Abstract

Aldehyde dehydrogenases (ALDHs) are overexpressed in various tumor types including prostate cancer and considered a potential target for therapeutic intervention. 4-(Diethylamino)benzaldehyde (DEAB) has been extensively reported as a pan-inhibitor of ALDH isoforms, and here, we report on the synthesis, ALDH isoform selectivity, and cellular potencies in prostate cancer cells of 40 DEAB analogues; three analogues (14, 15, and 16) showed potent inhibitory activity against ALDH1A3, and two analogues (18 and 19) showed potent inhibitory activity against ALDH3A1. Significantly, 16 analogues displayed increased cytotoxicity (IC50 = 10–200 μM) compared with DEAB (>200 μM) against three different prostate cancer cell lines. Analogues 14 and 18 were more potent than DEAB against patient-derived primary prostate tumor epithelial cells, as single agents or in combination treatment with docetaxel. In conclusion, our study supports the use of DEAB as an ALDH inhibitor but also reveals closely related analogues with increased selectivity and potency.
Introduction
The aldehyde dehydrogenase (ALDH) superfamily of 19 different human isoforms is a group of NAD(P)+-dependent enzymes that catalyze several cellular processes, including detoxification of endogenous and exogenous aldehydes and biosynthesis of retinoic acid (RA), which is a modulator of stem cell (SC) differentiation.1−3 Recent interest in ALDHs emerges from their roles linked to cancer cell proliferation, differentiation, and survival4,5 and potential as markers of tumor-initiating cells or cancer SCs (CSCs).6
Identification of CSCs is frequently carried out using the Aldefluor assay, which includes the addition of 4-(diethylamino)benzaldehyde (DEAB), a small-molecule inhibitor that is used as a control to identify subpopulations of cells with high ALDH expression (ALDHhigh) and with SC-like properties.4 DEAB is a pan-ALDH inhibitor, which has been found to delay differentiation of CSCs,7,8 also showing potential in combination treatments with other drugs.9−13 Early work showed that 4-(dipropylamino)benzaldehyde was more potent than DEAB as a reversible inhibitor against mouse and human ALDH1, with variable inhibitory effects according to the selected substrate.14 In the same study, 4-(dimethylamino)benzaldehyde and 4-(dibutylamino)benzaldehyde showed low binding affinity to ALDH1 but provided no information on isoform selectivity.14 Many studies indicate that DEAB is a reversible and broad inhibitor of several ALDH isoforms, which is a confounding factor when using the Aldefluor assay.15,16 The complexity of the ALDH interaction is evident by recent work, which has shown that DEAB is also a substrate for ALDH1A1 and ALDH3A1,15 an irreversible inhibitor for ALDH1A2 and ALDH2, and neither a substrate nor inhibitor for ALDH1L1 and ALDH4A1, while it has been shown to be metabolized by ALDH1A3, ALDH1B1, and ALDH5A1.17 Additionally, DEAB has been shown to covalently bind to ALDH7A1, in which the DEAB-enzyme complex was successfully cocrystallized with the cofactor NAD+.18 It has been proposed that the structural features of the amino acid residues at the ALDH active sites are the main factors determining whether DEAB is a substrate or an inhibitor.17 Moreover, the ALDH–DEAB binary structure has been hypothesized to be stabilized by resonance arrangement, which is initiated and supported by the amine electron donors at the para position to the carbonyl group.17
ALDH1A1 and 1A3 are highly expressed in SC-like subpopulations and several cancer types.8,19 We have recently reported ALDH1A1 and 1A3 isoform expression in different prostatic tissue-derived cell lines (normal, benign, and malignant) and patient-derived primary prostate tumor epithelial cells and shown potential in inhibiting these for therapeutic intervention.20 Given the utility of DEAB in both identifying CSC populations21 and potential in treating such ALDHhigh-expressing populations,8 we decided to generate a new library of analogues that could be used to explore the DEAB scaffold to unravel key features. Here, we report on the synthesis, biological evaluation against ALDH1A1, 1A3, and 3A1, and antiproliferative activity as single agents against a panel of prostate cancer (PCa) cell lines. We also assessed selected agents for their potential in combination treatment with docetaxel.
Results and Discussion
Chemistry
DEAB analogues were synthesized using either aliphatic or aromatic nucleophilic substitution one-step reactions. The nucleophilic aromatic substitution of fluorine by the desired secondary amine is shown in Scheme 1A. The presence of the aldehyde linked para to the fluorine group is likely facilitating the nucleophilic attack through electronic arrangement within the aromatic ring.22−24 The aldehyde is chemically reactive toward amines; however, as imines are less likely to be produced by secondary amines, the products were generally afforded in good yields (33–100%). Compounds 16, 33, and 35 were synthesized by the aliphatic nucleophilic substitution reaction between the phenolic hydroxyl group and isopropyl iodide, as shown in Scheme 1B.25 All DEAB analogues were synthesized, purified, and characterized by 1H NMR, 13C NMR, and HRMS as described in the Experimental Section.
Scheme 1. Nomenclature of DEAB Analogues and General Synthesis Scheme; Reaction Starts by a Nucleophilic Substitution at the Fluorine (A) or Hydroxyl Group (B) of the Substituted Benzaldehyde.
Standard conditions: (a) secondary amine, DMF, K2CO3, 25–100 °C and (b) 2-iodopropane, DMF, K2CO3, 25–90 °C, 6 h.
Inhibitory Effect of DEAB Analogues against ALDH Isoforms
Previous studies have established the importance of ALDH1 isoforms: 1A126 and 1A327 in CSCs and 3A128 in drug resistance. Accordingly, we decided to explore a new library of DEAB analogues for interaction with these isoforms by generating structure–activity relationships (SAR) useful in informing DEAB properties critical to biological activity.
The inhibition screening and IC50 values were assayed using the conditions described in the Experimental Section. The IC50 value for compound 7 was measured at a saturating substrate concentration with ALDH3A1 because the same remaining activity was observed in inhibition screening both near the Km value and at the saturating substrate concentration. Experimental values are shown as the mean ± SE. IC50 values and represent the concentration of the compound that decreases 50% the enzyme activity determined in the absence of the inhibitor. ND = not determined.
ALDH1A1
Given the presence of the aldehyde group and the evidence that DEAB has been reported to be a slow ALDH1A1 substrate, we decided to perform both inhibition and substrate studies. Most compounds displayed lower inhibitory potency against ALDH1A1 compared with DEAB and were evaluated at an initial dose of 10 μM (Figure S1). Due to the significant substrate activity shown with many compounds, even much higher than that with DEAB (Figure S2), it was difficult to assess their inhibitory properties.
Accordingly, we captured full data analysis for compounds 14, 26, 29, and DEAB, and IC50 values were calculated for the best fits, demonstrating the latter two compounds to be nearly equipotent with DEAB (IC50 = 0.48 ± 0.06 μM), while compound 14 was ∼15-fold less effective in inhibiting ALDH1A1 activity (Table 1). In general, those compounds with a better inhibition profile had dipropyl, diethyl, or a pyrrolidine as an R group, the latter being the best, especially compared to those with other groups such as morpholine or methyl-piperazine. Among those compounds that provided satisfactory results, unsubstituted analogues at the meta position (X group) yielded better inhibition results (measured as remaining activity at 10 μM compound) than those substituted with halogen atoms, that is, DEAB (m-H, 2%) versus 13 (m-Br, 24%), pyrrolidine 4 (m-H, 10%) versus 12 (m-Cl, 17%), or dipropylamine 1 (m-H, 12%) versus 14 (m-Br, 45%) (Figure S1). In fact, 14 yielded an IC50 of 7.08 ± 0.70 μM, approximately 15-fold less active when compared to DEAB as an inhibitor for ALDH1A1 (IC50 = 0.48 ± 0.06 μM). Contrary to ALDH1A3 and 3A1, the most potent ALDH1A1 inhibitors were analogues bearing an electron-donating group: 26 (m-CH3, IC50 = 0.80 ± 0.16 μM) and 29 (m-OCH3, IC50 = 0.88 ± 0.05 μM) (Table 1 1).
Table 1. IC50 Values of the Most Potent DEAB Analogues against ALDH1A1, ALDH1A3, and ALDH3A1 Isoforms.

ALDH1A3
From the initial one-dose (10 μM) evaluation (Figure S1), the selection of compounds was based not only on inhibitory properties but also on their ability to act as substrates for the other two ALDH isoforms. Some compounds were shown to display very low IC50 values regarding ALDH1A3 (Table 1), yet its high percentage of substrate activity for ALDH1A1 and/or ALDH3A1 would complicate their further investigation and use as inhibitors. For example, 16 displayed the lowest IC50 value of all compounds in the library for ALDH1A3 (0.26 μM), yet it was shown to be a good ALDH3A1 substrate (Figure S2). Similarly, compounds 7 and 14 revealed comparable IC50 values (0.55 and 0.66 μM, respectively); however, 7 presented a significantly higher ALDH1A1 substrate activity compared to that of 14 (17.9 vs 3.14%). This result may be due to chlorine being a smaller atom compared to bromine, and thus, steric hindrance might explain why compound 7 can be better accommodated into the active site. Nonetheless, ALDH1A3 studies revealed many compounds to be superior to DEAB in inhibiting ALDH1A3 enzymatic activity with hexanal as a substrate (Table 1 and Figure S1).
There are several interesting SARs that reveal trends of inhibitory properties. Analogues bearing a methyl group at the meta position to the aldehyde and an aliphatic moiety at the para position reveal that the modulation of the latter from diethyl (DEAB) or dipropyl (1) to constrained heterocycles based on pyrrolidine (26) or piperidine (15) rings increases the inhibitory effect: 15 (IC50 = 0.29 μM) > 26 (IC50 = 1.15 μM) > DEAB (IC50 = 10.4 μM) ≈ 1 (only tested at 10 μM, Figure S1). Analogues incorporating para-positioned piperazine 24 and especially morpholine 27 also showed inhibitory capacity decreasing ALDH1A3 activity down to 45 and 10% at 10 μM dose, respectively. Pyrrolidine 12 with a chlorine installed at the meta position to the aldehyde was almost twice as potent (IC50 = 0.31 μM) as the propyl analogue 7 (IC50 = 0.55 μM), further substantiating the presence of a heterocycle at the para position for enhanced potent ALDH1A3 inhibitory activity. Interestingly, 34 was the only compound with an NO2 electron-withdrawing group that appeared effective at inhibiting ALDH1A3, with an IC50 = 0.27 μM. Comparison of propyl analogues 7 (m-Cl) and 14 (m-Br) revealed similar IC50 values (0.55 and 0.63 μM, respectively), indicating that the atomic volume and electronegativity of these halogens are not critical for ALDH1A3 inhibition.
Notably, comparison of diethyl 13 (IC50 = 10.7 μM) and diisopropyl 14 (IC50 = 0.63 μM) compounds revealed approximately 17-fold difference in capacity to inhibit ALDH1A3. This result suggests that the extra two methyl groups may provide additional nonpolar interactions with the active-site pocket, enough to strengthen binding. Figure S6 depicts the comparison of docking poses of 13 with 14 and DEAB within the ALDH1A3 binding pocket. Analogue 13 demonstrated a similar binding orientation as DEAB, establishing H-bonds with Cys313, Cys314, and Thr315. Both the N,N-diethyl side chains of 13 and DEAB established van der Waals contacts with Ile132 and Leu185 in the binding site of ALDH1A3, whereas analogue 14 was able to form only one H-bond with Thr315 and maintained van der Waals contacts with Ile132 and Leu185. Interestingly, the two extra methyl groups at the side chain of 14 were accommodated between Trp189 and Leu471 residues, favoring stronger van der Waals contacts than 13 and DEAB, thus improving its binding to ALDH1A3. Therefore, these findings may explain why 13 acted as an ALDH1A1 substrate with almost 60% activity relative to hexanal, whereas 14 only yielded 4% activity (Figure S2).
From all the compounds with a CH3 group at the meta position, compound 15 displayed the most potent inhibitory properties. Compound 16, containing an isopropoxy group at the para position, was found to be one of the most potent compounds under the conditions investigated (IC50 = 0.26 μM). Significantly, several new analogues exhibited up to 100-fold higher potency in inhibiting ALDH1A3 as compared to DEAB (IC50 = 10.20 ± 2.15 μM) when using hexanal as a substrate.
The type of inhibition and Ki values determined for compounds 14, 15, and 16 against ALDH1A3 are shown in Figures 1, S4, and S5, respectively. It can be observed that when increasing the inhibitor concentration, the Km value tends to increase, whereas the Vmax value barely changes. This behavior best fits with a competitive type of inhibition. For these compounds, Ki values are all below 1 μM, in good agreement with the IC50 values. The calculated value for 14 was Ki = 0.46 ± 0.15 μM, which indicates that this compound is an excellent inhibitor for ALDH1A3.
Figure 1.
Inhibition kinetics of ALDH1A3 by compound 14 at various concentrations of inhibitor: −○– 0 μM; −●– 0.25 μM; −□– 2.5 μM; and −■– 5 μM. Hexanal was used as the substrate. The values of the kinetic parameters calculated from a fit to the competitive inhibition equation are Vmax = 0.37 ± 0.01 U/mg; Km = 16.1 ± 4 μM; and Ki = 0.46 ± 0.15 μM. Results are the mean ± SE of duplicate experiments.
With all observations taken together, it can be concluded that compound 14 yielded the most promising inhibition parameters while also displaying very low activity as a substrate for the ALDH1A1 and ALDH3A1 isoforms (lower than 5% at 10 μM for both isoforms).
ALDH3A1
Compounds 14, 18, and 19 were shown to display IC50 values below 10 μM, with both 18 (IC50 = 1.61 μM) and 19 (IC50 = 1.29 μM) approximately threefold more potent than DEAB (IC50 = 5.67 μM). The latter two compounds share similar chemical structures, and the inhibitory activity is most likely related to three structural features: the lipophilicity of the para-substituted functional groups, the side chain flexibility involving free rotations, and the meta-substituted functional group. They differ in the para substitution groups diethylamine and dipropylamine. Compared to other analogues, these groups appear to be better accommodated in the active site and show more activity than those compounds with a similar m-NO2 group but with p-substituted groups of lower lipophilicity (i.e., morpholine 32 or pyrrolidine 34).
