Abstract
Multiple myeloma (MM) patients have an 11-fold increased risk of developing myeloid neoplasms compared to the general population; however, acute lymphoblastic leukemia (ALL) is rarely observed. Given that both MM and the majority of ALL are of B cell origin, this raises the question of whether ALL in patients with MM arises from the same clone. We report 13 cases of B-cell ALL following therapy for MM. The interval from MM diagnosis to ALL onset was 5.4 years (range, 3.3–10). The median age at the time of ALL diagnosis was 60 years (range, 43–67). MM therapy included immunomodulatory agents in all patients and autologous hematopoietic cell transplantation in 10 (77%) patients preceding ALL diagnosis. ALL genetics showed a normal karyotype, TP53 mutation/deletion, and monosomy 7 or 7q deletion in 5, 3, and 2 cases, respectively. Analysis of paired samples of MM and ALL using whole exome sequencing demonstrated that the malignancies arose from different clones. Thus, ALL as a second primary malignancy following MM is not clonally related but could potentially represent a therapy-related leukemia.
Second primary malignancies (SPM) remain a concern for patients with multiple myeloma (MM), especially as MM survival improves.1 Large population-based studies suggest that the risk of an SPM is 26% higher in MM patients compared with the general population.2 The etiology of these SPMs remains unclear, but theories implicate host, genetic, and MM treatment factors.1
The incidence of hematologic malignancies as an SPM in MM is 0.8–3.1%.3–5 The majority of cases are myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML), but acute lymphoblastic leukemia (ALL) has been occasionally reported.6–8 The IFM2005 study and the CALGB100104 trial of lenalidomide maintenance following hematopoietic cell transplantation (HCT) reported three cases and one case of ALL, respectively.6, 7 However, no details on their clonal origin were reported. Because both B-cell ALL and MM originate from abnormal post-germinal center B-cells, this fact raises the question of whether post-MM ALL represents a clonal dedifferentiation from a more indolent MM to an aggressive ALL, or a therapy-related leukemia triggered by the genotoxic effect of MM therapy.8, 9 The transformation of mature B-cell lymphomas to ALL has been reported, with related clones in follicular lymphoma and unrelated clones in chronic lymphocytic leukemia.10, 11 Other possibilities include the coincident development of two unrelated malignancies or the presence of a germline mutation predisposing the patient to several malignancies as noted in MM with other malignancies.12
Herein, we describe a single-institution series of ALL cases as SPM after MM, and we examine the clonal relationship between these two B-cell malignancies in a subset of cases to further explore their relationship.
We retrospectively reviewed all consecutive cases of adult ALL treated at City of Hope between 2000 and 2017, and we identified cases that had an antecedent MM diagnosis. This study was approved by the Institutional Review Board.
Six paired MM and ALL samples were obtained when available, and whole exome sequencing was performed to assess the clonal relationship between the two malignancies. Pre-autologous HCT mobilized stem cells and a bone marrow biopsy at remission were used as a surrogate for germline controls in 4 and 1 cases, respectively. These were the only available samples from the deceased subjects in the study. Genomic DNA was extracted with QIAamp DNA Micro Kit (Qiagen; Venlo, Netherlands) according to manufacturer instructions. The libraries were prepared with TruSeq Exome Library Prep Kit (Illumina; San Diego, CA, USA) and 100 ng of genomic DNA. Two or three libraries were pooled together and sequenced on Illumina HiSeq 4000 in high-output mode with 100 paired-end reads to obtain a minimum coverage of 100X per sample. Sequencing data were analyzed using the pipelines of the Broad Institute of Harvard and MIT (Firehose, www.broadinstitute.org/cancer/cga), resulting in BAM files aligned to hg19 with calibrated quality scores.13 We used MuTect within the Firehose framework to call somatic mutations in tumor aspirates.13 We filtered out potential artefactual