Abstract
Transition from maternal to embryonic transcriptional control is crucial for embryogenesis. However, alternative splicing regulation during this process remains understudied. Using transcriptomic data from human, mouse, and cow preimplantation development, we show that the stage of zygotic genome activation (ZGA) exhibits the highest levels of exon skipping diversity reported for any cell or tissue type. Much of this exon skipping is temporary, leads to disruptive noncanonical isoforms, and occurs in genes enriched for DNA damage response in the three species. Two core spliceosomal components, Snrpb and Snrpd2, regulate these patterns. These genes have low maternal expression at ZGA and increase sharply thereafter. Microinjection of Snrpb/d2 messenger RNA into mouse zygotes reduces the levels of exon skipping at ZGA and leads to increased p53-mediated DNA damage response. We propose that mammalian embryos undergo an evolutionarily conserved, developmentally programmed splicing failure at ZGA that contributes to the attenuation of cellular responses to DNA damage.
Alternative splicing temporarily disrupts genes involved in DNA damage response during maternal-to-zygotic transition.
INTRODUCTION
Preimplantation embryonic development follows a morphogenetically similar path in all placental mammals. It progresses from an unfertilized oocyte to a fertilized zygote through fusion with sperm, followed by symmetric cell divisions, morula compaction, establishment of the first cell lineages, and formation of a blastocyst that implants into the uterine wall. A distinctive control over the cell cycle and DNA damage response (DDR) has been observed during the first cell divisions (1, 2), a moment in which the embryo needs to ensure cell cycle progression while preserving genome integrity, as any unrepaired damage will be inherited by all embryo lineages. These preimplantation events are mirrored by large epigenetic and transcriptomic changes. Arguably, the most studied aspect is the transition from maternal to embryonic transcriptional control at the stage of zygotic genome activation (ZGA). Here, the maternal mRNA contribution is cleared both actively and passively (3), and the zygotic genome activates in different waves (4). The relative timing of the major wave differs between species, disconnecting morphogenetic and transcriptomic events. For example, while morula compaction occurs roughly at the eight-cell (8C) stage in both human and mouse embryos, the major wave of ZGA also takes place at the 8C stage in human but at the 2C stage in mouse. These differences, added to the difficulty of obtaining reliable transcriptomic data from preimplantation embryos from multiple mammalian species, complicate transcriptomic evolutionary comparisons between species. Thus, despite some initial reports based on microarrays [e.g., (5, 6)], in-depth investigation of transcriptome-wide remodeling had remained elusive until the advent of single-cell and low-input RNA sequencing (RNA-seq). Using these techniques, several studies have undertaken large transcriptomic analyses of preimplantation development in human (7–9), mouse (3, 7, 10–13), and cow (14–16). These studies have confirmed some previous findings (e.g., the different timing of the ZGA in each species) and provided comparative insights. However, they focused nearly exclusively on variations in the steady-state levels of protein-coding genes.
Alternative splicing (AS) is the process by which different pairs of splice sites are selected in precursor mRNAs leading to different combinations of exons in the final mature mRNA. It is responsible for greatly expanding the functional and regulatory capacity of eukaryotic genomes (17), potentially generating numerous transcript and protein products from a single gene. Over half of human protein–coding genes produce multiple transcript isoforms that are widely regulated across cell and tissue types (18), with particularly high prevalence in brain and testis (19, 20). These AS events may generate distinct functional protein isoforms or lead to unproductive mRNA products that are degraded by nonsense-mediated decay (NMD), thereby contributing to the modulation of gene expression. Although proper AS regulation is crucial for postimplantation mammalian embryo development (21), only a handful of studies have descriptively assessed isoform diversity during preimplantation stages, concluding that hundreds of genes dynamically express different transcripts in human and mouse, particularly at the maternal-to-zygotic transition (8, 12, 22–25). However, the regulatory mechanisms, evolutionary conservation, and physiological implications of these patterns are unknown.
Here, we generated a comprehensive dataset of AS quantifications for preimplantation development of human, mouse, and cow. We found that the blastomeres undergoing the major wave of ZGA show the highest levels of exon skipping reported so far for any cell or tissue type. In most cases, this exon skipping was temporary and restored soon after ZGA. These AS events often disrupt the open reading frame (ORF) and are enriched in genes involved in DDR in the three studied species. We identified the Sm ring components Snrpb and Snrpd2 as major regulators of these patterns and showed that induced expression of these factors before the major wave of ZGA leads to reduced splicing disruption and increased DDR upon etoposide treatment at this developmental point.
RESULTS
AS profiles in early embryo development of human, mouse, and cow
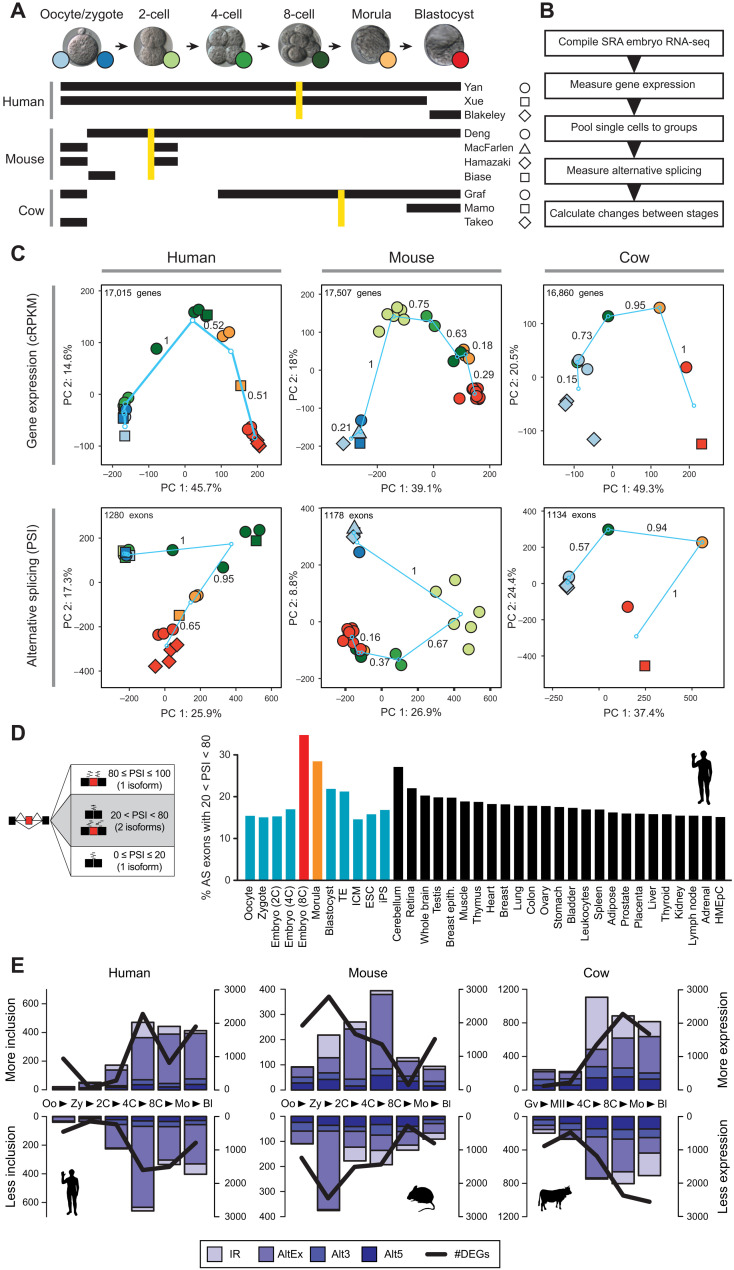
To investigate AS during preimplantation development, we took advantage of the abundant publicly available RNA-seq datasets (Fig. 1A and table S1). These data comprised samples from oocyte to blastocyst stage embryos from multiple studies obtained from either single blastomere or bulk embryo RNA-seq. To confidently estimate AS levels across the time courses, we performed the following steps (Fig. 1B; see Materials and Methods for details). We first measured gene steady-state mRNA levels [hereafter referred to as gene expression (GE)] genome-wide for each sample and clustered these samples using hierarchical clustering (figs. S1 to S3). Samples largely grouped by stage and not by experiment and outlier cells were removed from further study (table S1 for details). On the basis of this information, we merged single cells/samples into pools of ~160 million reads on average (table S1) to acquire high coverage on exon-exon junctions and improve quantifications of AS. Principal components analyses (PCAs) of GE measurements for the pooled groups showed a V-shaped temporal profile for PCs 1 and 2, with the largest difference between consecutive stages occurring at the time of the major wave of ZGA in the three species [Fig. 1C; between the 4C and 8C stages in human, zygote and 2C stage in mouse, and 4C and 8C + morula in cow]. For simplicity, throughout the manuscript, we refer to the stages immediately before and immediately after the major ZGA wave as the “pre-ZGA” and “ZGA” stages.
Fig. 1. AS profiles during early development reveal the highest exon skipping diversity at ZGA.
(A) Summary of datasets used in this study for each species. Golden lines indicate the major wave of ZGA for each species. (B) Schematic view of the methodological steps taken to process the RNA-seq data and obtain AS quantifications with vast-tools. SRA, Short Read Archive. (C) PCA for the merged groups of samples per stage for GE {top; cRPKMs [corrected (for mappability) reads per kilobase of transcript per million mapped reads]} and AS (bottom; exons with sufficient read coverage in at least 80% of the samples). Color codes and shapes for each stage as depicted in (A). Turquoise circles represent the centroids of each stage, and lines show the change between consecutive stages, with values representing the relative change respect to the largest change (=1). PSI, percent-spliced-in. (D) Percentage of exons with 20 < PSI < 80 (i.e., generating two substantial isoforms) in each sample was calculated for every stage and differentiated tissue, showing the highest relative exon skipping diversity for the ZGA stage. Equivalent plots for mouse and cow are shown in fig. S5A. Trophectoderm (TE), Inner Cell Mass (ICM); Embryonic Stem Cells (ESC); induced Pluripotent Stem cells (iPS); Human Mammary Epithelial Cells (HMEpC). (E) Numbers of alternative exons (AltEx), alternative 3/5′ splice sites (Alt3/5) and retained introns (IR) with increased/decreased inclusion levels in consecutive pairwise transitions (bars, left y axes) or genes with higher/lower expression (DEGs) (line, right y axes). Each data point represents the change in inclusion/expression between the stages at each side of the arrowhead in the x axis (e.g., Oo > Zy). Oo, oocyte; Zy, zygote; Mo, morula; Bl, blastocyst; Gv, oocyte GV; and MII, oocyte MII.
We then used vast-tools (18, 26) to quantify alternative sequence inclusion levels for alternative exons, alternative donor/acceptor sites, and retained introns. We used the percent-spliced-in (PSI) metric, which gives a value between 0 and 100 corresponding to the percentage of expressed transcripts from the host gene that include the alternative sequence; therefore, the PSI metric is independent of the expression level of the host gene (fig. S4A). In addition, to ensure that early embryo–specific exons were not missed from vast-tools annotations, we conducted a de novo search for cassette exons using a custom pipeline (see Materials and Methods for details; table S2). Similar to GE, PCAs of exon PSIs separated embryos by cell stage and not experiment, showed V-shaped temporal dynamics, and had the largest difference between consecutive stages at the major ZGA wave for the three species (Fig. 1C). To facilitate the access of this large resource to the research community, we have provided AS and GE plots as special datasets in VastDB (http://vastdb.crg.eu).
ZGA stage embryos show the highest levels of exon skipping diversity of any cell and tissue type
To have a first assessment of the contribution that AS has to diversify transcriptomes at each developmental stage, we used a simple measure of diversity where alternative exons with sufficient read coverage were classified as either producing one main isoform (PSI ≤ 20 or PSI ≥ 80) or two (20 < PSI < 80) (Fig. 1D). Remarkably, the stage undergoing the ZGA showed the highest levels of exon skipping diversity in the three studied species (8C in human, 2C in mouse, and 8C in cow), returning to the preceding lower levels in the subsequent stages (Fig. 1D and fig. S5A). Furthermore, the level of AS of cassette exons upon ZGA was not matched by any differentiated cell or tissue type, including neural, muscle, and testis (Fig. 1D and fig. S5A). The increased level of AS among alternative exons was robust to different cutoffs of PSI range and read coverage (fig. S5B), also observed at the single-blastomere level (fig. S5C), and was not found for intron retention or alternative 3′/5′ splice site choices (fig. S5B). Together, these results reveal that transcriptome diversification driven by exon skipping reaches its maximum in mammals only for a brief moment in life during ZGA.
Changes in GE and AS are maximal at ZGA
We next measured the number of AS events that change between each pair of consecutive stages (hereafter, stage transitions) in our time course (fig. S4B and Materials and Methods). In total, we found 2711, 1828, and 4748 unique AS events of all types with differential regulation in 2058, 1350, and 2735 genes for human, mouse, and cow, respectively (Fig. 1E). For comparison, we also calculated the numbers of differentially expressed genes (fig. S4C) and found 6545, 8895, and 8118 genes in each respective species that showed differential expression in at least one transition. Consistent with the PCA results, we found the largest number of changes for both AS and GE at the major ZGA wave in the three species (Fig. 1E), with only small effects observed for the minor ZGA wave. For example, in human, 1130 of 2711 (41.7%) of AS events with differential regulation and 3826 of 6545 (58.5%) of genes differentially expressed were observed at ZGA (4C-8C transition). Moreover, a clear bias was observed in the direction of regulation for alternative exons and intron retention in the three species at this transition: Whereas most exons showed increased skipping at ZGA, most differentially spliced introns had increased retention (Fig. 1E). Despite AS and GE changes mostly occurring at ZGA, the specific genes with AS and GE changes did not significantly overlap for nearly all transitions, a pattern that was consistent for intron retention and exon skipping separately (fig. S6, A to C). In other words, changes in GE and AS at ZGA occur largely independently. Given that a large number of genes become transcribed at this stage, these results imply that, in many cases, the maternally inherited transcript isoform is (partially) substituted by a new isoform without significantly altering the overall mRNA steady state level of the gene.