It can be concluded that the NO2 group at the meta position seems to be providing these compounds with higher inhibitory activity. The docking analysis suggested that Tyr115, one of the ALDH3A1 residues that normally interacts with the CHO part of the molecule, presents some interaction with the NO2 group as well. This additional binding might be related to the better inhibitory capacity.
The only structural difference among 18 and 19 is the N-substituted group at the para position to the aldehyde, and although the results of the ALDH3A1 inhibition are very similar, their ability to act as substrates differs, especially with ALDH1A1 (17% of 18 vs 69% of 19, Figure S2). Other para-substituted dipropylamine analogues (1, 7, 14, 17, 18, and 38) were evaluated, but only those with electron-withdrawing groups demonstrated the capacity to significantly inhibit ALDH3A1 activity (below 15% remaining activity). Selectivity might be linked to a Gln122 residue, as a site for providing selectivity for ALDH3A1 only.29
Kinetic evaluation of both 18 and 19 revealed them as excellent candidates for ALDH3A1 inhibition. Both compounds were further characterized as ALDH3A1 inhibitors by determination of the type of inhibition and Ki value. Data are shown in Figures 2 and 3. Compounds were best fitted to a competitive type of inhibition. The calculated value for Ki was 0.30 ± 0.06 μM for 18 and 0.24 ± 0.04 μM for 19, which indicates that compounds 18 and 19 are excellent inhibitors of ALDH3A1 functional activity.
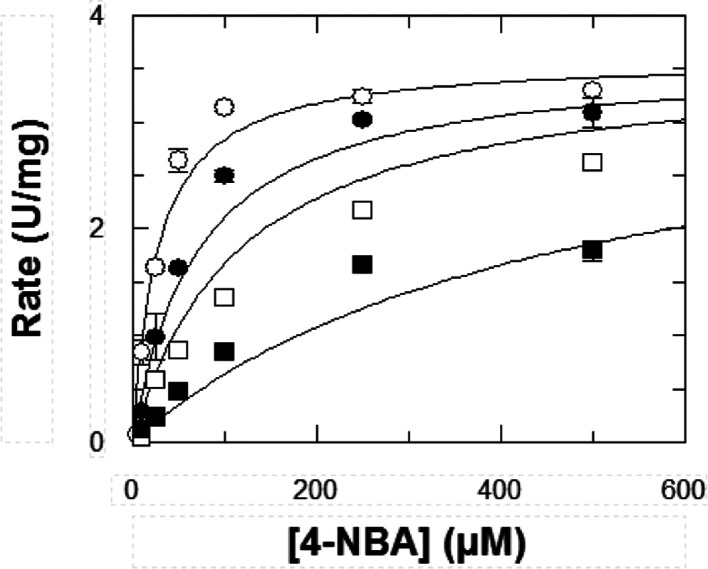
Figure 2.
Inhibition kinetics of ALDH3A1 by compound 18 at various concentrations of inhibitor: −○– 0 μM; −●– 0.5 μM; −□– 1 μM; and −■– 5 μM. 4-Nitrobenzaldehyde (4-NBA) was used as the substrate. The values of the kinetic parameters calculated from a fit to the competitive inhibition equation are Vmax = 3.59 ± 0.16 U/mg; Km = 26.92 ± 5.41 μM; and Ki = 0.30 ± 0.06 μM. Results are the mean ± SE of duplicate experiments.
Figure 3.
Inhibition kinetics of ALDH3A1 by compound 19 at various concentrations of inhibitor: −○– 0 μM; −●– 0.5 μM; −□– 1 μM; and −■– 5 μM. 4-NBA was used as the substrate. The values of the kinetic parameters calculated from a fit to the competitive inhibition equation are Vmax = 3.69 ± 0.16 U/mg; Km = 27.81 ± 5.36 μM; and Ki = 0.24 ± 0.04 μM. Results are the mean ± SE of duplicate experiments.
Additionally, we decided to further investigate the role of 18 as an ALDH3A1 substrate (4.21% with 10 μM, Figure S2) to assess whether it would interfere with the inhibition experiments. Kinetic analysis of the saturation profile for 18 with ALDH3A1 was best fitted to the Michaelis–Menten equation modified for substrate inhibition (Figure 4), producing a Km of 2.82 ± 0.35 μM and a Ksi of 113 ± 25 μM. For comparison, the Km of the best ALDH3A1 substrate, 4-NBA, is 10-fold higher, with a Km of 31.0 ± 4.9 μM. Regarding kcat, 4-NBA has a 26-fold higher turnover number (359 ± 14 min–1) than compound 18, which has a kcat value of 13.4 ± 0.7 min–1. These findings suggest that 18 performs better as an inhibitor than as an ALDH3A1 substrate. We also performed two different IC50 calculations for compound 18. The aim was to check whether 18 was acting as a substrate during the standard 5-min preincubation time, when NADP+ and the enzyme were added but with no standard substrate in the reaction mixture. One IC50 was calculated by incubating with the cofactor and the other was calculated without it, and no differences were observed (Figure S6), indicating that the function of compound 18 as an inhibitor was not being affected by its role as a substrate under the assayed conditions.
Figure 4.
Km value for ALDH3A1 using compound 18 as a substrate. Experimental values were fitted to the substrate inhibition equation and the kinetic values were Km = 2.82 ± 0.35 μM, Ksi = 113 ± 25 μM, and kcat of 13.4 ± 0.7 min–1. Data were the result of duplicate experiments and expressed as the mean ± SE.
Antiproliferative Activity of DEAB Analogues in Prostate Cancer Cell Lines
Several members of the ALDH family of enzymes have been shown to be expressed in PCa,30−34 and ALDH1A1 and 1A3 isoforms have been reported to be expressed at higher levels in tumor tissues compared to benign prostatic hyperplasia and normal prostate.35 ALDHs have also been acknowledged to promote clonogenic and migration cell capabilities in vitro and enhance the metastatic potential in vivo,34,36 while the expression correlates with a higher Gleason score (G8-9) in vivo.20,31,35 We have previously shown the importance of the RA pathway in PCa37,38 and ALDHs as a potential target,20 and in this context, it was suitable to explore the new DEAB library of compounds. Due to their ALDH expression profile (Figure 5), we chose to use a small panel consisting of the three prostate cancer cell lines PC3, DU145, and LNCaP.
Figure 5.

Immunoblot analysis was carried out using ALDH1A1, 1A3, and 3A1 specific antibodies in a panel of immortalized PCa cell lines (PC-3, LNCaP, and DU145) and using GAPDH as a control.
All compounds were initially assessed using the MTT assay in an initial two-dose point screen (96 h exposure) to identify hit compounds. The results revealed clear dose-dependent trends (Figure S14), which, together with observations from the biochemical screening, led to 17 compounds including DEAB to be further investigated in a five-dose screen.
All DEAB analogues displayed IC50 values in the micromolar range (10–200 μM) as measured using the MTT assay. Most compounds showed equipotent antiproliferative activity in PC3 (expressing ALDH1A1 and 3A1) and DU145 (expressing ALDH1A1) cell lines and increased potency against LNCaP (expressing ALDH1A3) cells (Table 2); the cellular potency is likely to be multifactorial and not just a direct correlation of the ALDH isoform expression. Nonetheless, the dipropyl moiety appeared to provide compound 14 with a higher antiproliferative potency than diethyl-based analogue 13 and piperidine analogue 23 across all three cell lines. These findings were also found for compound 18 with the dipropyl moiety, which showed higher antiproliferative activity than diethyl analogue 19. Therefore, analogues 14 and 18 were shown to be the most promising compounds in agreement with data obtained from the biochemical screening. In addition, compound 14 showed the highest antiproliferative activity in both DU145 and PC3 cell lines with IC50 values of 61 and 47 μM, respectively; this correlated with the biochemical assay IC50 data obtained for ALDH1A1 (7.08 μM), ALDH1A3 (0.63 μM), and ALDH3A1 (8.00 μM).
Table 2. Antiproliferative Effect of DEAB Analogues Against PC3, LNCaP, and DU145 PCa Cell Lines.
| IC50 (μM) |
|||
|---|---|---|---|
| Cmpd | PC3 | LNCaP | DU145 |
| 1 | >200 | 73 ± 14 | 190 ± 25 |
| 6 | 172 ± 30 | 137 ± 14 | 133 ± 11 |
| 7 | 77 ± 17 | 10 ± 3 | 90 ± 27 |
| 8 | 130 ± 9 | 37 ± 1.4 | 110 ± 3 |
| 13 | 166 ± 41 | 47 ± 0.6 | 123 ± 12 |
| 14 | 47 ± 6 | 25 ± 1 | 61 ± 5 |
| 15 | 123 ± 29 | 61 ± 14 | 106 ± 13 |
| 16 | >200 | >200 | >200 |
| 17 | 106 ± 25 | 61 ± 18 | 103 ± 4 |
| 18 | 98 ± 12 | 31 ± 6 | 100 ± 13 |
| 19 | >200 | 136 ± 13 | >200 |
| 21 | >200 | 179 ± 14 | 187 ± 14 |
| 22 | 196 ± 4 | 82 ± 10 | 155 ± 32 |
| 23 | 84 ± 6 | 46 ± 10 | 84 ± 12 |
| 26 | >200 | 172 ± 8 | >200 |
| 38 | 75 ± 10 | 50 ± 3 | 71 ± 11 |
| DEAB | >200 | >200 | >200 |
IC50 values represent the concentration of the compound that decreases cell survival by 50%. Results are expressed in μM ± SD and derived from at least three independent experiments.
Cell line sensitivity after treatment with ALDH-targeting compounds might not only be a reflection of the ALDH expression and functional activity. Previous reports have demonstrated that there is a striking difference in the metabolic phenotypes of the three cell lines used in this study.39 Due to a mitochondrial dysfunction, PC3 and DU145 cells have been shown to have an increased glycolytic reliance, unlike LNCaP which is highly oxidative, which might contribute to ALDH activity and compound sensitivity. Resistance mechanisms might contribute to compound sensitivity. For example, the PC3 cell line represents the type of cancer that is difficult to treat as it is an androgen-independent metastatic prostate cancer type.20 Nevertheless, apart from compound 16, it was noticeable that all analogues selected for chemosensitivity screening displayed more potent antiproliferative effects when compared with DEAB.
ALDHs have also been linked to chemo- and radioresistance in cancer therapy.40,41 An increased expression of ALDH can have a chemoprotective effect on cells due to their metabolic and detoxifying abilities [reviewed in ref (41)]. The taxanes paclitaxel and docetaxel have been reported to be less effective in ALDH-expressing cells.42−44 Given that docetaxel is the most commonly used cytotoxic drug for the treatment of advanced PCa, we next decided to evaluate three compounds (14, 18, and DEAB) as single agents and in combination with docetaxel against five patient samples (four cancers with Gleason score 7 and one BPH) to investigate the potential of such a combination in clinical samples. The compounds were evaluated using two different concentrations (50 and 200 μM), and results revealed dose-dependent reduction in percentage cell viability of primary prostate epithelial cultures (Figure 6A–C). In line with the PCa cell line data, analogues 14 and 18 were more potent than DEAB, a pattern also evident in the combination with docetaxel (1 nM). To gain further insights into the observations obtained from both the biochemical assays and chemosensitivity experiments, docking studies were performed for compounds 14 and 18, as they showed promising results.
Figure 6.

Cell viability of primary cells following treatment with 14 (A), 18 (B), and DEAB (C) as a single or combination treatment with docetaxel. Patient samples assessed included BPH sample H415/15 and cancer samples H568/15 RM, H431/14 LM, H488/14 RM, and H517/15 RM. The experiment was carried out in triplicate and values are represented as the mean. Statistical significance was calculated using a paired two-tailed Student’s t-test, in which the mean of untreated cells was compared with the mean of treated cells, *p < 0.05, **p < 0.01, and ***p < 0.001, combination treatment with docetaxel.
Docking Studies of Compounds 14 and 18 on ALDH1A1, ALDH1A3, and ALDH3A1 Isoforms
Analogues 14 and 18 were docked into the three investigated isoforms (ALDH1A1, 1A3, and 3A1), with all analysis shown in Figures S7–S13. In addition, binding modes with DEAB were included to compare its binding affinity to these three analogues, as shown in Figure 7. From the docking study of analogue 14 with the ALDH1A1 binding site (Figure 7A), the aldehyde oxygen of 14 was found to form an H-bond with Tyr297, and the phenyl ring established face-to-face π–π stacking with Phe171. The electron-rich m-Br group was found to be in close proximity of Phe171, Tyr297, and Ile304, capable of constituting van der Waals contacts with these residues. One of the propyl groups of the N-substituted side chain established van der Waals contacts with Trp178. Compound 14 was found to superimpose on DEAB, maintaining the abovementioned protein–ligand interactions. Compound 18 (Figure 7B) displayed similar protein–ligand interactions to 14 and DEAB, in the binding pocket of ALDH1A1. However, the presence of m-NO2 in the place of an m-Br group did not result in an H-bond interaction within the binding site.
Figure 7.
Molecular docking of compounds 14 (green) and 18 (magenta) into ALDH1A1 (A,B, PDB ID: 4WPN), ALDH1A3 (C,D, PDB ID: 5FHZ), and ALDH3A1 (E,F, PDB ID: 4H80) binding sites. Best fit binding modes are compared to DEAB (orange).
As depicted in Figure 7C, analogue 14 occupied the binding pocket of ALDH1A3 with high affinity through several intermolecular forces: the aldehyde oxygen formed a H-bond contact with Thr315, the m-Br phenyl ring established van der Waals interactions with Phe182, Trp189, and Leu471, and the electronic interactions between bromine and both Phe308 and Ile132 residues significantly contributed to enhanced binding affinity with the target, which may explain the results obtained in the ALDH1A3 biochemical studies (Table 1). In addition, compound 14 was found to be superimposed with the binding disposition of DEAB, altogether contributing to the improved binding affinity of 14 for ALDH1A3. Analogue 18, as shown in Figure 7D, did not properly occupy the ALDH1A3 binding site, justifying the poor/inactive potencies against the ALDH1A3 isoform obtained from the biochemical studies. The presence of the m-NO2 group in 18, as an H-bond acceptor, completely altered the binding disposition of this analogue within the enzyme binding site, by forming an H-bond network at the entrance of the ALDH1A3 binding pocket with Arg139 and Ala473 residues, thus restricting their deeper entry into the active site.
