Abstract
Replication fork stalling occurs when the replisome encounters a barrier to normal fork progression. Replisome stalling events are common during scheduled DNA synthesis, but vary in their severity. At one extreme, a lesion may induce only temporary pausing of a DNA polymerase; at the other, it may present a near-absolute barrier to the replicative helicase and effectively block fork progression. Many alternative pathways have evolved to respond to these different types of replication stress. Among these, the homologous recombination (HR) pathway plays an important role, protecting the stalled fork and processing it for repair. Here, we review recent advances in our understanding of how blocked replication forks in vertebrate cells can be processed for recombination and for replication restart.
Keywords: Fanconi anemia, homologous recombination, replication restart, tandem duplication, structural variation
Introduction
Damaged DNA can present a barrier to replication, stalling either individual DNA polymerases or the replicative helicase and thereby the entire replisome. Stalling lesions include certain types of base damage, DNA adducts, DNA-protein crosslinks (DPCs) and inter-strand crosslinks (ICLs). Fork stalling can also occur at hairpin structures and G4 quadruplexes in undamaged DNA [1, 2], or from collisions between replication and transcription [3, 4]. The ICL, which can be formed from endogenous aldehydes [5], is the most formidable of fork barriers, since it covalently binds the two parental DNA strands. Unless it can be disrupted or bypassed, the ICL is an absolute barrier to the replicative helicase. In higher eukaryotes, ICLs encountered during replication can be processed by the Fanconi anemia (FA) pathway, a tightly choreographed, multi-step pathway that processes the ICL-blocked fork for repair by homologous recombination (HR) [6, 7]. The FA pathway has been the focus of intense study because of its pivotal role in preventing genomic instability in cycling cells. The mitotic recombinase Rad51 (eukaryotic homolog of bacterial RecA) has canonical roles in the repair of double-strand breaks (DSBs) by HR. It also has non-canonical roles at stalled forks, where it enables fork reversal and protects nascent strands from nucleolytic attack. These non-canonical functions of Rad51 have been reviewed recently, as have non-recombinational pathways of replication-coupled repair [8, 9].
In the repair of a replication-independent DSB, HR can have either error-free or error-prone outcomes. The latter entail aberrant replicative responses, exemplified by the phenomenon of break-induced replication (BIR) in yeast [10]. In BIR, Rad51-mediated strand exchange of a one-ended break establishes stable DNA synthesis, efficiently copying to the end of the donor chromosome at a rate slower than conventional DNA synthesis [11]. DSB-induced BIR entails conservative DNA synthesis; both strands of the BIR tract are newly synthesized by Polδ through a ‘bubble migration’ mechanism [12–14]. Leading and lagging strand synthesis may become uncoupled during BIR, provoking mutagenesis [15]. In contrast, conventional DNA replication is semi-conservative; leading and lagging strand synthesis (by Polε and Polδ respectively) are coordinated at the unperturbed fork. In Saccharomyces cerevisiae, BIR is mediated by the specialized helicase Pif1 and requires the non-essential DNA polymerase subunit Pol32 [12, 16]. Replicative HR responses are also seen in mammalian cells, although they appear to be less robust than yeast BIR [17–21]. Replication across a nicked DNA template induces one-ended breaks and loss of the replicative CMG helicase, potentially triggering BIR-like fork restart [17, 22, 23]. Recent work, discussed below, suggests that BIR-like responses can also occur at blocked replication forks, where they may either limit or promote genomic instability.
In this review, we will assess recent advances in our understanding of the FA pathway. We will also review current models of replication fork restart, based in part on work using site-specific replication fork barriers (RFBs).
Conservative HR at stalled forks: the Fanconi anemia pathway
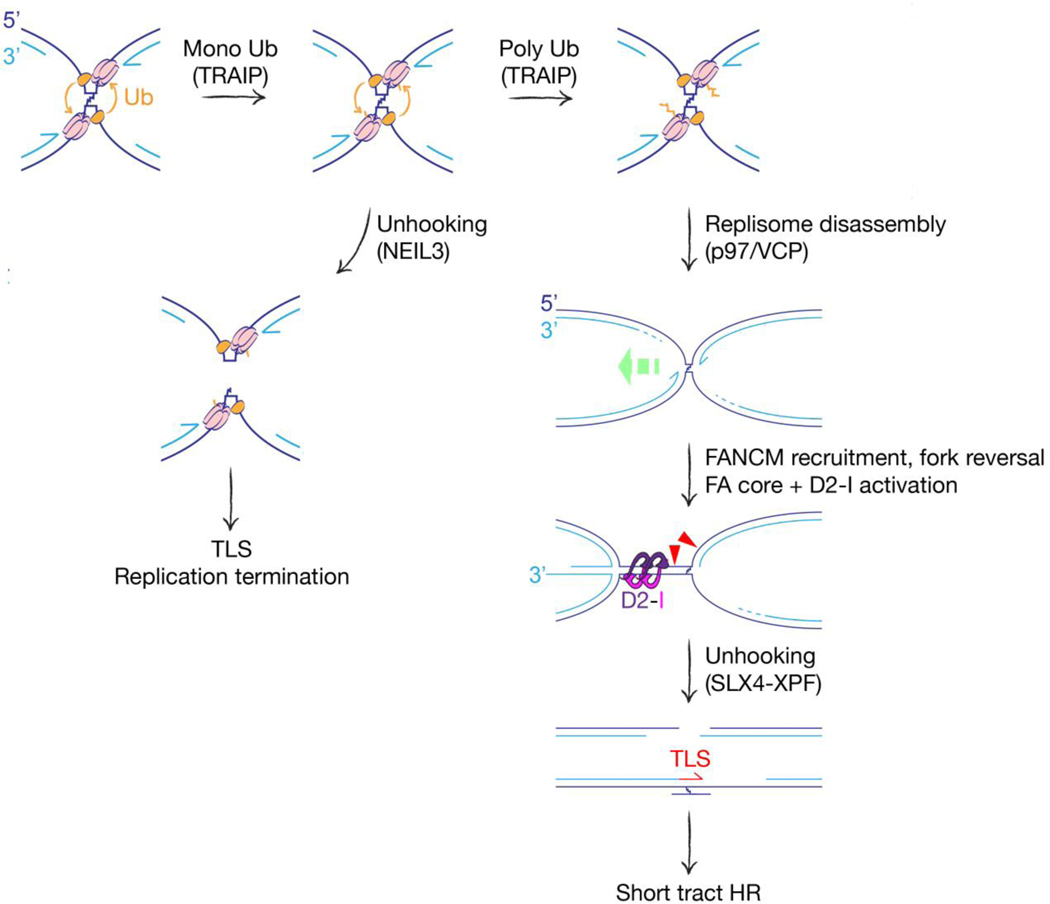
Fanconi anemia is a rare autosomal recessive or X-linked syndrome characterized by developmental abnormalities, bone marrow failure and cancer predisposition. At least 23 FA genes define a pathway of replication-coupled ICL repair [6, 24]. Much of our knowledge of the molecular steps of the FA pathway comes from analysis of ICL repair in plasmids replicating in frog egg extracts. An orchestrated series of steps, initially triggered by the stalling of two opposing forks at the ICL (‘bidirectional’ fork stalling), leads to scheduled incisions either side of the ICL on one sister chromatid, thereby ‘unhooking’ the ICL and generating a two-ended DSB intermediate (Figure 1). Translesion synthesis (TLS) DNA polymerases fill the gap opposite the unhooked ICL and the two-ended DSB is repaired by conservative ‘short tract’ HR. This pathway effectively transforms a highly dangerous lesion—the ICL—into a more benign lesion, at the cost of localized mutation. A recent review has addressed each of these steps in detail, including discussion of the caveats of studying ICL repair in small replicating plasmids [24]. Some ICLs can be hydrolyzed by endogenous enzymes, converting them to less deleterious lesions. These include psoralen-UV-induced crosslinks, which can be hydrolyzed by the NEIL3 glycosylase [25], and acetaldehyde-induced crosslinks, which can be reversed by an as yet uncharacterized mechanism [26]. Each of these repair pathways requires bidirectional fork stalling at the ICL. A pathway of ICL ‘traverse’, mediated by the DNA translocase FANCM, enables the CMG helicase to bypass some ICLs [27, 28]. Thus, the ‘decision tree’ of ICL repair is complex and lesion-specific.
