ABSTRACT
Long noncoding RNAs (lncRNAs) widely exist in the cells and play important roles in various biological processes. The role of lncRNAs in immunity remains largely unknown. lncRNA BST2-2 (lncBST2-2) was upregulated upon viral infection and dependent on the interferon (IFN)/JAK/STAT signaling pathway. There was no coding potential found in the lncBST2-2 transcript. Overexpression of lncBST2-2 inhibited the replication of hepatitis C virus (HCV), Newcastle disease virus (NDV), vesicular stomatitis virus (VSV), and herpes simplex virus (HSV), while knockdown of lncBST2-2 facilitated viral replication. Further studies showed that lncBST2-2 promoted the phosphorylation, dimerization, and nuclear transport of IRF3, promoting the production of IFNs. Importantly, lncBST2-2 interacted with the DNA-binding domain of IRF3, which augmented TBK1 and IRF3 interaction, thereby inducing robust production of IFNs. Moreover, lncBST2-2 impaired the interaction between IRF3 and PP2A-RACK1 complex, an essential step for the dephosphorylation of IRF3. These data shown that lncBST2-2 promotes the innate immune response to viral infection through targeting IRF3. Our study reveals the lncRNA involved in the activation of IRF3 and provides a new insight into the role of lncRNA in antiviral innate immunity.
IMPORTANCE Innate immunity is an important part of the human immune system to resist the invasion of foreign pathogens. IRF3 plays a critical role in the innate immune response to viral infection. In this study, we demonstrated that lncBST2-2 plays an important role in innate immunity. Virus-induced lncBST2-2 positively regulates innate immunity by interacting with IRF3 and blocking the dephosphorylation effect of RACK1-PP2A complex on IRF3, thus inhibiting viral infection. Our study provides a new insight into the role of lncBST2-2 in the regulation of IRF3 signaling activation.
KEYWORDS: innate immunity, viral infection, lncBST2-2, interferons, IRF3, PP2A, RACK1
INTRODUCTION
As the first line of host defense, the innate immune system deploys pattern recognition receptors (PRRs) to detect and respond to the presence of pathogens (1). There are four main types of PRRs, cytosolic retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), membrane-bound toll-like receptors (TLRs), nucleotide-binding domain and leucine-rich repeat containing molecules (NLRs), and C-type lectin receptors(CLRs) (2–4). RLRs consist of three RNA helicases, retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2) (1, 2). Once viral RNA is recognized, RIG-I and MDA5 undergo a conformational change, which exposes the N-terminal CARD domains to interact with the CARD domain of the mitochondrial adaptor protein (MAVS) (5). Then MAVS recruits and activates TBK1, subsequently phosphorylating IRF3, causing the translocation of IRF3 into the nucleus (6). Finally, interferons (IFNs) are induced, stimulating antiviral IFN-stimulated genes (ISGs) through the JAK/STAT signaling pathway (7).
With the development of high-throughput sequencing, more than 98% of DNA transcripts are noncoding RNAs (ncRNAs) with very low or no coding ability (8). According to the length of ncRNAs, ncRNAs are divided into small ncRNAs and long ncRNAs (lncRNAs), the latter of which are more than 200 nucleotides (nt) (9). Their modes of actions depend on the specific sequence and natural structure. By forming RNA-RNA, RNA-DNA, and RNA-protein complexes, lncRNAs regulate transcription, splicing, nucleic acid degradation, chromatin modification, and translation (10–12). Recently, numerous studies have found that lncRNAs play an important regulatory role in immunity (13–16). LncMxA binds to IFN-β promoter and inhibits the binding of IRF3 to IFN-β promoter. LncSros1 binds to microRNA miR-1 and induces the degradation of lncSros1, thereby stabilizing Stat1 mRNA (16). Lnczc3h7a promotes RIG-I antiviral innate immune response by targeting TRIM25 (17).
As IRF3 is an essential transcription factor regulating IFN production, IRF3’s expression and activity directly affect the activation of innate immune signals. RAUL, FoxO1, RBCK1, and MID1 promote the degradation of IRF3 to inhibit innate immunity (18–21). MKP5, RACK1, PP2A, and PP1 inhibit the phosphorylation of IRF3, thereby negatively regulating innate immunity (22–24). While there are many reports on host factors regulating the degradation and activity of IRF3, few studies are investigating the effect of lncRNAs on IRF3. Here, we identify that lncRNA BST2-2 (lncBST2-2) (NONHSAT062886; chr19:17419305–17419774), which is involved in viral infection, can promote innate immunity by targeting IRF3. Our data showed that lncBST2-2 promotes the expression of IFNs and ISGs and inhibits the replication of hepatitis C virus (HCV), Newcastle disease virus (NDV), vesicular stomatitis virus (VSV), and herpes simplex virus (HSV). LncBST2-2 interacts with IRF3 and augments the interaction between IRF3 and TBK1. Moreover, lncBST2-2 inhibits the binding of RACK1-PP2A complex to IRF3, thereby promoting the phosphorylation of IRF3. LncBST2-2 is a positive regulator of innate immune responses and plays a regulatory role in resisting the invasion of foreign pathogens.
RESULTS
LncBST2-2 is identified as an lncRNA involved in viral infection.
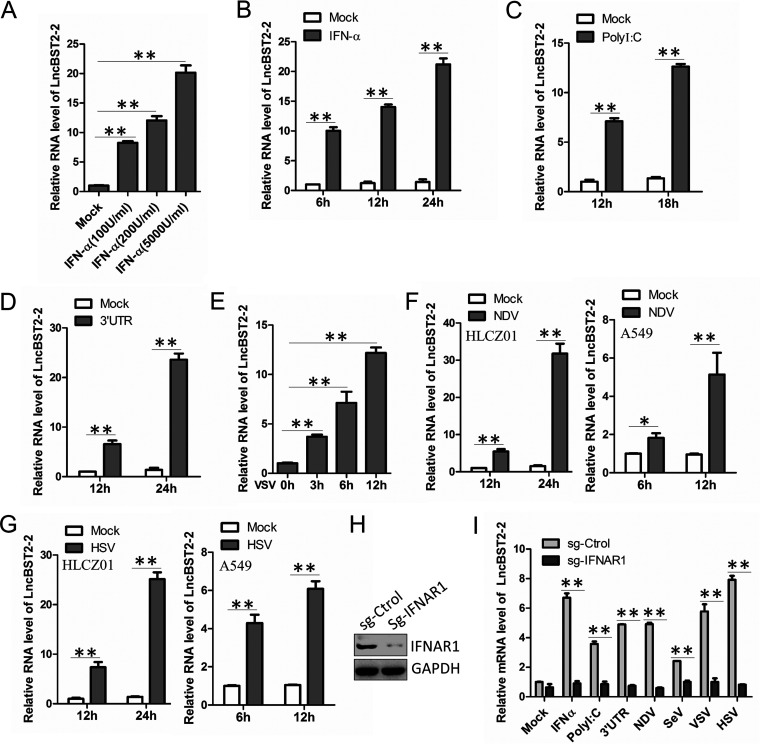
To evaluate the activity of lncBST2-2 during viral infection, we first examined the expression level of lncBST2-2 upon IFN-α stimulation in human hepatocytes. HLCZ01 cells mount an innate immune response to RNA virus infection (25). Reverse transcription-quantitative PCR (qRT-PCR) data showed that lncBST2-2 was upregulated in HLCZ01 cells in both dose- and time-dependent manners (Fig. 1A and B). HCV 3′ untranslated region (UTR) RNA and poly(I·C) can be recognized by RIG-I or MDA5, activating IFN signals (26). HCV 3′ UTR RNA and poly(I·C) also induce the expression of lncBST2-2 (Fig. 1C and D). These results showed that lncBST2-2 can be induced by IFN or PRR agonists. Under viral infection, the host will have an obvious IFN response. To investigate the association between lncBST2-2 and viral infection, we infected HLCZ01 cells with VSV, NDV, or HSV and detected the expression level of lncBST2-2. As shown in Fig. 1E to G, VSV, NDV, and HSV infection upregulated the expression of lncBST2-2. To rule out cell specificity, we infected A549 cells with NDV or HSV and found that the expression of lncBST2-2 was also upregulated in A549 cells upon NDV or HSV infection (Fig. 1F and G). Viral infection induces the expression of IFNs. Then, IFNs are recognized by the IFN receptors and activate the JAK-STAT signaling pathway to induce the expression of ISGs. To determine whether the induction of LncBST2-2 depends on the JAK-STAT signaling pathway, we tested the expression of LncBST2-2 in IFNAR1-knockdown HLCZ01 cells. The silenced effect was shown in Fig. 1H. As expected, knockdown of IFNAR1 attenuated the induction of lncBST2-2 by IFN-α, poly(I·C), HCV 3′ UTR RNA, NDV, Sendai virus (SeV), VSV, or HSV (Fig. 1I). These data supported that viral infection induces lncBST2-2 and that the induction of lncBST2-2 depends on the JAK/STAT signaling pathway activated by IFN.
FIG 1.
LncBST2-2 is identified as an lncRNA involved in viral infection. (A) HLCZ01 cells were treated with different doses of IFN-α for 6 h. LncBST2-2 level was analyzed by qRT-PCR. (B) HLCZ01 cells were treated with IFN-α (200 U/mL) for indicated time points. qRT-PCR was performed to determine the lncBST2-2 level. (C and D) HLCZ01 cells were transfected with high-molecular-weight (HMW) poly(I·C) (C) or HCV 3′ UTR (D) for indicated time points. LncBST2-2 level was analyzed by qRT-PCR. (E) HLCZ01 cells were infected with VSV for indicated time points. qRT-PCR was performed to determine the lncBST2-2 level. (F and G) HLCZ01 cells or A549 cells were infected with NDV (multiplicity of infection [MOI] = 0.1) (F) or HSV (MOI = 0.5) (G) for indicated time points. LncBST2-2 level was analyzed by qRT-PCR. (H) The protein level of IFNAR1 in HLCZ01 cells or IFNAR1-knockdown HLCZ01 cells was assayed by Western blotting. (I) IFNAR1-knockdown HLCZ01 cells or control cells were treated with IFN-α (500 U/mL) for 6 h; transfected with HMW poly(I·C) (500 ng) or HCV 3′ UTR RNA (500 ng) for 16 h; or treated with NDV (MOI = 0.1), SeV (MOI = 0.1), VSV (MOI = 0.1), or HSV (MOI = 0.5) for 9 h. LncBST2-2 level was analyzed by qRT-PCR. Experiments were independently repeated two or three times, with similar results. The results are presented as means ± standard deviations. *, P ≤ 0.05; **, P ≤ 0.01 versus control.
LncBST2-2 inhibits viral replication.
