Abstract
Introduction
Gut microbiota is associated with host characteristics such as age, sex, immune condition or frailty and is thought to be a key player in numerous human diseases. Nevertheless, its association with outcome in critically ill patients has been poorly investigated. The aim of this study is to assess the association between gut microbiota composition and Day-28 mortality in critically ill patients.
Methods
Rectal swab at admission of every patient admitted to intensive care unit (ICU) between October and November 2019 was frozen at − 80 °C. DNA extraction was performed thanks to QIAamp® PowerFecal® Pro DNA kit (QIAgen®). V3–V4 regions of 16SRNA and ITS2 coding genes were amplified by PCR. Sequencing (2x250 bp paired-end) was performed on MiSeq sequencer (Illumina®). DADA2 pipeline on R software was used for bioinformatics analyses. Risk factors for Day-28 mortality were investigated by logistic regression.
Results
Fifty-seven patients were consecutively admitted to ICU of whom 13/57 (23%) deceased and 44/57 (77%) survived. Bacteriobiota α-diversity was lower among non-survivors than survivors (Shannon and Simpson index respectively, p < 0.001 and p = 0.001) as was mycobiota α-diversity (respectively p = 0.03 and p = 0.03). Both gut bacteriobiota and mycobiota Shannon index were independently associated with Day-28 mortality in multivariate analysis (respectively OR: 0.19, 97.5 CI [0.04–0.60], p < 0.01 and OR: 0.29, 97.5 CI [0.09–0.75], p = 0.02). Bacteriobiota β-diversity was significantly different between survivors and non-survivors (p = 0.05) but not mycobiota β-diversity (p = 0.57). Non-survivors had a higher abundance of Staphylococcus haemolyticus, Clostridiales sp., Campylobacter ureolyticus, Akkermansia sp., Malassezia sympodialis, Malassezia dermatis and Saccharomyces cerevisiae, whereas survivors had a higher abundance of Collinsella aerofaciens, Blautia sp., Streptococcus sp., Faecalibacterium prausnitzii and Bifidobacterium sp.
Conclusion
The gut bacteriobiota and mycobiota α diversities are independently associated with Day-28 mortality in critically ill patients. The causal nature of this interference and, if so, the underlying mechanisms should be further investigated to assess if gut microbiota modulation could be a future therapeutic approach.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13054-022-03980-8.
Keywords: Microbiota, Mycobiota, Intensive care unit
Introduction
The emergence of next-generation sequencing during the last decade has allowed the exploration of the gut microbiota role in human health and diseases. The gut microbiota is composed of microbes from different kingdoms (bacteria, fungus, archae, virus). The number of microbes composing this microbiota is approximately equivalent to the number of our own human cells [1]. Critical illness is known to have profound effects on gut microbiota with extreme dysbiosis of gut microbiota identified in critically ill patients [2, 3]. Despite a large interpersonal variation in gut microbiota dysregulation, critically illness is consistently associated with decreased diversity of Actinobacteria, decreased abundance of butyrate-producing bacteria and commensal Firmicutes and Bacteroidetes, while the abundance of opportunistic pathogens is increased [4]. Host impact on gut microbiota during critical illness is mediated by both endogenous and external factors [5]. Endogenous factors include increased production of opioids and catecholamines, decreased bile-salt concentration, gastrointestinal dysmotility and loss of epithelial integrity in the intestine. External factors include antibiotics, proton pump inhibitor, enteral/parenteral feeding, sedatives, opioids and catecholamines. Extreme dysbiosis of gut microbiota during critical illness is thought to have numerous and profound effects on host metabolism including decreased systemic short-chain fatty acid levels, impaired immune condition and to increase the risk of super-infection, of acute kidney injury (AKI) or of muscle wasting [5]. Despite those data derived from in vitro and animal studies and trials focused on non-critical human diseases, only one study assessed the association between gut microbiota and death in critically ill patients. It suggested that bacteriobiota α-diversity and the presence of Enterococcaceae family could be associated with death in critically ill patients [6]. The aim of this study was thus to assess the association between gut microbiota composition and Day-28 mortality in critically ill patients.
Patients and methods
Patients inclusion and data collection
Every consecutive patient older than 18 years of age admitted to the medical intensive care unit (ICU) at Bordeaux University Hospital in October and November 2019 was prospectively screened to participate to Microbe study (NCT04131569) and included in this ancillary analysis.
Data were prospectively recorded by physicians in charge of the patient by questioning the patients, patients’ family and patients’ general practitioners. Electronic worksheet was completed by two medical intensive care residents. Comorbidities were defined as follows: chronic obstructive pulmonary disease and asthma were defined according to lung function testing. Chronic heart failure was defined according to transthoracic echocardiography and chronic coronary disease based on stress test or percutaneous coronary intervention. Other comorbidities included history of chronic kidney disease (glomerular filtration rate < 60 mL/min/1.73 m2), immunosuppression (drugs, haematological disease, blood marrow transplantation, solid organ transplantation, plasma exchanges indicated by autoimmune disorders, human immunodeficiency virus infection) and the worse simplified acute physiology score II (SAPSII) within the first 24 h following admission. Acute respiratory distress syndrome was defined according to Berlin’s criteria [7], septic shock according to Sepsis-3 definition [8] and AKI to KDIGO guidelines [9].