Figure 1. Pathway choice in replication-coupled ICL repair.
Bidirectional fork stalling activates TRAIP (orange), which ubiquitinates CMG components in trans (MCM subunits shown in pink). Short ubiquitin chains recruit NEIL3 glycosylase, providing an opportunity for direct unhooking of ICL. TLS: Translesional synthesis. Long ubiquitin chains recruit the p97/VCP ATPase, which extracts replisome components and disassembles the replisome. FANCM recognizes the collapsed fork and recruits the FA core complex, which activates FANCD2-I by monoubiquitination. Green dashed arrow: fork reversal. Activated FANCD2-I forms a sliding clamp on dsDNA—possibly cloaking the reannealed parental strands of the reversed fork, as shown. Fork reversal and D2-I monoubiquitination are both required for activation of SLX4-XPF. XPF-mediated incisions (red triangles) unhook the ICL, setting up gap filling by TLS and repair of the two-ended DSB by conservative ‘short tract’ HR.
One of the earliest steps of the FA pathway is the disassembly of replisome components of the stalled fork (‘fork collapse’). Recent work has identified the E3 ubiquitin ligase TRAIP as a key mediator of this step [29]. TRAIP travels with the replisome and ubiquitylates in trans MCM components of the opposing CMG helicase, which are then extracted by the p97/VCP ATPase for proteasomal degradation. TRAIP itself regulates the selection of ICL processing pathways. TRAIP-mediated MCM monoubiquitination recruits NEIL3, providing an opportunity for ICL hydrolysis, while TRAIP-mediated MCM polyubiquitination recruits p97/VCP, thereby channeling repair towards the FA pathway (Figure 1). Recognition of the stalled fork by FANCM and its associated proteins enables recruitment of the FA core complex—an E3 ubiquitin ligase that monoubiquitinates and activates the FANCD2/FANCI heterodimer [24]. In parallel with these events, the bidirectionally stalled fork undergoes asymmetric fork reversal, converting the X-shaped bidirectional stall site to a ‘chicken foot’ on one side of the ICL and a simple stalled fork on the other [30] (Figure 1). Fork reversal and monoubiquitination of FANCD2/I are each required for the incision/unhooking step, which is orchestrated by SLX4/FANCP and the associated endonuclease XPF/FANCQ [31, 32]. XPF-mediated ICL unhooking requires fork reversal [30]. Additional nucleases including SNM1A and FAN1 may also participate in the unhooking mechanism [33, 34].
The identity of the motor protein(s) that mediate fork reversal in the FA pathway remains unclear. FANCM itself is a candidate, since its motor function can reverse replication forks in vitro [35]. Our work on the Tus/Ter site-specific RFB has shown that conservative, two-ended ‘short tract’ HR triggered by fork stalling at Tus/Ter is specifically mediated by the FA pathway [36, 37]. FANCM motor function is required for efficient Tus/Ter-induced HR, further underscoring FANCM as a candidate mediator of fork reversal/remodeling in the FA pathway. However, the stalled fork can recruit numerous additional motor proteins, including known fork reversal enzymes SMARCAL1, ZRANB3, HLTF and FBH1, as well as the RecQ helicases BLM (product of the Bloom’s syndrome gene) and RECQ1 [38–42]. Some combination of these enzymes might be required for efficient fork remodeling during FA pathway activation.
Recent cryo-electron microscopy structural studies have provided important insights into the mechanisms of action of the multi-subunit FA core complex and its target, FANCD2-FANCI [43–46]. The FA core complex forms an extended, asymmetric dimer in which all FA core components are represented twice, with the exception of FANCC, FANCE and FANCF. The binding of the CEF subcomplex to one FANCL subunit inactivates it as an E3 ubiquitin ligase, leaving the second FANCL subunit available to bind the E2 UBE2T and to monoubiquitinate FANCD2-I. Structural studies of monoubiquitinated FANCD2-I heterodimers produced additional surprises. FANCD2-I monoubiquitination remodels the heterodimer, creating a channel that encircles double stranded DNA [43, 45]. Activated D2-I is therefore a sliding DNA clamp. Ubiquitin locks the complex in this configuration, enabling D2-I to form filamentous arrays on dsDNA in vitro, but monoubiquitinated D2-I does not directly recruit SLX4 [47]. These new findings establish new modes of action of key players in the FA pathway. They also raise a host of new questions. For example, which tracts of dsDNA near the stalled fork are clamped by activated FANCD2-I? One possibility is that activated D2-I encircles the re-annealed parental duplex produced by fork reversal, generating a specialized, D2-I-cloaked nucleosome-free zone that facilitates the action of SLX4/XPF [24] (Figure 1).