Conserved and species-specific changes in AS
We next asked whether changes in AS and GE were conserved across species or were specific to individual species. For exons changing in each transition for a given species, we assessed in each other species: (i) whether the exons were present in their genome [“genome-conserved” (27)], (ii) whether these orthologous exons had sufficient read coverage, and, if so, (iii) whether they changed (|ΔPSI| > 15) in the same direction at any transition (fig. S7, A to F; see Materials and Methods for details). This analysis showed that most AS events changing in two species change at ZGA but highlighted overall low levels of conservation. For instance, only 3.6 and 5.1% (31 and 44 of 859) of ZGA-regulated exons in human also change their inclusion in the same direction at the mouse or cow ZGA, respectively. These percentages increase up to 15.8 and 30.1%, respectively, for exons with an ortholog and sufficient read coverage at the ZGA in the other species, a higher fraction than expected by chance (P = 0.024 and P = 3.7 × 10−8, respectively, one-sided Fisher’s exact test). In the case of GE, 27.2 and 35.0% of genes with differential expression at ZGA in human overlapped with those changing at ZGA in mouse and cow, respectively, although a high level of heterochronies was also observed (fig. S8), as previously reported (5). To further identify exons that were dynamically regulated during preimplantation development across mammals, we next searched for orthologous exons whose inclusion levels were different between any two stages in the three species (see Materials and Methods). This revealed 259 exons (fig. S7G and table S3), which were significantly enriched for genes involved in key signaling pathways (e.g., Wnt pathway), transcription and chromatin modifiers (e.g., Tcerg1, Dnmt3b, and Ezh2), and genes related to morphogenesis (e.g., cadherin binding) (fig. S7H).
Peak profiles dominate AS dynamics
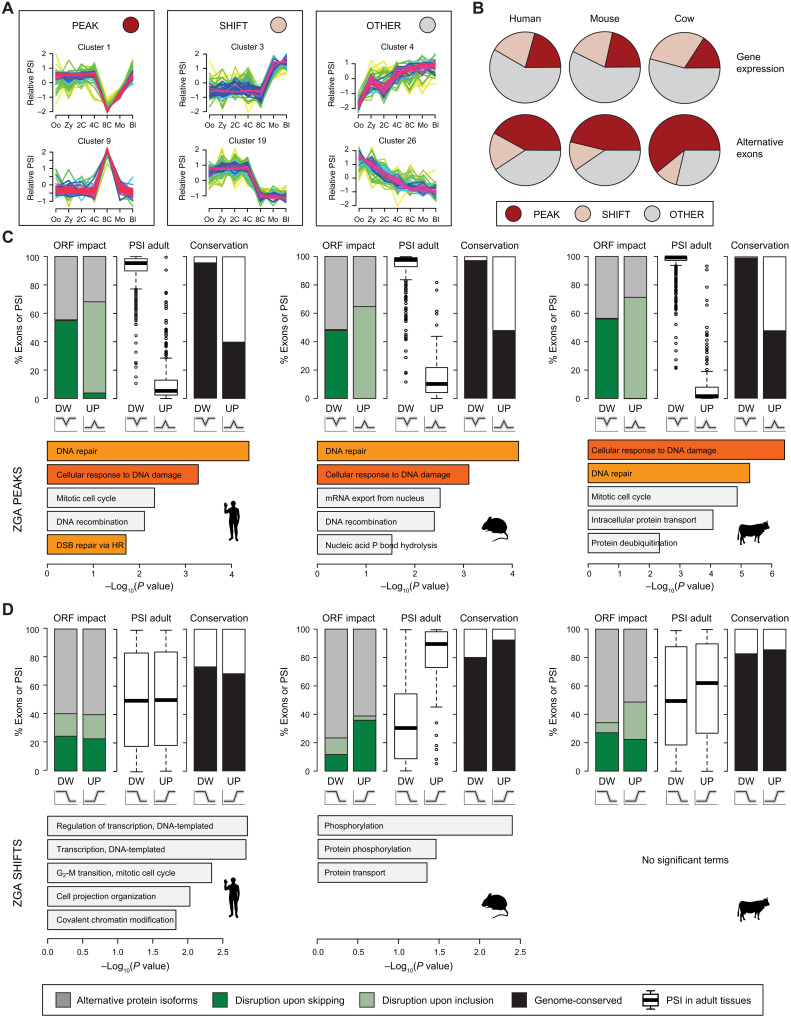
To further characterize the temporal dynamics of AS, we used Mfuzz (28) to cluster alternative exons according to their coregulated inclusion levels throughout the time course. We obtained 28, 18, and 22 exon clusters in human, mouse, and cow, respectively (figs. S9 to S11 and table S4). Most of these clusters could be broadly classified into three general patterns: (i) peak-like regulation, in which the exon is highly included or skipped only in a given stage, quickly returning to the initial levels (hereafter, “peak exons”); (ii) shift-like regulation, in which the exon goes from high to low inclusion at a specific transition, or vice versa, but does not return to the initial level (“shift exons”); and (iii) other regulation, those that do not fit the previous descriptions (Fig. 2A; see Materials and Methods for precise definitions). The peak behavior was the most common regulation among alternative exons (Fig. 2B; e.g., 42.1% versus 17.3% of shift exons in human). This contrasted with the patterns of Mfuzz clusters based on GE, which were more represented by shift or other behaviors (Fig. 2B). These results were consistent with the V-shape patterns in the PCA (Fig. 1C) and the asymmetric patterns observed for exon skipping and intron retention at ZGA, which were inverted in the post-ZGA transition in the three species (Fig. 1E), suggesting that a large proportion of changes at this time are temporary. Peak profiles were particularly enriched among AS events changing at ZGA (fig. S12); for instance, 428 of 750 (57%) human alternative exons with significant changes at ZGA covered in the Mfuzz cluster analysis showed peak behavior. Mfuzz clustering patterns were highly validated by reverse transcription polymerase chain reaction (RT-PCR) using RNA from independent pools of embryos, with 21 of 23 (91%) of peak, 11 of 11 (100%) of shift, and 19 of 19 (100%) of other alternative exons showing the expected temporal dynamics (fig. S13).
Fig. 2. Exons peaking at ZGA disrupt the ORFs and are enriched for DDR genes.
(A) Representative examples of the three main Mfuzz cluster types (Peak, Shift, and Other). (B) Proportions of each type of Mfuzz cluster for GE (top) and alternative exons with sufficient read coverage in all stages (bottom). (C and D) For each species, several features are shown for exons in Mfuzz clusters with Peak (C) or Shift (D) dynamics at ZGA, either with decreased (DW) or increased (UP) PSI. Top: Percentage of exons in coding sequences predicted to disrupt the ORF upon inclusion or skipping or to produce alternative protein isoforms (left), distribution of median PSIs in adult tissues (center), and percentage of exons with genomic conservation in any of the other two species (right). Bottom: Enriched Gene Ontology (GO) categories. Orange bars indicate DNA damage/repair–related categories. DSB, double-strand break.
Peak AS changes at ZGA often disrupt the ORF and are enriched for DDR genes
To begin elucidating the potential functional impact of AS changes during early embryogenesis, we investigated the predicted effect that alternative sequence inclusion or exclusion had on ORFs at each stage. Although the fractions of AS events that are predicted to alter the ORF were similar across stage transitions and species (fig. S14A), we found strong biases in the direction of ORF disruption depending on the transition: The vast majority of non–frame-preserving AS events changing at ZGA disrupt the ORF specifically at that stage in the three species (fig. S14B). That is, these alternative sequences were more included at ZGA when the inclusion was predicted to disrupt the ORF, and vice versa for exclusion. Consistent with these predictions, isoforms predicted to disrupt the ORF at ZGA showed strong up-regulation upon NMD disruption through UPF1 knockdown in HR1 cells (fig. S14C). Moreover, the disruptive impact on ORFs seems further strengthened by a global differential engagement of isoforms in translating ribosomes. Comparison of RNA-seq data from high and low polysome fractions from human embryonic stem cells (29) or human embryonic kidney (HEK) 293 cells (30) revealed a strong bias for ORF-disrupting isoforms at the ZGA to be less engaged by translating ribosomes, whereas the opposite was true for the few ORF-recovering isoforms (fig. S14, D and E).
Analysis of the subsets of exons that belonged to different Mfuzz clusters changing at ZGA further informed these patterns. Most peak exons disrupted the ORF at the peaking stage in the three species, whereas shift exons more often generated alternative protein isoforms (Fig. 2, C and D, top left). In addition, peak-down exons were usually constitutively or highly included in differentiated tissues, whereas peak-up exons were normally not or only lowly included in differentiated tissues (Fig. 2C, top middle). The biased inclusion levels in peak exons occurred for both ORF-preserving and ORF-disrupting events, underscoring a widespread change from canonical to rare isoforms at this stage. In line with this, ZGA isoforms were found to be up-regulated upon NMD depletion and depleted in the high polysome fraction in both embryonic stem cells and 293 T cells, irrespective of the predicted ORF impact (fig. S14, F to H). Two additional lines of evidence supported the opposite nature of peak-down and peak-up exons. First, each cluster type had distinct profiles of overlap with transposable elements: Peak-down exons were strongly depleted for transposable elements, whereas peak-up exons were enriched for these genetic elements (fig. S15). Second, peak-down exons had the highest levels of genome conservation among mammals, whereas most peak-up exons were species specific (Fig. 2C, top right). Therefore, together, these patterns are consistent with most peak-down exons being constitutive exons of major coding relevance and whose skipping at ZGA leads to nonfunctional isoforms, whereas peak-up exons are cryptic-like exons that do not encode important protein domains and often disrupt the gene’s canonical isoforms when included.
Despite the low level of regulatory conservation of individual events (see above), exons in clusters peaking at ZGA were altogether significantly enriched for genes involved in cellular response to DNA damage and DNA repair in human, mouse, and cow (Fig. 2C, bottom). These Gene Ontology (GO) enrichments contrast sharply with those for genes with shift events, with terms related to transcription or protein phosphorylation, and whose regulated exons had more intermediate inclusion levels across adult cell and tissues (Fig. 2D). In summary, these results show that a large number of peak exons are likely to disrupt protein function temporarily upon ZGA, affecting genes involved in DDR and DNA repair.
Reduced DDR to etoposide during ZGA
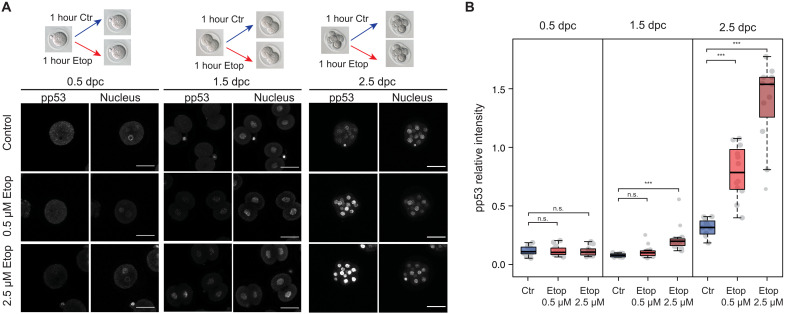
The predicted impact of AS on the function of proteins involved in DDR suggests that this process may be affected during ZGA. DDR and DNA repair pathways operate during the early stages of mammalian development (31–33). However, several studies in different mammalian species indicate that cleavage stage embryos are particularly resistant to certain DNA damage–inducing agents (34–38), suggesting that at least some DDR pathways may not be fully functional during ZGA. To gain further insights into DDR regulation at this stage, we first treated mouse embryos with the topoisomerase inhibitor etoposide at different developmental points: before, during, and after the major ZGA wave (1C, late 2C, and 8C-16C stages, respectively). Etoposide induces DNA double-strand breaks, which primarily get resolved through the activation of the Ataxia-telangiectasia mutated (ATM) pathway (39) followed by the phosphorylation of p53, one of its main downstream targets (40). We therefore used the levels of p53 phosphorylation at Ser15, as well as phospho-ATM (Ser1981), to ask whether embryos at the time of ZGA display a correct activation of the ATM pathway in response to etoposide. After treatment of embryos for 1 hour with either 0.5 or 2.5 μM etoposide, the levels of phospo-p53 and phospho-ATM were significantly higher in treated morulas in both conditions, whereas they remained low in treated 1C embryos and were only mildly induced with the higher concentration in 2C embryos (Fig. 3 and fig. S16, A to C). However, the basal levels of phospho-p53, but not of phospho-ATM, were lower in 2C embryos when compared to 1C or early morula (fig. S16, D and E), indicating a singular regulation of p53 phosphorylation during the major ZGA wave. Consequently, only a very high concentration of etoposide (10 μM) induced levels of p53 phosphorylation in 2C embryos, equivalent to those seen with 0.5 μM in morulas (fig. S16F), suggesting that much higher levels of DNA damage are tolerated in early embryos before ATM-p53 activation.
Fig. 3. Mouse embryos at ZGA have a lower response to DNA damage induced by etoposide.
(A) Scheme of etoposide treatment and immunostaining for phospho-p53 (Ser15; pp53). Scale bars, 50 μm. (B) Quantification of nuclear phospho-p53 (Ser15) levels from embryos is shown for each stage. Each dot represents the average relative intensity of all cells of a single embryo. Box plots show staining levels of embryos from one representative experiment. Two more experiments are shown in fig. S16C. Number of embryos: n = 12 (control, 1C), n = 14 (0.5 μM etoposide, 1C), n = 14 (2.5 μM etoposide, 1C), n = 14 (control, 2C), n = 15 (0.5 μM etoposide, 2C), n = 15 (2.5 μM etoposide, 2C), n = 11 (control, morula), n = 12 (0.5 μM etoposide, morula), and n = 11 (2.5 μM etoposide, morula). ***P < 0.001 based on Wilcoxon rank sum tests. n.s., not significant. days post coitum (dpc).
