Abstract
Obesity is the leading cause of health-related diseases in the United States and World. Previously, we reported that obesity can change gut microbiota using the Zucker rat model. Metformin is an oral anti-hyperglycemic agent approved by the FDA to treat type 2 diabetes (T2D) in adults and children older than 10 years of age. The correlation of short-term metformin treatment and specific alterations to the gut microbiota in obese models is less known. Short-term metformin has been shown to reduce liver steatosis. Here we investigate the effects of short-term metformin treatment on population of gut microbiota profile in an obese rat model. Five week old obese (n = 12) female Zucker rats after 1 week of acclimation, received AIN-93 G diet for 8 weeks and then rats were randomly assigned into two groups (6 rats/group): (1) obese without metformin (ObC), or (2) obese with metformin (ObMet). Metformin was mixed with AIN-93G diet at 1,000 mg/kg of diet. Rats were weighed twice per week. All rats were sacrificed at the end of metformin treatment at 10 weeks and fecal samples were collected and kept at −80°C. Total microbial DNA was collected directly from the fecal samples used for shotgun-metagenomics sequencing and subsequently analyzed using MetaPlAn and HUMAnN. After stringent data filtering and quality control we found significant differences (p = 0.0007) in beta diversity (Aitchison distances) between the ObC vs. ObMet groups. Supervised and unsupervised analysis of the log-ratios Bacteroides dorei and B. massiliensis vs. all other Bacteroides spp., revealed that B. dorei and B. massiliensis were enriched in the ObMet group, while the remaining Bacteroides spp. where enriched in the ObC group (p = 0.002). The contributional diversity of pathways is also significantly associated by treatment group (p = 0.008). In summary, in the obese Zucker rat model, short-term metformin treatment changes the gut microbiota profile, particularly altering the composition Bacteroides spp. between ObC and ObMet.
Keywords: obesity, metformin, Bacteroides dorei, Zucker rat, gut microbiota
Introduction
Obesity has been an epidemic in the United States (US) and the rate of adult obesity continues to grow. Data from the Centers for Disease Control and Prevention (CDC) indicated that more than one-third of United States adults are obese (Ogden et al., 2014). However, recent data from CDC indicate that In 2017–2018, the age-adjusted prevalence of obesity in adults was 42.4%, and there were no significant differences between men and women among all adults or by age group. The age-adjusted prevalence of severe obesity in adults was 9.2% and was higher in women than in men (Flegal et al., 2016). Obesity is associated with several health problems such as type 2 diabetes, cardiovascular disease, liver disease and certain types of cancers (Hakkak et al., 2012). Childhood obesity has more than doubled in children, and obesity has quadrupled in adolescents in the past 30 years (Angulo, 2007; Ogden et al., 2008; Fryar et al., 2021). In the United States, the prevalence of obesity is 19.3% and affects about 14.4 million children and adolescents. Obesity prevalence was 13.4% among 2- to 5-year-olds, 20.3% among 6- to 11-year-olds, and 21.2% among 12- to 19-year-olds (Fryar et al., 2021). Obesity is often associated with an increased risk of non-alcoholic fatty liver disease (NAFLD), in which liver steatosis is commonly observed (Fabbrini et al., 2010).
Non-alcoholic fatty liver disease is the leading cause of liver disease in adolescents in the United States and world, and the risk has increased with the rise of obesity (Lazo et al., 2013; Welsh et al., 2013). Data from our animal studies using the obese Zucker (fa/fa) rat model reported that obesity increases fatty liver (steatosis) and that obese Zucker rats can develop fatty liver by the starting age of 8 weeks (Hakkak et al., 2018, 2021).
The effects of obesity on composition and metabolic activity of the intestinal microbiota is an active area of study (Ley et al., 2005; Tremaroli and Bäckhed, 2012; Devaraj et al., 2013; Harsch and Konturek, 2018; Pascale et al., 2019; Cao et al., 2020; Zhang and Hu, 2020). Several studies have identified some differences between the microbiota populations in lean and obese subjects (Tremaroli and Bäckhed, 2012). Mice homozygous for the leptin receptor mutation that results in the development of obesity show a reduction in Bacteroidetes and an increase in Firmicutes compared to their wild-type siblings when fed the same diet. This effect is not limited to animals with a genetic predisposition to obesity. Diet-induced obesity is also linked to changes in the intestinal microbiota in mice (Turnbaugh et al., 2008). This connection between adiposity and the gut microbial ecology appears to apply to humans as well (Turnbaugh et al., 2009). It is clear that both community composition and discrete bacterial species can exert either pathogenic effects that encourage disease development or probiotic effects that maintain health status (Devaraj et al., 2013).
We recently reported the effects of obesity on gut microbiota using a Zucker rat model via amplicon sequencing of the 16S rRNA gene (Hakkak et al., 2017). Several groups of bacteria were differentially abundant between lean and obese rats after 60 days. Furthermore, we found that principal coordinate analysis (PCoA) plots of beta diversity, and LEfSe analysis (Segata et al., 2011) suggested differences in intestinal microbiota populations associated with both time point, and lean or obese status, within the Zucker rat model for obesity (Hakkak et al., 2017). The scientific community emphasizes the need to investigate the effects of metformin in conjunction with gut microbiota (McCreight et al., 2016; Rodriguez et al., 2018; Pascale et al., 2019; Zhang and Hu, 2020). The gut microbiota contains a diverse population of obligate and facultative anaerobic microorganisms that contribute a broad range of metabolic activities. These microorganisms usually exist in a symbiotic relationship with the host and are important in the digestion of dietary components and the metabolism of nutrients and drugs (Ley et al., 2006; Vázquez-Baeza et al., 2018; Weersma et al., 2020). The specific population of organisms comprising the intestinal microbiota in an individual is relatively stable under normal conditions, but several factors, such as diet, disease state, antimicrobial use, etc., can cause changes in the distribution of different bacterial groups (Turnbaugh et al., 2009; Tagliabue and Elli, 2013). These population changes can affect the metabolic capabilities of the total microbiota population, which can affect the health of the host (Rodriguez et al., 2018; Vázquez-Baeza et al., 2018; Zimmermann et al., 2019).
Members of the genus of Bacteroides (e.g., B. vulgatus and B. dorei) are mostly gram-negative anaerobic organisms. Bacteroides spp. are often characterized as a predominant gut bacterial species (Wexler and Goodman, 2017). Bacteroides dorei, was recently isolated and distinguished from Bacteroides vulgatus (Pedersen et al., 2013). Prior to this, both species were difficult to disambiguate until the advent of 16S rRNA gene amplicon sequencing methods (Pedersen et al., 2013). Although very short-read high-throughput sequencing of the 16S rRNA gene is unable to differentiate among the two taxa, success can be achieved by targeting the longer V3V4 region of the 16S rRNA gene (Davis-Richardson et al., 2014). This potentially explains seemingly contradictory results across ampicon-based microbiome surveys regarding the identity of Bacteroides spp. Prior studies have shown that these organisms might improve the enteric environment and reduce bacterial lipopolysaccharide (LPS) production (Yoshida et al., 2018). LPS is confirmed as a potent inducer of hepatic inflammation like NAFLD in obese subjects (Fukunishi et al., 2014).
Metformin is an oral anti-hyperglycemic agent approved by the FDA to treat type 2 diabetes (T2D) in adults and children older than 10 years of age. Several clinical trials have identified modest improvements following metformin treatment in insulin sensitivity in obese children with normal glucose tolerance (Srinivasan et al., 2006; Burgert et al., 2008; Love-Osborne et al., 2008; Brufani et al., 2013; McDonagh et al., 2014; Lentferink et al., 2018; Raman and Foster, 2021), as well as a decrease in the BMI of obese adolescents (Wilson et al., 2010). In addition, metformin appears to improve lipid profiles in obese children (Kay et al., 2001; Atabek and Pirgon, 2008). Furthermore, metformin is not only used for the treatment of diabetes, but also for various other diseases including cancer, cardiovascular diseases, and liver steatosis (Lin et al., 2000; Foretz et al., 2014; Madsen et al., 2015; Wang et al., 2016; Fujita and Inagaki, 2017; Lv and Guo, 2020; Hakkak et al., 2021).