OxoG mutations as well as somatic single nucleotide variations (SSNVs) and indels present in a panel of normal samples. To estimate somatic copy number alteration, we used ReCapSeg (http://gatkforums.broadinstitute.org/categories/recapseg-documentation), which calculated proportional coverage for each target region and then normalized each segment using the median proportional coverage in a panel of normal (PON) samples sequenced with the same capture technology. The sample was projected to a hyperplane defined by the PON, and the tumor copy-ratio was estimated. These copy-ratio profiles were segmented with CBS.14 To estimate allelic copy number, germline heterozygous sites in the normal sample were called via GATK Haplotype Caller.15, 16 Then, the contribution of each homologous chromosome was assessed via reference and alternate read counts at the germline heterozygous sites. Finally, we segmented the allele specific copy ratios using PSCBS.14 Copy ratios and the force-called SSNVs and indels were used to estimate the sample purity, ploidy, and cancer cell fraction by ABSOLUTE.13 To assess mutation clonality in matched samples, we used PHYLOGIC to perform clustering of ABSOLUTE CCFs, as previously described.13
Demographic, disease, and treatment characteristics were reported using descriptive statistics; median and range for continuous variables; and frequency and percent for categorical variables. Leukemia free survival (LFS) and overall survival (OS) were calculated from the date of ALL diagnosis using the Kaplan-Meier product-limit method. All analyses were performed using SAS version 9.4 (SAS Institute; Cary, NC, USA). Data were locked for analysis on January 8, 2018.
We identified 13 (1.3%) cases of ALL preceded by MM out of 1022 adults diagnosed with ALL; all had a B-cell phenotype. The median age at the time of ALL diagnosis was 60 years (range 43–67), and 62% were male. The median interval of latency from MM diagnosis to ALL diagnosis was 5.4 years (range, 3.3–10.0), and the interval from the date of autologous HCT to ALL diagnosis was 4.3 years (range, 2.7–6.0) (Table 1).
Table 1.
Patient characteristics
| Category | Number (percent or range). N=13. |
|---|---|
| Median age at ALL diagnosis | 60 (43–67) |
| Sex | |
| Female | 5 (38.5) |
| Male | 8 (61.5) |
| Race | |
| White | 5 (38.5) |
| Hispanic | 6 (46.1) |
| Other/unknown | 2 (15.4) |
| MM subtype | |
| IgG | 6 (46.2) |
| IgM | 0 (0) |
| IgA | 4 (30.8) |
| Light chain MM | 3 (23.1) |
| MM cytogenetics | |
| Unavailable | 5 (38.5) |
| Normal karyotype | 6 (46.2) |
| TP53 deletion | 1 (7.7) |
| Monosomy 13 and trisomy 4, 5, 9 and 15 | 1 (7.7) |
| MM therapy | |
| IMiDs | 13 (100) |
| bortezomib | 7 (53.8) |
| chemotherapy | 12 (92.3) |
| radiation | 3 (23.1) |
| Maintenance therapy for MM | |
| Yes | 11 (84.6) |
| No | 2 (15.4) |
| IMiDs for MM | |
| thalidomide | 8 (61.5) |
| lenladomide | 6 (46.2) |
| AutoHCT for MM | |
| Yes | 10 (76.9) |
| No | 3 (23.1) |
| MM relapsed | |
| Yes | 6 (46.2) |
| No | 7 (53.8) |
| Median time of latency | |
| From MM diagnosis | 5.4 (3.3–10.0) |
| From autoHCT | 4.3 (2.7–6.0) |
| MM active at the time of ALL | |
| Yes | 3 (23.1) |
| No | 10 (76.9) |
| WBC at ALL presentation | 2 (1.2–29) |
| ALL phenotype | |
| B-cell | 13 (100) |
| T-cell | 0 (0) |
| ALL cytogenetics | |
| Normal karyotype | 5 (38.5) |
| TP53 deletion/mutation | 3 (23.1) |
| Monosomy 7 or 7q deletion | 2 (15.4) |
| Trisomy 8 | 1 (7.7) |
| Hyperdiploidy | 1 (7.7) |
| add(9)(q34),del(14)(q22q32),inv(20)(q13) | 1 (7.7) |
ALL, acute lymphoblastic leukemia; autoHCT, autologous hematopoietic cell transplantation; IMID, immunomodulatory drug; MM, multiple myeloma; WBC, white blood cell
All patients received immunomodulatory drugs (IMiDs) as part of MM therapy before ALL onset, including six patients who were treated with lenalidomide and eight who were treated with thalidomide. Bortezomib was administered to 7 (54%) patients, chemotherapy (either standard dose or high dose [n=10, 77%]) was given to 12 (92%) patients, and 3 (23%) patients had prior involved field radiation. Eleven patients received maintenance therapy, the majority with lenalidomide. Three (23%) patients had evidence of active MM at the time of ALL diagnosis.
