Abstract
Swimming motility is a flagellum-dependent form of movement observed in the Gram-negative bacterium Pseudomonas aeruginosa. Swimming motility is defined as the movement in liquid or low-viscosity conditions (up to 0.3 % agar concentration). Unlike swarming motility, swimming motility requires a functional flagellum, but neither quorum sensing (QS) systems nor biosurfactants. While swimming motility can also be observed via microscopy, here we describe a reproducible plate-based method.
Keywords: Swimming motility, Flagellum, Chemotaxis
1. Introduction
Pseudomonas aeruginosa has a polar flagellum that it uses to swim in liquid-to-low viscosity environments. This flagellum is composed of numerous proteins including the cap, filament, hook, and the basal body. Collectively, the hook and the filament are capable of both clockwise (CW) and counterclockwise (CCW) rotation to propel the bacterium, while the basal body (stator and motor parts combined) hold the flagellum in place, as well as conductions necessary to power this machine [1]. Similar to ATP synthesis via oxidative phosphorylation, conductance of cations (H+ for most neutrophiles, and Na+ for alkalophiles and Vibrio species) through the basal body fuels flagellar motility [2–4].
Swimming motility is a unicellular behavior, and requires a functional polar flagellum with its motor-stator complex. The plate-based swimming motility assay described below measures the degree of this flagellar-dependent motility of the bacterium. As a consequence, a mutant lacking a properly functioning flagellum demonstrates decreased motility compared to its wild-type counterpart (see Fig. 1). It is important to note that the extent of the bacterium’s swimming motility is scored based on its radial migration through the agar.
Fig. 1.
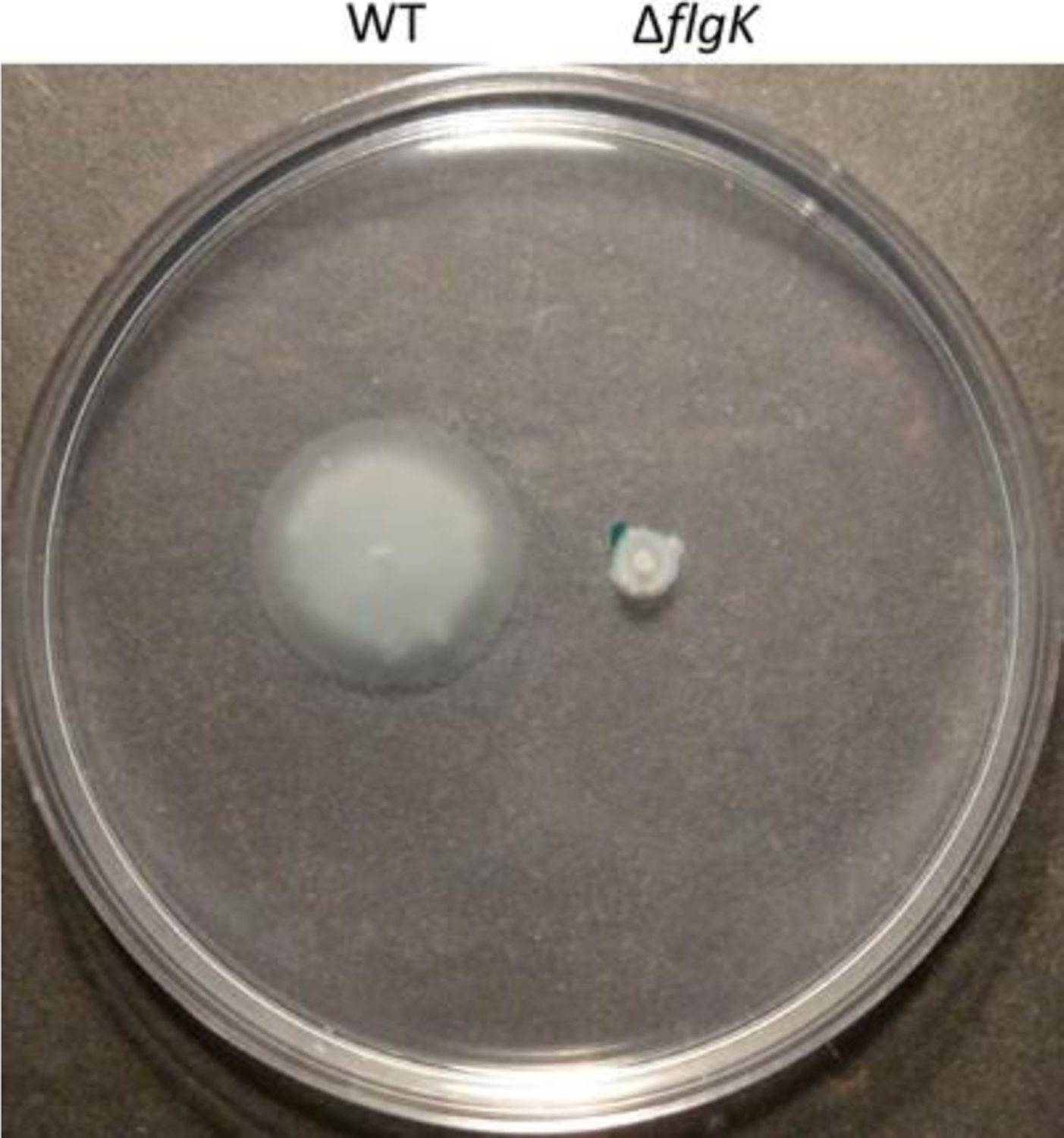
Swimming motility of Pseudomonas aeruginosa PA14 wild type vs. ΔflgK mutant
It should also be noted that in addition to a functioning flagellum and the basal body, chemotaxis machinery is also important for proper bacterial motility. Loss of chemotaxis results in a phenotype similar to loss of flagellar function, and thus, this plate-based assay does not readily distinguish between flagellar and chemotactic defects. Therefore, should the investigator be interested in chemotaxis-related phenotypes, microscopy-based observations are a recommended alternative approach to this plate-based assay.