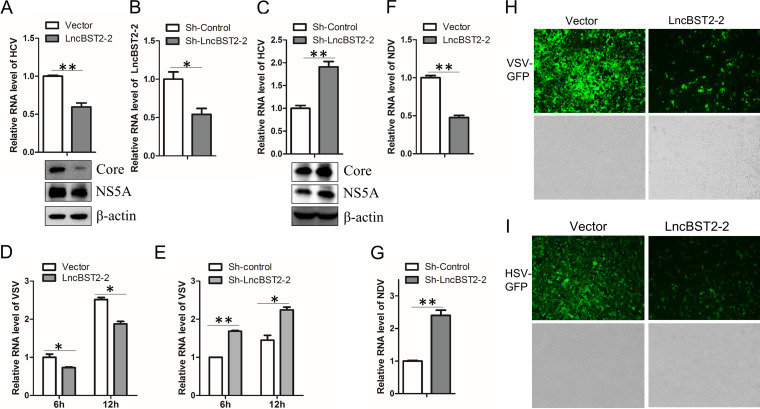
BST2 is recognized as an antiviral gene that shares a bidirectional promoter with lncBST2-2 (27, 28). Given that the above data showed that viral infection induces lncBST2-2, we hypothesized that lncBST2-2 might have antiviral function. Overexpression of lncBST2-2 decreased the level of intracellular HCV RNA and viral core and NS5A protein (Fig. 2A). To determine the effect of endogenous lncBST2-2 on HCV replication, we knocked down lncBST2-2 in HLCZ01 cells (Fig. 2B). Silencing of lncBST2-2 increased the level of HCV RNA and viral protein (Fig. 2C). To further verify whether lncBST2-2 has broad-spectrum antiviral function, we included NDV, VSV, and HSV in our study. Overexpression or silencing of lncBST2-2 inhibited or enhanced the replication of VSV (Fig. 2D and E) or NDV (Fig. 2F and G), respectively. Fluorescence staining results further confirmed the antiviral role of lncBST2-2 in green fluorescent protein (GFP)-VSV or GFP-HSV infection (Fig. 2H and I). These data demonstrated that lncBST2-2 inhibits viral replication.
FIG 2.
LncBST2-2 inhibits viral replication. (A) HLCZ01 cells were infected with HCV for 12 h before lncBST2-2 plasmid transfection for 48 h. HCV RNA was examined by qRT-PCR (top), and viral proteins were measured by Western blotting (bottom). (B) The knockdown effect of shRNA plasmid targeting lncBST2-2 was examined by qRT-PCR. (C) HLCZ01 cells stably expressing shRNA plasmid targeting lncBST2-2 were infected with HCV for 72 h. HCV RNA was examined by qRT-PCR (top), and viral proteins were measured by Western blotting (bottom). (D) HLCZ01 cells were transfected with pcDNA3.1a-lncBST2-2 for 48 h and infected with VSV for indicated time points. VSV RNA level was analyzed by qRT-PCR. (E) HLCZ01 cells were transfected with sh-lncBST2-2 for 48 h and infected with VSV for indicated time points. VSV RNA level was analyzed by qRT-PCR. (F) Huh7 cells infected with NDV for 24 h after overexpression of lncBST2-2. NDV RNA level was analyzed by qRT-PCR. (G) Huh7 cells infected with NDV for 24 h after transfection of sh-lncBST2-2 plasmid for 24 h. qRT-PCR was performed to determine NDV RNA level. (H and I) Microscopy imaging of lncBST2-2-overexpressing A549 cells infected with VSV-GFP (MOI = 0.01) (H) or HSV-GFP (MOI = 0.01) (I) for 12 h. Experiments were independently repeated two or three times, with similar results. The results are presented as means ± standard deviations. *, P ≤ 0.05; **, P ≤ 0.01 versus control.
LncBST2-2 promotes virus-induced innate immune response.
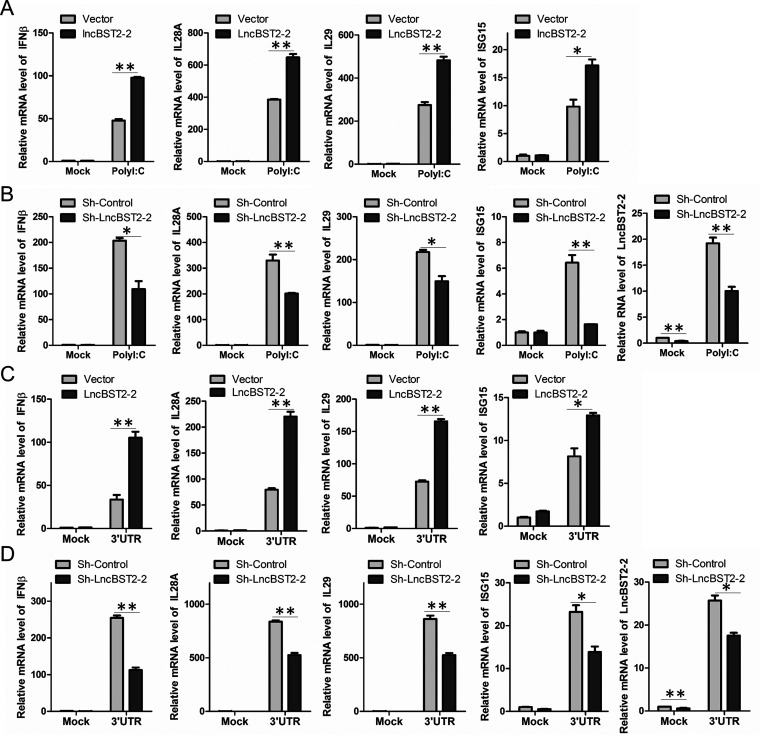
The innate immune response is critical for the host to resist viral invasion. Since our above data showed that viral infection induces lncBST2-2 expression and lncBST2-2 inhibits viral infection, we assumed that lncBST2-2 might play an important role in the innate immune response to viral infection. To test the hypothesis, we first explored the effect of lncBST2-2 on cytokine expression triggered by RNA nucleic acid mimics in HLCZ01 cells. Overexpression of lncBST2-2 enhanced poly(I·C) and HCV 3′ UTR RNA-triggered expression of IFN-β, interleukin-28A (IL-28A), IL-29, and ISG15 (Fig. 3A and C). Silencing of lncBST2-2 in HLCZ01 cells suppressed the expression of IFN-β, IL-28A, IL-29, and ISG15 induced by poly(I·C) and HCV 3′ UTR RNA (Fig. 3B and D). These data suggested that lncBST2-2 promotes the innate immune response to RNA nucleic acid mimics.
FIG 3.
LncBST2-2 positively regulates innate immune response to RNA nucleic acid mimics. (A and C) HLCZ01 cells were transfected with the lncBST2-2 plasmid for 40 h and then were transfected with 500 ng poly(I·C) (A) or 500 ng 3′ UTR (C) for 9 h. The levels of IFN-β, IL-28A, IL-29, and ISG15 mRNA were examined by qRT-PCR. (B and D) LncBST2-2-silenced HLCZ01 cells were transfected with 500 ng poly(I·C) (B) or 500 ng HCV 3′ UTR RNA (D) for 9 h. IFN-β, IL-28A, IL-29, ISG15, and lncBST2-2 levels were examined by qRT-PCR. Experiments were independently repeated two or three times, with similar results. The results are presented as means ± standard deviations. *, P ≤ 0.05; **, P ≤ 0.01 versus control.
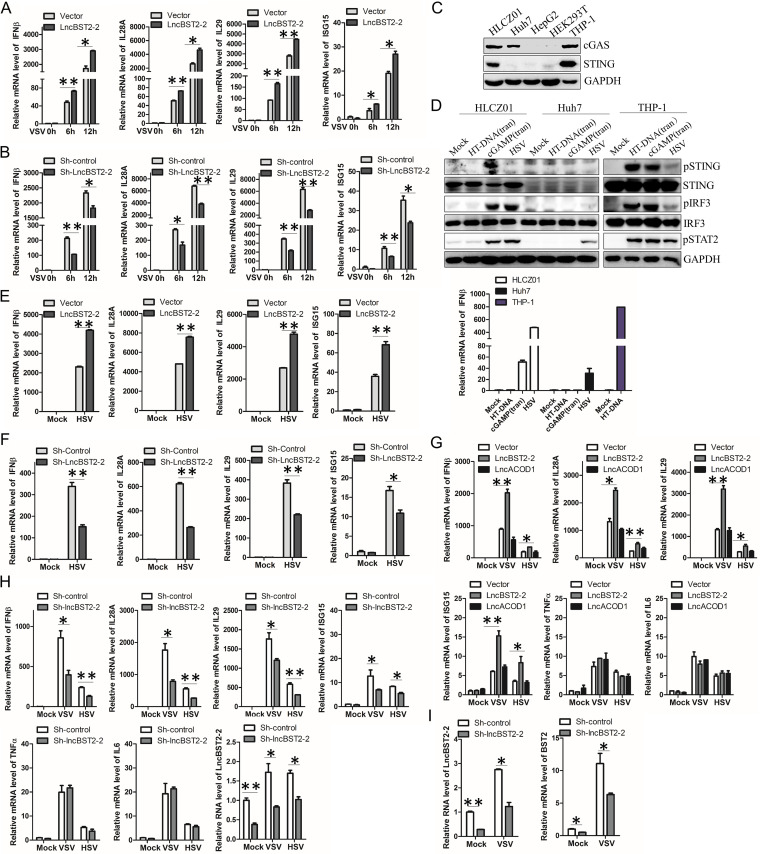
Viral infection leads to the activation of the host’s innate immune response. To determine the role of lncBST2-2 in viral infection, we infected HLCZ01 cells with an RNA virus, VSV. Ectopic expression of lncBST2-2 enhanced VSV-induced expression of IFN-β, IL-28A, IL-29, and ISG15 (Fig. 4A). Knockdown of lncBST2-2 attenuated the expression of IFNs and ISG triggered by VSV infection (Fig. 4B). To determine whether lncBST2-2 has an effect on DNA virus-induced IFN responses, we examined the expression of cGAS and STING in HLCZ01 cells. Both cGAS and STING were expressed in HLCZ01 cells (Fig. 4C). To test whether cGAS and/or STING is functional in HLCZ01 cells, we treated the cells with herringtestis DNA(HT-DNA), cGAMP, or HSV. cGAMP activated STING and IRF3 in the cells, indicating that STING is functional in HLCZ01 cells. To our surprise, HT-DNA could not activate STING in the cells (Fig. 4D). The activated IRF3 and STAT2 were triggered by HSV infection in HLCZ01 cells. Consistently, HSV induced the expression of IFN-β in the cells (Fig. 4D). Overexpression of lncBST2-2 enhanced the expression of IFN-β, IL-28A, IL-29, and ISG15 triggered by HSV in HLCZ01 cells (Fig. 4E). Knockdown of lncBST2-2 suppressed HSV-induced cytokine expression (Fig. 4F). And similar results were observed in VSV- or HSV-infected A549 cells. Overexpression or silencing of lncBST2-2 augmented or impaired the expression of IFN-β, IL-28A, IL-29, and ISG15 induced by VSV or HSV infection, respectively, while lncBST2-2 had no effect on the expression of tumor necrosis factor alpha (TNF-α) and IL-6 (Fig. 4G and H). These data indicated that lncBST2-2 may not affect NF-κB signaling. Previous studies showed that another long noncoding RNA BST2 (BISPR) (NONHSAT062871; chr19:17405694–17415736), which is also adjacent to the gene BST2, can regulate the expression of BST2 (29). Similarly, silencing of lncBST2-2 suppressed BST2 expression (Fig. 4I). These data suggested that lncBST2-2 promotes the innate immune response to viral infection.