Samples collection and preparation for microbiota analysis
The rectal swab (Transport Swab VWR, Copan®) performed for faecal ESBL-E carriage screening at admission before administration of antimicrobial agents was collected and frozen at − 80 °C. DNA extraction was performed by QIAamp® PowerFaecal® Pro DNA kit (QIAgen®). A step of mechanical lysis (2 cycles of 30 s at 7000 rpm on Precellys evolution) was added just after the chemical lysis of the kit. V3–V4 regions of 16SRNA coding gene and ITS2 were amplified by PCR as previously reported [10]. Sequencing (2x250 bp paired-end) was performed on MiSeq sequencer (Illumina®) at the Bordeaux Transciptome Genome platform (INRAe, France).
Bioinformatics analysis
DADA2 pipeline on R software was used for bioinformatics analyses [11]. DADA2 pipeline was preferred as it allows inter-studies comparison (if identical primers are used for amplification) [11] and is more accurate for mycobiota analysis [12]. We defined bacteriobiota as the bacterial kingdom of the microbiota and mycobiota as the fungal kingdom of the microbiota. Gut bacteriobiota and mycobiota α-diversity was expressed by Shannon index, Simpson index and evenness. Between sample beta-diversity differences (measured using Bray Curtis dissimilarity) were tested using a permutational multivariate ANOVA (Permanova) from “vegan” package with 10,000 permutations, while accounting for individual identity as a covariate. Gut bacteriobiota and mycobiota α- and β-diversities were compared thanks to “Phyloseq” package on R software v3.6.0. Linear discriminant analysis (LDA) effect size (LefSe) analysis was performed from microbiomeMarker package. We used mock communities to avoid a non-efficient sequencing experiment, and negative controls to identify and remove potential reagent contaminants of bacterial and fungal microbiota with the microDecon R package [13]. Comparison of β-diversity between negative control, mock community and samples is available in Additional file 1: Figs. S1 and S2 for bacteriobiota and mycobiota respectively). The final average read counts were 66,434 (standard deviation ± 17,634) for 1285 bacterial ASVs and 3647 (standard deviation ± 1267) for 361 fungal ASVs. The 16S rRNA gene and ITS2 sequences have been submitted to the European Nucleotide Archive (Accession N◦ ERP134948).
Statistical analysis
No statistical sample size calculation was performed a priori, and sample size was equal to the number of patients admitted to ICU with available rectal swab during the study period. Quantitative variables are presented as median and interquartile range (IQR) and compared by use of the Mann–Whitney Wilcoxon rank-sum test. Categorical variables are expressed as number of patients (percentage) and compared by use of the Chi-square or Fisher’s test. Risk factors for Day-28 mortality were investigated by logistic regression. First, a univariate analysis was performed. Only variables with a p value < 0.10 were included in the multivariate analysis. Variables comprised in SAPSII such as malignancy, sepsis (blood pressure), ARDS (Pa02/FiO2 ratio) and acute kidney injury (urine output, serum urea level and kaliemia) were not included in the analysis as it is already known that they are associated with SAPSII.
All statistical tests were 2-tailed, and statistical significance was defined as p < 0.05. Statistical analyses were assessed by the R 3.6.0 statistical software (R foundation for Statistical Computing Vienna, Austria).
Ethics
According to French law and the French Data Protection Authority, the handling of these data for research purposes was declared to the Data Protection Officer of the Bordeaux University Hospital. The study obtained the approval of the Institutional Review Board of the Bordeaux University Hospital (declaration number CER BDX-2021-36). Patients (or their relatives, if any) were notified about the anonymized use of their healthcare data via the department's booklet.
Results
Flow chart and patients’ characteristics
Ninety-three patients were prospectively screened during the study period. Twenty-one of them (22.6%) did not have any swab available either due to lack of screening or swab without any stool on it, 9 declined to participate (9.7%), and 6 (6.5%) had an estimated length of stay of less than 48 h (Fig. 1). Among the 57 patients with rectal swabs available for microbiota analysis, 13 (23%) deceased within the first 28 days following admission to ICU and 44 (77%) survived. Patients’ characteristics are summarized in Table 1. Non-survivors were more often male (12/13 (92.9%) vs. 27/44 (61.4%), p = 0.04) with active solid cancer (4/13 (28.6%) vs. 2/44 (4.5%), p = 0.02) and higher SAPSII score (78 [75–87] vs. 61 [46–74], p < 0.01).
Fig. 1.
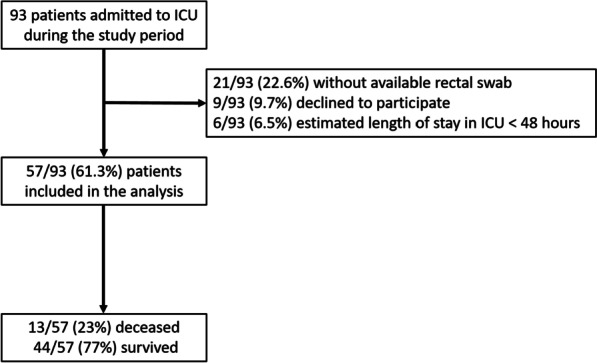
Flow chart. ICU intensive care unit
Table 1.
