Abstract
Automated segmentation of grey matter (GM) and white matter (WM) in gigapixel histopathology images is advantageous to analyzing distributions of disease pathologies, further aiding in neuropathologic deep phenotyping. Although supervised deep learning methods have shown good performance, its requirement of a large amount of labeled data may not be cost-effective for large scale projects. In the case of GM/WM segmentation, trained experts need to carefully trace the delineation in gigapixel images. To minimize manual labeling, we consider semi-surprised learning (SSL) and deploy one state-of-the-art SSL method (FixMatch) on WSIs. Then we propose a two-stage scheme to further improve the performance of SSL: the first stage is a self-supervised module to train an encoder to learn the visual representations of unlabeled data, subsequently, this well-trained encoder will be an initialization of consistency loss-based SSL in the second stage. We test our method on Amyloid-β stained histopathology images and the results outperform FixMatch with the mean IoU score at around 2% by using 6,000 labeled tiles while over 10% by using only 600 labeled tiles from 2 WSIs.
Clinical relevance—
This work minimizes the required labeling efforts by trained personnel. An improved GM/WM segmentation method could further aid in the study of brain diseases, such as Alzheimer’s disease.
I. INTRODUCTION
Alzheimer’s disease, the sixth leading cause of death, resulted in nearly 122,019 deaths in 2018 and the number of patients is expected to rise to 13.8 million in U.S. by mid-century [1]. To comprehensively study this disease, neuropathologists assess histopathology images to identify extracellular Amyloid-β plaques [2], which have different distributions in grey matter (GM) and white matter (WM) [3]. To determine the density and distribution of these plaques in the two regions, it is imperative to segment GM and WM in histopathology images. Many image processing-based methods have been proposed for histopathology image segmentation, such as [4], [5]. Although these methods are computationally efficient, the inter and intra-variations in staining and color contrast could significantly impair the performances of these methods on a hold-out test set [6].
Recently convolutional neural networks (CNN) have also gained wide popularity in medical segmentation problems. Among these methods, FCN [7] and U-Net [6] based architectures are the predominant choices [8], [9]. In [10], [11], they developed an automated GM and WM segmentation pipeline with promising results and compared different deep learning methods: FCN [7], U-Net [6], ResNet-Patch and ResNet-NCRF. However, these CNNs show their performance through supervised learning, which heavily relies on a large labeled dataset. For example, a recent study [12] claimed that it requires more than 30,000 labeled tiles from gigapixel WSIs to achieve the well-defined performance of CNNs, which requires labor-intensive labeling [13]. Furthermore, the labeling cost could be much higher when annotations must be done by experts (for example, doctors required for medical problems) [14]. Therefore, these challenges in procuring a sufficiently large dataset with annotations limit the wide-adoption of deep learning-based methods in realworld medical problems [15].
As such, it is vital to design an algorithm that not only automates histopathology segmentation but minimizes manual labeling. Semi-supervised learning (SSL) is one that train models without requiring heavy annotations combining a small set of labeled samples with a large amount of unlabeled samples [16]. Consistency loss-based SSL methods involve both pseudo labels and data augmentation, showing their powerful performance on CIFAR-10 [14]. One drawback of these consistency loss-based SSL methods is that the imperfect class conditional distribution is used to generate pseudo labels and the over-reliance on pseudo labels make it difficult to correctly update the class conditional distribution [17]. For example, a recent study [18] applied FixMatch [14] and Mix-Match [16] to a histology dataset and showed that the performance of these state-of-the-art SSL methods are limited due to the above drawback.
To deal with the above issue, inspired by [15] who claims that pre-training a classifier and then transferring it has the potential to outperform SSL in some settings (using 4000 labeled labeled points from CIFAR-10), we design a novel two-stage SSL, SIM-FixMatch, to further reduce the labeling cost when labeled data is too rare for transfer learning. Our first stage is to employ self-supervised learning [19] for learning visual representations, which plays a similar role to pre-training in transfer learning but requires no labels. After the first stage, a pretrained encoder will be fed into the standard consistency loss-based SSL models. We employ our proposed scheme to segment GM and WM in Amyloid-β stained histopathology images. To our best knowledge, our work is the first to tackle this task with minimal labeling cost and our proposed method outperforms FixMatch [14] when the amount of labeled data is much reduced (e.g., to 0.1% of total tiles from WSIs).