Both ligands constituted the fundamental H-bonds with Cys243 and Asn114 of ALDH3A1 (Figure 7E,F). In particular, compound 18 exhibited a preference binding for ALDH3A1 when compared with 1A1 and 1A3 isoforms. Apart from H-bond contacts, both analogues displayed significant van der Waals interactions in the ALDH3A1 binding pocket; the phenyl ring of 14 was observed to establish an edge-to-face π–π stacking with Tyr115 and van der Waals contact with the Ile394 side chain. The m-Br group of 14 was found to be in proximity of Ile391, thus favoring the electronic interaction, and one of the propyl of the N-substituted side chain formed a hydrophobic contact with Tyr65. Figure 7F also shows that the phenyl ring of ligand 18 formed a face-to-face π–π stacking with Tyr315 and the N-substituted side chain provided a van der Waals contact with Tyr65. In addition, the docking results of 14 and 18 were compared with those for DEAB in the ALDH3A1 binding pocket (Figure 7E,F). DEAB was superimposed on the binding orientation of 14 within the ALDH3A1 binding site, exhibiting identical H-bond and hydrophobic interactions. The phenyl ring of DEAB formed an edge-to-face π–π stacking with Tyr115 and van der Waals contacts with Ile394, whereas the ethyl group from one of the N-ethyl side chains of DEAB established another van der Waals contact with Tyr65. The phenyl ring of 18 was oriented differently relative to the phenyl ring of 14.
Conclusions
DEAB has been shown to be effective in treating aggressive ALDHhigh-expressing subpopulations with SC-like propensity in vitro and features as a key component of the Aldefluor assay used to identify cancer cells with SC properties. In regard to the former, it is possible that the inclusion of a therapeutic agent aimed at eradicating prostate CSCs could increase the overall survival rate.19,45−47 In PCa, patients who no longer respond to androgen therapy (ADT) develop an aggressive disease known as castrate-resistant prostate cancer (CRPC), which has high propensity for metastasis and short median survival rates ranging from 12.1 to 27.0 months.48−50 Currently, drugs that are used to treat advanced PCa include small-molecule-based therapies such as the androgen receptor (AR) inhibitor enzalutamide,51 the CYP17A1 inhibitor abiraterone acetate,51 and the taxanes docetaxel52 and cabazitaxel.53 Unfortunately, these therapies are rarely curative, necessitating the identification of new molecular targets and/or development of therapeutic strategies to treat aggressive PCa. In this study, we wanted to (i) generate a DEAB library to help understand ALDH isoform selectivity and (ii) investigate the potential of treating PCa cells as single agents and combination with docetaxel. Accordingly, we synthesized a small library of 40 compounds with a benzaldehyde scaffold and evaluated them against three ALDH isoforms—1A1, 1A3, and 3A1—known to be expressed in PCa. The results showed that an electron-donating group (amine or oxygen) at the para position to the aldehyde group is important for activity, particularly when connecting with a lipophilic carbon chain that participates in van der Waals interactions with ALDH active sites. In addition, the presence of an electron-withdrawing group at the meta position to the aldehyde also was found to increase affinity to the binding site. The synthesized compounds showed promising inhibitory properties, notably identifying compounds 14–16, 18, and 19 with superior activity against ALDH1A3 and 3A1 isoforms when compared with DEAB.
The antiproliferative activity of the compounds was also evaluated in three PCa cell lines, and interestingly, most compounds outperformed DEAB in cellular potency. We further investigated DEAB and analogues 14 and 18 in combination with docetaxel in primary PCa cells from tumors with a high Gleason score of 7. Although the results are only indicative, they do suggest that docetaxel treatment of aggressive primary PCa cells might benefit from the inclusion of an ALDH inhibitor such as compound 14 or 18. In uterine endometrial cancer, paclitaxel treatment has been shown to increase the proportion of ALDHhigh cells in clinical samples and in spheroids.54 Spheroids are enriched in CSCs, which mainly depend on an enhanced glycolytic metabolic pathway for their proliferation and survival. This glycolytic activation is mediated by the expression of ALDH and its crucial downstream effector GLUT1. Interestingly, the combination of ALDH/GLUT1 inhibitors with paclitaxel has been shown to suppress the proliferation of endometrial cancer cells in a synergistic manner, indicating that ALDH-dependent GLUT activation might be relevant for the maintenance of chemoresistance of CSCs.
In conclusion, further mechanistic studies are required to fully understand how inhibition of ALDH activity might be linked to potentiation of docetaxel treatment in PCa. Given the well-established capacity of DEAB to inhibit ALDHhigh-expressing CSCs, it also remains to be explored whether analogues from our DEAB library, such as 14 and 18, can be used as chemical probes to further unravel the significance of ALDH expression in CSCs and possible link to taxane resistance. An additional outcome of the present work might indicate that the Aldefluor assay specificity could be improved using compounds 14 and 15 to label ALDH1A3-expressing cells or 18 and 19 for ALDH3A1-expressing cells, given their increased selectivity as compared with DEAB.
Experimental Section
General Procedures
All materials and reagents were used as received with no further purification. 4-Fluoro-3-nitrobenzaldehyde, 3-cyano-4-fluorobenzaldehyde, 2-bromo-4-fluorobenzaldehyde, and 4-fluoro-2-methoxybenzaldehyde were purchased from fluorochem, 3-bromo-4-benzaldehyde, 4-fluorobenzaldehyde, 4-fluoro-3-methoxybenzaldehyde, 3-chloro-4-fluorobenzaldehyde 4-fluoro-3-methylbenzaldehyde, diethylamine, dipropylamine, morpholine, piperidine, pyrrolidine, DMSO, and 4-diethylaminobenzaldehyde (DEAB) were purchased Sigma-Aldrich; 1-methylpiperazine and 5-nitrovanillin were purchased from Acros Organics. Chemical reactions were monitored by analytical thin-layer chromatography using Merck 9385 silica gel 60 F254 aluminum-backed plates through visualizing the spotted plates under ultraviolet (UV) at 254 and 366 nm. Intermediates and final products were purified by column chromatography using silicagel 60A 40–63 μm. Proton and carbon NMR spectra were analyzed for all intermediates and final products on a Bruker AMX400 (400 MHz) nuclear magnetic resonance spectrometer. Chemical shifts were reported in parts per million (δ, ppm) downfield from internal TMS. Coupling constants (J) were expressed in Hertz (Hz). High-resolution mass spectra were obtained by the Engineering and Physical Sciences Research Council (EPSRC) mass spectrometry service, Swansea. Melting points were measured with a Gallenkamp melting point apparatus. All compounds biologically evaluated were >95% pure by HRMS/HPLC analysis except analogue 40 (82%); HPLC traces for all compounds can be found in the Supporting Information.
General Procedures for the Synthesis of Compounds 1–15, 17–32, 34, 36–38
To a stirred solution of starting aldehyde (1 equiv) in DMF (10 mL), the corresponding amine (6.0 equiv) and K2CO3 (2.0 equiv) were added and stirred at (25–100) °C for hours–days. The reaction was then cooled to room temperature, and DMF volume was reduced by evaporation under vacuum. Water (30 mL) was then added to the mixture and stirred for 30 min. The mixture was then extracted with ethyl acetate (EA) (3 × 20 mL), and the organic fractions were combined, washed with water, and dried with MgSO4. EA was then evaporated under vacuum to give the product as crude, which was purified by silica gel column chromatography, affording the desired targeted compounds.
General Procedures for the Synthesis of Compounds 16, 33, and 35
To a stirred solution of the starting aldehyde (1 equiv) in DMF (10 mL), iodopropane (4.0 equiv) and K2CO3 (2 equiv) were added and stirred at 90 °C for 6 h. The reaction was then cooled to room temperature, and DMF volume was reduced by evaporation under vacuum. Water (30 mL) was then added to the mixture and stirred for 30 min. The mixture was then extracted with EA (2 × 20 mL), and the organic fractions were combined, washed with water, and dried with MgSO4. EA then evaporated under vacuum to give the product as crude, which was then purified by silica gel column chromatography, affording the desired targeted compounds.
4-(Dipropylamino)benzaldehyde (1)
Starting reagents were 4-fluorobenzaldehyde and dipropylamine, processed at 100 °C for 48 h. The crude was purified by column chromatography using 5% EA in petroleum ether (PE), affording 56% of the title product as a yellow oil. Rf = 0.26 (EA/PE, 1:10). 1H NMR (400 MHz, CDCl3): δ 9.72 (s, 1H), 7.72 (d, 2H, J = 8.9 Hz), 6.67 (d, 2H, J = 8.9 Hz), 3.40–3.25 (t, 4H, J = 7.2 Hz), 1.78–1.53 (m, 4H), 0.98 (t, 6H, J = 7.4 Hz). 13C NMR (101 MHz, CDCl3): δ 190.0, 152.5, 132.2, 124.8, 111.0, 53.0, 20.3, 11.4. HRMS (ESI): calcd for C13H19NO [M + H]+, 206.1539; found, 206.1535.
4-(Piperidin-1-yl)benzaldehyde (2)
Starting reagents were 4-fluorobenzaldehyde and piperidine, processed at 100 °C for 24 h. The crude was purified by column chromatography using EA/PE (0.05:10) to afford 66.7% of the title product as a yellow solid. Rf = 0.13 (EA/PE, 1:10). mp 70 ± 1 °C. 1H NMR (400 MHz, CDCl3): δ 9.66 (s, 1H), 7.64 (d, 2H, J = 8.9 Hz), 6.81 (d, 2H, J = 8.9 Hz), 3.32 (m, 4H), 1.59 (m, 6H). 13C NMR (101 MHz, CDCl3): δ 190.3, 155.1, 132.0, 126.2, 113.3, 48.4, 25.3, 24.3. HRMS (ESI): calcd for C12H15NO [M + H]+, 190.1226; found, 190.1221.
4-Morpholinobenzaldehyde (3)
Starting reagents were 4-fluorobenzaldehyde and morpholine, processed at 100 °C for 48 h. The crude was purified by column chromatography using 10% EA in petroleum ether, affording 56.6% of the title product as a yellow solid. Rf = 0.47 (EA/PE, 1:1). mp 63 ± 1 °C. 1H NMR (400 MHz, CDCl3): δ 9.73 (s, 1H), 7.70 (d, 2H, J = 8.9 Hz), 6.85 (d, 2H, J = 8.9 Hz), 3.78 (t, 4H, J = 4.9 Hz), 3.27 (t, 4H, J = 4.9 Hz). 13C NMR (101 MHz, CDCl3): δ 190.6, 155.2, 131.8, 127.6, 113.5, 66.5, 47.3. HRMS (ESI): calcd for C11H13NO2 [M + H]+, 192.1019; found, 192.1015.
4-(Pyrrolidin-1-yl)benzaldehyde (4)
Starting reagents were 4-fluorobenzaldehyde and pyrrolidine, processed at 100 °C for 48 h. The crude was purified by column chromatography using EA/PE (0.5:10) to afford 87.5% of the title product as a yellow solid. Rf = 0.31 (EA/PE, 2:10). mp 92 ± 1 °C. 1H NMR (400 MHz, CDCl3): δ 9.73 (s, 1H), 7.74 (d, 2H, J = 8.8 Hz), 6.60 (d, 2H, J = 8.8 Hz), 3.40 (t, 4H, J = 6.6 Hz), 2.07 (t, 4H, J = 6.6 Hz). 13C NMR (101 MHz, CDCl3): δ 190.3, 151.9, 132.2, 124.9, 111.4, 47.8, 25.4. HRMS (ESI): calcd for C11H13NO [M + H]+, 176.1070; found, 176.1065.
4-(4-Methylpiperazin-1-yl)benzaldehyde (5)
Starting reagents were 4-fluorobenzaldehyde and 1-methylpiperazine, processed at 90 °C for 24 h. The crude was purified by column chromatography using (CH3OH/CH2Cl2, 0.5:10) to afford 96.7% of the title product as a yellow solid. Rf = 0.49 (CH3OH/CH2Cl2, 1:10). mp 74 ± 1 °C. 1H NMR (400 MHz, CDCl3): δ 9.73 (s, 1H), 7.70 (d, 2H, J = 8.9 Hz), 6.87 (d, 2H, J = 8.9 Hz), 3.40–3.31 (m, 4H), 2.58–2.45 (m, 4H), 2.31 (s, 3H). 13C NMR (101 MHz, CDCl3): δ 190.3, 155.0, 131.8, 127.0, 113.5, 54.6, 47.0, 46.1. HRMS (ESI): calcd for C12H16N2O [M + H]+, 205.1335; found, 205.1331.
4-(1H-Imidazole-1-yl)benzaldehyde (6)
Starting reagents were 4-fluorobenzaldehyde and 1H-imidazole, processed at 100 °C for 24 h. The crude was purified by column chromatography using (CH3OH/CH2Cl2, 0.1:10) to afford 38.9% of the title product as a pale-yellow solid. Rf = 0.29 (CH3OH/CH2Cl2, 0.05:10). mp 161 ± 1 °C. 1H NMR (400 MHz, CDCl3): δ 10.05 (s, 1H), 8.05–7.99 (m, 3H), 7.59 (d, 2H, J = 8.5 Hz), 7.38 (s, 1H), 7.26 (s, 1H). 13C NMR (101 MHz, CDCl3): δ 190.6, 141.7, 135.4, 135.0, 131.6, 131.2, 121.1, 117.7. HRMS (ESI): calcd for C10H8N2O [M + H]+, 173.0709; found, 173.0705.
3-Chloro-4-(dipropylamino)benzaldehyde (7)
Starting reagents were 3-chloro-4-fluorobenzaldehyde and dipropylamine, processed at 100 °C for 24 h. The crude was purified by column chromatography using EA/PE (0.1:10) to afford 33.1% of the title product as a yellow oil. Rf = 0.42 (EA/PE, 1:10). 1H NMR (400 MHz, CDCl3): δ 9.73 (s, 1H), 7.75 (d, 1H, J = 1.9 Hz), 7.58 (dd, 1H, J = 8.4, 1.9 Hz), 6.99 (d, 1H, J = 8.4 Hz), 3.21–3.05 (m, 4H), 1.58–1.38 (m, 4H), 0.80 (t, 6H, J = 7.4 Hz). 13C NMR (101 MHz, CDCl3): δ 189.7, 153.8, 132.9, 130.1, 128.7, 127.7, 121.2, 53.8, 20.6, 11.4. HRMS (ESI): calcd for C13H18ClNO [M + H]+, 240.1150; found, 240.1150.
