ABSTRACT
Binding to the receptor, CD4, drives the pretriggered, “closed” (State-1) conformation of the human immunodeficiency virus (HIV-1) envelope glycoprotein (Env) trimer ([gp120/gp41]3) into more “open” conformations. HIV-1 Env on the viral membrane is maintained in a State-1 conformation that resists binding and neutralization by commonly elicited antibodies. Premature triggering of Env before the virus engages a target cell typically leads to increased susceptibility to spontaneous inactivation or ligand-induced neutralization. Here, we showed that single amino acid substitutions in the gp41 membrane-proximal external region (MPER) of a primary HIV-1 strain resulted in viral phenotypes indicative of premature triggering of Env to downstream conformations. Specifically, the MPER changes reduced viral infectivity and globally increased virus sensitivity to poorly neutralizing antibodies, soluble CD4, a CD4-mimetic compound, and exposure to cold. In contrast, the MPER mutants exhibited decreased sensitivity to the State 1-preferring inhibitor, BMS-806, and to the PGT151 broadly neutralizing antibody. Depletion of cholesterol from virus particles did not produce the same State 1-destabilizing phenotypes as MPER alterations. Notably, State 1-stabilizing changes in Env distant from the MPER could minimize the phenotypic effects of MPER alteration but did not affect virus sensitivity to cholesterol depletion. Thus, membrane-proximal gp41 elements contribute to the maintenance of the pretriggered Env conformation. The conformationally disruptive effects of MPER changes can be minimized by distant State 1-stabilizing Env modifications, a strategy that may be useful in preserving the native pretriggered state of Env.
IMPORTANCE The pretriggered shape of the human immunodeficiency virus (HIV-1) envelope glycoprotein (Env) is a major target for antibodies that can neutralize many strains of the virus. An effective HIV-1 vaccine may need to raise these types of antibodies, but this goal has proven difficult. One reason is that the pretriggered shape of Env is unstable and dependent on interactions near the viral membrane. Here, we showed that the membrane-proximal external region (MPER) of Env plays an important role in maintaining Env in a pretriggered shape. Alterations in the MPER resulted in global changes in Env conformation that disrupted its pretriggered shape. We also found that these disruptive effects of MPER changes could be minimized by distant Env modifications that stabilized the pretriggered shape. These modifications may be useful for preserving the native shape of Env for structural and vaccine studies.
KEYWORDS: HIV-1 Env, MPER, native conformation, State 1, stabilization, cholesterol, membrane, triggerability
INTRODUCTION
The human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) trimer mediates virus entry into host cells (1). HIV-1 Env is a Class I viral fusion protein composed of three gp120 exterior subunits and three gp41 transmembrane subunits (1–4). Env is synthesized in the rough endoplasmic reticulum where signal peptide cleavage, high-mannose glycan modification, and trimerization take place (5–7). This trimeric Env precursor (gp160) then traffics to the cell surface either through the conventional secretory pathway via the Golgi apparatus, where furin-mediated cleavage and modification with complex glycans occur, or through a pathway that bypasses the Golgi compartment (8–12). Envs transported to the cell surface through the Golgi apparatus are selectively incorporated into virions (8). On virions, Env samples at least three conformational states, reflecting its dynamic nature (13). Envs of primary HIV-1 strains mainly occupy a pretriggered, “closed” (State-1) conformation that resists the binding of most antibodies elicited during natural infection (13–17). More rarely elicited broadly neutralizing antibodies recognize conserved elements of the State-1 Env conformation (13–15, 18). Binding to the first receptor, CD4, triggers major conformational changes in Env, leading initially to a default intermediate conformation (State 2) and then to the full CD4-bound conformation (State 3) (19–25). The CD4-bound conformation (State 3) consists of a prehairpin intermediate in which the gp41 heptad repeat (HR1) region is exposed (26–28). Subsequent binding of the State-3 Env to the second receptor, either CCR5 or CXCR4, leads to the formation of an energetically stable gp41 six-helix bundle, a process that results in fusion of the viral and target cell membranes (29–42).
The gp41 transmembrane subunits anchor Env to the lipid membrane on the surface of infected cells and virus particles (43–45). The gp41 glycoprotein is composed of an N-terminal fusion peptide, a fusion peptide-proximal region (FPPR), two heptad repeat regions (HR1 and HR2), a membrane-proximal external region (MPER), a transmembrane region, and a long cytoplasmic tail (43, 46). The gp41 MPER (residues ∼659 to 683) plays different roles during the distinct stages of HIV-1 entry, which may be reflected in its dynamic structure. In current high-resolution structures of soluble or detergent-solubilized HIV-1 Env trimers, the MPER has been removed or is disordered (47–53). Lower-resolution tomograms of virion Envs have provided different views of the MPER conformation, from a stalk tightly organized near the trimer axis to a tripod with various degrees of splay (21, 54–57). Synthetic MPER peptides partition into membranes and form α-helical structures. Peptides corresponding to the gp41 MPER-transmembrane region have been suggested to form trimers in bicelles, but other studies of similar peptides in lipid bilayers or nanodiscs found little evidence of trimers (58–64). These discrepancies may reflect the dynamic nature of the MPER and the dependence of its structure on interactions with both the viral membrane and the rest of the Env ectodomain.
Conserved elements of the HIV-1 gp41 MPER can be targeted by broadly neutralizing antibodies (65). The antibodies generally recognize the MPER in conjunction with the lipid membrane and, in some cases, have been shown to bind a tilted Env and to extract the peptide epitope from the membrane (63, 65–71). Most MPER-directed neutralizing antibodies recognize the CD4-bound Env better than the pretriggered Env, although the 10E8 antibody can bind the functional virion Env spike in the absence of CD4 (72–77). These observations are consistent with the induction of changes in MPER conformation and/or exposure because of Env-receptor interactions.
The HIV-1 gp41 MPER has been suggested to play a role in virus entry by promoting the late stages of Env-mediated membrane fusion. Alteration of a conserved tryptophan-rich MPER motif disrupted the ability of Env to expand the initial fusion pore (78, 79). The MPER includes a potential cholesterol-binding motif and some changes in this element diminish Env function (80–84). Finally, functional links between the FPPR and MPER during the formation of the gp41 six-helix bundle have been suggested (85).
Some changes in the gp41 MPER can result in increased HIV-1 sensitivity to neutralization by antibodies directed against gp120 (86–92). The increased neutralization sensitivity resulting from these MPER changes is, in some cases, enhanced by additional gp41 or gp120 changes (86, 90, 91). We hypothesize that the observed increases in neutralization sensitivity result from a loss of MPER-mediated stabilization of the pretriggered (State-1) Env conformation. A corollary of this hypothesis is that the MPER functions to maintain a higher activation barrier between State 1 and downstream conformations of Env. Therefore, Env mutants whose MPER integrity is compromised are more triggerable and predisposed to sampling downstream conformations. A stabilized Env with a larger activation barrier between State 1 and downstream conformations might be expected to resist the disruptive effects of MPER changes. Here, we characterized in detail the effects of MPER changes in the Env of a Tier-2 primary virus, HIV-1AD8. We tested the above hypothesis by evaluating whether State 1-stabilizing changes in the rest of the Env ectodomain could revert the phenotypes that resulted from alteration of the MPER. The phenotypic consequences of cholesterol depletion on HIV-1 infectivity and neutralization sensitivity were compared with those of MPER alteration.
RESULTS
Effects of MPER changes on the HIV-1AD8 Env.
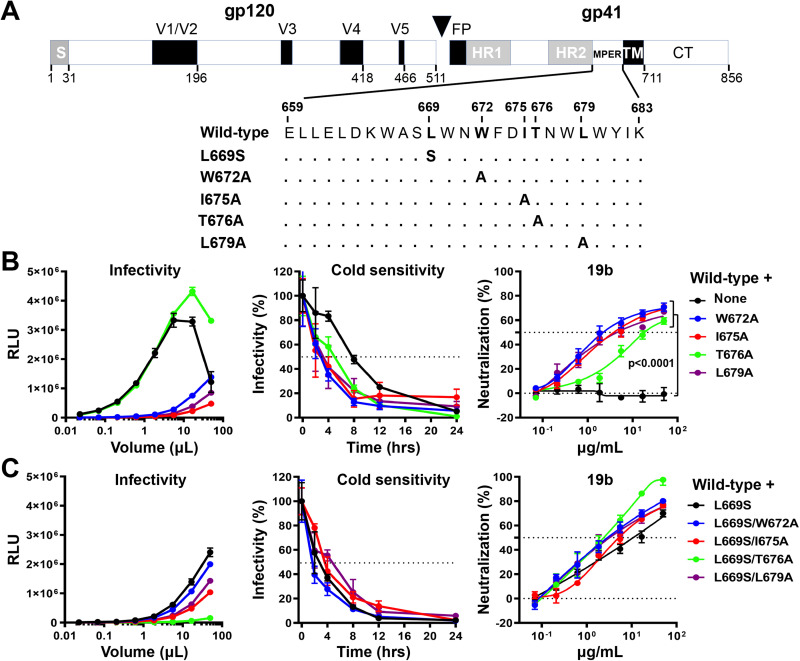
We evaluated the effects of MPER alterations on the phenotypes associated with the HIV-1AD8 Env. The wild-type HIV-1AD8 Env used in this work contained a signal peptide and part of the gp41 cytoplasmic tail from the HIV-1HXBc2 Env. Viruses with this chimeric Env exhibit a degree of CD4 dependence and an antibody neutralization profile consistent with those of a primary, Tier-2 HIV-1 (14, 86). To alter the MPER of the HIV-1AD8 Env, we individually changed Leu 669, Trp 672, Ile 675, Thr 676, and Leu 679 (Fig. 1A). These MPER residues are more than 98% conserved in HIV-1 strains except for Thr 676, which naturally tolerates substitutions of serine residues in ∼45% of HIV-1 strains (93). Alteration of the individual MPER residues reduced virus infectivity in all cases except for the T676A change, which supported HIV-1 infection comparably to the wild-type HIV-1AD8 Env (Fig. 1B). To evaluate the sensitivity of the variant Envs to cold, pseudoviruses were incubated on ice for different lengths of time and their infectivity was then measured. Compared to the wild-type HIV-1AD8 virus, the mutant viruses exhibited increased sensitivity to cold (Fig. 1B). Most of the MPER-modified viruses lost 50% of their infectivity after 4 h on ice. The T676A virus exhibited a 5-h half-life on ice, whereas the wild-type HIV-1AD8 required 8 h on ice to lose half of its infectivity. Consistent with its Tier-2 phenotype, the wild-type HIV-1AD8 was not neutralized by the poorly neutralizing 19b antibody, which is directed against the gp120 V3 loop (94) (Fig. 1B). In contrast, the MPER mutant viruses were neutralized by 19b. The T676A virus was less sensitive to 19b neutralization than the other mutant viruses. No additivity or synergy between the MPER changes was observed. Double mutants containing L669S and one of the other MPER changes displayed virus sensitivity to cold and 19b neutralization comparable to those of the L669S single mutant (Fig. 1C). Overall, these results supported our hypothesis that altered MPER integrity could result in more triggerable Envs that are more prone to making transitions from State 1 to downstream conformations. A more comprehensive and detailed characterization of the effects of MPER changes on the Env ectodomain is presented below.