FIG 4.
LncBST2-2 promotes virus-induced innate immune response. (A) HLCZ01 cells were transfected with the lncBST2-2 plasmid for 40 h and then infected with VSV (MOI = 0.01) for 6 h and 12 h. The level of IFN-β, IL-28A, IL-29, and ISG15 mRNA was examined by qRT-PCR. (B) LncBST2-2-silenced HLCZ01 cells were infected with VSV (MOI = 0.01) for 6 h and 12 h. IFN-β, IL-28A, IL-29, ISG15, were examined by qRT-PCR. (C) The expression of cGAS and STING in different hepatocytes. (D) HLCZ01, Huh7, and THP-1 cells were treated with 4 μg/mL of HT-DNA (transfection), 4 μg/mL of cGAMP (transfection), or HSV (MOI = 0.5) for 12 h. Western blotting was performed with antibodies against the indicated proteins (upper panel), and the mRNA level of IFN-β was measured by qPCR (lower panel). (E) HLCZ01 cells were transfected with the lncBST2-2 plasmid for 40 h and then infected with HSV (MOI = 0.5) for 12 h. The level of IFN-β, IL-28A, IL-29, and ISG15 mRNA was examined by qRT-PCR. (F) LncBST2-2-silenced HLCZ01 cells were infected with HSV (MOI = 0.5) for 12 h. IFN-β, IL-28A, IL-29, ISG15 were examined by qRT-PCR. (G) A549 cells were transfected with the lncBST2-2 plasmid for 40 h and then infected with VSV (MOI = 0.01) or HSV (MOI = 0.05) for 6 h. The mRNA level of IFN-β, IL-28A, IL-29, ISG15, TNF-α, and IL-6 was examined by qRT-PCR. lncACOD1 was used as a native control. (H) A549 cells were transfected with sh-lncBST2-2 plasmid for 40 h and then infected with VSV (MOI = 0.01) or HSV (MOI = 0.05) for 6 h. The mRNA level of IFN-β, IL-28A, IL-29, ISG15, TNF-α, IL-6, and lncBST2-2 was examined by qRT-PCR. (I) HLCZ01 cells were transfected with sh-lncBST2-2 plasmid for 40 h and then infected with VSV (MOI = 0.01) for 12 h. The mRNA level of lncBST2-2 and BST2 was examined by qRT-PCR. Experiments were independently repeated two or three times, with similar results. The results are presented as means ± standard deviations. *, P ≤ 0.05; **, P ≤ 0.01 versus control.
Identification of LncBST2-2 as a noncoding transcript.
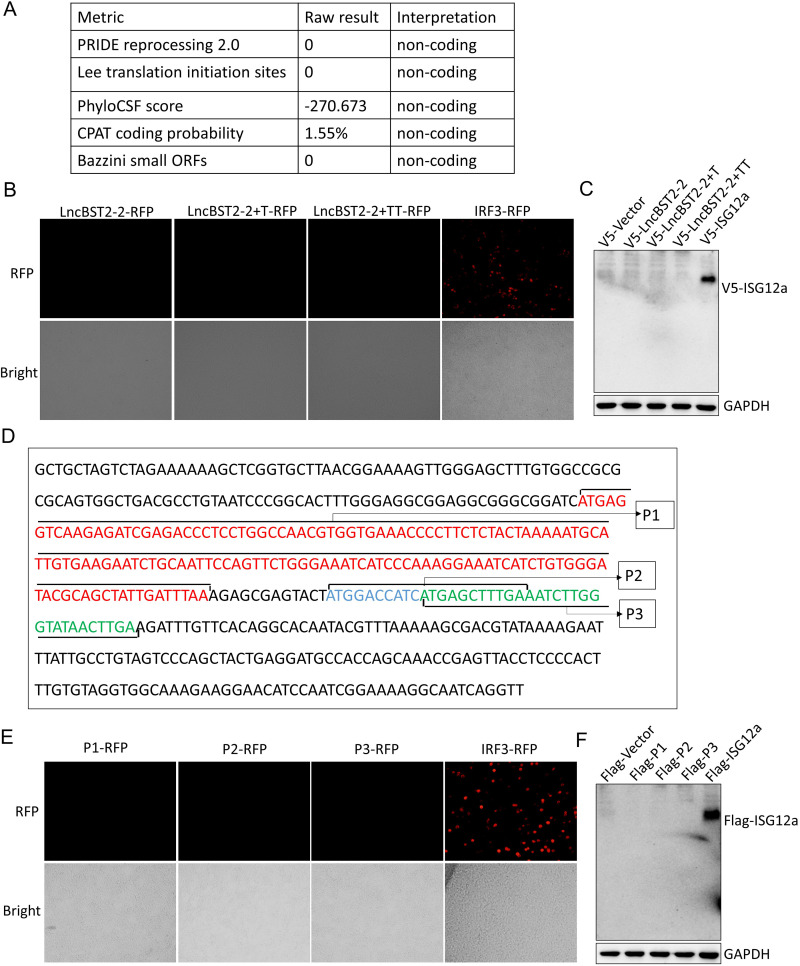
Although lncRNAs are generally non-protein-coding transcripts, recent studies have shown that some noncoding RNAs have coding capabilities (30–32). To investigate whether there is an lncBST2-2-coding peptide, we obtained the coding potential scores by different coding potential calculators, and all of them indicated that lncBST2-2 cannot encode peptides (Fig. 5A) (33). To further confirm the noncoding ability of lncBST2-2, we added T or TT to the end of lncBST2-2 transcript and cloned the constructs into pcDNA3.1a vector and pRed-max-N1 vector. If the transcript of lncRNA is encoded, the corresponding tag protein can be expressed. Consistent with the calculation, no red fluorescence was observed in HEK293T cells transfected with lncBST2-2-RFP (red fluorescent protein), lncBST2-2T-RFP, or lncBST2-2TT-RFP plasmids and no band was detected, while the expressions of positive controls, IRF3-RFP and V5-ISG12a, were clearly detectable (Fig. 5B and C). A previous study shows that lncRNAs may contain complete open reading frames (ORFs), and some ORFs have coding functions (30). The complete ORF has a termination codon. To exclude the premature termination of translation and the inability to express the tag protein, we checked the sequence of lncBST2-2 for possible ORFs. Interestingly, we found three possible ORFs in lncBST2-2, which have the initiation codon sequence, termination codon sequence, and the number of nucleobases in multiples of three (Fig. 5D). To identify the coding ability of these ORFs (P1, P2, P3), we cloned the corresponding sequence into pcDNA3.1a vector and pRed-max-N1 vector. Still, no red fluorescence or band was observed (Fig. 5E and F). All the data supported that lncBST2-2 is a noncoding transcript.
FIG 5.
LncBST2-2 is identified as a noncoding transcript. (A) The coding potential score of LncBST2-2 was obtained from the coding potential calculators. (B) HEK293T cells were transfected with lncBST2-2-RFP, lncBST2-2+T-RFP, lncBST2-2+TT-RFP, or IRF3-RFP plasmids for 48 h. The RFP-IRF3 plasmid served as a positive control. Images were taken under a fluorescence microscope. (C) HEK293T cells were transfected with the pcDNA3.1a vector, pcDNA3.1a-lncBST2-2, pcDNA3.1a-lncBST2-2+T, pcDNA3.1a-lncBST2-2+TT, or pcDNA3.1a-ISG12a for 48 h, followed by Western blotting. (D) Full sequence of LncBST2-2 and small open reading frames of LncBST2-2, P1, P2, and P3. (E) HEK293T cells were transfected with P1-RFP, P2-RFP, P3-RFP, or IRF3-RFP plasmids for 48 h. The RFP-IRF3 plasmid served as a positive control. Images were taken under a fluorescence microscope. (F) HEK293T cells were transfected with the p3×Flag-CMV vector, p3×Flag-CMV-P1, p3×Flag-CMV-P2, p3×Flag-CMV-P3, or p3×Flag-CMV-ISG12a for 48 h, followed by Western blotting.
LncBST2-2 promotes the activation of IRF3 signaling but not NF-κB signaling.
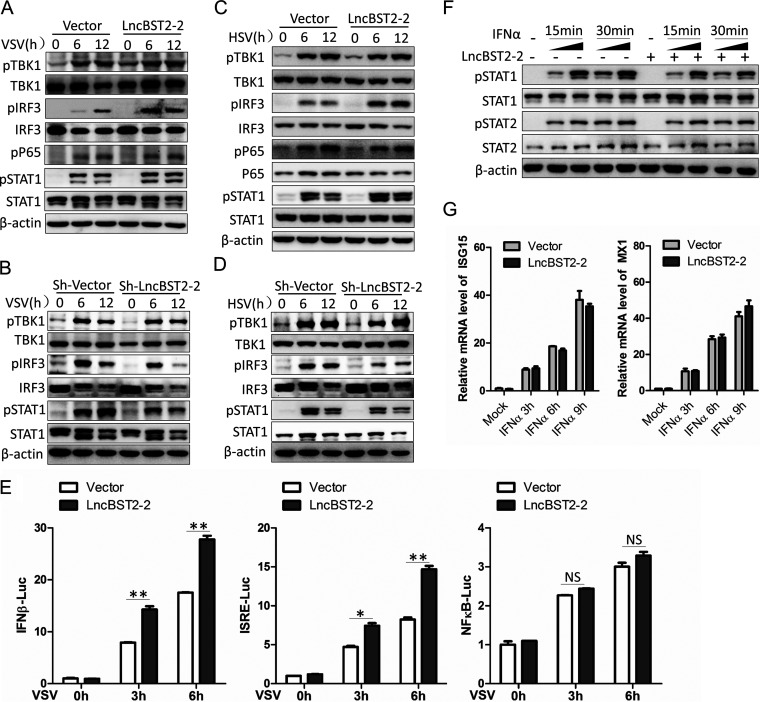
The above data showed that lncBST2-2 promotes the expression of IFNs triggered by viral infection. Upon viral infection, two major pathways responsible for inducing antiviral genes are activated. One is the activation of NF-κB signaling pathway, which phosphorylates P65 to activate downstream signals (34). The other is the activation of TBK1-IRF3 signal axis. TBK1 phosphorylates IRF3 to induce the expression of IFNs. We speculated that lncBST2-2 might affect the activation of both IRF3 signal and NF-κB signal. Overexpression of lncBST2-2 indeed augmented the phosphorylation of IRF3 and downstream phosphorylated STAT1 triggered by VSV or HSV, but not P65 phosphorylation (Fig. 6A and C). Correspondingly, silencing of lncBST2-2 impaired the phosphorylation of IRF3 and STAT1 triggered by VSV or HSV (Fig. 6B and D). Luciferase assay demonstrated that lncBST2-2 augmented the activation of IFN-β and IFN-sensitive response element (ISRE) promoters induced by VSV while it did not affect the NF-κB promoter activation (Fig. 6E). Considering the role of lncBST2-2 in the induction of IFNs by viral infection, we treated A549 cells with IFN-α to determine whether lncBST2-2 has a direct effect on the activation of JAK/STAT signal by IFN. As shown in Fig. 6F and G, overexpression of lncBST2-2 did not affect the phosphorylation of STAT1 and STAT2 triggered by IFN-α. Quantitative PCR (qPCR) analysis showed that lncBST2-2 did not change the expression of ISG15 and Mx1 induced by IFN-α. All the data supported that lncBST2-2 promotes the activation of IRF3 signaling but not NF-κB signaling upon viral infection.
