Abstract
The Cryptococcus neoformans PMA1 gene, encoding a plasma membrane H+-ATPase, was isolated from a genomic DNA library of serotype A strain ATCC 6352. An open reading frame of 3,380 nucleotides contains six introns and encodes a predicted protein consisting of 998 amino acids with a molecular mass of approximately 108 kDa. Plasma membranes were isolated, and the H+-ATPase was shown by sodium dodecyl sulfate-polyacrylamide gel electrophoresis to be slightly larger than the S. cerevisiae H+-ATPase, consistent with its predicted molecular mass. The plasma membrane-bound enzyme exhibited a pH 6.5 optimum for ATP hydrolysis, Km and Vmax values of 0.5 mM and 3.1 μmol mg−1 min−1, respectively, and an apparent Ki for vanadate inhibition of 1.6 μM. ATP hydrolysis in plasma membranes and medium acidification by whole cells were inhibited by ebselen, a nonspecific H+-ATPase antagonist which was also fungicidal. The predicted C. neoformans protein is 35% identical to proton pumps of both pathogenic and nonpathogenic fungi but exhibits more than 50% identity to PMA1 genes from plants. Collectively, this study provides the basis for establishing the Cryptococcus H+-ATPase as a viable target for antifungal drug discovery.
The opportunistic pathogen Cryptococcus neoformans causes pulmonary and central nervous system disease in immunocompromised individuals and is known to produce life-threatening meningoencephalitis in 5 to 10% of AIDS patients (1). Treatment of cryptococossis typically involves combined and sequential therapy with the polyene antibiotic amphotericin and azole-based drugs such as fluconazole. However, such therapy presents clinical problems, since amphotericin treatment results in a high degree of nephrotoxicity and azole resistance has emerged as a complicating factor, especially in AIDS patients with repeated or chronic exposure to fluconazole (4). As is the case with other fungal pathogens, the development of newer antifungal drugs with alternative sites of action that can reduce toxicity and be used in combination to minimize resistance is an important goal.
The plasma membrane H+-ATPase is a high-capacity proton pump that plays a critical role in fungal cell physiology by helping to regulate intracellular pH and maintain transmembrane electrochemical proton gradients necessary for nutrient uptake (26). The H+-ATPase has been characterized biochemically from various fungi in which it is known to be a predominant membrane protein comprised of a single 100-kDa subunit that contains both a membrane-spanning transport domain and a cytoplasmically located catalytic ATP hydrolysis domain. The gene encoding this enzyme, PMA1, has been cloned from diverse fungi and has been shown to be highly conserved (31). Gene-disruption experiments in Saccharomyces cerevisiae have confirmed the essential nature of this gene product (27). The H+-ATPase has recently been proposed as a target for antifungal drug development, largely because of its well-characterized biochemical and genetic properties (20) and the availability of several high-throughput screens that target unique functional properties of the enzyme (24). In addition, the H+-ATPase is a typical member of the P-type family of ion translocating enzymes (16), which serve as selective targets for cardiac glycosides and antiulcer therapeutics (22).
Presently there is little information about H+-ATPases from pathogenic fungi. In this study, we report the cloning and characterization of the plasma membrane H+-ATPase from a serotype A strain of C. neoformans. This work should serve as a basis for novel antifungal drug development.
MATERIALS AND METHODS
Strains and cell culture.
Escherichia coli BB4 (supF58 supE44 hsdR514 galK2 galT22 trpR55 metB1 tonA ΔlacU169 [F′ proAB+ laqIq lacZΔM15 Tn10 (Tetr)]) and Epicurian coli XL1-Blue MRF′ (Δ(mcrA) 183 Δ(mcrCB-hsdSMR-mrr) 173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIq ZΔM15 Tn10 (Tetr)]) were used for library propagation and cloning, respectively. C. neoformans serotype A strains ATCC 6352 and H99 and S. cerevisiae strain GW201 (32) were used in this study. Yeast cells were grown at 30°C in YPD medium (1% yeast extract, 2% peptone, 2% dextrose).
Identification of the C. neoformans PMA1 gene from genomic DNA.
DNA was extracted from C. neoformans strain ATCC 6352 as described by Fujimura and Sakuma (13). Primers (5′-TTGTGTTCCGACAAAACCGGTACTTTG ACC and 5′-TGGAGCATCGTTAACACCATCACCAGTCAT) from conserved regions of PMA genes from S. cerevisiae and other fungi were initially used to amplify a 0.8-kb fragment of C. neoformans PMA1 from the genomic DNA preparation. Following DNA sequence analysis of this fragment, two new C. neoformans-specific primers (5′-GCGTACCGAGATCACTTACCG and 5′-CGGTAAGTGATCTCGGTACGC) were used to amplify this fragment. The amplification was carried out using TaqPlus Long DNA polymerase with the low-salt buffer provided with the enzyme (Stratagene), and the nucleotide sequence of the amplified product was determined. The GenBank accession numbers for the mRNA and DNA sequences are AF217201 and AF21702, respectively.
Screening of the genomic library.