FIG 1.
Effects of MPER changes in the wild-type HIV-1AD8 Env. (A) A schematic representation of the HIV-1AD8 Env glycoprotein is shown with the gp120-gp41 cleavage site depicted as a black triangle. S, signal peptide; V1 to V5, gp120 hypervariable regions; FP, fusion peptide; HR, heptad repeat region; MPER, membrane-proximal external region; TM, transmembrane region; CT, cytoplasmic tail. The MPER amino acid changes studied herein are listed. Standard numbering of HIV-1 Env amino acid residues is used (140). (B and C) HEK293T cells were transfected with pNL4-3.Luc.R-E-vector and a plasmid expressing the wild-type or MPER-modified HIV-1AD8 Env. All Env mutants in C contain the L669S change either alone or in combination with the other MPER changes. Forty-eight hours later, the cell supernatants were collected, clarified, and added to TZM-bl cells in the presence of 20 μg/mL DEAE-Dextran. Forty-eight hours later, the infected cells were lysed, and the luciferase activity (in relative light units [RLU]) was measured as an indicator of virus infectivity (left). In the cold sensitivity assay, viruses were incubated on ice for the indicated times, after which the virus infectivity was measured (middle). In the neutralization assay with the 19b poorly neutralizing antibody, viruses were incubated with serial dilutions of 19b for 1 h at 37°C before TZM-bl cells were added and infectivity was measured. The results for cold sensitivity and 19b neutralization are normalized to those obtained in the absence of virus exposure to cold or the 19b antibody. The results shown are representative of those obtained in two independent experiments, expressed as means and standard deviations from triplicate luciferase readings. The significance of the difference in the sensitivity of the HIV-1AD8 Env variants to 19b neutralization was evaluated by a Student’s paired t test.
Effects of the MPER changes are minimized in the 2-4 RM6 AE Env.
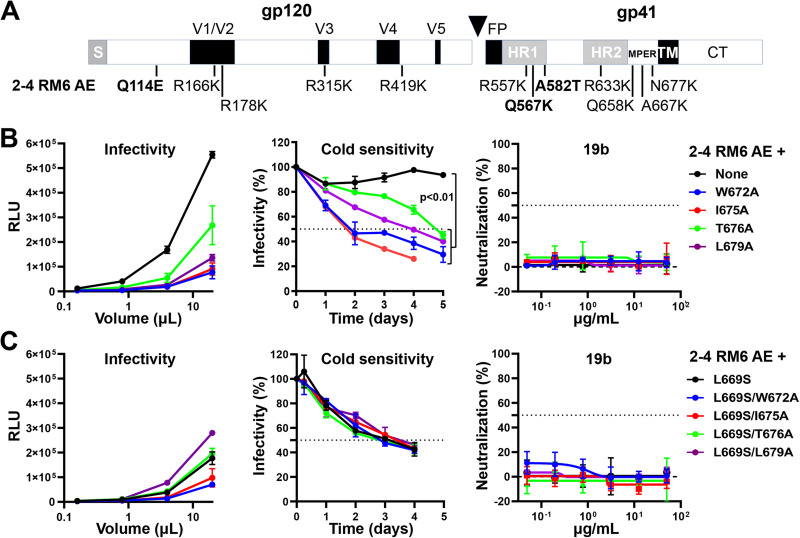
In a previous study, we identified several lysine-rich and well-cleaved HIV-1AD8 Env variants with phenotypes consistent with State-1 stabilization (95). Compared to the wild-type HIV-1AD8, viruses with these variant Envs were significantly more resistant to cold, soluble CD4, the CD4-mimetic compound BNM-III-170 (96), and gp120-trimer dissociation in detergent. One such Env variant, 2-4 RM6 AE, was found to tolerate the State 1-destabilizing effects of changes (R542A, I595F, and L602H) in or near the gp41 HR1 region (95). Here, we introduced the MPER changes into the 2-4 RM6 AE Env to test whether the phenotypes indicative of State-1 disruption would be less pronounced in the 2-4 RM6 AE Env background than in the context of the wild-type HIV-1AD8 Env (Fig. 2A). The MPER mutants exhibited decreases in infectivity relative to the 2-4 RM6 AE parent virus (Fig. 2B). Strikingly, the MPER-mediated phenotypes associated with State-1 destabilization were minimized or completely reverted in the 2-4 RM6 AE Env background. While the 2-4 RM6 AE viruses with MPER changes were more sensitive to cold relative to the parental 2-4 RM6 AE virus, they were significantly more cold-resistant than wild-type HIV-1AD8. The half-lives on ice of the 2-4 RM6 AE MPER mutants were at least 2 days, compared to the 8-h half-life of wild-type HIV-1AD8 (compare Fig. 2B, middle with Fig. 1B, middle). Likewise, the MPER modifications in the 2-4 RM6 AE Env did not result in increased sensitivity to neutralization by the 19b anti-V3 antibody (Fig. 2B). The 2-4 RM6 AE Env with the L669S change tolerated additional MPER changes without effects on cold sensitivity or sensitivity to 19b neutralization (Fig. 2C). These results support the hypothesis that some phenotypic effects of MPER alterations are minimized in the context of an Env that exhibits a more stable pretriggered (State-1) conformation.
FIG 2.
Effects of MPER changes in the State 1-stabilized 2-4 RM6 AE Env. (A) The 2-4 RM6 AE Env is an HIV-1AD8 Env variant in which the pretriggered (State-1) conformation is stabilized, compared to the wild-type HIV-1AD8 Env (95). Of the 12 changes in the 2–4 RM6 AE Env relative to the wild-type HIV-1AD8 Env, the three highlighted changes (Q114E, Q567K, and A582T) are the key determinants of the viral phenotypes associated with State-1 stabilization (95). (B, C) The experiments were carried out as described in the legend to Fig. 1. Briefly, cell supernatants containing recombinant luciferase-expressing viruses were collected from transiently transfected HEK293T cells, clarified, and either used directly for infection (left) or incubated with the 19b antibody for 1 h at 37°C (right) before TZM-bl target cells were added. After 48 to 72 h, luciferase activity (RLU) was measured. In the cold sensitivity assay, viruses were incubated on ice for the indicated times before virus infectivity was measured (middle). Note the cold resistance of the 2-4 RM6 AE variants, compared with that of wild-type HIV-1AD8 in Fig. 1B The results shown are representative of those obtained in two independent experiments, expressed as means and standard deviations from triplicate luciferase readings. The significance of the difference in the cold sensitivity of the 2-4 RM6 AE Env variants was evaluated by a Student’s paired t test.
State 1-stabilizing Env changes lessen the phenotypic effects of MPER alteration.
The 2-4 RM6 AE Env differs from the wild-type HIV-1AD8 Env by 12 amino acid residues (Fig. 2A). Three substitutions, Q114E, Q567K, and A582T, were identified as key determinants of the State 1-stabilizing phenotypes of the 2-4 RM6 AE Env (95). To test our hypothesis that State 1-stabilizing Env changes could revert the State 1-destabilizing effects of MPER changes, we introduced the MPER changes into HIV-1AD8 Envs with Q114E/Q567K and Q114E/Q567K/A582T alterations. Because the effects of these alterations are additive, the viral phenotypes associated with State-1 stabilization are more robust for the Q114E/Q567K/A582T mutant than for the Q114E/Q567K mutant (95). The MPER changes exerted only modest effects on the infectivity of the Q114E/Q567K and Q114E/Q567K/A582T viruses (Fig. 3A and B). The effects of the MPER changes on virus sensitivity to cold and 19b antibody neutralization were decreased in the Q114E/Q567K background compared with those seen in the context of the wild-type HIV-1AD8 (compare Fig. 3A with Fig. 1B). The half-lives in the cold of the MPER mutants increased from 4 to 5 h in the wild-type HIV-1AD8 background to around 24 h (or more for the T676A mutant) in the Q114E/Q567K virus background. Likewise, even at the highest concentrations tested, the 19b antibody only partially inhibited infection of the Q114E/Q567K MPER mutant viruses. The effects of the MPER changes on virus sensitivity to cold and 19b neutralization were further minimized by the addition of the A582T change, a gp41 HR1 substitution that stabilizes the pretriggered (State-1) conformation of Env (97–100) (Fig. 3B). Nearly all the resistance to the phenotypic effects of MPER changes displayed by the 2–4 RM6 AE Env could be accounted for by the Q114E/Q567K/A582T changes (compare Fig. 3B with Fig. 2B). These results support the hypothesis that the phenotypic effects of MPER alterations can be reverted by State 1-stabilizing changes in the Env ectodomain outside the MPER.
FIG 3.
Effects of MPER changes in HIV-1AD8 Envs with State 1-stabilizing changes. The indicated MPER changes were introduced into HIV-1AD8 Envs containing Q114E/Q567K (A) or Q114E/Q567K/A582T substitutions (B) that have previously been shown to stabilize a pretriggered (State-1) conformation (95). The experiments were carried out as described in the legend to Fig. 1. Briefly, recombinant viruses produced from HEK293T cells transfected with pNL4-3.Luc.R-E- and an Env-expressing plasmid were incubated with TZM-bl target cells for 48 h, after which luciferase activity was measured (left). In the cold sensitivity assay, the virus was incubated on ice before addition to target cells (middle). In the neutralization assay, the virus was preincubated with the 19b antibody for 1 h at 37°C before TZM-bl cells were added to the mixture (right). The results shown are representative of those obtained in two independent experiments, expressed as means and standard deviations from triplicate luciferase readings. The significance of the difference in the cold sensitivity of the Env variants was evaluated by a Student’s paired t test.
Overall effects of the MPER changes in different Env backgrounds.
The above findings suggested that MPER changes can promote Env transitions from its pretriggered conformation to downstream states. Furthermore, Envs exhibiting phenotypes indicative of increased State-1 stability were less susceptible to some of the consequences of MPER alteration. To obtain a more complete picture, we evaluated the range of Env phenotypes that potentially might be affected by MPER modification. We compared the MPER mutant phenotypes in the backgrounds of the wild-type HIV-1AD8 Env and the State 1-stabilized Envs.