With respect to ALL, the median white blood cell (WBC) count at the time of diagnosis was 2000/μL (range, 1.2k-29k). The hyperCVAD regimen was used as ALL initial induction in the majority of patients (n=12, 92%), and 85% of all patients achieved complete remission (CR). Eight patients (62%) subsequently underwent allogeneic HCT. The median follow-up for all patients was 16.2 months (range, 5.7–118.0). One-year event free survival and OS from the time of ALL diagnosis were both 77% (95% CI: 44%–92%).
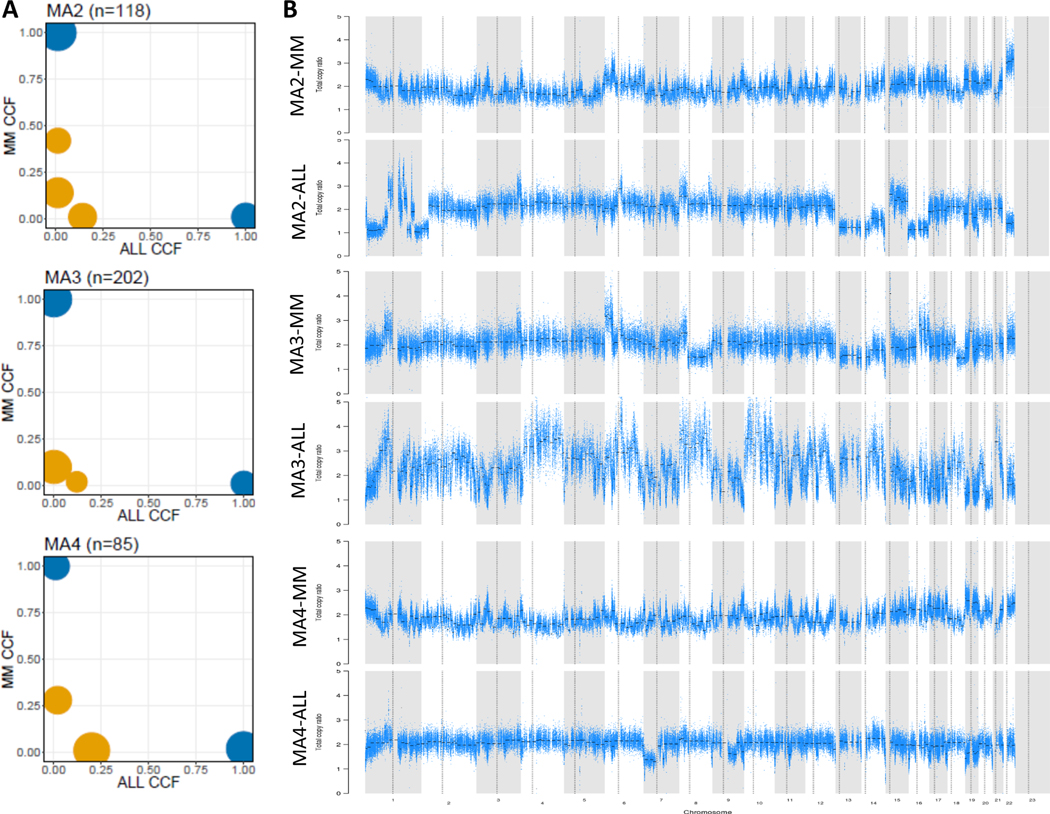
Six paired MM and ALL DNA samples, as well as germline controls for all the cases but one, were available and underwent whole exome sequencing. One paired sample was excluded from downstream analysis because of lack of germline control. The mean coverage was 229X for all sequenced samples. We investigated the number and spectrum of somatic mutations and copy number alterations (CNA) in both neoplasms, and we did not identify any differences compared to publicly available datasets. Only 3 matched samples were further analyzed by PHYLOGIC due to low purity (<15%) for two of the matched samples. Phylogenetic analysis clearly indicated that the two neoplasms are clonally unrelated, with clonal and subclonal mutations and CNAs unique to each malignancy (Figure 1).