The ATCC 6352 genomic library constructed in λGEM-11 vector was provided by Silvia Spitzer (SUNY at Stony Brook). The library was propagated in E. coli strain BB4 and plated on eight 100-mm-diameter Luria-Bertani agar plates. The plates were divided into quadrants, and the plaques were pooled. DNA from the phage pools was extracted using LambdaSorb phage adsorbent (Promega). Mixed phage containing the C. neoformans PMA1 gene were identified by PCR amplification of a 0.8-kb fragment of the gene using the above primers on DNA obtained from phage pools. Positive pools were replated, and new plaque pools were evaluated. After several rounds of enrichment, single-plaque DNA was extracted by heating plaque-containing agar plugs in 71 μl of H2O at 94°C for 7 min. PMA1 was amplified from the enriched phage DNA using the two C. neoformans PMA1-specific primers and primers designed to target the T7 and SP6 promoter regions of the λGEM-11 vector.
cDNA preparation and primer extension studies.
Total RNA was extracted from ATCC 6352 using the RNeasy Mini Kit (Qiagen). cDNA was obtained by reverse transcriptase PCR using the RETROscript kit (Ambion). Fragments of C. neoformans PMA1 were amplified using TaqPlus Long as described above using primers derived from the DNA sequence. The sequences of the PCR products were determined and compared with the genomic sequence. Primer extension was carried out using an N-terminal reverse primer, 5′-GGAGTTCTGAGAATTAGGTG GAGG. The primer was end labeled using [32P]dATP and T4 polynucleotide kinase (10 U; New England Biolabs). Total RNA (50 μg) diluted in a solution containing 100 mM KCl and 50 mM Tris-Cl (pH 8.3) was used for hybridization to labeled primer (12 ng) at 4°C following a denaturation step at 60°C for 2 min. The reverse transcriptase reaction was carried out at 47°C for 1 h using AMV reverse transcriptase (10 U; New England Biolabs). The product of the primer extension reaction was analyzed alongside a sequence analysis of the genomic DNA initiated by the same primer.
Disruption of the C. neoformans PMA1 gene.
The C. neoformans PMA1 coding region was amplified from total genomic DNA of the ATCC 6352 strain using the following primers: 5′-CCAACTCTTAGTTTTAGC and 5′-GCGGGTGATAATACGGGGG. The resulting 3.5-kb fragment was cloned into the pPCR-Script cloning vector (Stratagene). A disruption construct was made by inserting the Cryptococcus actin promoter fused to the hygromycin gene from E. coli into the C. neoformans PMA1 gene. The actin-hygromycin construct was amplified from the pCnTEL-Act:Hyg plasmid (9) using Pfu polymerase (Stratagene) and primers 5′-GCTATTGTCCAGGCTGCG and 5′-CCAATCGGCAGGCACGGGCGGCG. The 2.1-kb actin-hygromycin PCR fragment was ligated into the single HpaI site in the PMA1 gene. The plasmid was linearized with ApaI, precipitated with 95% ethanol, and introduced by electroporation transformation into strains ATCC 6352 and H99 (21). Stable transformants were selected on YPD plates containing 200 μg of hygromycin B per ml, and genomic DNA was isolated as described above. The region of PMA1 containing the actin-hygromycin B construct was amplified using primers 5′-CCTCTCTCCTGGGTCATGGAG and 5′-CGGGAAGAGACTCGCC and 50 ng of genomic DNA from hygromycin B-resistant transformants. PCR products were analyzed by agarose gel electrophoresis.
Purification of plasma membrane H+-ATPase and ATP hydrolysis measurements.
Plasma membranes from C. neoformans strain ATCC 6352 and S. cerevisiae strain GW201 (32) were isolated from mid-log-phase cells by the procedure described previously (28). ATP hydrolysis assays were performed in triplicate in 96-well microplates as described in Wang et al. (32). Inorganic phosphate released was determined by measuring the absorbance at 660 nm in a microtiter plate reader (Tecan SLT Instruments) after a 10-min incubation at 22°C. The optimal pH for ATP hydrolysis was determined in a standard reaction medium with the pH adjusted to 5.0 to 8.0. Km and Vmax were determined by measuring ATP hydrolysis with equimolar concentrations of ATP and MgSO4 from 0 to 15 mM. Vanadate sensitivity was assayed by measuring ATP hydrolysis in the presence of 0 to 100 μM sodium vanadate. Inhibition of ATP hydrolysis by the inhibitor ebselen (Astra-Zeneca) was determined by preincubating the membranes with 0 to 25 μM ebselen for 30 min at room temperature.
Glucose-dependent medium acidification.
Glucose-dependent medium acidification was monitored by a modification of a procedure described previously (23). Cultures of C. neoformans strain ATCC 6352 (50 ml) were grown to mid-log phase and harvested by centrifugation for 10 min at 3,000 × g. The pellets were washed by resuspension in 50 ml of 100 mM KCl, pH 5.0, and centrifugation as described above. The pellets were resuspended in 10 ml of the KCl solution and incubated with shaking at room temperature for 1 h. Samples were stored at 4°C for 16 h. Prior to use, cells were concentrated by centrifugation as described above and adjusted to give a final A590 of ∼2.3. Cells (20 μl) were incubated for 30 min at room temperature with 0 to 50 μM ebselen in 155 μl of bromophenol blue (50 μg/ml) in 100 mM KCl, pH 5.0. A 20-μl aliquot of 20% (wt/vol) glucose was added to initiate the reaction. Medium acidification was monitored at 590 nm over a period of 4 h (with a data point taken every 5 min) in a microtiter plate reader (Tecan SLT Instruments).
Growth inhibition.
C. neoformans cultures were grown for 16 h at 30°C. Cells (2.0 × 106) were diluted into 250 μl of YPD containing 0, 10, or 25 μM ebselen and incubated for 0 to 24 h at room temperature. The treated cells were diluted with YPD and plated on YPD agar. The number of colonies on each plate was determined after 48 h at 37°C.
