facile assays of serological assays of total immunoglobin G (IgG) antibodies [49]. The faster speed and capability of surface regeneration represent a step closer than many other studies for rapid analyses of numerous clinical samples. To the best of our knowledge, SPR and other related detection variants have not been used for rapid and high-throughput mass testing of samples for infectious diseases. However, three additional challenges are inherent in the serum assays for SARS-CoV-2 antibodies. First, the antibody concentrations of SARS-CoV-2 are in the low to mid-μg mL−1 range (vide supra), more than three orders of magnitude lower than the total IgG-type antibodies in serum (5.6–17.7 mg mL−1 [50]). The second difficulty confronting the serological antibody testing for SARS-CoV-2 is sample throughput. If an SPR sensor needs to be regenerated after only one or a few measurements, the sample throughput is undoubtedly compromised. Finally, sample delivery, sensor regeneration, and data collection should be as automated as possible. This will allow unattended measurements to be performed, affording not only high throughputs, but also accurate and reproducible results. We resolved these problems by using four analysis channels sequentially over a single tris-NTA sensor immobilized with a S1 protein. Using this method, at least 20 cycles of surface generations can be achieved on a single sensor for assaying at least 800 serum samples (the exact number of assays is dependent on the antibody concentrations in sera). With 96 titer plates preloaded with samples and placed in a pre-programmed autosampler, continuous analyses and surface regenerations can be realized in an unattended manner for days. In addition to the small channel-to-channel variability (1.9%), the variability between different sensors is less than 3%. The average time for assaying each sample is 6 min (including times needed for sample delivery and measurements and sensor regeneration in between) and the cost for each sample testing is less than 60 cents. The excellent correlation between our method and a commercial ELISA kit in assaying donors inoculated with different vaccines indicates that our method is accurate and reproducible. Thus, our approach represents a more rapid and cost-effective alternative to the current ELISA and CLIA methods and stands to offer important information for studies of the durability of vaccination, immunity, and suitability of convalescent serum samples for therapeutic treatments of patients.
2. Experimental section
2.1. Reagents and materials
N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), and ethanolamine hydrochloride were acquired from Sigma (St. Louis, MO, USA). K2HPO4, KH2PO4, NaCl, NaOH, NiCl2, ethylenediaminetetraacetic acid (EDTA), Tween-20 and heparin were obtained from Macklin Biochemical Technology (Shanghai, China). His-tagged SARS-CoV-2 S1 protein and its polyclonal antibody (anti-SARS-CoV-2, 95% purity, type rabbit) were purchased from Sangon Biotech (Shanghai, China). CM-dextran sensor chips were obtained from Biosensing Instrument Inc. (Tempe, AZ, USA). Tris-NTA sensor was prepared as described previously [49]. Other reagents were of analytical purity and used as received. Deionized water was purified by a Millipore system (Simplicity 185, Millipore Corp, Billerica, MA).
2.2. Serum sample collection
The blood samples from two unvaccinated and 113 vaccinated donors were collected by the Hospital of The University of Jinan. These samples were centrifuged for 10 min at 2000 rpm after clotting, and the supernatants were individually collected as serum samples. The 113 immunized donors all received two doses of vaccines made by Sinopharm Group (China), Sinovac Biotech Co., Ltd. (Beijing, China), or Moderna Therapeutics (Cambridge, MA, USA). The current study was in accordance with the Helsinki declaration and approved by the Medical Ethics Committee of the Hospital affiliated with The University of Jinan.
2.3. Surface plasmon resonance
All experiments were conducted on a five-channel instrument (SPR-4500, Biosensing Instrument Inc.). A programmable autosampler (Spark 7800, AJ Emmen Inc., Netherlands) that can accommodate two deep 96-well microplates (VWR International LLC, Radnor, PA) was used in conjunction with the SPR instrument. The running buffer was 10 mM PBS (10 mM KH2PO4/K2HPO4, 150 mM NaCl, 0.005% Tween-20, pH 7.4) containing 50 μM EDTA.
For assays of the anti-SARS-CoV-2 antibody in serum samples, the His-tagged S1 protein was immobilized onto each tris-NTA sensor to 1.000 ng mm−2 by injecting 250 μL of 5 μg mL−1 solution at 10 μL min−1, using a procedure similar to that described in our previous work [51]. A series of anti-SARS-CoV-2 antibody standards with varying concentrations (0.5, 1.0, 2.0, 4.0, 8.0, 16.0, 32.0, and 96.0 μg mL−1) were injected at 80 μL min−1 for 60 s to construct the calibration curve. Serum samples were diluted 2- to 1000-fold, and 60-μL aliquots of diluted samples were injected at 80 μL min−1. Each sample was analyzed at least three times, and 20 mM NaOH was used to dissociate preformed bioconjugates for sensor regeneration.
2.4. ELISA for antibody levels in serum samples
An ELISA kit for quantifying the anti-SARS-CoV-2 antibody was purchased from T & J Biomedical (Beijing, China) and used according to the manufacturer's instruction. Serum samples (100 μL each), pipetted into wells of a 96-well titer plate, were incubated at 37 °C for 60 min. Afterwards, the content in each well was decanted and each well was washed three times with PBS. Horseradish peroxidase (HRP)-conjugated secondary antibody (50 μL) was then added into each well. Following incubation at 37 °C for 30 min, each well was again rinsed with PBS to remove the unbound secondary antibody. Then 100 μL of 3,3′,5,5′-tetramethylbenzidine (TMB) solution was added into each well and the resulting mixture was incubated at room temperature for 15 min. The UV–vis absorbance was recorded at 450 nm with a multimode microplate reader (TECAN, Spark, Männedorf, Switzerland).