In this section, we describe a reproducible, plate-based method to measure swimming motility in P. aeruginosa. Although some reports have documented swimming motility assays conducted on rich media, e.g., lysogeny broth (LB) agar plates [5, 6], we have used and found M8-supplemented plates to be both consistent and reliable. And while we aim for uniformity in these assays, our general protocol should help produce reproducible results regardless of the chosen medium.
2. Materials
Prepare all solutions using ultrapure, distilled deionized water. All solutions used in this assay are sterilized (autoclaved at least 45 min at 121 °C, 15 psi for 1 L solution) prior to use and are kept at room temperature (unless indicated otherwise). Follow all waste disposal procedures when disposing of contaminated solutions at the completion of the experiments.
Tube(s) of P. aeruginosa strain(s) grown overnight at 37 °C in lysogeny broth (LB: 0.5 g yeast extract, 1 g tryptone, 0.5 g NaCl in 100 mL water) from a fresh streak plate, <7 days old.
Petri plates: sterile, 100 mm × 15 mm.
Autoclavable bottle: 1 L.
Autoclavable flask: 2 L.
Automatic pipettor.
Disposable, sterile serological pipettes: 10, 25, 50 mL.
20 % Casamino acids solution in water (see Note 1).
20 % Glucose solution in water.
1 M MgSO4 solution in water.
Pipette: P1000.
Agar.
1.5 mL disposable, sterile microcentrifuge tubes.
Sterile wooden toothpicks or sterile yellow plastic disposable pipette tips.
5× M8 solution: dissolve 64 g Na2HPO4·7H2O, or alternatively, 30 g Na2HPO4; 15 g KH2PO4; 2.5 g NaCl in water, and bring the final volume to 1 L (see Note 2).
3. Methods
All procedures are performed at room temperature (RT) unless specified otherwise. The following protocol is scaled to make 1 L batch of agar. This protocol for 1 L batch should yield roughly 30–36 swimming motility plates. For a different volume, scale accordingly to maintain the appropriate concentration for each component.
Add 3 g (for final concentration of 0.3 %) of agar to 800 mL of water. Autoclave sufficiently in 2 L flask to yield a sterile, homogeneous agar suspension; 45 min cycle is sufficient for a 1 L batch (see Note 3).
Autoclave 5× M8 solution in 1 L bottles. Add 200 mL of 5× M8 solution to the melted agar (see Note 4).
Add 10 mL of 20 % glucose to the melted agar; final concentration: 0.2 % glucose.
Add 25 mL of 20 % casamino acids to the melted agar; final concentration: 0.5 % casamino acids.
Add 1 mL of 1 M MgSO4 to the melted agar; final concentration: 1 mM MgSO4.
Mix the agar medium and cool prior to pouring onto petri plates (see Note 5).
Pour thick plates (~25 mL/plate) and let plates solidify at room temperature (RT) for a few hours (see Note 6).
Pipet a small volume (~100 μL) of the overnight bacterial culture into a 1.5 mL microcentrifuge tube.
Using a sterile toothpick (or a sterile, disposable yellow pipette tip), dip it in the overnight culture. Use the same toothpick (or pipette tip) to stab into the agar layer of the plate, but not all the way to the base of the petri plate (see Notes 7 and 8).
Incubate plate(s) upright at 37 °C for 16–24 h (see Note 9) and observe the phenotype. For semi-quantitative analysis, software like ImageJ can be used to compare radial growth of swimming phenotype between strains (see Note 10).
Fig. 2.