Our prior research has also shown that short-term dietary effects can be observed when obese Zucker rats are fed a diet of soy protein with high isoflavones that can protect against liver steatosis. Although these rats have gained more weight compared to obese casein-fed rats, they had lower liver steatosis and contained lower blood serum levels aspartate aminotransferase AST, alanine aminotransferase (Hakkak et al., 2018). Similar results were observed for obese Zucker rats fed an AIN-93 G diet during metformin treatment (Hakkak et al., 2021). Prior research within murine models and human subjects has shown that short-term metformin treatment was sufficient to reduce liver steatosis (Lin et al., 2000; Madsen et al., 2015; Wang et al., 2016; Hakkak et al., 2021).
Although various mechanisms by which metformin acts are still being investigated, the last decade of research has led to some insightful discoveries on how the gut microbiome responds to and contributes to the altered metabolic landscape driven by metformin treatment, as reviewed by Sanz et al. (2015), McCreight et al. (2016), and Pascale et al. (2019). The ability of the microbiome to affect many other therapeutic treatments is well known and of increasing interest (Vázquez-Baeza et al., 2018; Zimmermann et al., 2019). Investigations on the effects of metformin on the gut microbiota have shown increasing evidence that a key factor of metformin action involves the gut microbiome (Pastor-Villaescusa et al., 2016; de la Cuesta-Zuluaga et al., 2017; Pascale et al., 2019). The effects of metformin on murine models through the use of high-fat diet-induced obesity (Zhang and Hu, 2020) revealed that metformin had significant effects and changes on the composition of the gut microbiota (Lee and Ko, 2014; Shin et al., 2014; Cao et al., 2020). However, the correlation of short-term metformin treatment and specific alterations to the gut microbiota in obese models and liver steatosis is less known.
We have previously shown that short-term (10 weeks) metformin treatment is a useful model for early adolescent obesity related diseases, using obese Zucker rat model (Hakkak et al., 2021). In this model, we were able to show that this short-term metformin treatment can protect against NAFLD. However, the possible mechanisms of this protection is less known. Herein we extend our investigations of the gut microbiome by focusing on the effects of short-term metformin treatment on obese Zucker rats using shotgun metagenomics to better resolve the species and strain-level identification of microbial taxa (Hong et al., 2009; Jovel et al., 2016; Wasimuddin et al., 2020).
Materials and Methods
Experimental Design
All animal care and procedures were approved by the University of Arkansas for Medical Sciences/Arkansas Children’s Research Institute Institutional Animal Care and Use Committee and adhered to the institutional policies and procedures. The guidelines of the United States Department of Agriculture (USDA) Animal Welfare Act were followed to ensure that the care and use of animals were appropriate and humane.
Sampling and Storage
A total of 12 five-week-old female obese (fa/fa) Zucker rats were purchased from Envigo, (Indianapolis, IN, United States), as they are sexually mature by this age (Sengupta, 2013). Female obese Zucker (fa/fa) rats are often used for non-diabetic obesity studies because they are highly resistant to developing diabetes unless fed a high fat diet (Corsetti et al., 2000; Gustavsson et al., 2011), whereby we can investigate obesity and liver steatosis without the confounding effects of a diabetic phenotype. We have also shown that both obese male and female rats will develop obesity and liver steatosis at the same rates and that there is no difference on between both sexes (Hakkak et al., 2012, 2015, 2018). Rats were housed in an Association for Assessment and Accreditation of Laboratory Animal Care approved animal facility that is registered with the USDA and has a fully approved Letter of Assurance on file with the Office of Laboratory Animal Welfare of the National Institutes of Health. Rats were housed one per cage in 12-h light-dark cycles and had ad libitum access to feed and water. After 1 week of acclimation (age 42 days), rats had ad libitum access to water on semi-purified AIN-93G diet (Envigo, Indianapolis, IN) for 8 weeks to mimic obese adolescents (Hakkak et al., 2012, 2015, 2018). Rats were weighed twice weekly. After 8 weeks on AIN 93-G diet, obese rats were randomly assigned into two groups (6 rats/group): (1) obese without metformin (ObC), or (2) obese with metformin (ObMet), and maintained for 10 weeks. Metformin was mixed with AIN-93G diet at 1,000 mg/kg of diet (Envigo, Indianapolis, IN, United States). We used a modified formula of the Reagan-Shaw approach for obesity to calculate the maximum dose of metformin in the proposed experiment (Food and Drug Administration, 2005). All rats were euthanized after treatment in the 10th week. Fecal samples were collected over a 12-h period a day before the metformin treatment diet and at the end of the experiment. Fecal samples were stored at −80°C until analysis.
DNA Extraction and Sequencing
Total microbial DNA was collected directly from the fecal samples using a PowerSoil ® DNA isolation kit (MoBio Laboratories, Inc., Carlsbad, CA, United States). Isolated DNA was used for shotgun-metagenomics data collection using Illumina NextSeq 500 (Supplementary Figure 1). Minimum Information about a Metagenomic Sequence (MIMS) compliant data (Field et al., 2007, 2008) are available from the National Center for Biotechnology Information Sequence Read Archive, under BioProject PRJNA770726.
Metagenomic Sequencing and Analyses
Shotgun metagenomic reads were first processed through the metaWRAP (Uritskiy et al., 2018) read_qc module which wraps FastQC (Andrews, 2010), Trim Galore (Martin, 2011; Krueger, 2015), and BMTagger (Rotmistrovsky and Agarwala, 2017), to filter and trim low-quality reads, and remove potential mammalian host sequences downloaded from GenBank (Benson et al., 2005), i.e., human (Homo sapiens; hg38), mouse (Mus musculus; mm10), rat (Rattus norvegicus; rn6), and pig (Sus scrofa; 10.2). The resulting reads were then processed via MetaPhlAn and HUMAnN via the bioBakery suite (Beghini et al., 2020) to determine microbial taxonomy and functional potential. Data preparation, as well as unsupervised and supervised microbial compositional analysis were performed using DEICODE (Martino et al., 2019), songbird (Morton et al., 2019), Qurro (Fedarko et al., 2020) and Emperor (Vázquez-Baeza et al., 2013; Supplementary Figure 2). The data were formatted for these analyses as outlined in Baker et al. (2021).
A taxonomic abundance table (feature-table) was generated from MetaPhlAn and subsequently filtered to contain only those bacteria that were identified at the species-level, and which were present in at least 50% of the samples (6 of 12), and contained at least 10,000 reads. This helps avoid spurious log ratios, as reviewed in Baker et al. (2021). Beta-diversity was calculated with DEICODE, which uses Robust Aitchison PCA, and visualized via Emperor. DEICODE was run with the following settings: min-sample-count 500, min-feature-count 1000, min-feature-frequency 0, max-iterations 10. Beta-diversity significance testing was performed through PERMANOVA. Upon visual inspection of the PCA plot, the feature-loadings of Axis 1 (which appeared to separate the control and metformin treatment groups), were visualized in Qurro. Either log-ratios of specific microbes (e.g., Bacteroides spp.), or those ranked above 0 and those equal or less than 0, were exported for post-hoc significance testing to determine microbiota associated with treatment group separation. This comprised our unsupervised analysis of microbial taxa.
The same taxonomy table was then analyzed via Songbird to rank microbial species that are associated with our metformin treatment through the use of reference frames. The following parameters were used: batch-size 3, num-random-test-examples 3, learning-rate 0.0001, epochs 50000, differential-prior 0.5, min-feature-count 6, summary-interval 1. This comprised our supervised analysis of microbial taxa.
A functional pathway table was generated with HUMAnN, which contains the functional potential of each metagenome sample. This data was analyzed similarly as the MetaPhlAn data above except in this case DEICODE was used to ordinate the samples with respect to functional pathway composition as they relate to the treatment groups, while Songbird was used to rank the pathways themselves as they relate to metformin treatment. The HUMAnN pathway table was filtered to keep pathways that were present in at least 50% of the samples (6 of 12) and contained at least 1,000 reads. The settings used for DEICODE: min-sample-count 500, min-feature-count 1000, min-feature-frequency 0, max-iterations 10. The setting used for Songbird: batch-size 3, num-random-test-examples 3, learning-rate 0.0001, epochs 100,000, differential-prior 0.5, min-feature-count 6, summary-interval 1.
Results
Body Weight
The final body weights for ObC vs. ObMet groups was 597.5 ± 41.4 g vs. 573.1 ± 48.1 g, respectively and not significantly different (P = 0.20).