3. Results and discussion
3.1. SPR assay development
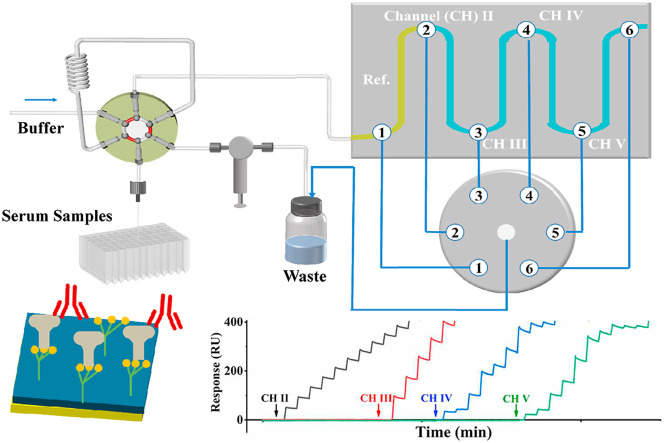
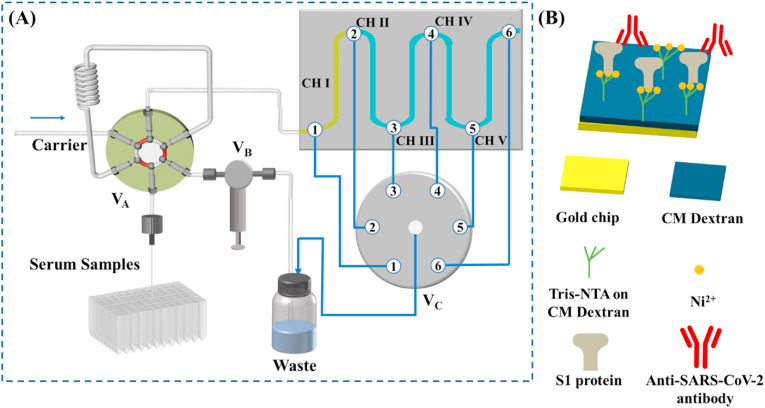
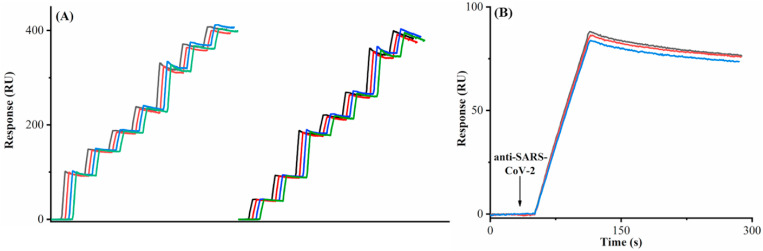
Fig. 1A illustrates the sequential use of four fluidic channels for detecting the anti-SARS-CoV-2 antibody captured by the His-tagged S1 protein (scheme shown in Fig. 1B), with the first channel serving as the reference. The programmable autosampler is used in conjunction with a six-port injector valve (VA) and a check valve (VB) for delivering samples from a deep 96-well plate to the sensor surface. In a typical configuration, channel I (CH I), without immobilized S1 protein, serves as the reference for background subtraction, whereas CH II−CH V are pre-immobilized with S1 protein. When the center port of the selector valve (VC) is connected to port 3, samples are injected from CH I to CH II. CH II is continuously used until the SPR signal reaches a threshold whereat analytes can no longer be captured quantitatively (cf. the determination of the threshold described below). At this moment, the program connects the center port of VC to port 4, opening CH III for quantitative analyses of the next samples. This process continues until all four channels have reached or surpassed the threshold value. For antibody measurements, with the center port staying connected to port 6, a single injection of 20 mM NaOH simultaneously regenerates all four channels and the sensor can be used in the next assay cycle.
Fig. 1.
(A) Schematic of the five-channel SPR, in conjunction with an autosampler, for detecting the anti-SARS-CoV-2 antibody in serum samples. (B) Captures of the anti-SARS-CoV-2 antibody by the His-tagged S1 protein pre-immobilized onto the CM-dextran-based tris-NTA sensor.
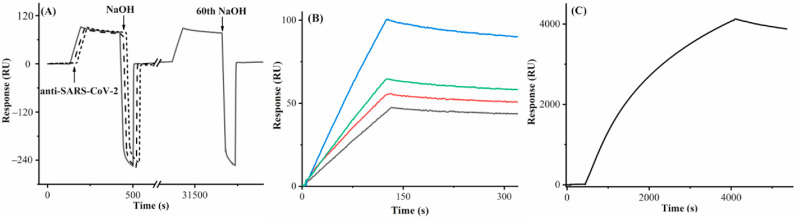
We first examined the effectiveness of sensor regeneration after the antibody capture. In Fig. 2 A, the first three consecutive injections of 20 mM NaOH all completely dissociated the conjugates formed between the antibody and S1 protein, as reflected by the full recovery of the original baseline. We found that after 60 regenerations the sensorgram signal for the same antibody concentration varied by less than 5%. Consequently, highly reproducible sensorgrams can be repeatedly collected. The antibody binding signal increased with the S1 protein density between 0.500 and 1.000 ng mm−2, but decreased beyond 1.000 ng mm−2. This trend suggests that 1.000 ng mm−2 is the optimal immobilization density that offers the greatest number of binding sites without imposing steric hindrance to the antibody binding (Fig. 2B). The sensorgram in Fig. 2C was collected from a continuous injection of a relatively high antibody concentration (20 μg mL−1) for 1 h. Although the signal is quite high, a steady-state signal was not obtained, indicating that concentration determination using SPR signals at the equilibrium phase is indeed time- and sample-consuming and often impractical. Thus, our method, by measuring the SPR signals at the earlier stage of the association phase, shortens the analysis time and cuts down the sample consumption. Adopting this facile method is particularly advantageous for relatively weak biomolecular interactions whose equilibria are too long to attain [48,49]. Based on the two criteria for constructing calibration curves [49], we found that 25 s is the right time to obtain the following calibration curve: R = 4.726[C] + 0.022, with a coefficient of determination R 2 of 0.9999 (Fig. S1). The relative standard deviations (RSDs) of signals in Fig. S1 range from 0.2% to 4.1%, indicative of high reproducibility. The detection limit of our method was estimated to be 0.057 μg mL−1 (S/N = 3), a value more than adequate for quantifying the anti-SARS-CoV-2 antibody in sera of convalescent patients [13,14,27].
Fig. 2.