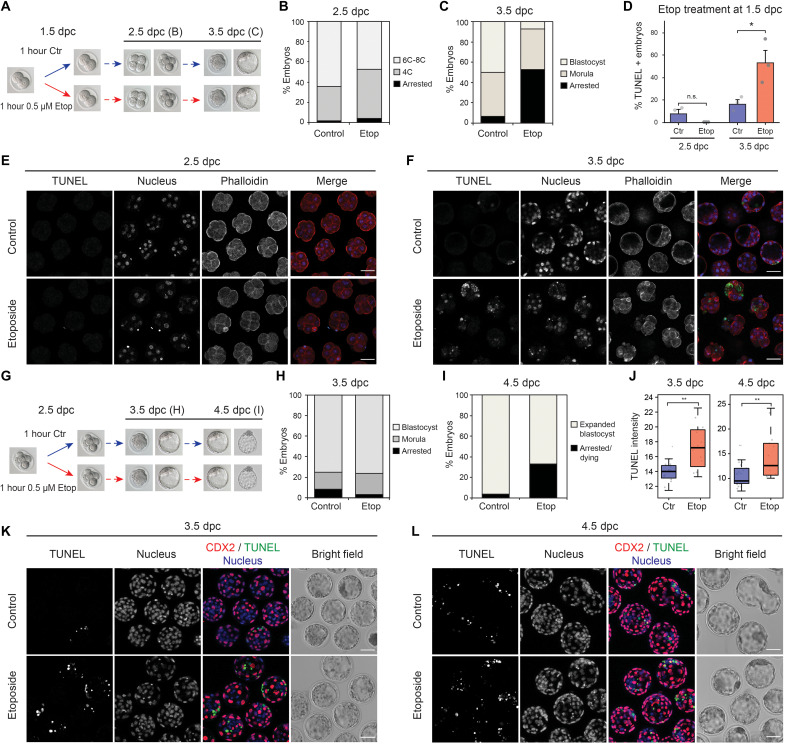
To assess the impact that this reduced response has on embryo development, we treated late 2C embryos with the lower dose (0.5 μM) of etoposide for 1 hour and analyzed the effect on their developmental progression for further 48 hours (Fig. 4A). Etoposide-treated embryos progressed to the 8C stage at the same rate as control embryos (Fig. 4B and fig. S17A) and became arrested after early morula compaction (Fig. 4C and fig. S17A). In addition, terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL) staining showed that cell death was suppressed until morula stage (48 hours after treatment), when a large proportion of etoposide-treated embryos were TUNEL positive (Fig. 4, D to F). When the same experiment was performed on early morulas (Fig. 4G), which show a strong immediate activation of the DDR (Fig. 3), a high proportion of treated embryos left to recover developed to form expanded blastocysts 48 hours after treatment (Fig. 4, H and I, and fig. S17B). Cell death was induced in treated embryos by 3.5 days post coitum (dpc) (24 hours after treatment), and no further increase was seen at 4.5 dpc (Fig. 4, J to L, and fig. S17, C and D). Overall, this suggests that the inability of 2C embryos to fully activate an immediate DDR (Fig. 3, A and B) leads to a high rate of developmental arrest at later stages. In contrast, proper activation of the DDR and DNA repair pathways in early morulas likely leads to either the repair or elimination of damaged cells, resulting in a lower developmental arrest rate than that observed for embryos treated at 2C stage.
Fig. 4. Mouse embryos treated with etoposide at ZGA present a delayed DDR that results in a high rate of developmental arrest.
(A) Scheme of etoposide treatment: 2C embryos were left untreated or treated with 0.5 μM etoposide for 1 hour and then left to recover for further 48 hours. (B and C) Proportion of embryos found arrested or developed to the stated stage 24 hours (B) or 48 hours (C) after treatment. Average of six independent experiments. Total number of embryos: n = 105 (control) and n = 113 (treated). (D) Percentage of embryos showing positive TUNEL staining at each developmental stage. Average of three independent experiments is shown. Total number of embryos: n = 72 (control, 2.5 dpc), n = 63 (etoposide, 2.5 dpc), n = 67 (control, 3.5 dpc), and n = 50 (etoposide, 3.5 dpc). *P = 0.036, two-sided Student’s t test. Error bars represent SEM. (E and F) TUNEL staining of embryos in (B) and (C). (G) Scheme of etoposide treatment: Early morula embryos were left untreated or treated with 0.5 μM etoposide for 1 hour and then left to recover for further 48 hours. (H and I) Proportion of embryos found arrested or developed to the stated stage 24 hours (H) or 48 hours (I) after treatment. Average of two independent experiments. Total number of embryos: n = 45 (control) and n = 44 (treated). (J) Quantification of TUNEL staining intensity in morphologically normal blastocysts from (H) and (I). Each dot represents one embryo. A representative experiment is shown. Total number of embryos: n = 16 (control, 3.5 dpc), n = 12 (etoposide, 3.5 dpc), n = 26 (control, 4.5 dpc), and n = 13 (etoposide, 4.5 dpc). Another experiment is shown in fig. S17D. **P < 0.01 based on Wilcoxon rank sum tests. (K and L) Double staining of TUNEL and the trophectoderm marker CDX2 of embryos in (H) and (I). In (B) and (C), “Arrested” embryos represent those not reaching the expected developmental stage because of either cell death or cell cycle arrest. Bar plots corresponding to (B), (C), (H), and (I) are shown in fig. S17 (A and B). Scale bars, 50 μm (E, F, K, and L).
Last, we asked whether other DDR pathways were also dampened during ZGA. For this, we treated embryos with aphidicolin, which induces replication stress and activates the Ataxia telangiectasia and Rad3 related (ATR) pathway (41). When 2C embryos were treated with aphidicolin (0.25 μg/ml) for 16 hours and then left to recover for further 8 hours, the vast majority arrested at the 4C stage (fig. S18A). However, when the same treatment was done at the early morula stage (8C-16C), treated embryos formed blastocysts at a similar rate to controls (fig. S18B). This shows that embryos around ZGA are able to activate an early response to replication stress that differs from that observed at morula stage.
These results thus indicate that not all DDR pathways are equally active during early development and demonstrate that sensitivity of ATM-p53–mediated DDR to double-strand breaks is low before and during major ZGA and only becomes fully active from the early morula stage, when the embryos can resolve the accumulated damage by inducing cell cycle arrest and death. Mechanistically, AS-mediated protein disruption at ZGA could lower the overall functional levels of some newly produced DDR proteins at this developmental stage, contributing to maintaining a reduced response to certain DNA lesions until early morula stage.
Peak exons are sensitive to SNRPB levels
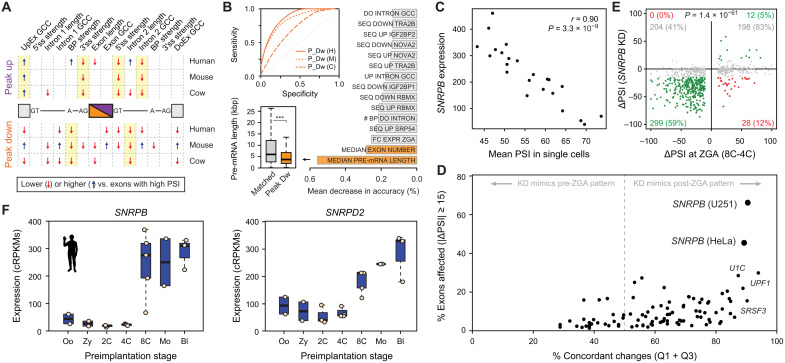
To gain insights into the characteristics and regulation of peak exons at ZGA, we first evaluated multiple exonic and intronic features associated with exon skipping using Matt (42). This analysis revealed some common patterns across the three species, including weaker branch points and shorter downstream introns for exons peaking down at ZGA and weaker 5′ and 3′ splice sites and differences in GC content for those peaking up (Fig. 5A and file S1). A Random Forest classifier based on multiple genomic and transcriptomic features was able to discriminate human and mouse peak exons from sets of exons with matched pre-ZGA PSIs with high sensitivity and specificity [e.g., Area Under the ROC curve (AUC) = 0.852 and 0.885 for human peak-down and peak-up exons, respectively; Fig. 5B and fig. S19A]. Investigation of the features that contributed the most to this discrimination revealed the length of pre-mRNA and intron number as top-ranking characteristics for peak-down exons, which were located in significantly shorter genes in the three species (Fig. 5B; fig. S19, B and C; and file S1).
Fig. 5. Snrpb and Snrpd2 are activated during ZGA and are associated with peak exons.
(A) Intron-exon features associated with ZGA peak-down or peak-up exons. Blue/red arrows indicate features with statistically significantly higher/lower median values compared to exons with high PSI (two-sided Mann-Whitney U tests). Full comparisons with exons with high/low PSI and control sets with matched pre-ZGA PSIs and value distributions are in file S1. UpEx, Upstream Exon; GC Content, GCC; BP, Branch Point; DoEx, Downstream Exon. (B) Top inset: Receiver Operating Characteristic (ROC) curves for Random Forest classifiers for peak-down exons (H: human, AUC = 0.852; M: mouse, AUC = 0.818; C: cow, AUC = 0.678). Bar plot shows top-ranking features based on mean decrease in accuracy averaged for the three species. Bottom inset: Distribution of pre-mRNA lengths for human peak-down exons and a control set of exons with matched pre-ZGA PSI distributions. ***P < 0.001 based on Wilcoxon rank sum tests. (C) Correlation at the single-cell level between human SNRPB expression and the mean PSI for peak-down exons. (D) Associations between human splicing factor knockdowns (KD) and exons from Mfuzz clusters that peak at ZGA (up and down). Y axis shows percentage of exons affected by the knockdown (|ΔPSI| ≥ 15). X axis shows concordance of the direction of change in the knockdown respect to the change at ZGA [in Cartesian axes as per (E), those in quadrants 1 and 3 (Q1 + Q3)]. (E) Scatter plot showing ΔPSI of ZGA peak exons upon SNRPB knockdown in U251 cells (y axis) versus ΔPSI at ZGA (8C-4C; x axis). Exons with |ΔPSI| ≥15 upon knockdown are green/red; gray, exons with no change. Percentages are given respect to exons with increased or decreased PSI at ZGA. P value: two-sided binomial test between Q1 + Q3 versus Q2 + Q4. U251 data from (72). (F) Expression of SNRPB and SNRPD2 in human early development. Mouse and cow plots in fig. S20B.
We next investigated which splicing regulators may be responsible for the specific temporal dynamics of peak exons at ZGA. For this purpose, we took several complementary approaches (see Materials and Methods for details). First, for each species, we looked for enrichment of known binding motifs for RNA binding proteins (RBPs) (43) in the exonic and neighboring intronic regions for exons from Mfuzz clusters peaking up or down at ZGA. Although a few significantly enriched motifs were found for individual species, the associated RBPs did not show changes in GE at ZGA and/or the enrichments were not evolutionarily conserved (table S5). Second, we correlated average PSIs of peak-down or peak-up exons with GE levels of known splicing regulators at the ZGA at the single-cell level (table S6). This identified multiple significant correlations at each transition, including several AS factors (e.g., SRSF2 and TIAL1) and core spliceosomal components (e.g., SNRPB; Fig. 5C). Third, because these correlations may be indirect, we collected RNA-seq data from 119 available experimental perturbations for 84 unique splicing regulators in different human cell or tissue types (table S1). For each regulator and experiment, we calculated the average change in PSI (or ΔPSI) between knockdown and control for all alternative exons and overlapped those with significant changes with exons peaking at ZGA (table S7). Notably, exons changing upon SNRPB knockdown showed the strongest association with ZGA peak exons in two independent available experiments (Fig. 5D). Noticeably, most overlapping exons corresponded to peak-down exons that are skipped upon SNRPB depletion (Fig. 5E, lower left quadrant). Moreover, a similar pattern was observed upon knockdown of Snrpb in mouse embryonic stem cells (fig. S20A).
Peak exon behavior depends on SNRPB and SNRPD2 developmental dynamics
Given this strong association, we decided to investigate the potential role of SNRPB during ZGA in more detail. SNRPB, also known as SmB, is part of the Sm heptameric ring, which is required for the biogenesis of the U1, U2, U4/U6, and U5 small nuclear ribonucleoprotein molecules on the pre-mRNA (44), and its knockdown had been previously shown to result mainly in skipping of alternative exons (45). SNRPB was very lowly expressed at pre-ZGA stages and showed a sharp increase in expression at the ZGA stage in human, mouse, and cow (Fig. 5F and fig. S20B). In addition, another gene encoding a subunit of the Sm ring, SNRPD2, had similar activation patterns at ZGA in the three species (Fig. 5F and fig. S20B), and its knockdown in mouse embryonic stem cells similarly resulted in widespread down-regulation of peak-down exons (fig. S20C). Misregulated exons that were predicted to cause ORF disruption upon either Snrpb or Snrpd2 knockdown in embryonic stem cells were enriched in genes related to DDR and DNA repair (fig. S20, D and E).
Considering all these data together, we hypothesized the following scenario (Fig. 6A, left). Genes with peak-down SNRPB/D2-sensitive exons become actively transcribed at ZGA when the maternal and zygotic levels of the two Sm proteins are low. This results in the production of transcripts that skip these exons, leading to a net decrease in their inclusion levels compared to pre-ZGA stages. As development proceeds, the burst of de novo transcription of SNRPB/D2 genes at ZGA eventually leads to a subsequent increase in Sm protein levels, which is naturally delayed with respect to the increase in mRNA levels. This protein increase, in turn, allows the correct inclusion of peak exons in nascent transcripts at post-ZGA stages, thereby eventually restoring pre-ZGA high inclusion levels. In this way, the dynamic production and turnover of transcripts from genes containing SNRPB/D2-sensitive exons, together with the expression dynamics of the SNRPB/D2 genes themselves, is predicted to result in temporary peak-like patterns of exon skipping around ZGA stages.