Replication restart at stalled forks: BIR and its relatives
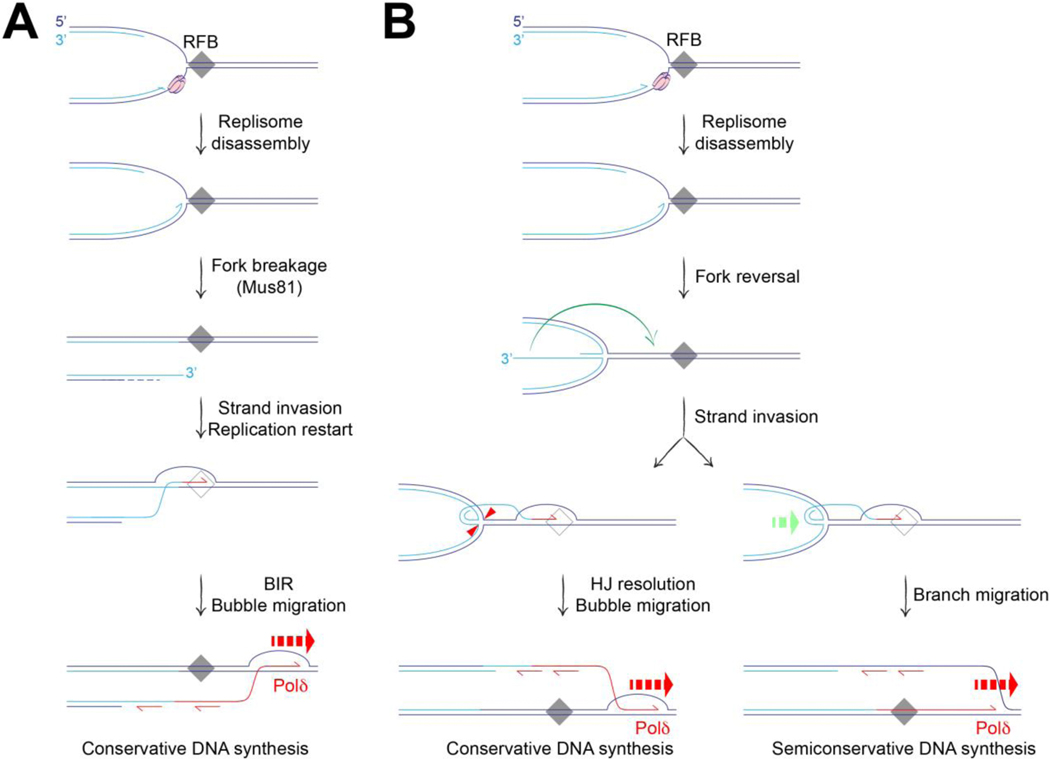
The resumption of replication at sites of fork stalling can be mediated by several different mechanisms. At its simplest (for example, following removal of a transient fork barrier), fork restart can entail the resumption of normal semiconservative DNA synthesis, supported by the CMG helicase. The CMG helicase can bypass DPCs and ICLs under some circumstances [27, 48], raising the possibility that it might sometimes be retained on chromatin during stalled fork remodeling. However, following its disassembly or loss, CMG reloading is not thought to be possible until the G1 phase of the following cell cycle. Therefore, a collapsed fork that is also broken, for example by the Mus81 nuclease, could only be restarted by non-canonical mechanisms, such as BIR (Figure 2A). An assumption underlying some models of fork restart (e.g., Figure 2A) is that certain RFBs are not absolute blocks, but are permeable under some circumstances. DNA synthesis through the RFB might require a switch in DNA polymerase or helicase use—as would occur during BIR-mediated restart—or the activation of mechanisms that degrade the RFB. High affinity protein-DNA complexes, DPCs and DNA:RNA hybrids (R-loops) exemplify this type of robust but permeable RFB. In Schizosaccharomyces pombe, aberrant replication restart at the RTS1 RFB (a protein-DNA complex) occurs by ‘homologous recombination–restarted replication’ (HoRReR) [49]. HoRReR entails a semi-conservative copying mechanism in which Polδ mediates both leading and lagging strand synthesis. The semi-conservative mechanism distinguishes HoRReR from BIR, and raises questions of how it is initiated at the stalled fork. One possibility is that Rad51 mediates invasion of a DNA end formed at the reversed fork into parental duplex, facilitating resumption of DNA synthesis (Figure 2B). How the Holliday junction (HJ) at the reversed fork is processed would determine the subsequent copying mechanism. HJ resolution could establish conservative synthesis (BIR-type copying), whereas HJ dissolution by branch migration would favor semi-conservative synthesis (HoRReR) (Figure 2B) [50]. These models illustrate the ‘topological alchemy’ that can occur when classical DSB repair mechanisms interact with pre-existing branched DNA structures at the stalled fork.
Figure 2. Aberrant fork restart initiated by a strand invasion step.
A. Break-induced replication can be triggered by fork breakage, either from collision of the fork with a nicked DNA template (not shown), or following replisome disassembly/fork collapse at a replication fork barrier (RFB). Fork collapse may expose the stalled fork to nucleases such as Mus81, leading to fork breakage and formation of a one-ended break. Strand invasion initiates BIR (conservative Polδ-mediated synthesis by bubble migration) at the site of fork breakage. B. HoRReR (semi-conservative Polδ-mediated DNA replication) can restart synthesis at an RFB. Initiation might occur following HR-dependent strand invasion of the parental duplex by the HoRReR (semi-conservative Polδ-mediated DNA replication) can restart synthesis at an RFB. Initiation might occur following HR-dependent strand invasion of the parental duplex by the solitary DNA end of a reversed fork (green arrow). Depending on how the Holliday junction (HJ) at the reversed fork is processed, the copying mechanism could be either conservative or semi-conservative, as shown. Red arrowheads: incisions of HJ resolution. Green dashed arrow: branch migration mediates HJ dissolution.