(A) A sensorgram showing captures of the anti-SARS-CoV-2 antibody (8 μg mL−1) by the His-tagged S1 protein immobilized at a density of 1.000 ng mm−2 after multiple regenerations by 20 mM NaOH. The flow rate was 80 μL min−1. The arrows denote the time of antibody and NaOH injections. For better visual comparison, the 1st, 2nd, and 3rd antibody captures and NaOH regenerations, depicted in solid, dashed and dotted line curves, were overlaid with a small offset, and shown with the 60th antibody capture/regeneration cycle. (B) Sensorgrams showing captures of the antibody (5 μg mL−1) by the His-tagged S1 protein immobilized at densities of 0.500 (black), 0.750 (red), 1.000 (blue) and 1.250 (green) ng mm−2. The flow rate used was 60 μL min−1. (C) A sensorgram showing a continuous injection of 20 μg mL−1 antibody into a channel covered with 1.000 ng mm−2 His-tagged S1 protein at 5 μL min−1. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Method establishment using antibody-spiked serum samples
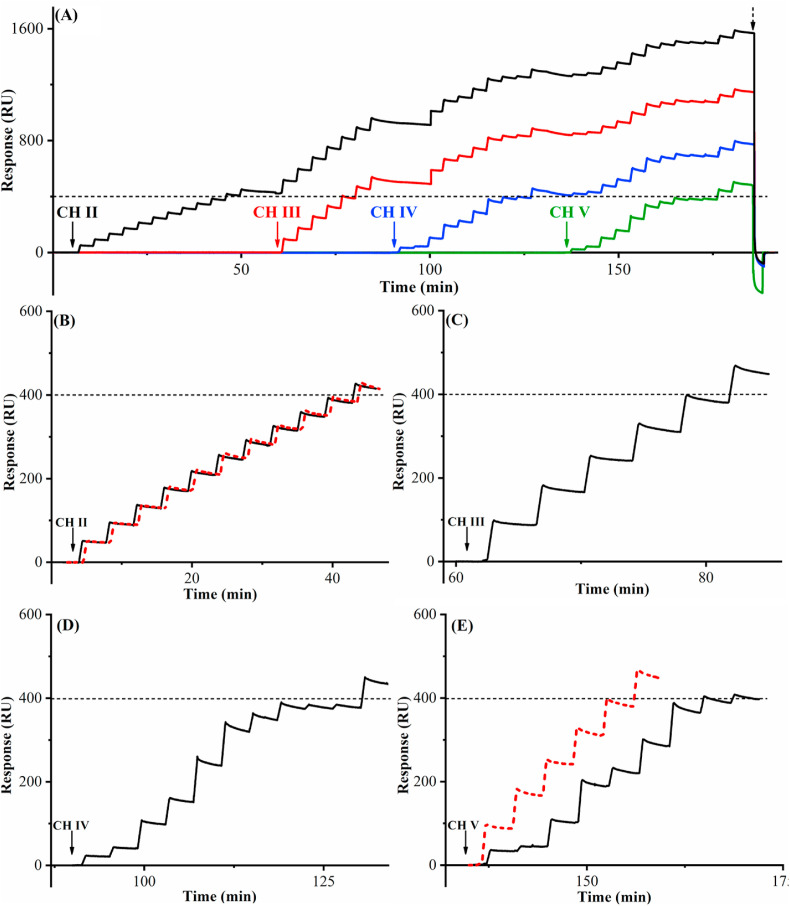
As mentioned in the Introduction, a viable assay of the anti-SARS-CoV-2 antibody in serum must be rapid, reproducible, and of high throughput. While the feasibility of regenerating the sensor surface in a single channel is apparent in Fig. 2A, the throughput will be much greater if all four channels are highly comparable and can be regenerated simultaneously. Along this line, we immobilized CHs II−V with the same S1 protein density (1.000 ng mm−2), with CH I serving as the reference channel for background subtraction. Fig. 3 A illustrates the entire measurement sequence comprising 41 injections. To obtain these sensorgrams, the center port of VC was first connected to port 3 (cf. Fig. 1), opening CH II for assays while preserving the other three channels for samples to be analyzed later. After ten consecutive injections of an 8.0-μg mL−1 antibody standard, the cumulative SPR signal reached 400 RU where many of the immobilized S1 protein molecules have captured the antibody (Fig. 3B). The next injection, above 400 RU, led to a binding signal that is more than 5% lower than the expected value (see also Table S1 in the Supplementary Information). Such a deviation indicates that the binding sites in CH II have become limited once the threshold value is reached. Instead of regenerating the channel right away, we programmed VC (cf. Fig. 1) to connect its center port to port 4 so that CH III was open for accurate measurements of additional samples. Because CHs II−V are connected in series, the downstream channels display similar signals to the upstream one(s), with only those below the threshold value being accurate. Fig. 3C shows that five consecutive injections of 16.0-μg mL−1 antibody into CH III resulted in a cumulative SPR signal of 400 RU, beyond which the accuracy of the sixth measurement deviates from the true value by more than 5% (cf. Table S1). Because the antibody concentration injected into CH III is doubled, the number of injections to reach this threshold value is halved when compared to that analyzed by CH II. Two points are worth mentioning: The amounts of antibody captured in the upstream channel hardly changes the antibody concentration in the bulk and does not affect the quantitative measurements in the downstream channels, a fact elucidated in our previous work [51]. The uniformity of the immobilized S1 protein in CH II is the same as that in CH III. When the same threshold in CHs IV and V is reached, the subsequent measurements also become less reliable (cf. Fig. 3D and E as well as Table S1). Thus, 33 injections (i.e., 10, 5, 10, and 8 measurements in CHs II, III, IV, and V) generated signals below the threshold. After signal in the last channel (CH V) reached 400 RU, 20 mM NaOH was injected to regenerate CHs II−V, reverting the signals of all four channels back to the original baseline.
Fig. 3.