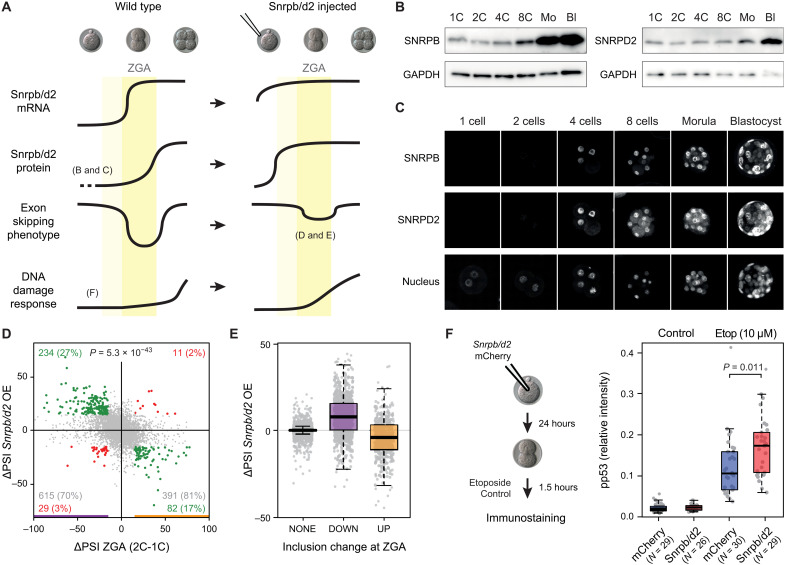
Fig. 6. Snrpb and Snrpd2 are partly responsible for peak-down exons affecting DDR at ZGA.
(A) Model for peak-down exon regulation during ZGA. Left (Wild type): Snrpb and Snrpd2 have low maternal mRNA and protein levels. At ZGA, mRNA levels increase, but the rise in protein levels is delayed, leading to exclusion of sensitive exons in newly transcribed genes. This situation starts being restored after ZGA, when SNRPB and SNRPD2 protein levels reach sufficient amounts. Aberrant exclusion of exons in DDR genes contributes to a delay in DDR at ZGA. Right: Induced expression of Snrpb/d2 before ZGA should prevent the abovementioned exon skipping in DDR genes and therefore enable an earlier stronger DDR. (B and C) Western blot and immunostaining showing SNRPB and SNRPBD2 levels during mouse development. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (D) Changes in exon PSIs at 2C stage upon early induced expression of Snrpb/d2 (y axis) respect to changes at ZGA (2C-1C; x axis). Exons with |ΔPSI| ≥ 15 or |ΔPSI| < 15 upon Snrpb/d2 expression and at ZGA are highlighted in green/red or gray. Percentages among ZGA-changing exons are indicated. P value: two-sided binomial test (Q2 + Q4 versus Q1 + Q3). OE, overexpression. (E) ΔPSIs upon Snrpb/d2-induced expression for all exons with increased (UP), decreased (DOWN), or no change (NONE) in PSI at ZGA. (F) Pronuclear stage embryos were injected with Snrpb/d2 or mCherry mRNA and left to develop to the 2C stage, when they were treated for 1.5 hours with 10 μM etoposide or left untreated. Box plots show nuclear phospho-p53 (Ser15) relative intensity. Each dot represents the average relative intensity of both cells of a single embryo. Data from three independent experiments (individual experiments in fig. S23). P value calculated from the differences between least squares means based on a generalized linear mixed model that incorporates the batch as a random effect.
To assess this hypothesis, we tested the major predictions made by this model. First, we checked whether or not Snrpb and Snrpd2 were depleted at the protein level at pre-ZGA stages. Western blot and immunostaining at different stages confirmed very low levels of both proteins until the 4C stage, with a sharp increase by the 8C/morula stage (Fig. 6, B and C). Although the precise regulation of the timing of SNRPB and SNRPD2 protein production remains to be elucidated, this mRNA-versus-protein lag is consistent with the overall poor temporal correlation between transcriptome and proteome previously observed for the early mouse embryo (46). Next, to investigate how such low levels of these two Sm ring subunits in particular were achieved during oogenesis, we looked at the expression of all the subunits in the transition from Germinal Vesicle (GV) to Metaphase II (MII) stage in five independent RNA-seq experiments. Whereas all other Sm ring subunits maintained similar levels in GV and MII oocytes, Snrpb and Snrpd2 showed a sharp decrease (fig. S21A). This result was confirmed through quantitative PCR (qPCR) assays on independent RNA samples (fig. S21A). During the GV-to-MII transition, the oocyte is transcriptionally inactive; therefore, these results suggest that Snrpb and Snrpd2 mRNAs are differentially degraded during the last steps of oocyte maturation. This is not likely to be due to general differences in mRNA stability of the different Sm ring subunits, which have similar half-lives in somatic cells (fig. S21B; as it is also the case for their protein half-lives, fig. S21C).
Next, we evaluated the effect that increasing SNRPB and SNRPD2 levels before ZGA had on exons changing at this particular stage (Fig. 6A, right). For this, we injected in vitro–transcribed mRNA from either Snrpb and Snrpd2 together (Snrpb/d2) or mCherry, as a control, into pronuclear stage embryos and sequenced polyadenylated [poly (A)+] mRNA from zygotes (5 hours after mRNA injection) and 2C stage embryos (24 hours after mRNA injection). We identified 878 and 484 alternative exons decreasing and increasing their inclusion levels (|ΔPSI| > = 15), respectively, between our zygote and 2C stage embryos, which largely recapitulated the inclusion patterns of peak-up and peak-down exons (fig. S22A). Of these, 234 of 878 (26.7%) exons with decreased PSI were less skipped after Snrpb/d2 overexpression compared to the control condition (in contrast to only 29 with further skipping). Moreover, 82 of 484 (16.9%) exons that increased their inclusion from 1C to 2C decrease their PSI in Snrpb/d2-injected embryos (compared to 11 in the opposite direction) (Fig. 6, D and E). The negative association between the changes at ZGA and upon Snrpb/d2 overexpression is highly significant (P = 1.1 × 10−30, binomial test), suggesting that higher Snrpb/d2 levels before ZGA maintain alternative exon patterns in a more pre-ZGA state, a trend that was also observed for all exons together (fig. S22B). A similar, but milder, reversion of ZGA AS patterns was observed upon early expression of Snrpb or Snrpd2 alone (fig. S22, C and D). Moreover, this pattern was also observed for intron retention (fig. S22E) but, importantly, not for GE (fig. S22F), for which very few changes were identified upon Snrpb/d2 overexpression [29 of 5048 (0.58%) of ZGA changing genes], indicating that the reversion was specific to AS.
Earlier Snrpb/d2 expression leads to increased DDR in 2C embryos
Our results show that peak-like exon skipping at ZGA can be, in part, prevented by combined overexpression of Snrpb and Snrpd2 (Fig. 6, D and E). Given that these exons are predicted to substantially affect the function of proteins involved in DDR (Fig. 2C), we next evaluated the effect of Snrpb/d2 overexpression on the ability of the 2C embryo to respond to DNA damage induced by etoposide. For this, we injected mRNA from Snrpb/d2 or mCherry into pronuclear stage embryos. Injected embryos were left to develop in culture up to the 2C stage and then treated for 1.5 hours with a high dose of etoposide (10 μM), which ensures induction of DDR even at this early stage (fig. S16F). Activation of DDR was evaluated by phospho-p53 (Ser15) immunostaining. Notably, while Snrpb/d2 overexpression did not induce a significant change on phospho-p53 levels on basal conditions, these levels were significantly higher after etoposide treatment in Snrpb/d2-injected embryos (Fig. 6F and fig. S23, A and B). This suggests that the temporal skipping of exons sensitive to Snrpb/d2, at least in part, attenuates p53-mediated DDR occurring during ZGA, potentially contributing to the low response observed at this stage.
DISCUSSION
We have combined multiple datasets and applied a strict quality control to generate a comprehensive and highly validated atlas of AS events during preimplantation development in three mammalian species. Previous studies revealed that AS is very dynamic during mouse preimplantation development, with most changes in isoform usage occurring at the ZGA (12, 22–24). Our transcriptomic analysis confirmed this observation not only for mouse but also for human and cow, despite the marked differences in the relative timing of ZGA in the three species (7, 15). Notably, blastomeres undergoing ZGA in the three species showed the highest levels of isoform diversity generated by exon skipping so far reported for any other tissue, cell type, or developmental stage. This is particularly unexpected given the low morphological complexity of these cells, especially when compared to complex organs as intricate as the brain, which had the previously highest levels of AS (20, 47). Intriguingly, this exceptional exon skipping complexity was observed during only one or two stages, lasting 24 to 48 hours in development, and was due to hundreds of alternative exons whose inclusion levels displayed a sharp peak-like temporal behavior and were not significantly associated with changes in the expression of the host genes. There are at least two potential nonmutually exclusive explanations for these patterns. First, the default hypothesis is that the high exon skipping levels are trivially due to splicing noise associated with novel transcription at ZGA. In this scenario, the relatively simple blastomeres could tolerate better than other cell types such temporary transcriptomic noise. However, peak exons share some remarkable patterns in the three studied species: They have an equivalent molecular impact (specific ORF disruption at ZGA), are in genes enriched for similar gene functions (DDR), and are likely regulated by the same factors (Sm ring components). Therefore, a second possibility is that at least part of these temporary AS patterns are the result of an evolutionarily conserved and developmentally programmed splicing failure specific to ZGA, which contributes to the attenuation of ATM-p53–mediated DDR during this stage. A recent study has shown that inducing a splicing failure in mouse embryonic stem cells, either chemically or by knocking down specific spliceosomal components (including Snrpb and Snrpd2), induces a sharp reprogramming toward totipotent cells. These cells have a bona fide 2C molecular profile (48), including an enrichment for DDR functions among genes with exons causing ORF disruption upon Snrpb or Snrpd2 depletion (fig. S20, D and E). These highly complementary findings suggest that the importance of the splicing failure we report here at ZGA may be even more widespread. Therefore, given the potential biological interest of the possibility that this splicing failure is developmentally programmed, and although we acknowledge that further work will be needed to prove this alternative hypothesis, we discuss it in detail below from the mechanistic and physiological viewpoints.
Mechanistically, the conserved and unique developmental dynamics of two Sm ring components would be, at least in part, behind the programmed splicing failure. Unlike most adult and embryonic cells, pre-ZGA blastomeres inherit remarkably low mRNA and protein levels of SNRPB and SNRPD2, leading to skipping of sensitive exons in genes that are transcribed at ZGA. Because the SNRPB/D2 genes themselves are strongly transcribed during the major ZGA wave, their mRNA levels increase quickly, which is followed by a gradual rise in protein levels that prompts high exon inclusion in the subsequent stages, restoring the canonical splicing patterns. Consistent with this model, zygotic injection of Snrpb/d2 mRNA led to a partial “rescue” of the splicing failure observed at ZGA. Only a subset of all exons is sensitive to low SNRPB/D2 levels, which is enriched among ZGA peak exons. These exons showed common genomic features and could be accurately discriminated from exons with matched pre-ZGA inclusion levels by a Random Forest model.
As a programmed splicing failure, the pattern that we see is reminiscent of the widespread intron detention reported during mouse spermatogenesis (49). In that case, an excess of transcription was proposed to overload the available spliceosomal machinery, leading to reduced splicing efficiency of a subset of introns with weak splice sites, which are then properly spliced and translated at later stages. Moreover, mouse embryos before ZGA have been shown to have highly inefficient pre-mRNA processing through additional mechanisms to those described here to avoid precocious spurious gene expression (50). Therefore, despite their different molecular mechanisms, associated targets, and physiological consequences, these processes suggest that developmentally programmed specific splicing failures may be exploited by multicellular organisms as a strategy to regulate their development and physiology more often than previously anticipated.
Although DDR and DNA repair pathways do operate in very early mammalian embryos (31–33), the activity of some of them appears to be reduced during cleavage stages. DDR to etoposide (Fig. 3) and to irradiation (36, 37) increases during mouse preimplantation development, and high genome instability is observed and tolerated in embryos from human and farmed animals (51–53). The mechanisms behind this change in sensitivity to DNA damage during very early development are mostly unknown. The enrichment for DDR and DNA repair functions among genes with peak exons at ZGA in the three studied species might provide insights to help in understanding the special regulation of these pathways during early mammalian embryogenesis. Here, we have shown that mouse embryos treated with low doses of etoposide before or during ZGA do not substantially activate a p53-mediated response and that embryo arrest is delayed up until early morula stage. This points to the ATM pathway not being fully active during early cleavage stages, in line with previous observations (54, 55). Partly reverting the peak-like exon skipping pattern at ZGA by Snrpb/d2 injection led to a mild, but significant, increase in phospho-p53 levels in response to etoposide at this stage (Fig. 6F). However, the levels of γ-H2AX (Histone H2A family member X), a mark common for the activation of different DDR pathways, increased to a similar extent upon etoposide treatment in control and Snrpb/d2-injected embryos (fig. S23, C and D). It is known that H2AX phosphorylation can happen independently of ATM (54, 56); therefore, these results suggest that Snrpb/d2-dependent exon skipping occurring at ZGA has a significant impact on DDR occurring through the ATM-p53 pathway, although other pathways are likely not affected by this type of regulation. Indeed, Atm itself has an exon that gets temporarily skipped at ZGA and whose exclusion is predicted to disrupt the ORF (fig. S13). Although the precise mechanisms responsible for the reduced DDR before ZGA remain unknown, our results show a role for AS in maintaining such low levels during ZGA, therefore delaying a full response to DNA damage up until morula stage.