FIG 6.
LncBST2-2 promotes the activation of IRF3 signaling but not NF-κB signaling. (A and B) HLCZ01 cells were transfected with pcDNA3.1a-lncBST2-2 (A) or shRNA that targets lncBST2-2 (B) for 40 h and then infected with VSV (MOI = 0.01) for the indicated times. Immunoblot assays were performed with the indicated antibodies. (C and D) A549 cells were transfected with pcDNA3.1a-LncBST2-2 (C) or shRNA that targets lncBST2-2 (D) for 40 h and then infected with HSV (MOI = 0.05) for the indicated times. Immunoblot assays were performed with the indicated antibodies. (E) Luciferase activity of lysates of HEK293T cells cotransfected with IFN-β-Luc, ISRE-Luc, or NFKB-Luc and pcDNA3.1a vector or pcDNA3.1a-LncBST2-2 for 16 h followed by VSV infection for 8 h. (F) A549 cells were transfected with pcDNA3.1a-LncBST2-2 for 40 h and then treated with IFN-α (100 U/mL [lane 2,4,7,9] and 200 U/mL [lane 3,5,8,10]) for the indicated time points. Immunoblot assays were performed with the indicated antibodies. (G) A549 cells were transfected with pcDNA3.1a-LncBST2-2 for 40 h and then treated with IFN-α (100 U/mL) for the indicated times. ISG15 and MX1 were examined by qRT-PCR. Experiments were independently repeated two or three times, with similar results. The results are presented as means ± standard deviations. *, P ≤ 0.05; **, P ≤ 0.01 versus control; NS, not significant.
LncBST2-2 targets IRF3 and triggers the nucleus translocation of IRF3.
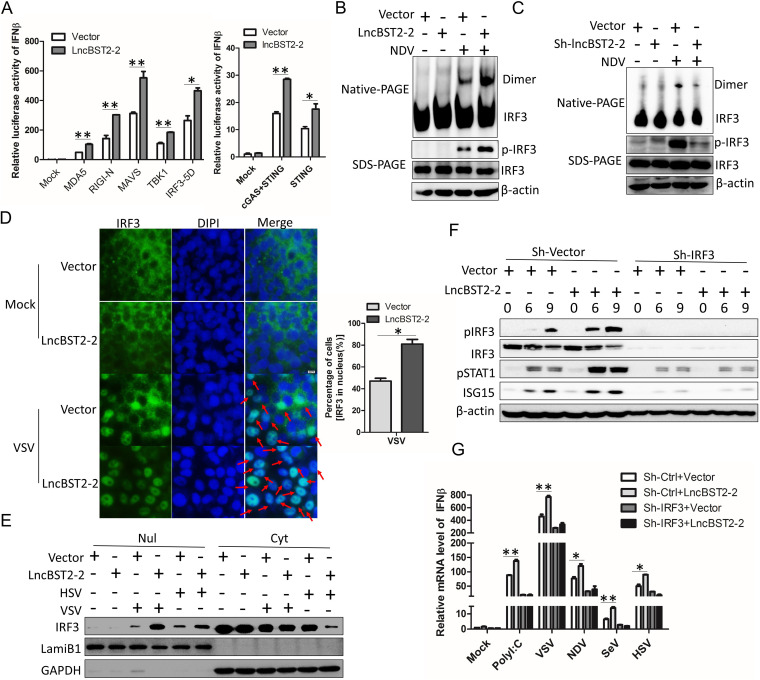
To explore which component(s) of IFN signaling pathway can be targeted by lncBST2-2, we cotransfected lncBST2-2 with various candidates of IFN signaling pathway in a reporter assay system. Overexpression of lncBST2-2 strengthened the activation of IFN-β promoter triggered by IRF3-5D and its upstream molecules, cGAS, STING, N-RIG-I, MDA5, MAVS, or TBK1 (Fig. 7A). Moreover, lncBST2-2 augmented the phosphorylation of IRF3 but not the phosphorylation of TBK1 triggered by VSV or HSV (Fig. 6A to D). These data indicated that lncBST2-2 might target IRF3. IRF3 is an essential transcription factor in IFN signaling pathway. Upon activation, IRF3 undergoes C-terminal phosphorylation and dimerization and translocates to the nucleus (6). Therefore, we adopted SDS-PAGE and native PAGE to detect the status of IRF3 phosphorylation and dimerization, respectively. HLCZ01 cells were transfected with pcDNA3.1a-LncBST2-2 and then infected by NDV. Overexpression of lncBST2-2 augmented the phosphorylation and dimerization of IRF3 (Fig. 7B). Oppositely, lncBST2-2 knockdown impaired the phosphorylation and dimerization of IRF3 (Fig. 7C). The entry of IRF3 into the nucleus is a key step in the activation of IFN signals. Here, immunofluorescence assays and nucleus and cytoplasm separation analysis results showed that lncBST2-2 promoted the nucleus translocation of IRF3 upon VSV or HSV infection (Fig. 7D and E). To further confirm that lncBST2-2 targets IRF3, we detected the activation of IFN pathway in IRF3-silenced HLCZ01 cells. Overexpression of lncBST2-2 failed to activate the phosphorylation of IRF3 and STAT1 and induce ISG expression in IRF3-silenced cells (Fig. 7F). Furthermore, knockdown of IRF3 abolished the production of IFN-β triggered by poly(I·C), VSV, NDV, SeV, or HSV (Fig. 7G). All these data suggested that lncBST2-2 targets IRF3 and promotes the nucleus translocation of IRF3.
FIG 7.
LncBST2-2 targets IRF3 and promotes the nucleus translocation of IRF3. (A) HEK293T cells were cotransfected with lncBST2-2, IFN-β luciferase, and pRL-CMV plasmids and cGAS, STING, MDA5, N-RIG-I, MAVS, TBK1, and IRF3-5D plasmids for 24 h, respectively. IFN-β luciferase assay was performed. (B) HLCZ01 cells were transfected with pcDNA3.1a-lncBST2-2 for 48 h and then infected with NDV (MOI = 0.1) for 24 h. Immunoblot assays were performed with the indicated antibodies. (C) HLCZ01 cells were transfected with shRNA that targets lncBST2-2 for 48 h and then infected with NDV (MOI = 0. 1) for 24 h. Immunoblot assays were performed with the indicated antibodies. (D) A549 cells were transfected with pcDNA3.1a-lncBST2-2 for 42 h and then infected with VSV (MOI = 0.05) for 6 h. The subcellular localization of endogenous IRF3 was analyzed by fluorescence microscopy. (E) A549 cells were transfected with pcDNA3.1a-lncBST2-2 for 42 h and then infected with VSV (MOI = 0.05) or HSV (MOI = 0.1) for 6 h, and nuclear (nul) and cytoplasmic (cyt) portions were extracted for immunoblot analysis. (F) IRF3 stable knockdown HLCZ01 cells were transfected with pcDNA3.1a-lncBST2-2 or an empty vector. The cells were infected with VSV (MOI = 0.05) for indicated times. Immunoblot assays were performed with the indicated antibodies. (G) IRF3 stable knockdown HLCZ01 cells were transfected with pcDNA3.1a-lncBST2-2 or an empty vector. The cells were treated with poly(I·C) or infected with VSV, NDV, SeV, and HSV for 9 h. The level of IFN-β was examined by real-time PCR. Experiments were independently repeated two or three times, with similar results. The results are presented as means ± standard deviations. *, P ≤ 0.05; **, P ≤ 0.01 versus control.
LncBST2-2 interacts with IRF3 and enhances the interaction between TBK1 and IRF3.
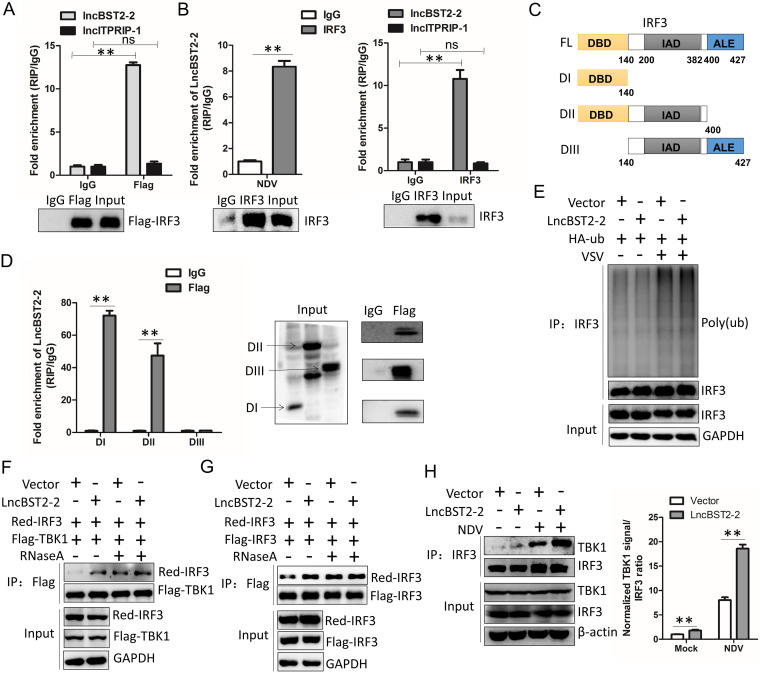
lncRNAs may function through forming RNA-protein complex (34). We speculate that lncBST2-2 may affect the activation of IRF3 by interacting with IRF3. To prove this speculation, we delivered pcDNA3.1a-LncBST2-2 or pcDNA3.1a-lncITPRIP-1 and pFlag-IRF3 into HEK293T cells. Then, RNA-immunoprecipitation (RIP) analysis of the interaction between lncBST2-2 and Flag-IRF3 was performed. LncITPRIP-1 served as a negative control. As expected, Flag-IRF3 interacted with lncBST2-2, but not lncITPRIP-1 (Fig. 8A). Moreover, lncBST2-2 also interacted with endogenous IRF3 under NDV infection (Fig. 8B). IRF3 has three main structures, the DNA-binding domain (DBD), the IRF association domain (IAD), and the autoinhibition element (AIE) domain. To explore which domain(s) of IRF3 interact(s) with lncBST2-2, we cloned three IRF3 truncations. DI contains the DNA-binding domain (DBD). DII contains the DBD domain and the IRF association domain (IAD). DIII contains the IAD domain and the autoinhibition element (AIE) (Fig. 8C). RIP assays showed that DI, the DNA-binding domain of IRF3, played a key role in the association between IRF3 and lncBST2-2 (Fig. 8D). These data suggested that lncBST2-2 interacts with IRF3.