Other procedures.
PCRs were carried out in a PTC-150 MiniCycler (MJ Research). PCR products were purified prior to sequence analysis using the Wizard PCR Preps DNA purification system (Promega). Genomic and cDNA sequences were analyzed at the New York University Medical Center and the Queens University DNA Sequencing Facilities. Sequence analysis for primer extension was performed using the Sequenase PCR product sequencing kit (Amersham) and a 6% polyacrylamide precast sequencing gel (Stratagene). Protein concentrations were determined by a modified Lowry assay (17). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using precast 10% minigels (Novex). Hydropathy profiles were generated using the Kyte-Doolittle method (15).
RESULTS
Cloning of C. neoformans PMA1.
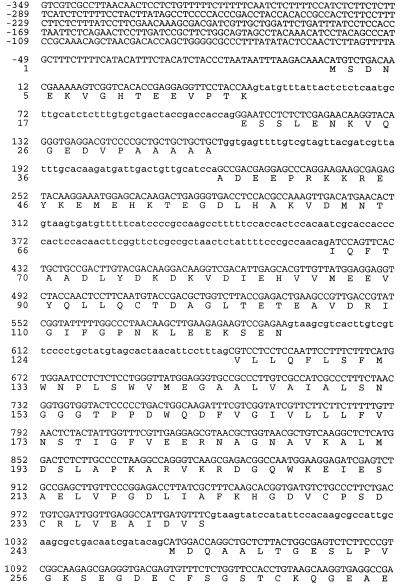
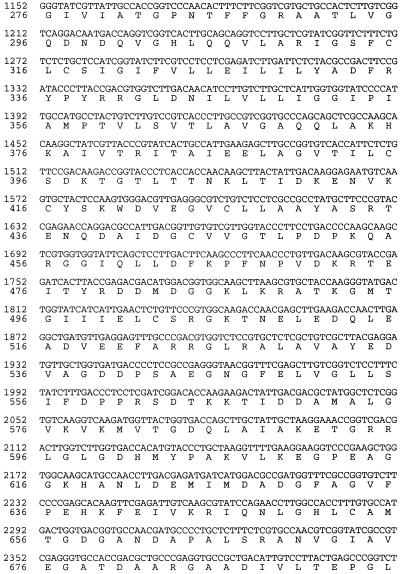
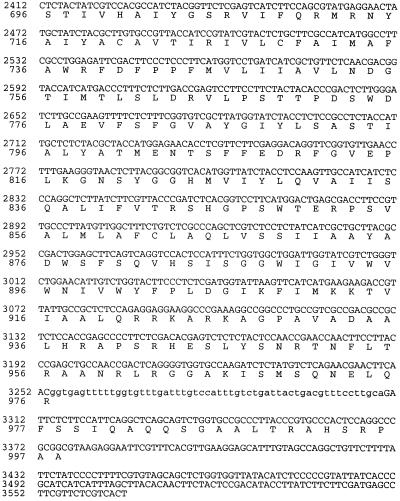
A set of primers representing a highly conserved region of the PMA1 gene from S. cerevisiae was used to amplify a 0.8-kb PCR product from genomic DNA of C. neoformans strain ATCC 6352. BLAST analysis of this product indicated a high level of similarity with other fungal and plant PMA genes. Screening of a phage library of genomic DNA fragments from this strain by PCR amplification produced the same 0.8-kb product. A clone with a 9-kb insert containing the entire coding region of C. neoformans PMA1 was obtained. Total RNA extracted from ATCC 6352 was used to prepare cDNA, and C. neoformans PMA1 sequences were amplified using primers derived from the DNA sequence. The nucleotide sequence and predicted amino acid sequence for C. neoformans PMA1 are shown in Fig. 1. Comparison of the genomic DNA sequence and the cDNA sequence indicated that the C. neoformans PMA1 gene contains six introns, five located in the N-terminal half and one near the C terminus. Primer extension studies revealed the presence of two transcriptional start sites, which yield 283- and 300-nucleotide (nt) leader sequences. The relatively large leader sequence is comparable in size to leader sequences from PMA1 genes from other fungi, including Candida albicans (18) and S. cerevisiae (6). The coding region initiates with an ATG and terminates with a TAA stop codon. The predicted protein consists of 998 amino acids and has a predicted molecular mass of 108,469 Da.
FIG. 1.
Nucleotide and predicted protein sequences of the C. neoformans PMA1 gene. Translated DNA sequences and protein sequence are indicated in capital letters. Untranslated regions and intron sequences are shown in lowercase letters.
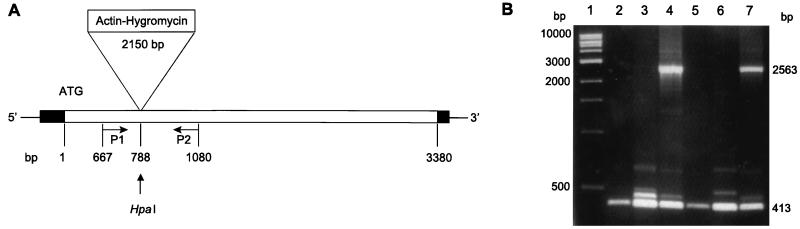
Disruption of the PMA1 gene.