Patients’ characteristics and comparison between survivors and non-survivors
| Total (n = 57) | Non-survivors (n = 13) | Survivors (n = 44) | p value | |
|---|---|---|---|---|
| Characteristics at admission to ICU | ||||
| Age | 75 [65–79] | 73 [65–79] | 76 [64–79] | 0.99 |
| Sex (male) | 39 (68.4%) | 12 (92.9%) | 27 (61.4%) | 0.04 |
| SAPSII | 68 [49–78] | 78 [75–87] | 61 [46–74] | < 0.01 |
| Septic shock | 20 (35.1%) | 7 (50%) | 13 (29.5%) | 0.18 |
| ARDS | 8 (14%) | 4 (28.6%) | 4 (9.1%) | 0.07 |
| Acute kidney injury | 32 (56.1%) | 12 (85.7%) | 20 (45.5%) | < 0.01 |
| Comorbidities | ||||
| Chronic respiratory disease | 24 (42.1%) | 5 (35.7%) | 19 (43.2%) | 0.52 |
| COPD | 13 (22.8%) | 3 (21.4%) | 10 (22.7%) | 1.00 |
| Asthma | 6 (10.5%) | 1 (7.1%) | 5 (11.4%) | 1.00 |
| Chronic heart failure | 28 (49.1%) | 9 (64.3%) | 19 (43.2%) | 0.12 |
| Chronic coronary disease | 22 (38.6%) | 7 (50%) | 15 (34.1%) | 0.22 |
| Chronic kidney disease | 13 (22.8%) | 4 (28.7%) | 9 (20.5%) | 0.47 |
| Immunosuppression | 12 (21%) | 3 (21.4%) | 9 (20.5%) | 1.00 |
| Active solid cancer | 6 (10.5%) | 4 (28.6%) | 2 (4.5%) | 0.02 |
| Proton pump inhibitor | 15 (26.3%) | 3 (21.4%) | 12 (27.3%) | 1.00 |
| Metformin | 6 (10.5%) | 4 (28.6%) | 6 (13.6%) | 0.21 |
| Antimicrobial treatment during the 3 previous months | 19 (33.3%) | 3 (21.4%) | 16 (36.4%) | 0.51 |
| Treatment | ||||
| Mechanical ventilation | 32 (56.1%) | 10 (71.4%) | 22 (50%) | 0.12 |
| Renal replacement therapy | 10 (17.5%) | 6 (42.9%) | 4 (9.1%) | < 0.01 |
Results are presented as proportion for categorical variables and median [interquartile range] for continuous variables
p values are for comparison between survivors and non-survivors. Threshold for statistical significance: p = 0.05
ARDS acute respiratory distress syndrome, ICU intensive care unit, SAPS simplified acute physiology score II
Gut bacteriobiota and mycobiota α-diversities are significantly decreased in non-survivors compared with survivors
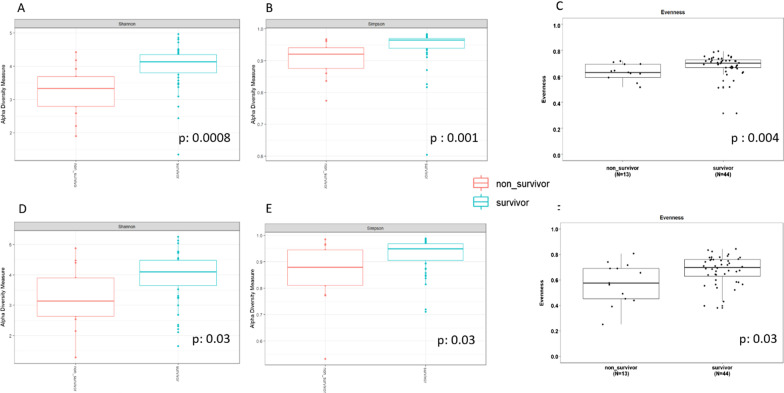
Non-survivors had lower gut bacteriobiota α-diversity (expressed by Shannon or Simpson index and evenness respectively, p < 0.001, p = 0.001 and p < 0.01) (Fig. 2A, B, C) than survivors as well as well as lower gut mycobiota α-diversity (expressed by Shannon or Simpson index and evenness, p = 0.03 for all those 3 index) (Fig. 2D, E, F).
Fig. 2.
Comparison of gut microbiota α-diversities between survivors (in blue) and non-survivors (in red). Gut bacteriobiota α-diversity according to Shannon index (A), Simpson index (B) and evenness (C). Gut mycobiota α diversity according to Shannon index (D), Simpson index (E) and evenness (F). Threshold for statistical significance: p = 0.05
Both gut bacteriobiota and mycobiota α-diversities are independently associated with Day-28 mortality in critically ill patients
After univariate analysis, gut bacteriobiota Shannon index, gut mycobiota Shannon index, sex, SAPSII, history of atrial fibrillation, current malignancy, sepsis at admission to ICU, ARDS at admission to ICU and acute kidney injury at admission to ICU were associated with Day-28 mortality (Table 2). Current malignancy, sepsis at admission to ICU, ARDS at admission to ICU and acute kidney injury at admission to ICU were not included in the multivariate analysis as they are included in SAPSII items.