FIG 8.
LncBST2-2 interacts with IRF3 and promotes the interaction between TBK1 and IRF3. (A) HEK293T cells were cotransfected with pcDNA3.1a-lncBST2-2, pcDNA3.1a-lncITPRIP-1, and Flag-IRF3 for 48 h, and then an RIP experiment was performed. lncITPRIP-1 was used as a native control. (B) Huh7 cells were transfected with pcDNA3.1a-lncBST2-2 for 24 h and then were infected with NDV (MOI = 0.1) for 24 h, and then an RIP experiment was performed with anti-IRF3 (left). HLCZ01 cells were infected with VSV (MOI = 0.05) for 12 h, and then an RIP experiment was performed with anti-IRF3. lncITPRIP-1 was used as a native control (right). (C) Schematic illustration of IRF3 truncations. (D) HEK293T cells were cotransfected with lncBST2-2 plasmid and truncations of IRF3 plasmid for 48 h, and then an RIP experiment was performed. (E) HEK293T cells were cotransfected with pcDNA3.1a-LncBST2-2 or HA-Ub for 40 h and then infected with VSV (MOI = 0.05) for 6 h. Ubiquitination and immunoblotting assays were performed with the indicated antibodies. (F) HEK293T cells were cotransfected with pcDNA3.1a-lncBST2-2, Red-IRF3, and Flag-TBK1 for 48 h. Proteins were lysed by IP lysis buffer and treated with or without RNase A (100 μg/mL). IP assays were performed. (G) HEK293T cells were cotransfected with pcDNA3.1a-lncBST2-2, Red-IRF3, and Flag-IRF3 for 48 h. Proteins were lysed by IP lysis buffer and treated with or without RNase A (100 μg/mL). IP assays were performed. (H) HLCZ01 cells were transfected with lncBST2-2 for 24 h and then infected with NDV for 30 h, and immunoprecipitation assays were performed. Experiments were independently repeated two or three times, with similar results. The results are presented as means ± standard deviations. *, P ≤ 0.05; **, P ≤ 0.01 versus control; ns, not significant.
Ubiquitination plays an essential role in the activation of IRF3 (35). Next, we investigated whether lncBST2-2 affects the ubiquitination of IRF3. LncBST2-2 did not affect the ubiquitination of IRF3 in resting state or upon VSV infection (Fig. 8E). The activation of IRF3 depends on the phosphorylation of IRF3 by TBK1, in which the interaction between TBK1 and IRF3 plays a critical role. IP experiments showed that lncBST2-2 enhanced the interaction between Flag-TBK1 and Red-IRF3 (Fig. 8F). The interaction between endogenous IRF3 and TBK1 triggered by NDV was also strengthened by lncBST2-2 (Fig. 8H). Interestingly, the effect of lncBST2-2 on the interaction of TBK1 with IRF3 disappeared after RNase A treatment of the protein lysates. However, the interaction of TBK1 with IRF3 was enhanced under RNase A treatment. A similar phenomenon was observed for the interaction of Red-IRF3 with Flag-IRF3 (Fig. 8F and G). Previous studies have shown that RNase L can cleave RNA into small fragments of RNA, making it better recognized by RNA PRRs such as RIG-I and activating IFN signaling (36). After RNase A treatment, the RNA in the lysates was cleaved and then recognized by RNA PRRs, which promoted the interaction between TBK1 and IRF3, and IRF3 dimer formation. These data suggested that lncBST2-2 promotes the innate immune response by enhancing the interaction between TBK1 and IRF3.
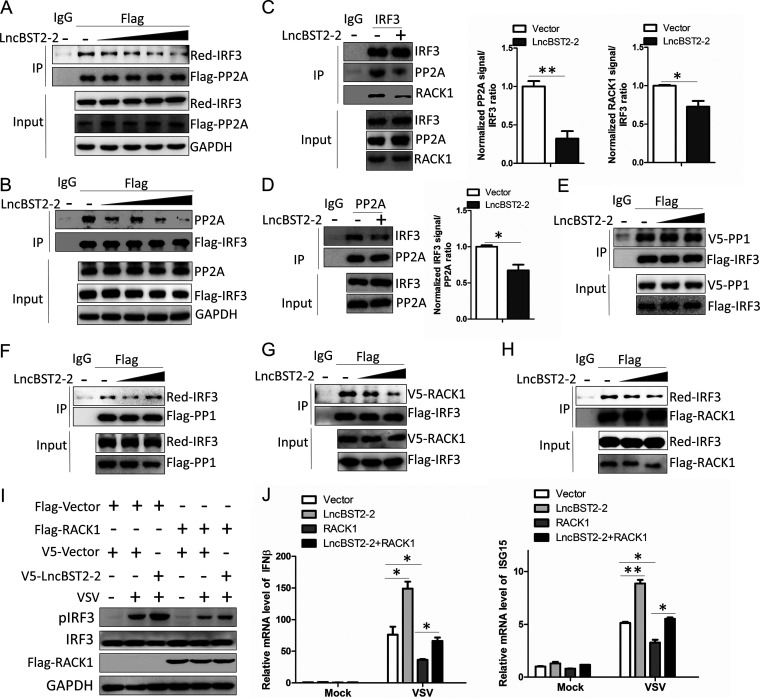
LncBST2-2 inhibits the interaction between IRF3 and RACK1, thereby inhibiting the recruitment of PP2A by IRF3.
PP2A and PP1 are important phosphatases that affect the phosphorylation of IRF3 (23, 24). PP2A or PP1 interacts with IRF3 and then dephosphorylates IRF3, inhibiting IRF3 signaling activation. Then, we explored whether lncBST2-2 is involved in the interaction between IRF3 and its phosphatases. We first examined whether lncBST2-2 affects the interaction between IRF3 and PP2A. As shown in Fig. 9A and B, lncBST2-2 impaired the interaction between exogenous IRF3 and PP2A in a dose-dependent manner in HEK293T cells. LncBST2-2 also inhibited the interaction between endogenous IRF3 and PP2A (Fig. 9C and D). However, lncBST2-2 had little effect on the interaction between IRF3 and PP1 (Fig. 9E and F). These data indicated that lncBST2-2 may specifically inhibit the effect of the phosphatase PP2A on IRF3. Previous studies have shown that RACK1 mediates the formation of the IRF3-RACK1-PP2A complex. Knocking down RACK1 impairs the interaction between IRF3 and PP2A, promoting the activation of IRF3 (24). We speculated that lncBST2-2 might inhibit the recruitment of PP2A by IRF3 through impairing the interaction between RACK1 and IRF3. Consistent with our speculation, lncBST2-2 did inhibit the interaction between IRF3 and RACK1 (Fig. 9C, G, and H). Moreover, overexpression of lncBST2-2 reversed the inhibitory effect of RACK1 on IRF3 phosphorylation, IFN production, and ISG expression triggered by VSV (Fig. 9I and J). Taken together, our data reveal that lncBST2-2 blocks the formation of IRF3-RACK1-PP2A complex, promotes the activation of IRF3, and thereby enhances the innate immune response to viral infection.
FIG 9.
LncBST2-2 inhibits the interaction between IRF3 and RACK1, thereby inhibiting the recruitment of PP2A by IRF3. (A) HEK293T cells were cotransfected with Flag-PP2A, Red-IRF3, and a gradient concentration of pcDNA3.1a-lncBST2-2 for 48 h. IP and immunoblot assays were performed with the indicated antibodies. (B) HEK293T cells were cotransfected with Flag-IRF3 and a gradient concentration of pcDNA3.1a-lncBST2-2 for 48 h. IP and immunoblot assays were performed with the indicated antibodies. (C and D) HLCZ01 cells were transfected with pcDNA3.1a-lncBST2-2 for 48 h. Immunoprecipitation was performed with anti-IRF3 (C) or anti-PP2A (D), and immunoblot assays were performed with the indicated antibodies. (E and F) HEK293T cells were cotransfected with pcDNA3.1a-lncBST2-2 and Flag-IRF3 and V5-PP1 (E) or Flag-PP1 and Red-IRF3 (F) for 48 h. IP and immunoblot assays were performed with the indicated antibodies. (G and H) HEK293T cells were cotransfected with pcDNA3.1a-lncBST2-2 and Flag-IRF3 and V5-RACK1 (G) or Flag-RACK1 and Red-IRF3 (H) for 48 h. IP and immunoblot assays were performed with the indicated antibodies. (I and J) HLCZ01 cells were transfected with the indicated plasmids for 40 h and then infected with VSV for 8 h. (I) Immunoblot assays were performed with the indicated antibodies. (J) The levels of IFN-β and ISG15 mRNAs were examined by real-time PCR. Experiments were independently repeated two or three times, with similar results. The results are presented as means ± standard deviations. *, P ≤ 0.05; **, P ≤ 0.01 versus control.
DISCUSSION
lncRNAs widely exist in cells and play important roles in various biological processes (10, 37, 38). Recently, some studies have reported that lncRNAs play a regulatory role in the host’s resistance to viral invasion (14, 39–41). Lnczc3h7a binds to TRIM25 and promotes RIG-I-mediated antiviral innate immune responses (17). LncEDAL inhibits the replication of multiple neurotropic viruses by promoting EZH2 lysosomal degradation (15). Nevertheless, some lncRNAs assist viruses to evade the host immune response (17, 42, 43). For example, lncRNA-IFI6 increases HCV replication by inhibiting IFI6 expression (14). lncRNA ACOD1 promotes viral infection by modulating cellular metabolism (44). PSMB8-AS1 promotes influenza virus replication (40). LncDLEU2 binds with hepatitis B virus (HBV) protein HBx to sustain HBV covalently closed circular DNA (cccDNA) (45). Our previous study shows that lncITPRIP-1 can be induced by HCV in JAK/STAT signal-dependent and independent ways. LncITPRIP-1 positively regulates the innate immune response through promoting oligomerization and activation of MDA5 (43). Here, in this study, we found that lncBST2-2 plays an important role in the innate immune response to viral infection.
Importantly, our data reveal that lncBST2-2 functions as an antiviral factor against viral infection. Overexpression of lncBST2-2 inhibits the replication of HCV, NDV, VSV, and HSV. Upon viral infection, lncBST2-2 promotes the activation of the IRF3 signaling pathway by targeting IRF3, enhancing the expression of IFNs and ISGs.