Intriguingly, a delay in the full activation of ATM-p53–mediated DDR would seemingly make early preimplantation embryos more sensitive to DNA damage, as it implies the accumulation of DNA lesions that will not be resolved until morula stage. In line with our observations (Fig. 4), DNA damage produced by double-strand breaks at these early stages has been associated to lower embryo survival than when induced at blastocyst stage (57). Is this beneficial for embryo development or simply a quasi-neutral molecular context that early embryos can tolerate? Although there could be many nonmutually exclusive explanations for the former, we could envision a scenario in which severe DNA damage occurring during the first cell divisions is not resolved and instead gets amplified to provoke embryo arrest and death before implantation, lowering the tolerance for DNA lesions at a time when any mutation will be transmitted to all embryo lineages. However, not all DDR pathways can be lower at this developmental point, as genome stability must be ensured during ZGA, a particularly disruptive stage at the molecular level, with global transcription occurring in a largely epigenetically naive and distinct context (58), and with transcription itself producing DNA breaks (59). In particular, because ATR responds to replication stress and is essential for coordinating replication and transcription, it should be fully implemented in early embryos to be able to cope with the activation of the genome. Consistently, we have seen that mouse embryos are particularly sensitive to the replication stress agent aphidicolin during ZGA (fig. S18), which goes in line with previous observations (60). Moreover, ATR, but not ATM, was recently shown to be required for the conversion of mouse embryonic stem cells into 2C-like cells, which are characterized by the activation of various ZGA markers (41). Thus, hampering DDR through ATM but not ATR pathway could allow lowering the tolerance for DNA damage while ensuring genome stability during mouse ZGA. Although the early tolerance for DNA damage and the delay in the apoptotic response seem to be commonalities to different mammalian species, it is important to note that genome stability is likely to be regulated differently in early mouse and human embryos, as the rate of aneuploidies is very high in human and minimum in mouse (53). It is thus possible that AS affects other aspects of DDR and DNA repair apart from the ATM response, and it remains an open question whether it could contribute to the high genome instability observed in the human embryo.
In summary, our results provide evidence for a specific programmed splicing failure dependent on the levels of Sm proteins that results in extensive protein disruption at the time of ZGA and contributes to the attenuation of the ATM-p53–mediated response to DNA damage early during development. Further research will clarify the full biological relevance of this type of regulation for embryo development.
MATERIALS AND METHODS
RNA-seq datasets
Publicly available Illumina RNA-seq samples from oocyte, zygote, 2C, 4C, 8C, 16C/morula, and blastocyst, were downloaded from National Center for Biotechnology Information Short Read Archive. All samples and associated information are provided in table S1. The selected datasets comprise three, four, and three independent experiments with single cells or bulk embryo samples in human, mouse, and cow, respectively (Fig. 1A). To ensure the selection of high-quality data representative of each stage and with sufficient read coverage to measure AS, we performed the following filtering steps (the reason for exclusion of each sample is detailed in table S1): (i) Only RNA-seq runs of at least 50 nucleotides (nt) were used and (ii) individual cells (“outliers”) were discarded from the analyses if they did not cluster with other samples of the expected stage based on GE clustering profiles (see below; figs. S1 to S3). In particular, mouse 2C and 4C samples from (10) were excluded as they did not cluster with those from (3) likely due to slightly different timings, and cow 8C from (14) were discarded since they did not yet undergo ZGA; (iii) experiments for which pooling replicates together did not provide sufficient reads for each stage were discarded (see below); (iv) samples with strong 3′ bias, as assessed by vast-tools align through mapping to the five 3′-most 500-nt segments of mRNAs longer than 2500 nt (18), were also removed; and (v) other miscellaneous reasons stated in table S1. In total, we selected 135, 183, and 28 individual single-cell or bulk embryo samples in human, mouse, and cow, respectively. Last, to assess relative AS levels (Fig. 1E and fig. S5A) or PSIs in differentiated adult cell and tissue types (Fig. 2, C and D), we compiled another set of samples for the three species from VastDB (http://vastdb.crg.eu/; table S1).
Quantification of AS and GE levels
To calculate the percent of inclusion for a given alternative sequence (either an exon, an intron, or an exon truncation/extension due to alternative 3′ or 5′ splice site choices), we used vast-tools v1 (18). vast-tools relies on a comprehensive database of exon-exon and exon-intron junctions for the identification and quantification of different types of AS events and has been used to quantify inclusion levels in multiple species with high validation rates [e.g., (18, 61–63)]. vast-tools provides a table with percent inclusion levels (using the metrics PSI) for each AS event and sample, as well as a series of quality scores on the reliability of the estimate. In this study, we have used a minimum read coverage of VLOW for all event types [for details, see (18)] and also filtered out intron retention events with a significant read imbalance at the two retention junctions (option --p_IR). For each species, we have used the following VASTDB libraries: human (hg19, hsa.16.02.18), mouse (mm9, mmu.16.02.18), and cow (bosTau6, bta.20.12.19). To measure GE of each gene in each cell or pool, we also used the align module of vast-tools, which provides a normalized count measure for each gene {cRPKM [corrected (for mappability) reads per kilobase of transcript per million mapped reads]; see (64)} and raw read counts for each gene.
De novo exon skipping events
To ensure that preimplantation-specific exons could also be quantified by vast-tools, we conducted a de novo search for alternative cassette exons for each species using our early development RNA-seq data (table S1) and created an additional vast-tools library with the exon-exon junctions from those exons. For this purpose, we mapped these RNA-seq samples to their respective genomes (hg19, mm9, and bosTau6 assemblies) using tophat2 and built gene models through cufflinks (65). The resulting Gene Transfer Format (GTF) files were merged using cuffmerge for each species and processed using SUPPA (66) to extract all identified cassette exons. Custom scripts were then used to detect previously unidentified exons, which corresponded to internal alternative exons that were not present in vast-tools and had at least one upstream (C1) or downstream (C2) exon annotated in Ensembl. Using this approach, we identified 3468 new alternative exons not present in vast-tools v1 for human (508 not annotated in Ensembl v60), 2206 (267 not annotated v62) for mouse, and 727 (180 not annotated v76) for cow (reported in table S2). Next, for each species, we created a library with exon-exon junctions for these exons by joining the upstream and alternative exons (inclusion, C1A), the alternative and the downstream exons (inclusion, AC2), and the upstream and downstream exons (skipping, C1C2), with a minimum of eight mapping positions from each exon for 50-nt reads [for details, see (67)]. This additional library was incorporated into the vast-tools workflow as an additional module.
Determination of cell identities, merging of samples, and PCAs
GE values for individual cells were normalized with DESeq (68) and clustered using heatmap.2 with default parameters. Raw counts obtained from vast-tools align were normalized using size factors and a variance stabilizing transformation (“blind” option) before plotting the top 500 most variable genes as a heatmap with z scores for rows (file S1). As stated above, individual cells that did not fit into their expected stage and may represent dying, damaged, or mislabeled cells were removed from further study (table S1). Next, because single-cell libraries have low molecular complexity and are often sequenced at low depth, resulting in low coverage across exon-exon junctions, we created pools of samples representative of specific stages, aiming at generating pools of samples with a total of >150 million reads, when possible. In all cases, cells from the same embryo were kept in the same pool, and where two embryos needed to be merged, this was based on the hierarchical clustering results. The pooling of samples was performed with the merge module of vast-tools, and the composition of the pools is described in table S1.
PCA was conducted in R using the function princomp on either normalized gene expression raw counts (for GE) or PSIs (for AS). For AS, all exons with sufficient read coverage (VLOW or higher) in >80% of the merged samples and SD ≥5 were considered. GE measurements took all genes with SD ≥5 across merged samples as input.
Estimation of AS complexity
We used a simple measure of the transcriptional complexity generated by AS at each developmental stage or differentiated tissue. For those AS events with sufficient read coverage (VLOW or higher) in at least 50% of all the compared samples, we calculated for each stage or tissue the fraction of AS events with sufficient read coverage in that sample whose PSI was 20 < PSI < 80 (i.e., was predicted to generate two prevalent isoforms). Modifying the range of PSI to define prevalent isoforms, the minimum coverage per event, and the minimum fraction of samples with coverage or using individual single cells/samples (instead of the pooled samples) did not qualitatively change the results (fig. S5, B and C).
Definition of differentially spliced AS events and expressed genes at consecutive stages
Given that the number of replicates per stage and species was relatively low and uneven, we made the following definitions to call differentially spliced AS events at each developmental transition (i.e., two consecutive developmental stages) based on differences in mean PSI between stages (fig. S4B):
1) Both consecutive stages must have at least two samples with sufficient read coverage (VLOW or higher) for human and mouse, or one for cow.
2) In all cases, the overlap between the PSI distributions of the two compared stages had to be ≤10 (i.e., “range diff” ≥ −10).
3a) Depending on the intrastage PSI range (i.e., the difference in PSI between the sample with the highest PSI and the one with the lowest for a given stage), a minimum mean change in PSI (∆PSI) between the two stages was required:
3a.i) If the PSI range in both stages was ≤30, then |∆PSI| ≥20, else
3a.ii) if the PSI range in both stages was ≤50 but >30, then |∆PSI| ≥30, else
3a.iii) if the PSI range in any stage was >50, then |∆PSI| ≥55.
3b) In addition, if the mean PSI of any of the two stages was ≥99.5 or ≤0.5 (i.e., either near complete inclusion or skipping), then |∆PSI| ≥10.
All exons that were differentially spliced at any transition are provided in table S4. For a more direct comparison with these differentially spliced definitions, we called differentially expressed genes between consecutive transitions also using qualitative definitions based on fold changes between mean cRPKM values for each stage (fig. S4C). First, we filtered out genes with low expression in both stages (mean cRPKM <2). Then, for genes with mean expression cRPKM <10 in both stages, we required that one stage had a mean cRPKM <1 and another ≥5 to consider it as differentially expressed. Last, for genes with mean expression cRPKM ≥10 in at least one stage, we imposed an absolute fold change ≥2 for a gene to be considered differentially expressed.
To assess the overlap between differentially spliced AS events and differentially expressed genes per transition (fig. S6, A to C), we categorized each differentially spliced AS event based on whether the host gene was up- or down-regulated at the GE level, not differentially expressed or had too low expression, as defined above. To obtain the expected overlap between both types of transcriptomic change at each transition (triangles in fig. S6, A to C), we calculated the fraction of differentially expressed genes among those hosting AS events that fulfil the minimum coverage criteria in both consecutive stages (see above). Statistical significance of the overlap was calculated through two-sided Fisher’s exact tests using a contingency table for differentially spliced and differentially expressed genes of the total number of genes tested in both analyses.
Clustering AS and GE by temporal dynamics and functional enrichment analysis
We used the soft clustering algorithm Mfuzz (28) to group alternative exons and genes based on their PSI or expression profiles. For exons, we first selected those that had sufficient read coverage (VLOW or higher) in at least one sample for each time point and that were differentially spliced (as described above) between any pair of stages (including nonconsecutive stages). This yielded a total of 1904, 968, and 1083 exons for human, mouse, and cow, respectively. Next, we provided a mean PSI per stage for each valid exon as input for Mfuzz. Default settings were used, and the optimum number of clusters was automatically determined for each species using the Dmin function. Next, we selected those exons that were differentially spliced between any pair of stages and that had sufficient read coverage in all but one or two stages (for human and mouse) or all but one stage (for cow), and imputed the missing values in two ways: (i) If the missing point is at the beginning or end of the time course, then it was assigned a value identical to the second or previous value, respectively; (ii) if the missing point is in between known values, then it was assigned the mean between the two neighboring values. These additional exons (1050, 689, and 649 exons for human, mouse, and cow, respectively) were then assigned to the previously defined Mfuzz clusters for exons with complete coverage using the “Mfuzz: membership” function. It should be noted that exons in the original clusters might be reassigned to other clusters through this process. Mfuzz clusters were also generated for mean GE values in the same way that was described for alternative exons with complete coverage.
Mfuzz profiles were then classified into “Peak,” “Shift,” and “Other” on the basis of the following definitions. First, for each exon or gene, the values across the time course were classified as being in the upper, mid, and lower tercile given segments of size = (Max–Min)/3, producing an array of seven elements, one per stage (e.g., 1113111; six for cow). A profile was classified as Other if it had either: (i) two or more values in the mid tercile, (ii) a change from upper to lower tercile in the first or last transition (or vice versa), in which case, potential peak and shift patterns cannot be discriminated, (iii) the first or last value in the mid tercile; or (iv) a profile gradually changing from high to low PSI (or vice versa), defined as having the first stage in the first tercile and the last stage in the third tercile (or vice versa), and the maximum change in PSI at any consecutive stage divided by the total PSI range < 0.4; as Peak if its first and last values were both in the first (peak-up) or third (peak-down) tercile and it was not defined as Other; and Shift for any other profile. Profiles of each cluster were then manually inspected to ensure the accurateness of these definitions. Last, to calculate the fraction of alternative exons that belonged to each type of cluster (Fig. 2B), only exons with coverage in all time points were considered. All exons that were included in any Mfuzz cluster and their associated features are provided in table S4.