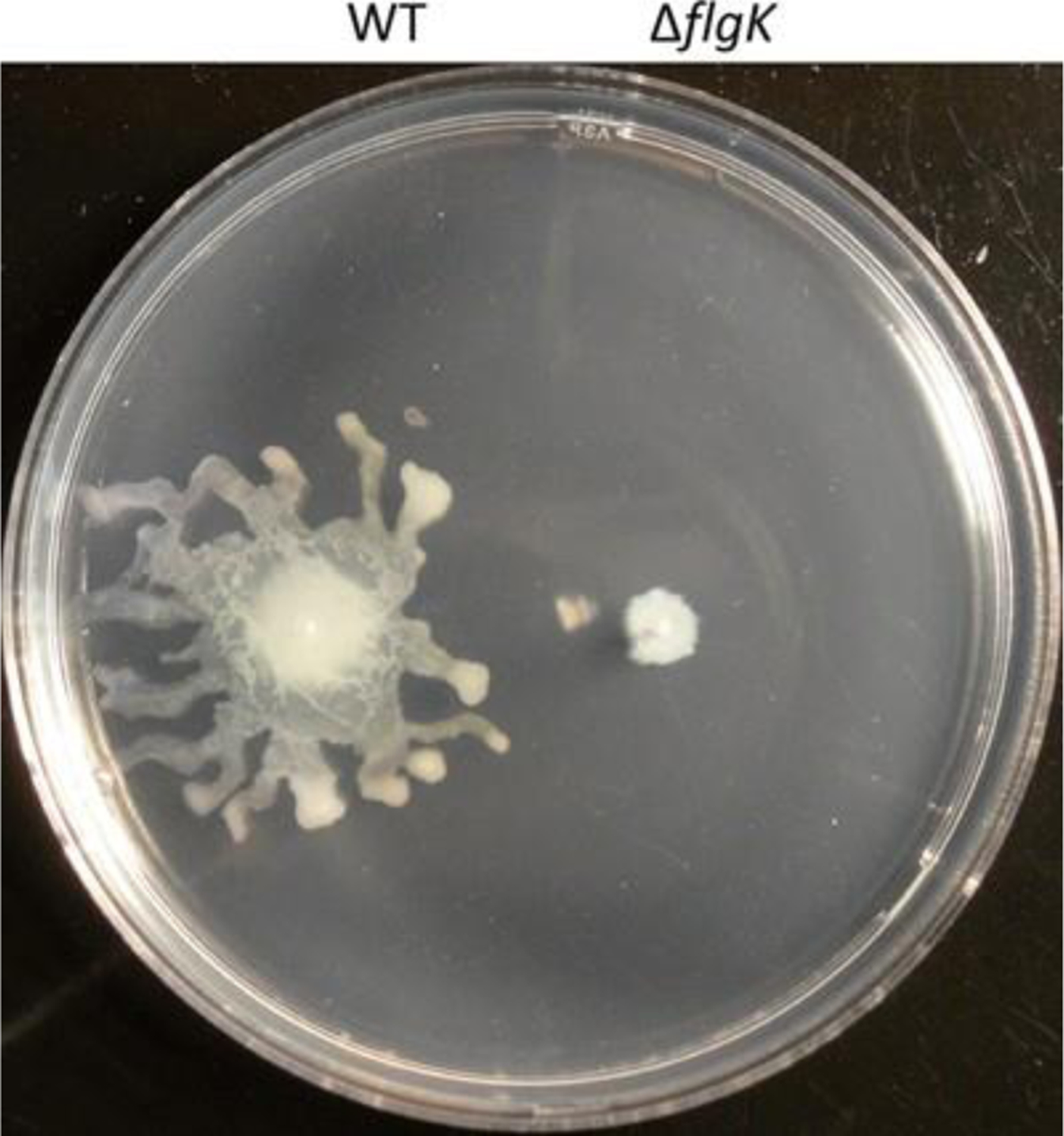
Occasional swarming motility may interfere with interpreting swimming motility
Acknowledgement
This work was supported by the NIH grant R01A1003256 to G.A.O. and the Rosaline Borison predoctoral fellowship awarded to D.G.H.
4 Notes
Since 5× M8 solution lacks any source of nitrogen, supplementation with a nitrogen source is key in a swimming motility assay. Therefore, casamino acids can be substituted with a different source of nitrogen depending on the investigator’s experimental question. As for 20 % stock solution of casamino acids, precipitates do accrue in older solutions at RT. In our experience, while these precipitates do not redissolve with increased temperature (~50 °C), they do not impact the final outcome of swimming motility phenotypes.
Completely dissolving Na2HPO4 (both for heptahydrate- and anhydrous-forms) into water takes some time; however, warming the solution is unnecessary. Add reagents to 800 mL of water before bringing up the final volume to 1 L.
We find it best to prepare a fresh batch of molten agar on the day of the assay. Addition of a magnetic stir bar to the agar is recommended, as it will assist in mixing other components.
