Abstract
Objectives
Electronic health records (EHRs) contain a large quantity of machine-readable data. However, institutions choose different EHR vendors, and the same product may be implemented differently at different sites. Our goal was to quantify the interoperability of real-world EHR implementations with respect to clinically relevant structured data.
Materials and Methods
We analyzed de-identified and aggregated data from 68 oncology sites that implemented 1 of 5 EHR vendor products. Using 6 medications and 6 laboratory tests for which well-accepted standards exist, we calculated inter- and intra-EHR vendor interoperability scores.
Results
The mean intra-EHR vendor interoperability score was 0.68 as compared to a mean of 0.22 for inter-system interoperability, when weighted by number of systems of each type, and 0.57 and 0.20 when not weighting by number of systems of each type.
Discussion
In contrast to data elements required for successful billing, clinically relevant data elements are rarely standardized, even though applicable standards exist. We chose a representative sample of laboratory tests and medications for oncology practices, but our set of data elements should be seen as an example, rather than a definitive list.
Conclusions
We defined and demonstrated a quantitative measure of interoperability between site EHR systems and within/between implemented vendor systems. Two sites that share the same vendor are, on average, more interoperable. However, even for implementation of the same EHR product, interoperability is not guaranteed. Our results can inform institutional EHR selection, analysis, and optimization for interoperability.
Keywords: electronic health records, information storage and retrieval, data aggregation, data management, common data elements, data warehousing
INTRODUCTION
Interoperability is the “degree to which two or more systems, products or components can exchange information and use the information that has been exchanged.”1 The widespread implementation of electronic health records (EHRs) has focused attention on the ability to share structured (clinical) data between sites; or, to be more precise, between EHR implementations.2,3 However, interoperability is often discussed as a binary property without consideration of any particular task or purpose. Clearly, it is possible for 2 systems to be able to share some data types (eg, demographics), but not others (eg, laboratory results). In addition, the sharing of a particular data type may be suitable for one purpose but not another. Thus, interoperability is a continuum based on context, rather than an absolute.
Interoperability can be subtyped even further. Interoperability can be assessed between specific EHR implementations (inter-system interoperability), answering the question: Can sites A and B exchange information? Interoperability can also be assessed between EHR vendors, answering the question: Can sites that implement a particular vendor’s product share information? Intra-vendor interoperability refers to the ability to share information between instances of the same vendor’s product (eg, Epic ↔ Epic). Inter-vendor interoperability refers to the ability to share information between instances of different vendor products (eg, Epic ↔ Cerner). The latter question is relevant to sites that are choosing to implement a new or replacement EHR system. Will choosing the same EHR vendor as a likely partner increase interoperability?
In previous work, we found that standards were used consistently for diagnoses (ICD-9/104) and encounter data (CPT5) often for race, and rarely for medications (RxNorm6), laboratory tests (Logical Observation Identifiers Names and Codes [LOINC]7), or biomarkers.8,9 However, selection of EHR vendor is sometimes made on the basis of perceived interoperability. For example, 2 collaborating entities may choose the same EHR vendor in hopes of being able to share data more easily, even if the data do not necessarily conform to a particular standard. To assess inter- and intra-EHR interoperability, we propose an approach to quantitating interoperability and report results based on a real-world sample of 68 sites.
METHODS
Data source
In 2014, the American Society of Clinical Oncology (ASCO), through its wholly owned nonprofit subsidiary CancerLinQ LLC, developed and implemented the health technology platform CancerLinQ to help improve the quality of cancer care and to generate new insights, by delivering a suite of electronic clinical measures and dashboards to oncologists and by enabling the creation of de-identified secondary-use real-world data sets, respectively. CancerLinQ extracts, aggregates, and normalizes structured data sourced primarily from the EHRs of participating centers.10 This presents the opportunity to evaluate the standards in use for structured data across a sample of oncology practices as well as the degree to which the EHRs implemented at these practices are interoperable with respect to structured data.