Table 2.
Factors associated with Day-28 mortality in critically ill patients
| Variables | Univariate analysis OR | 97.5 CI | p value |
|---|---|---|---|
| Shannon 16S rDNA | 0.26 | [0.09–0.62] | < 0.01 |
| Shannon ITS2 | 0.47 | [0.23–0.91] | 0.03 |
| Age | 1.02 | [0.97–1.08] | 0.50 |
| Sex (male) | 7.56 | [1.3–144] | 0.06 |
| SAPSII | 1.07 | [1.02–1.14] | 0.01 |
| Chronic pulmonary disease | 0.82 | [0.22–2.87] | 0.76 |
| COPD | 1.02 | [0.20–4.15] | 0.98 |
| Chronic heart disease | 2.96 | [0.83–12.3] | 0.11 |
| Atrial fibrillation | 3.33 | [0.88–12.7] | 0.07 |
| Coronary disease | 2.26 | [0.64–8.21] | 0.20 |
| Chronic kidney disease | 1.73 | [0.40–6.76] | 0.44 |
| Malignancy | 13.1 | [2.39–104] | < 0.01 |
| Immunosuppression | 1.17 | [0.23–4.83] | 0.84 |
| Long-term proton pump inhibitor | 0.89 | [0.18–3.59] | 0.87 |
| Long-term metformin | 1.01 | [0.98–1.03] | 0.99 |
| Antibiotics within the past 3 months | 0.53 | [0.11–2.07] | 0.39 |
| Septic shock at admission | 2.78 | [0.78–10.3] | 0.11 |
| Acute respiratory distress syndrome at admission | 4.44 | [0.90–22.4] | 0.06 |
| Acute kidney injury at admission | 14.4 | [2.51–274] | 0.01 |
| Digestive infection within the past 3 months | 1.75 | [0.08–19.8] | 0.66 |
| Variables | Multivariate analysis OR | 97.5 CI | p value |
|---|---|---|---|
| Shannon 16S rDNA | 0.19 | [0.04–0.60] | < 0.01 |
| SAPSII | 1.08 | [1.01–1.17] | 0.04 |
| Sex (male) | 14.4 | [1.45–516] | 0.05 |
| Atrial fibrillation | 5.03 | [0.76–41.1] | 0.10 |
| Variables | Multivariate analysis OR | 97.5CI | p value |
|---|---|---|---|
| Shannon ITS2 | 0.29 | [0.09–0.75] | 0.02 |
| SAPSII | 1.08 | [1.02–1.17] | 0.02 |
| Sex (male) | 18.8 | [1.84–582] | 0.04 |
| Atrial fibrillation | 4.53 | [0.07–33.4] | 0.11 |
Bold in univariate analysis: variables assessed for inclusion in the multivariate analysis. Bold in multivariate analysis: variables independently associated with Day-28 mortality
ICU intensive care unit, ITS2 internal transcribed spacer 2, OR odds ratio, SAPSII simplified acute physiology score II, 16S rDNA DNA region coding for ribosomal 16S RNA subunit, 97.5 CI 97.5% confidence interval
After multivariate analysis, both gut bacteriobiota and mycobiota Shannon index remain independently associated with Day-28 mortality (respectively OR: 0.19, 97.5 CI [0.04–0.60], p < 0.01 and OR: 0.29, 97.5 CI [0.09–0.75], p = 0.02) (Table 2).
Non-survivors and survivors have significantly dissimilar bacteriobiota but not mycobiota
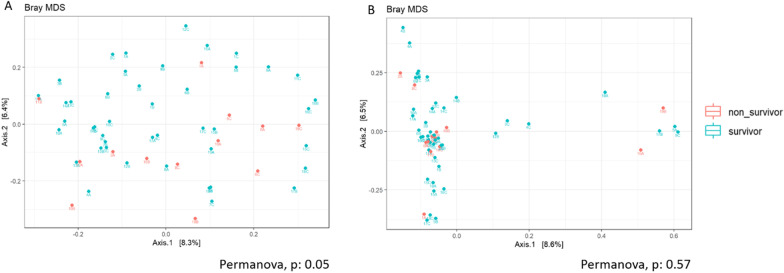
Gut bacteriobiota β-diversity was significantly different between survivors and non-survivors in ICU (Permanova, p = 0.05) but not mycobiota β-diversity (Permanova, p = 0.57) (Fig. 3A, B).
Fig. 3.