The important role that H+-ATPases play in lower-eukaryote cell physiology suggests that they should be essential to the cell. Gene disruption experiments have confirmed this for S. cerevisiae (27). To assess gene essentiality in C. neoformans, PMA1 was cloned into the pPCR-Script vector and disrupted by insertion of a construct containing hygromycin B gene under control of the actin promoter (9) into the middle of the gene (Fig. 2). A linearized construct was used to transform C. neoformans strains ATCC 6352 and H99, and transformants were selected on the basis of resistance to hygromycin B. Approximately 500 transformants were selected following electroporation, and genomic DNA was isolated. In every case, PCR analysis of genomic DNA using C. neoformans PMA1-specific primers indicated that the PMA1 disruption allele had not replaced the wild-type homologue of PMA1. Rather, both the wild-type copy of PMA1 along with the disrupted copy remained. The failure to obtain transformants displaying only a disrupted copy of PMA1 is consistent with this gene being essential to the cell. Validation of this assertion will require conditional expression of C. neoformans PMA1 under control of a conditional promoter. Unfortunately, a suitable conditional promoter has not been identified at this time.
FIG. 2.
(A) C. neoformans PMA1 disruption construct. Shown is a diagram of the PMA1 disruption construct. The actin-hygromycin insert was placed in the single HpaI site in the Cryptococcus PMA1 gene. (B) PCR analysis of hygromycin-resistant transformants. PCR products were amplified from wild-type and transformed strains using primers 1 and 2, which flank the actin-hygromycin insert and yield a fragment of 2,563 bp. Lanes: 1, molecular weight marker; 2, PCR product from the wild-type ATCC 6352; 3 and 4, PCR products from ATCC 6352 transformed with the disruption construct; 5, PCR product from the wild-type H99; 6 and 7, PCR products from H99 transformed with the disruption construct.
Biochemical properties of C. neoformans plasma membrane H+-ATPase.
Plasma membranes were purified from C. neoformans strain ATCC 6352 to determine the biochemical properties of the H+-ATPase. SDS-PAGE (Fig. 3) indicated that the C. neoformans H+-ATPase is slightly larger than the S. cerevisiae H+-ATPase, consistent with the predicted molecular mass of ∼108 kDa, 8 kDa larger than the S. cerevisiae H+-ATPase. Polyclonal antibodies prepared against the S. cerevisiae proton pump showed significant but weak cross-reaction with the C. neoformans protein (not shown). The C. neoformans enzyme was found to have a pH optimum for ATP hydrolysis of pH 6.5 and displayed Km and Vmax values of 0.5 mM and 3.1 μmol of Pi release per mg of protein per min, respectively (Table 1). An apparent Ki of 1.6 μM was found for the mechanism-specific inhibitor vanadate, which classically inhibits P-type enzymes (5). Thus, the Cryptococcus enzyme showed kinetic properties similar to those seen with the Saccharomyces protein.
FIG. 3.
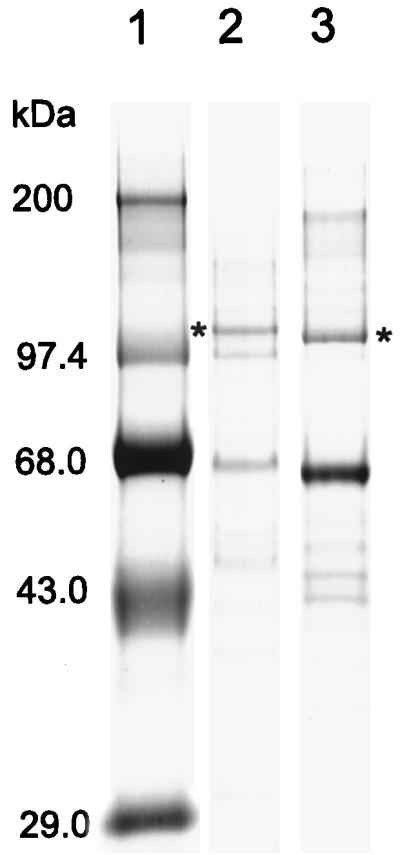
Isolation of C. neoformans plasma membrane H+-ATPase. Plasma membranes from C. neoformans and S. cerevisiae were purified as described in Materials and Methods. The position of the PMA1p band in each membrane preparation is indicated by an asterisk. Lanes: 1, molecular weight marker; 2, C. neoformans plasma membranes; 3, S. cerevisiae plasma membranes.
TABLE 1.
Kinetic properties of the C. neoformans plasma membrane H+-ATPasea
| Organism | Km (mM) | Vmax (μmol of Pi mg−1 min−1) | Ki of vanadate (μM) | pH optimum |
|---|---|---|---|---|
| C. neoformans (ATCC 6352) | 0.5 | 3.1 | 1.6 | 6.5 |
| S. cerevisiae (GW201) | 1.5 | 3.7 | 0.9 | 6.5 |
Values are the averages of triplicate assays, which were all within 5% error.
Effect of H+-ATPase inhibitor ebselen on the plasma membrane pump.