Sites
For purposes of this analysis a “site” is a CancerLinQ subscriber site, that is, an entity with a Business Associate Agreement, plus an EHR software system. Thus an organization with multiple physical locations or multiple instances of the same EHR vendor system (eg, 2 Epic instances) is considered one site. Conversely, a single subscriber with multiple EHR vendor systems (eg, an instance of Epic and an instance of Cerner) is represented as multiple sites. We included only EHR vendors (eg, Epic, Cerner) for which there were 3 or more sites. We masked EHR vendor names (in part to avoid revealing the identities of sites) but the 5 systems chosen (labeled A–E in the figures) were a subset of the CancerLinQ supported EHR list of: Allscripts (Allscripts, Chicago, IL), Aria (Varian Medical Systems, Palo Alto, CA), Centricity (General Electric, Boston, MA), Cerner (Cerner, North Kansas City, MO), Clarity (Epic Systems, Verona, WI), Intellidose (IntrinsiQ, LLC, Burlington, MA), MOSAIQ (Elekta, Stockholm, Sweden), NextGen (NextGen Healthcare, Irvine, CA), and OncoEMR (Flatiron Health, New York, NY). Using these criteria, 68 sites were analyzed.
Data element selection
We previously found that procedure and diagnosis codes (eg, CPT5 and ICD-9/104]) were consistently used, most likely for billing reasons.8 For this analysis, we selected a set of common laboratory tests and medications (Table 1). We focused on laboratory tests and medications because these data types are associated with standards recommended by the Office of the National Coordinator for Health Information Technology (ONC).11 Specifically, the ONC recommends using LOINC for laboratory results and RxNorm for medications.
Table 1.
Set of data elements used to determine interoperability
| Category | Value | Code system | Coded value |
|---|---|---|---|
| Medication | Acetaminophen | RxNorm | 161 |
| Medication | Cisplatin | RxNorm | 2555 |
| Medication | Dexamethasone | RxNorm | 3264 |
| Medication | Carboplatin | RxNorm | 40 048 |
| Medication | Trastuzumab | RxNorm | 224 905 |
| Medication | Nivolumab | RxNorm | 1 597 876 |
| Lab test | Hemoglobin (mass/volume) in blood | LOINC | 718-7 |
| Lab test | Alanine aminotransferase (enzymatic activity/volume) in serum or plasma | LOINC | 1742-6 |
| Lab test | Creatinine (Mass/volume) in serum or plasma | LOINC | 2160-0 |
| Lab test | Urea nitrogen (Mass/volume) in serum or plasma | LOINC | 3094-0 |
| Lab test | Hematocrit (volume fraction) of blood | LOINC | 20 570-8 |
| Lab test | Leukocytes (#/volume) in blood | LOINC | 26 464-8 |
Abbreviation: LOINC: Logical Observation Identifiers Names and Codes.
To assess medication variability, we selected widely prescribed drugs such as acetaminophen and dexamethasone as well as common cancer therapeutic agents such as carboplatin, trastuzumab, and nivolumab. These selections were based upon consensus of the authors, informed by their domain knowledge and experience with interoperability and health data standards.
For medications, we used both ordered and administered medications, which CancerLinQ manages in separate tables, given the variability that we see in the use of these concepts at various sites. We restricted our analysis to native EHR data, excluding cancer registry data and any data abstracted from clinical notes obtained through a manual curation pipeline.10 We only examined lab and medication name original values and did not assess categorical responses, units, etc.
Variability assessment
CancerLinQ receives data from EHRs as original values, that is, the representation of the value in the EHR. These are transformed by CancerLinQ into codified values, that is, the same content but coded to a controlled biomedical terminology (see Potter et al10 for a description of this process) via specific rules. These transformations are both syntactic (ie, structural) and semantic (ie, related to meaning). For example, a cancer stage group listed as “2A” in the EHR is transformed from whatever table and variable name is used locally into a triplet in the CancerLinQ procedure_performed table. For our example, the triplet comprises the stagegroup_codesystem that defined the specific controlled terminology used for that particular datum (ie, SNOMED CT12) a stagegroup_code, the concept unique identifier (CUI) from the code system that represents the value “2A” (SNOMED CUI 261614003); and a stagegroup_codelabel that has a human readable form of the coded value (“Stage 2A”).