The noncovalent association of gp120 with detergent-solubilized Env trimers is prone to disruption, providing information about the stability of the Env complex. The gp120-trimer association was evaluated by a Ni-NTA precipitation assay (101). For this assay, HOS cells transiently expressing Envs with C-terminal His6 tags were lysed in buffers containing the Cymal-5 detergent. The clarified lysates were incubated with Ni-NTA beads, and the precipitated Envs as well as an “input” sample from the cell lysates obtained before incubation with the Ni-NTA beads were analyzed by Western blot. The results of the assays conducted on the Env variants at 25°C and 4°C are shown in Fig. 4A and B, respectively. Comparison of the gp120 and gp160 bands in the “input” samples indicates that the MPER changes did not detectably affect either the proteolytic processing or the subunit association of the Env variants. As previously observed (95), the processing of the 2-4 RM6 AE, Q114E/Q567K, and Q114E/Q567K/A582T Envs was at least as efficient as that of the wild-type HIV-1AD8 Env. The gp120-trimer association in detergent lysates is reflected in the gp120:gp160 ratio in the Ni-NTA precipitates compared with the gp120:gp160 ratio in the input samples. As previously observed (101), the gp120-trimer association of the wild-type HIV-1AD8 Env in detergent lysates was lower at 25°C than at 4°C (Fig. 4A and B). Also consistent with a previous study (95), the Env variants with the State 1-stabilizing modifications (2-4 RM6 AE, Q114E/Q567K, and Q114E/Q567K/A582T) displayed a stronger gp120-trimer association in detergent lysates than the wild-type HIV-1AD8 Env. The MPER changes did not significantly affect gp120-trimer association in any of the Env backgrounds. In other assays, the MPER changes in the wild-type HIV-1AD8 background did not detectably affect spontaneous gp120 shedding from cell-surface Envs or Env incorporation into virus particles (data not shown). These results indicate that the general structural integrity of the Env trimer is maintained in the MPER mutants.
FIG 4.
Effects of the MPER changes on gp120-trimer association in detergent. HOS cells transiently expressing Env glycoproteins tagged at the C terminus with His6 were lysed and incubated with Ni-NTA beads for 1 h at 25°C (A) or 4°C (B). Total cell lysates (Input) and Ni-NTA precipitates were Western blotted with a goat anti-gp120 antibody. The gp120-trimer association index for each Env variant was calculated from the gp120:gp160 ratio in the Ni-NTA precipitates divided by the gp120:gp160 ratio in the Input samples. The gp120-trimer association indices of the parental and MPER mutants Envs are shown in the bar graphs (right). The results shown are representative of those obtained in two independent experiments.
To examine more thoroughly the effects of the MPER alterations on the conformation of the functional Envs, we carried out virus neutralization assays using monoclonal antibodies and inhibitors targeting different gp120 and gp41 regions (Fig. 5). Poorly neutralizing antibodies directed against gp120 included F105 against the CD4 binding site (CD4bs) (102), 17b against a CD4-induced (CD4i) epitope (103), and 19b against the V3 region (94). The gp120-directed broadly neutralizing antibodies included VRC03 against the CD4bs (104), PG9, and PGT145 against quaternary V2 epitopes (105, 106) and PGT151 against the gp120-gp41 interface (107). Virus entry inhibitors directed against gp120 included soluble CD4-Ig (sCD4-Ig), the small-molecule CD4-mimetic compound (CD4mc) BNM-III-170 (96), and the State 1-preferring small molecule, BMS-806 (108). The gp41-directed ligands included the T20 peptide, which mimics the HR2 region and targets the HR1 coiled-coil (109), and the broadly neutralizing 10E8 antibody against the MPER (60, 68, 77).
FIG 5.
Phenotypes of viruses with Envs containing MPER changes. Pseudoviruses containing the indicated Env variants were tested for sensitivity to cold and to antibodies/compounds directed against gp120 or gp41. Poorly neutralizing antibodies and broadly neutralizing antibodies are indicated. Cold sensitivity is reported as the half-life of infectious virus after incubation on ice. The 50% inhibitory concentrations (IC50 values) of the Env ligands are reported in μg/mL except for BNM-III-170 (in μM) and BMS-806 (in nM). Some of the phenotypes were not determined (ND) due to the low infectivity of the viruses. Values are colored based on their fold increase in sensitivity (green) or resistance (red) compared to the relevant parental virus (labeled “None”). The parental and associated MPER mutant viruses were compared in parallel experiments. The results shown are means and standard deviations from two independent experiments. Additional experiments validated the relative IC50 values of the parental viruses for sCD4-Ig, BNM-III-170, BMS-806, or cold. The following mean IC50 values of T20 and 10E8.v4 were obtained in a side-by-side comparison of the four parental viruses (wild-type HIV-1AD8, 2-4 RM6 AE, Q114E/Q567K and Q114E/Q567K/A582T): 0.56, 0.64, 2.5 and 0.89 μg/mL of T20, respectively; 1.7, 0.11, 2.4 and 2.0 μg/mL of 10E8.v4, respectively. Note that the increased 10E8.v4 sensitivity of the parental 2–4 RM6 AE virus relative to the wild-type HIV-1AD8 likely results from some of the lysine changes in the 2–4 RM6 AE Env; viruses containing only the key State 1-stabilizing changes (Q114E/Q567K/A582T) do not exhibit increased 10E8.v4 sensitivity.
The phenotypes resulting from the MPER changes depended upon the Env background, as shown in Fig. 5 and summarized schematically in Fig. 6. In the background of the wild-type HIV-1AD8 Env, the MPER mutants were more sensitive to neutralization by the poorly neutralizing F105, 17b, and 19b antibodies, and by sCD4 and the CD4mc BNM-III-170. These MPER mutants also exhibited modest increases in sensitivity to T20 compared with the wild-type HIV-1AD8 parent. Along with increased cold sensitivity, these MPER mutant phenotypes are consistent with the destabilization of the pretriggered (State-1) conformation. Relative to wild-type HIV-1AD8, the MPER mutants were significantly more resistant to the broadly neutralizing PGT151 antibody. Some MPER mutants also exhibited slight resistance to VRC03 and BMS-806, two State-1 preferring Env ligands (13, 110–112). No significant changes were detected in the sensitivity of the MPER mutants to neutralization by the PG9 and PGT145 antibodies against V2 quaternary epitopes. The MPER mutants were more sensitive to neutralization by the 10E8 antibody, except for W672A. The W672A change affects the MPER epitope for this antibody (77). In summary, multiple single-residue changes in the MPER of the wild-type HIV-1AD8 Env led to phenotypes indicative of global disruption of the pretriggered (State-1) conformation (Fig. 6).
FIG 6.
Summary of the phenotypic effects of the MPER and State 1-stabilizing changes on virus sensitivity to Env ligands or cold exposure. The overall phenotypic effects of MPER changes and State 1-stabilizing changes in Env on virus sensitivity to Env ligands and cold exposure (from Fig. 5 and additional side-by-side experiments comparing the four parental Envs) are summarized here. On a schematic view of the HIV-1 Env trimer, the approximately positioned ligands are colored according to increased sensitivity (green) or increased resistance (red). In each case, the phenotypes of the Env variant following the arrow are compared with those of the Env variant preceding the arrow. For example, the phenotypes of wild-type HIV-1AD8 Envs without and with MPER changes can be compared in (A and B), respectively. For the depictions of the State 1-stabilized Envs in C, D and E, the data from the Q114E/Q567K and Q114E/Q567K/A582T Envs were used, to avoid potential effects of the other changes in the 2-4 RM6 AE Env unrelated to State-1 stabilization. Examination of (B and D) allows a comparison of the effects of the MPER changes in the context of the wild-type HIV-1AD8 Env and the State 1-stabilized Envs, respectively. The phenotypes of the wild-type HIV-1AD8 Env, the HIV-1AD8 Env with MPER changes, and the State 1-stabilized Envs with MPER changes can be compared between in (A, B and E), respectively.
As expected (95), compared to the wild-type HIV-1AD8, viruses with the 2-4 RM6 AE, Q114E/Q567K, and Q114E/Q567K/A582T Envs were significantly resistant to cold, sCD4-Ig, and BNM-III-170 (Fig. 5), phenotypes consistent with State-1 stabilization. The IC50 values of the other Env ligands tested are not directly comparable for these four parental Envs (rows labeled “None” in Fig. 5) because they were not tested in parallel. However, the relative sensitivity of viruses pseudotyped with the wild-type HIV-1AD8, Q114E/Q567K, and Q114E/Q567K/A582T Envs to these other ligands was evaluated in separate assays and found not to differ significantly, as summarized in Fig. 6A and C. The relative resistance of the Q114E/Q567K and Q114E/Q567K/A582T pseudoviruses specifically to sCD4-Ig, CD4mc, and cold is indicative of the higher activation barrier of these Envs to CD4-induced transitions from State 1 to downstream conformations. When the MPER changes were introduced into these Env backgrounds, two major differences in the resulting viral phenotypes were observed compared to those seen in the wild-type HIV-1AD8 background. First, the MPER mutants remained resistant to poorly neutralizing antibodies. Second, relative to the parent viruses, the MPER mutants generally did not display resistance to PGT151, VRC03, or BMS-806 (Compare Fig. 6C and D). In all Env backgrounds, relative to the parent virus, alteration of the MPER led to enhanced viral sensitivity to cold, sCD4-Ig, BNM-III-170, T20, and 10E8 (Compare Fig. 6A and B; Fig. 6C and D). In contrast to the globally “opened” Env resulting from the introduction of MPER changes into the wild-type HIV-1AD8 background (Fig. 6B), viruses with both MPER and State 1-stabilizing Env changes differ minimally from wild-type HIV-1AD8 (Compare Fig. 6A and E). The viruses with combined MPER and State 1-stabilizing Env changes displayed an increased sensitivity to 10E8 neutralization, possibly reflecting local effects on the MPER, but exhibited none of the phenotypes associated with open, State 1-destabilized Envs.
The above findings suggest that Envs with MPER alterations are more prone to make transitions from the pretriggered (State-1) conformation to more open downstream conformations. We evaluated whether infection by wild-type and MPER mutant viruses was activated with different levels of efficiency by sCD4-Ig and the CD4mc, BNM-III-170. In this assay, viruses were spinoculated on CD4-negative, CCR5-expressing Cf2Th-CCR5 cells, after which sCD4-Ig or BNM-III-170 was added. Forty-eight hours later, virus infection was measured. In the absence of sCD4-Ig or BNM-III-170, a very low background was observed in this assay. Wild-type HIV-1AD8 infection of the Cf2Th-CCR5 cells was significantly enhanced by sCD4-Ig and BNM-III-170 (Fig. 7A). Infection by the T676A mutant virus was activated at lower doses of sCD4-Ig and BNM-III-170 than wild-type HIV-1AD8 infection. Infection by the other MPER mutant viruses was not detectably enhanced by sCD4-Ig or BNM-III-170, perhaps due to their intrinsically low infectivity. Infection by viruses with the 2-4 RM6 AE Env was activated only at the highest BNM-III-170 concentration tested, consistent with the lower expected triggerability of this State 1-stabilized variant (95). Infection by viruses with MPER-altered 2-4 RM6 AE Envs was not detectably activated by either sCD4-Ig or BNM-III-170. These observations are consistent with the T676A change in the MPER increasing the responsiveness of the HIV-1AD8 Env to CD4 and a CD4mc.