II. METHODS
A. SIM-FixMatch Pipeline
In this section, we will introduce SIM-FixMatch, a two-stage SSL approach. In the first stage, we utilize the self-supervised module to pretrain an encoder that learns the visual representations from unlabeled set. Then, we use this encoder as the input into a standard consistency loss-based SSL to leverage the information from both labeled and unlabeled set. Fig. 1 shows the overall architecture.
Fig. 1.
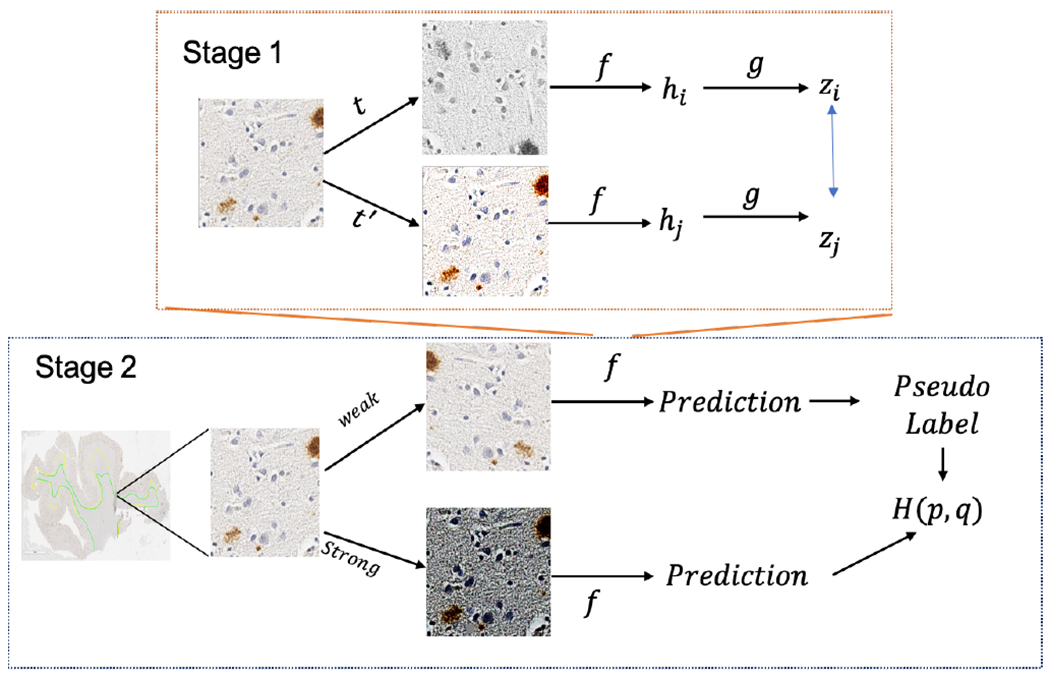
Two-stage Sim-FixMatch pipeline where encoder f in the 2nd stage is self-trained using the 1st stage.
1). First Stage - Self-supervised Pre-training:
SimCLR [19] is a simple self-supervised framework for contrastive learning of visual representations on unlabeled images. As shown in Fig. 1, an unlabeled image undergoes two random data augmentation operations t and t′ and produces outputs hi and hj after going through the encoder network f (·). g(·) is a projection head (multilayer perceptron with one hidden layer) to get zi = g(hi). f (·) and g(·) are trained to maximize the agreement using the contrastive loss function
where N is the size of a batch (two separate augmentation operators result in 2N data points), is an indicator function, which is 1 if k ≠ i, sim(·,·) is cosine similarity, and τ is a temperature parameter.