Some data elements, such as laboratory tests, medication orders, and administration events were constructed from multiple triplets. In these cases, the full semantics of the data element are captured by their presence in the specific table plus a series of triplets. For a laboratory test that would involve a labtestname triplet and either a labtestvalue numeric field and labtestunits triplet or a labtestvalue triplet for tests with categorical values (eg, positive/negative, 1+, 2+, 3+, etc.). For a medication order, the element is constructed from the presence in the order table and a set of triplets that describe medicationname, medicationroute, medicationunits, etc. In addition to the triplets, CancerLinQ retains the “original value,” making each triplet a quadruplet, although these values are neither presented to users nor included in secondary-use de-identified datasets for researchers.
Data are frequently received from EHRs in inconsistent ways. For example, laboratory tests from the same system can be received as LOINC7 coded values with the local “original values” attached, or just as the “original value” string. Some systems provide only the “original value” strings, and even where the test is coded, the coding can represent different levels of abstraction. For example, a test for hematocrit can be coded as a hematocrit or as a Complete Blood Count, meaning that the original value string is often key to understanding what is transferred. For this reason, we used these “original value” strings to assess the degree of variability.
Initial assessments of variability were determined by simply counting the number of distinct inbound original values that were coded to the same codified value with larger numbers indicating greater variability. That is, we counted the number of distinct original values from an EHR that were mapped to the same value. We focused on the name of the medication or test rather than the results (which would have undoubtedly added additional variability to the equation). Further, mapping requires clinical judgment (eg, should venous hematocrit be mapped the same value as arterial hematocrit). We did not analyze mapping accuracy and assumed that judgments made by human analysts to be correct.
Note that we defined “distinct” to mean string variants. This included names with variant capitalization (eg, Hematocrit vs hematocrit) or medications that attach dosage units or have different spacing. In addition, we coded to ingredient levels for medications (ie, any RxNorm6 variant or text string representing acetaminophen, including dose variants and brand vs generic medications). This approach is important because meaning can depend on capitalization, for example, estimated Glomerular Filtration Rate (eGFR) versus Epidermal Growth Factor Receptor (EGFR) and because machines do not apply clinical knowledge.
Interoperability assessment
We developed an objective measure of the likelihood that an unmodified record transmitted from one site to another can be understood. For this purpose, we define “understood” to mean that the specific text string used to define the meaning of the data element was used in the recipient site. This presupposes that the data item is meaningful in the recipient site, but we view this as reasonable given that it suggests that the data are comparable with data already existing in the recipient system (eg, can count the number of patients with a hematocrit < 30%).
We first created a series of matrices that represent the variability for each of the selected lab tests and medications within each site. These matrices, designated OV[name] had n rows each representing a site with a particular EHR and m columns with each representing one of the inbound original values that was mapped to a specific LOINC code or RxNorm ingredient code across the entire database. Each cell O[name]i, j contained the fraction of inbound values at site i that were represented using form (representation) j. Thus, if the jth original value for hematocrit was “Hct,” then O[name]i, j would represent the fraction of hematocrit values from site i that were indicated by an entry of “Hct.” Similarly if the kth original value was “hematocrit,” then O[name]i, k would represent the fraction of hematocrit values from site i that were indicated by an entry of “hematocrit.” The sum of each row equals 1 as each cell is a fraction of the data that was recorded using the particular original value. The maximum value for the sum of a column is m if and only if all sites used the same representation for that test or variable.
Calculation of interoperability for a data element value pair
We calculated probability matrices (P) that a value sent from site i would be understood at site j. The matrices each have n rows and columns, with each row and column representing a site, and each cell Pi, j representing the probability that a value transmitted from site i to site j (as an original value) had been seen before. The probability was calculated as follows:
For example, consider a data type with 4 original values, W, X, Y, and Z. Site A is distributed 33% W, 33% X, and 34% Y. Site B is distributed 25% each for W, X, Y and Z. P1,2 will be 1.0, since all variants from site A are seen in site B (calculation is 0.33(1)+0.33(1)+0.33(1)+0(0)). The measure is not symmetric. PB, A = 0.75 as 25% of the values at site B are not present at site A. The diagonal is always 1.
The calculation yielded a 68 by 68 matrix where each row represents a sending site and each column a receiving site. The diagonal represents site A versus site A and so it has the value of 1. The matrix is not symmetric across the diagonal. For example, consider 2 sites (sites A and B) with 2 common representations (ie, both A and B have these 2 representations) and one additional representation at the second site (ie, site B has one additional representation, thus 3 different representations at site B, but only 2 at site A). Since all of the representations from site A (2 representations) will be understandable at site B (3 representations), but the reverse is not true.