IRF3 is a critical transcription factor for IFN production. When IFN signaling pathway is activated, TBK1 or IKKε phosphorylates IRF3 (46). Phosphorylated IRF3 undergoes conformational changes that expose the DBD and IAD through the disassociation of the C-terminal autoinhibition domain. Then, phosphorylated IRF3 forms homodimers through the IAD and translocates to the nucleus (35). In our study, lncBST2-2 promotes the formation of IRF3 dimers, promoting the nucleus translocation of IRF3. The activation of IRF3 is regulated by multiple protein modifications, methylation, acetylation, ubiquitination, etc. The methyltransferase NSD3 directs lysine methylation of IRF3 and then promotes antiviral innate immunity (47). KAT8 mediates the acetylation of IRF3, inhibiting IRF3 recruitment to IFN gene promoters (48). Reports have shown that protein ubiquitination affects the degradation of IRF3. MID1, Ro52, RBCK1, and RAUL negatively regulate innate immunity by promoting K48-linked ubiquitination degradation of IRF3 (18, 20, 21, 49). TRIM21 catalyzes the K27-linked ubiquitination of IRF3 to promote its autophagic degradation (50). However, we found that lncBST2-2 has no significant influence on the ubiquitination of IRF3. LncBST2-2 interacts with the DBD of IRF3 and promotes the interaction between IRF3 and TBK1, the key step for the phosphorylation of IRF3.
Phosphorylation of IRF3 is also strictly regulated by phosphatases. PP2A and PP1 are important phosphatases for IRF3. PP2A and PP1 negatively regulate innate immunity by dephosphorylating IRF3 (23, 24). RACK1 is critical for IRF3 recruiting of PP2A. Knockdown of RACK1 inhibits the interaction of PP2A and IRF3 (24). In our study, lncBST2-2 interacts with IRF3, inhibits the binding of RACK1-PP2A complex to IRF3, suppresses the dephosphorylation of IRF3, and subsequently promotes the innate immune response to viral infection. In addition to the role in innate immunity, IRF3 also functions as an antitumor factor (51–54). There is a report that the expression of IRF3 suppresses gastric cancer and improves prognosticated patient survival (52). In colorectal cancer, IRF3 prevents colorectal tumorigenesis by inhibiting Wnt signaling and activation of IRF3 relieves its inhibition on Wnt signaling (53). Many reports show that lncRNAs also play an important role in the development of tumors (55, 56). We found that lncBST2-2 is expressed at low levels in some tumor samples (data not shown), which implies that it may also be involved in tumorigenesis. Therefore, we speculate that the low expression of LncBST2-2 in tumors is related to the inhibitory effect of IRF3 on tumor progression. This needs further exploring.
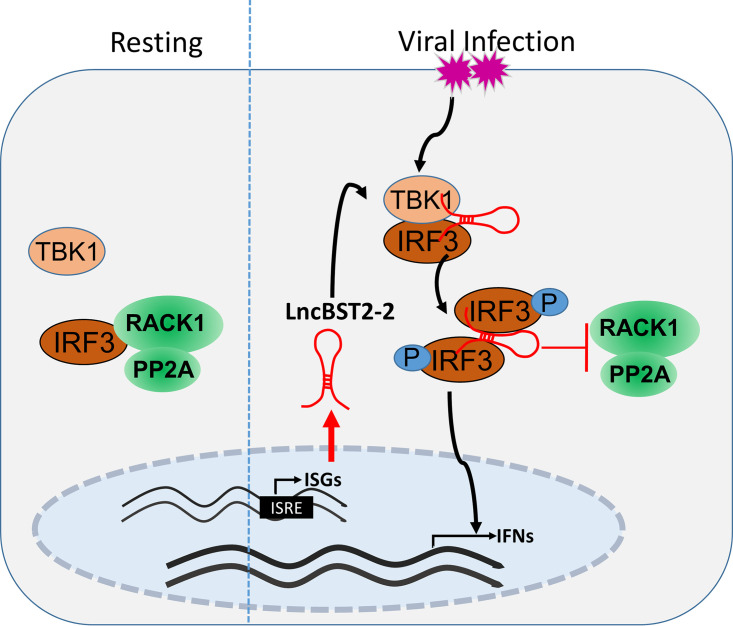
In summary, this study demonstrates the role of lncBST2-2 in the innate immune response to viral infection. Viruses induce the expression of lncBST2-2. LncBST2-2 promotes the innate immune response by interacting with IRF3, enhances the interaction between IRF3 and TBK1, and inhibits the formation of IRF3-RACK1-PP2A complex, thereby inhibiting viral replication (Fig. 10). Collectively, our study provides new insights into lncRNAs in the regulation of IRF3 signaling.
FIG 10.
Working model of lncBST2-2 in the regulation of the innate immune response to viral infection. Viruses induce the expression of lncBST2-2. LncBST2-2 enhances the interaction between IRF3 and TBK1 and inhibits the formation of IRF3-RACK1-PP2A complex, promoting the innate immune response to viral infection.
MATERIALS AND METHODS
Cell lines.
HLCZ01 cells, a novel hepatoma cell line supporting the entire life cycle of HBV and HCV, were established in our laboratory (25). Huh7 cells were purchased from the American Type Culture Collection. Huh7.5 cells were kindly provided by Charles M. Rice (Rockefeller University, New York, NY). HEK293T cells were purchased from Boster. HLCZ01 cells were cultured in collagen-coated tissue culture plates and cultured with Dulbecco’s modified Eagle’s medium (DMEM)–F-12 medium supplemented with 10% (vol/vol) fetal bovine serum (FBS) (Gibco), 40 ng/mL of dexamethasone (Sigma), insulin-transferrin-selenium (ITS) (Lonza), penicillin, and streptomycin. Huh7.5 cells, Huh7 cells, HEK293T cells, and A549 cells were propagated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS, l-glutamine, nonessential amino acid, penicillin, and streptomycin.
Antibodies.
Mouse monoclonal anti-NS5A and anti-Core antibodies were gifts from Chen Liu (Yale University, CT). Monoclonal antibodies against β-actin and Flag tag were purchased from Sigma. Anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) antibody and goat anti-mouse IgG-horseradish peroxidase (HRP) secondary antibody were obtained from Millipore. Anti-V5 antibody was obtained from Invitrogen. Antihemagglutinin (anti-HA) antibody was purchased from Abcam. Anti-TBK1, anti-pTBK1, anti-IRF3, anti-p-IRF3, anti-STAT1, anti-pSTAT1, anti-LMNB1, anti-PP2A, anti-PP1, and goat anti-rabbit IgG-HRP secondary antibodies were purchased from Cell Signaling Technology (CST). Anti-RACK1 antibody was obtained from Santa Cruz.
Viruses.
The protocol for the production of HCV in Huh7.5 cells was described previously (43). VSV, SeV, and HSV-1 were kindly provided by Xuetao Cao (Second Military Medical University, Shanghai, China). The NDV D817 strain was kindly provided by Joseph Peiris (University of Hong Kong, Hong Kong, China).
Plasmid construction.
LncBST2-2 was synthesized from total RNA isolated from HLCZ01 cells and cloned into the pcDNA3.1a vector. Short hairpin RNAs (shRNAs) targeting lncBST2-2 were cloned into pSilencer-neo plasmid (Ambion). The target sequence of lncBST2-2 shRNA was 5′-GAATCTGCAATTCCAGTTCTG-3′. The IFN-β-luciferase and pRL-CMV plasmids were purchased from InvivoGen. The plasmids pFlag-cGAS and pFlag-STING were kindly provided by Hongbing Shu (Wuhan University, Wuhan, China). Flag-MDA5 plasmids were kindly provided by Jianguo Wu (Wuhan University, Wuhan, China). MAVS was synthesized from total cellular RNA isolated from HLCZ01 cells by standard RT-PCR and was subsequently cloned into p3×FLAG-CMV (cytomegalovirus) vector. Flag-TBK1 plasmid was kindly provided by Zhengfan Jiang (Peking University, Beijing, China). Flag-IRF3-5D plasmid was a gift from Deyin Guo (Wuhan University, Wuhan, China). Multiple truncation domains of IRF3 were amplified from the full-length templates of IRF3 and were cloned into the p3×FLAG-CMV vector. LncBST2-2 was synthesized from total RNA isolated from HLCZ01 cells and cloned into the pcDNA3.1a vector and the pRed-max-N1 vector. Primers for the constructed plasmids are listed in Table 1.
TABLE 1.
Primers for the constructed plasmids
| Primer | Sequence |
|---|---|
| pcDNA3.1a-lncBST2-2(F) | 5′-GGGGTACCGCTGCTAGTCTAGAAAAAAGCTCGG-3′ |
| pcDNA3.1a-lncBST2-2(R) | 5′-CGGGATCCAACCTGATTGCCTTTTCCG-3′ |
| pcDNA3.1a-lncBST2-2+T(F) | 5′-GGGGTACCGCTGCTAGTCTAGAAAAAAGCTCGG-3′ |
| pcDNA3.1a-lncBST2-2+T(R) | 5′-CGGGATCCAAACCTGATTGCCTTTTCCG-3′ |
| pcDNA3.1a-lncBST2-2+TT(F) | 5′-GGGGTACCGCTGCTAGTCTAGAAAAAAGCTCGG-3′ |
| pcDNA3.1a-lncBST2-2+TT(R) | 5′-CGGGATCCAAAACCTGATTGCCTTTTCCG-3′ |
| pRed-Max-N1-lncBST2-2(F) | 5′-GGGGTACCGCTGCTAGTCTAGAAAAAAGCTCGG-3′ |
| pRed-Max-N1-lncBST2-2(F) | 5′-CGGGATCCAACCTGATTGCCTTTTCCG-3′ |
| pRed-Max-N1-lncBST2-2+T(F) | 5′-GGGGTACCGCTGCTAGTCTAGAAAAAAGCTCGG-3′ |
| pRed-Max-N1-lncBST2-2+T(F) | 5′-CGGGATCCAAACCTGATTGCCTTTTCCG-3′ |
| pRed-Max-N1-lncBST2-2+TT(F) | 5′-GGGGTACCGCTGCTAGTCTAGAAAAAAGCTCGG-3′ |
| pRed-Max-N1-lncBST2-2+TT(F) | 5′-CGGGATCCAAACCTGATTGCCTTTTCCG-3′ |
| pcDNA3.1a-P1(F) | 5′-GGGGTACCATGAGGTCAAGAGATCGAGACCCT-3′ |
| pcDNA3.1a-P1(R) | 5′-GCTCTAGAAATCAATAGCTGCGTATCCCACAGATG-3′ |
| pcDNA3.1a-P2(F) | 5′-CATGGACCATCATGAGCTTT-3′ |
| pcDNA3.1a-P2(R) | 5′-CTAGAAAGCTCATGATGGTCCATGGTAC-3′ |
| pcDNA3.1a-P3(F) | 5′-CATGAGCTTTGAAATCTTGGGTATAACTT-3′ |
| pcDNA3.1a-P3(R) | 5′-CTAGAAGTTATACCCAAGATTTCAAAGCTCATGGTAC-3′ |
| pRed-Max-N1-P1(F) | 5′-GGGGTACCATGAGGTCAAGAGATCGAGACCCT-3′ |
| pRed-Max-N1-P1(R) | 5′-GCTCTAGAAATCAATAGCTGCGTATCCCACAGATG-3 |
| pRed-Max-N1-P2(F) | 5′-CATGGACCATCATGAGCTTT-3′ |
| pRed-Max-N1-P2(R) | 5′-CTAGAAAGCTCATGATGGTCCATGGTAC-3′ |
| pRed-Max-N1-P3(F) | 5′-CATGAGCTTTGAAATCTTGGGTATAACTT-3′ |
| pRed-Max-N1-P3(R) | 5′-CTAGAAGTTATACCCAAGATTTCAAAGCTCATGGTAC-3′ |
IFNAR1 knockdown HLCZ01 cell line.