Comparison of gut microbiota similarity (β-diversities) between survivors and non-survivors. Metric Bray–Curtis analysis of β-diversity for gut bacteriobiota (A) and gut mycobiota (B). Red: non-survivors. Green: survivors. Threshold for statistical significance: p = 0.05
Identification of bacterial and fungal species associated with survival
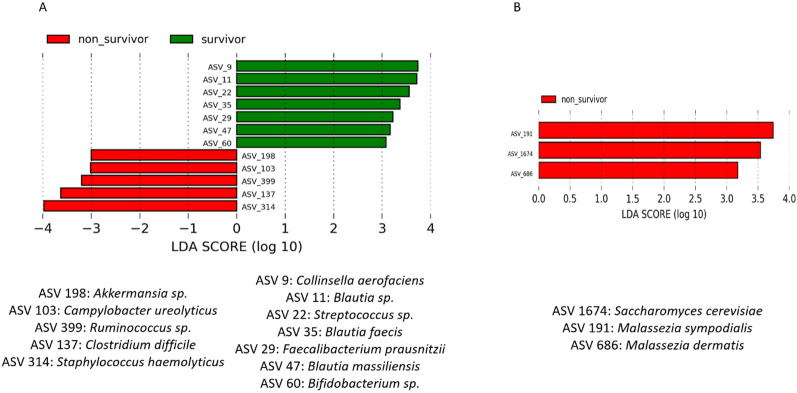
Non-survivors had a higher abundance (LDA > 3log) in gut microbiota of Staphylococcus haemolyticus, Clostridiales sp., Campylobacter ureolyticus, Akkermansia sp., Malassezia sympodialis, Malassezia dermatis and Saccharomyces cerevisiae whereas survivors had a higher abundance of Blautia sp., Streptococcus sp., Faecalibacterium prausnitzii and Bifidobacterium sp. (Fig. 4A, B).
Fig. 4.
Microbial species associated with mortality. LefSe analysis with linear discriminant analysis (LDA) for bacterial species (A) and fungal species (B). Threshold for statistical significance: LDA > 3log. ASV amplicon sequence variant
Discussion
In critically ill patients, gut bacteriobiota and mycobiota α diversities are decreased in non-survivors compared to survivors. Gut bacteriobiota was dissimilar (β-diversity) between non-survivors and survivors but not mycobiota. Moreover, our study is the first to assess the association of both gut bacteriobiota and mycobiota α-diversities with fatal outcome in ICU patients and both are independently associated with Day-28 mortality. This result is not surprising as commensal fungi have been demonstrated to recapitulate the protective benefits of intestinal bacteria. In fact, mannans, a highly conserved component class of fungal cell walls, stimulate local and systemic immunity and protect mice depleted of commensal bacteria from colitis and influenza A virus infection [14]. Fungal kingdom of microbiota should not be under-investigated in future studies.
Few studies investigated the impact of gut microbiota on ICU patients’ outcome and mostly focused on identifying biomarkers instead of analysing α diversity. In addition, they focused on the bacterial kingdom (bacteriobiota) exclusively. A first cohort study of 98 neurocritically ill patients indicates that the gut bacteriobiota composition differs significantly from that in a healthy population. In 50 neurocritically ill patients of this cohort, the magnitude of dysbiosis increased during the first week in the neurological ICU with an increase in gut Enterobacteriales burden which was associated with a 92% increased risk of mortality at Day 180 [15]. Similarly, gut bacteriobiota in sepsis and septic shock patients had an increased abundance of microbes tightly associated with inflammation, such as Parabacteroides, Fusobacterium and Bilophila species and evidenced a remarkable loss of microbial diversity during the ICU stay. The increase in abundance of pathogenic species, such as Enterococcus spp., was differentially increased in sepsis patients who died [16]. Other biomarkers derived from gut microbiota could be the abundance of Bifidobacterium in gut bacteriobiota [17], a higher abundance being associated with survival, or the progression of imbalance in the ratio of Bacteroidetes to Firmicutes within the first 7 days [18].
Three studies investigated the role of lung microbiota in ICU patients, still focusing on bacteriobiota. A first study with 29 endotracheal aspirates demonstrated a negative correlation between lung bacteriobiota α-diversity and APACHE II score [19]. A second study demonstrated that lung bacterial burden, but not bacteriobiota α-diversity, was associated with ventilator-free days in 91 critically ill patients receiving mechanical ventilation [20]. The last study included 36 mechanically ventilated patients with extra-pulmonary sepsis (thus excluding patients with lung infections) and suggested that lung bacterial α-diversity could predict ICU mortality [21].
A major limitation is to know whether there is any causal inference in this association between gut and lung microbiota and clinical outcomes in critically ill patients, but several data suggest that there could be a causal relationship. In fact, gut microbiota dysbiosis is known to have various deleterious effects on the host. Locally, it could increase microbial virulence and favour microbial translocation in systemic and lymphatic circulation [5]. Antibiotic-induced changes in the gut microbiota have been demonstrated to be associated with decreased neutrophils maturation in the bone marrow, decreased splenic B1 B lymphocytes production and IgM production, decreased dendritic cells migration to the lungs and other numerous systemic immunity impairment [22]. Thus, the absence of some protective microbial species or the expansion of some deleterious microbial species in gut microbiota associated with ICU stay could worsen host condition.
Interestingly, we found that the abundance of Bifidobacterium sp. was positively associated with survival, as previously discussed [17]. We found several other microbial species to be associated with survival that have been described to have anti-inflammatory properties. For instance, Blautia faecis, C. aerofaciens and F. prausnitzii are butyrate-producing bacteria that alleviate inflammatory disease and are associated with clinical remission in ulcerative colitis or Crohn’s disease patients [23–26]. The two latter ones (C. aerofaciens and F. prausnitzii) are also associated with clinical response to immunotherapy (anti-CTLA-4 or anti-PD-1 treatments) in cancer patients [27, 28]. C. aerofaciens is also associated with faecal microbiota transplantation efficacy to treat recurrent Clostridioides difficile infection [29], a bacteria known to cause potentially lethal colitis which occurs in frailer patients and abundance of which was associated with mortality in this study.