Microbial Diversity
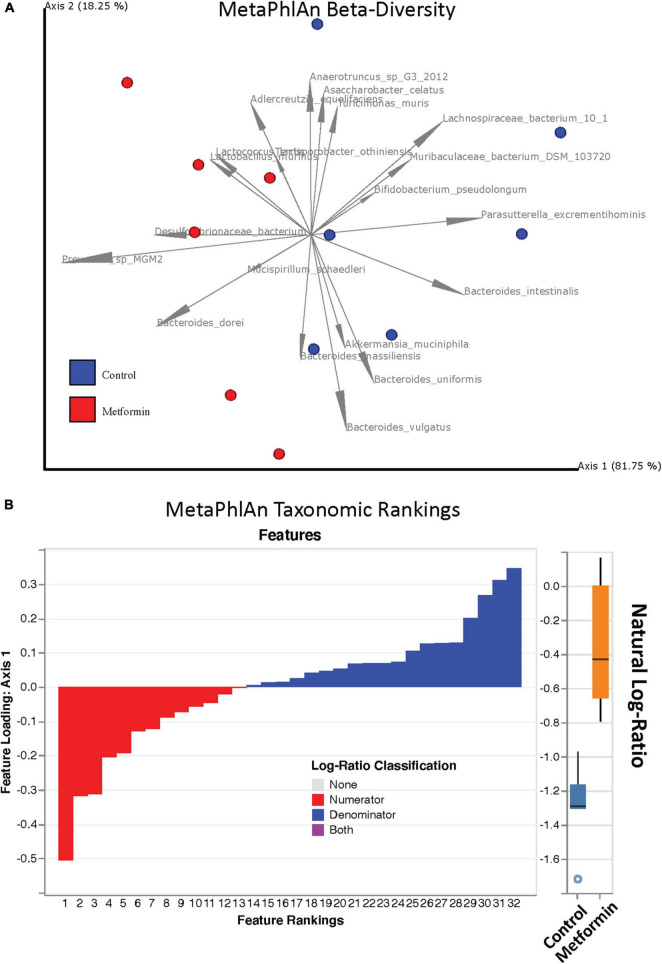
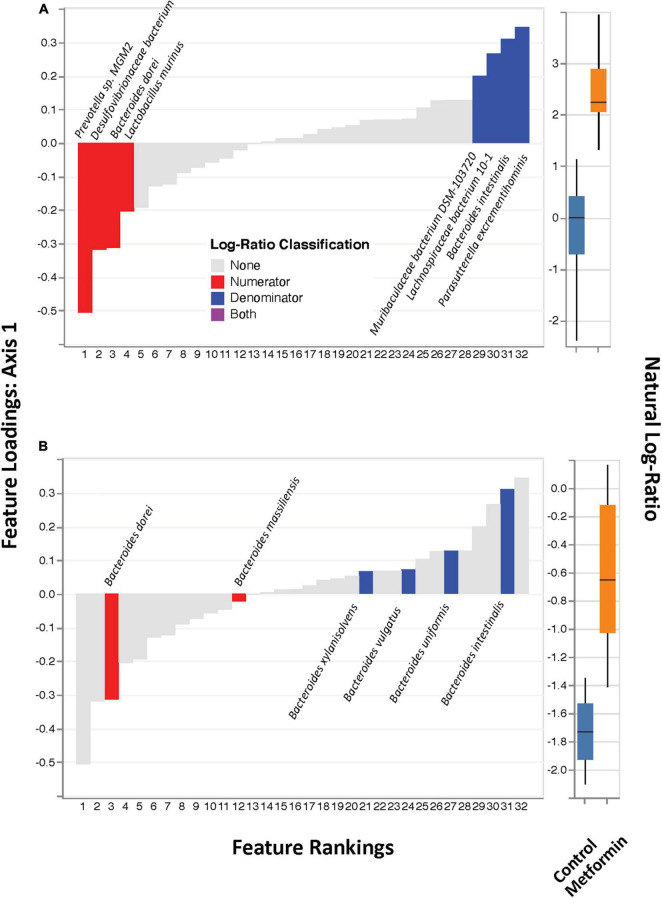
No significant differences in alpha diversity were found between the control and metformin treated groups. However, significant differences in beta diversity were observed between these groups, as measured by Bray-Curtis (p < 0.011) and DEICODE (Figure 1; p < 0.002), but not Jaccard. DEICODE was further used as an unsupervised approach to detect differentially abundant ratios of microbiota. The log-ratio rankings of individual bacterial taxa were found to be significantly different (p < 0.0025) between the control and metformin groups after post-hoc analysis (Figure 1). This signal was still present when either the top feature-rankings were selected (Figure 2A), or only the feature rankings of Bacteroides taxa (Figure 2B). The most notable finding is the differential ratios of Bacteroides species present in each of the treatment groups, i.e., increased B. dorei and B. massiliensis (numerator taxa) in the metformin group relative to the control group, while the opposite is observed for (denominator taxa) B. xylanisolvens, B. vulgatus, B. uniformis, and B. intestinalis (Figure 2B). The above patterns were also observed through a supervised approach with Songbird (Supplementary Figure 3).
FIGURE 1.
(A) Robust Aitchison PCA plot of metagenome samples processed through MetaPhlAn 3 and DEICODE and visualized with qurro. Bacterial species not present in at least 50% of samples were removed from the analysis. Separation of Control and Metformin treated groups was significant (p-value 0.002). (B) Corresponding log-ratio rankings for individual bacterial taxa oriented to Axis 1 (right). Numerator (red) / Denominator (blue) ratios of these taxa were computed to generate a box-whisker plot (left), p-value 0.0025. These results were corroborated with songbird analyses.
FIGURE 2.
DEICODE feature rankings. (A) Same plot as in Figure 1, with only the top-feature rankings selected (left), and their ratios plotted as a box-whisker-plot (right). (B) Only Bacteroides spp. selected (left) and their ratios plotted as box-whisker plot (right).
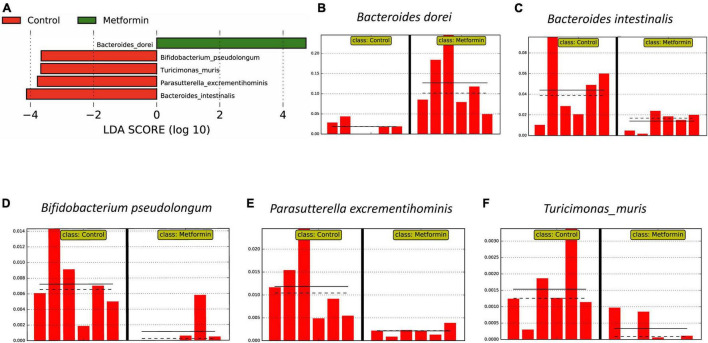
These results were generally consistent with LEfSe analyses (strict all-vs.-all setting; Figure 3), B. dorei is enriched in ObMet (Figures 3A,B) while B. intestinalis is enriched in the ObC (Figures 3A,C), with other taxa also enriched in the control group (Figures 3A,D–F).
FIGURE 3.
Differentially abundant bacterial taxa as determined by LEfSe analysis on MetaPhlAn 3 output. (A) Linear discriminant analysis (LDA) score plot of differential taxa associated with the Control and Metformin groups. Differential feature plots for the respective taxa are shown across samples: (B) Bacteroides dorei, (C) Bacteroides intestinalis, (D) Bifidobacterium pseudolongum, (E) Parasutterella excrementihominis, and (F) Turicimonas muris.