3-Chloro-4-(piperidin-1-yl)benzaldehyde (8)
Starting reagents were 3-chloro-4-fluorobenzaldehyde and piperidine, processed at 100 °C for 24 h. The crude was purified by column chromatography using EA/PE (0.2:10) to afford 65.0% of the title product as a yellow oil. Rf = 0.08 EA/PE (0.2:10). 1H NMR (400 MHz, CDCl3): δ 9.84 (s, 1H), 7.86 (d, 1H, J = 1.9 Hz), 7.71 (dd, 1H, J = 8.2, 1.9 Hz), 7.11 (d, 1H, J = 8.2 Hz), 3.22–3.08 (m, 4H), 1.86–1.70 (m, 4H), 1.65 (m, 2H). 13C NMR (101 MHz, CDCl3): δ 189.9, 155.6, 132.1, 130.9, 129.6, 128.3, 120.0, 52.2, 25.9, 24.1. HRMS (ESI): calcd for C12H14ClNO [M + H]+, 224.0837; found, 224.0832.
3-Chloro-4-morpholinobenzaldehyde (9)
Starting reagents were 3-chloro-4-fluorobenzaldehyde and morpholine, at processed 100 °C for 24 h. The crude was purified by column chromatography using EA/PE (0.5:10) to afford 87% of the title product as a yellow solid. Rf = 0.24 (EA/PE, 2:10). mp 83 ± 1 °C. 1H NMR (400 MHz, CDCl3): δ 9.88 (s, 1H), 7.89 (d, 1H, J = 1.9 Hz), 7.76 (dd, 1H, J = 8.3, 1.9 Hz), 7.12 (d, 1H, J = 8.3 Hz), 3.98–3.80 (m, 4H), 3.27–3.12 (m, 4H). 13C NMR (101 MHz, CDCl3): δ 189.9, 154.2, 132.1, 131.7, 129.7, 128.5, 119.9, 66.8, 51.1. HRMS (ESI): calcd for C11H12ClNO2 [M + H]+, 226.0629; found, 226.0630.
3-Chloro-4-(4-methylpiperazin-1-yl)benzaldehyde (10)
Starting reagents were 3-chloro-4-fluorobenzaldehyde and N-methylpiperazine, processed at 100 °C for 24 h. The crude was purified by column chromatography starting with CH2Cl2 and gradually increasing CH3OH to final mixture of CH3OH/CH2Cl2 (0.5:10), affording 80.5% of the title product as a yellow solid. Rf = 0.20 (CH3OH/CH2Cl2, 0.5:10). mp 42 ± 1 °C. 1H NMR (400 MHz, CDCl3): δ 9.76 (s, 1H), 7.77 (d, 1H, J = 1.9 Hz), 7.63 (dd, 1H, J = 8.3, 1.9 Hz), 7.02 (d, 1H, J = 8.3 Hz), 3.15 (m, 4H), 2.55 (m, 4H), 2.29 (s, 3H). 13C NMR (101 MHz, CDCl3): δ 189.8, 154.5, 132.0, 131.4, 129.7, 128.3, 120.0, 54.9, 50.6, 46.0. HRMS (ESI): calcd for C12H15ClN2O [M + H]+, 239.0944; found, 239.0946.
3-Chloro-4-(diethylamino)benzaldehyde (11)
Starting reagents were 3-chloro-4-fluorobenzaldehyde and diethylamine, processed at 55 °C for 48 h. The crude was purified by column chromatography using EA/PE (0.1:10) to afford 65.9% of the title product as a yellow oil. Rf = 0.60 (EA/PE, 2:10). 1H NMR (400 MHz, CDCl3): δ 9.73 (s, 1H), 7.76 (d, 1H, J = 1.9 Hz), 7.59 (dd, 1H, J = 8.5, 1.9 Hz), 6.99 (d, 1H, J = 8.5 Hz), 3.24 (q, 4H, J = 7.1 Hz), 1.05 (t, 6H, J = 7.1 Hz). 13C NMR (101 MHz, CDCl3): δ 189.8, 153.5, 132.7, 130.2, 128.8, 127.8, 121.0, 45.7, 12.5. HRMS (ESI): calcd for C11H14ClNO [M + H]+, 212.0837; found, 212.0835.
3-Chloro-4-(pyrrolidin-1-yl)benzaldehyde (12)
Starting reagents were 3-chloro-4-fluorobenzaldehyde and pyrrolidine, processed at 60 °C for 24 h. The crude was purified by column chromatography using EA/PE (0.5:10) to yield 81.9% of the title product as a yellow oil. Rf = 0.54 (EA/PE, 2:10). 1H NMR (400 MHz, CDCl3): δ 9.61 (s, 1H), 7.66 (d, 1H, J = 1.9 Hz), 7.50 (dd, 1H, J = 8.6, 1.9 Hz), 6.66 (d, 1H, J = 8.6 Hz), 3.52 (t, 4H, J = 6.6 Hz), 1.97–1.77 (m, 4H). 13C NMR (101 MHz, CDCl3): δ 189.3, 150.7, 133.9, 129.5, 127.1, 119.5, 115.3, 51.2, 25.8. HRMS (ESI): calcd for C11H12ClNO [M + H]+, 210.0680; found, 210.0678.
3-Bromo-4-(diethylamino)benzaldehyde (13)
Starting reagents were 3-bromo-4-fluorobenzaldehyde and diethylamine, processed at 55 °C for 48 h. The crude was purified by column chromatography using EA/PE (0.1:10) to yield 72.6% of the title product as a yellow oil. Rf = 0.43 (EA/PE, 1:10). 1H NMR (400 MHz, CDCl3): δ 9.74 (s, 1H), 7.98 (d, 1H, J = 2.0 Hz), 7.65 (dd, 1H, J = 8.3, 2.0 Hz), 7.02 (d, 1H, J = 8.3 Hz), 3.20 (q, 1H, J = 7.1 Hz), 1.03 (t, 1H, J = 7.1 Hz). 13C NMR (101 MHz, CDCl3): δ 189.8, 155.1, 136.0, 131.2, 129.2, 122.2, 119.2, 46.0, 12.4. HRMS (ESI): calcd for C11H14BrNO [M + H]+, 256.0332; found, 256.0333, calculated for C11H14BrNO [M + H+2]+, 258.0311; found, 258.0309.
3-Bromo-4-(dipropylamino)benzaldehyde (14)
Starting reagents were 3-bromo-4-fluorobenzaldehyde and dipropylamine, processed at 80 °C for 24 h. The crude was purified by column chromatography using EA/PE (0.05:10) to yield 48.5% of the title product as a yellow oil. Rf = 0.45 (EA/PE, 1:10). 1H NMR (400 MHz, CDCl3): δ 9.73 (s, 1H), 7.97 (d, 1H, J = 1.9 Hz), 7.64 (dd, 1H, J = 8.3, 1.9 Hz), 7.02 (d, 1H, J = 8.3 Hz), 3.17–3.04 (m, 4H), 1.54–1.38 (m, 4H), 0.79 (t, 6H, J = 7.4 Hz). 13C NMR (101 MHz, CDCl3): δ 189.8, 155.4, 136.2, 131.0, 129.2, 122.2, 118.8, 54.0, 20.4, 11.5. HRMS (ESI): calcd for C13H18BrNO [M + H]+, 284.0645; found, 284.0645, calculated for C13H18BrNO [M + H+2]+, 286.0624; found, 286.0620.
3-Methyl-4-(piperidin-1-yl)benzaldehyde (15)
Starting reagents were 4-fluoro-3-methylbenzaldehyde and piperidine, processed at 100 °C for 24 h. The crude was purified by column chromatography using EA/PE (0.1:10) to yield 64.7% of the title product as a yellow oil. Rf = 0.46 (EA/PE, 1:10). 1H NMR (400 MHz, CDCl3): δ 9.77 (s, 1H), 7.60 (s, 1H) 7.58–7.53 (m, 1H), 6.94 (d, 1H, J = 8.1 Hz), 2.95–2.78 (m, 4H), 2.25 (s, 3H), 1.71–1.59 (m, 4H), 1.53 (dt, 2H, J = 10.9, 5.6 Hz). 13C NMR (101 MHz, CDCl3): δ 191.5, 158.6, 132.5, 132.0, 130.4, 129.3, 118.5, 52.5, 26.3, 24.3, 18.6. HRMS (ESI): calcd for C13H17NO [M + H]+, 204.1383; found, 204.1381.
4-Isopropoxybenzaldehyde (16)
Starting reagents were 4-hydroxybenzaldehyde and 2-iodopropane. The crude was purified by column chromatography using EA/PE (0.1:10) to yield quantitatively the title compound as a pale-yellow oil. Rf = 0.38 (EA/PE, 1:10). 1H NMR (400 MHz, CDCl3): δ 9.87 (s, 1H), 7.95–7.68 (m, 2H), 7.09–6.84 (m, 2H), 4.68 (Sep, 1H, J = 6.1 Hz), 1.38 (d, 6H, J = 6.1 Hz). 13C NMR (101 MHz, CDCl3): δ 190.8, 163.2, 132.1, 129.5, 115.6, 70.3, 21.9. HRMS (ESI): calcd for C10H12O2 [M + H]+, 165.0910; found, 165.0906.
4-(Dipropylamino)-3-methoxybenzaldehyde (17)
Starting reagents were 4-fluoro-3-methoxybenzaldehyde and dipropylamine, processed at 100 °C for 5 days. The crude was purified by column chromatography using EA/PE (0.1:10) to yield 65.9% of the title compound as a yellow oil. Rf = 0.12 (EA/PE, 0.5:10). 1H NMR (400 MHz, CDCl3): δ 9.68 (s, 1H), 7.28 (dd, 1H, J = 8.7, 1.7 Hz), 7.26 (d, 1H, J = 1.7 Hz), 6.75 (d, 1H, J = 8.7 Hz), 3.80 (s, 3H), 3.25–3.09 (m, 4H), 1.59–1.40 (m, 4H), 0.80 (t, 6H, J = 7.4 Hz). 13C NMR (101 MHz, CDCl3): δ 190.5, 151.4, 146.4, 128.5, 126.7, 116.9, 109.9, 55.6, 54.2, 20.8, 11.5. HRMS (ESI): calcd for C14H21NO2 [M + H]+, 236.1645; found, 236.1640.
4-(Dipropylamino)-3-Nitrobenzaldehyde (18)
Starting reagents were 4-fluoro-3-nitrobenzaldehyde and dipropylamine, processed at 25 °C for 1 h. The crude was purified by column chromatography using CH2Cl2 to yield 96% of the title product as a dark yellow solid. Rf = 0.44 (EA/PE, 2:10). mp 42 ± 0.5 °C. 1H NMR (400 MHz, CDCl3): δ 9.78 (s, 1H), 8.18 (d, 1H, J = 2.0 Hz), 7.84 (dd, 1H, J = 8.9, 2.0 Hz), 7.12 (d, 1H, J = 8.9 Hz), 3.28–3.12 (m, 4H), 1.71–1.52 (m, 4H), 0.87 (t, 6H, J = 7.4 Hz). 13C NMR (101 MHz, CDCl3): δ 188.7, 148.9, 139.0, 132.2, 130.4, 125.8, 119.7, 53.7, 20.7, 11.2. HRMS (ESI): calcd for C13H18N2O3 [M + H]+, 251.1390; found, 251.1388.
4-(Diethylamino)-3-Nitrobenzaldehyde (19)
Starting reagents were 4-fluoro-3-nitrobenzaldehyde and dirthylamine, processed at 25 °C for 1 h. The crude was purified by column chromatography using CH2Cl2 to yield 91.7% of the title product as a dark yellow solid. Rf = 0.29 (EA/PE, 2:10). mp 50 ± 0.5 °C. 1H NMR (400 MHz, CDCl3): δ 9.77 (s, 1H), 8.14 (d, 1H, J = 2.0 Hz), 7.83 (dd, 1H, J = 8.9, 2.0 Hz), 7.10 (d, 1H, J = 8.9 Hz), 3.32 (q, 4H, J = 7.1 Hz), 1.20 (t, 6H, J = 7.1 Hz). 13C NMR (101 MHz, CDCl3): δ 188.7, 148.1, 138.9, 132.3, 130.2, 125.8, 119.2, 46.0, 12.4. HRMS (ESI): calcd for C11H14N2O3 [M + H]+, 223.1077; found, 223.1074.
4-(Diethylamino)-3-methoxybenzaldehyde (20)
Starting reagents were 4-fluoro-3-methoxybenzaldehyde and diethylamine, processed at 100 °C for 5 days. The crude compound was purified by column chromatography starting with EA/PE (0.1:10) and gradually increasing to EA/PE (0.5:10) to yield 60.4% of the title compound as a yellow oil. Rf = 0.26 (EA/PE, 1:10). 1H NMR (400 MHz, CDCl3): δ 9.70 (s, 1H), 7.30 (dd, 1H, J = 7.9, 1.5 Hz), 7.27 (d, 1H, J = 1.5 Hz), 6.80 (d, 1H, J = 7.9 Hz), 3.81 (s, 3H), 3.26 (q, 4H, J = 7.0 Hz), 1.05 (t, 6H, J = 7.1 Hz). 13C NMR (101 MHz, CDCl3): δ 190.6, 151.7, 146.1, 128.9, 126.6, 117.3, 109.6, 55.6, 45.7, 12.7. HRMS (ESI): calcd for C12H17NO2 [M + H]+, 208.1332; found, 208.1330.
3-Bromo-4-(4-methylpiperazin-1-yl)benzaldehyde (21)
Starting reagents were 3-bromo-4-fluorobenzaldehyde and N-methylpiperazine, processed at 100 °C for 24 h. The crude was purified by column chromatography starting with CH2Cl2 and followed by a gradual increase of polarity to (CH3OH/CH2Cl2, 0.5:10) to yield 93.0% of the title product as a yellow oil, which was solidified under vacuum. Rf = 0.13 (CH3OH/CHCl2, 0.5:10). mp 55 ± 1 °C. 1H NMR (400 MHz, CDCl3): δ 9.74 (s, 1H), 7.95 (d, 1H, J = 1.9 Hz), 7.67 (dd, 1H, J = 8.3, 1.9 Hz), 7.02 (d, 1H, J = 8.3 Hz), 3.21–3.10 (m, 4H), 2.61–2.49 (m, 4H), 2.29 (s, 3H). 13C NMR (101 MHz, CDCl3): δ 189.6, 155.8, 135.3, 131.9, 130.2, 120.4, 118.7, 54.8, 51.0, 46.0. HRMS (ESI): calcd for C12H15BrN2O [M + H]+, 283.0441; found, 283.0443. [M+2 + H]+, 285.0420; found, 285.0419.