Figure 1: ALL as a second primary malignancy is clonally unrelated to preceding MM.
A) Cancer cell fraction (CCF) for clusters of SSNVs detected in MM and ALL samples from the same patient. Mutations were clustered by CCF for each pair of samples using PHYLOGIC. Clonal (navy blue) SSNVs were defined as events having ≥0.9 CCF in both samples. Subclonal (yellow) SSNVs were defined as events having <0.9 CCF in samples. Size of circles indicated the fraction of SSNVs. Mutations having ≥0.9 detection power in both samples are shown, and clusters with <3 mutations are excluded. At the top of each plot are reported the ID of the patient and the number of mutations considered in the plot. In the x axis are represented clonal (navy blue) and subclonal (yellow) mutations identified in ALL, while on the y axis are shown mutations found in MM; B) Copy number alteration in paired samples showing distinctive variations. The panel of each sample visualizes the total copy number ratio of each chromosome.
Secondary hematologic malignancies are a known risk in patients with myeloma, and they mainly comprise cases of AML or MDS.17 Originally, this finding was ascribed to the use of melphalan as a mainstay of MM therapy, and more recently, the use of lenalidomide, especially following autologous HCT.7 On the other hand, ALL has rarely been noted.
Here, we have described a case series of ALL as an SPM after MM, encompassing the clinical and genetic features of this cohort. We have shown that all patients in this cohort were treated with IMiDs for their myeloma and the majority also underwent autologous HCT before ALL onset.
Both MM and the majority of ALL are B-cell malignancies, and their sequential development in the same patient may suggest an MM clonal dedifferentiation into a more aggressive form of B-cell malignancy, such as B-cell ALL. Nonetheless, our exome sequencing analysis of paired DNA samples from 6 cases of this cohort illustrated that the neoplasms were clonally unrelated, contradicting this hypothesis. On the other hand, therapy-related ALL has been described as a complication of exposure to cytotoxic therapies in the course of unrelated primary malignancies.8, 9 Our ALL cohort was enriched with the TP53 mutation/deletion, as well as other genetic features observed frequently in therapy-related myeloid neoplasms, such as complex karyotype, monosomal karyotype and 7q deletion.18 Therefore, we postulate that a subset of these cases could be therapy-related leukemia as the result of prior MM treatment, presenting as a B-cell ALL phenotype rather than the more frequently seen myeloid phenotype. Further evaluation of a larger cohort is required, and it would be of interest to expand this study to therapy-related acute myeloid leukemia and myelodysplastic syndrome.
Acknowledgments
The Biostatistics and Molecular Pathology Cores at City of Hope were supported by the National Institutes of Health under award number P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosure of conflict of interest
I.A. serves on the speakers’ bureau for Jazz Pharmaceuticals. A.K. is a consultant and serves on the speakers’ bureau for Janssen, Celgene, Takeda, and Onyx. The other authors have no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication. Springer Nature are providing this early version of the manuscript as a service to our customers. The manuscript will undergo copyediting, typesetting and a proof review before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers apply.