Depending on the temperature of the molten agar, addition of the 5× M8 solution could either prematurely solidify the agar, or speed up the cooling process. We find that the best approach is to pre-warm the 5× M8 solution in a warm water bath (~55 °C) prior to adding into the molten agar. If working on a shorter time frame, however, 5× M8 solution at RT can be added to a hot molten agar to decrease the time needed to cool the molten agar prior to pouring plates.
While swimming motility measures bacterial movements within the agar layer, and not on top, for clear and easy to image swim plates, we recommend pouring a smooth agar layer. Mix the added components using a magnetic stir bar; aggressive mixing tends to create unnecessary foam. Once poured, any remaining bubbles/foam can be removed by gently running a flame over a poured plate (not recommended on batch volumes >1 L, as the plates begin solidifying rather quickly once poured). Agar suspension that is cool to the touch (5 s to the touch without burning one’s hand) is when we generally start pouring plates. We feel this temperature is warm enough to prevent any premature solidifying of the agar, but also cool enough to pour manually without feeling too hot, nor melting the plastic petri plates.
Plate thickness does not have to be exact, but the general rule of thumb is that thicker plates will produce better results. The reason being that thicker agar layer retains moisture longer, and prevents premature drying of the plate. Since swimming motility occurs through the agar medium, retention of moisture ensures a constant viscosity. After the plates have been poured, we find it best to solidify them either as single plates or in small stacks (<4 plates/stack) of plates for approximately 3–4 h, as plates on the upper level of the stack tend to dry slower than those at the bottom. When plates are dry, make sure to remove any condensation that has accumulated on the underside of the petri plate cap. Overnight incubation at 37 °C will further increase condensation, which may fall back onto the agar.
Whereas swimming motility occurs in the agar medium, any residual inoculum on the surface of the agar can result in unwanted swarming motility. Tendril projections of swarming motility may interfere with estimating the radial growth representing swimming motility (see Fig. 2). In order to minimize residual inoculum on the agar surface, we find it best to preferentially use a pointy-ended toothpick or pipet tip, and also to dip less (do not dunk) into the overnight culture. Prior to inoculating the agar medium, briefly check the toothpick or pipet tip end to ensure there is not any excess inoculum on the sides. Furthermore, it is helpful to keep the toothpick or pipet tip perpendicular to the agar medium to prevent any unintentional inoculation on the agar surface.
Should the inoculum pass through the entire layer of the agar medium and come in contact with the basal petri plate surface, twitch motility may occur and may interfere with the interpretation of the radial growth depicting swimming motility. It is recommended that the investigator simply poke part way into the agar, rather than applying force to stab into the agar medium. Considering the possibility of these unintentional errors, we recommend replicate plates per strain tested (at least 3 per strain).
Even though the agar solidifies after 3–4 h on the benchtop, due to its low viscosity (0.3 % agar), the plate should be maintained in an upright position. This applies to both during- and post-incubation at 37 °C. Failure to do so and inverting the plate for any extended period of time (>10 min) may result in loss of the integrity of the agar medium, allowing it to detach from the base of the petri plate. Due to lower viscosity and easier detachment of the agar, it is also recommended that the investigator take extra caution when disposing of these plates.
While this plate-based assay can be scored qualitatively, software such as ImageJ can aid in semi-quantitatively measuring the swimming motility of each strain. Simply measure the area covered by each strain’s swimming motility zone (as depicted by a radial growth), and draw a comparison with other strains in question. In case of an obstructed view due to swarming phenotype, omit the plate from consideration, and if necessary, repeat the assay.
References
- 1.Blair DF (2003) Flagellar movement driven by proton translocation. FEBS Lett 545: 86–95 [DOI] [PubMed] [Google Scholar]
- 2.Larsen SH, Adler J, Gargus JJ, Hogg RW (1974) Chemomechanical coupling without ATP: the source of energy for motility and chemotaxis in bacteria. Proc Natl Acad Sci USA 71:1239–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manson MD, Tedesco P, Berg HC, Harold FM, Van der Drift C (1977) A protonmotive force drives bacterial flagella.Proc Natl Acad Sci USA 74:3060–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirota N, Imae Y (1983) Na+-driven flagellar motors of an alkalophilic Bacillus strain YN-1. J Biol Chem 258:10577–10581 [PubMed] [Google Scholar]
- 5.Murray TS, Ledizet M, Kazmierczak BI (2010) Swarming motility, secretion of type 3 effectors and biofilm formation phenotypes exhibited within a large cohort of Pseudomonas aeruginosa clinical isolates. J Med Microbiol 59:511–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soscia C, Hachani A, Bernadac A, Filloux A, Bleves S (2007) Cross talk between type III secretion and flagellar assembly systems in Pseudomonas aeruginosa. J Bacteriol 189:3124–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]