The eventual goal for this stage is to train the encoder f(·) for learning visual representations from unlabeled dataset. In our experiment, we use ResNet [20] for the encoder.
2). Second Stage - Standard FixMatch:
In this study, we mainly adopt FixMatch [14], which generates artificial labels using both pseudo-labeling and consistency regularization. Specifically, the pseudo label is generated based on a weakly-augmented unlabeled image (weak), which will be the target to compare with the output of the model on a strong-augmented version of the same unlabeled image (strong) as shown in Fig. 1. As pseudo-labels generated here could be hurt by the imperfect class conditional distribution, we use the encoder f(·) pretrained on unlabeled data from the first stage to provide an initialization for FixMatch. For the optimizer, instead of using standard SGD reported to have the best performance in [14], our experiments show Adam [21] performs better for our WSI dataset. The “strong” augmentation operations include RandAugment and CTAugment [14] while the “weak” includes standard flip-and-shift augmentation.
B. Datasets
1). Overview:
In this study, we utilize 30 Whole Slide Images (WSIs) of 5um formalin fixed paraffin embedded sections of human temporal cortex stained with an Amyloid-β antibody (4G8, recognizing residues 17–24, dilution 1:1600, BioLegend (formally Covance) catalog number SIG-39200). These slides were scanned and digitized with an Aperio AT2 at up to 20× magnification, resulting in the average resolution at nearly 60,000 × 50,000 pixels each. Among these 30 WSIs, 18 slides (from 10 males and 8 females with an average age at death of 84 ± 7 years) from deceased patients pathologically diagnosed as Alzheimer’s disease, and will be referred to as AD cases; the remaining 12 slides lacked a pathological diagnosis of Alzheimer’s disease, referred as NAD cases. Among these 12 NAD cases, one had a diagnosis of metastatic carcinoma, and five with cerebrovascular disease. The Ethnoracial make up of the cohort was 22 non-Hispanic White (73%) descendants, 5 African Americans (17%), and 3 Hispanics (10%). To further protect data confidentiality, we refer to the AD cases as WSI-1 to WSI-18 and NAD cases as WSI-19 to WSI-30.
2). Training Data Preparation:
As downsampling [13] may lose medical features, we follow a patch-based method in [11] to divide WSIs into 256 × 256 patches to cope with the ultra-high resolution. In this paper, 20 WSIs (12 AD cases and 8 NAD cases) were randomly selected for training and validation while the remaining 10 WSIs (6 AD cases and 4 NAD cases) were used for hold-out testing and inference. From the 20 WSIs, we selected one AD case and NAD case that have highest inter-rater agreement as the source of labeled patches while we kept the remaining 18 WSIs for generating unlabeled patches (around 600,000 patches). In our setting, we first generated 6,000 labeled patches (nearly 1% proportion of all patches) from 2 labeled WSIs, then we generated 6,00 labeled patches (nearly 0.1%) to further verify the effectiveness of our proposed method.
III. RESULTS
A. Ablation Study
To verify the effectiveness of our first stage, which is to learn visual representations on the unlabeled set and provide an encoder for the second stage, we visualized the training process of the second stage by using our method and baseline FixMatch. We trained both of them over 40 epochs and found that they converged quickly within 15 epochs. As shown in Fig. 2, with the first stage, the model will be well-trained after only 3 epochs in the second stage, while it takes almost 20 epochs without the first stage. Besides, our proposed method starts from nearly 50% higher accuracy after the first epoch compared with original FixMatch, which shows the effectiveness of our proposed first stage on learning the representations via contrastive learning.
Fig. 2.