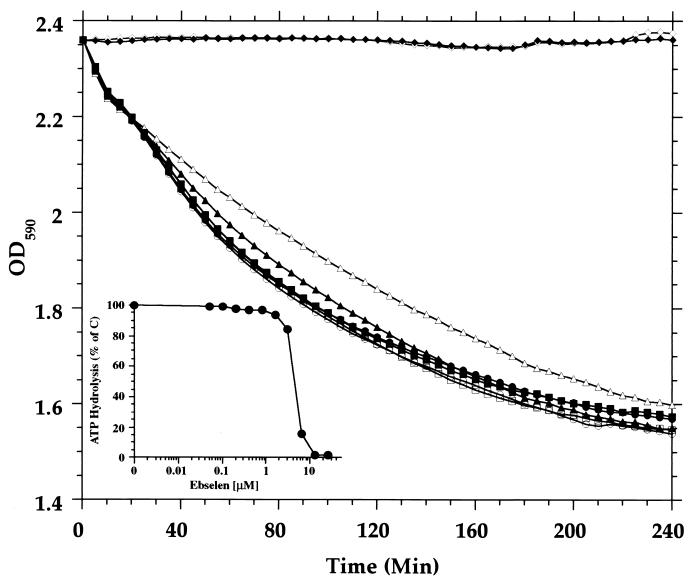
The seleno-organic drug ebselen [2-phenyl-1,2-benzoisoselenazol-3(2H)-one] is a thiol-reactive reagent that inhibits a number of enzymes, including protein kinases and the gastric H+,K+-ATPase (25), and an analog displays antifungal behavior (3). It inhibits H+-ATPases from Candida and Saccharomyces with a 50% inhibitory concentration in the micromolar range (D. S. Perlin, unpublished data). Figure 4 (inset) shows that ATP hydrolysis by the H+-ATPase in isolated plasma membranes was inhibited by ebselen (IC50, 4.5 μM). To assess the effect of ebselen proton efflux by the H+-ATPase in whole cells, a medium acidification assay was employed (23). Carbon-starved cells were preincubated for 30 min with 0 to 50 μM ebselen. Proton pumping was assessed by observing the ability of the cells to acidify the medium in response to glucose. In the medium acidification assay proton pumping was totally inhibited in the presence of a 25 μM concentration of the compound. The approximately threefold-higher 50% inhibitory concentration for proton pumping relative to ATP hydrolysis either reflects low permeability and/or the prevalence of free sulfhydryl groups in the cell wall that effectively reduce the level of inhibitor at the plasma membrane.
FIG. 4.
Effect of ebselen on activity of C. neoformans plasma membrane H+-ATPase. C. neoformans cells were incubated with ebselen for 30 min, and proton efflux by the H+-ATPase was monitored by medium acidification. Symbols: ●, 0 μM; ○, 0.5 μM; ■, 1 μM; □, 2.5 μM; ▴, 5 μM; ▵, 10 μM; ⧫, 25 μM; ◊, 50 μM. (Inset) Plasma membranes were incubated for 30 min with increasing concentrations of ebselen, and the rate of ATP hydrolysis measured. The control value was taken as the activity in the absence of drug.
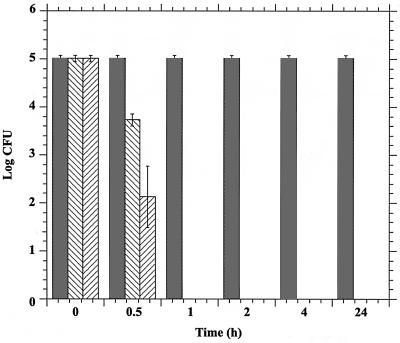
Due to the likely essential nature of the H+-ATPase, it is expected that inhibition of proton transport would be lethal. Figure 5 shows that inhibition of the H+-ATPase is fungicidal in C. neoformans. Cells (2.0 × 106) were incubated with 10 or 25 μM ebselen for 0 to 24 h, diluted, and plated on YPD agar. The number of colonies formed on each plate (CFU) was determined after 48 h. Following treatment of the cells for 30 min, the number of CFU was reduced 1.2 and 3 log orders at 10 and 25 μM, respectively. No viable cells were detected after treatment for 60 min with either 10 or 25 μM ebselen. This result suggests that inhibition of the H+-ATPase is sufficient to block cell growth, as has been observed in other fungi (8), and that the inhibition is fungicidal. This observation reflects the critical role of the H+-ATPase in cellular physiology.
FIG. 5.
Fungicidal properties of ebselen. C. neoformans cells were incubated with ebselen for the times indicated, diluted in YPD, and plated on YPD agar. After 48 h the number of colonies resulting from each treatment was determined. Bars:  , 0 μM; ▧, 10 μM; ▨, 25 μM. Error bars, standard deviations.
, 0 μM; ▧, 10 μM; ▨, 25 μM. Error bars, standard deviations.
DISCUSSION
The C. neoformans PMA1 gene encoding the plasma membrane H+-ATPase was cloned from a genomic library derived from C. neoformans strain ATCC 6352, serotype A. It has a coding region of 3,380 nt and a transcript containing an upstream untranslated sequence of 283 or 300 nt. Relatively large 5′ untranslated regions have been observed in fungal genes, including those from S. cerevisiae, C. albicans, and C. neoformans (6, 10, 19, 30, 33). Two putative CAAT boxes are found upstream of the transcriptional start sites at −314 and −336 nt. Analysis of the C. neoformans LAC1 gene has suggested that the transcriptional regulation of some C. neoformans genes appears more similar to those of mammalian and plant systems than to those of other fungi with multiple DNA binding sites distributed over a relatively large upstream region (33). The C. neoformans PMA1 gene contains six introns, five located within the N-terminal half and one near the C terminus. An open reading frame of 998 amino acids encodes a predicted protein with a molecular mass of 108,469 Da (Fig. 1). SDS-PAGE confirmed that the Cryptococcus H+-ATPase was somewhat larger than its counterpart from S. cerevisiae, which has a known molecular mass of 99,572 Da (27) (Fig. 3).