Calculation of inter-site interoperability
Intuitively, we considered inter-institutional interoperability to be a function of the data element pair interoperability (described above). In other words, institutional EHR implementations were interoperable to the extent that they could meaningfully exchange data element pairs. We created a virtual 3-dimensional matrix consisting of the interoperability matrices described above. Functionally, the x and y axes are sites as before, and the z axis moves along the elements listed in Table 1. We then calculated the mean for each x and y pair across the z dimension, yielding an inter-site interoperability score matrix. This is another 68 by 68 matrix with a diagonal that is filled with 1s, and that is not symmetric across the diagonal.
Conceptually, we took the probability matrices for each of the tested values and created a 3-dimensional n × n × v matrix where v is the number of laboratory results or medications examined. Each of the first 2 dimensions represents an EHR vendor plus site combination and the third dimension represents a score for one of the measured data elements. We then calculate an inter-site interoperability score for site i and site j by calculating the mean interoperability score for all of the individual measures for those 2 sites as below:
Intuitively, we are taking a slice out of the z-dimension of the 3-dimensional matrix for each value in the x- and y-dimensions and calculating the mean value in that slice, and then placing this value in a new 2-dimensional matrix I of dimension n × n with each row and column representing a site. The maximum value for each cell is 1 and the minimum is 0. The diagonal is defined to be 1; however, in this matrix we set the Ii, i to null where i = j to support our final analysis. Each data type is weighted equally (ie, interoperability of acetaminophen is equally important as interoperability of hematocrit).
Calculation of inter- versus intra-electronic health records vendor interoperability
Intuitively, inter-vendor interoperability reflects the ability of implementations of different vendor products to meaningfully share data (eg, Epic ↔ Cerner). Similarly, intra-vendor interoperability reflects the ability of same-vendor implementations to meaningfully share data (eg, Epic ↔ Epic). To determine inter-vendor interoperability, we computed the average of the interoperability scores of each EHR vendor versus other EHR vendors. Functionally, we calculated the mean of a series of regions representing each EHR. Intra-vendor interoperability regions are focused around the diagonal and inter-vendor interoperability scores are off of the diagonal. We did not include the values along the diagonal in the calculation of the intra-vendor interoperability because the diagonal represents interoperability within a specific instance of an EHR system (ie, the site exchanging data with itself, which is by definition 1.0 and would artificially inflate the interoperability score). This yields a 5 by 5 matrix that provides a rough measure of interoperability between implemented EHR vendor products, as opposed to between 2 specific EHR instances.
RESULTS
We analyzed 12 data elements at 68 sites. Thus, these calculations yielded a series of 12 matrices of size 68 by n where n is the number of distinct inbound values for each individual data element. As an example, consider the hypothetical element that indicates smoking status (not evaluated in this work) that is labeled “Smoking” in 20 EHR implementations, “Smoking Status” in 20 EHR implementations, “Smoke Stat” in 20 EHR implementations, and “Tobacco Use” in 30 EHRs. The matrix for this data element would have 68 rows and 4 columns. If EHR implementation number 1 had 95% of its smoking status data coded to “Smoking” and 5% to “Tobacco Use,” the first row of the matrix would be [0.95,0,0,0.05]. The rest of the matrix would be calculated similarly for each of the other EHR installations. The key is that the number of columns in the matrix defines the totality of all representations of the same data element across all EHRs examined.