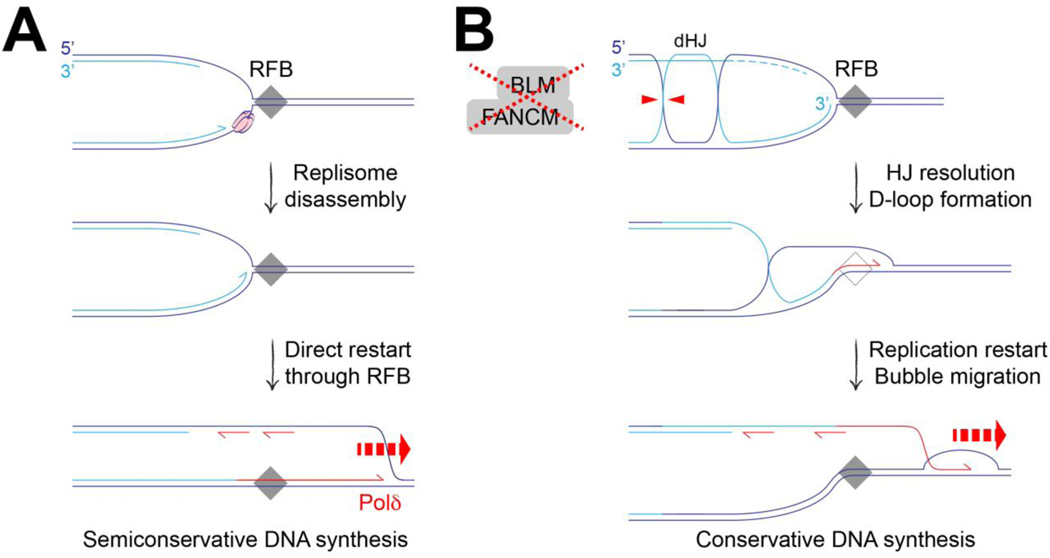
BIR-like or HoRReR-like replication restart could also occur in the absence of an initiating strand exchange event. Simple restart following replisome disassembly could produce HR-independent HoRReR-like copying, provided that the original RFB is an not absolute block (Figure 3A). Our work on tandem duplications (TDs) led us to consider how BIR-like restart might occur without an initiating strand exchange step [36, 51]. Small ~10 kb TDs (‘Group 1’ TDs) form specifically in BRCA1-linked breast and ovarian cancers, and the Tus/Ter system recapitulates this process [51–53]. Tus/Ter-induced TDs in BRCA1 mutants form by a replication restart/replication bypass mechanism. FANCM and BLM synergize with BRCA1 to suppress TDs and, perhaps related to this synergy, BRCA1 mutation is synthetic lethal with FANCM loss [36, 51, 54]. The roles of FANCM and BLM in TD suppression potentially implicate a BIR-like mechanism of fork restart. Notably, the TD mechanism and, hence, the underlying replication restart process, is HR-independent. Conceivably, the processing of postreplicative HJs or hemicatenanes at the blocked fork in FANCM/BLM-defective cells might establish a D-loop in the absence of an initiating DSB or strand exchange step, thereby priming BIR-type copying (Figure 3B) [36].
Figure 3. Aberrant fork restart in the absence of an initiating strand invasion step.
A. Polδ-mediated leading strand synthesis could restart collapsed forks at a replication fork barrier (RFB) without an initiating strand exchange step. B. BIR-type copying could restart stalled forks in the absence of an initiating DSB or strand invasion step. In FANCM/BLM-defective cells, post-replicative double Holliday junctions (dHJ) might persist and be channeled towards HJ resolution, potentially leaving a D-loop at the site of stalling, as shown. Loss of FANCM/BLM would also allow persistence of the D-loop, favoring replication restart by a BIR-like bubble migration mechanism.
Another example of possible BIR-related fork restart is the phenomenon of mitotic DNA synthesis (MiDAS) [55]. MiDAS completes DNA synthesis at origin-poor chromosomal regions known as ‘common fragile sites’ during mitosis, and is mediated by Rad52, SLX4, RTEL1, Mus81 and Polδ [56]. The involvement of Mus81 suggests that MiDAS is initiated at stalled, Mus81-cleaved forks as part of a post-replicative salvage pathway. Half of the MiDAS replication tracts visualized were detected on only one sister chromatid, suggesting a conservative mechanism of DNA synthesis; the remaining tracts involved both sisters or were complex [55]. MiDAS proceeds from the border of the unreplicated tract towards the center of the fragile site [57, 58]. In some cell lines, leading and lagging strand synthesis are uncoupled during MiDAS. These observations are suggestive of a BIR mechanism. Indirect support for MiDAS as a break-induced phenomenon came from analysis of the impact of mitotic CDK activity on ICL-stalled forks in frog egg extracts [59]. Unlike the S phase environment, where TRAIP acts only in trans, mitotic CDKs license TRAIP-dependent CMG ubiquitination in cis. In this setting, CMG ubiquitination in cis triggers replisome disassembly and breakage of solitary stalled forks, forming one-ended breaks—lesions that would be conducive to BIR (Figure 2A). Although this example of MiDAS portrays it as a pathway for limiting genomic instability, MiDAS may also promote catastrophic genomic instability, as part of a cascade of cumulative damage triggered by chromosome bridges formed in a previous cell cycle [60].
Breakage-fusion, Microhomology-mediated BIR or replication bypass?
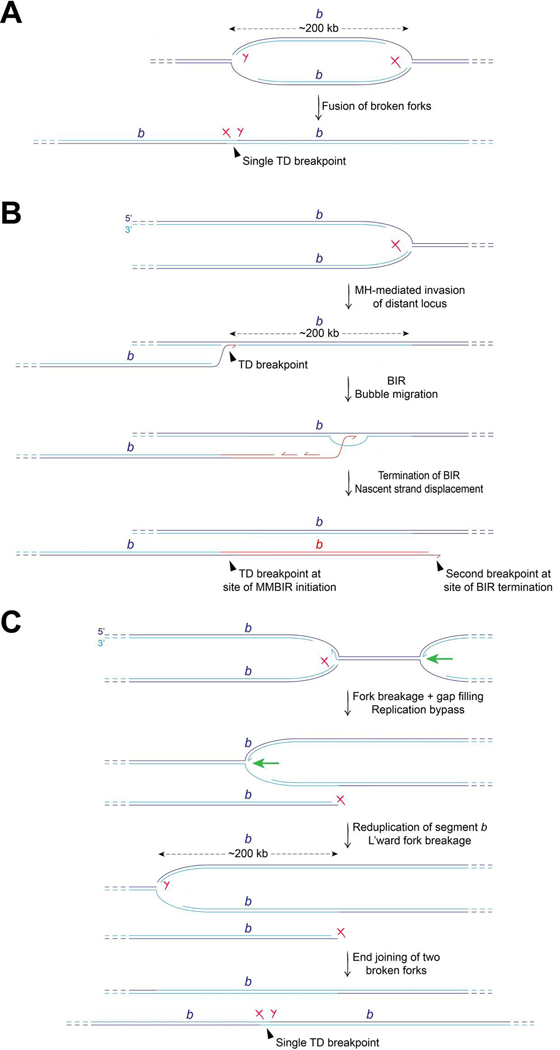
Tandem duplications (TDs) are important drivers in the evolution of species and are the most common form of structural variation in the cancer genome. TDs are characterized by a single non-homologous breakpoint at the boundary between the two copies of the duplicated segment. The most common type of cancer-associated TDs has a median span size of ~200 kb (‘Group 2’ TDs) and is strongly associated with Cyclin E overexpression, which may promote fork breakage [61]. Group 2 TDs might arise by the fusion of two broken forks (Figure 4A). In this model, no localized new synthesis beyond conventional replication is required to form the TD. Alternative models propose that localized re-replication (reduplication) of a chromosome segment drives TD formation. One such model invokes a ‘microhomology (MH)-mediated BIR’ (MMBIR) mechanism, in which the duplicated segment is synthesized by BIR [20] (Figure 4B). In this model, BIR-type copying of ≥200 kb is initiated following MH-mediated invasion of a broken fork into a non-homologous locus ~200 kb from the site of breakage. The site of MH-mediated invasion and BIR initiation would define the TD breakpoint and, hence, the TD span size. Some problems associated with this model are currently unresolved. First, the mechanisms that might promote MH-mediated invasions preferentially at sites ~200 kb upstream of the site of breakage are unclear. Second, current mammalian examples of MMBIR entail synthesis tracts of only a few hundred base pairs in length. Third, the MMBIR mechanism would be expected to generate a second breakpoint at the site of BIR termination (Figure 4A).