3-Bromo-4-morpholinobenzaldehyde (22)
Starting reagents were 3-bromo-4-fluorobenzaldehyde and morpholine, processed at 100 °C for 48 h. The crude was purified by column chromatography using CH2Cl2 to yield 71.5% of the title product as a yellow solid. Rf = 0.20 (Ch3OH/CH2Cl2, 0.5:10). mp 96 ± 1 °C. 1H NMR (400 MHz, CDCl3): δ 9.78 (s, 1H), 8.00 (d, 1H, J = 1.8 Hz), 7.71 (dd, 1H, J = 8.2, 1.9 Hz), 7.03 (d, 1H, J = 8.3 Hz), 3.93–3.71 (m, 4H), 3.18–3.02 (m, 4H). 13C NMR (101 MHz, CDCl3): δ 189.8, 155.6, 135.5, 132.3, 130.2, 120.4, 118.9, 66.8, 51.5. HRMS (ESI): calcd for C11H12BrNO2 [M + H]+, 270.0124; found, 270.0126. [M+2 + H]+, 272.0104; found, 272.0103.
3-Bromo-4-(piperidin-1-yl)benzaldehyde (23)
Starting reagents were 3-bromo-4-fluorobenzaldehyde and piperidine, processed at 100 °C for 24 h. The crude was purified by column chromatography using EA/PE (0.1:10) to yield 84.6% of the title product as a yellow oil. Rf = 0.33 (EA/PE, 1:10). 1H NMR (400 MHz, CDCl3): δ 9.81 (s, 1H), 8.03 (d, 1H, J = 1.9 Hz), 7.73 (dd, 1H, J = 8.3, 1.9 Hz), 7.07 (d, 1H, J = 8.3 Hz), 3.18–2.99 (m, 4H), 1.85–1.68 (m, 4H), 1.62 (m, 2H). 13C NMR (101 MHz, CDCl3): δ 189.8, 157.1, 135.4, 131.5, 130.1, 120.4, 118.9, 52.7, 25.9, 24.0. HRMS (ESI): calcd for C12H14BrNO [M + H]+, 268.0332; found, 268.0333. [M+2 + H]+, 270.0311; found, 270.0310.
3-Methyl-4-(4-methylpiperazin-1-yl)benzaldehyde (24)
Starting reagents were 4-fluoro-3-methylbenzaldehyde and N-methylpiperazine, processed at 100 °C for 6 days. The crude was purified by column chromatography starting with CH2Cl2 and gradually increasing polarity to CH3OH/CH2Cl2 (0.5:10) to yield 71.4% of the title product as a pale yellow solid. Rf = 0.420 (CH3OH/CH2Cl2, 1:10). mp 68 ± 0.5 °C. 1H NMR (400 MHz, CDCl3): δ 9.82 (s, 1H), 7.63 (s, 1H), 7.60 (m, 1H), 7.02 (d, 1H, J = 8.0 Hz), 2.98 (t, 4H, J = 4.78 Hz), 2.56 (s, 4H), 2.33 (s, 3H), 2.30 (s, 3H). 13C NMR (101 MHz, CDCl3): δ 191.32, 157.19, 132.43, 131.97, 130.90, 129.31, 118.54, 55.21, 50.92, 46.06, 18.52. HRMS (ESI): calcd for C13H18N2O [M + H]+, 219.1495; found, 219.1497.
4-(4-Methylpiperazin-1-yl)-3-nitrobenzaldehyde (25)
Starting reagents were 4-fluoro-3-nitrobenzaldehyde and N-methylpiperazine, processed at 25 °C for 1 h. The crude was purified by column chromatography using CH2Cl2 followed by (CH3OH/CH2Cl2, 0.1:10) to yield quantitatively the title compound as a yellow solid. Rf = 0.24 (CH3OH/CH2Cl2, 0.5:10). mp 101 ± 1 °C. 1H NMR (400 MHz, CDCl3): δ 9.82 (s, 1H), 8.23 (d, 1H, J = 2.0 Hz), 7.90 (dd, 1H, J = 8.7, 2.0 Hz), 7.13 (d, 1H, J = 8.7 Hz), 3.32–3.16 (m, 4H), 2.64–2.47 (m, 4H), 2.34 (s, 3H). 13C NMR (101 MHz, CDCl3): δ 188.7, 149.4, 139.8, 133.2, 129.7, 127.7, 119.9, 54.4, 50.5, 45.9. HRMS (ESI): calcd for C12H15N3O3 [M + H]+, 250.1191; found, 250.1192.
3-Methyl-4-(pyrrolidin-1-yl)benzaldehyde (26)
Starting reagents were 4-fluoro-3-methylbenzaldehyde and pyrrolidine, processed at 100 °C for 48 h. The crude was purified by column chromatography using first EA/PE (0.1:10) before increasing to EA/PE (0.2:10) to yield 70.4% of the title compound as a dark yellow oil. Rf = 0.10 (EA/PE, 0.5:10). 1H NMR (400 MHz, CDCl3): δ 9.64 (s, 1H), 7.51–7.48 (m, 1H), 7.48–7.45 (m, 1H), 6.64 (d, 1H, J = 9.0 Hz), 3.44–3.28 (m, 4H), 2.34 (s, 3H), 1.94–1.80 (m, 4H). 13C NMR (101 MHz, CDCl3): δ 191.5, 154.4, 134.1, 129.9 127.1, 124.7, 113.8, 51.0, 25.7, 22.0. HRMS (ESI): calcd for C12H15NO [M + H]+, 190.1226; found, 190.1221.
3-Methyl-4-morpholinobenzaldehyde (27)
Starting reagents were 4-fluoro-3-methylbenzaldehyde and morpholine, at 100 °C for 6 days. The crude was purified by column chromatography using first EA/PE (0.5:10) followed by EA/PE (1:10) to yield 45.9% of the title product as a yellow solid. Rf = 0.08 (EA/PE, 1:10). mp 67 ± 1 °C. 1H NMR (400 MHz, CDCl3): δ 9.81 (s, 1H), 7.62–7.59 (m, 1H), 7.58–7.62 (m, 1H), 7.00 (d, 1H, J = 7.9 Hz), 3.91–3.64 (m, 4H), 3.02–2.82 (m, 4H), 2.29 (s, 3H). 13C NMR (101 MHz, CDCl3): δ 191.4, 156.9, 132.6, 132.2, 131.3, 129.4, 118.5, 67.1, 51.6, 18.5. HRMS (ESI): calcd for C12H15NO2 [M + H]+, 206.1176; found, 206.1173.
3-Methoxy-4-(piperidin-1-yl)benzaldehyde (28)
Starting reagents were 4-fluoro-3-methoxybenzaldehyde and piperidine, processed at 100 °C for 48 h. The crude was purified by column chromatography using first EA/PE (0.1:10) followed with gradual increase up to EA/PE (2:10) to yield 70.4% of the title compound as a yellow oil. Rf = 0.50 (EA/PE, 2:10). 1H NMR (400 MHz, CDCl3): δ 9.75 (s, 1H), 7.32 (dd, 1H, J = 8.1, 1.8 Hz), 7.28 (d, 1H, J = 1.8 Hz), 6.89 (d, 1H, J = 8.1 Hz), 3.84 (s, 3H), 3.11–2.96 (m, 4H), 1.74–1.60 (m, 4H), 1.53 (m, 2H). 13C NMR (101 MHz, CDCl3): δ 191.0, 152.2, 148.6, 130.4, 126.6, 117.4, 109.1, 55.6, 51.5, 26.0, 24.3. HRMS (ESI): calcd for C13H17NO2 [M + H]+, 220.1332; found, 220.1330.
3-Methoxy-4-(pyrrolidin-1-yl)benzaldehyde (29)
Starting reagents were 4-fluoro-3-methoxybenzaldehyde and pyrrolidine, processed at 100 °C for 48 h. The crude was purified by column chromatography using first EA/PE (0.1:10) and then EA/PE (0.5:10) to yield 57.9% of the title compound as a yellow solid. Rf = 0.25 (EA/PE, 0.1:10). mp 38 ± 0.5 °C. 1H NMR (400 MHz, CDCl3): δ 9.60 (s, 1H), 7.23 (dd, 1H, J = 8.0, 1.7 Hz), 7.20 (d, 1H, J = 1.8 Hz), 6.50 (d, 1H, J = 8.0 Hz), 3.74 (s, 3H), 3.44 (t, 4H, J = 6.7 Hz), 1.84 (t, 4H, J = 6.7 Hz). 13C NMR (101 MHz, CDCl3): δ 190.0, 148.7, 145.1, 128.1, 126.4, 112.6, 109.7, 55.7, 50.6, 25.5. HRMS (ESI): calcd for C12H15NO2 [M + H]+, 206.1176; found, 206.1174.
3-Methoxy-4-(4-methylpiperazin-1-yl)benzaldehyde (30)
Starting reagents were 4-fluoro-3-methoxybenzaldehyde and N-methylpiperazine, processed at 100 °C for 48 h. The crude was purified by column chromatography using first CH3OH/CH2Cl2 (0.1:10) with gradual increase of CH3OH to CH3OH/CH2Cl2 (0.5:10) to yield 66.9% of the title product as a yellow solid. Rf = 0.28 (CH3OH/CH2Cl2, 0.5:10). mp 78 ± 1 °C. 1H NMR (400 MHz, Acetone): δ 9.85 (s, 1H), 7.47 (dd, 1H, J = 8.1, 1.7 Hz), 7.40 (d, 1H, J = 1.7 Hz), 7.03 (d, 1H, J = 8.1 Hz), 3.93 (s, 3H), 3.21 (t, 4H, J = 4.7 Hz), 2.51 (t, 4H, J = 4.7 Hz), 2.27 (s, 3H). 13C NMR (101 MHz, acetone): δ 191.2, 153.1, 148.3, 131.7, 126.5, 118.1, 110.8, 56.0, 55.9, 50.6, 46.4. HRMS (ESI): calcd for C13H18N2O2 [M + H]+, 235.1441; found, 235.1439.
3-Methoxy-4-morpholinobenzaldehyde (31)
Starting reagents were 4-fluoro-3-methoxybenzaldehyde and morpholine, processed at 100 °C for 48 h. The crude was purified by column chromatography using first EA/PE (0.5:10) then gradually increase of EA to EA/PE (2:10) to yield 68.4% of the title compound as a pale-yellow solid. Rf = 0.59 (EA/PE, 1:1). mp 96 ± 1 °C. 1H NMR (400 MHz, acetone): δ 9.72 (s, 1H), 7.35 (dd, 1H, J = 8.1, 1.8 Hz), 7.27 (d, 1H, J = 1.8 Hz), 6.90 (d, 1H, J = 8.1 Hz), 3.79 (s, 3H), 3.62 (t, 4H, J = 4.6 Hz), 3.13 (t, 4H, J = 4.6 Hz). 13C NMR (101 MHz, acetone): δ 191.3, 153.1, 148.0, 132.0, 126.4, 118.0, 110.8, 67.4, 56.0, 51.2. HRMS (ESI): calcd for C12H15NO3 [M + H]+, 222.1125; found, 222.1123.
4-Morpholino-3-Nitrobenzaldehyde (32)
Starting reagents were 4-fluoro-3-nitrobenzaldehyde and morpholine, processed at 25 °C for 1 h. The crude was purified by column chromatography using CH2Cl2 and gradual increase to CH3OH/CH2Cl2 (0.1:10) to yield quantitatively the title compound as a dark yellow oil. Rf = 0.35 (EA/PE, 4:10). 1H NMR (400 MHz, Acetone): δ 9.93 (s, 1H), 8.31 (d, 1H, J = 1.9 Hz), 8.03 (dd, 1H, J = 8.7, 1.9 Hz), 7.42 (d, 1H, J = 8.7 Hz), 3.80 (t, 4H, J = 4.7 Hz), 3.26 (t, 4H, J = 4.7 Hz). 13C NMR (101 MHz, CDCl3): δ 189.9, 150.0, 141.1, 134.0, 129.7, 129.2, 121.1, 66.9, 51.6. HRMS (ESI): calcd for C11H12N2O4 [M + H]+, 237.0875; found, 237.0879.
4-Isopropoxy-3-methoxybenzaldehyde (33)
Starting reagents were 4-hydroxy-3-methoxybenzaldehyde and 2-iodopropane. The crude was purified by column chromatography using CH2Cl2 to yield quantitatively the title compound as a yellow oil. Rf = 0.18 (CH2Cl2). 1H NMR (400 MHz, Acetone): δ 9.86 (s, 1H), 7.50 (dd, 1H, J = 8.2, 1.9 Hz), 7.43 (d, 1H, J = 1.9 Hz), 7.12 (d, 1H, J = 8.2 Hz), 4.75 (sept, 1H, J = 6.1 Hz), 3.89 (s, 3H), 1.36 (d, 6H, J = 6.1 Hz). 13C NMR (101 MHz, Acetone): δ 191.2, 154.0, 151.5, 131.1, 126.6, 114.4, 111.0, 71.7, 56.1, 22.2. HRMS (ESI): calcd for C11H14O3 [M + H]+, 195.1016; found, 195.1013.
3-Nitro-4-(pyrrolidin-1-yl)benzaldehyde (34)
Starting reagents were 4-fluoro-3-nitrobenzaldehyde and pyrrolidine, processed at 25 °C for 1 h. The crude was purified by column chromatography using CH2Cl2 to yield quantitatively the title compound as a yellow solid. Rf = 0.20 (EA/PE, 2:10). mp 118 ± 1 °C. 1H NMR (400 MHz, Acetone): δ 9.81 (s, 1H), 8.19 (d, 1H, J = 2.0 Hz), 7.87 (dd, 1H, J = 8.9, 2.0 Hz), 7.13 (d, 1H, J = 8.9 Hz), 3.33 (t, 4H, J = 6.5 Hz), 2.05 (t, 4H, J = 6.5 Hz). 13C NMR (101 MHz, acetone): δ 189.3, 146.5, 137.3, 132.6, 130.7, 125.4, 117.5, 51.5, 26.2. HRMS (ESI): calcd for C11H12N2O3 [M + H]+, 221.0926; found, 221.0932.