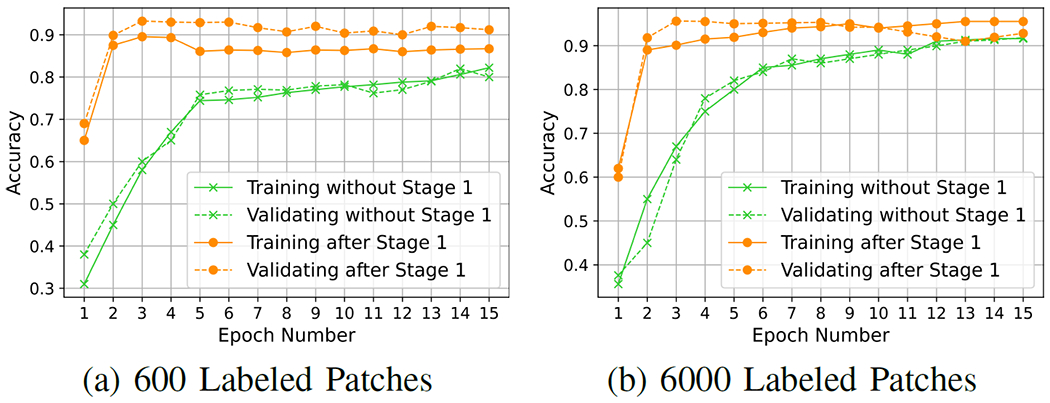
Trends of training and validation during the training process of FixMatch with or without the 1st stage.
B. Quantitative Results
IoU and STD.
We first use a standard segmentation metric — Intersection over Union (IoU) to compare the masks from our proposed method and original FixMatch. IoU score is designed for measuring the overlapping degree between two masks. And we also use standard deviation (STD) to evaluate how consistent and robust of our methods across different hold-out test slides. The results of both IoU scores and STD are summarized in Table. I.
TABLE I.
Pixel-wise IoU Scores for AD, NAD, and overall test set
| 2 Labeled WSIs | 0.1% Labeled | 1% Labeled | ||||
|---|---|---|---|---|---|---|
| FCN | U-Net | FixMatch | Proposed | FixMatch | Proposed | |
| AD Back ± STD | 61.04 ± 5.44 | 59.74 ± 13.9 | 93.15 ± 2.41 | 94.10 ± 2.21 | 96.59 ± 1.04 | 96.33 ± 1.05 |
| AD GM ± STD | 46.98 ± 2.78 | 37.16 ± 9.93 | 78.57 ± 3.87 | 84.59 ± 3.27 | 87.12 ± 3.46 | 88.21 ± 3.84 |
| AD WM ± STD | 27.75 ± 5.50 | 7.57 ± 6.02 | 56.66 ± 16.4 | 74.31 ± 3.36 | 73.94 ± 6.89 | 76.33 ± 4.77 |
| AD Mean ± STD | 45.26 ± 3.55 | 35.40 ± 7.12 | 76.13 ± 5.89 | 84.34 ± 1.88 | 85.88 ± 3.09 | 86.95 ± 2.72 |
| NAD Back ± STD | 66.66 ± 5.17 | 78.46 ± 18.5 | 97.07 ± 0.31 | 96.70 ± 0.73 | 97.71 ± 0.86 | 97.63 ± 0.88 |
| NAD GM ± STD | 50.15 ± 0.49 | 59.59 ± 13.6 | 83.97 ± 7.76 | 86.58 ± 5.44 | 90.01 ± 4.02 | 90.93 ± 4.12 |
| NAD WM ± STD | 19.72 ± 13.6 | 3.02 ± 3.09 | 22.72 ± 19.0 | 62.17 ± 7.04 | 59.71 ± 9.99 | 68.79 ± 7.22 |
| NAD Mean ± STD | 45.51 ± 3.29 | 47.02 ± 10.9 | 67.92 ± 6.53 | 81.82 ± 2.15 | 82.47 ± 2.59 | 85.78 ± 2.00 |
| Test Back ± STD | 63.29 ± 5.81 | 68.28 ± 17.2 | 94.72 ± 2.71 | 95.14 ± 2.17 | 97.04 ± 1.08 | 96.85 ± 1.15 |
| Test GM ± STD | 48.25 ± 2.66 | 46.13 ± 15.8 | 80.73 ± 6.01 | 85.39 ± 4.10 | 88.27 ± 3.78 | 89.30 ± 3.98 |
| Test WM ± STD | 24.54 ± 9.80 | 5.75 ± 5.37 | 43.08 ± 24.0 | 69.45 ± 7.88 | 68.25 ± 10.7 | 73.31 ± 6.72 |
| Test Mean ± STD | 45.36 ± 3.26 | 40.05 ± 10.2 | 72.84 ± 7.18 | 83.33 ± 2.28 | 84.52 ± 3.26 | 86.48 ± 2.41 |
AD is the average results on the 6 Alzheimer’s disease cases in hold-out test set. NAD is the average results on the 4 non-Alzheimer’s disease cases in test set. Test is the average results on all 10 WSIs.