(A) Background-subtracted sensorgrams corresponding to assays of multiple samples containing same (CH II and CH III) and different (CH IV and CH V) antibody concentrations. The black, red, blue, and green sensorgrams represent signals obtained from CHs II, III, IV, and V. The dashed line is the threshold at which the next channel should be open for further measurements. The vertical arrows indicate the opening of each channel. The antibody molecules captured in the four channels were simultaneously removed with a single injection of 20 mM NaOH (the inverted peak after the dashed arrow). The flow rate was 80 μL min−1. (B) The sensorgram of the first eleven injections (solid black) of 8.0-μg mL−1 antibody shown in Fig. 3A, overlaid with a replicate performed after regeneration of CH II (dotted red). (C) The sensorgram of the first six injections of 16.0-μg mL−1 antibody solution into CH III, as shown in Fig. 3A. (D) The sensorgram showing two replicate injections of five different antibody concentrations (4.0, 4.0, 10.0, 10.0, 16.0, 16.0, 8.0, 8.0, 2.0 and 2.0 μg mL−1) below the threshold in CH IV and an injection of 12.0 μg mL−1 antibody solution afterwards. (E) The sensorgram (solid black) shows the injections of randomized antibody concentrations (6.0, 2.0, 10.0, 16.0, 8.0, 14.0, 16.0, 8.0, and 4.0 μg mL−1) into CH V as in Fig. 3A, and the dotted red sensorgram denotes six injections of 16.0 μg mL−1 into this channel after sensor regeneration. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The regeneration is indeed effective and not perturbative to the S1 protein conformation, as evidenced by Fig. 3B and E. In the former, the sensorgrams before (solid black curve) and after (dashed red) the regeneration are almost congruent, while in the latter, after analyses of samples of randomized antibody concentrations, six injections of 16.0 μg mL−1 produced a sensorgram that is essentially identical to that recorded in CH III. We found that the channel-to-channel variability is only 1.2%. This is consistent with the small RSD values manifested in Fig. S1. The 33 accurate measurements displayed in Fig. 3A (see the measured values in Table S1) require only 185 min. Thus, each sample can be analyzed in 5.6 min, and the total time per sample is 6 min if the time for sensor regeneration is included, a time even shorter than that required for the qualitative LFA method (∼15 min) [52,53].
Before we applied our method to the analysis of the anti-SARS-CoV-2 antibody in real samples, we conducted a recovery experiment to ensure that the matrix of the serum samples does not affect the assay accuracy. Table 1 shows the actual amounts of different antibody standards spiked into serum samples of uninfected and unvaccinated donors, along with values measured with our method. With a serum sample diluted 10-fold by the running buffer, the differences between the spiked and measured values are exceedingly small and the recovery values are satisfactory. Moreover, the anti-SARS-CoV-2 antibody measurement is not affected by the presence of heparin (cf. Fig. S2) [54,55], based on the exceedingly small variance (<1%). As heparin is commonly used in treating Covid-19 patients, this small variance ensures accurate quantification of antibody levels in serum samples of patients under treatments. Thus, the matrix effects are essentially negligible, due to the excellent anti-fouling properties of the dextran-based tris-NTA sensor surface [49].
Table 1.
Antibody recovery in serum samples.
| Samples | Antibody spiked (μg mL−1) | Antibody measured (μg mL−1) | Recovery (%) |
|---|---|---|---|
| Serum | 0.20 | 0.21 | 105.0 |
| 1.00 | 1.01 | 101.0 | |
| 5.00 | 5.20 | 104.0 | |
| 10.00 | 10.34 | 103.4 | |
| 40.00 | 40.07 | 100.2 | |
| 45.00 | 44.75 | 99.4 |
We also found that our sensor can be continuously used for at least 20 assay cycles of serum samples, as evidenced by the high comparability (RSD% less than 1.9%) between the sensorgrams of the same samples measured in the same channel and that between the sensorgrams of samples measured in different channels subject to different regeneration steps (Fig. 4A). The four channels of this sensor were used uninterruptedly for a total of 877 assays in 87.6 h or 3.65 days. If the same approach is extended to SPR instruments possessing parallel channels [56] or related techniques designed for simultaneous analyses of different samples, the throughput will be even greater. For example, SPR imaging (SPRi) [[57], [58], [59]], an SPR variant, is capable of monitoring reactions occurring simultaneously in 96 channels [60]. We should also note that, in addition to the excellent channel-to-channel consistency, the variability between different sensors is also quite small. In Fig. 4B, the RSD% of the signals at 25 s of the three sensorgrams, collected separately from three sensors, is 2.1%.
Fig. 4.
(A) Overlaid sensorgrams that were selected from a continuous assay of 877 serum samples. The left half is the overlaid sensorgrams from the 1st (black curve), 6th (red), 14th (blue) and 17th (green) cycles in CH II. The antibody concentrations were measured to be 20.2, 9.2, 6.3, 8.1, 20.2, 9.2 and 6.3 μg mL−1. The right half is the overlaid sensorgrams from different cycles in different channels, with the black, red, blue and green curves corresponding to sensorgrams from the 2nd cycle in CH II, the 6th cycle in CH III, the 13th cycle in CH IV and the 18th cycle in CH V. The antibody concentrations in serum samples were 6.3, 9.2, 20.2, 6.3, 9.2, 20.2, and 8.1 μg mL−1. (B) Sensorgrams showing binding of 8.0 μg mL−1 anti-SARS-CoV-2 antibody to three different tris-NTA sensors immobilized with 1.000 ng mm−2 His-tagged S protein. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Assays of clinical serum samples
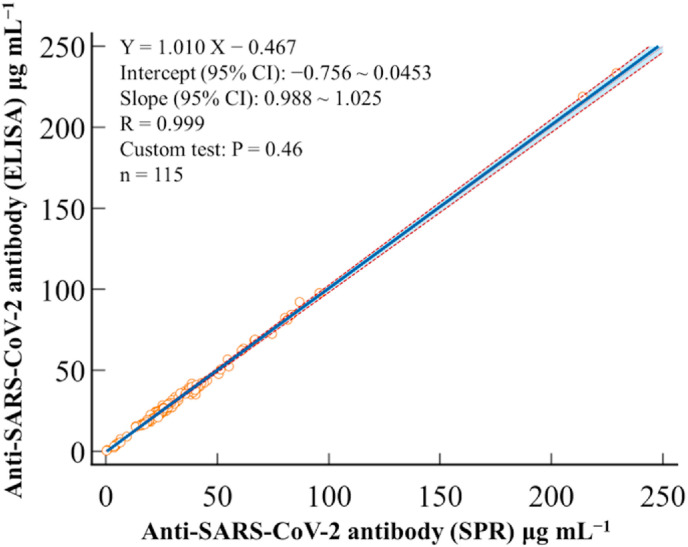
With the method established for serum sample analysis, we programed the instrument for automated analyses of the serum antibody levels from donors who have received two doses of different vaccines, as well as those from donors who were unvaccinated but not infected by Covid-19 (confirmed by the PCR tests of their oropharyngeal swab samples). The SPR assays of the 115 clinical samples were performed parallelly with ELISA in a double-blind fashion. The results displayed in Table S2 are in excellent agreement with each other. The Passing−Bablok regression plot comparing SPR and ELISA method afforded a slope of 1.010 and an intercept of −0.467 μg mL−1, whose 95% confidence interval (CI) encompasses values 1 and 0, respectively (Fig. 5 ). This plot indicates that proportional and constant differences are insignificant between SPR and ELISA [61]. Moreover, the Bland–Altman plot revealed a mean bias of 0.0 μg mL−1, further indicating that the two methods are equivalent (Fig. S3A). Between these two methods, only four outliers were observed within ±1.96 SD (Fig. S3A), consistent with the normal error (Gaussian) distribution for a large yet finite number of measurements, as shown by the Shapiro–Wilk test (Fig. S3B). As can be seen from Table S2, all of the vaccinated donors have antibody levels substantially greater than the two unvaccinated donors. Moreover, the test results are considered to be antibody-positive because their concentrations are all greater than the cut-off value, 1.0 μg mL−1 [[24], [25], [26],62].