In addition to Clostridioides difficile, several microbial species known to have pro-inflammatory properties were positively associated with mortality. In fact, Staphylococcus haemolyticus abundance in gut microbiota is increased in patients with active coeliac disease compared to control subjects [30]. Many species belonging to Ruminococcus genus are associated with intestinal inflammation in ulcerative colitis [31]; Campylobacter ureolyticus is associated with acute and prolonged gastroenteritis and is implicated in the development of inflammatory bowel diseases [32]. Malassezia sp. is associated with cystic fibrosis lung exacerbation [33] and with intestinal inflammation in Crohn’s disease patients [34]. Saccharomyces cerevisiae is also associated with intestinal inflammation in celiac disease [35].
The main limitation of this study is the relative small number of patients and its monocentric design. The possible prognostic value of both gut bacteriobiota and mycobiota in critically ill patients should be addressed in large multicentre cohort studies to assess if it could be of interest alongside of SAPSII score which is much more convenient to perform than microbiome sequencing. Furthermore, our analysis assessed for linear but not for nonlinear association of variables with mortality outcome. Thanks to the development of machine learning, complex logistic regression model or different models tested in ensemble modelling could address non-linearity automatically without pre-specification [36]. Another limitation of our study is the lack of causality demonstration as stated for previous studies discussed above. In fact, gut microbiota composition is highly dependent on host condition—including age, sex, immune condition, frailty—[3, 37] which could be confounding factors underlying correlation but not causality. To get beyond the association links provided in this study, animal or organoïd models are needed to decipher the causality between gut microbiota and mortality through the modulation of the host condition. Confirmation of a causal link would enhance the hypothesis that gut microbiota modulation could be a therapeutic approach in ICU patients. If so, in vitro studies will be also required to identify the underlying mechanisms as concerns exist about the translocation of probiotics given to ICU patients who often have increased gut permeability [38].
Conclusion
The gut bacteriobiota and mycobiota α diversities of critically ill patients are significantly decreased in non-survivors than in survivors and are independently associated with Day-28 mortality. The causal nature of this interference and, if so, the underlying mechanisms should be further investigated to assess if gut microbiota modulation could be a future therapeutic approach in critically ill patients.
Supplementary Information
Additional file 1. Supplemental Figure 1. Non metric Bray-curtis analysis of β-diversity of the V3-V4 sequencing run. XXA, B, and C samples: gut bacteriobiota samples. GXX: lung bacteriobiota samples. BlancV3-V4: negative control. Mock: mock community. Supplemental Figure 2. Non metric Bray-curtis analysis of β-diversity of ITS2 sequencing run. XXA, B, and C samples: gut mycobiota samples. GXX: lung mycobiota samples. Blanc1 and blanc2: negative controls. Mock: mock community.
Acknowledgements
We would like to thank Fabien Beaufils for his participation in the “Bordeaux Micro-Mycobiota Study Group”. We would like to thank Erwan Guichoux and Marie Massot for technical assistance.
Abbreviations
- AKI
Acute kidney injury
- ARDS
Acute respiratory distress syndrome
- AUC
Area under the curve
- ICU
Intensive care unit
- IQR
Interquartile range
- ITS
Internal transcribed spacer 2
- LDA
Linear discriminant analysis
- LefSe
Linear discriminant analysis size effect
- NPV
Negative predictive value
- OR
Odds ratio
- PPV
Positive predictive value
- ROC
Receiver operating characteristic
- SAPSII
Simplified acute physiology score II
- Se
Sensitivity
- Sp
Specificity
- 16S rDNA
DNA region coding for ribosomal 16S RNA subunit
Author contributions
RP, PB, LD, AB and DG contributed to the conception and design of the study. AO contributed to the acquisition of data. RP and AC performed DNA extraction. RP and RE performed bioinformatics and statistical analysis. Each author drafted or provided critical revision of the article and provided final approval of the version submitted for publication. All authors read and approved the final manuscript.
Funding
Funded by a grant from “Fédération Girondine de Lutte contre les Maladies Respiratoires». RP received a personal salary grant (MD/PhD program) from CHU de Bordeaux.
Availability of data and materials
The 16S rRNA gene and ITS2 sequences have been submitted to the European Nucleotide Archive (Accession N◦ ERP134948).
Code availability
The scripts used for bioinformatics analysis during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The study was approved by the Research Ethics Committee of Bordeaux University Hospital (CER BDX-2021-36) and performed according to The Code of Ethics of the World Medical Association (Declaration of Helsinki). An informed consent was obtained from all patients or from their legal representatives.
Consent for publication
Not applicable.