Figure 4. Models of Group 2 (~200 kb) Tandem Duplication formation.
A. Breakage-fusion model. Rejoining of two broken forks of the same replicon forms the TD. B. MMBIR model. Breakage of one fork (marked with red ‘X’) liberates a solitary DNA end that invades the chromosome ~200 kb upstream by a strand exchange mechanism involving minimal microhomology (MH). BIR, extending ≥200 kb, reduplicates chromosome segment b (marked in red). Note that this model predicts the formation of two breakpoints: the TD breakpoint at the site of MH invasion/BIR initiation; and a second breakpoint formed at the point of termination of BIR. C. Replication bypass model. Breakage of the rightward fork (marked with red ‘X’) liberates a DNA end, and the residual gap at the site of fork breakage on the intact sister chromatid is rapidly filled. The opposing leftward fork (green arrow) passes through the site of fork breakage, reduplicating chromosome segment b by conventional DNA synthesis. Subsequent breakage of the leftward fork ~200kb downstream generates a second DNA end (marked with red ‘Y’). Rejoining of the two DNA ends of the broken forks generates the solitary TD breakpoint (marked with red ‘XY’). The DNA end of the first broken fork (X) might engage in futile cycles of BIR (not shown) during replication bypass.
A third model proposes that Group 2 TDs arise by replication bypass, in which a conventional replication fork reduplicates the ~200 kb segment (Figure 4B). In this model, residual ssDNA gaps on the unbroken sister chromatid at the site of fork breakage are sealed before the arrival of the opposing fork. As a result, the opposing replication fork encounters no termination signal, enabling it to reduplicate the chromosome segment previously copied by the broken fork. Replication bypass continues until the overshooting fork itself is broken, and the single TD breakpoint forms by end joining. A problem with the replication bypass model is the fate of the original broken fork, the DNA end of which must remain unrepaired for an extended period while replication bypass occurs. Conceivably, this broken fork might be occupied by futile cycles of BIR, mediated by sister chromatid recombination, regenerating a free DNA end periodically as the BIR nascent strand is displaced. A defined mammalian model system is needed that recapitulates Group 2 TD formation in mammalian cells and is capable of distinguishing between these alternative hypotheses.
Concluding remarks
Recombination at stalled forks includes the conservative FA pathway and a diverse set of replication restart mechanisms. Aberrant fork restart may protect under-replicated loci in mitosis, but it can also drive structural variation in developmental disorders and cancer [62]. The ability to recapitulate specific types of structural variation in model systems will make it possible to define underlying mechanisms. One reward for this type of mechanistic enquiry will be the identification of new molecular targets for therapy in human disease, as exemplified by the synthetic lethal interaction between mutations of BRCA1 and FANCM [36].
Acknowledgements
We thank Drs. Johannes Walter, David Pellman, David Cortez, Andrew Deans and Eli Rothenberg for helpful discussions. This work was supported by NIH grants R01CA217991 and R01GM134425 (to R.S.) and by AACR fellowship 19–40-12-PAND (to A.P.).
Footnotes
Declaration of Interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tubbs A, Sridharan S, van Wietmarschen N, Maman Y, Callen E, Stanlie A, et al. Dual Roles of Poly(dA:dT) Tracts in Replication Initiation and Fork Collapse. Cell. 2018;174(5):1127–42 e19. Epub 2018/08/07. doi: 10.1016/j.cell.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Wietmarschen N, Sridharan S, Nathan WJ, Tubbs A, Chan EM, Callen E, et al. Repeat expansions confer WRN dependence in microsatellite-unstable cancers. Nature. 2020;586(7828):292–8. Epub 2020/10/02. doi: 10.1038/s41586-020-2769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crossley MP, Bocek M, Cimprich KA. R-Loops as Cellular Regulators and Genomic Threats. Mol Cell. 2019;73(3):398–411. Epub 2019/02/09. doi: 10.1016/j.molcel.2019.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.García-Muse T, Aguilera A. R Loops: From Physiological to Pathological Roles. Cell. 2019;179(3):604–18. Epub 2019/10/15. doi: 10.1016/j.cell.2019.08.055. [DOI] [PubMed] [Google Scholar]
- 5.Garaycoechea JI, Crossan GP, Langevin F, Mulderrig L, Louzada S, Yang F, et al. Alcohol and endogenous aldehydes damage chromosomes and mutate stem cells. Nature. 2018;553(7687):171–7. Epub 2018/01/13. doi: 10.1038/nature25154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niraj J, Färkkilä A, D’Andrea AD. The Fanconi Anemia Pathway in Cancer. Annual review of cancer biology. 2019;3:457–78. Epub 2019/03/19. doi: 10.1146/annurev-cancerbio-030617-050422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor AMR, Rothblum-Oviatt C, Ellis NA, Hickson ID, Meyer S, Crawford TO, et al. Chromosome instability syndromes. Nature reviews Disease primers. 2019;5(1):64. Epub 2019/09/21. doi: 10.1038/s41572-019-0113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortez D Replication-Coupled DNA Repair. Mol Cell. 2019;74(5):866–76. Epub 2019/06/08. doi: 10.1016/j.molcel.2019.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berti M, Cortez D, Lopes M. The plasticity of DNA replication forks in response to clinically relevant genotoxic stress. Nat Rev Mol Cell Biol. 2020;21(10):633–51. Epub 2020/07/03. doi: 10.1038/s41580-020-0257-5. [DOI] [PubMed] [Google Scholar]