References
- 1.Razavi P, Rand KA, Cozen W, Chanan-Khan A, Usmani S, Ailawadhi S. Patterns of second primary malignancy risk in multiple myeloma patients before and after the introduction of novel therapeutics. Blood cancer journal 2013. Jun 28; 3: e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jonsdottir G, Lund SH, Bjorkholm M, Turesson I, Hultcrantz M, Porwit A, et al. The impact of prior malignancies on second malignancies and survival in MM patients: a population-based study. Blood advances 2017. Nov 28; 1(25): 2392–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barlogie B, Tricot G, Haessler J, van Rhee F, Cottler-Fox M, Anaissie E, et al. Cytogenetically defined myelodysplasia after melphalan-based autotransplantation for multiple myeloma linked to poor hematopoietic stem-cell mobilization: the Arkansas experience in more than 3,000 patients treated since 1989. Blood 2008. Jan 01; 111(1): 94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Usmani SZ, Sexton R, Hoering A, Heuck CJ, Nair B, Waheed S, et al. Second malignancies in total therapy 2 and 3 for newly diagnosed multiple myeloma: influence of thalidomide and lenalidomide during maintenance. Blood 2012. Aug 23; 120(8): 1597–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rollison DE, Komrokji R, Lee JH, Hampras S, Fulp W, Fisher K, et al. Subsequent primary malignancies among multiple myeloma patients treated with or without lenalidomide. Leukemia & lymphoma 2017. Mar; 58(3): 560–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Attal M, Lauwers-Cances V, Marit G, Caillot D, Moreau P, Facon T, et al. Lenalidomide maintenance after stem-cell transplantation for multiple myeloma. The New England journal of medicine 2012. May 10; 366(19): 1782–1791. [DOI] [PubMed] [Google Scholar]
- 7.McCarthy PL, Owzar K, Hofmeister CC, Hurd DD, Hassoun H, Richardson PG, et al. Lenalidomide after stem-cell transplantation for multiple myeloma. N Engl J Med 2012. May 10; 366(19): 1770–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aldoss I, Dagis A, Palmer J, Forman S, Pullarkat V. Therapy-related ALL: cytogenetic features and hematopoietic cell transplantation outcome. Bone marrow transplantation 2015. May; 50(5): 746–748. [DOI] [PubMed] [Google Scholar]
- 9.Tang G, Zuo Z, Thomas DA, Lin P, Liu D, Hu Y, et al. Precursor B-acute lymphoblastic leukemia occurring in patients with a history of prior malignancies: is it therapy-related? Haematologica 2012. Jun; 97(6): 919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kishimoto W, Shirase T, Chihara D, Maeda T, Arimoto-Miyamoto K, Takeoka T, et al. Double-hit lymphoma with a feature of follicular lymphoma concurrent with clonally related B lymphoblastic leukemia : a preference of transformation for the bone marrow. Journal of clinical and experimental hematopathology : JCEH 2012; 52(2): 113–119. [DOI] [PubMed] [Google Scholar]
- 11.Chakhachiro Z, Yin CC, Abruzzo LV, Aladily TN, Barron LL, Banks HE, et al. B-Lymphoblastic Leukemia in Patients With Chronic Lymphocytic Leukemia: A Report of Four Cases. American journal of clinical pathology 2015. Aug; 144(2): 333–340. [DOI] [PubMed] [Google Scholar]
- 12.Dilworth D, Liu L, Stewart AK, Berenson JR, Lassam N, Hogg D. Germline CDKN2A mutation implicated in predisposition to multiple myeloma. Blood 2000. Mar 1; 95(5): 1869–1871. [PubMed] [Google Scholar]
- 13.Manier S, Park J, Capelletti M, Bustoros M, Freeman S, Ha G, et al. Whole-exome sequencing of cell-free DNA and circulating tumor cells in multiple myeloma. Nature communications 2018; In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics 2007. Mar 15; 23(6): 657–663. [DOI] [PubMed] [Google Scholar]
- 15.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011. May; 43(5): 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010. Sep; 20(9): 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landgren O, Thomas A, Mailankody S. Myeloma and second primary cancers. N Engl J Med 2011. Dec 08; 365(23): 2241–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith SM, Le Beau MM, Huo D, Karrison T, Sobecks RM, Anastasi J, et al. Clinical-cytogenetic associations in 306 patients with therapy-related myelodysplasia and myeloid leukemia: the University of Chicago series. Blood 2003. Jul 1; 102(1): 43–52. [DOI] [PubMed] [Google Scholar]