Contributional Pathway Diversity
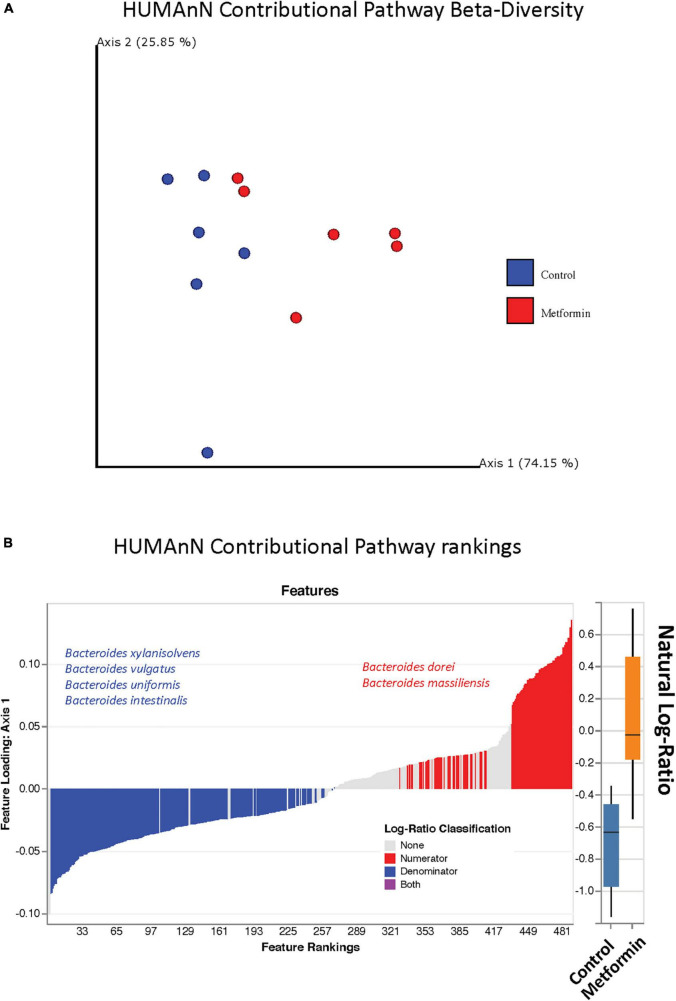
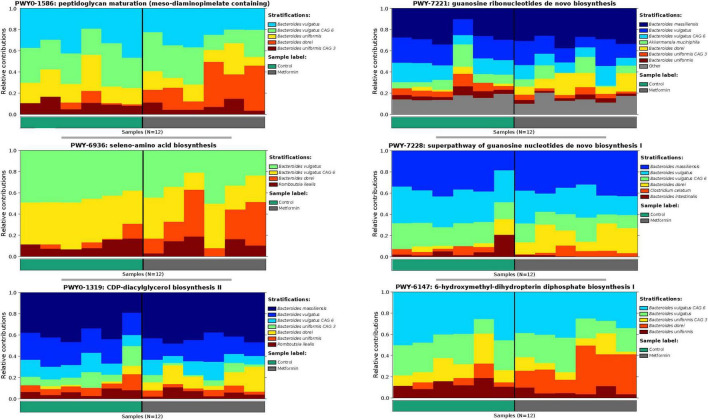
The observed contributional diversity of microbial taxa was also significantly associated with the treatment group (p < 0.012), as observed through unsupervised analysis via DEICODE (Figure 4A). The majority of all pathways were contributed by Bacteroides (Figure 4B). Although identical pathways can be contributed by all the Bacteroides, the specific species from which they are contributed, differs between the metformin and control groups. This can be exemplified through the differential contributional pathway diversity plots generated by HUMAnN, using some of the top-ranked pathways as determined by DEICODE (Figure 5 and Supplementary Figure 4). These results were confirmed using the supervised approach of Songbird (Supplementary Figure 5).
FIGURE 4.
(A) Robust Aitchison PCA plot of metagenome samples processed through HUMAnN and DEICODE and visualized with qurro. Pathways not present in at least 50% of samples were removed from the analysis. These values Separation of Control and Metformin treated groups was significant (p-value 0.012). (B) Corresponding log-ratio rankings for Pathways oriented to Axis 1 (right), displaying only pathways contributed by Bacteroides spp. Numerator (red)/Denominator (blue) ratios of these pathways were computed to generate a box-whisker plot (left), p-value 0.008. These results were corroborated with songbird analyses.
FIGURE 5.
Differential contributional pathway diversity of top-ranked pathways, determined by DEICODE and qurro from HUMAnN analysis.
Discussion
In this study, we examined the role of metformin, an anti-hyperglycemic agent, on the gut microbiome of obese female Zucker rats (fa/fa) during short-term treatment. This animal model is the most widely used model for non-diabetic obesity related research, unlike the Zucker Diabetic Fatty rat model. The primary cause for obesity in Zucker (fa/fa) rats is due to the mutation in the leptin receptor gene (fa) which is inherited by the rats as an autosomal recessive trait. The rats become noticeably obese by the age of 3–5 weeks, and by 14 weeks, >40% of their body is composed of lipids (Zucker, 1972; Bray et al., 1989). Furthermore, clinical studies and murine model systems have shown that short-term metformin treatment was sufficient to reduce liver steatosis (Lin et al., 2000; Madsen et al., 2015; Wang et al., 2016; Hakkak et al., 2021). Zucker rat models have also significantly contributed to the study of the function and role of microbiota in the gastrointestinal tract and its association with diseases such as metabolic disorder besides obesity (Gawler et al., 1989; Hakkak et al., 2017; Sui et al., 2019).
It is well established that the gut microbiota co-evolves with the host and the modified gut microbiome has been strongly linked with host obesity and associated therapeutics (Devaraj et al., 2013; Harsch and Konturek, 2018; Whang et al., 2019). Studies have shown that a high-fat diet for obesity, facilitates metabolic disorders, like Type 2 diabetes mellitus (T2DM) which may alter the gut microbiome (Pascale et al., 2019; Zhang and Hu, 2020). We reported the effects of obesity on the gut microbiome of Zucker rats using amplicon sequencing of the 16S rRNA gene. We found that obese rats showed higher ratios of the Firmicutes to Bacteroidetes phyla than the lean rats within a given timeframe, even though the rats were fed same high-fat diet. Gut microbiota populations appeared to shift with both the time point and observed phenotype (Hakkak et al., 2017).
Metformin is primarily administered alone or in combination with other hypoglycemic drugs to effectively control the blood glucose level of diabetic individuals, particularly to obese or overweight patients (Maruthur et al., 2016; Thomas and Gregg, 2017). Subsequently, evidence from multiple studies underscores the ability of metformin to reshape the gut microbiome in rat models induced by high-fat diet and in T2DM individuals (Zhang et al., 2015; Zhang and Hu, 2020). One of the studies reported that metformin exerts hypoglycemic effects by affecting the gut microbiome, which maintains intestinal barrier function, increases the production of short chain fatty acids, regulates bile acid metabolism, and affects glucose homeostasis. Hence, it is important to understand the relationships between obesity and the intestinal microbiota in correlation with the effects of metformin in obese Zucker rats significantly in their widespread use as a model disease system (Zhang et al., 2015).
Our study herein, PCA plots of beta-diversity showed the separation of each test group (ObC and ObMet) into different bacterial populations by unsupervised clustering through Aitchison distances. The dominating taxa that are associated with the separation of the metagenomic samples include Bacteroides, Akkermansia, Bifidobacterium (Figure 1). Previous studies related to metformin treatment on high-fat diet induced obese rats have also reported the alterations of similar taxa, particularly Bacteroides, Bifidobacterium (Zhang et al., 2019; Zhang and Hu, 2020). In one such study, Zhang and Hu (2020) has shown that metformin modulates the gut microbiota by enriching short chain fatty acid (SCFA) producing bacteria, like Bacteroides spp. We’ve observed similar taxonomic alterations through the analysis of log-ratios, based on MetaPhlAn output. Not only are Bacteroides spp. enriched HFD Zucker rats in general, but our shotgun metagenomics analysis revealed differences in the Bacteroides spp. that are differentially enriched between the treatment groups. For example, we observed that B. dorei, and B. massiliensis are enriched in ObMet, while B. vulgatus, B. uniformis, B. intestinalis, and B. xylanisolvens are enriched in ObC. The enrichment of D. dorei in the metformin-treated Zucker rats was also observed in rats with T2D (Yoshida et al., 2018; Zhang et al., 2020). Other studies in murine and human models often observe the co-enrichment of B. vulgatus and B. dorei across various disease states (Yoshida et al., 2018). However, we observe that B. dorei is enriched in ObMet while B. vulgatus is enriched in ObC (Figure 2 and Supplementary Figure 3). This pattern, where the ratio of B. dorei to B. vulgatus differs with respect to treatment groups, was also observed in blockade treatments in metastatic melanoma (Usyk et al., 2021) and with host epigenomic alterations of inflammatory bowel disease (Ryan et al., 2020). It was proposed by Ryan et al. (2020), that this pattern may be indicative of colonic-crypt species-specific colonization and competition (Sonnenburg et al., 2010; Lee et al., 2013) due to differences in metabolic capabilities (Gutiérrez and Garrido, 2019). Whether this, or other similar phenomenon, is occurring in our study system remains to be determined.