Competing interests
On behalf of all authors, the corresponding author states that there are no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164:337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 2.Zaborin A, Smith D, Garfield K, Quensen J, Shakhsheer B, Kade M, et al. Membership and behavior of ultra-low-diversity pathogen communities present in the gut of humans during prolonged critical illness. MBio. 2014;5:e01361–01314. doi: 10.1128/mBio.01361-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonald D, Ackermann G, Khailova L, Baird C, Heyland D, Kozar R, et al. Extreme dysbiosis of the microbiome in critical illness. mSphere. 2016;1:e00199–16. doi: 10.1128/mSphere.00199-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lankelma JM, van Vught LA, Belzer C, Schultz MJ, van der Poll T, de Vos WM, et al. Critically ill patients demonstrate large interpersonal variation in intestinal microbiota dysregulation: a pilot study. Intensive Care Med. 2017;43:59–68. doi: 10.1007/s00134-016-4613-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haak BW, Wiersinga WJ. The role of the gut microbiota in sepsis. Lancet Gastroenterol Hepatol. 2017;2:135–143. doi: 10.1016/S2468-1253(16)30119-4. [DOI] [PubMed] [Google Scholar]
- 6.Rubio E, Vergara A, Aziz F, Narváez S, Cuesta G, Hernández M, et al. Changes in the gut microbiota and risk of colonization by multidrug-resistant bacteria, infection and death in critical care patients. Clin Microbiol Infect. 2022;S1198-743X(22)00024-6. [DOI] [PubMed]
- 7.ARDS Definition Task Force. Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, Caldwell E, et al. Acute respiratory distress syndrome: the Berlin Definition. JAMA. 2012;307:2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 8.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The Third International Consensus Definitions for sepsis and septic shock (sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.On behalf of the Acute Disease Quality Initiative Workgroup 16. Chawla LS, Bellomo R, Bihorac A, Goldstein SL, Siew ED, et al. Acute kidney disease and renal recovery: consensus report of the Acute Disease Quality Initiative (ADQI) 16 Workgroup. Nat Rev Nephrol. 2017;13:241–257. doi: 10.1038/nrneph.2017.2. [DOI] [PubMed] [Google Scholar]
- 10.Vandenborght L-E, Enaud R, Urien C, Coron N, Girodet P-O, Ferreira S, et al. Type 2-high asthma is associated with a specific indoor mycobiome and microbiome. J Allergy Clin Immunol. 2021;147:1296–1305.e6. doi: 10.1016/j.jaci.2020.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pauvert C, Buée M, Laval V, Edel-Hermann V, Fauchery L, Gautier A, et al. Bioinformatics matters: the accuracy of plant and soil fungal community data is highly dependent on the metabarcoding pipeline. Fungal Ecol. 2019;41:23–33. doi: 10.1016/j.funeco.2019.03.005. [DOI] [Google Scholar]
- 13.McKnight DT, Huerlimann R, Bower DS, Schwarzkopf L, Alford RA, Zenger KR. microDecon: a highly accurate read-subtraction tool for the post-sequencing removal of contamination in metabarcoding studies. Environ DNA. 2019;1:14–25. doi: 10.1002/edn3.11. [DOI] [Google Scholar]
- 14.Jiang TT, Shao T-Y, Ang WXG, Kinder JM, Turner LH, Pham G, et al. Commensal fungi recapitulate the protective benefits of intestinal bacteria. Cell Host Microbe. 2017;22:809–816.e4. doi: 10.1016/j.chom.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu R, Tan C, Zhu J, Zeng X, Gao X, Wu Q, et al. Dysbiosis of the intestinal microbiota in neurocritically ill patients and the risk for death. Crit Care. 2019;23:195. doi: 10.1186/s13054-019-2488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agudelo-Ochoa GM, Valdés-Duque BE, Giraldo-Giraldo NA, Jaillier-Ramírez AM, Giraldo-Villa A, Acevedo-Castaño I, et al. Gut microbiota profiles in critically ill patients, potential biomarkers and risk variables for sepsis. Gut Microbes. 2020;12:1707610. doi: 10.1080/19490976.2019.1707610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei R, Chen X, Hu L, He Z, Ouyang X, Liang S, et al. Dysbiosis of intestinal microbiota in critically ill patients and risk of in-hospital mortality. Am J Transl Res. 2021;13:1548–1557. [PMC free article] [PubMed] [Google Scholar]
- 18.Ojima M, Shimizu K, Motooka D, Ishihara T, Nakamura S, Shintani A, et al. Gut dysbiosis associated with antibiotics and disease severity and its relation to mortality in critically ill patients. Dig Dis Sci. 2021;3:1–13. doi: 10.1007/s10620-021-07000-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamarche D, Johnstone J, Zytaruk N, Clarke F, Hand L, Loukov D, et al. Microbial dysbiosis and mortality during mechanical ventilation: a prospective observational study. Respir Res. 2018;19:245. doi: 10.1186/s12931-018-0950-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickson RP, Schultz MJ, van der Poll T, Schouten LR, Falkowski NR, Luth JE, et al. Lung microbiota predict clinical outcomes in critically ill patients. Am J Respir Crit Care Med. 2020;201:555–563. doi: 10.1164/rccm.201907-1487OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sepsis Lung Microbiome Study Group Could lung bacterial dysbiosis predict ICU mortality in patients with extra-pulmonary sepsis? A proof-of-concept study. Intensive Care Med. 2020;46:2118–2120. doi: 10.1007/s00134-020-06190-4. [DOI] [PubMed] [Google Scholar]