FIG 7.
CD4- and CD4mc-mediated activation of infection of CD4-negative cells by HIV-1 Env variants. Wild-type HIV-1AD8 (A) and 2-4 RM6 AE viruses (B) containing the indicated MPER changes were spinoculated on CD4-negative, CCR5-expressing Cf2Th-CCR5 cells, after which serial dilutions of sCD4-Ig or BNM-III-170, a CD4-mimetic compound, were added. Note that a lower range of sCD4-Ig and BNM-III-170 was used in the bottom section of (A). Forty-eight hours later, the cells were lysed, and luciferase activity was measured. The results shown are representative of those obtained in two independent experiments, expressed as means and standard deviations from triplicate luciferase readings.
Effect of membrane cholesterol on HIV-1 Env phenotypes.
The MPER contains a conserved cholesterol recognition motif spanning residues 679 to 683 (80–83). Changes in this motif can result in viruses with impaired replicative ability and increased sensitivity to cold, poorly neutralizing antibodies and CD4mc (84, 87, 88, 92). Depletion of cholesterol from the virus particles of certain HIV-1 strains produces similar effects (92, 113–118). We asked whether changes that stabilize the State-1 conformation would alter virus susceptibility to decreases in membrane cholesterol. Methyl-β-cyclodextrin (MBCD) was used to deplete cholesterol from the viral membrane (119). The wild-type HIV-1AD8, Q114E/Q567K, Q114E/Q567K/A582T, and 2-4 RM6 AE viruses were all inhibited comparably by MBCD (unpublished data). These results indicate that stabilization of the pretriggered (State-1) Env conformation does not bypass or diminish the requirement of HIV-1 for cholesterol in the membrane.
Next, we asked whether cholesterol depletion would result in State 1-destabilized phenotypes similar to those resulting from MPER changes. The viruses were incubated with MBCD at a 50% inhibitory concentration for 1 h and then tested for sensitivity to neutralization by antibodies and BNM-III-170. As seen in Fig. 8, no difference was observed in the sensitivity of wild-type HIV-1AD8, Q114E/Q567K, Q114E/Q567K/A582T, and 2-4 RM6 AE viruses to poorly neutralizing antibodies (19b, 17b, F105), the broadly neutralizing antibody PGT151, or the CD4mc BNM-III-170. However, the MBCD-treated viruses were slightly more sensitive than the untreated control viruses to neutralization by 10E8.v4, an optimized version of the 10E8 antibody against the MPER (120). This observation verifies that viral components near or within the membrane were affected by MBCD treatment. Some local effects of cholesterol removal from the viral membrane may resemble those resulting from MPER changes, which also result in increased sensitivity to 10E8 antibody neutralization (Fig. 5 and 6). However, the more global effects of MPER changes on HIV-1 Env conformation are distinct from the effects of cholesterol removal from the viral membrane.
FIG 8.
Effects of cholesterol depletion on virus sensitivity to neutralization. The wild-type HIV-1AD8 (A), 2-4 RM6 AE (B), Q114E/Q567K (C), and Q114E/Q567K/A582T (D) viruses were incubated with methyl-β-cyclodextrin (MBCD) at the predetermined IC50 concentration for each virus for 1 h at 37°C. Then the indicated antibodies or BNM-III-170 were added and the mixture was incubated for another hour. The mixture was added to TZM-bl target cells and 48 h later, luciferase activity was measured. For each Env variant, the results were normalized to those obtained for viruses that were not incubated with MBCD, antibodies, or BNM-III-170. The results shown are representative of those obtained in two independent experiments, expressed as means and standard deviations from triplicate luciferase readings. The significance of the difference in virus sensitivity to 10E8.v4 neutralization with and without MBCD was evaluated by a Student’s paired t test.
DISCUSSION
Changes introduced into several highly conserved amino acid residues in the HIV-1 gp41 MPER resulted in remarkably similar viral phenotypes. This observation suggests that each of these changes exerted a significant impact on MPER integrity. Only the T676A mutant, affecting a residue that naturally tolerates Thr/Ser polymorphism (93), exhibited phenotypes intermediate between those of the parental viruses and the other mutants. Despite efficient processing and subunit association, the MPER mutants exhibited modest decreases in infectivity, in keeping with the role proposed for this gp41 region in membrane fusion and virus entry (78–85). Our results highlight another key role played by the gp41 MPER, namely, the regulation of the conformation of the HIV-1 Env ectodomain. MPER alteration led to both local and distant conformational effects. In gp41, MPER changes resulted in enhanced sensitivity to the 10E8 antibody against the MPER and the T20 peptide against HR1 (Fig. 6), which has been previously observed (121). The increased sensitivity of some MPER mutants to other MPER-directed antibodies, 2F5 and 4E10, has been suggested to result from enhanced/prolonged exposure of the MPER on virus particles (87–89, 122, 123). At a distance, in gp120, the MPER changes resulted in increased sensitivity to sCD4-Ig, a CD4-mimetic compound, BNM-III-170, and several poorly neutralizing antibodies. This pattern of enhanced virus sensitivity to ligands that prefer State-2/3 Env conformations along with increased virus inactivation in the cold is strongly associated with destabilization of the pretriggered (State-1) Env conformation (14, 86–92, 95, 110, 124–138). Infection of CD4-negative, CCR5-expressing cells by the T676A mutant, which retains the highest level of infectivity among the MPER mutants, was also more easily activated by sCD4-Ig and BNM-III-170 than the wild-type HIV-1AD8. Together, these findings support the hypothesis that the MPER changes destabilize State 1, predisposing Env to assume downstream conformations.
To test this hypothesis, we examined whether the viral phenotypes associated with the MPER changes would be mitigated in the context of State 1-stabilized Envs. Viruses with State 1-stabilized Envs retained the resistance to the poorly neutralizing antibodies seen for the Tier-2 parental virus, HIV-1AD8, but were relatively resistant to CD4-Ig, the CD4mc BNM-III-170, and exposure to cold Fig. 6A and C) (95). Remarkably, in the context of State 1-stabilized Envs, the multiple phenotypic effects of MPER changes on wild-type HIV-1AD8 were nullified, except for increased sensitivity to the 10E8 antibody against the MPER (Compare Fig. 6B and E). The State 1-destabilizing effects of the MPER changes can be balanced by the State 1-stabilizing changes present in the 2-4 RM6 AE, Q114E/Q567K, and Q114E/Q567K/A582T Envs. This mechanism may underlie the HIV-1 strain dependence of the phenotypes observed for some MPER changes (89, 92), predicting lower penetrance (less apparent phenotypes) in the context of Envs with lower triggerability (more stabilized State-1 conformations). For example, the introduction of the L669S change into the Tier-2 HIV-1AD8 Env in our study generated a virus that exhibited increased sensitivity to multiple poorly neutralizing antibodies (Fig. 6A and B). In contrast, the same L669S change in a Tier-3 HIV-1253-11 Env resulted in more than 10-fold increases in sensitivity to the MPER antibodies 2F5, 4E10, and 10E8, but no differences in sensitivity to the F105, 17b, and PGT151 antibodies (89). In the latter instance, the phenotypic effects of the L669S MPER change may have been offset by the low triggerability and strong State-1 stability of the Tier-3 HIV-1 Env.
The mechanism by which the MPER contributes to the stability of the State-1 Env conformation is unknown, but potentially involves direct interactions between the MPER and the viral membrane and/or other gp41 elements, as well as indirect effects on gp120. Based on studies of MPER peptides, the MPER may be partly submerged in the lipid environment of the viral membrane (58–64). Such membrane interactions could stabilize the position and orientation of the Env spike, influencing trimer symmetry and packing of the Env protomers. Changes in MPER conformation may be allosterically coupled to entry-related conformational changes in the rest of the Env ectodomain. Indeed, destabilization of the pretriggered (State-1) Env conformation by CD4 binding or alteration of “restraining residues” in gp120 can lead to increased MPER exposure and enhanced HIV-1 sensitivity to neutralization by MPER-directed antibodies (72–77, 110, 133). Such allosteric coupling between gp120 and gp41 conformations has been suggested by smFRET studies (111) and helps explain the observed synergy between gp120 and gp41 changes in allowing HIV-1 to become CD4-independent (86).
The effects of cholesterol removal from the viral membrane mediated by MBCD differed from the effects of MPER changes. As expected (113–118), MBCD treatment resulted in a decrease in HIV-1 infectivity. However, the apparent requirement of HIV-1 infectivity for membrane cholesterol was not bypassed or diminished by the State 1-stabilizing Env modifications. At MBCD concentrations that reduced HIV-1 infectivity by 50%, global effects on virus sensitivity to neutralizing antibodies were not detected. However, increased sensitivity to 10E8 antibody neutralization was observed, suggesting that cholesterol removal from the viral membrane can increase the efficacy of an anti-MPER antibody. It has been proposed that the 10E8 antibody extracts its peptide epitope from the viral membrane, a process that may be facilitated in a membrane lacking cholesterol (60, 62, 63, 68, 71). In summary, although local effects of MBCD on the MPER were observed, the global effects of MPER changes on HIV-1 Env conformation are distinct from the effects of cholesterol removal from the viral membranes.
The sensitivity of the State-1 HIV-1 Env conformation to MPER disruption has implications for structural and vaccine studies. In most preparations of soluble or detergent-solubilized HIV-1 Env trimers, the MPER has been deleted or is disordered (24, 25, 47–53, 107). Single-molecule FRET studies indicate that the State-1 conformation is not well represented in these Env preparations (111, 139). Thus, detailed information about the State-1 Env conformation, the major target for broadly neutralizing antibodies (13–15, 18, 111, 112), is currently unavailable. Remedying this knowledge gap may require approaches to maintain MPER integrity in Env preparations or minimizing the impact of MPER disruption on Env conformation. The State 1-stabilized Env designs described here offset many of the effects of MPER changes on functional Env conformation. These Env variants could assist efforts to characterize the elusive State-1 conformation and to maintain this native conformation in vaccine immunogens.
MATERIALS AND METHODS
Cell lines.
HEK293T, TZM-bl and HOS (ATCC) were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 μg/mL penicillin-streptomycin (Life Technologies). CD4 negative, CCR5-expressing Cf2Th-CCR5 cells were cultured in the same medium supplemented with 400 μg/mL G418. Expi293 cells (Invitrogen) were maintained in suspension culture in Expi293TM Expression Medium, supplemented with 50 units/mL penicillin and 50 μg/mL streptomycin.