Statistical significance of GO term enrichments was calculated using proportion tests (prop.test in R) given a foreground and a background list of genes. GO annotations for the three species were downloaded from Biomart (Ensembl v91), and GO terms from each species were transferred to the orthologs in the other species to standardize the comparisons and improve cow annotations. In particular, the genes belonging to enriched DDR-related categories (Fig. 2) are provided in table S8. For the GO enrichment analysis of exons in different Mfuzz clusters changing at ZGA (either Peak or Shift), we used as background all multiexonic genes that had at least one AS event (of any type) that changed in any pairwise comparison (a total of 11,096, 9550, and 12,229 genes for human, mouse, and cow, respectively). To test the GO enrichments associated with the exons that cause ORF disruption upon Snrpb or Snrpd2 knockdown, we downloaded the RNA-seq data from (48) and run vast-tools v2.5.1 for mm9. Differentially regulated AS events were identified with vast-tools compare using standard parameters (--min_dPSI 15), and genes with up-/down-regulated exons predicted to cause ORF disruption upon inclusion/skipping, respectively, were selected (1087 and 1197 genes for Snrpb and Snrpd2, respectively). The background for GO analysis was obtained using the option --GO from vast-tools compare and includes all genes with at least one AS event with the required read coverage (10,103 and 9984 genes for Snrpb and Snrpd2, respectively).
Assessment of evolutionary conservation
To compare each set of exons changing at a given transition in one species (species 1) against another (species 2), we performed the following steps to assess the evolutionary conservation at different levels, as previously described (27, 62, 69). First, to find which exons are conserved at the genomic level, we use the liftOver tool (70) with -minMatch=0.10 -multiple -minChainT=200 -minChainQ=200 parameters. Next, for those coordinates lifted to species 2, we extracted the two neighboring dinucleotides. Lifted exons with at least one canonical 5′ or 3′ splice site (GT/C or AG) were considered Genome-conserved. Then, we matched Genome-conserved exons to vast-tools v1 identifiers based on coordinate overlap and selected those that had sufficient read coverage in at least the equivalent transition of species 2 with respect to the ZGA. Exons were considered to have a “PSI change in the same direction” if they displayed a |∆PSI| > 15 in the same direction as those in species 1 in at least one transition. The equivalence between transitions in species 1 and 2 was displayed using alluvial plots for each pair of species (fig. S7, A to F). For species 2, only the transition with the largest ∆PSI in the same direction was selected.
In addition, we performed the following steps to identify a set of orthologous exons that were regulated during preimplantation development of the three studied mammalian species (fig. S7G). First, for each species, we identified differentially spliced exons between any pair of developmental stages (whether consecutive or not) as described above (fig. S4B). From these, we identified 93 orthologous exons that were differentially spliced in the three species. Furthermore, for those identified as differentially spliced in two species, we then asked whether there was an ortholog exon in the third species and, if so, whether it had an average |∆PSI| > 15 between any pair of stages. This resulted in a total of 259 orthologous exons (table S3). To assess the enrichment of GO terms among the genes hosting these exons, we used as background multiexonic genes with 1:1:1 orthologs in the three species and with at least one exon skipping event with coverage in two stages in any species (15,132 genes).
Last, to assess regulatory conservation of genes differentially expressed at each transition in a given species (species 1) against another (species 2) (fig. S8), we first identified one-to-one orthologs based on Ensembl-BioMart information and then checked whether any differentially up- or down-regulated gene in species 1 was also identified as up- or down-regulated in species 2 (“GE change same direction”). The equivalence between transitions was shown using alluvial plots, in which only the transition with the largest fold change in the same direction was selected for species 2.
Predicted impact on ORFs, NMD, and ribosome-engagement analyses
The predicted impact on the ORF of the inclusion/exclusion of each alternatively spliced sequence was obtained from VastDB (release 3), and it was inferred essentially as described in (26). Four major categories are reported: (i) AS events that are predicted to generate alternative protein isoforms both upon inclusion and skipping of the alternative sequence [i.e., are located in the coding sequence (CDS) and maintain the ORF and/or are located toward the end of the CDS and are not predicted to trigger NMD or to create a large protein truncation (>20% of the reference isoform and/or > 300 amino acids)]; (ii) AS events that disrupt the ORF upon sequence inclusion (e.g., most introns, exons that are usually not included and whose length is not multiple of three and/or contain in-frame stop codons); (iii) AS events that disrupt the ORF upon sequence exclusion (e.g., exons that are normally constitutive and whose length is not multiple of three); and (iv) AS events in the 5′ or 3′ untranslated regions. Comparisons among ZGA Mfuzz clusters (Fig. 2, C and D) included only those exons that were labeled as (i) alternative protein isoform, (ii) disruptive upon inclusion, or (iii) disruptive upon exclusion.
To compare the relative engagement of the inclusion and exclusion isoforms on ribosomes for different exon subsets (fig. S14, D, E, G, and H), we used Transcript Isoforms in Polysomes sequencing data for human embryonic stem cells (29) or HEK 293 T cells (30). We used vast-tools to obtain PSI values for each exon in the cytosolic and high-polysome fractions and calculated the ΔPSI between the two fractions, which gave a measure of the difference in ribosomal engagement (positive ΔPSI means higher engagement upon inclusion and the opposite for negative ΔPSI and exclusion). For each exon category, we plotted events with sufficient read coverage (VLOW or higher) in both the cytosolic and high-polysome fractions. For differentially spliced exons at the ZGA transition (fig. S14, D and E), we plotted the ΔPSI with respect to the ZGA isoform grouped by the predicted impact on the ORF at ZGA (i.e., positive/negative ΔPSI values imply that the ZGA isoform is more/less engaged). For each type of temporal dynamics with change at ZGA (fig. S14, G and H), we plotted directly the ΔPSI between cytosolic and high-polysome fractions (i.e., a negative/positive ΔPSI implies that the inclusion/skipping isoform, which is the ZGA isoform for peak-up/peak-down exons, respectively, is less engaged). In addition, to assess the impact of NMD disruption on these sets of exons (fig. S14, C and F), we performed an equivalent analysis calculating the ΔPSI upon UPF1 knockdown in HR1 cells for each category [data from (71)]. In these box plots and others throughout the manuscript, center line represents the median, box limits the upper and lower quartiles, and whiskers show the 1.5× interquartile range.
Last, to study the enrichment or depletion of transposable elements in different Mfuzz exon clusters (fig. S15), we overlapped the coordinates of these elements for each species as defined by RepeatMasker (excluding simple repeats) and of the alternative exons and neighboring intronic regions (500 base pairs upstream and downstream the exon) using bedtools intersect and counted the number of exons with at least 1-nt overlap with any transposable element family. For each Mfuzz cluster, we plotted the relative fraction of exons overlapping transposable elements with respect to the fraction of all exons included in Mfuzz clusters (“ALL”).
Evaluation of intron-exon features of ZGA-Peak exons
We used Matt v1.3.0 (42) to compare exon and intron features associated with splicing regulation between exons with Peak dynamics at ZGA and different reference exon sets. For each group of exons being compared, Matt cmpr_exons automatically extracts and compares 69 genomic features associated with AS regulation, including exon and intron length and GC content, splice site strength, branch point number, strength and distance to the 3′ splice site using different predictions, length and position of the polypyrimidine tract, etc. For the calculation of splice site and branch point strength, we used the available human models. Comparisons among groups are performed using Mann-Whitney U tests and visualized using box plots (file S1). For this analysis, we defined the following six exon sets for each species (table S9):
(i) P_Dw: exons with a peak-down profile at ZGA, sufficient read coverage (VLOW or higher) in the pre-ZGA and ZGA stages, and a ΔPSI ≤−10 at the ZGA transition. Given the broader time of ZGA in cow, the ZGA ΔPSI was defined as the largest ΔPSI from either 4C-8C or 4C-Morula [also in (ii)]. N of exons: human = 526, mouse = 345, and cow = 646.
(ii) P_Up: exons with a peak-up profile at ZGA, sufficient read coverage in the pre-ZGA and ZGA stages, and a ΔPSI ≥10 at the ZGA transition. N of exons: human = 236, mouse = 32, and cow = 152.
(iii) HIGH_PSI: exons with PSI >90 across both preimplantation and differentiated samples with sufficient read coverage. N of exons: human = 11,878, mouse = 6768, and cow = 5108.
(iv) LOW_PSI: exons with PSI <10 across both preimplantation and differentiated samples with sufficient read coverage. N of exons: human = 4077, mouse = 1213, and cow = 955.
In addition, we extracted 33,710, 16,999, and 9228 background exons with sufficient read coverage in the pre-ZGA and ZGA stages, |ΔPSI| < 5 at the ZGA transition, and that are not part of the foreground sets (i) and (ii) for human, mouse, and cow, respectively. Because the pre-ZGA PSI distribution of these background exons differ from those of (i) and (ii), we constructed two stratified background sets. For this purpose, for each exon of each foreground set (i and ii), we randomly selected one background exon whose pre-ZGA PSI deviates from the pre-ZGA PSI of the foreground exon by no more than 5. Selected background exons were then excluded from the pool of background exons, and the process was repeated a total of four times. The stratified background exon sets after the four iterations [(v) Bg_Dwstrat and (vi) Bg_Upstrat] contained 2104/1224/2584 and 944/128/604 for sets (i) and (ii) for human/mouse/cow and matched at least 89% of the foreground exons.
Classification and feature analysis with Random Forest
To consolidate and extend results obtained by Matt’s discriminative feature analysis, we applied a Random Forest model to the classification of peak-down exons versus a matched background and peak-up versus a matched background for each of the three species and extracted the variable importance with the goal to identify features most relevant for these classifications. First, we constructed a comprehensive set of 746 features for the exons as follows: (i) All 69 intron-exon–related genomic features extracted with function get_efeatures of splicing toolkit Matt v1.3.0, as mentioned above; (ii) GE fold change at ZGA (ZGA/pre-ZGA) of the gene the exon belongs to; and (iii) for each 338 regular-expression RNA binding motifs from CisBP-RNA v0.6, we scanned the 200-nt upstream and 200-nt downstream (150 nt intronic + 50 exonic) sequences for each exon for motif hits and added the number of hits as individual features.
We then used the R package randomForest v4.6-12 to train models for each foreground exon set. Because the foreground and background sets differ in size by at least one order of magnitude, we downsampled the background exon set anew for each Random Forest and trained multiple models in an iterative manner. In each iteration, we randomly chose for each foreground exon one background exon with |ΔPSI| <5 at ZGA, ensuring that the characteristics of the foreground and background exons have similar weight when training Random Forests. We held fixed the Random Forest parameters ntry = 25 and nodesize = 1, whose recommended values for classification tasks are sqrt(N features) and 1. We optimized the number of trees (ntree) by iterating it from 100 to 3100 by 300. For each ntree value, we trained 100 Random Forest and determined average classification accuracies measures by comparing the out of bag (OOB) classification votes to the ground truth. We chose the ntree value that gave overall best average OOB error, area under Receiver Operating Characteristic (ROC) and precision recall curves, and average sensitivities for specificities 90% and 95%. At the same time, we chose ntree as small as possible to guard against overfitting effects. Last, we trained 1000 Random Forests with the optimal ntree value and with different downsampled stratified background sets in each run and reported average ROC and precision recall curves, as well rankings of the features according to their average feature importance (i.e., mean decrease of accuracy as reported by the Random Forest model).
Identification of potential regulators of exons with peak dynamics
To identify potential regulators of exons with peak-up and peak-down Mfuzz dynamics at ZGA in the three species, we took three main approaches:
Sequence motif enrichment analyses
From the foreground sets (i, P_Dw) and (ii, P_Up) described above, we extracted 300 nt of the upstream and of the downstream intronic regions as well as the exon sequences and used Matt v1.3 (42) to test enrichment of all available motifs for RBPs in CisBP-RNA (43) in each of three regions separately. The background set corresponded to those exons that have sufficient read coverage in all or all but one stage and that were not part of P_Dw or P_Up sets (human: 34,452 exons, mouse: 20,232 exons, and cow: 11,177 exons).
Correlation between RBP expression and exon PSIs at the single-cell level
For each foreground exon set in human and in mouse, we correlated (Pearson) at the single-cell level the GE values of 196 manually curated splicing factors (table S6) with the mean PSIs of those peak-down or peak-up exons (i and ii sets above) with sufficient read coverage in each blastomere of the ZGA stage (8C in human and 2C in mouse). This was performed using corr.test from the psych package in R.
Overlap with specific splicing factor–dependent exons
To identify peak exons whose inclusion levels were dependent on specific splicing factors, we first collected publicly available RNA-seq data from specific splicing factor depletion experiments (knockdown or knockout) in any cell or tissue type (table S1). In total, we compiled data for 119 available experimental perturbations for 84 splicing factors comprising 64 independent studies. These data were processed using vast-tools to obtain ΔPSIs for each experiment and splicing factor for all exons with sufficient read coverage in control and experimental condition. Next, for each splicing factor and experiment, we scored the number of ZGA peak exons (i and ii sets above) that show a |ΔPSI| ≥15 upon splicing factor depletion. To identify the most promising splicing factors (Fig. 5D), we calculated: (i) the percentage of exons with sufficient read coverage affected by the depletion and (ii) the consistency of these changes (e.g., down-regulation of peak-down and up-regulation of peak-up), as estimated by the percentage of exons in quadrants Q1 and Q3 and evaluated using a two-sided binomial test (table S7).