- 10.Kockler ZW, Osia B, Lee R, Musmaker K, Malkova A Repair of DNA Breaks by Break-Induced Replication. Annu Rev Biochem. 2021. Epub 2021/04/02. doi: 10.1146/annurev-biochem-081420-095551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu L, Yan Z, Osia BA, Twarowski J, Sun L, Kramara J, et al. Tracking break-induced replication shows that it stalls at roadblocks. Nature. 2021. Epub 2021/01/22. doi: 10.1038/s41586-020-03172-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, et al. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502(7471):389–92. Epub 2013/09/13. doi: 10.1038/nature12584nature12584 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donnianni RA, Symington LS. Break-induced replication occurs by conservative DNA synthesis. Proc Natl Acad Sci U S A. 2013;110(33):13475–80. Epub 2013/07/31. doi: 10.1073/pnas.1309800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donnianni RA, Zhou ZX, Lujan SA, Al-Zain A, Garcia V, Glancy E, et al. DNA Polymerase Delta Synthesizes Both Strands during Break-Induced Replication. Mol Cell. 2019;76(3):371–81.e4. Epub 2019/09/10. doi: 10.1016/j.molcel.2019.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, Mathur R, et al. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9(2):e1000594. Epub 2011/02/25. doi: 10.1371/journal.pbio.1000594. PubMed PMID: 21347245; PubMed Central PMCID: PMC3039667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lydeard JR, Jain S, Yamaguchi M, Haber JE Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448(7155):820–3. PubMed PMID: 17671506. [DOI] [PubMed] [Google Scholar]
- 17.Li S, Wang H, Jehi S, Li J, Liu S, Wang Z, et al. PIF1 helicase promotes break-induced replication in mammalian cells. Embo j. 2021;40(8):e104509. Epub 2021/01/21. doi: 10.15252/embj.2020104509. PubMed PMID: 33470420; PubMed Central PMCID: PMCPMC8047440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. Embo J. 2000;19(13):3398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagaraju G, Odate S, Xie A, Scully R. Differential regulation of short- and long-tract gene conversion between sister chromatids by Rad51C. Mol Cell Biol. 2006;26(21):8075–86. PubMed PMID: ; PubMed Central PMCID: PMC1636746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, et al. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343(6166):88–91. doi: 10.1126/science.1243211. PubMed PMID: 24310611; PubMed Central PMCID: PMC4047655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puget N, Knowlton M, Scully R Molecular analysis of sister chromatid recombination in mammalian cells. DNA Repair (Amst). 2005;4(2):149–61. PubMed PMID: 15590323; PubMed Central PMCID: PMC2967438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayle R, Campbell IM, Beck CR, Yu Y, Wilson M, Shaw CA, et al. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science. 2015;349(6249):742–7. doi: 10.1126/science.aaa8391. PubMed PMID: 26273056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vrtis KB, Dewar JM, Chistol G, Wu RA, Graham TGW, Walter JC. Single-strand DNA breaks cause replisome disassembly. Mol Cell. 2021;81(6):1309–18.e6. Epub 2021/01/24. doi: 10.1016/j.molcel.2020.12.039. PubMed PMID: 33484638; PubMed Central PMCID: PMCPMC7979477. • Using a model system in frog egg extracts, the authors show that nick-induced fork breakage causes CMG loss. The mechanisms of CMG loss differ, depending on whether the nick is on the leading or lagging strand.
- 24.Semlow DR, Walter JC. Mechanisms of Vertebrate DNA Interstrand Cross-Link Repair. Annu Rev Biochem. 2021. Epub 2021/04/22. doi: 10.1146/annurev-biochem-080320-112510. PubMed PMID: 33882259. [DOI] [PubMed] [Google Scholar]
- 25.Semlow DR, Zhang J, Budzowska M, Drohat AC, Walter JC. Replication-Dependent Unhooking of DNA Interstrand Cross-Links by the NEIL3 Glycosylase. Cell. 2016;167(2):498511 e14. Epub 2016/10/04. doi: 10.1016/j.cell.2016.09.008. PubMed PMID: 27693351; PubMed Central PMCID: PMCPMC5237264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hodskinson MR, Bolner A, Sato K, Kamimae-Lanning AN, Rooijers K, Witte M, et al. Alcohol-derived DNA crosslinks are repaired by two distinct mechanisms. Nature. 2020;579(7800):603–8. Epub 2020/03/07. doi: 10.1038/s41586-020-2059-5. PubMed PMID: 32132710; PubMed Central PMCID: PMCPMC7116288. • The authors report that acetaldehyde-induced ICLs can be processed by the FA pathway, or can be directly unhooked by an as yet unidentified mechanism.
- 27.Huang J, Liu S, Bellani MA, Thazhathveetil AK, Ling C, de Winter JP, et al. The DNA translocase FANCM/MHF promotes replication traverse of DNA interstrand crosslinks. Mol Cell. 2013;52(3):434–46. doi: 10.1016/j.molcel.2013.09.021. PubMed PMID: 24207054; PubMed Central PMCID: PMC3880019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang J, Zhang J, Bellani MA, Pokharel D, Gichimu J, James RC, et al. Remodeling of Interstrand Crosslink Proximal Replisomes Is Dependent on ATR, FANCM, and FANCD2. Cell reports. 2019;27(6):1794–808.e5. Epub 2019/05/09. doi: 10.1016/j.celrep.2019.04.032. PubMed PMID: 31067464; PubMed Central PMCID: PMCPMC6676478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu RA, Semlow DR, Kamimae-Lanning AN, Kochenova OV, Chistol G, Hodskinson MR, et al. TRAIP is a master regulator of DNA interstrand crosslink repair. Nature. 2019;567(7747):267–72. Epub 2019/03/08. doi: 10.1038/s41586-019-1002-0. PubMed PMID: 30842657; PubMed Central PMCID: PMCPMC6417926. •• This paper identified the E3 ubiquitin ligase TRAIP as a key regulator of pathway choice in replication-coupled ICL repair. In S phase extracts, TRAIP ubiquitinates the CMG helicase complex in trans. Short ubiquitin chains on CMG recruit the NEIL3 glycosylase to promote direct unhooking of the ICL. Longer ubiquitin chains program CMG for extraction by the p97/VCP ATPase, disassembling the replisome and enabling subsequent steps of the Fanconi anemia pathway.