Lenti-CRISPR-v2 vector encoding Cas9 and IFNAR1 single guide RNA (sgRNA) (TTCCATCAGATGCTTGTACGCGG) was transfected into HLCZ01 cells. The cells were selected with puromycin (1 mg/mL) for 1 month.
Real-time PCR assay.
Total cellular RNA was extracted in TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. RNA was reverse transcribed by the PrimeScript RT reagent kit gDNA Eraser (Perfect Real Time) (TaKaRa). The SYBR RT-PCR kit (Roche) was used for real-time qRT-PCR analysis. Gene expression was normalized to GAPDH. Primers for related genes are listed in Table 2.
TABLE 2.
Primers for real-time PCR
| Primer | Sequence |
|---|---|
| IFN-β-(F) | 5′-CAGCATTTTCAGTGTCAGAAGC-3′ |
| IFN-β-(R) | 5′-TCATCCTGTCCTTGAGGCAGT-3′ |
| IL-28A (F) | 5′-GCCTCAGAGTTTCTTCTGC-3′ |
| IL-28A (R) | 5′-AAGGCATCTTTGGCCCTCTT-3′ |
| IL-29 (F) | 5′-CGCCTTGGAAGAGTCACTCA-3′ |
| IL-29 (R) | 5′-GAAGCCTCAGGTCCCAATTC-3′ |
| GAPDH (F) | 5′-AATGGGCAGCCGTTAGGAAA-3′ |
| GAPDH (R) | 5′-GCGCCCAATACGACCAAATC-3′ |
| NDV (F) | 5′-TCACAGACTCAACTCTTGGG-3′ |
| NDV (R) | 5′-CAGTATGAGGTGTCAAGTTCTTC-3′ |
| ISG15 (F) | 5′-CACCGTGTTCATGAATCTGC-3′ |
| ISG15 (R) | 5′-CTTTATTTCCGGCCCTTGAT-3′ |
| VSV (F) | 5′-CAAGTCAAAATGCCCAAGAGTCACA-3′ |
| VSV (R) | 5′-TTTCCTTGCATTGTTCTACAGATGG-3′ |
| MX1(F) | 5′-CAACCTGTGCAGCCAGTATGA-3′ |
| MX1(R) | 5′-AGCCCGCAGGGAGTCAAT-3′ |
| LncBST2-2(F) | 5′-GGGGTACCCCAGGATGGTCTT-3′ |
| LncBST2-2(R) | 5′-CGGGATCCCCAGCCTGGGTG-3′ |
| IL6-(F) | 5′-CTCAATATTAGAGTCTCAACCCCCA-3′ |
| IL6-(R) | 5′-GAGAAGGCAACTGGACCGAA-3′ |
| TNF-α-(F) | 5′-AGAACTCACTGGGGCCTACA-3′ |
| TNF-α-(R) | 5′-GCTCCGTGTCTCAAGGAAGT-3′ |
| BST2-(F) | 5′-AAGTACTACCCCAGCTCCCA-3′ |
| BST2-(R) | 5′-TTCCAAGATGTGCCAGCTTCC-3′ |
Western blotting.
Cells were washed with phosphate-buffered saline (PBS) and lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (ThermoFisher) supplemented with protease and phosphatase inhibitor cocktail. The protein was resolved by SDS-PAGE, transferred to the polyvinylidene difluoride (PVDF) membrane at a constant voltage of 100 V for 90 min, and probed with appropriate primary and secondary antibodies. The bound antibodies were detected by SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL).
Luciferase assay.
Luciferase assays were performed with a luciferase assay kit (Promega) according to the manufacturer’s instructions.
Immunofluorescence staining.
The A549 cells that were seeded in a glass-bottom dish were fixed with 4% formaldehyde for 15 min at 25°C. The supernatant was removed, and cells were washed with PBS to remove residual formaldehyde. Cells were blocked with goat serum (diluted in PBS to 1:50) for 60 min and then incubated with rabbit monoclonal anti-IRF3 overnight at 4°C. The cells were washed three times with PBS, 5 min each time, and stained with fluorescence-labeled secondary antibodies (diluted in PBS to 1:400; Invitrogen) for 90 min. Finally, the dish was washed with PBS three times and counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Fluorescent images were obtained under a fluorescence microscope (Olympus).
Immunoprecipitation and RNA immunoprecipitation.
The protocol for immunoprecipitation was described previously (43). For RNA immunoprecipitation, IRF3 and truncations of IRF3-bound lncBST2-2 were quantified relative to IgG. Cells were collected, washed gently in 1 mL PBS, and pelleted at 3,000 × g for 1 min. Then, the cells were resuspended in 1 mL 1% formaldehyde and allowed to stand for 15 min at room temperature. Cells were pelleted at 3,000 × g for 1 min, resuspended in 500 μL 0.25 M glycine solution (diluted in PBS), and allowed to stand for 10 min at room temperature again. Cells were pelleted and washed with 500 μL PBS and then lysed with RIPA buffer supplemented with protease inhibitor cocktail and RNase inhibitor (Roche) on ice for 30 min. And subsequent IP was carried out as described above. Protein-RNA complexes binding to beads were eluted in PBS at 70°C for 45 min. The eluted supernatant was lysed in an ice-cold TRIzol reagent for RT-PCR.
Native PAGE.
The protocol for native PAGE was described previously (57).
Nuclear and cytoplasmic extraction.
The protocol for nuclear and cytoplasmic extraction was described previously (57).
Statistical analysis.
All results are presented as means ± standard deviations (SD). Comparisons between two groups were performed using Student’s t test.
ACKNOWLEDGMENTS
We thank Charles M. Rice for the Huh7.5 cell lines, Takaji Wakita for pJFH1 and pJFH1/GND, and Xuetao Cao for VSV and HSV. We also appreciate Chen Liu, Jianguo Wu, Zhengfan Jiang, Deyin Guo, and Hongbing Shu for kindly sharing research materials.
This work was supported by the National Natural Science Foundation of China (81730064, 82072269, 81571985), the National Science and Technology Major Project (2017ZX10202201-005), and the Hunan Natural Science Foundation (2018JJ3090).
Contributor Information
Haizhen Zhu, Email: zhuhaizhen69@yahoo.com.
J.-H. James Ou, University of Southern California
REFERENCES
- 1.Wu J, Chen ZJ. 2014. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol 32:461–488. 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 2.Luecke S, Sheu KM, Hoffmann A. 2021. Stimulus-specific responses in innate immunity: multilayered regulatory circuits. Immunity 54:1915–1932. 10.1016/j.immuni.2021.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zindel J, Kubes P. 2020. DAMPs, PAMPs, and LAMPs in immunity and sterile inflammation. Annu Rev Pathol 15:493–518. 10.1146/annurev-pathmechdis-012419-032847. [DOI] [PubMed] [Google Scholar]
- 4.Fitzgerald KA, Kagan JC. 2020. Toll-like receptors and the control of immunity. Cell 180:1044–1066. 10.1016/j.cell.2020.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ren Z, Ding T, Zuo Z, Xu Z, Deng J, Wei Z. 2020. Regulation of MAVS expression and signaling function in the antiviral innate immune response. Front Immunol 11:1030. 10.3389/fimmu.2020.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, Chen ZJ. 2015. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347:aaa2630. 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 7.Villarino AV, Kanno Y, O’Shea JJ. 2017. Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat Immunol 18:374–384. 10.1038/ni.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aune TM, Spurlock CF, III.. 2016. Long non-coding RNAs in innate and adaptive immunity. Virus Res 212:146–160. 10.1016/j.virusres.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong CM, Tsang FH, Ng IO. 2018. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol 15:137–151. 10.1038/nrgastro.2017.169. [DOI] [PubMed] [Google Scholar]
- 10.Liu SJ, Lim DA. 2018. Modulating the expression of long non-coding RNAs for functional studies. EMBO Rep 19:e46955. 10.15252/embr.201846955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nair L, Chung H, Basu U. 2020. Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery. Nat Rev Mol Cell Biol 21:123–136. 10.1038/s41580-019-0209-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palazzo AF, Koonin EV. 2020. Functional long non-coding RNAs evolve from junk transcripts. Cell 183:1151–1161. 10.1016/j.cell.2020.09.047. [DOI] [PubMed] [Google Scholar]
- 13.Chai W, Li J, Shangguan Q, Liu Q, Li X, Qi D, Tong X, Liu W, Ye X. 2018. Lnc-ISG20 inhibits influenza A virus replication by enhancing ISG20 expression. J Virol 92:e00539-18. 10.1128/JVI.00539-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Duan X, Holmes JA, Li W, Lee SH, Tu Z, Zhu C, Salloum S, Lidofsky A, Schaefer EA, Cai D, Li S, Wang H, Huang Y, Zhao Y, Yu ML, Xu Z, Chen L, Hong J, Lin W, Chung RT. 2019. A long noncoding RNA regulates hepatitis C virus infection through interferon alpha-inducible protein 6. Hepatology 69:1004–1019. 10.1002/hep.30266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sui B, Chen D, Liu W, Wu Q, Tian B, Li Y, Hou J, Liu S, Xie J, Jiang H, Luo Z, Lv L, Huang F, Li R, Zhang C, Tian Y, Cui M, Zhou M, Chen H, Fu ZF, Zhang Y, Zhao L. 2020. A novel antiviral lncRNA, EDAL, shields a T309 O-GlcNAcylation site to promote EZH2 lysosomal degradation. Genome Biol 21:228. 10.1186/s13059-020-02150-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu H, Jiang Y, Xu X, Su X, Liu Y, Ma Y, Zhao Y, Shen Z, Huang B, Cao X. 2019. Inducible degradation of lncRNA Sros1 promotes IFN-gamma-mediated activation of innate immune responses by stabilizing Stat1 mRNA. Nat Immunol 20:1621–1630. 10.1038/s41590-019-0542-7. [DOI] [PubMed] [Google Scholar]
- 17.Lin H, Jiang M, Liu L, Yang Z, Ma Z, Liu S, Ma Y, Zhang L, Cao X. 2019. The long noncoding RNA Lnczc3h7a promotes a TRIM25-mediated RIG-I antiviral innate immune response. Nat Immunol 20:812–823. 10.1038/s41590-019-0379-0. [DOI] [PubMed] [Google Scholar]
- 18.Yu Y, Hayward GS. 2010. The ubiquitin E3 ligase RAUL negatively regulates type I interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity 33:863–877. 10.1016/j.immuni.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lei CQ, Zhang Y, Xia T, Jiang LQ, Zhong B, Shu HB. 2013. FoxO1 negatively regulates cellular antiviral response by promoting degradation of IRF3. J Biol Chem 288:12596–12604. 10.1074/jbc.M112.444794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Xu Y, Tu W, Huang F, Zuo Y, Zhang H-G, Jin L, Feng Q, Ren T, He J, Miao Y, Yuan Y, Zhao Q, Liu J, Zhang R, Zhu L, Qian F, Zhu C, Zheng H, Wang J. 2021. Ubiquitin E3 ligase MID1 inhibits the innate immune response by ubiquitinating IRF3. Immunology 163:278–292. 10.1111/imm.13315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M, Tian Y, Wang RP, Gao D, Zhang Y, Diao FC, Chen DY, Zhai ZH, Shu HB. 2008. Negative feedback regulation of cellular antiviral signaling by RBCK1-mediated degradation of IRF3. Cell Res 18:1096–1104. 10.1038/cr.2008.277. [DOI] [PubMed] [Google Scholar]