4-Isopropoxy-3-methoxy-5-Nitrobenzaldehyde (35)
Starting reagents were 4-hydroxy-3-methoxy-5-nitrobenzaldehyde and 2-iodopropane. The crude was purified by column chromatography using EA/PE (0.5:10) with a gradual increase to EA/PE (3:10) to yield 25.8% of the title compound as a pale yellow solid. Rf = 0.41 (EA/PE, 3:10). mp 71 ± 0.5 °C. 1H NMR (400 MHz, Acetone): δ 9.85 (s, 1H), 7.76 (d, 1H, J = 1.8 Hz), 7.63 (d, 1H, J = 1.8 Hz), 4.88–4.76 (sept, 1H, J = 6.1 Hz), 3.93 (s, 3H), 1.15 (d, 6H, J = 6.1 Hz). 13C NMR (101 MHz, Acetone): δ 190.4, 155.3, 146.9, 145.5, 132.6, 118.7, 114.8, 78.1, 57.2, 22.6. HRMS (ESI): calcd for C11H13NO5 [M + H]+, 240.0872; found, 240.0869.
3-Nitro-4-(piperidin-1-yl)benzaldehyde (36)
Starting reagents were 4-fluoro-3-nitrobenzaldehyde and piperidine, processed at 25 °C for 1 h. The crude was purified by column chromatography using CH2Cl2 to yield quantitatively the title compound as a yellow oil. Rf = 0.31 (EA/PE, 2:10). 1H NMR (400 MHz, acetone): δ 9.75 (s, 1H), 8.12 (d, 1H, J = 2.0 Hz), 7.82 (dd, 1H, J = 8.7, 2.0 Hz), 7.23 (d, 1H, J = 8.7 Hz), 3.14–3.07 (m, 4H), 1.67–1.44 (m, 6H). 13C NMR (101 MHz, acetone): δ 189.7, 150.6, 140.7, 133.6, 130.0, 128.1, 121.1, 52.4, 26.4, 24.4. HRMS (ESI): calcd for C12H14N2O3 [M + H]+, 235.1083; found, 235.1084.
4-(Diethylamino)-2-methoxybenzaldehyde (37)
Starting reagents were with 4-fluoro-2-methoxybenzaldehyde and diethylamine, processed at 100 °C for 48 h. The crude was purified by column chromatography using EA/PE (1:10) to yield 59.4% of the title compound as yellow solid. Rf = 0.20 (EA/PE, 2:10). mp 101 ± 1 °C. 1H NMR (400 MHz, Acetone): δ 9.98 (s, 1H), 7.45 (d, 1H, J = 8.9 Hz), 6.23 (dd, 1H, J = 8.9, 2.1 Hz), 6.10 (d, 1H, J = 2.1 Hz), 3.78 (s, 3H), 3.37 (q, 4H, J = 7.1 Hz), 1.07 (t, 6H, J = 7.1 Hz). 13C NMR (101 MHz, acetone) δ 186.0, 165.0, 154.8, 130.3, 115.0, 105.1, 93.6, 55.7, 45.2, 12.8. HRMS (ESI): calcd for C12H17NO2 [M + H]+, 208.1332; found, 208.1329.
4-(Dipropylamino)-2-methoxybenzaldehyde (38)
Starting reagents were 4-fluoro-2-methoxybenzaldehyde and dipropylamine, processed at 100 °C for 48 h. The crude was purified by column chromatography using EA/PE (1:10) to yield 43.7% of the title compound as yellow solid. Rf = 0.38 (EA/PE, 2:10). mp 92 ± 1 °C. 1H NMR (400 MHz, acetone) δ 9.97 (s, 1H), 7.44 (d, 1H, J = 8.9 Hz), 6.22 (dd, 1H, J = 8.9, 2.1 Hz), 6.08 (d, 1H, J = 2.1 Hz), 3.77 (s, 3H), 3.28 (t, 4H, J = 7.6 Hz), 1.62–1.43 (sextet, 4H, J = 7.5 Hz), 0.81 (t, 6H, J = 7.4 Hz). 13C NMR (101 MHz, acetone) δ 186.0, 164.9, 155.3, 130.3, 115.0, 105.3, 93.8, 55.7, 53.2, 21.2, 11.5. HRMS (ESI): calcd for C14H21NO2 [M + H]+, 236.1645; found, 236.1643.
tert-Butyl 4-(4-formyl-2-nitrophenyl)piperazine-1-carboxylate (39)
To a solution of 4-fluoro-3-nitrobenzaldehyde (1 equiv) in acetone (20 mL), pyridine (1 mL) and 1-Boc-piperazine (1.2 equiv) were added and stirred at room temperature for 4 h. The solvent was then evaporated under vacuum to yield a dark-yellow oil, which was purified by column chromatography using EA/PE (1:10) to afford 39.4% of the title compound as a yellow solid. Rf = 0.19 (EA/PE, 2:10). mp 141 ± 1 °C. 1H NMR (400 MHz, acetone): δ 9.94 (s, 1H), 8.33 (d, 1H, J = 2.0 Hz), 8.03 (dd, 1H, J = 8.9, 2.0 Hz), 7.44 (d, 1H, J = 8.9 Hz), 3.76–3.43 (m, 4H), 3.43–3.15 (m, 4H), 1.48 (s, 9H). 13C NMR (101 MHz, acetone): δ 189.91, 155.04, 150.11, 141.03, 133.88, 129.70, 129.18, 121.35, 80.08, 51.03, 44.23, 28.52. HRMS (ESI): calcd for C16H21N3O5 [M + H]+, 336.1559; found, 336.1560.
3-Nitro-4-(piperazin-1-yl)benzaldehyde (40)
To a solution of 39 (1 equiv) in CH2Cl2 (15 mL), trifluoroacetic acid (5 mL) was added and stirred at room temperature for 2 h. CH2Cl2 was then evaporated under vacuum to give a dark-yellow oil, which was purified by column chromatography using (CH3OH/CH2Cl2, 0.5:10) to yield quantitatively the title compound as a yellow-orange solid (82%). Rf = 0.38 (CH3OH/CH2Cl2, 1:10). mp 144 ± 1 °C. 1H NMR (400 MHz, acetone) δ 9.98 (s, 1H), 8.38 (d, 1H, J = 1.9 Hz), 8.10 (dd, 1H, J = 8.6, 1.9 Hz), 7.56 (d, 1H, J = 8.6 Hz), 3.71–3.61 (m, 4H), 3.61–3.50 (m, 4H), 2.10 (s, 1H). 13C NMR (101 MHz, acetone) δ 190.19, 149.65, 141.82, 134.47, 130.55, 129.34, 122.25, 48.60, 43.97. HRMS (ESI): calcd for C11H13N3O3 [M + H]+, 236.1030; found, 236.1031. Compound 40 was obtained in 82% purity.
Purification of Recombinant Human ALDHs and Enzymatic Assays
Human ALDH1A1, ALDH1A3, and ALDH3A1 were cloned and recombinantly expressed from the pET-30 Xa/LIC vector. Protein purification was achieved by affinity chromatography on a nickel-charged chelating Sepharose Fast Flow 5-mL column (His Trap column, Cytiva), which specifically binds the protein due to its N-terminal (His)6 tag, using an ÄKTA FPLC system (Cytiva), as previously described.55 Enzymes including the His tag were stored at −80 °C in 20 mM Tris/HCl and 0.5 M NaCl, pH 8.0, until use. Reaction buffers were as follows: ALDH1A1 was assayed in 50 mM HEPES, 0.5 mM EDTA, and 0.5 mM DTT, pH 8.0; ALDH1A3 was assayed in 50 mM HEPES, 30 mM MgCl2, and 5 mM DTT, pH 8.0; and ALDH3A1 was assayed in 50 mM Tris–HCl and 5 mM DTT, pH 8.0. Activity under standard conditions was measured fluorimetrically at 25 °C using a Cary Eclipse (Varian) fluorimeter to follow the purification procedure and to check the enzyme concentration before each kinetic experiment. The fluorescence of NADH was characterized at 460 nm with excitation at 340 nm, and 5 μM NAD(P)H was added as an internal standard to obtain the absolute reaction rates.55 Standard activity was measured at saturating concentrations of substrate using 30 μM hexanal (ALDH1A1), 250 μM hexanal (ALDH1A3), or 250 μM 4-NBA (ALDH3A1). NAD+ was 500 μM for ALDH1A1 and ALDH1A3, and NADP+ was 1 mM for ALDH3A1.
Inhibition Screening
All compounds tested were dissolved in DMSO and assayed at a final concentration of 1% (v/v) DMSO. Single-point measurements of enzymatic activity at 10 μM inhibitor were performed for the 40 DEAB analogues against the three isoforms (ALDH1A1, ALDH1A3, and ALDH3A1). For the initial screening, the enzymatic activity was measured in 96-well plates (final volume of 200 μL) in a Victor 3 Multilabel Plate Reader (Perkin Elmer), by monitoring the fluorescence of the NAD(P)H produced during the reaction (excitation at 340 nm and emission at 460 nm). Alternatively, the enzymatic activity was measured in a Varian Cary 400 UV–vis spectrophotometer, by monitoring the increase in the absorbance of NAD(P)H at 340 nm (ε = 6.22 mM–1·cm–1) or in a Cary Eclipse Varian fluorimeter, as described above. The inhibition screening was preferably performed at two substrate concentrations (near the Km value and at a saturating substrate concentration), except for ALDH1A1, which was only tested at substrate saturation due to a lack of sensitivity when its low substrate concentration was used in the assay. For ALDH1A1, 5 μM hexanal (Sigma) was used. For ALDH1A3, 10 and 250 μM hexanal (Sigma) were used. For ALDH3A1, 4-NBA (Sigma) was used at 31 and 250 μM. All substrates were prepared in the corresponding assay buffer at a concentration of 2 mM and further diluted to reach the final concentrations required per experiment. The concentration of the enzyme was kept from 50- to 100-fold lower than that of the substrate for all enzymatic assays.
Enzyme Kinetics with DEAB Analogues as Substrates
According to their structure with a carbonyl group, compounds were tested for their substrate properties against ALDH1A1, ALDH1A3, and ALDH3A1. For this assay, the conditions were the same as previously described except using the DEAB analogues as substrates instead of hexanal or 4-NBA at a concentration of 10 μM. Results are expressed as the percentage of activity at 10 μM versus the activity at 10 μM of their standard substrate (hexanal or 4-NBA). Values are expressed as the mean ± SE.
To calculate the Km value of compound 18 as a substrate for
ALDH3A1, several concentrations of compound 18 were used.
Experimental values were fitted to the adaptation of the Michaelis–Menten
equation for substrate inhibition  and shown as the mean ± SE.
and shown as the mean ± SE.
Determination of the Kinetic Constants (IC50 and Ki)
In order to determine the IC50 values,
reaction rates were determined at various concentrations
of inhibitor at a fixed concentration of substrate. As substrates
for ALDH1A1, ALDH1A3, and ALDH3A1, 5 μM hexanal, 6 μM
hexanal, and 31 μM 4-NBA, respectively, were used. The IC50 values were calculated by nonlinear fitting of the obtained
data to a sigmoidal plot using GraFit 5.0 (Erithacus software), with
the following 4-parameter equation  , where y is the specific
activity, x is the inhibitor concentration, background
is the minimum y value, range is the fitted uninhibited
value minus the background, and s is the slope factor.
Values are expressed as the mean ± SE.
, where y is the specific
activity, x is the inhibitor concentration, background
is the minimum y value, range is the fitted uninhibited
value minus the background, and s is the slope factor.
Values are expressed as the mean ± SE.
Activity assays
to determine the type of inhibition and Ki value were performed using various substrate and inhibitor concentrations
maintaining the same conditions as for the IC50 experiments
and using GraFit 5.0 for data processing. The data of enzymatic activities
at different inhibitor concentrations were fitted to the Michaelis–Menten
equation to determine the values of Km and Vmax. Next, results were fitted
to the equations for competitive, given by  ; noncompetitive, given by
; noncompetitive, given by  ; uncompetitive, given by
; uncompetitive, given by  ; and mixed,
given by
; and mixed,
given by  .56 The type
of inhibition was selected based on the lowest error value. Parameters
were expressed as the mean ± SE.
.56 The type
of inhibition was selected based on the lowest error value. Parameters
were expressed as the mean ± SE.
Docking studies of compounds 14 and 18 on ALDH1A3 and ALDH3A1 isoforms.
(a) Protein and ligand preparation: The three X-ray complexes ALDH1A3, ALDH3A1, and ALDH1A1 were downloaded from the Protein Data Bank with their PBD IDs 5FHZ,584H80,59 and 4WPN,60 respectively. The cocrystallized ligands, ions, and water molecules were removed from the X-ray complexes and H-bonds; missing residues were added to the protein with the aid of protein preparation wizard of Maestro. All the compounds were drawn using the Build panel of Maestro and subjected to a conformational search of 1000 steps in a water environment (using the generalized-Born/surface-area model) through Macromodel software. A Monte Carlo algorithm with the MMFF and a distance-dependent dielectric constant of 1.0 was applied while using Macromodel.61
(b) Molecular docking: All docking calculations were carried out on the X-ray structure of human ALDH1A1 in complex with selective inhibitor 1-{[1,3-dimethyl-7-(3-methylbutyl)-2,6-dioxo-2,3,6,7-tetrahydro-1H-purin-8-yl]methyl}piperidine-4-carboxamide (PDB ID: 4WPN), X-ray structure of human ALDH1A3 in complex with RA and NAD+ (PDB ID: 5FHZ), and X-ray structure of human ALDH3A1 in complex with selective inhibitor—N-[4-(4-methylsulfonyl-2-nitroanilino)phenyl]acetamide (PDB ID: 4H80). Glide 5.0 with the standard precision (SP) method62 was used for docking of all compounds on the three X-ray structures.