- 30.Amunugama R, Willcox S, Wu RA, Abdullah UB, El-Sagheer AH, Brown T, et al. Replication Fork Reversal during DNA Interstrand Crosslink Repair Requires CMG Unloading. Cell reports. 2018;23(12):3419–28. doi: 10.1016/j.celrep.2018.05.061. PubMed PMID: 29924986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoogenboom WS, Boonen R, Knipscheer P. The role of SLX4 and its associated nucleases in DNA interstrand crosslink repair. Nucleic Acids Res. 2019;47(5):2377–88. Epub 2018/12/24. doi: 10.1093/nar/gky1276. PubMed PMID: 30576517; PubMed Central PMCID: PMCPMC6411836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein Douwel D, Boonen RA, Long DT, Szypowska AA, Raschle M, Walter JC, et al. XPF-ERCC1 acts in Unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell. 2014;54(3):460–71. Epub 2014/04/15. doi: 10.1016/j.molcel.2014.03.015. PubMed PMID: ; PubMed Central PMCID: PMCPMC5067070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang AT, Sengerová B, Cattell E, Inagawa T, Hartley JM, Kiakos K, et al. Human SNM1A and XPF-ERCC1 collaborate to initiate DNA interstrand cross-link repair. Genes Dev. 2011;25(17):1859–70. Epub 2011/09/08. doi: 10.1101/gad.15699211. PubMed PMID: 21896658; PubMed Central PMCID: PMCPMC3175721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thongthip S, Bellani M, Gregg SQ, Sridhar S, Conti BA, Chen Y, et al. Fan1 deficiency results in DNA interstrand cross-link repair defects, enhanced tissue karyomegaly, and organ dysfunction. Genes Dev. 2016;30(6):645–59. Epub 2016/03/17. doi: 10.1101/gad.276261.115. PubMed PMID: 26980189; PubMed Central PMCID: PMCPMC4803051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci U S A. 2008;105(42):16107–12. doi: 10.1073/pnas.0804777105. PubMed PMID: 18843105; PubMed Central PMCID: PMCPMC2570989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Panday A, Willis NA, Elango R, Menghi F, Duffey EE, Liu ET, et al. FANCM regulates repair pathway choice at stalled replication forks. Mol Cell. 2021. Epub 2021/04/22. doi: 10.1016/j.molcel.2021.03.044. PubMed PMID: 33882298. • Using the Tus/Ter site-specific RFB to directly measure stalled fork repair, we show that FANCM’s role in the FA pathway is genetically separable from its role in suppressing fork restart. We identify a synthetic lethal interaction between BRCA1 and FANCM mutation, which might be exploitable for therapy of BRCA1-linked breast and ovarian cancer.
- 37.Willis NA, Chandramouly G, Huang B, Kwok A, Follonier C, Deng C, et al. BRCA1 controls homologous recombination at Tus/Ter-stalled mammalian replication forks. Nature. 2014;510(7506):556–9. doi: 10.1038/nature13295. PubMed PMID: 24776801; PubMed Central PMCID: PMC4118467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pike AC, Gomathinayagam S, Swuec P, Berti M, Zhang Y, Schnecke C, et al. Human RECQ1 helicase-driven DNA unwinding, annealing, and branch migration: insights from DNA complex structures. Proc Natl Acad Sci U S A. 2015;112(14):4286–91. Epub 2015/04/02. doi: 10.1073/pnas.1417594112. PubMed PMID: 25831490; PubMed Central PMCID: PMCPMC4394259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Popuri V, Bachrati CZ, Muzzolini L, Mosedale G, Costantini S, Giacomini E, et al. The Human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J Biol Chem. 2008;283(26):17766–76. Epub 2008/05/02. doi: 10.1074/jbc.M709749200. PubMed PMID: 18448429. [DOI] [PubMed] [Google Scholar]
- 40.Whelan DR, Lee WTC, Marks F, Kong YT, Yin Y, Rothenberg E. Super-resolution visualization of distinct stalled and broken replication fork structures. PLoS genetics. 2020;16(12):e1009256. Epub 2020/12/29. doi: 10.1371/journal.pgen.1009256. PubMed PMID: 33370257; PubMed Central PMCID: PMCPMC7793303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quinet A, Lemaçon D, Vindigni A. Replication Fork Reversal: Players and Guardians. Mol Cell. 2017;68(5):830–3. Epub 2017/12/09. doi: 10.1016/j.molcel.2017.11.022. PubMed PMID: 29220651; PubMed Central PMCID: PMCPMC5895179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu W, Krishnamoorthy A, Zhao R, Cortez D. Two replication fork remodeling pathways generate nuclease substrates for distinct fork protection factors. Sci Adv. 2020;6(46). Epub 2020/11/15. doi: 10.1126/sciadv.abc3598. PubMed PMID: 33188024; PubMed Central PMCID: PMCPMC7673757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang R, Wang S, Dhar A, Peralta C, Pavletich NP. DNA clamp function of the monoubiquitinated Fanconi anaemia ID complex. Nature. 2020;580(7802):278–82. Epub 2020/04/10. doi: 10.1038/s41586-020-2110-6. PubMed PMID: 32269332; PubMed Central PMCID: PMCPMC7398534. •• These reports revealed the structures of the FA core complex and of activated (monoubiquitinated) FANCD2-FANCI. Notably, activated D2-I forms a sliding clamp around dsDNA, providing new insights into the mechanism of FA pathway activation.
- 44. Wang S, Wang R, Peralta C, Yaseen A, Pavletich NP. Structure of the FA core ubiquitin ligase closing the ID clamp on DNA. Nature structural & molecular biology. 2021;28(3):300–9. Epub 2021/03/10. doi: 10.1038/s41594-021-00568-8. PubMed PMID: 33686268. •• These reports revealed the structures of the FA core complex and of activated (monoubiquitinated) FANCD2-FANCI. Notably, activated D2-I forms a sliding clamp around dsDNA, providing new insights into the mechanism of FA pathway activation.
- 45. Alcón P, Shakeel S, Chen ZA, Rappsilber J, Patel KJ, Passmore LA. FANCD2-FANCI is a clamp stabilized on DNA by monoubiquitination of FANCD2 during DNA repair. Nature structural & molecular biology. 2020;27(3):240–8. Epub 2020/02/19. doi: 10.1038/s41594-020-0380-1. PubMed PMID: 32066963; PubMed Central PMCID: PMCPMC7067600. •• These reports revealed the structures of the FA core complex and of activated (monoubiquitinated) FANCD2-FANCI. Notably, activated D2-I forms a sliding clamp around dsDNA, providing new insights into the mechanism of FA pathway activation.
- 46. Shakeel S, Rajendra E, Alcón P, O’Reilly F, Chorev DS, Maslen S, et al. Structure of the Fanconi anaemia monoubiquitin ligase complex. Nature. 2019;575(7781):234–7. Epub 2019/11/02. doi: 10.1038/s41586-019-1703-4. PubMed PMID: 31666700; PubMed Central PMCID: PMCPMC6858856. •• These reports revealed the structures of the FA core complex and of activated (monoubiquitinated) FANCD2-FANCI. Notably, activated D2-I forms a sliding clamp around dsDNA, providing new insights into the mechanism of FA pathway activation.