- 22.Becattini S, Taur Y, Pamer EG. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med. 2016;22:458–478. doi: 10.1016/j.molmed.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferreira-Halder CV, de Sousa Faria AV, Andrade SS. Action and function of Faecalibacterium prausnitzii in health and disease. Best Pract Res Clin Gastroenterol. 2017;31:643–648. doi: 10.1016/j.bpg.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 24.Qin P, Zou Y, Dai Y, Luo G, Zhang X, Xiao L. Characterization of a novel butyric acid-producing bacterium Collinsella aerofaciens subsp. shenzhenensis subsp. nov. Microorganisms. 2019;7:E78. doi: 10.3390/microorganisms7030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quagliariello A, Del Chierico F, Reddel S, Russo A, Onetti Muda A, D’Argenio P, et al. Fecal microbiota transplant in two ulcerative colitis pediatric cases: gut microbiota and clinical course correlations. Microorganisms. 2020;8:E1486. doi: 10.3390/microorganisms8101486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi K, Nishida A, Fujimoto T, Fujii M, Shioya M, Imaeda H, et al. Reduced abundance of butyrate-producing bacteria species in the fecal microbial community in Crohn’s disease. Digestion. 2016;93:59–65. doi: 10.1159/000441768. [DOI] [PubMed] [Google Scholar]
- 27.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre M-L, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller PL, Carson TL. Mechanisms and microbial influences on CTLA-4 and PD-1-based immunotherapy in the treatment of cancer: a narrative review. Gut Pathog. 2020;12:43. doi: 10.1186/s13099-020-00381-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mullish BH, McDonald JAK, Pechlivanis A, Allegretti JR, Kao D, Barker GF, et al. Microbial bile salt hydrolases mediate the efficacy of faecal microbiota transplant in the treatment of recurrent Clostridioides difficile infection. Gut. 2019 doi: 10.1136/gutjnl-2018-317842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sánchez E, Ribes-Koninckx C, Calabuig M, Sanz Y. Intestinal Staphylococcus spp. and virulent features associated with coeliac disease. J Clin Pathol. 2012;65:830–834. doi: 10.1136/jclinpath-2012-200759. [DOI] [PubMed] [Google Scholar]
- 31.Baumgartner M, Lang M, Holley H, Crepaz D, Hausmann B, Pjevac P, et al. Mucosal biofilms are an endoscopic feature of irritable bowel syndrome and ulcerative colitis. Gastroenterology. 2021;S0016–5085(21):03138–3143. doi: 10.1053/j.gastro.2021.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serichantalergs O, Ruekit S, Pandey P, Anuras S, Mason C, Bodhidatta L, et al. Incidence of Campylobacter concisus and C. ureolyticus in traveler’s diarrhea cases and asymptomatic controls in Nepal and Thailand. Gut Pathog. 2017;9:47. doi: 10.1186/s13099-017-0197-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soret P, Vandenborght L-E, Francis F, Coron N, Enaud R, Avalos M, et al. Respiratory mycobiome and suggestion of inter-kingdom network during acute pulmonary exacerbation in cystic fibrosis. Sci Rep. 2020;10:3589. doi: 10.1038/s41598-020-60015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Limon JJ, Tang J, Li D, Wolf AJ, Michelsen KS, Funari V, et al. Malassezia is associated with Crohn’s disease and exacerbates colitis in mouse models. Cell Host Microbe. 2019;25:377–388.e6. doi: 10.1016/j.chom.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El Mouzan M, Al-Hussaini A, Fanelli B, Assiri A, AlSaleem B, Al Mofarreh M, et al. Fungal dysbiosis in children with celiac disease. Dig Dis Sci. 2021;67:216–223. doi: 10.1007/s10620-021-06823-8. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Z, Chen L, Xu P, Hong Y. Predictive analytics with ensemble modeling in laparoscopic surgery: a technical note. Laparosc Endosc Robot Surg. 2022;5:25–34. doi: 10.1016/j.lers.2021.12.003. [DOI] [Google Scholar]
- 37.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 38.Yelin I, Flett KB, Merakou C, Mehrotra P, Stam J, Snesrud E, et al. Genomic and epidemiological evidence of bacterial transmission from probiotic capsule to blood in ICU patients. Nat Med. 2019;25:1728–1732. doi: 10.1038/s41591-019-0626-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplemental Figure 1. Non metric Bray-curtis analysis of β-diversity of the V3-V4 sequencing run. XXA, B, and C samples: gut bacteriobiota samples. GXX: lung bacteriobiota samples. BlancV3-V4: negative control. Mock: mock community. Supplemental Figure 2. Non metric Bray-curtis analysis of β-diversity of ITS2 sequencing run. XXA, B, and C samples: gut mycobiota samples. GXX: lung mycobiota samples. Blanc1 and blanc2: negative controls. Mock: mock community.
Data Availability Statement
The 16S rRNA gene and ITS2 sequences have been submitted to the European Nucleotide Archive (Accession N◦ ERP134948).
The scripts used for bioinformatics analysis during the current study are available from the corresponding author on reasonable request.