We selected the most updated version of FCN [7] and U-Net [6] as the supervised learning (SL) baselines for our comparison. Both of them are trained on only 2 labeled slides (1 AD case + 1 NAD case). Compared to the results reported in [10] that are trained on 20 labeled slides, their performance drastically deteriorates with reduced labeled WSIs from 20 to 2. The mean IoU scores for these two methods are only around 40%. FixMatch and our proposed method are trained on labeled patches (600 and 6000) from the same 2 labeled WSIs while the unlabeled patches are from other 18 unlabeled WSIs. For 6000 labeled patches setting, the labeled ratio is only 1%. FixMatch could achieve 84.52% of mean IoU while our proposed SIM-FixMatch is around 2% higher and has better performance in almost all classes, especially for the WM region in NAD cases (9.08% of improvement). Besides, our proposed method achieves 2.28% lower in terms of STD compared to original FixMatch. To further stress-test of our proposed method, we consider an extreme situation by using only 600 labeled patches (the labeled ratio is down to 0.1%). The improvement of SIM-FixMatch is significant, almost 40% of increase in terms of the WM region in NAD cases and 10.49% of increase in the mean IoU while the STD is still close to original FixMatch.
DICE coefficient.
Besides IoU, we also use DICE coefficient [22] to further evaluate the proposed methods. When only 600 patches are labeled, the DICE coefficient of WM increases from 61.32% (FixMatch) to 82.67% (proposed). And when 6000 patches are labeled, it also gains 3% of improvement in WM.
C. Segmentation Visualization
Fig. 3 shows the segmentation visualization of SL methods (FCN [7], U-Net [6]) trained on the same 2 labeled WSIs and SSL methods ( original FixMatch [14] and our proposed method) trained using 600 labeled patches from the same two labeled slides and unlabeled patches from the other 18 WSIs (the labeled ratio is only 0.1%). The masks of U-Net (Fig. 3 the 2nd column) indicates that U-Net is unable to distinguish the WM from the GM; the masks of FCN (Fig. 3 the 3rd column) have better visualization than U-Net but there are still many incorrectly labeled regions. FixMatch (Fig. 3 the 4th column) is able to find the rough boundary between GM and WM but there are noisy pixels within WM, indicating it wrongly predicts some WM pixels as GM in the WM region. Our proposed method (Fig. 3 the 5th column) could provide more distinguishable boundary for each region and the masks are the closest to the ground truth masks.
Fig. 3.