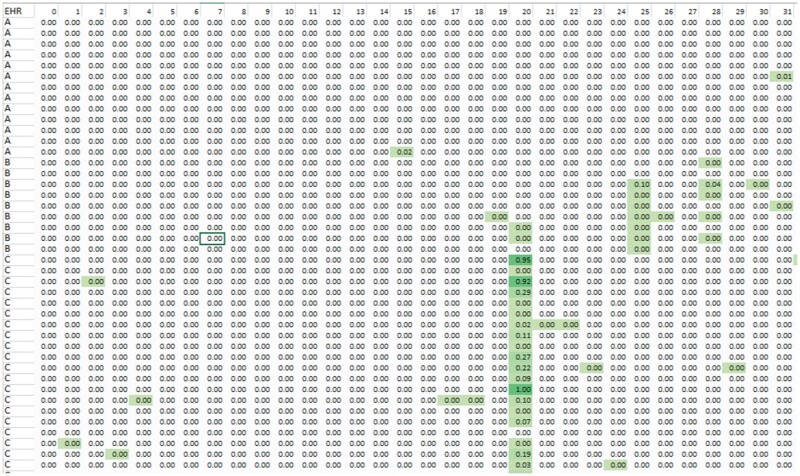
A sample portion of such a matrix is shown in Figure 1. Notably, even standard data elements such as hematocrit had many different representations. As indicated, the matrix values are the fractions of each row that is represented as this original value, and each row sums to 1.0. Generally, highly consistent representations of data internal to a single implementation of an EHR yield rows with single cells with high scores (eg, the cells in Figure 1 with values of 0.95 or 1.0). Internally inconsistent representations will have values distributed across multiple cells (eg, a number of EHR “C” installations represent this data element using representation 20 for this data element from 19% to 27%, with other values comprising the remainder of the 73% to 81% of the values.)
Figure 1.
Representation matrix. Rows are instances of electronic health records vendor products (eg, implementation of vendor A, B, etc.), columns are distinct representations of a particular data item (eg, hematocrit). Rows do not sum to 1.0 because we are showing only a portion of the entire matrix. The cells are color-coded by value (darker green = larger fraction of data following that representation).
Interoperability for a data element value pair
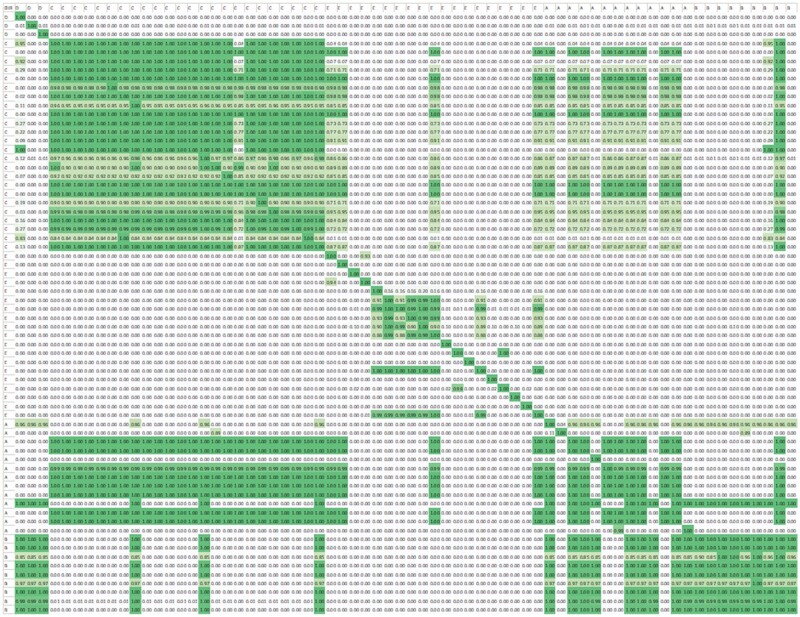
Figure 2 shows the 68 by 68 matrix. Each row represents a sending site and each column a receiving site. Visually, it is apparent that sites that share the same vendor tend to be relatively interoperable, but this is not universally true. For example, although sites implementing vendor C are relatively interoperable with other sites implementing vendor C, they are also interoperable with some, but not all sites implementing vendor A.
Figure 2.
Interoperability matrix for an example data element. Each column row represents a vendor product implemented at a site for an example data element.