Antibodies and sCD4-Ig.
Poorly neutralizing antibodies (F105, 17b and 19b) and broadly neutralizing antibodies (VRC03, PG9, PGT145, PGT151, 10E8 and 10E8.v4) against the HIV-1 Env were used in this study. Antibodies were produced by cotransfecting paired heavy- and light-chain antibody-expressing plasmids at a ratio of 1:1 using FectoPRO DNA transfection reagent (VWR) into Expi293 cells. Antibodies were purified from the cell medium 5 days after transfection by affinity chromatography using protein A columns (Thermo Fisher Scientific). Soluble CD4-Ig (sCD4-Ig) encoding the first two domains of human CD4 fused to an antibody Fc was expressed and purified as described above for the antibodies.
Plasmids.
The wild-type HIV-1AD8, 2-4 RM6 AE, Q114E/Q567K, and Q114E/Q567K/A582T Envs were cloned into the pSVIIIenv expression plasmid, using the Kpn I and BamHI sites, as described previously (95). The signal peptide/N terminus (residues 1–33) and the cytoplasmic tail C terminus (residues 751 to 856) of these Envs are derived from the HIV-1HXBc2 Env (86). All the Envs contain a His6 tag (GGHHHHHH or GGGHHHHHH) at the carboxyl terminus. The MPER changes (L669S, W672A, I675A, T676A, and L679A) were introduced using the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies). All the mutations were confirmed by DNA sequencing. Antibody-expressing plasmids were obtained from Dennis Burton (Scripps), Peter Kwong and John Mascola (NIH Vaccine Research Center), Barton Haynes (Duke University), Hermann Katinger (Polymun), James Robinson (Tulane University), Marshall Posner (Mount Sinai Medical Center), David Ho (Columbia University Vagelos College of Physicians and Surgeons), and the NIH HIV Reagent Program.
Virus infectivity and cold sensitivity.
Pseudoviruses bearing the wild-type or variant HIV-1 Envs were produced by cotransfecting the Env-expressing pSVIIIenv plasmid and the luciferase-encoding pNL4-3.Luc.R-E-vector (NIH HIV Reagent Program) at a 1:1 ratio into HEK293T cells using polyethyleneimine (Polysciences). The cell supernatants containing pseudoviruses were harvested 48 h later, clarified by low-speed centrifugation (2000 rpm for 10 min), aliquoted, and either used directly to measure virus infectivity or stored at −80°C until use. To compare the infectivity of the Env variants, freshly prepared pseudoviruses were serially diluted and incubated with TZM-bl cells in the presence of 20 μg/mL DEAE-dextran. TZM-bl cells were supplied by John C. Kappes and Xiaoyun Wu (University of Alabama at Birmingham) through the NIH HIV Reagent Program. Forty-eight hours later, the luciferase activity in TZM-bl cells was measured using a luminometer.
Previously frozen pseudoviruses were used to evaluate the sensitivity of the Env variants to cold inactivation, antibody neutralization, and inhibition by virus entry blockers. For these assays, similar TCID50 (50% tissue culture infectious dose) levels of the pseudoviruses were incubated with TZM-bl cells in the presence of 20 μg/mL DEAE-dextran. To evaluate the cold sensitivity of viruses with different Envs, pseudoviruses were incubated on ice for different lengths of time and virus infectivity was subsequently measured, as described above.
Virus neutralization assay.
Approximately 100 to 200 TCID50 of pseudovirus was incubated with serial dilutions of purified antibodies, sCD4-Ig, T20, BNM-III-170, or BMS-806 in triplicate wells of 96-well plates at 37°C for 1 h. Then approximately 2 × 104 TZM-bl cells in 20 μg/mL DEAE-dextran in the medium were added to each well and the mixture was incubated at 37°C/5%CO2 for 48 to 72 h, after which luciferase activity was measured. The concentrations of antibodies and other inhibitors that inhibit 50% of infection (the IC50 values) were determined by fitting the data in five-parameter dose-response curves using GraphPad Prism 8.
Env expression and gp120-trimer association.
HOS cells in 6-well plates were transfected with plasmids encoding His6-tagged Env variants and Tat at a ratio of 8:1, using the Effectene transfection reagent (Qiagen) according to the manufacturer’s instructions. Forty-eight hours after transfection, cells were lysed in buffer containing 1.5% Cymal-5. Clarified lysates were prepared, and a portion was saved as the “Input” sample. The remainder of the clarified lysate was incubated with Ni-NTA beads at room temperature or 4°C for 1 h. The beads were washed, boiled, and analyzed by Western blotting with 1:2,000 goat anti-gp120 antibody (Invitrogen) and 1:3,000 HRP-conjugated rabbit anti-goat antibody (Invitrogen). The intensity of the gp120 and gp160 bands on the Western blot was quantified using the Bio-Rad Image Lab program. The gp120-trimer association index was calculated by dividing the gp120:gp160 ratio in the Ni-NTA precipitates by the gp120:gp160 ratio in the Input samples.
Cholesterol depletion.
To determine the IC50 of methyl-β-cyclodextrin (MBCD), approximately 100 to 200 TCID50 of pseudovirus was incubated with serial dilutions of MBCD at 37°C for 1 h. TZM-bl cells were then added and luciferase activity was measured 48 h later. The IC50 value of MBCD for each virus was then determined using GraphPad Prism 8, as described above. To determine the sensitivity of cholesterol-depleted viruses to antibodies and BNM-III-170, pseudoviruses were treated with MBCD at the IC50 for 1 h at 37°C, after which the neutralization assay was carried out as described above.
Activation of virus infection by sCD4-Ig and a CD4-mimetic compound, BNM-III-170.
Pseudoviruses were incubated with CD4-negative, CCR5-expressing Cf2Th-CCR5 cells in 96-well plates. The plates were centrifuged at 1,800 rpm for 30 min at 21°C. Medium containing serial dilutions of sCD4-Ig or BNM-III-170 was then added. Forty-eight hours later, the luciferase activity was measured.
ACKNOWLEDGMENTS
We thank Elizabeth Carpelan for her help with manuscript preparation.
This work was supported by grants from the National Institutes of Health (AI145547, AI124982, and AI150471) and by a gift from the late William F. McCarty-Cooper.
We declare no conflict of interest.
Contributor Information
Hanh T. Nguyen, Email: hanht_nguyen@dfci.harvard.edu.
Joseph G. Sodroski, Email: joseph_sodroski@dfci.harvard.edu.
Viviana Simon, Icahn School of Medicine at Mount Sinai.
REFERENCES
- 1.Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888. 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 2.Harrison SC. 2005. Mechanism of membrane fusion by viral envelope proteins. Adv Virus Res 64:231–261. 10.1016/S0065-3527(05)64007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robey WG, Safai B, Oroszlan S, Arthur LO, Gonda MA, Gallo RC, Fischinger PJ. 1985. Characterization of envelope and core structural gene products of HTLV-III with sera from AIDS patients. Science 228:593–595. 10.1126/science.2984774. [DOI] [PubMed] [Google Scholar]
- 4.Allan JS, Coligan JE, Barin F, McLane MF, Sodroski JG, Rosen CA, Haseltine WA, Lee TH, Essex M. 1985. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science 228:1091–1094. 10.1126/science.2986290. [DOI] [PubMed] [Google Scholar]
- 5.Willey RL, Bonifacino JS, Potts BJ, Martin MA, Klausner RD. 1988. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc Natl Acad Sci USA 85:9580–9584. 10.1073/pnas.85.24.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Luo L, Thomas DY, Kang CY. 2000. The HIV-1 Env protein signal sequence retards its cleavage and down-regulates the glycoprotein folding. Virology 272:417–428. 10.1006/viro.2000.0357. [DOI] [PubMed] [Google Scholar]
- 7.Earl PL, Moss B, Doms RW. 1991. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J Virol 65:2047–2055. 10.1128/JVI.65.4.2047-2055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang S, Nguyen HT, Ding H, Wang J, Zou S, Liu L, Guha D, Gabuzda D, Ho DD, Kappes JC, Sodroski JG. 2021. Dual pathways of human immunodeficiency virus (HIV-1) envelope glycoprotein trafficking modulate the selective exclusion of uncleaved trimers from virions. J Virol 95:e01369-20. 10.1128/JVI.01369-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doores KJ, Bonomelli C, Harvey DJ, Vasiljevic S, Dwek RA, Burton DR, Crispin M, Scanlan CN. 2010. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc Natl Acad Sci USA 107:13800–13805. 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Go EP, Herschhorn A, Gu C, Castillo-Menendez L, Zhang S, Mao Y, Chen H, Ding H, Wakefield JK, Hua D, Liao H-X, Kappes JC, Sodroski J, Desaire H. 2015. Comparative analysis of the glycosylation profiles of membrane-anchored HIV-1 envelope glycoprotein trimers and soluble gp140. J Virol 89:8245–8257. 10.1128/JVI.00628-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merkle RK, Helland DE, Welles JL, Shilatifard A, Haseltine WA, Cummings RD. 1991. gp160 of HIV-I synthesized by persistently infected Molt-3 cells is terminally glycosylated: evidence that cleavage of gp160 occurs subsequent to oligosaccharide processing. Arch Biochem Biophys 290:248–257. 10.1016/0003-9861(91)90616-q. [DOI] [PubMed] [Google Scholar]
- 12.Brakch N, Dettin M, Scarinci C, Seidah NG, Di Bello C. 1995. Structural investigation and kinetic characterization of potential cleavage sites of HIV GP160 by human furin and PC1. Biochem Biophys Res Commun 213:356–361. 10.1006/bbrc.1995.2137. [DOI] [PubMed] [Google Scholar]