Table 2 shows an amino acid comparison of the C. neoformans H+-ATPase to other fungal and plant plasma membrane ATPases. The protein is 37% identical to the S. cerevisiae plasma membrane H+-ATPase and shows similar levels of identity with H+-ATPases from pathogenic and nonpathogenic fungi. This relatively low level of identity helps explain why polyclonal antibodies to the H+-ATPase from S. cerevisiae cross-react weakly with the C. neoformans enzyme (not shown). Interestingly, C. neoformans PMA1 shows more than 50% similarity to PMA genes from plants.
TABLE 2.
Similarity of the C. neoformansa H+-ATPase to other H+-ATPases
| Category and organism | % Similarity | % Identity | Accession no. |
|---|---|---|---|
| Pathogenic fungi | |||
| Cryptococcus neoformans (B-3501) | 99 | 98 | AF077766 |
| Histoplasma capsulatum | 45 | 35 | L07305 |
| Pneumocystis carinii | 45 | 34 | U65004 |
| Candida albicans | 44 | 35 | P28877 |
| Nonpathogenic fungi | |||
| Dictyostelium discoideum | 51 | 42 | P54679 |
| Saccharomyces cerevisiae | 46 | 37 | P05030 |
| Zygosaccharomyces rouxii | 46 | 36 | P24545 |
| Neurospora crassa | 45 | 37 | M14085 |
| Aspergillus nidulans | 44 | 35 | AF036763 |
| Schizosaccharomyces pombe | 44 | 34 | P09627 |
| Plants | |||
| Nicotiana plumbaginifolia | 63 | 54 | Q08435 |
| Lycopersicon esculentum | 62 | 53 | P22180 |
| Zea mays | 62 | 52 | X85805 |
| Arabidopsis thaliana | 60 | 52 | P20649 |
Compared with a C. neoformans serotype D sequence that was recently placed in GenBank (accession number AF077766), the DNA and protein sequences were found to be 96 and 98% identical, respectively. In the coding region, 112 nucleotide differences were found, with the majority of nucleotide changes occurring in the third codon position. At the protein level, serotypes A and D differ by 22 amino acids, though 14 of these are conservative substitutions. The large number of nucleotide substitutions is consistent with allelic variation that has been seen in other Cryptococcus genes (7, 12). In fact, the large number of nucleotide differences in the URA5 gene between serotypes A and D has led to the suggestion that they be considered different varieties of C. neoformans (12).
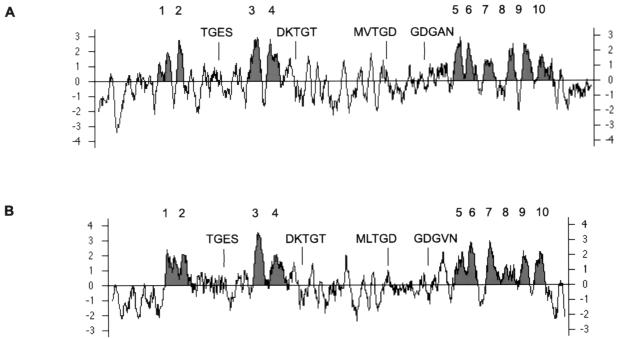
Members of the family of P-type enzymes contain a number of conserved sequence motifs essential to catalysis (14, 16). Highly conserved regions such as the TGES, CSDKTG, MXTDG, and GDGXNDXP motifs were all completely conserved in the H+-ATPase from C. neoformans (Fig. 6). Hydropathy profiles of the C. neoformans and S. cerevisiae enzymes were similar, with the only significant differences being the longer length of the N and C termini in the Cryptococcus enzyme (Fig. 7).
FIG. 6.
Alignment of conserved regions of P-type ATPases. The consensus sequences TGES, CSDKTG, MXTDG, and GDGXNDXP and their locations in the proteins are indicated. Abbreviations: C.a., C. albicans; S.c., S. cerevisiae; C.n., C. neoformans; N.p., Nicotiana plumbaginifolia; R.n., Rattus norvegicus; O.c., Oryctolagus cuniculus.
FIG. 7.
Hydropathy profiles for plasma membrane H+-ATPase from C. neoformans and S. cerevisiae. Several conserved regions of each protein, including the region around the phosphorylation site, are indicated. Transmembrane segments 1 to 10 are numbered and shaded.
The kinetic properties of the plasma membrane-bound H+-ATPase are indicative of a high-capacity proton pump with a high catalytic turnover number, as has been observed for other members of this family (19, 29). This property is critical to the essential role that the H+-ATPase plays in cell physiology by establishing ion gradients and regulating intracellular pH. The essentiality of C. neoformans PMA1 was assessed with a gene disruption strategy involving homologous recombination between the chromosomal wild-type gene and a disrupted allele containing the selectable marker hygromycin B (Fig. 2). In all transformants, an intact copy of C. neoformans PMA1 was observed, which is consistent with the notion the H+-ATPase is critical for cell survival but does not prove the case. A more definitive test of gene essentiality was presented recently by Del Poeta et al. (11), whereby a second copy of the gene is inserted in the chromosome. Unfortunately, it has not yet been possible to introduce a second copy of the gene (P. Soteropoulos and D. S. Perlin, unpublished data). This may reflect induced lethality from gene dosage effects that are commonly observed with PMA1 in S. cerevisiae. A confirmation of PMA1 essentiality in Cryptococcus will have to await the development of a more suitable conditional expression system, as was demonstrated for PMA1 in S. cerevisiae (27).