- 22.James SJ, Jiao H, Teh HY, Takahashi H, Png CW, Phoon MC, Suzuki Y, Sawasaki T, Xiao H, Chow VTK, Yamamoto N, Reynolds JM, Flavell RA, Dong C, Zhang Y. 2015. MAPK phosphatase 5 expression induced by influenza and other RNA virus infection negatively regulates IRF3 activation and type I interferon response. Cell Rep 10:1722–1734. 10.1016/j.celrep.2015.02.030. [DOI] [PubMed] [Google Scholar]
- 23.Hu X, Wang B, Feng H, Zhou M, Lin Y, Cao H. 2020. Protein phosphatase PP1 negatively regulates IRF3 in response to GCRV infection in grass carp (Ctenopharyngodon idella). Front Immunol 11:609890. 10.3389/fimmu.2020.609890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long L, Deng Y, Yao F, Guan D, Feng Y, Jiang H, Li X, Hu P, Lu X, Wang H, Li J, Gao X, Xie D. 2014. Recruitment of phosphatase PP2A by RACK1 adaptor protein deactivates transcription factor IRF3 and limits type I interferon signaling. Immunity 40:515–529. 10.1016/j.immuni.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 25.Yang D, Zuo C, Wang X, Meng X, Xue B, Liu N, Yu R, Qin Y, Gao Y, Wang Q, Hu J, Wang L, Zhou Z, Liu B, Tan D, Guan Y, Zhu H. 2014. Complete replication of hepatitis B virus and hepatitis C virus in a newly developed hepatoma cell line. Proc Natl Acad Sci USA 111:E1264–E1273. 10.1073/pnas.1320071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue B, Li H, Guo M, Wang J, Xu Y, Zou X, Deng R, Li G, Zhu H. 2018. TRIM21 promotes innate immune response to RNA viral infection through Lys27-linked polyubiquitination of MAVS. J Virol 92:e00321-18. 10.1128/JVI.00321-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong N, Shan T, Wang H, Jiao Y, Zuo Y, Li L, Tong W, Yu L, Jiang Y, Zhou Y, Li G, Gao F, Yu H, Zheng H, Tong G. 2020. BST2 suppresses porcine epidemic diarrhea virus replication by targeting and degrading virus nucleocapsid protein with selective autophagy. Autophagy 16:1737–1752. 10.1080/15548627.2019.1707487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.OhAinle M, Helms L, Vermeire J, Roesch F, Humes D, Basom R, Delrow JJ, Overbaugh J, Emerman M. 2018. A virus-packageable CRISPR screen identifies host factors mediating interferon inhibition of HIV. Elife 7:e39823. 10.7554/eLife.39823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barriocanal M, Carnero E, Segura V, Fortes P. 2014. Long non-coding RNA BST2/BISPR is induced by IFN and regulates the expression of the antiviral factor tetherin. Front Immunol 5:655. 10.3389/fimmu.2014.00655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsumoto A, Pasut A, Matsumoto M, Yamashita R, Fung J, Monteleone E, Saghatelian A, Nakayama KI, Clohessy JG, Pandolfi PP. 2017. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 541:228–232. 10.1038/nature21034. [DOI] [PubMed] [Google Scholar]
- 31.Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng X, Xiong F, Guo C, Wu X, Li Y, Li X, Li G, Zeng Z, Xiong W. 2020. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol Cancer 19:22. 10.1186/s12943-020-1147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang J-Z, Chen M, Chen D, Gao X-C, Zhu S, Huang H, Hu M, Zhu H, Yan G-R. 2017. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell 68:171–184.e6. 10.1016/j.molcel.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 33.Volders PJ, Anckaert J, Verheggen K, Nuytens J, Martens L, Mestdagh P, Vandesompele J. 2019. LNCipedia 5: towards a reference set of human long non-coding RNAs. Nucleic Acids Res 47:D135–D139. 10.1093/nar/gky1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun SC. 2017. The non-canonical NF-kappaB pathway in immunity and inflammation. Nat Rev Immunol 17:545–558. 10.1038/nri.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwanke H, Stempel M, Brinkmann MM. 2020. Of keeping and tipping the balance: host regulation and viral modulation of IRF3-dependent IFNB1 expression. Viruses 12:733. 10.3390/v12070733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malathi K, Dong B, Gale M, Jr, Silverman RH. 2007. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 448:816–819. 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agliano F, Rathinam VA, Medvedev AE, Vanaja SK, Vella AT. 2019. Long noncoding RNAs in host-pathogen interactions. Trends Immunol 40:492–510. 10.1016/j.it.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu S, Wang Q, Kang Y, Liu J, Yin Y, Liu L, Wu H, Li S, Sui S, Shen M, Zheng W, Pang D. 2020. Long noncoding RNAs control the modulation of immune checkpoint molecules in cancer. Cancer Immunol Res 8:937–951. 10.1158/2326-6066.CIR-19-0696. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Cen S. 2020. Roles of lncRNAs in influenza virus infection. Emerg Microbes Infect 9:1407–1414. 10.1080/22221751.2020.1778429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.More S, Zhu Z, Lin K, Huang C, Pushparaj S, Liang Y, Sathiaseelan R, Yang X, Liu L. 2019. Long non-coding RNA PSMB8-AS1 regulates influenza virus replication. RNA Biol 16:340–353. 10.1080/15476286.2019.1572448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Unfried JP, Fortes P. 2020. LncRNAs in HCV infection and HCV-related liver disease. Int J Mol Sci 21:2255. 10.3390/ijms21062255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Wang Y, Luo W, Song X, Huang L, Xiao J, Jin F, Ren Z, Wang Y. 2020. Roles of long non-coding RNAs and emerging RNA-binding proteins in innate antiviral responses. Theranostics 10:9407–9424. 10.7150/thno.48520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie Q, Chen S, Tian R, Huang X, Deng R, Xue B, Qin Y, Xu Y, Wang J, Guo M, Chen J, Tang S, Li G, Zhu H. 2018. Long noncoding RNA ITPRIP-1 positively regulates the innate immune response through promotion of oligomerization and activation of MDA5. J Virol 92:e00507-18. 10.1128/JVI.00507-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang P, Xu J, Wang Y, Cao X. 2017. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science 358:1051–1055. 10.1126/science.aao0409. [DOI] [PubMed] [Google Scholar]
- 45.Salerno D, Chiodo L, Alfano V, Floriot O, Cottone G, Paturel A, Pallocca M, Plissonnier ML, Jeddari S, Belloni L, Zeisel M, Levrero M, Guerrieri F. 2020. Hepatitis B protein HBx binds the DLEU2 lncRNA to sustain cccDNA and host cancer-related gene transcription. Gut 69:2016–2024. 10.1136/gutjnl-2019-319637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yum S, Li M, Fang Y, Chen ZJ. 2021. TBK1 recruitment to STING activates both IRF3 and NF-kappaB that mediate immune defense against tumors and viral infections. Proc Natl Acad Sci USA 118:e2100225118. 10.1073/pnas.2100225118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang C, Wang Q, Xu X, Xie B, Zhao Y, Li N, Cao X. 2017. The methyltransferase NSD3 promotes antiviral innate immunity via direct lysine methylation of IRF3. J Exp Med 214:3597–3610. 10.1084/jem.20170856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huai W, Liu X, Wang C, Zhang Y, Chen X, Chen X, Xu S, Thomas T, Li N, Cao X. 2019. KAT8 selectively inhibits antiviral immunity by acetylating IRF3. J Exp Med 216:772–785. 10.1084/jem.20181773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oke V, Wahren-Herlenius M. 2012. The immunobiology of Ro52 (TRIM21) in autoimmunity: a critical review. J Autoimmun 39:77–82. 10.1016/j.jaut.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 50.Wu Y, Jin S, Liu Q, Zhang Y, Ma L, Zhao Z, Yang S, Li YP, Cui J. 2021. Selective autophagy controls the stability of transcription factor IRF3 to balance type I interferon production and immune suppression. Autophagy 17:1379–1392. 10.1080/15548627.2020.1761653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown MC, Mosaheb MM, Mohme M, McKay ZP, Holl EK, Kastan JP, Yang Y, Beasley GM, Hwang ES, Ashley DM, Bigner DD, Nair SK, Gromeier M. 2021. Viral infection of cells within the tumor microenvironment mediates antitumor immunotherapy via selective TBK1-IRF3 signaling. Nat Commun 12:1858. 10.1038/s41467-021-22088-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiao S, Guan J, Chen M, Wang W, Li C, Wang Y, Cheng Y, Zhou Z. 2018. Targeting IRF3 as a YAP agonist therapy against gastric cancer. J Exp Med 215:699–718. 10.1084/jem.20171116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tian M, Wang X, Sun J, Lin W, Chen L, Liu S, Wu X, Shi L, Xu P, Cai X, Wang X. 2020. IRF3 prevents colorectal tumorigenesis via inhibiting the nuclear translocation of beta-catenin. Nat Commun 11:5762. 10.1038/s41467-020-19627-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding C, He J, Zhao J, Li J, Chen J, Liao W, Zeng Y, Zhong J, Wei C, Zhang L, Zhou M, Jia Z, Zhang Y, Li H, Zhou Y, Xiao X, Han D, Li C, Zhu Z, Xia Z, Peng J. 2018. Beta-catenin regulates IRF3-mediated innate immune signalling in colorectal cancer. Cell Prolif 51:e12464. 10.1111/cpr.12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ni W, Zhang Y, Zhan Z, Ye F, Liang Y, Huang J, Chen K, Chen L, Ding Y. 2017. A novel lncRNA uc.134 represses hepatocellular carcinoma progression by inhibiting CUL4A-mediated ubiquitination of LATS1. J Hematol Oncol 10:91. 10.1186/s13045-017-0449-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qin Y, Hou Y, Liu S, Zhu P, Wan X, Zhao M, Peng M, Zeng H, Li Q, Jin T, Cui X, Liu M. 2021. A novel long non-coding RNA lnc030 maintains breast cancer stem cell stemness by stabilizing SQLE mRNA and increasing cholesterol synthesis. Adv Sci (Weinh) 8:2002232. 10.1002/advs.202002232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J, Li H, Xue B, Deng R, Huang X, Xu Y, Chen S, Tian R, Wang X, Xun Z, Sang M, Zhu H. 2020. IRF1 promotes the innate immune response to viral infection by enhancing the activation of IRF3. J Virol 94:e01231-20. 10.1128/JVI.01231-20. [DOI] [PMC free article] [PubMed] [Google Scholar]