- 13.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB, Kwong PD, Blanchard SC, Mothes W. 2014. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346:759–763. 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haim H, Salas I, McGee K, Eichelberger N, Winter E, Pacheco B, Sodroski J. 2013. Modeling virus- and antibody-specific factors to predict human immunodeficiency virus neutralization efficiency. Cell Host Microbe 14:547–558. 10.1016/j.chom.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678–682. 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 16.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312. 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 17.Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med 201:1407–1419. 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guttman M, Cupo A, Julien JP, Sanders RW, Wilson IA, Moore JP, Lee KK. 2015. Antibody potency relates to the ability to recognize the closed, pre-fusion form of HIV Env. Nat Commun 6:6144. 10.1038/ncomms7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman JC, Montagnier L. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767–768. 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 20.Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763–767. 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113. 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma X, Lu M, Gorman J, Terry DS, Hong X, Zhou Z, Zhao H, Altman RB, Arthos J, Blanchard SC, Kwong PD, Munro JB, Mothes W. 2018. HIV-1 Env trimer opens through an asymmetric intermediate in which individual protomers adopt distinct conformations. Elife 7:e34271. 10.7554/eLife.34271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khasnis MD, Halkidis K, Bhardwaj A, Root MJ. 2016. Receptor activation of HIV-1 Env leads to asymmetric exposure of the gp41 trimer. PLoS Pathog 12:e1006098. 10.1371/journal.ppat.1006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Cohen AA, Galimidi RP, Gristick HB, Jensen GJ, Bjorkman PJ. 2016. Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop. Proc Natl Acad Sci USA 113:E7151–E7158. 10.1073/pnas.1615939113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozorowski G, Pallesen J, de Val N, Lyumkis D, Cottrell CA, Torres JL, Copps J, Stanfield RL, Cupo A, Pugach P, Moore JP, Wilson IA, Ward AB. 2017. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature 547:360–363. 10.1038/nature23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furuta RA, Wild CT, Weng Y, Weiss CD. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol 5:276–279. 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 27.Koshiba T, Chan DC. 2003. The prefusogenic intermediate of HIV-1 gp41 contains exposed C-peptide regions. J Biol Chem 278:7573–7579. 10.1074/jbc.M211154200. [DOI] [PubMed] [Google Scholar]
- 28.He Y, Vassell R, Zaitseva M, Nguyen N, Yang Z, Weng Y, Weiss CD. 2003. Peptides trap the human immunodeficiency virus type 1 envelope glycoprotein fusion intermediate at two sites. J Virol 77:1666–1671. 10.1128/jvi.77.3.1666-1671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. 1995. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science 270:1811–1815. 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 30.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661–666. 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 31.Feng Y, Broder CC, Kennedy PE, Berger EA. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872–877. 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 32.Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955–1958. 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 33.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135–1148. 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 34.Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, Peiper SC, Parmentier M, Collman RG, Doms RW. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149–1158. 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 35.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667–673. 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 36.Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179–183. 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 37.Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184–187. 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 38.Chan DC, Fass D, Berger JM, Kim PS. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263–273. 10.1016/S0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 39.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426–430. 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 40.Lu M, Blacklow SC, Kim PS. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol 2:1075–1082. 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 41.Melikyan GB, Markosyan RM, Hemmati H, Delmedico MK, Lambert DM, Cohen FS. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J Cell Biol 151:413–423. 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilen CB, Tilton JC, Doms RW. 2012. Molecular mechanisms of HIV entry. Adv Exp Med Biol 726:223–242. 10.1007/978-1-4614-0980-9_10. [DOI] [PubMed] [Google Scholar]
- 43.Weiss CD. 2003. HIV-1 gp41: mediator of fusion and target for inhibition. AIDS Rev 5:214–221. [PubMed] [Google Scholar]
- 44.Gabuzda D, Olshevsky U, Bertani P, Haseltine WA, Sodroski J. 1991. Identification of membrane anchorage domains of the HIV-1 gp160 envelope glycoprotein precursor. J Acquir Immune Defic Syndr (1988) 4:34–40. [PubMed] [Google Scholar]
- 45.Nguyen HT, Madani N, Ding H, Elder E, Princiotto A, Gu C, Darby P, Alin J, Herschhorn A, Kappes JC, Mao Y, Sodroski JG. 2017. Evaluation of the contribution of the transmembrane region to the ectodomain conformation of the human immunodeficiency virus (HIV-1) envelope glycoprotein. Virol J 14:33. 10.1186/s12985-017-0704-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White JM, Delos SE, Brecher M, Schornberg K. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol 43:189–219. 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. 2014. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514:455–461. 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JH, Ozorowski G, Ward AB. 2016. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351:1043–1048. 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–1490. 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bartesaghi A, Merk A, Borgnia MJ, Milne JL, Subramaniam S. 2013. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol 20:1352–1357. 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torrents de la Pena A, Rantalainen K, Cottrell CA, Allen JD, van Gils MJ, Torres JL, Crispin M, Sanders RW, Ward AB. 2019. Similarities and differences between native HIV-1 envelope glycoprotein trimers and stabilized soluble trimer mimetics. PLoS Pathog 15:e1007920. 10.1371/journal.ppat.1007920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pan J, Peng H, Chen B, Harrison SC. 2020. Cryo-EM structure of full-length HIV-1 Env bound with the Fab of antibody PG16. J Mol Biol 432:1158–1168. 10.1016/j.jmb.2019.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zanetti G, Briggs JA, Grunewald K, Sattentau QJ, Fuller SD. 2006. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog 2:e83. 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu P, Liu J, Bess J, Jr., Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. 2006. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441:847–852. 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 56.Li Z, Li W, Lu M, Bess J, Jr., Chao CW, Gorman J, Terry DS, Zhang B, Zhou T, Blanchard SC, Kwong PD, Lifson JD, Mothes W, Liu J. 2020. Subnanometer structures of HIV-1 envelope trimers on aldrithiol-2-inactivated virus particles. Nat Struct Mol Biol 27:726–734. 10.1038/s41594-020-0452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.White TA, Bartesaghi A, Borgnia MJ, Meyerson JR, de la Cruz MJ, Bess JW, Nandwani R, Hoxie JA, Lifson JD, Milne JL, Subramaniam S. 2010. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog 6:e1001249. 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schibli DJ, Montelaro RC, Vogel HJ. 2001. The membrane-proximal tryptophan-rich region of the HIV glycoprotein, gp41, forms a well-defined helix in dodecylphosphocholine micelles. Biochemistry 40:9570–9578. 10.1021/bi010640u. [DOI] [PubMed] [Google Scholar]
- 59.Suarez T, Nir S, Goni FM, Saez-Cirion A, Nieva JL. 2000. The pre-transmembrane region of the human immunodeficiency virus type-1 glycoprotein: a novel fusogenic sequence. FEBS Lett 477:145–149. 10.1016/s0014-5793(00)01785-3. [DOI] [PubMed] [Google Scholar]
- 60.Sun ZY, Oh KJ, Kim M, Yu J, Brusic V, Song L, Qiao Z, Wang JH, Wagner G, Reinherz EL. 2008. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity 28:52–63. 10.1016/j.immuni.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 61.Fu Q, Shaik MM, Cai Y, Ghantous F, Piai A, Peng H, Rits-Volloch S, Liu Z, Harrison SC, Seaman MS, Chen B, Chou JJ. 2018. Structure of the membrane proximal external region of HIV-1 envelope glycoprotein. Proc Natl Acad Sci USA 115:E8892–E8899. 10.1073/pnas.1807259115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kwon B, Lee M, Waring AJ, Hong M. 2018. Oligomeric structure and three-dimensional fold of the HIV gp41 membrane-proximal external region and transmembrane domain in phospholipid bilayers. J Am Chem Soc 140:8246–8259. 10.1021/jacs.8b04010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y, Kaur P, Sun ZJ, Elbahnasawy MA, Hayati Z, Qiao ZS, Bui NN, Chile C, Nasr ML, Wagner G, Wang JH, Song L, Reinherz EL, Kim M. 2019. Topological analysis of the gp41 MPER on lipid bilayers relevant to the metastable HIV-1 envelope prefusion state. Proc Natl Acad Sci USA 116:22556–22566. 10.1073/pnas.1912427116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carravilla P, Chojnacki J, Rujas E, Insausti S, Largo E, Waithe D, Apellaniz B, Sicard T, Julien JP, Eggeling C, Nieva JL. 2019. Molecular recognition of the native HIV-1 MPER revealed by STED microscopy of single virions. Nat Commun 10:78. 10.1038/s41467-018-07962-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu H, Su X, Si L, Lu L, Jiang S. 2018. The development of HIV vaccines targeting gp41 membrane-proximal external region (MPER): challenges and prospects. Protein Cell 9:596–615. 10.1007/s13238-018-0534-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ofek G, Tang M, Sambor A, Katinger H, Mascola JR, Wyatt R, Kwong PD. 2004. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol 78:10724–10737. 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Irimia A, Sarkar A, Stanfield RL, Wilson IA. 2016. Crystallographic identification of lipid as an integral component of the epitope of HIV broadly neutralizing antibody 4E10. Immunity 44:21–31. 10.1016/j.immuni.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Irimia A, Serra AM, Sarkar A, Jacak R, Kalyuzhniy O, Sok D, Saye-Francisco KL, Schiffner T, Tingle R, Kubitz M, Adachi Y, Stanfield RL, Deller MC, Burton DR, Schief WR, Wilson IA. 2017. Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: insights for vaccine and therapeutic design. PLoS Pathog 13:e1006212. 10.1371/journal.ppat.1006212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanchez-Martinez S, Lorizate M, Hermann K, Kunert R, Basanez G, Nieva JL. 2006. Specific phospholipid recognition by human immunodeficiency virus type-1 neutralizing anti-gp41 2F5 antibody. FEBS Lett 580:2395–2399. 10.1016/j.febslet.2006.03.067. [DOI] [PubMed] [Google Scholar]
- 70.Lorizate M, Gomara MJ, de la Torre BG, Andreu D, Nieva JL. 2006. Membrane-transferring sequences of the HIV-1 Gp41 ectodomain assemble into an immunogenic complex. J Mol Biol 360:45–55. 10.1016/j.jmb.2006.04.056. [DOI] [PubMed] [Google Scholar]