Fig. 5.
Comparison between the SPR and ELISA methods. Passing–Bablok regression depicted by the solid blue line and 95% confidence interval (CI) for the regression represented by the dashed orange lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We should note that the cost for our homemade tris-NTA sensor is about $200, which constitutes the bulk of the expense for screening. Thus, each measurement costs about 25 cents or less. Even if the commercially available sensor chip (∼$500) [[63], [64], [65]] were used and costs of other reagents were included, the overall cost would be still quite low (<60 cents/sample). An ELISA kit based on the platform of 96-well plates ranges from $420 to $1100 [66], and the cost for testing one sample corresponds to a range between $4.4 and $11.4. Therefore, our SPR method is much more cost-effective than ELISA. In addition, to minimize trial-to-trial and plate-to-plate variations, constructing a new calibration curve for a different 96-well plate is generally recommended [67]. All the procedures and requirements of ELISA decrease its throughput. Our method obviates the use of enzymes and the incubation/washing steps, and the programmable autosampler and regenerable sensor chips facilitate uninterrupted assays for days (cf. Fig. 4A).
4. Conclusions
Towards the objective of rapid, accurate, and high-throughput screening of the anti-SARS-CoV-2 antibody, we coupled a programmable autosampler with a five-channel SPR instrument in conjunction with a regenerable S1 protein sensor surface for continuous and automated analyses of many serum samples. Different from commercial ELISA that requires enzyme-linked antibodies, our method is label-free and uses signals of the early association phase of the biomolecular interaction to reduce analysis time and enhance the throughput. Consequently, hundreds of samples can be analyzed on a single, renewable sensor, and the high channel-to-channel and sensor-to-sensor comparability ensures data quality and assay fidelity. It is worth noting that SPR has been used for detections of viral particles such as H1N1, H7N9, and even SARS-CoV-2 [68,69]. Therefore, by immobilizing different proteins on different channels of a single sensor, serological testing can provide additional information about immunity, convalescence, and immunization efficacy. As a result, SPR is a versatile and powerful tool for studying the prevalence and pathogenicity of SARS-CoV-2 and other related infectious diseases.
CRediT authorship contribution statement
Meng Jiang: Formal analysis, Investigation. Tianbao Dong: Investigation, Data curation. Chaowei Han: Investigation. Luyao Liu: Investigation. Tiantian Zhang: Resources. Qing Kang: Supervision, Funding acquisition. Pengcheng Wang: Investigation, Supervision, Writing – original draft, Writing – review & editing, Funding acquisition. Feimeng Zhou: Conceptualization, Methodology, Supervision, Writing – original draft, Writing – review & editing, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Nature Science Foundation of China (No. 21906065 and 21802051), the Natural Science Foundation of Shandong Province of China (No. ZR2019QB007), and the Shandong Provincial Program of Talent-Leading Teams.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aca.2022.339830.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395:689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hui D.S., Azhar E.I., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., McHugh T.D., Memish Z.A., Drosten C., Zumla A., Petersen E. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - the latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization WHO coronavirus (COVID-19) dashboard. https://covid19.who.int/ accessed Mar 30 2022)
- 5.Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatmal M.m.M., Alshaer W., Al-Hatamleh M.A.I., Hatmal M., Smadi O., Taha M.O., Oweida A.J., Boer J.C., Mohamud R., Plebanski M. Comprehensive structural and molecular comparison of spike proteins of SARS-CoV-2, SARS-CoV and MERS-CoV, and their Interactions with ACE2. Cells. 2020;9:2638. doi: 10.3390/cells9122638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Standl F., Joeckel K.-H., Brune B., Schmidt B., Stang A. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 2021;21:e238–e244. doi: 10.1016/S1473-3099(20)30648-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansson M.A., Quandelacy T.M., Kada S., Prasad P.V., Steele M., Brooks J.T., Slayton R.B., Biggerstaff M., Butler J.C. SARS-CoV-2 transmission from people without COVID-19 symptoms. JAMA Netw. Open. 2021;4 doi: 10.1001/jamanetworkopen.2020.35057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuce M., Filiztekin E., Ozkaya K.G. COVID-19 diagnosis - a review of current methods. Biosens. Bioelectron. 2021;172:112752. doi: 10.1016/j.bios.2020.112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattioli I.A., Hassan A., Oliveira O.N., Jr., Crespilho F.N. On the challenges for the diagnosis of SARS-CoV-2 based on a review of current methodologies. ACS Sens. 2020;5:3655–3677. doi: 10.1021/acssensors.0c01382. [DOI] [PubMed] [Google Scholar]