- 47.Tan W, van Twest S, Leis A, Bythell-Douglas R, Murphy VJ, Sharp M, et al. Monoubiquitination by the human Fanconi anemia core complex clamps FANCI:FANCD2 on DNA in filamentous arrays. eLife. 2020;.9. Epub 2020/03/14. doi: 10.7554/eLife.54128. PubMed PMID: 32167469; PubMed Central PMCID: PMCPMC7156235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sparks JL, Chistol G, Gao AO, Raschle M, Larsen NB, Mann M, et al. The CMG Helicase Bypasses DNA-Protein Cross-Links to Facilitate Their Repair. Cell. 2019;176(12):167–81 e21. Epub 2019/01/01. doi: 10.1016/j.cell.2018.10.053. PubMed PMID: 30595447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyabe I, Mizuno K, Keszthelyi A, Daigaku Y, Skouteri M, Mohebi S, et al. Polymerase δ replicates both strands after homologous recombination-dependent fork restart. Nature structural & molecular biology. 2015;22(11):932–8. Epub 2015/10/06. doi: 10.1038/nsmb.3100. PubMed PMID: 26436826; PubMed Central PMCID: PMCPMC4655445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen MO, Jalan M, Morrow CA, Osman F, Whitby MC. Recombination occurs within minutes of replication blockage by RTS1 producing restarted forks that are prone to collapse. eLife. 2015;4:e04539. doi: 10.7554/eLife.04539. PubMed PMID: 25806683; PubMed Central PMCID: PMCPMC4407270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willis NA, Frock RL, Menghi F, Duffey EE, Panday A, Camacho V, et al. Mechanism of tandem duplication formation in BRCA1-mutant cells. Nature. 2017;551(7682):590–5. doi: 10.1038/nature24477. PubMed PMID: 29168504; PubMed Central PMCID: PMCPMC5728692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534(7605):47–54. doi: 10.1038/nature17676. PubMed PMID: 27135926; PubMed Central PMCID: PMCPMC4910866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Menghi F, Inaki K, Woo X, Kumar PA, Grzeda KR, Malhotra A, et al. The tandem duplicator phenotype as a distinct genomic configuration in cancer. Proc Natl Acad Sci U S A. 2016;113(17):E2373–82. doi: 10.1073/pnas.1520010113. PubMed PMID: 27071093; PubMed Central PMCID: PMCPMC4855596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scully R, Panday A, Elango R, Willis NA DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol. 2019;20(11):698–714. Epub 2019/07/03. doi: 10.1038/s41580-019-0152-0. PubMed PMID: 31263220; PubMed Central PMCID: PMCPMC7315405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhowmick R, Minocherhomji S, Hickson ID. RAD52 Facilitates Mitotic DNA Synthesis Following Replication Stress. Mol Cell. 2016;64(6):1117–26. doi: 10.1016/j.molcel.2016.10.037. PubMed PMID: 27984745. [DOI] [PubMed] [Google Scholar]
- 56.Wu W, Bhowmick R, Vogel I, Özer Ö, Ghisays F, Thakur RS, et al. RTEL1 suppresses G-quadruplex-associated R-loops at difficult-to-replicate loci in the human genome. Nature structural & molecular biology. 2020;27(5):424–37. Epub 2020/05/14. doi: 10.1038/s41594-020-0408-6. PubMed PMID: 32398827. [DOI] [PubMed] [Google Scholar]
- 57. Macheret M, Bhowmick R, Sobkowiak K, Padayachy L, Mailler J, Hickson ID, et al. High-resolution mapping of mitotic DNA synthesis regions and common fragile sites in the human genome through direct sequencing. Cell research. 2020;30(11):997–1008. Epub 2020/06/21. doi: 10.1038/s41422-020-0358-x. PubMed PMID: 32561860; PubMed Central PMCID: PMCPMC7784693. • The authors use high resolution mapping of DNA synthesis to analyze MiDAS. In some cell lines, leading and lagging strand synthesis were uncoupled, potentially suggestive of BIR as an underlying mechanism.
- 58.Ji F, Liao H, Pan S, Ouyang L, Jia F, Fu Z, et al. Genome-wide high-resolution mapping of mitotic DNA synthesis sites and common fragile sites by direct sequencing. Cell research. 2020;30(11):1009–23. Epub 2020/06/21. doi: 10.1038/s41422-020-0357-y. PubMed PMID: 32561861; PubMed Central PMCID: PMCPMC7785011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Deng L, Wu RA, Sonneville R, Kochenova OV, Labib K, Pellman D, et al. Mitotic CDK Promotes Replisome Disassembly, Fork Breakage, and Complex DNA Rearrangements. Mol Cell. 2019;73(5):915–29.e6. Epub 2019/03/09. doi: 10.1016/j.molcel.2018.12.021. PubMed PMID: ; PubMed Central PMCID: PMCPMC6410736. •• Using frog egg extracts re-programed with mitotic cyclin-dependent kinases, the authors show that the mitotic environment alters the activity of TRAIP, allowing it to ubiquitinate the CMG complex in cis. Replisome disassembly of isolated stalled forks leads to fork breakage, potentially promoting BIR at the site of breakage.
- 60. Umbreit NT, Zhang CZ, Lynch LD, Blaine LJ, Cheng AM, Tourdot R, et al. Mechanisms generating cancer genome complexity from a single cell division error. Science. 2020;368(6488). Epub 2020/04/18. doi: 10.1126/science.aba0712. PubMed PMID: 32299917; PubMed Central PMCID: PMCPMC7347108. • The authors identify a role for MiDAS in the generation of complex patterns of genomic instability driven by a chromosome bridge formed in the previous cell cycle. This paper shows that MiDAS can be an engine of genomic instability, in addition to the proposed protective role it plays at a CFS.
- 61.Menghi F, Barthel FP, Yadav V, Tang M, Ji B, Tang Z, et al. The Tandem Duplicator Phenotype Is a Prevalent Genome-Wide Cancer Configuration Driven by Distinct Gene Mutations. Cancer Cell. 2018;34(2):197–210 e5. Epub 2018/07/19. doi: 10.1016/j.ccell.2018.06.008. PubMed PMID: 30017478; PubMed Central PMCID: PMCPMC6481635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y, Roberts ND, Wala JA, Shapira O, Schumacher SE, Kumar K, et al. Patterns of somatic structural variation in human cancer genomes. Nature. 2020;578(7793):112–21. Epub 2020/02/07. doi: 10.1038/s41586-019-1913-9. PubMed PMID: 32025012; PubMed Central PMCID: PMCPMC7025897. [DOI] [PMC free article] [PubMed] [Google Scholar]