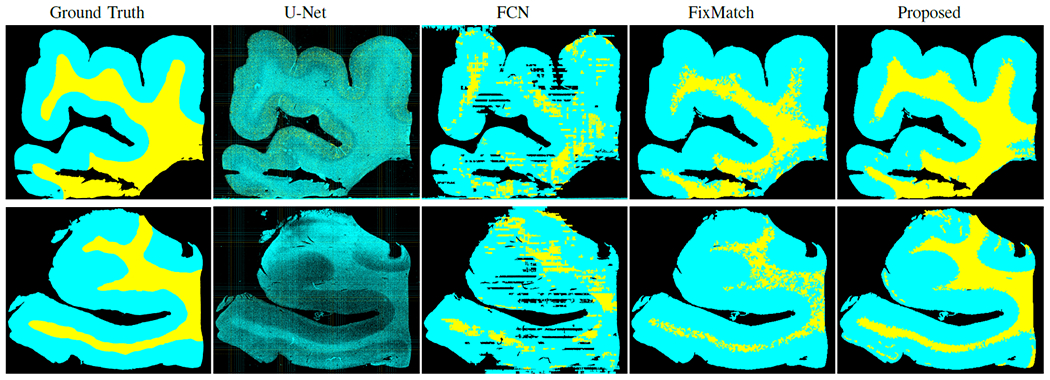
Segmentation masks visualization by only using 0.1% labeled patches from one AD case (top) and one NAD case (bottom). Both are from hold-out test set. Here GM, WM, and background are indicated by cyan, yellow, and black, respectively.
IV. conclusions
In this paper, we investigate the applicability of state-of-the-art semi-supervised learning in histology images and propose a two-stage approach to further improve the performance of SSL methods on Amyloid-β stained WSIs at gigapixel level with the minimal labeling efforts. In our two-stage method, we verify the effectiveness of the first stage (self-supervised pretraining) by providing an encoder that has learned adequate visual representations among unlabeled data. Our proposed method outperformed the original FixMatch, especially in the case where labeled tiles are extremely rare (0.1%). While we showed promising results by running our experiments using randomly selected two WSIs, we will evaluate the selection criteria of WSIs more systematically in our future work.
These techniques have the potential to be applied to other classification and segmentation problems in medical images to minimize the expensive labeling cost. In addition, it takes nearly 3 days for SimCLR to train a good representation. Consequently, our future direction involves developing a task-based architecture to accelerate this process.
ACKNOWLEDGMENT
The authors would like to thank the families and participants of the University of California, Davis Alzheimer’s Disease Research Center (UCD-ADRC) for their generous donations as well as the commitments of faculty and staff of the UCD-ADRC.
This work was supported by the NSF HDR:TRIPODS grant CCF-1934568 and the National Institute On Aging of the National Institutes of Health under Award Numbers P30AG010129, and AG062517, a research grant from the University of California office of the president (MRI-19-599956) and a research grant from the California Department Of Public Health (19-10611) with partial funding from the 2019 California Budget Act. The views and opinions expressed in this manuscript are those of the author and do not necessarily reflect the official policy or position of any public health agency of California or of the United States government.
References
- [1].A. Association et al. “2020 Alzheimer’s Disease facts and figures,” Alzheimer’s & Dementia, vol. 16, no. 3, pp. 391–460, 2020. [Google Scholar]
- [2].Dugger BN and Dickson DW, “Pathology of neurodegenerative diseases,” Cold Spring Harbor perspectives in biology, vol. 9, no. 7, p. a028035, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Iwamoto N, Nishiyama E, Ohwada J, and Arai H, “Distribution of amyloid deposits in the cerebral white matter of the alzheimer’s disease brain: relationship to blood vessels,” Acta Neuropathol, vol. 93, no. 4, pp. 334–340, 1997. [DOI] [PubMed] [Google Scholar]
- [4].Kłeczek P, Dyduch G, Jaworek-Korjakowska J, and Tadeusiewicz R, “Automated epidermis segmentation in histopathological images of human skin stained with hematoxylin and eosin,” in Medical Imaging 2017: Digital Pathology, vol. 10140. ISOP, 2017, p. 101400M. [Google Scholar]
- [5].Bug D, Feuerhake F, and Merhof D, “Foreground extraction for histopathological whole slide imaging,” in Bildverarbeitung für die Medizin 2015. Springer, 2015, pp. 419–424. [Google Scholar]
- [6].Oskal K, Risdal M, Janssen E, Undersrud E, and Gulsrud T, “A U-net based approach to epidermal tissue segmentation in whole slide histopathological images,” SN Appl. Sci, vol. 1, p. 672, 06 2019. [Google Scholar]