GLIDE 5.0
The binding site was defined by a rectangular box of 10 Å along the x, y, and z axes centered on the ligand. The option of imposing a maximum value to the number of atoms that a ligand may have (when docked) was deactivated. Thus, all the ligands were docked independently from the number of their atoms, whereas the GLIDE defaults were used for the remaining parameters. The GlideScore fitness function is based on Chemscore but includes a steric-clash term and adds buried polar terms to penalize electrostatic mismatches and modifications on other secondary terms. The docking analyses were carried out using the SP method. A total of 50 docking solutions were generated for each ligand, and the top-ranked docking pose was considered as the final pose.
The reliability of the docking program: GLIDE 5.0 was assessed by performing self-docking analysis and calculating the root-mean-square deviation (RMSD) between the crystallographic position of the ligand and the ligand’s disposition predicted by docking. The rms_analysis program of the Gold suite was used to calculate the RMSD difference, considering only the heavy atoms of the ligand. The docking method is able to produce a binding pose within 2.0 Å RMSD of the crystallographic disposition, therefore considered as reliable.63
Immunoblotting
The PCa cell lysates (DU145, LNCaP, and PC3) were prepared using the RIPA buffer. The lysates were briefly sonicated and centrifuged, and the protein concentrations were determined using the BCA protein assay kit (Thermo Fisher, CA, USA). A total of 40 μg of cell lysates was separated using 10% SDS-polyacrylamide gel electrophoresis and transferred on to Amersham Hybond ECL nitrocellulose membranes (Amersham, QC, Canada). The membranes were blocked using 5% skim milk in phosphate-buffered saline with 0.1% Tween-20 for 1 h and then incubated with specific primary antibodies overnight at 4 °C followed by incubation with HRP-conjugated secondary antibodies for 1 h at room temperature. Specific protein bands were visualized using an ECL detection kit (Amersham). The primary and secondary antibodies used were rabbit anti-ALDH1A1 (D9J7R, Cell signaling Technology, UK), mouse anti-GAPDH (6C5, Abcam, Cambridge, UK), rabbit anti-ALDH1A3 (N2C2, GeneTex, California, USA), mouse anti-ALDH3A1 (G-2, Santa Cruz Biotechnology, CA), anti-rabbit HRP (Dako), and goat anti-mouse HRP (Abcam, Cambridge, UK).
Chemosensitivity Studies
MTT (3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2H-tetrazolium bromide) assay was used to determine cell viability.64 Immortalized PCa cells (PC-3, Du-145, and LNCaP) were seeded in 96-well plates and allowed to adhere overnight at 37 °C, 5% CO2, and 100% humidity. The following day, cells were treated with the DEAB analogues at appropriate concentrations ranging from 12.5 to 200 μM. After 96 h of exposure, 200 μL of MTT (Sigma) solution (0.5 mg/mL) was added to each well and incubated at 37 °C for 4 h. The formazan crystals were dissolved in DMSO (Sigma), and the absorbance was read using a microplate reader (Multiskan EX; Thermo Fisher Scientific) at 540 nm. Data analysis was performed using Microsoft Excel 2013 and GraphPad Prism software.
Primary cells cultured at early passage from human prostate tissue biopsies included one BPH sample H415/15–BPH/PSA 0.61/age 74 and four PCa samples H568/15 RM–Gl7(3 + 4)/PSA 8.5/age 69, H431/14 LM–Gl7(3 + 4)/PSA 14/age 66, H488/14 RM–Gl7(3 + 4)/PSA 12.6/age 60, and H517/15 RM–Gl7(3 + 4)/PSA 4.4/age 65. Primary cells were obtained with ethical consent (REC ref 07/H1304/121) at radical prostatectomy (cancer) and transurethral resection (BPH) and were seeded as previously described37 in 96-well plates 5000 cells/well in 100 μL of SCM and incubated at 37 °C, 5% CO2 for 24 h. DEAB, 14, and 18 were prepared in DMSO at a stock concentration of 200 mM. Cells were treated with 100 μL of ALDH inhibitors at concentrations of 50 and 200 μM as single treatment. Combination treatments included 100 μL of 50 μM ALDH inhibitor + 1 nM docetaxel and 100 μL of 200 μM ALDH inhibitor + 1 nM docetaxel. Cells were also treated with 100 μL of 1 nM docetaxel only. 1 nM docetaxel was chosen since it was the IC50 when other primary samples were analyzed in our laboratory. Control wells included blank (media only) and untreated cells (DMSO only). Each experiment was performed in triplicate, and cell seeding density was the same in all wells; therefore, changes in treated cells were compared to untreated DMSO control cells. Plates were returned to the incubator for a 72 h incubation before further processing. Alamar Blue solution was added at 10% of total sample volume, and plates were incubated at 37 °C for 1–4 h before absorbance was analyzed in a plate reader.65 Alamar Blue has an excitation wavelength of 530–560 nm and an emission wavelength of 590 nm. Total % cell viability/cell survival was calculated by dividing the absorbance of the treated sample by the absorbance of control and multiplying it by 100.
Acknowledgments
We thank Maria Butjosa and Jan Julià for their assistance in performing inhibition screening and kinetic characterizations.
Glossary
Abbreviations
- ADT
androgen therapy
- ALDH
aldehyde dehydrogenase
- AR
androgen receptor
- BPH
benign prostate hyperplasia
- 13C NMR
carbon-13 nuclear magnetic resonance
- CH2Cl2
dichloromethane
- CH3OH
methanol
- CRPC
castrate-resistant prostate cancer
- CYP
cytochrome p450
- DEAB
4-(diethylamino)benzaldehyde
- DMF
dimethylformamide
- DMSO
dimethyl sulfoxide
- DPBA
4-(dipropylamino)benzaldehyde
- EA
ethyl acetate
- GLUT1
glucose transporter 1
- 1H NMR
proton nuclear magnetic resonance
- HRMS
high resolution mass spectrometry
- Hz
hertz
- IC50
half-maximal inhibitory concentration
- Ki
inhibitory constant
- mp
melting point
- MHz
megahertz
- NAD+
nicotinamide adenine dinucleotide
- NADH
nicotinamide adenine dinucleotide hydrogen
- 4-NBA
4-nitrobenzaldehyde
- PCa
prostate cancer
- PDB
protein data bank
- PE
petroleum ether
- RA
retinoic acid
- Rf
retention factor
- RMSD
root-mean-square deviation
- SCs
stem cells
- CSCs
cancer stem cells
- TLC
thin layer chromatography
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.1c01367.
HPLC traces for all compounds. PDB codes 4WPN (ALDH1A1), 5FHZ (ALDH1A3) and 4H80 (ALDH3A1) were used for docking studies of compounds 14 and 18, respectively (ZIP)
Molecular Formula Strings (CSV)
Additional data include results of inhibition screening as % of remaining ALDH activities in the presence of DEAB analogues, % of substrate activities of DEAB analogues,IC50 representations of 14 with ALDH1A3 and 18 with ALDH3A1, inhibition kinetics curves of ALDH1A3 by 15 and 16, IC50 representations of 18 with ALDH3A1 with and without NADP+, docking studies of compounds 14 and 18 on ALDH1A1, ALDH1A3 and ALDH3A1 isoforms and representations of the preliminary two-dose point antiproliferative data using PC3 cells and the MTT assay (PDF)
Author Contributions
A.I.M.I., J.F. and K.P. designed the study. A.I.M.I. performed all chemical experiments, V.J. and T.T. performed computational studies, E.B., R.J., R.P. and T.R. provided recombinant ALDH protein and performed biochemical studies, and S.S., M.S., F.F. and N.J.M. performed ALDH expression analysis and compound evaluation in PCa cell lines and primary cells. The first draft of the manuscript was co-written by A.I.M.I., E.B., J.F. and K.P. J.F., X.P. and K.P., funding acquisition. All authors have given approval to the final version of the manuscript. All authors have contributed to editorial improvements of the first draft.
This research was funded by Prostate Cancer UK (grant ref. no.: S12-027), the Spanish Ministerio de Ciencia e Innovación (PID2020-119424RB-I00), Al-Zaytoonah University of Jordan and UoB International Development Fund Scheme (PhD studentship to EBR). RJ is a recipient of a PIF predoctoral fellowship from Universitat Autònoma de Barcelona.
The authors declare no competing financial interest.
Supplementary Material
References
- Ginestier C.; Wicinski J.; Cervera N.; Monville F.; Finetti P.; Bertucci F.; Wicha M. S.; Birnbaum D.; Charafe-Jauffret E. Retinoid signaling regulates breast cancer stem cell differentiation. Cell Cycle 2009, 8, 3297–3302. 10.4161/cc.8.20.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson B.; Brocker C.; Thompson D. C.; Black W.; Vasiliou K.; Nebert D. W.; Vasiliou V. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum. Genom. 2011, 5, 283–303. 10.1186/1479-7364-5-4-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasimanickam V. Expression of retinoic acid-metabolizing enzymes, ALDH1A1, ALDH1A2, ALDH1A3, CYP26A1, CYP26B1 and CYP26C1 in canine testis during post-natal development. Reprod. Domest. Anim. 2016, 51, 901–909. 10.1111/rda.12756. [DOI] [PubMed] [Google Scholar]
- Moreb J. S.; Zucali J. R.; Ostmark B.; Benson N. A. Heterogeneity of aldehyde dehydrogenase expression in lung cancer cell lines is revealed by Aldefluor flow cytometry-based assay. Cytometry, Part B 2007, 72, 281–289. 10.1002/cyto.b.20161. [DOI] [PubMed] [Google Scholar]
- Muzio G.; Maggiora M.; Paiuzzi E.; Oraldi M.; Canuto R. A. Aldehyde dehydrogenases and cell proliferation. Free Radic. Biol. Med. 2012, 52, 735–746. 10.1016/j.freeradbiomed.2011.11.033. [DOI] [PubMed] [Google Scholar]
- Ma I.; Allan A. L. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev. 2011, 7, 292–306. 10.1007/s12015-010-9208-4. [DOI] [PubMed] [Google Scholar]
- Chute J. P.; Muramoto G. G.; Whitesides J.; Colvin M.; Safi R.; Chao N. J.; McDonnell D. P. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 11707–11712. 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P.; Joshi K.; Li J.; Kim S.-H.; Li P.; Santana-Santos L.; Luthra S.; Chandran U. R.; Benos P. V.; Smith L.; Wang M.; Hu B.; Cheng S.-Y.; Sobol R. W.; Nakano I. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 8644–8649. 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croker A. K.; Allan A. L. Inhibition of aldehyde dehydrogenase (ALDH) activity reduces chemotherapy and radiation resistance of stem-like ALDHhiCD44(+) human breast cancer cells. Breast Cancer Res. Treat. 2012, 133, 75–87. 10.1007/s10549-011-1692-y. [DOI] [PubMed] [Google Scholar]
- Awad O.; Yustein J. T.; Shah P.; Gul N.; Katuri V.; O’Neill A.; Kong Y.; Brown M. L.; Toretsky J. A.; Loeb D. M. High ALDH activity identifies chemotherapy-resistant Ewing’s sarcoma stem cells that retain sensitivity to EWS-FLI1 inhibition. PloS One 2010, 5, e13943 10.1371/journal.pone.0013943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Zheng S.; He H.; Ge H.; Saeed B. N,N-diethylaminobenzaldehyde targets aldehyde dehydrogenase to eradicate human pancreatic cancer cells. Exp. Ther. Med. 2020, 20, 662–670. 10.3892/etm.2020.8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. W.; Jung K.-H.; Lee J. H.; Moon S. H.; Cho Y. S.; Lee K.-H. Inhibition of aldehyde dehydrogenase 1 enhances the cytotoxic effect of retinaldehyde on A549 cancer cells. Oncotarget 2017, 8, 99382–99393. 10.18632/oncotarget.19544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez R.; Pequerul R.; Amor A.; Lorenzo J.; Metwally K.; Avilés F. X.; Parés X.; Farrés J. Inhibitors of aldehyde dehydrogenases of the 1A subfamily as putative anticancer agents: Kinetic characterization and effect on human cancer cells. Chem. Biol. Interact. 2019, 306, 123–130. 10.1016/j.cbi.2019.04.004. [DOI] [PubMed] [Google Scholar]
- Russo J.; Chung S.; Contreras K.; Lian B.; Lorenz J.; Stevens D.; Trousdell W. Identification of 4-(N,N-dipropylamino)benzaldehyde as a potent, reversible inhibitor of mouse and human class I aldehyde dehydrogenase. Biochem. Pharmacol. 1995, 50, 399–406. 10.1016/0006-2952(95)00138-p. [DOI] [PubMed] [Google Scholar]
- Zhou L.; Sheng D.; Wang D.; Ma W.; Deng Q.; Deng L.; Liu S. Identification of cancer-type specific expression patterns for active aldehyde dehydrogenase (ALDH) isoforms in ALDEFLUOR assay. Cell Biol. Toxicol. 2019, 35, 161–177. 10.1007/s10565-018-9444-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreb J. S.; Ucar D.; Han S.; Amory J. K.; Goldstein A. S.; Ostmark B.; Chang L.-J. The enzymatic activity of human aldehyde dehydrogenases 1A2 and 2 (ALDH1A2 and ALDH2) is detected by Aldefluor, inhibited by diethylaminobenzaldehyde and has significant effects on cell proliferation and drug resistance. Chem. Biol. Interact. 2012, 195, 52–60. 10.1016/j.cbi.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan C. A.; Parajuli B.; Buchman C. D.; Dria K.; Hurley T. D. N,N-diethylaminobenzaldehyde (DEAB) as a substrate and mechanism-based inhibitor for human ALDH isoenzymes. Chem. Biol. Interact. 2015, 234, 18–28. 10.1016/j.cbi.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M.; Gates K. S.; Henzl M. T.; Tanner J. J. Diethylaminobenzaldehyde is a covalent, irreversible inactivator of ALDH7A1. ACS Chem. Biol. 2015, 10, 693–697. 10.1021/cb500977q. [DOI] [PubMed] [Google Scholar]