An important benefit of the putative essentiality of the C. neoformans H+-ATPase is that it becomes an attractive target for antifungal drug discovery. Many of the important antifungal drugs in clinical use today are limited by their fungistatic growth properties which prevent additional growth of cells but have little affect on existing cell populations. Thus, a competent immune system is required to clear infections. Fungicidal agents which are able to kill existing cells are therefore desirable. The H+-ATPase is needed for both growth and stable cell maintenance. Due to these factors and its slow turnover in the membrane in other fungi (∼11 h) (2), it is likely that inhibitors of the H+-ATPase will be fungicidal, as was observed with compound ebselen (Fig. 5).
The fungal proton pumps share less than 30% sequence identity with P-type ATPases from animal cells. Clinically active therapeutics like cardiac glycosides and reversible antiulcer acid blockers can be selectively targeted to members of the P-type class. This well-documented therapeutic specificity should facilitate the development of highly selective antifungal drugs. In addition, H+-ATPase antagonists should display broad-spectrum activity on diverse pathogenic fungi due the high-degree of sequence similarity found among these enzymes.
Overall, the fungal plasma membrane H+-ATPase has well-defined properties that facilitate drug discovery. We have validated the H+-ATPase as an antifungal target by showing that the proton pump can be inhibited both in vivo and in vitro and that inhibition results in cell death. The enzyme is amenable to detailed genetic and biochemical analyses, which facilitate an evaluation of drug-target interactions. In addition, there are a variety of high-throughput screens that assess functional properties of the H+-ATPase in vitro and in whole cells (24). The cloning and characterization of the plasma membrane H+-ATPase from C. neoformans along with the other plasma membrane pumps already isolated will aid in the development of new antifungal agents.
ACKNOWLEDGMENTS
We acknowledge John Perfect for providing plasmid pCnTEL-Act:Hyg and for a critical review of the manuscript. We also thank Silvia and Eric Spitzer for providing the genomic library of C. neoformans, Carin Briving and Ingemar Starke of Astra Hässle, Sweden, for suggesting the use of Ebselen as an ATPase inhibitor, and Steven Park for technical assistance.
This work was partially supported by a grant from Astra Hässle AB (to D.S.P.).
REFERENCES
- 1.Aberg J A, Powderly W G. Cryptococcal disease: implications of recent clinical trials on treatment and management. AIDS Clin Rev. 1997;1997–1998:229–248. [PubMed] [Google Scholar]
- 2.Benito B, Moreno E, Lagunas R. Half-life of the plasma membrane ATPase and its activating system in resting yeast cells. Biochim Biophys Acta. 1991;1063:265–268. doi: 10.1016/0005-2736(91)90381-h. [DOI] [PubMed] [Google Scholar]
- 3.Bien M, Blaszczyk B, Kalinowska K, Mlochowski J, Inglot A D. Antifungal activity of 2-(4-chlorophenyl)-1,2-benzisoselenazol-3(2H)-one, the analog of Ebselen. Arch Immunol Ther Exp (Warsaw) 1999;47:185–193. [PubMed] [Google Scholar]
- 4.Bossche H V, Dromer F, Improvisi I, Lozano-Chiu M, Rex J H, Sanglard D. Antifungal drug resistance in pathogenic fungi. Med Mycol. 1998;36(Suppl. 1):119–128. [PubMed] [Google Scholar]
- 5.Bowman B J, Slayman C W. The effects of vanadate on the plasma membrane ATPase of Neurospora crassa. J Biol Chem. 1979;254:2928–2934. [PubMed] [Google Scholar]
- 6.Capieaux E, Vignais M L, Sentenac A, Goffeau A. The yeast H+-ATPase gene is controlled by the promoter binding factor TUF. J Biol Chem. 1989;264:7437–7446. [PubMed] [Google Scholar]
- 7.Casadevall A, Freundlich L F, Marsh L, Scharff M D. Extensive allelic variation in Cryptococcus neoformans. J Clin Microbiol. 1992;30:1080–1084. doi: 10.1128/jcm.30.5.1080-1084.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cid A, Perona R, Serrano R. Replacement of the promoter of the yeast plasma membrane ATPase gene by a galactose-dependent promoter and its physiological consequences. Curr Genet. 1987;12:105–110. doi: 10.1007/BF00434664. [DOI] [PubMed] [Google Scholar]
- 9.Cox G M, Toffaletti D L, Perfect J R. Dominant selection system for use in Cryptococcus neoformans. J Med Vet Mycol. 1996;34:385–391. [PubMed] [Google Scholar]
- 10.Cruz M C, Edlind T. β-Tubulin genes and the basis for benzimidazole sensitivity of the opportunistic fungus Cryptococcus neoformans. Microbiology. 1997;143:2003–2008. doi: 10.1099/00221287-143-6-2003. [DOI] [PubMed] [Google Scholar]