The most abundant bacteria reported in our study is Bacteroides dorei. Which is capable of metabolizing many drugs (Zimmermann et al., 2019), and could potentially be doing so with metformin. LEfSe analysis (Segata et al., 2011), also confirmed that Bacteroides dorei was enriched in metformin treated obese rats compared to the control group (Figure 3). Bacteroides intestinalis, Bifidobacterium pseudolongum, Parasutterella excrementihominis, and Turicimonas muris were found to be abundant in non-metformin treated obese rats.
A subsequent pathway ranking analysis revealed similar results about the taxa contributing functional pathways. The Bacteroides sp. described above were found to be contributing to the pathways from both test groups. Some taxa are reportedly found exclusively in mouse models like Turicimonas sp. (Lagkouvardos et al., 2016), while others may be present in both rat models and humans.
Recent evidence has shown that individuals who are genetically susceptible to various autoimmune diseases, like T2DM have substantial differences in gut microbial composition than non-genetically susceptible individuals (Bakir et al., 2006). Davis-Richardson et al. (2014) reported in their study about the role of Bacteroides dorei dominating the gut microbiome in Finnish children likely susceptible to type 1 diabetes mellitus (T1D). Significantly higher composition of B. dorei and B. vulgatus was found in the autoimmune susceptible children than in the control group with the help of metagenomic sequencing. The study also suggested the potential involvement of B. dorei in autoimmune disorders like T1D.
Bacteroides vulgatus has been associated with the reduced risk of immune-related adverse events (Usyk et al., 2021), including dermatological skin toxicity (where B. dorei anti-correlated) (Zhang et al., 2021). Furthermore, B. vulgatus has been shown to be protective against Escherichia coli induced colitis of interleukin-2-deficient gnotobiotic mice (Waidmann et al., 2003). In contrast, several studies revealed that B. vulgatus is generally viewed as a pathobiont (Ó Cuív et al., 2017), often associated with colitis in murine (Bloom et al., 2011) model systems, as well as human ulcerative colitis (Mills et al., 2022), irritable bowel disease (Ryan et al., 2020), and celiac disease (Schippa et al., 2010; Bloom et al., 2011; Wu et al., 2021). It has also been shown that B. vulgatus has an increased protease activity compared to other Bacteroides spp., and can cause barrier dysfunction (Riepe et al., 1980; Mills et al., 2022). We are unable to determine if this is at all related to the observed patterns described herein. This highlights the importance of ecological context when considering where a particular microbe lies on the “parasitism (pathogen)–mutualism” spectrum (Drew et al., 2021).
The major differences in this current study with previously related studies is the reliance of obese animals fed with same diets compared to high-fat diet for inducing obesity (Zhang et al., 2015). Also, the emphasis of metformin treatment and subsequent examination of the gut microbiota for compositional changes is less known in the scientific community. Thereby, the relationship between obesity, gut microbiome and the role of B. dorei demands further investigation in the Zucker rat model. Sample size is a major limiting factor in this study, a larger sample size applying short-term metformin treatment may strengthen the weight/power of the data.
Subsequently, the development of different chronic diseases is linked to obesity and also the host’s gut microbiome, where the relationship between the two plays a vital role in development of autoimmune diseases, cardiovascular disease, kidney disease and certain types of cancers (Wang et al., 2017; Vázquez-Baeza et al., 2018; Hobby et al., 2019; Kazemian et al., 2020; Sepich-Poore et al., 2021). Consequently, future investigations may be conducted on the relationship between obesity, metformin or other related drug, gut microbiota and several related diseases can be based on the standard data from provided by our study on the interaction and role of metformin correlated with obesity and the gut microbiome in Zucker rats.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA770726.
Ethics Statement
The animal study was reviewed and approved by the Association for Assessment and Accreditation of Laboratory Animal Care approved animal facility. This facility is registered with the USDA and has a fully approved Letter of Assurance on file with the Office of Laboratory Animal Welfare of the National Institutes of Health.
Author Contributions
RH conceived of the project, acquired the funding, and conducted laboratory experiments. CR, RH, and SB collected the data. MR analyzed the data. All authors contributed to the writing of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Funding
This research was supported, in part, by the Arkansas Children’s Research Institute and the Arkansas Biosciences Institute to RH.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.834776/full#supplementary-material
Overall study design.
Metagenomics workflow.
Songbird differential rankings processed through MetaPhlAn. (A) Same plot as in Figure 1B with all features above and below 0 selected, and their ratios plotted as a box-whisker-plot (right). (B) Same plot as with Figure 2A, with only the top-feature rankings selected (left), and their ratios plotted as a box-whisker-plot (right). (C) Same as Figure 2B, only Bacteroides spp. selected (left) and their ratios plotted as box-whisker plot (right).
Songbird differential rankings of HUMAnN pathways. These are shown in the HUMAnN contributional diversity plot of Figure 5.
Songbird differential rankings processed through HUMAnN. (A) Same plot as in Supplementary Figure 3A with all features above and below 0 selected, and their ratios plotted as a box-whisker-plot (right). (B) Same plot as with Supplementary Figure 3B, with only the top-feature rankings selected (left), and their ratios plotted as a box-whisker-plot (right). (C) Same as Supplementary Figure 3C, only Bacteroides spp. selected (left) and their ratios plotted as box-whisker plot (right).
References
- Andrews S. (2010). FastQC: A Quality Control Tool For High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. (accessed March 13, 2020). [Google Scholar]
- Angulo P. (2007). GI Epidemiology: nonalcoholic fatty liver disease. Aliment. Pharmacol. Ther. 25 883–889. 10.1111/j.1365-2036.2007.03246.x. [DOI] [PubMed] [Google Scholar]
- Atabek M. E., Pirgon O. (2008). Use of metformin in obese adolescents with hyperinsulinemia: a 6-month, randomized, double-blind, placebo-controlled clinical trial. J. Pediatr. Endocrinol. Metab. 21 339–348. 10.1515/jpem.2008.21.4.339 [DOI] [PubMed] [Google Scholar]
- Baker J. L., Morton J. T., Dinis M., Alvarez R., Tran N. C., Knight R., et al. (2021). Deep metagenomics examines the oral microbiome during dental caries, revealing novel taxa and co-occurrences with host molecules. Genome Res. 31 64–74. 10.1101/gr.265645.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakir M. A., Sakamoto M., Kitahara M., Matsumoto M., Benno Y. (2006). Bacteroides dorei sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 56 1639–1643. 10.1099/ijs.0.64257-0 [DOI] [PubMed] [Google Scholar]
- Beghini F., McIver L. J., Blanco-Míguez A., Dubois L., Asnicar F., Maharjan S., et al. (2020). Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife 10:e65088. 10.7554/eLife.65088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D. A., Karsch-Mizrachi I., Lipman D. J., Ostell J., Wheeler D. L. (2005). GenBank. Nucleic Acids Res. 33 D34–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom S. M., Bijanki V. N., Nava G. M., Sun L., Malvin N. P., Donermeyer D. L., et al. (2011). Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe 9 390–403. 10.1016/j.chom.2011.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray G. A., York D. A., Fisler J. S. (1989). Experimental obesity: a homeostatic failure due to defective nutrient stimulation of the sympathetic nervous system. Vitam. Horm. 45 1–125. 10.1016/s0083-6729(08)60393-3 [DOI] [PubMed] [Google Scholar]