- 11.Jayamohan H., Lambert C.J., Sant H.J., Jafek A., Patel D., Feng H., Beeman M., Mahmood T., Nze U., Gale B.K. SARS-CoV-2 pandemic: a review of molecular diagnostic tools including sample collection and commercial response with associated advantages and limitations. Anal. Bioanal. Chem. 2021;413:49–71. doi: 10.1007/s00216-020-02958-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubina R., Dziedzic A. Molecular and serological tests for COVID-19 a comparative review of SARS-CoV-2 coronavirus laboratory and point-of-care diagnostics. Diagnostics. 2020;10:434. doi: 10.3390/diagnostics10060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zarletti G., Tiberi M., De Molfetta V., Bossu M., Toppi E., Bossu P., Scapigliati G. A cell-based ELISA to improve the serological analysis of anti-SARS-CoV-2 IgG. Viruses. 2020;12:1274. doi: 10.3390/v12111274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan X., Krel M., Dolgov E., Park S., Li X., Wu W., Sun Y.-L., Zhang J., Oo M.K.K., Perlin D.S., Fan X. Rapid and quantitative detection of SARS-CoV-2 specific IgG for convalescent serum evaluation. Biosens. Bioelectron. 2020;169:112572. doi: 10.1016/j.bios.2020.112572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schasfoort R.B.M., van Weperen J., van Amsterdam M., Parisot J., Hendriks J., Koerselman M., Karperien M., Mentink A., Bennink M., Krabbe H., Terstappen L.W.M.M., Mulder A.H.L. Presence and strength of binding of IgM, IgG and IgA antibodies against SARS-CoV-2 during COVID-19 infection. Biosens. Bioelectron. 2021;183:113165. doi: 10.1016/j.bios.2021.113165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang R., Wu J., Ao H., Fu J., Qiao B., Wu Q., Ju H. A rolling circle-amplified G-quadruplex/hemin DNAzyme for chemiluminescence immunoassay of the SARS-CoV-2 protein. Anal. Chem. 2021;93:9933–9938. doi: 10.1021/acs.analchem.1c02229. [DOI] [PubMed] [Google Scholar]
- 17.Roberts A., Mahari S., Shahdeo D., Gandhi S. Label-free detection of SARS-CoV-2 Spike S1 antigen triggered by electroactive gold nanoparticles on antibody coated fluorine-doped tin oxide (FTO) electrode. Anal. Chim. Acta. 2021;1188 doi: 10.1016/j.aca.2021.339207. 339207-339207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y., Chen B., He M., Hu B. A homogeneous nucleic acid assay for simultaneous detection of SARS-CoV-2 and influenza A (H3N2) by single-particle inductively coupled plasma mass spectrometry. Anal. Chim. Acta. 2021;1186 doi: 10.1016/j.aca.2021.339134. 339134-339134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yelin I., Aharony N., Tamar E.S., Argoetti A., Messer E., Berenbaum D., Shafran E., Kuzli A., Gandali N., Shkedi O., Hashimshony T., Mandel-Gutfreund Y., Halberthal M., Geffen Y., Szwarcwort-Cohen M., Kishony R. Evaluation of COVID-19 RT-qPCR test in multi sample pools. Clin. Infect. Dis. 2020;71:2073–2078. doi: 10.1093/cid/ciaa531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Javier Candel F., Barreiro P., San Roman J., Carlos Abanades J., Barba R., Barberan J., Bibiano C., Canora J., Canton R., Calvo C., Carretero M., Cava F., Delgado R., Garcia-Rodriguez J., Gonzalez del Castillo J., Gonzalez de Villaumbrosia C., Hernandez M., Emilio Losa J., Javier Martinez-Peromingo F., Maria Molero J., Munoz P., Onecha E., Onoda M., Rodriguez J., Sanchez-Celaya M., Antonio Serra J., Zapatero A. Recommendations for use of antigenic tests in the diagnosis of acute SARS-CoV-2 infection in the second pandemic wave: attitude in different clinical settings. Rev. Esp. Quim. 2020;33:466–484. doi: 10.37201/req/120.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nolan T., Hands R.E., Bustin S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 22.Kucirka L.M., Lauer S.A., Laeyendecker O., Boon D., Lessler J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction-based SARS-CoV-2 tests by time since exposure. Ann. Intern. Med. 2020;173:262–267. doi: 10.7326/M20-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y., Yao L., Li J., Chen L., Song Y., Cai Z., Yang C. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J. Med. Virol. 2020;92:903–908. doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Y., Peng C., Yuan M., Huang J., Wang Z., Yu J., Gao X., Wang D., Yu X., Li L., Zhang J., Wu X., Li B., Xu Y., Chen W., Peng Y., Hu Y., Lin L., Liu X., Huang S., Zhou Z., Zhang L., Wang Y., Zhang Z., Deng K., Xia Z., Gong Q., Zhang W., Zheng X., Liu Y., Yang H., Zhou D., Yu D., Hou J., Shi Z., Chen S., Chen Z., Zhang X., Yang X. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. U.S.A. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Focosi D., Anderson A.O., Tang J.W., Tuccori M. Convalescent plasma therapy for COVID-19: state of the art. Clin. Microbiol. Rev. 2020;33 doi: 10.1128/CMR.00072-20. e00072-00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., Tang X., Yu J., Lan J., Yuan J., Wang H., Zhao J., Zhang S., Wang Y., Shi X., Liu L., Zhao J., Wang X., Zhang Z., Zhang L. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 27.Santiago L., Uranga-Murillo I., Arias M., Gonzalez-Ramirez A.M., Macias-Leon J., Moreo E., Redrado S., Garcia-Garcia A., Taleb V., Lira-Navarrete E., Hurtado-Guerrero R., Aguilo N., del Mar Encabo-Berzosa M., Hidalgo S., Galvez E.M., Ramirez-Labrada A., de Miguel D., Benito R., Miranda P., Fernandez A., Domingo J.M., Serrano L., Yuste C., Villanueva-Saz S., Pano-Pardo J.R., Pardo J. Determination of the concentration of IgG against the spike receptor-binding domain that predicts the viral neutralizing activity of convalescent plasma and serum against SARS-CoV-2. Biology. 2021;10:208. doi: 10.3390/biology10030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ong D.S.Y., Fragkou P.C., Schweitzer V.A., Chemaly R.F., Moschopoulos C.D., Skevaki C., European Society of Clinical M., Infectious D. V. Study Group for Respiratory, How to interpret and use COVID-19 serology and immunology tests. Clin. Microbiol. Infect. 2021;27:981–986. doi: 10.1016/j.cmi.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deeks J.J., Dinnes J., Takwoingil Y., Davenport C., Spijker R., Taylor-Phillips S., Adrianol A., Beesel S., Dretzkel J., di Ruffanol L.F., Harris I.M., Price M.J., Dittrich S., Emperador D., Hooft L., Leeflang M.M.G., Van den Bruel A., Cochrane C.-D.T. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst. Rev. 2020;6:CD013652. doi: 10.1002/14651858.CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long Q.X., Tang X.J., Shi Q.L., Li Q., Deng H.J., Yuan J., Hu J.L., Xu W., Zhang Y., Lv F.J., Su K., Zhang F., Gong J., Wu B., Liu X.M., Li J.J., Qiu J.F., Chen J., Huang A.L. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 31.Varona J.F., Madurga R., Penalver F., Abarca E., Almirall C., Cruz M., Ramos E., Maria Castellano-Vazquez J. Kinetics of anti-SARS-CoV-2 antibodies over time. Results of 10 month follow up in over 300 seropositive Health Care Workers. Eur. J. Intern. Med. 2021;89:97–103. doi: 10.1016/j.ejim.2021.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerhards C., Thiaucourt M., Kittel M., Becker C., Ast V., Hetjens M., Neumaier M., Haselmann V. Longitudinal assessment of anti-SARS-CoV-2 antibody dynamics and clinical features following convalescence from a COVID-19 infection. Int. J. Infect. Dis. 2021;107:221–227. doi: 10.1016/j.ijid.2021.04.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amanat F., Thapa M., Lei T., Ahmed S.M.S., Adelsberg D.C., Carreno J.M., Strohmeier S., Schmitz A.J., Zafar S., Zhou J.Q., Rijnink W., Alshammary H., Borcherding N., Reiche A.G., Srivastava K., Sordillo E.M., van Bakel H., Turner J.S., Bajic G., Simon V., Ellebedy A.H., Krammer F. SARS-CoV-2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2. Cell. 2021;184:3936–3948. doi: 10.1016/j.cell.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saad-Roy C.M., Wagner C.E., Baker R.E., Morris S.E., Farrar J., Graham A.L., Levin S.A., Mina M.J., Metcalf C.J.E., Grenfell B.T. Immune life history, vaccination, and the dynamics of SARS-CoV-2 over the next 5 years. Science. 2020;370:811–818. doi: 10.1126/science.abd7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galipeau Y., Greig M., Liu G., Driedger M., Langlois M.A. Humoral responses and serological assays in SARS-CoV-2 infections. Front. Immunol. 2020;11:610688. doi: 10.3389/fimmu.2020.610688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Younes N., Al-Sadeq D.W., Al-Jighefee H., Younes S., Al-Jamal O., Daas H.I., Yassine H.M., Nasrallah G.K. Challenges in laboratory diagnosis of the novel coronavirus SARS-CoV-2. Viruses. 2020;12:582. doi: 10.3390/v12060582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hosseini S., Vázquez-Villegas P., Rito-Palomares M., Martinez-Chapa S.O. Advantages, Disadvantages and Modifications of Conventional ELISA. Springer Singapore; Singapore: 2018. Enzyme-linked Immunosorbent Assay (ELISA): from A to Z; pp. 67–115. [Google Scholar]
- 38.Kricka L.J. In: Immunoassay. Diamandis E.P., Christopoulos T.K., editors. Academic Press; San Diego: 1996. 15 - chemiluminesence immunoassay; pp. 337–353. [Google Scholar]
- 39.Nimse S.B., Sonawane M.D., Song K.S., Kim T. Biomarker detection technologies and future directions. Analyst. 2016;141:740–755. doi: 10.1039/c5an01790d. [DOI] [PubMed] [Google Scholar]
- 40.Tang M.S., Case J.B., Franks C.E., Chen R.E., Anderson N.W., Henderson J.P., Diamond M.S., Gronowski A.M., Farnsworth C.W. Association between SARS-CoV-2 neutralizing antibodies and commercial serological assays. Clin. Chem. 2020;66:1538–1547. doi: 10.1093/clinchem/hvaa211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Elslande J., Houben E., Depypere M., Brackenier A., Desmet S., Andre E., Van Ranst M., Lagrou K., Vermeersch P. Diagnostic performance of seven rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin. Microbiol. Infect. 2020;26:1082–1087. doi: 10.1016/j.cmi.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaisocherova H., Brynda E., Homola J. Functionalizable low-fouling coatings for label-free biosensing in complex biological media: advances and applications. Anal. Bioanal. Chem. 2015;407:3927–3953. doi: 10.1007/s00216-015-8606-5. [DOI] [PubMed] [Google Scholar]
- 43.Hoang Hiep N., Park J., Kang S., Kim M. Surface plasmon resonance: a versatile technique for biosensor applications. Sensors. 2015;15:10481–10510. doi: 10.3390/s150510481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niu L., Zhang N., Liu H., Zhou X., Knoll W. Integrating plasmonic diagnostics and microfluidics. Biomicrofluidics. 2015;9 doi: 10.1063/1.4929579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niu L., Wohland T., Knoll W., Koper I. Interaction of a synthetic antimicrobial peptide with a model bilayer platform mimicking bacterial membranes. Biointerphases. 2017;12 doi: 10.1116/1.5001020. [DOI] [PubMed] [Google Scholar]
- 46.Zhao H., Boyd L.F., Schuck P. Measuring protein interactions by optical biosensors. Curr. Protoc. Protein Sci. 2017;88:20.22.21–20.22.25. doi: 10.1002/cpps.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKeating K.S., Hinman S.S., Rais N.A., Zhou Z., Cheng Q. Antifouling lipid membranes over protein A for orientation-controlled immunosensing in undiluted serum and plasma. ACS Sens. 2019;4:1774–1782. doi: 10.1021/acssensors.9b00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karlsson R., Roos H., Fägerstam L., Persson B. Kinetic and concentration analysis using BIA technology. Methods. 1994;6:99–110. [Google Scholar]
- 49.Liu L., Han C., Jiang M., Zhang T., Kang Q., Wang X., Wang P., Zhou F. Rapid and regenerable surface plasmon resonance determinations of biomarker concentration and biomolecular interaction based on tris-nitrilotriacetic acid chips. Anal. Chim. Acta. 2021;1170:338625. doi: 10.1016/j.aca.2021.338625. [DOI] [PubMed] [Google Scholar]
- 50.Petar P., Dubois D., Rabin B.S., Shurin M.R. In: Measuring Immunity. Lotze M.T., Thomson A.W., editors. Academic Press; London: 2005. Chapter 12 - immunoglobulin titers and immunoglobulin subtypes; pp. 158–171. [Google Scholar]
- 51.Wang X., Li Z., Nguyen L., Zhou F. One-step ligand immobilization and single sample injection for regeneration-free surface plasmon resonance measurements of biomolecular interactions. Anal. Chem. 2017;89:3261–3265. doi: 10.1021/acs.analchem.7b00121. [DOI] [PubMed] [Google Scholar]