- 11.Del Poeta M, Toffaletti D L, Rude T H, Dykstra C C, Heitman J, Perfect J R. Topoisomerase I is essential in Cryptococcus neoformans: role in pathobiology and as an antifungal target. Genetics. 1999;152:167–178. doi: 10.1093/genetics/152.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franzot S P, Fries B C, Cleare W, Casadevall A. Genetic relationship between Cryptococcus neoformans var. neoformans strains of serotypes A and D. J Clin Microbiol. 1998;36:2200–2204. doi: 10.1128/jcm.36.8.2200-2204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujimura H, Sakuma Y. Simplified isolation of chromosomal and plasmid DNA from yeasts. BioTechniques. 1993;14:538–540. [PubMed] [Google Scholar]
- 14.Goldshleger R, Karlish S J D. Fe-catalyzed cleavage of the a subunit of Na/K-ATPase: evidence for conformation-sensitive interactions between cytoplasmic domains. Proc Natl Acad Sci USA. 1997;94:9596–9601. doi: 10.1073/pnas.94.18.9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kyte J, Doolittle R F. A simple method for displaying the hydrophobic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 16.Lutsenko S, Kaplan J H. Organization of P-type ATPases: significance of structural diversity. Biochemistry. 1995;34:15607–15613. doi: 10.1021/bi00048a001. [DOI] [PubMed] [Google Scholar]
- 17.Markwell M A K, Haas S M, Bieber L L, Tolbert N E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 18.Monk B C, Kurtz M B, Marrinan J A, Perlin D S. The cloning and characterization of the plasma membrane H+-ATPase from Candida albicans. J Bacteriol. 1991;173:6826–6836. doi: 10.1128/jb.173.21.6826-6836.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monk B C, Niimi M, Shepherd M G. The Candida albicans plasma membrane and H+-ATPase during yeast growth and germ tube formation. J Bacteriol. 1993;175:5566–5573. doi: 10.1128/jb.175.17.5566-5574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monk B C, Perlin D S. Fungal plasma membrane proton pumps as promising new antifungal targets. Crit Rev Microbiol. 1994;20:209–223. doi: 10.3109/10408419409114555. [DOI] [PubMed] [Google Scholar]
- 21.Perfect J R, Lang S D R, Durack D T. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am J Pathol. 1980;101:177–194. [PMC free article] [PubMed] [Google Scholar]
- 22.Perlin, D. S. 15 August 1998, posting date. Ion pumps as targets for therapeutic intervention: old and new paradigms. Electron. J. Biotechnol. http://www.ejb.org/content/vol1/issue2/full/2/.
- 23.Perlin D S, Brown C L, Haber J E. Membrane potential defect in hygromycin B-resistant pma1 mutants of Saccharomyces cerevisiae. J Biol Chem. 1988;263:18118–18122. [PubMed] [Google Scholar]
- 24.Perlin D S, Seto-Young D, Monk B C. The plasma membrane H+-ATPase of fungi. A candidate drug target? Ann N Y Acad Sci. 1997;834:609–617. doi: 10.1111/j.1749-6632.1997.tb52330.x. [DOI] [PubMed] [Google Scholar]
- 25.Schewe T. Molecular actions of ebselen—an antiinflammatory antioxidant. Gen Pharmacol. 1995;26:1153–1169. doi: 10.1016/0306-3623(95)00003-j. [DOI] [PubMed] [Google Scholar]
- 26.Serrano R. Structure and function of proton translocating ATPase in plasma membranes of plants and fungi. Biochim Biophys Acta. 1988;947:1–28. doi: 10.1016/0304-4157(88)90017-2. [DOI] [PubMed] [Google Scholar]
- 27.Serrano R, Kielland-Brandt M C, Fink G R. Yeast plasma membrane ATPase is essential for growth and has homology with (Na++K+),K+- and Ca2+-ATPase. Nature. 1986;319:689–693. doi: 10.1038/319689a0. [DOI] [PubMed] [Google Scholar]
- 28.Seto-Young D, Monk B C, Perlin D S. Assessing hydrophobic regions of the plasma membrane H+-ATPase from Saccharomyces cerevisiae. Biochim Biophys Acta. 1992;1102:213–219. doi: 10.1016/0005-2728(92)90102-8. [DOI] [PubMed] [Google Scholar]
- 29.Slayman C L. The plasma membrane ATPase of Neurospora: a proton-pumping electroenzyme. J Bioenerg Biomembr. 1987;19:1–20. doi: 10.1007/BF00769728. [DOI] [PubMed] [Google Scholar]
- 30.Thornewell S J, Peery R B, Skatrud P L. Cloning and characterization of CneMDR1: a Cryptococcus neoformans gene encoding a protein related to multidrug resistance proteins. Gene. 1997;201:21–29. doi: 10.1016/s0378-1119(97)00421-6. [DOI] [PubMed] [Google Scholar]
- 31.Wach A, Schlesser A, Goffeau A. An alignment of 17 deduced protein sequences from plant, fungi, and ciliate H+-ATPase genes. J Bioenerg Biomembr. 1992;24:309–317. doi: 10.1007/BF00768851. [DOI] [PubMed] [Google Scholar]
- 32.Wang G, Tamas M J, Hall M J, Pascual-Ahuir A, Perlin D S. Probing conserved regions of the cytoplasmic LOOP1 segment linking transmembrane segments 2 and 3 of the Saccharomyces cerevisiae plasma membrane H+-ATPase. J Biol Chem. 1996;271:25438–25445. doi: 10.1074/jbc.271.41.25438. [DOI] [PubMed] [Google Scholar]
- 33.Zhang S, Varma A, Williamson P R. The yeast Cryptococcus neoformans uses ‘mammalian’ enhancer sites in the regulation of the virulence gene, CNLAC1. Gene. 1999;227:231–240. doi: 10.1016/s0378-1119(98)00590-3. [DOI] [PubMed] [Google Scholar]