Embryo collection and manipulation
All protocols were carried out in accordance to the European Community Council Directive 2010/63/EU and approved by the local Ethics Committee for Animal Experiments [Comitè Ètic d’Experimentació Animal–Parc de Recerca Biomèdica de Barcelona (CEEA-PRBB), CEEA number 9086]. All embryos were obtained from B6CBA F1 crosses. Zygotes were collected from the oviduct of superovulated females 20 hours after human chorionic gonadotropin (HCG) injection and cultured in EmbryoMax KSOM Mouse Embryo Media (Millipore) at 37°C, 5% CO2 up to 2C, 4C, 8C, morula, or blastocyst stages. Two-cell embryos were collected 46 to 48 hours after HCG injection to ensure that ZGA had taken place. The 4C, 8C, morula, and blastocyst stage embryos were collected 65, 72, 90, and 100 hours after HCG injection, respectively. For the experiments in Figs. 3 and 4 (G to L) and figs. S16, S17 (B to D), and S18B, early morulas (8 to 16 cells) were collected at 2.5 dpc (72 hours after HCG injection) by flushing of the oviduct in M2 media (Sigma-Aldrich). For GV isolation, 4- to 6-week-old females were injected with Pregnant Mare Serum Gonadotropin (PMSG) (5 IU), ovaries were dissected 48 hours after injection, and gently punctured with a 27G needle in L-15 Leibovitz media (Sigma-Aldrich) containing 10% fetal bovine serum (Thermo Fisher Scientific). Cumulus cells were removed from GV oocytes by pipetting. MII oocytes were collected from the oviducts of 4-week-old superovulated females 16 hours after HCG administration. Collected oocytes were directly placed into RNA extraction buffer from the RNeasy Micro Kit (Qiagen) that was further used for RNA isolation.
For the overexpression experiments (Fig. 6, D to F), Snrpb and Snrpd2 complementary DNA (cDNA) were cloned into a modified pCS2 + 8NmCherry vector lacking mCherry tag (Addgene) for their in vitro transcription. The mCherry used as control was transcribed from the pCS2 + 8NmCherry vector (Addgene). In vitro transcription was performed using the mMESSAGE mMACHINE SP6 Transcription Kit (Ambion) according to the manufacturer’s instructions. For all overexpression experiments, one-cell embryos at pronuclear stage were microinjected with mCherry mRNA (control, 300 ng/μl) or Snrpb mRNA (150 ng/μl) and Snrpd2 mRNA (150 ng/μl) (Snrpb/d2) following standard pronuclear injection procedures, where microinjection is assessed by around 50% pronulear swelling (=2pl).
Etoposide and aphidicolin treatment
For experiments in Figs. 3 and 4, embryos were either left untreated or treated for 1 hour with etoposide (Sigma-Aldrich) at the stated stage and concentration. Following treatment, embryos were either fixed directly for immunostaining or washed in Potassium Simplex Optimized Medium (KSOM) media and kept in culture to be fixed at the desired stage. For Snrpb and Snrpd2 overexpression experiments (Fig. 6F), injected one-cell embryos were left in culture and either treated with 10 μM etoposide for 1.5 hours or left untreated. Following the treatment, embryos were fixed for immunostaining. For experiments in fig. S18, 2C stage (50 hours after HCG injection) and early morula stage (72 hours after HCG injection) embryos were cultured in the presence of aphidicolin (0.25 μg/ml; Sigma-Aldrich) for 16 hours and then washed in KSOM media and kept in culture for further 8 hours, when developmental progress was assessed.
RT-PCR validation assays qPCR and RNA-seq experiments
RT-PCR assays for alternative exon validations were performed on pools of embryos at different developmental stages. The Arcturus Pico Pure RNA extraction kit (Thermo Fisher Scientific) was used for RNA extraction, and cDNA was transcribed with Superscript III Reverse Transcriptase (Thermo Fisher Scientific). RT-PCR primers can be found in table S10. PSI quantification from RT-PCR was performed using the Fiji software.
RNA extraction from GV and MII oocytes was performed using the RNeasy Micro Kit (Qiagen), and Superscript III Reverse Transcriptase (Thermo Fisher Scientific) was used for cDNA synthesis. qPCR was performed with SYBR Green Master Mix (Thermo Fisher Scientific). qPCR primers are listed in table S10.
For RNA-seq experiments following Snrpb and Snrpd2 overexpression, one-cell embryos were microinjected as described above and collected for RNA extraction at either 5 hours after injection (zygote stage) or 24 hours after injection (2C stage, 48 hours after HCG injection). For each condition, 40 embryos coming from three independent experiments were pooled to extract RNA for sequencing. RNA was extracted using the Qiagen RNeasy Micro Kit. SMARTer Stranded RNA-Seq Kit was used for library preparation before Illumina sequencing. Libraries were sequenced in a HiSeq 2500 machine, generating an average of ~69 million 125-nt paired-end reads per sample. Read numbers and mapping statistics are provided in table S11.
Snrpb and Snrpd2 overexpression RNA-seq analysis
RNA-seq data for control or Snrpb and/or Snrpd2 overexpressing embryos at 1C or 2C stages was processed using vast-tools. For both AS and GE analyses, control 1C and 2C embryos were compared to identify differentially spliced exons or expressed genes at ZGA, and 2C embryos overexpressing mRNA from Snrpb, Snrpd2, or both genes were compared with the control condition. For AS analyses (Fig. 6, C and D, and fig. S22, C to E), only exons or introns with sufficient read coverage in 1C and 2C controls as well as the tested experimental condition were used, and a cutoff of |ΔPSI| ≥15 was used. For GE analyses, cRPKM values were normalized using quantile normalization in R, and genes with fewer than 50 read counts or expression levels lower than cRPKM <5 in the three conditions (1C and 2C controls as well as the tested experimental condition) were discarded. To calculate log2 fold changes, 0.01 was added to each normalized cRPKM value, and a minimum fold change of |FC| ≥ 2 was set as a cutoff. Full PSI and normalized cRPKM values are provided as tables S12 and S13, respectively.
Embryo immunostaining, TUNEL staining, and Western blot
Embryos were fixed in 4% paraformaldehyde (PFA)/phosphate-buffered saline (PBS) for 10 min. Following fixation, they were permeabilized in 0.5% Triton X-100 for 15 min, blocked in 10% bovine serum albumin (BSA)/0.1% Triton X-100/PBS, and incubated overnight in primary antibody: anti-SNRPB (Thermo Fisher Scientific), anti-SNRPD2 (Thermo Fisher Scientific), anti–phospho-H2AX Ser139 (Cell Signaling Technology), anti–phospho-p53 Ser15 (Cell Signaling Technology), anti–phospho-ATM Ser1981 (Cell Signaling Technology), and anti-CDX2 clone EPR2764Y (Abcam). Hoechst was used for nuclear staining and CytoPainter Phalloidin-iFluor 647 Reagent (Abcam) for membrane staining. Imaging was conducted in a Leica SP8 inverted confocal microscope and images processed with Fiji software. For quantification, relative intensity represents the mean fluorescent intensity of the nucleus relative to nucleus area along the biggest nuclear section, measured with Fiji.
The Apop-Tag Fluorescein Kit (Millipore) was used for TUNEL staining according to the manufacturer’s instruction with minor modifications. Briefly, embryos were fixed in 4% PFA for 10 min at room temperature, washed in 0.1% Triton X-100/PBS, and permeabilized for 15 min in 0.4% Triton X-100/PBS. Following washes in PBS, embryos were equilibrated and stained according to the Apop-Tag kit’s protocol. In the case of CDX2 immunostaining, this was performed immediately after TUNEL staining starting from the blocking. Hoechst and Cytopainter Phalloidin-iFluor 647 Reagent (Abcam) were used for counterstaining following TUNEL staining. The quantification of TUNEL staining intensity shown in Fig. 4J and fig. S17D was performed for each blastocyst on the z-stack maximum projection image generated using the maximum intensity. Image quantification was performed with Fiji.
For Western blot, embryos at different developmental stages were collected as described above. For protein extraction, pools of embryos at each stage were collected in Laemmli Buffer. Pools of 150 or 60 embryos per stage were used for the SNRPB and SNRPD2 Western blots, respectively. Proteins were separated by 14% SDS–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Bio-Rad). Following blocking in 5% milk (Sigma-Aldrich) in Tris-Buffered Saline buffer supplemented with 0.1% Tween-20 (TBST), membranes were incubated overnight with the primary antibody in 5% BSA (Sigma-Aldrich) in TBST. The following antibodies and dilutions were used: anti-SNRPB (1:200; Thermo Fisher Scientific), anti SNRPD2 (1:2000; Abcam), and anti–glyceraldehyde-3-phosphate dehydrogenase (1:10,000; Abcam).
Acknowledgments
We thank J. Ule and R. Faraway for feedback on the analyses, J. Valcárcel for critical comments on the manuscript, and F. Mantica and L. P. Iñiguez for assistance on R plotting and statistical testing. We acknowledge the support of the CERCA Programme/Generalitat de Catalunya and of the Spanish Ministry of Economy, Industry and Competitiveness (MEIC) to the EMBL partnership.
Funding: This work was funded by the Spanish Ministerio de Ciencia grants BFU2014-55076-P, BFU2017-89201-P and PID2020-115040GB-I00 (M.I.), Marie Skłodowska-Curie actions grant H2020-MSCA-IF-2014_ST-656843 (B.P.), La Caixa PhD fellowship (C.D.R.W.), and “Centro de Excelencia Severo Ochoa 2013-2017” SEV-2012-0208 (CRG-MI).
Author contributions: Conceptualization: C.D.R.W., B.P., and M.I. Methodology: C.D.R.W., B.P., M.I., A.G., Q.R., S.B., M.C.S., and E.B. Investigation: C.D.R.W., B.P., M.I., A.G., M.M.-C., L.G., Q.R., M.C.S., E.B., and O.B. Visualization: C.D.R.W., B.P., M.I., and A.G. Supervision: B.P. and M.I. Writing—original draft: C.D.R.W., B.P., and M.I. Writing—review and editing: C.D.R.W., B.P., and M.I., with input from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq data generated for this study have been submitted to Gene Expression Omnibus (accession code: GSE163205). Public RNA-seq data are reported in table S1. Gene expression and AS profiles from these RNA-seq data are available in vastdb.crg.eu. All software used to analyze the data are publicly available and indicated in Materials and Methods.
Supplementary Materials
This PDF file includes:
Figs. S1 to S23
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S13
Supplementary file S1
REFERENCES AND NOTES
- 1.Palmer N., Kaldis P., Regulation of the embryonic cell cycle during mammalian preimplantation development. Curr. Top. Dev. Biol. 120, 1–53 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Kermi C., Aze A., Maiorano D., Preserving genome integrity during the early embryonic dna replication cycles. Genes (Basel) 10, 398 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng Q., Ramskold D., Reinius B., Sandberg R., Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343, 193–196 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Hamatani T., Carter M. G., Sharov A. A., Ko M. S. H., Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell 6, 117–131 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Xie D., Chen C.-C., Ptaszek L. M., Xiao S., Cao X., Fang F., Ng H. H., Lewin H. A., Cowan C., Zhong S., Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res. 20, 804–815 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kues W. A., Sudheer S., Herrmann D., Carnwath J. W., Havlicek V., Besenfelder U., Lehrach H., Adjaye J., Niemann H., Genome-wide expression profiling reveals distinct clusters of transcriptional regulation during bovine preimplantation development in vivo. Proc. Natl. Acad. Sci. U.S.A. 105, 19768–19773 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xue Z., Huang K., Cai C., Cai L., Jiang C.-y., Feng Y., Liu Z., Zeng Q., Cheng L., Sun Y. E., Liu J.-y., Horvath S., Fan G., Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500, 593–597 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J., Huang J., Li M., Wu X., Wen L., Lao K., Li R., Qiao J., Tang F., Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1131–1139 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Petropoulos S., Edsgärd D., Reinius B., Deng Q., Panula S. P., Codeluppi S., Reyes A. P., Linnarsson S., Sandberg R., Lanner F., Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 165, 1012–1026 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biase F., Cao X., Zhong S., Cell fate inclination within 2-cell and 4-cell mouse embryos revealed by single-cell RNA sequencing. Genome Res. 24, 1787–1796 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boroviak T., Loos R., Lombard P., Okahara J., Behr R., Sasaki E., Nichols J., Smith A., Bertone P., Lineage-specific profiling delineates the emergence and progression of naive pluripotency in mammalian embryogenesis. Dev. Cell 35, 366–382 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiao Y., Ren C., Huang S., Yuan J., Liu X., Fan J., Lin J., Wu S., Chen Q., Bo X., Li X., Huang X., Liu Z., Shu W., High-resolution annotation of the mouse preimplantation embryo transcriptome using long-read sequencing. Nat. Commun. 11, 2653 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan X., Zhang X., Wu X., Guo H., Hu Y., Tang F., Huang Y., Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 16, 148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeo S., Kawahara-Miki R., Goto H., Cao F., Kimura K., Monji Y., Kuwayama T., Iwata H., Age-associated changes in gene expression and developmental competence of bovine oocytes, and a possible countermeasure against age-associated events. Mol. Reprod. Dev. 80, 508–521 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Graf A., Krebs S., Zakhartchenko V., Schwalb B., Blum H., Wolf E., Fine mapping of genome activation in bovine embryos by RNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 111, 4139–4144 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mamo S., Mehta J. P., McGettigan P., Fair T., Spencer T. E., Bazer F. W., Lonergan P., RNA sequencing reveals novel gene clusters in bovine conceptuses associated with maternal recognition of pregnancy and implantation. Biol. Reprod. 