Inter-site interoperability
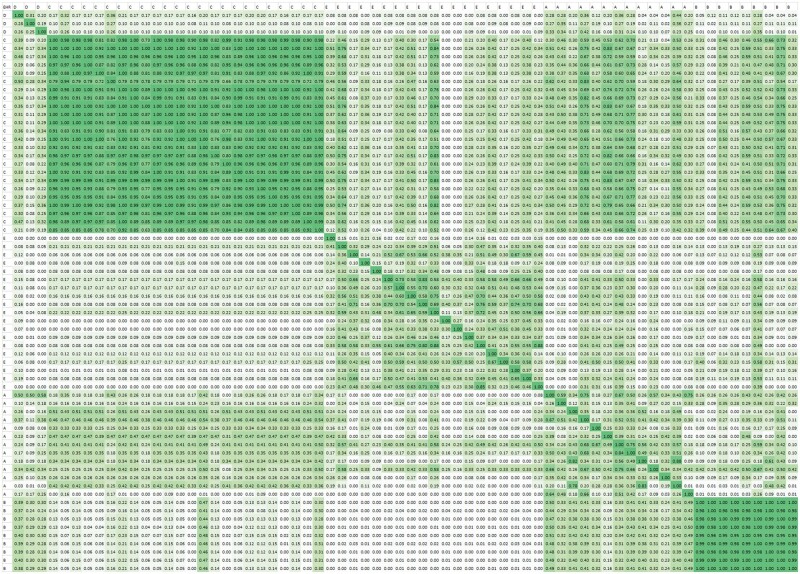
Figure 3 shows the resulting 68 by 68 matrix. Again, sites implementing the same vendor product tend to be more interoperable. However, this is neither guaranteed nor exclusive. For example, sites implementing vendor A vary with respect to their interoperability with each other and are somewhat interoperable with some, but not all sites implementing vendor C.
Figure 3.
Interoperability matrix for all data elements (inter-site interoperability). A–E represent 5 distinct vendors. Each cell represents the interoperability between products grouped by vendor on the X and Y axes.
Inter- versus intra-electronic health records vendor interoperability
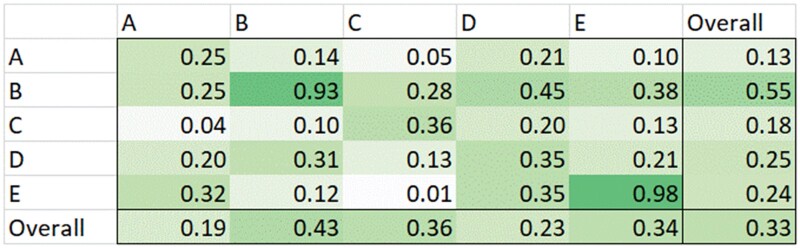
Figure 4 quantitatively summarizes the inter- and intra-vendor interoperability.
Figure 4.
Inter- versus intra-electronic health records interoperability.
The mean intra-vendor interoperability score was 0.68, compared to a mean of 0.22 for inter-vendor interoperability, when weighted by number of sites implementing the particular vendor’s product, and 0.57 and 0.20 when not weighting for the number of sites implementing that vendor’s product.
DISCUSSION
We proposed an approach to calculate objective interoperability scores and demonstrated this approach using data from 68 real-world EHR implementations from a variety of vendors. We found that inter-vendor interoperability was lower than intra-vendor interoperability (0.22 vs 0.68, respectively, if weighted by the number of implementing sites). Thus, 2 sites that implement the same vendor product are more likely to be able to share data (laboratory test results and medications). However, interoperability is far from perfect.
To our knowledge, this is the first attempt to define and demonstrate an objective approach to quantifying interoperability using real-world structured data within EHRs. A major strength of our study is the relatively large sample of 68 sites with real-world data that were collected for routine clinical care. Although we did not include all EHR vendors, our sample included 4 of the 7 most frequently implemented vendors based on expenditure-weighted Meaningful Use attestation rates of certified EHRs.13
We recognize that the specifics of our interoperability score represent a particular set of choices and that a community consensus might differ from these choices. Initially, we examined a measure based on vector distances, that is, we treated each row on the initial distribution matrix (O) as a vector, and constructed the equivalent of P using P(i, j) as the vector distance between rows i and j. Such a measure created a transposable matrix (in contrast to the measure we ultimately selected) that provided a good measure of similarity, but did not reflect whether the value had been seen, and therefore was likely to be understood, at the receiving site.