- 71.Rantalainen K, Berndsen ZT, Antanasijevic A, Schiffner T, Zhang X, Lee WH, Torres JL, Zhang L, Irimia A, Copps J, Zhou KH, Kwon YD, Law WH, Schramm CA, Verardi R, Krebs SJ, Kwong PD, Doria-Rose NA, Wilson IA, Zwick MB, Yates JR, 3rd, Schief WR, Ward AB. 2020. HIV-1 envelope and MPER antibody structures in lipid assemblies. Cell Rep 31:107583. 10.1016/j.celrep.2020.107583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dimitrov AS, Jacobs A, Finnegan CM, Stiegler G, Katinger H, Blumenthal R. 2007. Exposure of the membrane-proximal external region of HIV-1 gp41 in the course of HIV-1 envelope glycoprotein-mediated fusion. Biochemistry 46:1398–1401. 10.1021/bi062245f. [DOI] [PubMed] [Google Scholar]
- 73.de Rosny E, Vassell R, Jiang S, Kunert R, Weiss CD. 2004. Binding of the 2F5 monoclonal antibody to native and fusion-intermediate forms of human immunodeficiency virus type 1 gp41: implications for fusion-inducing conformational changes. J Virol 78:2627–2631. 10.1128/jvi.78.5.2627-2631.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rathinakumar R, Dutta M, Zhu P, Johnson WE, Roux KH. 2012. Binding of anti-membrane-proximal gp41 monoclonal antibodies to CD4-liganded and -unliganded human immunodeficiency virus type 1 and simian immunodeficiency virus virions. J Virol 86:1820–1831. 10.1128/JVI.05489-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harris AK, Bartesaghi A, Milne JL, Subramaniam S. 2013. HIV-1 envelope glycoprotein trimers display open quaternary conformation when bound to the gp41 membrane-proximal external-region-directed broadly neutralizing antibody Z13e1. J Virol 87:7191–7196. 10.1128/JVI.03284-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chakrabarti BK, Walker LM, Guenaga JF, Ghobbeh A, Poignard P, Burton DR, Wyatt RT. 2011. Direct antibody access to the HIV-1 membrane-proximal external region positively correlates with neutralization sensitivity. J Virol 85:8217–8226. 10.1128/JVI.00756-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. 2012. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491:406–412. 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Salzwedel K, West JT, Hunter E. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J Virol 73:2469–2480. 10.1128/JVI.73.3.2469-2480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Munoz-Barroso I, Salzwedel K, Hunter E, Blumenthal R. 1999. Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J Virol 73:6089–6092. 10.1128/JVI.73.7.6089-6092.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vincent N, Genin C, Malvoisin E. 2002. Identification of a conserved domain of the HIV-1 transmembrane protein gp41 which interacts with cholesteryl groups. Biochim Biophys Acta 1567:157–164. 10.1016/s0005-2736(02)00611-9. [DOI] [PubMed] [Google Scholar]
- 81.Greenwood AI, Pan J, Mills TT, Nagle JF, Epand RM, Tristram-Nagle S. 2008. CRAC motif peptide of the HIV-1 gp41 protein thins SOPC membranes and interacts with cholesterol. Biochim Biophys Acta 1778:1120–1130. 10.1016/j.bbamem.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schwarzer R, Levental I, Gramatica A, Scolari S, Buschmann V, Veit M, Herrmann A. 2014. The cholesterol-binding motif of the HIV-1 glycoprotein gp41 regulates lateral sorting and oligomerization. Cell Microbiol 16:1565–1581. 10.1111/cmi.12314. [DOI] [PubMed] [Google Scholar]
- 83.Vishwanathan SA, Thomas A, Brasseur R, Epand RF, Hunter E, Epand RM. 2008. Hydrophobic substitutions in the first residue of the CRAC segment of the gp41 protein of HIV. Biochemistry 47:124–130. 10.1021/bi7018892. [DOI] [PubMed] [Google Scholar]
- 84.Chen SS, Yang P, Ke PY, Li HF, Chan WE, Chang DK, Chuang CK, Tsai Y, Huang SC. 2009. Identification of the LWYIK motif located in the human immunodeficiency virus type 1 transmembrane gp41 protein as a distinct determinant for viral infection. J Virol 83:870–883. 10.1128/JVI.01088-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bellamy-McIntyre AK, Lay CS, Baar S, Maerz AL, Talbo GH, Drummer HE, Poumbourios P. 2007. Functional links between the fusion peptide-proximal polar segment and membrane-proximal region of human immunodeficiency virus gp41 in distinct phases of membrane fusion. J Biol Chem 282:23104–23116. 10.1074/jbc.M703485200. [DOI] [PubMed] [Google Scholar]
- 86.Haim H, Strack B, Kassa A, Madani N, Wang L, Courter JR, Princiotto A, McGee K, Pacheco B, Seaman MS, Smith AB, III, Sodroski J. 2011. Contribution of intrinsic reactivity of the HIV-1 envelope glycoproteins to CD4-independent infection and global inhibitor sensitivity. PLoS Pathog 7:e1002101. 10.1371/journal.ppat.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bradley T, Trama A, Tumba N, Gray E, Lu X, Madani N, Jahanbakhsh F, Eaton A, Xia SM, Parks R, Lloyd KE, Sutherland LL, Scearce RM, Bowman CM, Barnett S, Abdool-Karim SS, Boyd SD, Melillo B, Smith AB, III, Sodroski J, Kepler TB, Alam SM, Gao F, Bonsignori M, Liao HX, Moody MA, Montefiori D, Santra S, Morris L, Haynes BF. 2016. Amino acid changes in the HIV-1 gp41 membrane proximal region control virus neutralization sensitivity. EBioMedicine 12:196–207. 10.1016/j.ebiom.2016.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ringe R, Bhattacharya J. 2012. Association of enhanced HIV-1 neutralization by a single Y681H substitution in gp41 with increased gp120-CD4 interaction and macrophage infectivity. PLoS One 7:e37157. 10.1371/journal.pone.0037157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moyo T, Ereno-Orbea J, Jacob RA, Pavillet CE, Kariuki SM, Tangie EN, Julien JP, Dorfman JR. 2018. Molecular basis of unusually high neutralization resistance in Tier 3 HIV-1 strain 253-11. J Virol 92:e02261-17. 10.1128/JVI.02261-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blish CA, Nguyen MA, Overbaugh J. 2008. Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41. PLoS Med 5:e9. 10.1371/journal.pmed.0050009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lovelace E, Xu H, Blish CA, Strong R, Overbaugh J. 2011. The role of amino acid changes in the human immunodeficiency virus type 1 transmembrane domain in antibody binding and neutralization. Virology 421:235–244. 10.1016/j.virol.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Salimi H, Johnson J, Flores MG, Zhang MS, O'Malley Y, Houtman JC, Schlievert PM, Haim H. 2020. The lipid membrane of HIV-1 stabilizes the viral envelope glycoproteins and modulates their sensitivity to antibody neutralization. J Biol Chem 295:348–362. 10.1074/jbc.RA119.009481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Foley B, Leitner T, Apetrei C, Hahn B, Mizrachi I, Mullins J, Rambaut A, Wolinsky S, Korber B. 2014. HIV sequence compendium 2014. Los Alamos National Laboratory, Theoretical Biology and Biophysics, Los Alamos, New Mexico. [Google Scholar]
- 94.Moore JP, Trkola A, Korber B, Boots LJ, Kessler JA, 2nd, McCutchan FE, Mascola J, Ho DD, Robinson J, Conley AJ. 1995. A human monoclonal antibody to a complex epitope in the V3 region of gp120 of human immunodeficiency virus type 1 has broad reactivity within and outside clade B. J Virol 69:122–130. 10.1128/JVI.69.1.122-130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nguyen H, Qualizza A, Anang S, Zhao M, Zou S, Zhou R, Wang Q, Zhang S, Deshpande A, Ding H, Smith AB, III, Kappes J, Sodroski J. 2021. Functional and highly crosslinkable HIV-1 envelope glycoproteins enriched in a pretriggered conformation. J Virol. [DOI] [PMC free article] [PubMed]
- 96.Melillo B, Liang S, Park J, Schon A, Courter JR, LaLonde JM, Wendler DJ, Princiotto AM, Seaman MS, Freire E, Sodroski J, Madani N, Hendrickson WA, Smith AB III. 2016. Small-molecule CD4-mimics: structure-based optimization of HIV-1 entry inhibition. ACS Med Chem Lett 7:330–334. 10.1021/acsmedchemlett.5b00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wilson C, Reitz MS, Jr., Aldrich K, Klasse PJ, Blomberg J, Gallo RC, Robert-Guroff M. 1990. The site of an immune-selected point mutation in the transmembrane protein of human immunodeficiency virus type 1 does not constitute the neutralization epitope. J Virol 64:3240–3248. 10.1128/JVI.64.7.3240-3248.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reitz MS, Jr., Wilson C, Naugle C, Gallo RC, Robert-Guroff M. 1988. Generation of a neutralization-resistant variant of HIV-1 is due to selection for a point mutation in the envelope gene. Cell 54:57–63. 10.1016/0092-8674(88)90179-1. [DOI] [PubMed] [Google Scholar]
- 99.Thali M, Charles M, Furman C, Cavacini L, Posner M, Robinson J, Sodroski J. 1994. Resistance to neutralization by broadly reactive antibodies to the human immunodeficiency virus type 1 gp120 glycoprotein conferred by a gp41 amino acid change. J Virol 68:674–680. 10.1128/JVI.68.2.674-680.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pacheco B, Alsahafi N, Debbeche O, Prevost J, Ding S, Chapleau JP, Herschhorn A, Madani N, Princiotto A, Melillo B, Gu C, Zeng X, Mao Y, Smith AB, III, Sodroski J, Finzi A. 2017. Residues in the gp41 ectodomain regulate HIV-1 envelope glycoprotein conformational transitions induced by gp120-directed inhibitors. J Virol 91:e02219-16. 10.1128/JVI.02219-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zou S, Zhang S, Gaffney A, Ding H, Lu M, Grover JR, Farrell M, Nguyen HT, Zhao C, Anang S, Zhao M, Mohammadi M, Blanchard SC, Abrams C, Madani N, Mothes W, Kappes JC, Smith AB, III, Sodroski J. 2020. Long-acting BMS-378806 analogues stabilize the State-1 conformation of the human immunodeficiency virus type 1 envelope glycoproteins. J Virol 94:e00148-20. 10.1128/JVI.00148-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Posner MR, Hideshima T, Cannon T, Mukherjee M, Mayer KH, Byrn RA. 1991. An IgG human monoclonal antibody that reacts with HIV-1/GP120, inhibits virus binding to cells, and neutralizes infection. J Immunol 146:4325–4332. [PubMed] [Google Scholar]
- 103.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol 67:3978–3988. 10.1128/JVI.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, McLellan JS, Bailer RT, Dai K, Loesgen S, Louder MK, Staupe RP, Yang Y, Zhang B, Parks R, Eudailey J, Lloyd KE, Blinn J, Alam SM, Haynes BF, Amin MN, Wang LX, Burton DR, Koff WC, Nabel GJ, Mascola JR, Bewley CA, Kwong PD. 2013. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat Struct Mol Biol 20:804–813. 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Protocol GPI, Koff WC, Wilson IA, Burton DR, Poignard P, Protocol G Principal Investigators . 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, de la Pena AT, Cupo A, Julien JP, van Gils M, Lee PS, Peng W, Paulson JC, Poignard P, Burton DR, Moore JP, Sanders RW, Wilson IA, Ward AB. 2014. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity 40:669–680. 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang T, Zhang Z, Wallace OB, Deshpande M, Fang H, Yang Z, Zadjura LM, Tweedie DL, Huang S, Zhao F, Ranadive S, Robinson BS, Gong YF, Ricarrdi K, Spicer TP, Deminie C, Rose R, Wang HG, Blair WS, Shi PY, Lin PF, Colonno RJ, Meanwell NA. 2003. Discovery of 4-benzoyl-1-[(4-methoxy-1H- pyrrolo[2,3-b]pyridin-3-yl)oxoacetyl]-2- (R)-methylpiperazine (BMS-378806): a novel HIV-1 attachment inhibitor that interferes with CD4-gp120 interactions. J Med Chem 46:4236–4239. 10.1021/jm034082o. [DOI] [PubMed] [Google Scholar]