85, 1143–1151 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Irimia M., Blencowe B. J., Alternative splicing: Decoding an expansive regulatory layer. Curr. Opin. Cell Biol. 24, 323–332 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Tapial J., Ha K. C. H., Sterne-Weiler T., Gohr A., Braunschweig U., Hermoso-Pulido A., Quesnel-Vallières M., Permanyer J., Sodaei R., Marquez Y., Cozzuto L., Wang X., Gómez-Velázquez M., Rayon T., Manzanares M., Ponomarenko J., Blencowe B. J., Irimia M., An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res. 27, 1759–1768 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soumillon M., Necsulea A., Weier M., Brawand D., Zhang X., Gu H., Barthès P., Kokkinaki M., Nef S., Gnirke A., Dym M., de Massy B., Mikkelsen T. S., Kaessmann H., Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 3, 2179–2190 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Barbosa-Morais N. L., Irimia M., Pan Q., Xiong H. Y., Gueroussov S., Lee L. J., Slobodeniuc V., Kutter C., Watt S., Colak R., Kim T. H., Misquitta-Ali C. M., Wilson M. D., Kim P. M., Odom D. T., Frey B. J., Blencowe B. J., The evolutionary landscape of alternative splicing in vertebrate species. Science 338, 1587–1593 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Kalsotra A., Cooper T. A., Functional consequences of developmentally regulated alternative splicing. Nat. Rev. Genet. 12, 715–729 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen G., Chen J., Yang J., Chen L., Qu X., Shi C., Ning B., Shi L., Tong W., Zhao Y., Zhang M., Shi T., Significant variations in alternative splicing patterns and expression profiles between human-mouse orthologs in early embryos. Sci. China Life Sci. 60, 178–188 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Xing Y., Yang W., Liu G., Cui X., Meng H., Zhao H., Zhao X., Li J., Liu Z., Zhang M. Q., Cai L., Dynamic alternative splicing during mouse preimplantation embryo development. Front. Bioeng. Biotechnol. 8, 35 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian G. G., Li J., Wu J., Alternative splicing signatures in preimplantation embryo development. Cell Biosci. 10, 33 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng R., Zheng X., Wang Y., Wang M., Zhou C., Liu J., Zhang Y., Quan F., Liu X., Genome-wide analysis of alternative splicing differences between oocyte and zygote†. Biol. Reprod. 102, 999–1010 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Irimia M., Weatheritt R. J., Ellis J. D., Parikshak N. N., Gonatopoulos-Pournatzis T., Babor M., Quesnel-Vallières M., Tapial J., Raj B., ’Hanlon D. O., Barrios-Rodiles M., Sternberg M. J. E., Cordes S. P., Roth F. P., Wrana J. L., Geschwind D. H., Blencowe B. J., A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell 159, 1511–1523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irimia M., Rukov J. L., Roy S. W., Vinther J., Garcia-Fernandez J., Quantitative regulation of alternative splicing in evolution and development. Bioessays 31, 40–50 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Kumar L., Futschik M. E., Mfuzz: A software package for soft clustering of microarray data. Bioinformatics 2, 5–7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blair J. D., Hockemeyer D., Doudna J. A., Bateup H. S., Floor S. N., Widespread translational remodeling during human neuronal differentiation. Cell Rep. 21, 2005–2016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Floor S. N., Doudna J. A., Tunable protein synthesis by transcript isoforms in human cells. eLife 5, e10921 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fogarty N. M. E., Mc Carthy A., Snijders K. E., Powell B. E., Kubikova N., Blakeley P., Lea R., Elder K., Wamaitha S. E., Kim D., Maciulyte V., Kleinjung J., Kim J.-S., Wells D., Vallier L., Bertero A., Turner J. M. A., Niakan K. K., Genome editing reveals a role for OCT4 in human embryogenesis. Nature 550, 67–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H., Yang H., Shivalila C. S., Dawlaty M. M., Cheng A. W., Zhang F., Jaenisch R., One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daigneault B. W., Rajput S., Smith G. W., Ross P. J., Embryonic POU5F1 is required for expanded bovine blastocyst formation. Sci. Rep. 8, 7753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Middelkamp S., van Tol H. T. A., Spierings D. C. J., Boymans S., Guryev V., Roelen B. A. J., Lansdorp P. M., Cuppen E., Kuijk E. W., Sperm DNA damage causes genomic instability in early embryonic development. Sci. Adv. 6, eaaz7602 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hardy K., Spanos S., Becker D., Iannelli P., Winston R. M. L., Stark J., From cell death to embryo arrest: Mathematical models of human preimplantation embryo development. Proc. Natl. Acad. Sci. U.S.A. 98, 1655–1660 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adiga S. K., Toyoshima M., Shimura T., Takeda J., Uematsu N., Niwa O., Delayed and stage specific phosphorylation of H2AX during preimplantation development of gamma-irradiated mouse embryos. Reproduction 133, 415–422 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Adiga S. K., Toyoshima M., Shiraishi K., Shimura T., Takeda J., Taga M., Nagai H., Kumar P., Niwa O., p21 provides stage specific DNA damage control to preimplantation embryos. Oncogene 26, 6141–6149 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Yukawa M., Oda S., Mitani H., Nagata M., Aoki F., Deficiency in the response to DNA double-strand breaks in mouse early preimplantation embryos. Biochem. Biophys. Res. Commun. 358, 578–584 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Awasthi P., Foiani M., Kumar A., ATM and ATR signaling at a glance. J. Cell Sci. 128, 4255–4262 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Sun B., Ross S. M., Rowley S., Adeleye Y., Clewell R. A., Contribution of ATM and ATR kinase pathways to p53-mediated response in etoposide and methyl methanesulfonate induced DNA damage. Environ. Mol. Mutagen. 58, 72–83 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Atashpaz S., Shams S. S., Gonzalez J. M., Sebestyén E., Arghavanifard N., Gnocchi A., Albers E., Minardi S., Faga G., Soffientini P., Allievi E., Cancila V., Bachi A., Fernández-Capetillo Ó., Tripodo C., Ferrari F., López-Contreras A. J., Costanzo V., ATR expands embryonic stem cell fate potential in response to replication stress. eLife 9, e54756 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gohr A., Irimia M., Matt: Unix tools for alternative splicing analysis. Bioinformatics 35, 130–132 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Ray D., Kazan H., Cook K. B., Weirauch M. T., Najafabadi H. S., Li X., Gueroussov S., Albu M., Zheng H., Yang A., Na H., Irimia M., Matzat L. H., Dale R. K., Smith S. A., Yarosh C. A., Kelly S. M., Nabet B., Mecenas D., Li W., Laishram R. S., Qiao M., Lipshitz H. D., Piano F., Corbett A. H., Carstens R. P., Frey B. J., Anderson R. A., Lynch K. W., Penalva L. O. F., Lei E. P., Fraser A. G., Blencowe B. J., Morris Q. D., Hughes T. R., A compendium of RNA-binding motifs for decoding gene regulation. Nature 499, 172–177 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neuenkirchen N., Chari A., Fischer U., Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett. 582, 1997–2003 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Saltzman A. L., Pan Q., Blencowe B. J., Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev. 25, 373–384 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao Y., Liu X., Tang B., Li C., Kou Z., Li L., Liu W., Wu Y., Kou X., Li J., Zhao Y., Yin J., Wang H., Chen S., Liao L., Gao S., Protein expression landscape of mouse embryos during pre-implantation development. Cell Rep. 21, 3957–3969 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Grau-Bove X., Ruiz-Trillo I., Irimia M., Origin of exon skipping-rich transcriptomes in animals driven by evolution of gene architecture. Genome Biol. 19, 135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen H., Yang M., Li S., Zhang J., Peng B., Wang C., Chang Z., Ong J., Du P., Mouse totipotent stem cells captured and maintained through spliceosomal repression. Cell 184, 2843–2859.e20 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Naro C., Jolly A., di Persio S., Bielli P., Setterblad N., Alberdi A. J., Vicini E., Geremia R., de la Grange P., Sette C., An orchestrated intron retention program in meiosis controls timely usage of transcripts during germ cell differentiation. Dev. Cell 41, 82–93.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abe K., Yamamoto R., Franke V., Cao M., Suzuki Y., Suzuki M. G., Vlahovicek K., Svoboda P., Schultz R. M., Aoki F., The first murine zygotic transcription is promiscuous and uncoupled from splicing and 3′ processing. EMBO J. 34, 1523–1537 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vanneste E., Voet T., le Caignec C., Ampe M., Konings P., Melotte C., Debrock S., Amyere M., Vikkula M., Schuit F., Fryns J. P., Verbeke G., D’Hooghe T., Moreau Y., Vermeesch J. R., Chromosome instability is common in human cleavage-stage embryos. Nat. Med. 15, 577–583 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Destouni A., Esteki M. Z., Catteeuw M., Tšuiko O., Dimitriadou E., Smits K., Kurg A., Salumets A., Van Soom A., Voet T., Vermeesch J. R., Zygotes segregate entire parental genomes in distinct blastomere lineages causing cleavage-stage chimerism and mixoploidy. Genome Res. 26, 567–578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tšuiko O., Jatsenko T., Grace L. K. P., Kurg A., Vermeesch J. R., Lanner F., Altmäe S., Salumets A., A speculative outlook on embryonic aneuploidy: Can molecular pathways be involved? Dev. Biol. 447, 3–13 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Marangos P., Carroll J., Oocytes progress beyond prophase in the presence of DNA damage. Curr. Biol. 22, 989–994 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Wang H., Luo Y. B., Lin Z. L., Lee I. W., Kwon J., Cui X. S., Kim N. H., Effect of ATM and HDAC inhibition on etoposide-induced DNA damage in porcine early preimplantation embryos. PLoS One 10, e0142561 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koike M., Mashino M., Sugasawa J., Koike A., Histone H2AX phosphorylation independent of ATM after X-irradiation in mouse liver and kidney in situ. J. Radiat. Res. 49, 445–449 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Muller W. U., Streffer C., Pampfer S., The question of threshold doses for radiation damage: Malformations induced by radiation exposure of unicellular or multicellular preimplantation stages of the mouse. Radiat. Environ. Biophys. 33, 63–68 (1994). [DOI] [PubMed] [Google Scholar]
- 58.Dahl J. A., Jung I., Aanes H., Greggains G. D., Manaf A., Lerdrup M., Li G., Kuan S., Li B., Lee A. Y., Preissl S., Jermstad I., Haugen M. H., Suganthan R., Bjørås M., Hansen K., Dalen K. T., Fedorcsak P., Ren B., Klungland A., Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537, 548–552 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marnef A., Cohen S., Legube G., Transcription-coupled dna double-strand break repair: Active genes need special care. J. Mol. Biol. 429, 1277–1288 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Spindle A., Nagano H., Pedersen R. A., Inhibition of DNA replication in preimplantation mouse embryos by aphidicolin. J. Exp. Zool. 235, 289–295 (1985). [DOI] [PubMed] [Google Scholar]
- 61.Solana J., Irimia M., Ayoub S., Orejuela M. R., Zywitza V., Jens M., Tapial J., Ray D., Morris Q., Hughes T. R., Blencowe B. J., Rajewsky N., Conserved functional antagonism of CELF and MBNL proteins controls stem cell-specific alternative splicing in planarians. eLife 5, e16797 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burguera D., Marquez Y., Racioppi C., Permanyer J., Torres-Méndez A., Esposito R., Albuixech-Crespo B., Fanlo L., D’Agostino Y., Gohr A., Navas-Perez E., Riesgo A., Cuomo C., Benvenuto G., Christiaen L. A., Martí E., D’Aniello S., Spagnuolo A., Ristoratore F., Arnone M. I., Garcia-Fernàndez J., Irimia M., Evolutionary recruitment of flexible Esrp-dependent splicing programs into diverse embryonic morphogenetic processes. Nat. Commun. 8, 1799 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Torres-Méndez A., Bonnal S., Marquez Y., Roth J., Iglesias M., Permanyer J., Almudí I., O’Hanlon D., Guitart T., Soller M., Gingras A. C., Gebauer F., Rentzsch F., Blencowe B. J., Valcárcel J., Irimia M., A novel protein domain in an ancestral splicing factor drove the evolution of neural microexons. Nature Ecol. Evol. 3, 691–701 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Labbé R. M., Irimia M., Currie K. W., Lin A., Zhu S. J., Brown D. D. R., Ross E. J., Voisin V., Bader G. D., Blencowe B. J., Pearson B. J., A comparative transcriptomic analysis reveals conserved features of stem cell pluripotency in planarians and mammals. Stem Cells 30, 1734–1745 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., Pachter L., Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alamancos G. P., Pages A., Trincado J. L., Bellora N., Eyras E., Leveraging transcript quantification for fast computation of alternative splicing profiles. RNA 21, 1521–1531 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Han H., Irimia M., Ross P. J., Sung H. K., Alipanahi B., David L., Golipour A., Gabut M., Michael I. P., Nachman E. N., Wang E., Trcka D., Thompson T., O’Hanlon D., Slobodeniuc V., Barbosa-Morais N. L., Burge C. B., Moffat J., Frey B. J., Nagy A., Ellis J., Wrana J. L., Blencowe B. J., MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature 498, 241–245 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anders S., Huber W., Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Irimia M., Denuc A., Burguera D., Somorjai I., M.-Durán J. M., Genikhovich G., Jimenez-Delgado S., Technau U., Roy S. W., Marfany G., G.-Fernàndez J., Stepwise assembly of the nova-regulated alternative splicing network in the vertebrate brain. Proc. Natl. Acad. Sci. U.S.A. 108, 5319–5324 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hinrichs A. S., Karolchik D., Baertsch R., Barber G. P., Bejerano G., Clawson H., Diekhans M., Furey T. S., Harte R. A., Hsu F., Hillman-Jackson J., Kuhn R. M., Pedersen J. S., Pohl A., Raney B. J., Rosenbloom K. R., Siepel A., Smith K. E., Sugnet C. W., Sultan-Qurraie A., Thomas D. J., Trumbower H., Weber R. J., Weirauch M., Zweig A. S., Haussler D., Kent W. J., The UCSC genome browser database: Update 2006. Nucleic Acids Res. 34, D590–D598 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Attig J., de Los Mozos I. R., Haberman N., Wang Z., Emmett W., Zarnack K., König J., Ule J., Splicing repression allows the gradual emergence of new Alu-exons in primate evolution. eLife 5, e19545 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Correa B. R., de Araujo P. R., Qiao M., Burns S. C., Chen C., Schlegel R., Agarwal S., Galante P. A. F., Penalva L. O. F., Functional genomics analyses of RNA-binding proteins reveal the splicing regulator SNRPB as an oncogenic candidate in glioblastoma. Genome Biol. 17, 125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S23
Tables S1 to S13
Supplementary file S1