- Brufani C., Crinò A., Fintini D., Patera P. I., Cappa M., Manco M. (2013). Systematic review of metformin use in obese nondiabetic children and adolescents. Horm. Res. Paediatr. 80 78–85. 10.1159/000353760 [DOI] [PubMed] [Google Scholar]
- Burgert T. S., Duran E. J., Goldberg-Gell R., Dziura J., Yeckel C. W., Katz S., et al. (2008). Short-term metabolic and cardiovascular effects of metformin in markedly obese adolescents with normal glucose tolerance. Pediatr. Diabetes 9 567–576. 10.1111/j.1399-5448.2008.00434.x. [DOI] [PubMed] [Google Scholar]
- Cao T. T. B., Wu K.-C., Hsu J.-L., Chang C.-S., Chou C., Lin C.-Y., et al. (2020). Effects of non-insulin anti-hyperglycemic agents on gut microbiota: a systematic review on human and animal studies. Front. Endocrinol. 11:573891. 10.3389/fendo.2020.573891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsetti J. P., Sparks J. D., Peterson R. G., Smith R. L., Sparks C. E. (2000). Effect of dietary fat on the development of non-insulin dependent diabetes mellitus in obese Zucker diabetic fatty male and female rats. Atherosclerosis 148 231–241. 10.1016/s0021-9150(99)00265-8 [DOI] [PubMed] [Google Scholar]
- Davis-Richardson A. G., Ardissone A. N., Dias R., Simell V., Leonard M. T., Kemppainen K. M., et al. (2014). Bacteroides dorei dominates gut microbiome prior to autoimmunity in finnish children at high risk for type 1 diabetes. Front. Microbiol. 5:678. 10.3389/fmicb.2014.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cuesta-Zuluaga J., Mueller N. T., Corrales-Agudelo V., Velásquez-Mejía E. P., Carmona J. A., Abad J. M., et al. (2017). Metformin is associated with higher relative abundance of mucin-degrading akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care 40 54–62. 10.2337/dc16-1324 [DOI] [PubMed] [Google Scholar]
- Devaraj S., Hemarajata P., Versalovic J. (2013). The human gut microbiome and body metabolism: implications for obesity and diabetes. Clin. Chem. 59 617–628. 10.1373/clinchem.2012.187617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew G. C., Stevens E. J., King K. C. (2021). Microbial evolution and transitions along the parasite–mutualist continuum. Nat. Rev. Microbiol. 19 623–638. 10.1038/s41579-021-00550-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbrini E., Sullivan S., Klein S. (2010). Obesity and nonalcoholic fatty liver disease: biochemical, metabolic, and clinical implications. Hepatology 51 679–689. 10.1002/hep.23280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedarko M. W., Martino C., Morton J. T., González A., Rahman G., Marotz C. A., et al. (2020). Visualizing ’omic feature rankings and log-ratios using Qurro. NAR Genom. Bioinform. 2:lqaa023 10.1093/nargab/lqaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D., Garrity G., Gray T., Morrison N., Selengut J., Sterk P., et al. (2008). The minimum information about a genome sequence (MIGS) specification. Nat. Biotechnol. 26 541–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D., Garrity G., Gray T., Selengut J., Sterk P., Thomson N., et al. (2007). eGenomics: cataloguing our complete genome collection III. Comp. Funct. Genomics 2007 1–7. 10.1155/2007/47304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegal K. M., Kruszon-Moran D., Carroll M. D., Fryar C. D., Ogden C. L. (2016). Trends in obesity among adults in the United States, 2005 to 2014. JAMA 315 2284–2291. 10.1001/jama.2016.6458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Drug Administration (2005). Guidance for Industry: Estimating the Maximum Safe Starting Dose in Initial Clinical Trials for Therapeutics in Adult Healthy Volunteers. Beltsville, MD: Center for Drug Evaluation and Research (CDER), 7. [Google Scholar]
- Foretz M., Guigas B., Bertrand L., Pollak M., Viollet B. (2014). Metformin: from mechanisms of action to therapies. Cell Metab. 20 953–966. 10.1016/j.cmet.2014.09.018 [DOI] [PubMed] [Google Scholar]
- Fryar C. D., Carroll M. D., Afful J., and Division of Health and Nutrition Examination Surveys. (2021). Prevalence of Overweight, Obesity, and Severe Obesity Among Children and Adolescents Aged 2–19 Years: United States, 1963–1965 Through 2017–2018. National Center for Health Statistics: Hyattsville, MD. [Google Scholar]
- Fujita Y., Inagaki N. (2017). Metformin: new preparations and nonglycemic benefits. Curr. Diab. Rep. 17:5. 10.1007/s11892-017-0829-8 [DOI] [PubMed] [Google Scholar]
- Fukunishi S., Sujishi T., Takeshita A., Ohama H., Tsuchimoto Y., Asai A., et al. (2014). Lipopolysaccharides accelerate hepatic steatosis in the development of nonalcoholic fatty liver disease in Zucker rats. J. Clin. Biochem. Nutr. 54 39–44. 10.3164/jcbn.13-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawler D. J., Wilson A., Houslay M. D. (1989). Metformin treatment of lean and obese Zucker rats modulates the ability of glucagon and insulin to regulate hepatocyte adenylate cyclase activity. J. Endocrinol. 122 207–212. 10.1677/joe.0.1220207 [DOI] [PubMed] [Google Scholar]
- Gustavsson C., Soga T., Wahlström E., Vesterlund M., Azimi A., Norstedt G., et al. (2011). Sex-dependent hepatic transcripts and metabolites in the development of glucose intolerance and insulin resistance in Zucker diabetic fatty rats. J. Mol. Endocrinol. 47 129–143. 10.1530/JME-11-0007 [DOI] [PubMed] [Google Scholar]
- Gutiérrez N., Garrido D. (2019). Species deletions from microbiome consortia reveal key metabolic interactions between gut microbes. mSystems 4:e00185-19 10.1128/mSystems.00185-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkak R., Al-Dwairi A., Fuchs G. J., Korourian S., Simmen F. A. (2012). Dietary soy protein induces hepatic lipogenic enzyme gene expression while suppressing hepatosteatosis in obese female Zucker rats bearing DMBA-initiated mammary tumors. Genes Nutr. 7 549–558. 10.1007/s12263-012-0294-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkak R., Gauss C. H., Bell A., Korourian S. (2018). Short-term soy protein isolate feeding prevents liver steatosis and reduces serum ALT and AST levels in obese female zucker rats. Biomedicines 6:55 10.3390/biomedicines6020055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkak R., Korourian S., Foley S. L., Erickson B. D. (2017). Assessment of gut microbiota populations in lean and obese Zucker rats. PLoS One 12:e0181451. 10.1371/journal.pone.0181451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkak R., Rose S., Spray B., Kozaczek M., Korourian S. (2021). Effects of obesity and 10 weeks metformin treatment on liver steatosis. Biomed. Rep. 14 1–6. 10.3892/br.2021.1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkak R., Zeng H., Dhakal I. B., Korourian S. (2015). Short- and long-term soy diet versus casein protects liver steatosis independent of the arginine content. J. Med. Food 18 1274–1280. 10.1089/jmf.2015.0002 [DOI] [PubMed] [Google Scholar]
- Harsch I. A., Konturek P. C. (2018). The role of gut microbiota in obesity and type 2 and type 1 diabetes mellitus: new insights into “old” diseases. Med. Sci. 6:32 10.3390/medsci6020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobby G. P., Karaduta O., Dusio G. F., Singh M., Zybailov B. L., Arthur J. M. (2019). Chronic kidney disease and the gut microbiome. Am. J. Physiol. Renal Physiol. 316 F1211–F1217. 10.1152/ajprenal.00298.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Bunge J., Leslin C., Jeon S., Epstein S. S. (2009). Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J. 3 1365–1373. 10.1038/ismej.2009.89 [DOI] [PubMed] [Google Scholar]
- Jovel J., Patterson J., Wang W., Hotte N., O’Keefe S., Mitchel T., et al. (2016). Characterization of the gut microbiome using 16S or shotgun metagenomics. Front. Microbiol. 7:459. 10.3389/fmicb.2016.00459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay J. P., Alemzadeh R., Langley G., D’Angelo L., Smith P., Holshouser S. (2001). Beneficial effects of metformin in normoglycemic morbidly obese adolescents. Metabolism 50 1457–1461. 10.1053/meta.2001.28078 [DOI] [PubMed] [Google Scholar]
- Kazemian N., Mahmoudi M., Halperin F., Wu J. C., Pakpour S. (2020). Gut microbiota and cardiovascular disease: opportunities and challenges. Microbiome 8:36 10.1186/s40168-020-00821-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F. (2015). Trim galore. A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. Bioinformatics 516:517. [Google Scholar]