- 52.Wu J.L., Tseng W.P., Lin C.H., Lee T.F., Chung M.Y., Huang C.H., Chen S.Y., Hsueh P.R., Chen S.C. Four point-of-care lateral flow immunoassays for diagnosis of COVID-19 and for assessing dynamics of antibody responses to SARS-CoV-2. J. Infect. 2020;81:435–442. doi: 10.1016/j.jinf.2020.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicol T., Lefeuvre C., Serri O., Pivert A., Joubaud F., Dubee V., Kouatchet A., Ducancelle A., Lunel-Fabiani F., Le Guillou-Guillemette H. Assessment of SARS-CoV-2 serological tests for the diagnosis of COVID-19 through the evaluation of three immunoassays: two automated immunoassays (Euroimmun and Abbott) and one rapid lateral flow immunoassay (NG Biotech) J. Clin. Virol. 2020;129:104511. doi: 10.1016/j.jcv.2020.104511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cui J., Zang S., Shu W., Nie H., Jing J., Zhang X. Highly sensitive and selective detection of heparin in serum based on a long-wavelength tetraphenylethylene-cyanopyridine aggregation-induced emission luminogen. Anal. Chem. 2020;92:7106–7113. doi: 10.1021/acs.analchem.0c00496. [DOI] [PubMed] [Google Scholar]
- 55.Novelli C., Borotto E., Beverina I., Punzi V., Radrizzani D., Brando B. Heparin dosage, level, and resistance in SARS-CoV2 infected patients in intensive care unit. Int. J. Lab. Hematol. 2021;43:1284–1290. doi: 10.1111/ijlh.13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown M.E., Bedinger D., Lilov A., Rathanaswami P., Vasquez M., Durand S., Wallace-Moyer I., Zhong L., Nett J.H., Burnina I., Caffry I., Lynaugh H., Sinclair M., Sun T., Bukowski J., Xu Y., Abdiche Y.N. Assessing the binding properties of the anti-PD-1 antibody landscape using label-free biosensors. PLoS One. 2020;15 doi: 10.1371/journal.pone.0229206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bockova M., Slaby J., Springer T., Homola J. In: Advances in Surface Plasmon Resonance Imaging and Microscopy and Their Biological Applications. Bohn P.W., Pemberton J.E., editors. Annu. Rev. Anal. Chem.; 2019. pp. 151–176. [DOI] [PubMed] [Google Scholar]
- 58.Manuel G., Luptak A., Corn R.M. A microwell-printing fabrication strategy for the on-chip templated biosynthesis of protein microarrays for surface plasmon resonance imaging. J. Phys. Chem. C. 2016;120:20984–20990. doi: 10.1021/acs.jpcc.6b03307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malinick A.S., Lambert A.S., Stuart D.D., Li B., Puente E., Cheng Q. Detection of multiple sclerosis biomarkers in serum by ganglioside microarrays and surface plasmon resonance imaging. ACS Sens. 2020;5:3617–3626. doi: 10.1021/acssensors.0c01935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abdiche Y.N., Miles A., Eckman J., Foletti D., Van Blarcom T.J., Yeung Y.A., Pons J., Rajpal A. High-throughput epitope binning assays on label-free array-based biosensors can yield exquisite epitope discrimination that facilitates the selection of monoclonal antibodies with functional activity. PLoS One. 2014;9 doi: 10.1371/journal.pone.0092451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bilic-Zulle L. Comparison of methods: passing and bablok regression. Biochem. Med. 2011;21:49–52. doi: 10.11613/bm.2011.010. [DOI] [PubMed] [Google Scholar]
- 62.Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., Gong Y., Xiao H., Fan Z., Tan S., Wu G., Tan W., Lu X., Fan C., Wang Q., Liu Y., Zhang C., Qi J., Gao G.F., Gao F., Liu L. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi H., Tice C.M., Emert-Sedlak L., Chen L., Li W.F., Carlsen M., Wrobel J.E., Reitz A.B., Smithgall T.E. Tight-binding hydroxypyrazole HIV-1 Nef inhibitors suppress viral replication in donor mononuclear cells and reverse Nef-mediated MHC-I downregulation. ACS Infect. Dis. 2020;6:302–312. doi: 10.1021/acsinfecdis.9b00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bhandari D., Chen F.C., Hamal S., Bridgman R.C. Kinetic analysis and epitope mapping of monoclonal antibodies to Salmonella typhimurium flagellin using a surface plasmon resonance biosensor. Antibodies. 2019;8:22. doi: 10.3390/antib8010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dye J.M., Wu H., Hooper J.W., Khurana S., Kuehne A.I., Coyle E.M., Ortiz R.A., Fuentes S., Herbert A.S., Golding H., Bakken R.A., Brannan J.M., Kwilas S.A., Sullivan E.J., Luke T.C., Smith G., Glenn G., Li W., Ye L., Yang C., Compans R.W., Tripp R.A., Jiao J.A. Production of potent fully human polyclonal antibodies against Ebola Zaire virus in transchromosomal cattle. Sci. Rep. 2016;6:24897. doi: 10.1038/srep24897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fisher Scientific SARS-CoV-2 ELISA kit price. https://www.fishersci.com/us/en/catalog/search/products?keyword=SARS-CoV-2+ELISA
- 67.Jones G., Wortberg M., Kreissig S.B., Hammock B.D., Rocke D.M. Sources of experimental variation in calibration curves for enzyme-linked immunosorbent assay. Anal. Chim. Acta. 1995;313:197–207. [Google Scholar]
- 68.Chang Y.F., Wang W.H., Hong Y.W., Yuan R.Y., Chen K.H., Huang Y.W., Lu P.L., Chen Y.H., Chen Y.M.A., Su L.C., Wang S.F. Simple strategy for rapid and sensitive detection of avian Influenza A H7N9 virus based on intensity-modulated SPR biosensor and new generated antibody. Anal. Chem. 2018;90:1861–1869. doi: 10.1021/acs.analchem.7b03934. [DOI] [PubMed] [Google Scholar]
- 69.Huang L., Ding L., Zhou J., Chen S., Chen F., Zhao C., Xu J., Hu W., Ji J., Xu H., Liu G.L. One-step rapid quantification of SARS-CoV-2 virus particles via low-cost nanoplasmonic sensors in generic microplate reader and point-of-care device. Biosens. Bioelectron. 2021;171:112685. doi: 10.1016/j.bios.2020.112685. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.