- Pors K.; Moreb J. S. Aldehyde dehydrogenases in cancer: an opportunity for biomarker and drug development?. Drug Discov. Today 2014, 19, 1953–1963. 10.1016/j.drudis.2014.09.009. [DOI] [PubMed] [Google Scholar]
- Quattrini L.; Sadiq M.; Petrarolo G.; Maitland N. J.; Frame F. M.; Pors K.; La Motta C. Aldehyde Dehydrogenases and Prostate Cancer: Shedding Light on Isoform Distribution to Reveal Druggable Target. Biomedicines 2020, 8, 569. 10.3390/biomedicines8120569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcato P.; Dean C. A.; Giacomantonio C. A.; Lee P. W. K. Aldehyde dehydrogenase Its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle 2011, 10, 1378–1384. 10.4161/cc.10.9.15486. [DOI] [PubMed] [Google Scholar]
- Li D.; Zhang Q.; Wang X.; Li S.; Zhou H.; Wu J.; Tian Y. Self-assembly of a series of thiocyanate complexes with high two-photon absorbing active in near-IR range and bioimaging applications. Dyes Pigments 2015, 120, 175–183. 10.1016/j.dyepig.2015.03.039. [DOI] [Google Scholar]
- Canfield B. K.; Kuzyk M. G.; Hightower S. E.; Li A. D. Q. Use of quadratic electroabsorption for measurement of the hyperpolarizability ss of asymmetric molecules. J. Opt. Soc. Am. B 2005, 22, 723–734. 10.1364/josab.22.000723. [DOI] [Google Scholar]
- Yongpruksa N.; Pandey S.; Baker G. A.; Harmata M. Benzothiazines in organic synthesis. Synthesis of fluorescent 7-amino-2,1-benzothiazines. Org. Biomol. Chem. 2011, 9, 7979–7982. 10.1039/c1ob06051a. [DOI] [PubMed] [Google Scholar]
- Amslinger S.; Lindner S. Limno-CP: A Natural-Product-Inspired 5-Aryl-3(2H)-furanone as Scaffold for a Library of alpha-Modified Enones. Synthesis 2011, 2011, 2671–2683. 10.1055/s-0030-1260115. [DOI] [Google Scholar]
- Tomita H.; Tanaka K.; Tanaka T.; Hara A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget 2016, 7, 11018–11032. 10.18632/oncotarget.6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcato P.; Dean C. A.; Pan D.; Araslanova R.; Gillis M.; Joshi M.; Helyer L.; Pan L.; Leidal A.; Gujar S.; Giacomantonio C. A.; Lee P. W. K. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells 2011, 29, 32–45. 10.1002/stem.563. [DOI] [PubMed] [Google Scholar]
- Suwala A. K.; Koch K.; Rios D. H.; Aretz P.; Uhlmann C.; Ogorek I.; Felsberg J.; Reifenberger G.; Köhrer K.; Deenen R.; Steiger H.-J.; Kahlert U. D.; Maciaczyk J. Inhibition of Wnt/beta-catenin signaling downregulates expression of aldehyde dehydrogenase isoform 3A1 (ALDH3A1) to reduce resistance against temozolomide in glioblastoma. Oncotarget 2018, 9, 22703–22716. 10.18632/oncotarget.25210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parajuli B.; Fishel M. L.; Hurley T. D. Selective ALDH3A1 inhibition by benzimidazole analogues increase mafosfamide sensitivity in cancer cells. J. Med. Chem. 2014, 57, 449–461. 10.1021/jm401508p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milone M. R.; Pucci B.; Bifulco K.; Iannelli F.; Lombardi R.; Ciardiello C.; Bruzzese F.; Carriero M. V.; Budillon A. Proteomic analysis of zoledronic-acid resistant prostate cancer cells unveils novel pathways characterizing an invasive phenotype. Oncotarget 2015, 6, 5324–5341. 10.18632/oncotarget.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.; Su Y.; Mei Y.; Leng Q.; Leng B.; Liu Z.; Stass S. A.; Jiang F. ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients’ outcome. Lab. Invest. 2010, 90, 234–244. 10.1038/labinvest.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Zhou X.; Liang C.; Bao M.; Tian Y.; Zhu J.; Zhang T.; Yang J.; Wang Z. ALDH1A3 serves as a predictor for castration resistance in prostate cancer patients. BMC Cancer 2020, 20, 387. 10.1186/s12885-020-06899-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J.; De Melo J.; Cutz J.-C.; Aziz T.; Tang D. Aldehyde dehydrogenase 3A1 associates with prostate tumorigenesis. Br. J. Cancer 2014, 110, 2593–2603. 10.1038/bjc.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen C.; van der Horst G.; Cheung H.; Buijs J. T.; Pelger R. C. M.; van der Pluijm G. The aldehyde dehydrogenase enzyme 7A1 is functionally involved in prostate cancer bone metastasis. Clin. Exp. Metastasis 2011, 28, 615–625. 10.1007/s10585-011-9395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Magnen C.; Bubendorf L.; Rentsch C. A.; Mengus C.; Gsponer J.; Zellweger T.; Rieken M.; Thalmann G. N.; Cecchini M. G.; Germann M.; Bachmann A.; Wyler S.; Heberer M.; Spagnoli G. C. Characterization and clinical relevance of ALDHbright populations in prostate cancer. Clin. Cancer Res. 2013, 19, 5361–5371. 10.1158/1078-0432.ccr-12-2857. [DOI] [PubMed] [Google Scholar]
- van den Hoogen C.; van der Horst G.; Cheung H.; Buijs J. T.; Lippitt J. M.; Guzmán-Ramírez N.; Hamdy F. C.; Eaton C. L.; Thalmann G. N.; Cecchini M. G.; Pelger R. C. M.; van der Pluijm G. High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res. 2010, 70, 5163–5173. 10.1158/0008-5472.can-09-3806. [DOI] [PubMed] [Google Scholar]
- Frame F. M.; Pellacani D.; Collins A. T.; Maitland N. J. Harvesting Human Prostate Tissue Material and Culturing Primary Prostate Epithelial Cells. Methods Mol. Biol. 2016, 1443, 181–201. 10.1007/978-1-4939-3724-0_12. [DOI] [PubMed] [Google Scholar]
- Rivera-Gonzalez G. C.; Droop A. P.; Rippon H. J.; Tiemann K.; Pellacani D.; Georgopoulos L. J.; Maitland N. J. Retinoic acid and androgen receptors combine to achieve tissue specific control of human prostatic transglutaminase expression: a novel regulatory network with broader significance. Nucleic Acids Res. 2012, 40, 4825–4840. 10.1093/nar/gks143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins L. H.; Withers H. G.; Garbens A.; Love H. D.; Magnoni L.; Hayward S. W.; Moyes C. D. Hypoxia and the metabolic phenotype of prostate cancer cells. Biochim. Biophys. Acta 2009, 1787, 1433–1443. 10.1016/j.bbabio.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Cojoc M.; Peitzsch C.; Kurth I.; Trautmann F.; Kunz-Schughart L. A.; Telegeev G. D.; Stakhovsky E. A.; Walker J. R.; Simin K.; Lyle S.; Fuessel S.; Erdmann K.; Wirth M. P.; Krause M.; Baumann M.; Dubrovska A. Aldehyde Dehydrogenase Is Regulated by beta-Catenin/TCF and Promotes Radioresistance in Prostate Cancer Progenitor Cells. Cancer Res. 2015, 75, 1482–1494. 10.1158/0008-5472.CAN-14-1924. [DOI] [PubMed] [Google Scholar]
- Ibrahim A. I. M.; Sadiq M.; Frame F. M.; Maitland N. J.; Pors K. Expression and regulation of aldehyde dehydrogenases in prostate cancer. J. Cancer Metastasis Treat. 2018, 4, 44. 10.20517/2394-4722.2018.07. [DOI] [Google Scholar]
- Tanei T.; Morimoto K.; Shimazu K.; Kim S. J.; Tanji Y.; Taguchi T.; Tamaki Y.; Noguchi S. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin. Cancer Res. 2009, 15, 4234–4241. 10.1158/1078-0432.ccr-08-1479. [DOI] [PubMed] [Google Scholar]
- Ajani J. A.; Wang X.; Song S.; Suzuki A.; Taketa T.; Sudo K.; Wadhwa R.; Hofstetter W. L.; Komaki R.; Maru D. M.; Lee J. H.; Bhutani M. S.; Weston B.; Baladandayuthapani V.; Yao Y.; Honjo S.; Scott A. W.; Skinner H. D.; Johnson R. L.; Berry D. ALDH-1 expression levels predict response or resistance to preoperative chemoradiation in resectable esophageal cancer patients. Mol. Oncol. 2014, 8, 142–149. 10.1016/j.molonc.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott A. F.; Landis M. D.; Dontu G.; Griffith K. A.; Layman R. M.; Krop I.; Paskett L. A.; Wong H.; Dobrolecki L. E.; Lewis M. T.; Froehlich A. M.; Paranilam J.; Hayes D. F.; Wicha M. S.; Chang J. C. Preclinical and clinical studies of gamma secretase inhibitors with docetaxel on human breast tumors. Clin. Cancer Res. 2013, 19, 1512–1524. 10.1158/1078-0432.ccr-11-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitland N. J.; Collins A. T. Prostate cancer stem cells: a new target for therapy. J. Clin. Oncol. 2008, 26, 2862–2870. 10.1200/jco.2007.15.1472. [DOI] [PubMed] [Google Scholar]
- Carnero A.; Garcia-Mayea Y.; Mir C.; Lorente J.; Rubio I. T.; LLeonart M. E. The cancer stem-cell signaling network and resistance to therapy. Cancer Treat Rev. 2016, 49, 25–36. 10.1016/j.ctrv.2016.07.001. [DOI] [PubMed] [Google Scholar]
- Wang T.; Shigdar S.; Gantier M. P.; Hou Y.; Wang L.; Li Y.; Shamaileh H. A.; Yin W.; Zhou S.-F.; Zhao X.; Duan W. Cancer stem cell targeted therapy: progress amid controversies. Oncotarget 2015, 6, 44191–44206. 10.18632/oncotarget.6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso C. S.; Wu Y. M.; Robinson D. R.; Cao X.; Dhanasekaran S. M.; Khan A. P.; Quist M. J.; Jing X.; Lonigro R. J.; Brenner J. C.; Asangani I. A.; Ateeq B.; Chun S. Y.; Siddiqui J.; Sam L.; Anstett M.; Mehra R.; Prensner J. R.; Palanisamy N.; Ryslik G. A.; Vandin F.; Raphael B. J.; Kunju L. P.; Rhodes D. R.; Pienta K. J.; Chinnaiyan A. M.; Tomlins S. A. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012, 487, 239–243. 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris W. P.; Mostaghel E. A.; Nelson P. S.; Montgomery B. Androgen deprivation therapy: progress in understanding mechanisms of resistance and optimizing androgen depletion. Nat. Clin. Pract. Urol. 2009, 6, 76–85. 10.1038/ncpuro1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drazer M. W.; Stadler W. M. The Role of Testosterone in the Treatment of Castration-Resistant Prostate Cancer. Cancer J. 2016, 22, 330–333. 10.1097/ppo.0000000000000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L. J. Enzalutamide: A Review in Castration-Resistant Prostate Cancer. Drugs 2018, 78, 1913–1924. 10.1007/s40265-018-1029-9. [DOI] [PubMed] [Google Scholar]
- Tannock I. F.; de Wit R.; Berry W. R.; Horti J.; Pluzanska A.; Chi K. N.; Oudard S.; Théodore C.; James N. D.; Turesson I.; Rosenthal M. A.; Eisenberger M. A.; Investigators T. A. X. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N. Engl. J. Med. 2004, 351, 1502–1512. 10.1056/nejmoa040720. [DOI] [PubMed] [Google Scholar]
- Tsao C.-K.; Cutting E.; Martin J.; Oh W. K. The role of cabazitaxel in the treatment of metastatic castration-resistant prostate cancer. Ther. Adv. Urol. 2014, 6, 97–104. 10.1177/1756287214528557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y.; Yamawaki K.; Ishiguro T.; Yoshihara K.; Ueda H.; Sato A.; Ohata H.; Yoshida Y.; Minamino T.; Okamoto K.; Enomoto T. ALDH-Dependent Glycolytic Activation Mediates Stemness and Paclitaxel Resistance in Patient-Derived Spheroid Models of Uterine Endometrial Cancer. Stem Cell Rep. 2019, 13, 730–746. 10.1016/j.stemcr.2019.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pequerul R.; Vera J.; Giménez-Dejoz J.; Crespo I.; Coines J.; Porté S.; Rovira C.; Parés X.; Farrés J. Structural and kinetic features of aldehyde dehydrogenase 1A (ALDH1A) subfamily members, cancer stem cell markers active in retinoic acid biosynthesis. Arch. Biochem. Biophys. 2020, 681, 108256. 10.1016/j.abb.2020.108256. [DOI] [PubMed] [Google Scholar]
- Copeland R. A.Evaluation of Enzyme Inhibitors in Drug Discovery: A Guide for Medicinal Chemists and Pharmacologists, 2nd ed.; John Wiley & Sons, Inc., 2013; pp 57–124. [PubMed] [Google Scholar]
- Moretti A.; Li J.; Donini S.; Sobol R. W.; Rizzi M.; Garavaglia S. Crystal structure of human aldehyde dehydrogenase 1A3 complexed with NAD+ and retinoic acid. Sci. Rep. 2016, 6, 35710. 10.1038/srep35710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M.; Westbrook J.; Feng Z.; Gilliland G.; Bhat T. N.; Weissig H.; Shindyalov I. N.; Bourne P. E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan C. A.; Hurley T. D. Characterization of two distinct structural classes of selective aldehyde dehydrogenase 1A1 inhibitors. J. Med. Chem. 2015, 58, 1964–1975. 10.1021/jm501900s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger . MacroModel; Schrödinger, LLC: New York, NY, 2009.
- Halgren T. A.; Murphy R. B.; Friesner R. A.; Beard H. S.; Frye L. L.; Pollard W. T.; Banks J. L. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- Tuccinardi T.; Poli G.; Romboli V.; Giordano A.; Martinelli A. Extensive consensus docking evaluation for ligand pose prediction and virtual screening studies. J. Chem. Inf. Model. 2014, 54, 2980–2986. 10.1021/ci500424n. [DOI] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Hirst A. M.; Simms M. S.; Mann V. M.; Maitland N. J.; O’Connell D.; Frame F. M. Low-temperature plasma treatment induces DNA damage leading to necrotic cell death in primary prostate epithelial cells. Br. J. Cancer 2015, 112, 1536–1545. 10.1038/bjc.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.