The particular data elements that we selected to assess interoperability and the even weight given to these selections are an example, not a definitive recommendation for calculating a formal and reproducible interoperability score. Many important categories of clinical data were not included. Since our data were derived from oncology practices, we chose a small subset of medications, some used to treat a variety of conditions (eg, acetaminophen, dexamethasone) and some cancer-specific (eg, cisplatin, carboplatin). Similarly, we chose a small subset of laboratory test results. Less commonly prescribed medications and less frequently checked laboratory values are likely to be even less interoperable.
Another limitation is that we did not leverage EHR data extraction and export tools. For example, we did not make our assessment based on Clinical Data Architecture (CDA) extracts or translation layers that might be included in some EHR implementations but not others. Instead, we analyzed the data present in the EHR. The use of directly stored values could be an issue if the EHR stores coded information rather than as understandable text. For example, some EHRs code performance status based on the Eastern Cooperative Oncology Group scale as “2.” The meaning of “2” is driven by a combination of the context in which it is stored and the values that are used to represent the data element. Within the EHR (and potentially in a CDA extract) this information could be clearly understandable, but becomes functionally useless without that context.
Organizations with multiple instances of the same vendor product (eg, multiple Epic instances, but no Cerner instances) were considered one site. Thus, we were not able to calculate “within site” variability for organizations with multiple instances of the same vendor product. Due to restrictions on the data set imposed for confidentiality reasons, we cannot re-analyze the data to address this question, so it must be left to future work. However, we expect this situation to occur rarely, if ever, thus the impact on our results was likely small. Due to variable use of coded values, we measured interoperability using local “original values.” Over time, we saw substantial improvement in the capture of coded information, particularly in laboratory tests, but many key oncology tests remain uncoded in the EHR due to the mechanics of collecting the information. One prime example of this is estrogen and progesterone receptor and human epidermal growth factor receptor 2 (HER2) status in breast cancer. Because this information is manually transcribed from pathology reports by clinicians, it often comes with no coding, despite the fact that the actual value and methods (particularly for HER2) can be crucial for making clinical decisions.
We hope that the informatics community will take our work and expand upon it to develop a nuanced and relevant measure (or multiple measures) of interoperability. Multiple measures may be needed because some medical specialties, such as radiation oncology, may require assessment of data elements that are not relevant for general EHRs. Interoperability measures should also account for increased use of coded values, and whether the coding specifics improve or hinder interoperability.
An alternative to developing specialty-specific measures is to create an interoperability profile based on data types such as medications, laboratory results, risk assessments, etc. This approach has the advantage of allowing for reasonable interoperability measures with systems that support limited subsets of medical care (eg, ePrescribing or imaging). Thus, 2 systems may be more interoperable with respect to medications, for example, than with respect to laboratory test results. We expect that all such approaches will require careful selection of data elements and values to measure, and determinations of reasonable weights for each of the elements (eg, common medications may be weighted more than rare medications).
In practice, interoperability is determined by a combination of product features chosen by the vendor (eg, defaults) and implementation choices made by the institution (eg, how to represent a particular data type, legacy data, etc.). A potentially interesting consequence of implementing objective measures of interoperability is that vendors or institutions will focus interoperability efforts on the data elements chosen for assessment of interoperability. Just as in the economics domain where the Consumer Price Index can be manipulated to artificially reduce apparent inflation, it will be important to ensure that certification does not replace poor interoperability with poor interoperability except for a few chosen data elements.
In this article, we describe a quantitative approach to measuring interoperability using real-world data. We demonstrated our approach by assessing interoperability between sites and implementations of vendor products. Overall, interoperability was relatively poor with a maximum of 0.68. In the most favorable case (intra-vendor, weighted for the number of sites) approximately two-thirds of data types will be “understood” by a receiving site. Thus, institutions implementing products sold by the same vendor are likely to be more interoperable than institutions implementing products sold by different vendors. However, vendor choice does not ensure reliable interoperability. Reliable interoperability requires institutions to map their data to the same standards and ensure that mapping practices are consistent across institutions.
FUNDING
This work was supported in part by the National Center for Advancing Translational Sciences (NCATS) under awards UL1TR000371 and U01TR002393; the Cancer Prevention and Research Institute of Texas (CPRIT), under award RP170668, ASCO CancerLinQ, LLC and the Reynolds and Reynolds Professorship in Clinical Informatics.