- [7].Bándi P, van de Loo R, Intezar M, Geijs D, Ciompi F, van Ginneken B, van der Laak J, and Litjens G, “Comparison of different methods for tissue segmentation in histopathological whole-slide images,” in IEEE ISBI 2017, pp. 591–595. [Google Scholar]
- [8].Ronneberger O, Fischer P, and Brox T, “U-net: Convolutional networks for biomedical image segmentation,” in MICCAI. Springer, 2015, pp. 234–241. [Google Scholar]
- [9].Zhou Z, Siddiquee MMR, Tajbakhsh N, and Liang J, “Unet++: A nested u-net architecture for medical image segmentation,” in Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer, 2018, pp. 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lai Z, Guo R, Xu W, Hu Z, Mifflin K, DeCarli C, Dugger BN, Cheung S.-c., and Chuah C-N, “Automated segmentation of amyloid-β stained whole slide images of brain tissue,” bioRxiv 2020.11.13.381871, 2020. [Google Scholar]
- [11].Lai Z, Guo R, Xu W, Hu Z, Mifflin K, Dugger BN, Chuah C-N, and Cheung S.-c., “Automated grey and white matter segmentation in digitized aβ human brain tissue slide images,” in 2020 ICMEW. IEEE, 2020, pp. 1–6. [Google Scholar]
- [12].Tang Z, Chuang KV, DeCarli C, Jin L-W, Beckett L, Keiser MJ, and Dugger BN, “Interpretable classification of alzheimer’s disease pathologies with a convolutional neural network pipeline,” Nat. Commun, vol. 10, no. 1, pp. 1–14, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chen W, Jiang Z, Wang Z, Cui K, and Qian X, “Collaborative global-local networks for memory-efficient segmentation of ultra-high resolution images,” in IEEE CVPR, 2019, pp. 8924–8933. [Google Scholar]
- [14].Sohn K, Berthelot D, Li C-L, Zhang Z, Carlini N, Cubuk ED, Kurakin A, Zhang H, and Raffel C, “Fixmatch: Simplifying semi-supervised learning with consistency and confidence,” arXiv:2001.07685, 2020. [Google Scholar]
- [15].Oliver A, Odena A, Raffel CA, Cubuk ED, and Goodfellow I, “Realistic evaluation of deep semi-supervised learning algorithms,” Advances in Neural Information Processing Systems, vol. 31, pp. 3235–3246, 2018. [Google Scholar]
- [16].Berthelot D, Carlini N, Goodfellow I, Papernot N, Oliver A, and Raffel C, “Mixmatch: A holistic approach to semi-supervised learning,” arXiv:1905.02249, 2019. [Google Scholar]
- [17].Rizve MN, Duarte K, Rawat YS, and Shah M, “In defense of pseudo-labeling: An uncertainty-aware pseudo-label selection framework for semi-supervised learning,” arXiv:2101.06329, 2021. [Google Scholar]
- [18].Pulido JV, Guleria S, Ehsan L, Fasullo M, Lippman R, Mutha P, Shah T, Syed S, and Brown DE, “Semi-supervised classification of noisy, gigapixel histology images,” in 2020 BIBE. IEEE, 2020, pp. 563–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chen T, Kornblith S, Norouzi M, and Hinton G, “A simple framework for contrastive learning of visual representations,” in ICML. PMLR, 2020, pp. 1597–1607. [Google Scholar]
- [20].He K, Zhang X, Ren S, and Sun J, “Deep residual learning for image recognition,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 770–778. [Google Scholar]
- [21].Kingma DP and Ba J, “Adam: A method for stochastic optimization,” arXiv:1412.6980, 2014. [Google Scholar]
- [22].Zou KH, Warfield SK, Bharatha A, Tempany CM, Kaus MR, Haker SJ, Wells III WM, Jolesz FA, and Kikinis R, “Statistical validation of image segmentation quality based on a spatial overlap index1: scientific reports,” Academic radiology, vol. 11, no. 2, pp. 178–189, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