- 109.Matthews T, Salgo M, Greenberg M, Chung J, DeMasi R, Bolognesi D. 2004. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat Rev Drug Discov 3:215–225. 10.1038/nrd1331. [DOI] [PubMed] [Google Scholar]
- 110.Herschhorn A, Ma X, Gu C, Ventura JD, Castillo-Menendez L, Melillo B, Terry DS, Smith AII, Blanchard SC, Munro JB, Mothes W, Finzi A, Sodroski J. 2016. Release of gp120 restraints leads to an entry-competent intermediate state of the HIV-1 envelope glycoproteins. mBio 7:e01598-16. 10.1128/mBio.01598-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lu M, Ma X, Castillo-Menendez LR, Gorman J, Alsahafi N, Ermel U, Terry DS, Chambers M, Peng D, Zhang B, Zhou T, Reichard N, Wang K, Grover JR, Carman BP, Gardner MR, Nikić-Spiegel I, Sugawara A, Arthos J, Lemke EA, Smith AB, III, Farzan M, Abrams C, Munro JB, McDermott AB, Finzi A, Kwong PD, Blanchard SC, Sodroski JG, Mothes W. 2019. Associating HIV-1 envelope glycoprotein structures with states on the virus observed by smFRET. Nature 568:415–419. 10.1038/s41586-019-1101-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lu M, Ma X, Reichard N, Terry DS, Arthos J, Smith AII, Sodroski JG, Blanchard SC, Mothes W. 2020. Shedding-resistant HIV-1 envelope glycoproteins adopt downstream conformations that remain responsive to conformation-preferring ligands. J Virol 94:e00597-20. 10.1128/JVI.00597-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liao Z, Cimakasky LM, Hampton R, Nguyen DH, Hildreth JE. 2001. Lipid rafts and HIV pathogenesis: host membrane cholesterol is required for infection by HIV type 1. AIDS Res Hum Retroviruses 17:1009–1019. 10.1089/088922201300343690. [DOI] [PubMed] [Google Scholar]
- 114.Liao Z, Graham DR, Hildreth JE. 2003. Lipid rafts and HIV pathogenesis: virion-associated cholesterol is required for fusion and infection of susceptible cells. AIDS Res Hum Retroviruses 19:675–687. 10.1089/088922203322280900. [DOI] [PubMed] [Google Scholar]
- 115.Graham DR, Chertova E, Hilburn JM, Arthur LO, Hildreth JE. 2003. Cholesterol depletion of human immunodeficiency virus type 1 and simian immunodeficiency virus with beta-cyclodextrin inactivates and permeabilizes the virions: evidence for virion-associated lipid rafts. J Virol 77:8237–8248. 10.1128/jvi.77.15.8237-8248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guyader M, Kiyokawa E, Abrami L, Turelli P, Trono D. 2002. Role for human immunodeficiency virus type 1 membrane cholesterol in viral internalization. J Virol 76:10356–10364. 10.1128/jvi.76.20.10356-10364.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Campbell SM, Crowe SM, Mak J. 2002. Virion-associated cholesterol is critical for the maintenance of HIV-1 structure and infectivity. AIDS 16:2253–2261. 10.1097/00002030-200211220-00004. [DOI] [PubMed] [Google Scholar]
- 118.Kalyana Sundaram RV, Li H, Bailey L, Rashad AA, Aneja R, Weiss K, Huynh J, Bastian AR, Papazoglou E, Abrams C, Wrenn S, Chaiken I. 2016. Impact of HIV-1 membrane cholesterol on cell-independent lytic inactivation and cellular infectivity. Biochemistry 55:447–458. 10.1021/acs.biochem.5b00936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mahammad S, Parmryd I. 2015. Cholesterol depletion using methyl-beta-cyclodextrin. Methods Mol Biol 1232:91–102. 10.1007/978-1-4939-1752-5_8. [DOI] [PubMed] [Google Scholar]
- 120.Kwon YD, Georgiev IS, Ofek G, Zhang B, Asokan M, Bailer RT, Bao A, Caruso W, Chen X, Choe M, Druz A, Ko SY, Louder MK, McKee K, O'Dell S, Pegu A, Rudicell RS, Shi W, Wang K, Yang Y, Alger M, Bender MF, Carlton K, Cooper JW, Blinn J, Eudailey J, Lloyd K, Parks R, Alam SM, Haynes BF, Padte NN, Yu J, Ho DD, Huang J, Connors M, Schwartz RM, Mascola JR, Kwong PD. 2016. Optimization of the solubility of HIV-1-neutralizing antibody 10E8 through somatic variation and structure-based design. J Virol 90:5899–5914. 10.1128/JVI.03246-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zwick MB, Jensen R, Church S, Wang M, Stiegler G, Kunert R, Katinger H, Burton DR. 2005. Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J Virol 79:1252–1261. 10.1128/JVI.79.2.1252-1261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shen X, Parks RJ, Montefiori DC, Kirchherr JL, Keele BF, Decker JM, Blattner WA, Gao F, Weinhold KJ, Hicks CB, Greenberg ML, Hahn BH, Shaw GM, Haynes BF, Tomaras GD. 2009. In vivo gp41 antibodies targeting the 2F5 monoclonal antibody epitope mediate human immunodeficiency virus type 1 neutralization breadth. J Virol 83:3617–3625. 10.1128/JVI.02631-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shen X, Dennison SM, Liu P, Gao F, Jaeger F, Montefiori DC, Verkoczy L, Haynes BF, Alam SM, Tomaras GD. 2010. Prolonged exposure of the HIV-1 gp41 membrane proximal region with L669S substitution. Proc Natl Acad Sci USA 107:5972–5977. 10.1073/pnas.0912381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.O'Rourke SM, Schweighardt B, Scott WG, Wrin T, Fonseca DP, Sinangil F, Berman PW. 2009. Novel ring structure in the gp41 trimer of human immunodeficiency virus type 1 that modulates sensitivity and resistance to broadly neutralizing antibodies. J Virol 83:7728–7738. 10.1128/JVI.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.O'Rourke SM, Schweighardt B, Phung P, Mesa KA, Vollrath AL, Tatsuno GP, To B, Sinangil F, Limoli K, Wrin T, Berman PW. 2012. Sequences in glycoprotein gp41, the CD4 binding site, and the V2 domain regulate sensitivity and resistance of HIV-1 to broadly neutralizing antibodies. J Virol 86:12105–12114. 10.1128/JVI.01352-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kolchinsky P, Kiprilov E, Sodroski J. 2001. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J Virol 75:2041–2050. 10.1128/JVI.75.5.2041-2050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hoffman TL, LaBranche CC, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie JA, Doms RW. 1999. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc Natl Acad Sci USA 96:6359–6364. 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ringe R, Sharma D, Zolla-Pazner S, Phogat S, Risbud A, Thakar M, Paranjape R, Bhattacharya J. 2011. A single amino acid substitution in the C4 region in gp120 confers enhanced neutralization of HIV-1 by modulating CD4 binding sites and V3 loop. Virology 418:123–132. 10.1016/j.virol.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.O'Rourke SM, Schweighardt B, Phung P, Fonseca DP, Terry K, Wrin T, Sinangil F, Berman PW. 2010. Mutation at a single position in the V2 domain of the HIV-1 envelope protein confers neutralization sensitivity to a highly neutralization-resistant virus. J Virol 84:11200–11209. 10.1128/JVI.00790-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Powell RLR, Totrov M, Itri V, Liu X, Fox A, Zolla-Pazner S. 2017. Plasticity and Epitope Exposure of the HIV-1 Envelope Trimer. J Virol 91:e00410-17. 10.1128/JVI.00410-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Herschhorn A, Gu C, Moraca F, Ma X, Farrell M, Smith AII, Pancera M, Kwong PD, Schon A, Freire E, Abrams C, Blanchard SC, Mothes W, Sodroski JG. 2017. The beta20-beta21 of gp120 is a regulatory switch for HIV-1 Env conformational transitions. Nat Commun 8:1049. 10.1038/s41467-017-01119-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zolla-Pazner S, Cohen SS, Boyd D, Kong XP, Seaman M, Nussenzweig M, Klein F, Overbaugh J, Totrov M. 2016. Structure/function studies involving the V3 region of the HIV-1 envelope delineate multiple factors that affect neutralization sensitivity. J Virol 90:636–649. 10.1128/JVI.01645-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Guzzo C, Zhang P, Liu Q, Kwon AL, Uddin F, Wells AI, Schmeisser H, Cimbro R, Huang J, Doria-Rose N, Schmidt SD, Dolan MA, Connors M, Mascola JR, Lusso P. 2018. Structural constraints at the trimer apex stabilize the HIV-1 envelope in a closed, antibody-protected conformation. mBio 9:e00955-18. 10.1128/mBio.00955-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Keller PW, Morrison O, Vassell R, Weiss CD. 2018. HIV-1 gp41 Residues modulate CD4-induced conformational changes in the envelope glycoprotein and evolution of a relaxed conformation of gp120. J Virol 92:e00583-18. 10.1128/JVI.00583-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang P, Kwon AL, Guzzo C, Liu Q, Schmeisser H, Miao H, Lin Y, Cimbro R, Huang J, Connors M, Schmidt SD, Dolan MA, Armstrong AA, Lusso P. 2021. Functional anatomy of the trimer apex reveals key hydrophobic constraints that maintain the HIV-1 envelope spike in a closed state. mBio 12:e00090-21. 10.1128/mBio.00090-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stamatatos L, Cheng-Mayer C. 1998. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J Virol 72:7840–7845. 10.1128/JVI.72.10.7840-7845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mishra N, Sharma S, Dobhal A, Kumar S, Chawla H, Singh R, Das BK, Kabra SK, Lodha R, Luthra K. 2020. A rare mutation in an infant-derived HIV-1 envelope glycoprotein alters interprotomer stability and susceptibility to broadly neutralizing antibodies targeting the trimer apex. J Virol 94:e00814-20. 10.1128/JVI.00814-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Upadhyay C, Mayr LM, Zhang J, Kumar R, Gorny MK, Nadas A, Zolla-Pazner S, Hioe CE. 2014. Distinct mechanisms regulate exposure of neutralizing epitopes in the V2 and V3 loops of HIV-1 envelope. J Virol 88:12853–12865. 10.1128/JVI.02125-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang S, Wang K, Wang WL, Nguyen HT, Chen S, Lu M, Go EP, Ding H, Steinbock RT, Desaire H, Kappes JC, Sodroski J, Mao Y. 2021. Asymmetric structures and conformational plasticity of the uncleaved full-length human immunodeficiency virus envelope glycoprotein trimer. J Virol 95:e0052921. 10.1128/JVI.00529-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Korber B, Foley BT, Kuiken C, Pillai SK, Sodroski JG. 1998. Numbering positions in HIV relative to HBX2CG. Human Retroviruses and AIDS. 102–111. Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, New Mexico. [Google Scholar]