AUTHOR CONTRIBUTIONS
EVB, JLW, JCK, EA, WR, GK, RSM, and JLC conceived the study idea. EVB, RSM, and GK wrote the initial manuscript. GK performed the data analysis. All authors revised and expanded the manuscript. GK, RSM, WR provided the data. All authors reviewed and approved the manuscript prior to submission.
ACKNOWLEDGMENTS
Dr. Rubinstein participated in this work prior to joining the Food and Drug Administration. This work and related conclusions reflect the independent work of study authors and does not necessarily represent the views of the Food and Drug Administration or US government.
CONFLICT OF INTEREST STATEMENT
Wendy Rubinstein, George Komatsoulis, and Robert S. Miller made contributions to this study while employees of CancerLinQ, LLC.
DATA AVAILABILITY
The data underlying this article cannot be shared publicly due to CancerLinQ policies.
Contributor Information
Elmer V Bernstam, School of Biomedical Informatics, The University of Texas Health Science Center at Houston, Houston, Texas, USA; Department of Internal Medicine, McGovern Medical School, The University of Texas Health Science Center at Houston, Houston, Texas, USA.
Jeremy L Warner, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
John C Krauss, University of Michigan Medical School, Ann Arbor, Michigan, USA.
Edward Ambinder, The Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, New York, USA.
Wendy S Rubinstein, CancerLinQ LLC, American Society of Clinical Oncology, Alexandria, Virginia, USA.
George Komatsoulis, CancerLinQ LLC, American Society of Clinical Oncology, Alexandria, Virginia, USA.
Robert S Miller, CancerLinQ LLC, American Society of Clinical Oncology, Alexandria, Virginia, USA.
James L Chen, Division of Medical Oncology and Department of Biomedical Informatics, The Ohio State University, Columbus, Ohio, USA.
REFERENCES
- 1.ISO/IEC/IEEE. Systems and software engineering – Vocabulary 2017. https://standards.iso.org/ittf/PubliclyAvailableStandards/c071952_ISO_IEC_IEEE_24765_2017.zip (item 3.2089, p. 235). Accessed December 30, 2021.
- 2. Walker J, Pan E, Johnston D, et al. The value of health care information exchange and interoperability. Health Affairs 2005; 24 (Suppl1): W5-10–18. [DOI] [PubMed] [Google Scholar]
- 3. Halamka JD, Tripathi M. The HITECH era in retrospect. N Engl J Med 2017; 377 (10): 907–9. [DOI] [PubMed] [Google Scholar]
- 4.Classification of Diseases (ICD). World Health Organization. https://www.who.int/standards/classifications/classification-of-diseases. Accessed September 17, 2021.
- 5.Current Procedural Terminology (CPT). American Medical Association. https://www.ama-assn.org/amaone/cpt-current-procedural-terminology. Accessed September 17, 2021.
- 6.RxNorm. National Library of Medicine (National Institutes of Health). https://www.nlm.nih.gov/research/umls/rxnorm/index.html. Accessed September 17, 2021.
- 7. McDonald CJ, Huff SM, Suico JG, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem 2003; 49 (4): 624–33. [DOI] [PubMed] [Google Scholar]
- 8. Bernstam EV, Warner JL, Ambinder E, et al. Continuum of interoperability in oncology EHR implementations. In: Proceedings of the AMIA Fall Symposium; November 16–20, 2019, pp. 1304–5; Washington, DC.
- 9. Bernstam EV, Warner JL, Krauss JC, et al. Quantifying interoperability: an analysis of oncology practice electronic health record data variability. J Clin Oncol 2019; 37 (15_suppl): e18080. [Google Scholar]
- 10. Potter D, Brothers R, Kolacevski A, et al. Development of CancerLinQ, a health information learning platform from multiple electronic health record systems to support improved quality of care. J Clin Cancer Inform 2020; 4: 929–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Office of the National Coordinator for Health IT, 2020 Interoperability Standards Advisory, Reference Edition. Washington, DC.
- 12.SNOMED - Home | SNOMED International. https://www.snomed.org/. Accessed September 17, 2021.
- 13. Sorace J, Wong H-H, DeLeire T, et al. Quantifying the competitiveness of the electronic health record market and its implications for interoperability. Int J Med Inform 2020; 136: 104037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article cannot be shared publicly due to CancerLinQ policies.