- Lagkouvardos I., Pukall R., Abt B., Foesel B. U., Meier-Kolthoff J. P., Kumar N., et al. (2016). The Mouse Intestinal Bacterial Collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nat. Microbiol. 1:16131 10.1038/nmicrobiol.2016.131 [DOI] [PubMed] [Google Scholar]
- Lazo M., Hernaez R., Eberhardt M. S., Bonekamp S., Kamel I., Guallar E., et al. (2013). Prevalence of nonalcoholic fatty liver disease in the united states: the third national health and nutrition examination survey, 1988–1994. Am. J. Epidemiol. 178 38–45. 10.1093/aje/kws448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Ko G. (2014). Effect of metformin on metabolic improvement and gut microbiota. Appl. Environ. Microbiol. 80 5935–5943. 10.1128/AEM.01357-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. M., Donaldson G. P., Mikulski Z., Boyajian S., Ley K., Mazmanian S. K. (2013). Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501 426–429. 10.1038/nature12447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentferink Y. E., Knibbe C. A. J., van der Vorst M. M. J. (2018). Efficacy of metformin treatment with respect to weight reduction in children and adults with obesity: a systematic review. Drugs 78 1887–1901. 10.1007/s40265-018-1025-0 [DOI] [PubMed] [Google Scholar]
- Ley R. E., Backhed F., Turnbaugh P., Lozupone C. A., Knight R. D., Gordon J. I. (2005). Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. 102 11070–11075. 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. (2006). Microbial ecology: human gut microbes associated with obesity. Nature 444 1022–1023. 10.1038/4441022a [DOI] [PubMed] [Google Scholar]
- Lin H. Z., Yang S. Q., Chuckaree C., Kuhajda F., Ronnet G., Diehl A. M. (2000). Metformin reverses fatty liver disease in obese, leptin-deficient mice. Nat. Med. 6 998–1003. 10.1038/79697 [DOI] [PubMed] [Google Scholar]
- Love-Osborne K., Sheeder J., Zeitler P. (2008). Addition of metformin to a lifestyle modification program in adolescents with insulin resistance. J. Pediatr. 152 817–822. 10.1016/j.jpeds.2008.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Z., Guo Y. (2020). Metformin and its benefits for various diseases. Front. Endocrinol. 11:191. 10.3389/fendo.2020.00191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen A., Bozickovic O., Bjune J.-I., Mellgren G., Sagen J. V. (2015). Metformin inhibits hepatocellular glucose, lipid and cholesterol biosynthetic pathways by transcriptionally suppressing steroid receptor coactivator 2 (SRC-2). Sci. Rep. 5:16430. 10.1038/srep16430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 17 10–12. 10.1089/cmb.2017.0096 [DOI] [PubMed] [Google Scholar]
- Martino C., Morton J. T., Marotz C. A., Thompson L. R., Tripathi A., Knight R., et al. (2019). A novel sparse compositional technique reveals microbial perturbations. mSystems 4:e00016-19. 10.1128/mSystems.00016-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruthur N. M., Tseng E., Hutfless S., Wilson L. M., Suarez-Cuervo C., Berger Z., et al. (2016). Diabetes medications as monotherapy or metformin-based combination therapy for type 2 diabetes: a systematic review and meta-analysis. Ann. Intern. Med. 164 740–751. 10.7326/M15-2650 [DOI] [PubMed] [Google Scholar]
- McCreight L. J., Bailey C. J., Pearson E. R. (2016). Metformin and the gastrointestinal tract. Diabetologia 59 426–435. 10.1007/s00125-015-3844-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonagh M. S., Selph S., Ozpinar A., Foley C. (2014). Systematic review of the benefits and risks of metformin in treating obesity in children aged 18 years and younger. JAMA Pediatr. 168 178–184. 10.1001/jamapediatrics.2013.4200 [DOI] [PubMed] [Google Scholar]
- Mills R. H., Dulai P. S., Vázquez-Baeza Y., Sauceda C., Daniel N., Gerner R. R., et al. (2022). Multi-omics analyses of the ulcerative colitis gut microbiome link Bacteroides vulgatus proteases with disease severity. Nat. Microbiol. 7 262–276. 10.1038/s41564-021-01050-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton J. T., Marotz C., Washburne A., Silverman J., Zaramela L. S., Edlund A., et al. (2019). Establishing microbial composition measurement standards with reference frames. Nat. Commun. 10:2719. 10.1038/s41467-019-10656-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ó Cuív P., de Wouters T., Giri R., Mondot S., Smith W. J., Blottière H. M., et al. (2017). The gut bacterium and pathobiont Bacteroides vulgatus activates NF-κB in a human gut epithelial cell line in a strain and growth phase dependent manner. Anaerobe 47 209–217. 10.1016/j.anaerobe.2017.06.002 [DOI] [PubMed] [Google Scholar]
- Ogden C. L., Carroll M. D., Flegal K. M. (2008). High body mass index for age among US children and adolescents, 2003–2006. JAMA 299 2401–2405. 10.1001/jama.299.20.2401 [DOI] [PubMed] [Google Scholar]
- Ogden C. L., Carroll M. D., Kit B. K., Flegal K. M. (2014). Prevalence of childhood and adult obesity in the United States, 2011–2012. JAMA 311 806–814. 10.1001/jama.2014.732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascale A., Marchesi N., Govoni S., Coppola A., Gazzaruso C. (2019). The role of gut microbiota in obesity, diabetes mellitus, and effect of metformin: new insights into old diseases. Curr. Opin. Pharmacol. 49 1–5. 10.1016/j.coph.2019.03.011 [DOI] [PubMed] [Google Scholar]
- Pastor-Villaescusa B., Caballero-Villarraso J., Cañete M. D., Hoyos R., Maldonado J., Bueno G., et al. (2016). Evaluation of differential effects of metformin treatment in obese children according to pubertal stage and genetic variations: study protocol for a randomized controlled trial. Trials 17:323. 10.1186/s13063-016-1403-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen R. M., Marmolin E. S., Justesen U. S. (2013). Species differentiation of Bacteroides dorei from Bacteroides vulgatus and Bacteroides ovatus from Bacteroides xylanisolvens - back to basics. Anaerobe 24 1–3. 10.1016/j.anaerobe.2013.08.004 [DOI] [PubMed] [Google Scholar]
- Raman V., Foster C. M. (2021). Metformin treatment of pediatric obesity. Pediatrics 147:e2020044982 10.1542/peds.2020-044982. [DOI] [PubMed] [Google Scholar]
- Riepe S. P., Goldstein J., Alpers D. H. (1980). Effect of secreted Bacteroides proteases on human intestinal brush border hydrolases. J. Clin. Invest. 66 314–322. 10.1172/JCI109859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J., Hiel S., Delzenne N. M. (2018). Metformin: old friend, new ways of action-implication of the gut microbiome? Curr. Opin. Clin. Nutr. Metab. Care 21 294–301. 10.1097/MCO.0000000000000468 [DOI] [PubMed] [Google Scholar]
- Rotmistrovsky K., Agarwala R. (2017). BMTagger: Best Match Tagger for Removing Human Reads From Metagenomics Datasets. Available online at: ftp://ftp.ncbi.nlm.nih.gov/pub/agarwala/bmtagger/. (accessed March 9, 2020). [Google Scholar]
- Ryan F. J., Ahern A. M., Fitzgerald R. S., Laserna-Mendieta E. J., Power E. M., Clooney A. G., et al. (2020). Colonic microbiota is associated with inflammation and host epigenomic alterations in inflammatory bowel disease. Nat. Commun. 11:1512. 10.1038/s41467-020-15342-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz Y., Olivares M., Moya-Pérez Á., Agostoni C. (2015). Understanding the role of gut microbiome in metabolic disease risk. Pediatr. Res. 77 236–244. 10.1038/pr.2014.170 [DOI] [PubMed] [Google Scholar]
- Schippa S., Iebba V., Barbato M., Di Nardo G., Totino V., Checchi M. P., et al. (2010). A distinctive “microbial signature” in celiac pediatric patients. BMC Microbiol. 10:175. 10.1186/1471-2180-10-175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P. (2013). The laboratory rat: relating its age with human’s. Int. J. Prev. Med. 4 624–630. [PMC free article] [PubMed] [Google Scholar]
- Sepich-Poore G. D., Zitvogel L., Straussman R., Hasty J., Wargo J. A., Knight R. (2021). The microbiome and human cancer. Science 371:eabc4552. 10.1126/science.abc4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin N.-R., Lee J.-C., Lee H.-Y., Kim M.-S., Whon T. W., Lee M.-S., et al. (2014). An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63 727–735. 10.1136/gutjnl-2012-303839 [DOI] [PubMed] [Google Scholar]
- Sonnenburg E. D., Zheng H., Joglekar P., Higginbottom S. K., Firbank S. J., Bolam D. N., et al. (2010). Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141 1241–1252. 10.1016/j.cell.2010.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S., Ambler G. R., Baur L. A., Garnett S. P., Tepsa M., Yap F., et al. (2006). Randomized, controlled trial of metformin for obesity and insulin resistance in children and adolescents: improvement in body composition and fasting insulin. J. Clin. Endocrinol. Metab. 91 2074–2080. 10.1210/jc.2006-0241 [DOI] [PubMed] [Google Scholar]
- Sui Y., Kong X., Fan R., Ye Y., Mai H., Zhuo S., et al. (2019). Long-term treatment with metformin in the prevention of fatty liver in Zucker diabetic fatty rats. Diabetol. Metab. Syndr. 11:94. 10.1186/s13098-019-0491-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliabue A., Elli M. (2013). The role of gut microbiota in human obesity: recent findings and future perspectives. Nutr. Metab. Cardiovasc. Dis. 23 160–168. 10.1016/j.numecd.2012.09.002 [DOI] [PubMed] [Google Scholar]
- Thomas I., Gregg B. (2017). Metformin; a review of its history and future: from lilac to longevity. Pediatr. Diabetes 18 10–16. 10.1111/pedi.12473 [DOI] [PubMed] [Google Scholar]
- Tremaroli V., Bäckhed F. (2012). Functional interactions between the gut microbiota and host metabolism. Nature 489 242–249. 10.1038/nature11552 [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J., Bäckhed F., Fulton L., Gordon J. I. (2008). Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3 213–223. 10.1016/j.chom.2008.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ridaura V. K., Faith J. J., Rey F. E., Knight R., Gordon J. I. (2009). The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1:6ra14. 10.1126/scitranslmed.3000322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uritskiy G. V., DiRuggiero J., Taylor J. (2018). MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6:158. 10.1186/s40168-018-0541-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usyk M., Pandey A., Hayes R. B., Moran U., Pavlick A., Osman I., et al. (2021). Bacteroides vulgatus and Bacteroides dorei predict immune-related adverse events in immune checkpoint blockade treatment of metastatic melanoma. Genome Med. 13:160. 10.1186/s13073-021-00974-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Baeza Y., Callewaert C., Debelius J., Hyde E., Marotz C., Morton J. T., et al. (2018). Impacts of the human gut microbiome on therapeutics. Annu. Rev. Pharmacol. Toxicol. 58 253–270. 10.1146/annurev-pharmtox-042017-031849 [DOI] [PubMed] [Google Scholar]
- Vázquez-Baeza Y., Pirrung M., Gonzalez A., Knight R. (2013). EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2:16. 10.1186/2047-217X-2-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waidmann M., Bechtold O., Frick J.-S., Lehr H.-A., Schubert S., Dobrindt U., et al. (2003). Bacteroides vulgatus protects against Escherichia coli-induced colitis in gnotobiotic interleukin-2-deficient mice. Gastroenterology 125 162–177. 10.1016/s0016-5085(03)00672-3 [DOI] [PubMed] [Google Scholar]
- Wang B., Yao M., Lv L., Ling Z., Li L. (2017). The human microbiota in health and disease. Proc. Est. Acad. Sci. Eng. 3 71–82. [Google Scholar]
- Wang N., Zhang J., Wu Y., Liu J., Liu L., Guo X. (2016). Metformin improves lipid metabolism disorders through reducing the expression of microsomal triglyceride transfer protein in OLETF rats. Diabetes Res. Clin. Pract. 122 170–178. 10.1016/j.diabres.2016.10.006 [DOI] [PubMed] [Google Scholar]
- Wasimuddin, Schlaeppi K., Ronchi F., Leib S. L., Erb M., Ramette A. (2020). Evaluation of primer pairs for microbiome profiling from soils to humans within the One Health framework. Mol. Ecol. Resour. 20: 1558–1571. 10.1111/1755-0998.13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weersma R. K., Zhernakova A., Fu J. (2020). Interaction between drugs and the gut microbiome. Gut 69 1510–1519. 10.1136/gutjnl-2019-320204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J. A., Karpen S., Vos M. B. (2013). Increasing prevalence of nonalcoholic fatty liver disease among United States adolescents, 1988–1994 to 2007–2010. J. Pediatr. 162 496.e1–500.e1. 10.1016/j.jpeds.2012.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler A. G., Goodman A. L. (2017). An insider’s perspective: Bacteroides as a window into the microbiome. Nat. Microbiol. 2:17026. 10.1038/nmicrobiol.2017.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whang A., Nagpal R., Yadav H. (2019). Bi-directional drug-microbiome interactions of anti-diabetics. EBioMedicine 39 591–602. 10.1016/j.ebiom.2018.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. M., Abrams S. H., Aye T., Lee P. D. K., Lenders C., Lustig R. H., et al. (2010). Metformin extended release treatment of adolescent obesity: a 48-week randomized, double-blind, placebo-controlled trial with 48-week follow-up. Arch. Pediatr. Adolesc. Med. 164 116–123. 10.1001/archpediatrics.2009.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Qian L., Liu K., Wu J., Shan Z. (2021). Gastrointestinal microbiome and gluten in celiac disease. Ann. Med. 53 1797–1805. 10.1080/07853890.2021.1990392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N., Emoto T., Yamashita T., Watanabe H., Hayashi T., Tabata T., et al. (2018). Bacteroides vulgatus and Bacteroides dorei reduce gut microbial lipopolysaccharide production and inhibit atherosclerosis. Circulation 138 2486–2498. 10.1161/CIRCULATIONAHA.118.033714 [DOI] [PubMed] [Google Scholar]
- Zhang F., Ferrero M., Dong N., D’Auria G., Reyes-Prieto M., Herreros-Pomares A., et al. (2021). Analysis of the gut microbiota: an emerging source of biomarkers for immune checkpoint blockade therapy in non-small cell lung cancer. Cancers 13:2514 10.3390/cancers13112514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Feng R., Yang M., Qian C., Wang Z., Liu W., et al. (2019). Effects of metformin, acarbose, and sitagliptin monotherapy on gut microbiota in Zucker diabetic fatty rats. BMJ Open Diabetes Res. Care 7:e000717. 10.1136/bmjdrc-2019-000717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Hu N. (2020). Effects of metformin on the gut microbiota in obesity and type 2 diabetes mellitus. Diabetes Metab. Syndr. Obes. 13 5003–5014. 10.2147/DMSO.S286430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhao Y., Xu J., Xue Z., Zhang M., Pang X., et al. (2015). Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci. Rep. 5:14405 10.1038/srep14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Gu Y., Ren H., Wang S., Zhong H., Zhao X., et al. (2020). Gut microbiome-related effects of berberine and probiotics on type 2 diabetes (the PREMOTE study). Nat. Commun. 11:5015. 10.1038/s41467-020-18414-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M., Zimmermann-Kogadeeva M., Wegmann R., Goodman A. L. (2019). Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 570 462–467. 10.1038/s41586-019-1291-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker L. M. (1972). Fat mobilization in vitro and in vivo in the genetically obese Zucker rat “fatty.” J. Lipid Res. 13 234–243. 10.1016/s0022-2275(20)39417-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overall study design.
Metagenomics workflow.
Songbird differential rankings processed through MetaPhlAn. (A) Same plot as in Figure 1B with all features above and below 0 selected, and their ratios plotted as a box-whisker-plot (right). (B) Same plot as with Figure 2A, with only the top-feature rankings selected (left), and their ratios plotted as a box-whisker-plot (right). (C) Same as Figure 2B, only Bacteroides spp. selected (left) and their ratios plotted as box-whisker plot (right).
Songbird differential rankings of HUMAnN pathways. These are shown in the HUMAnN contributional diversity plot of Figure 5.
Songbird differential rankings processed through HUMAnN. (A) Same plot as in Supplementary Figure 3A with all features above and below 0 selected, and their ratios plotted as a box-whisker-plot (right). (B) Same plot as with Supplementary Figure 3B, with only the top-feature rankings selected (left), and their ratios plotted as a box-whisker-plot (right). (C) Same as Supplementary Figure 3C, only Bacteroides spp. selected (left) and their ratios plotted as box-whisker plot (right).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA770726.