Abstract
Neurofilaments (Nfs) are the major structural component of neurons. Their role as a potential biomarker of several neurodegenerative diseases has been investigated in past years with promising results. However, even under physiological conditions, little is known about the leaking of Nfs from the neuronal system and their detection in the cerebrospinal fluid (CSF) and blood. This study aimed at developing a mathematical model of Nf transport in healthy subjects in the 20–90 age range. The model was implemented as a set of ordinary differential equations describing the trafficking of Nfs from the nervous system to the periphery. Model parameters were calibrated on typical Nf levels obtained from the literature. An age‐dependent function modeled on CSF data was also included and validated on data measured in serum. We computed a global sensitivity analysis of model rates and volumes to identify the most sensitive parameters affecting the model’s steady state. Age, Nf synthesis, and degradation rates proved to be relevant for all model variables. Nf levels in the CSF and in blood were observed to be sensitive to the Nf leakage rates from neurons and to the blood clearance rate, and CSF levels were also sensitive to rates representing CSF turnover. An additional parameter perturbation analysis was also performed to investigate possible transient effects on the model variables not captured by the sensitivity analysis. The model provides useful insights into Nf transport and constitutes the basis for implementing quantitative system pharmacology extensions to investigate Nf trafficking in neurodegenerative diseases.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
The change of neurofilament (Nf) concentration in the cerebrospinal fluid and blood is a potential marker of several neurodegenerative diseases and brain injury.
WHAT QUESTION DID THIS STUDY ADDRESS?
Is it possible to build a mathematical model of Nf trafficking that describes the different Nf subunits in healthy subjects aged 20–90 years?
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
We introduce a mathematical model describing the transport of Nfs from neurons to the periphery in healthy conditions calibrated on previously reported Nf data in the literature. Model sensitivity analysis highlights the role of aging and the rates governing Nf leakage from the nervous system and blood clearance as main regulative players of Nf trafficking.
HOW MIGHT THIS CHANGE DRUG DISCOVERY, DEVELOPMENT, AND/OR THERAPEUTICS?
The model can be a valuable starting point for developing a quantitative system pharmacology model to analyze the role of Nf subunits as potential biomarkers of disease onset and treatment efficacy in several neurodegenerative diseases.
INTRODUCTION
Neurofilaments (Nfs) are type IV intermediate filaments, a major structural component of the neuronal cytoskeleton in the central nervous system (CNS) and peripheral nervous system (PNS), forming polymeric filaments that are fundamental to shape the morphology of the cell body, dendrites, and above all, axons. 1 Nfs are composed of three different subunits: light (NfL), medium (NfM), and heavy (NfH) chains, with molecular weights of 68, 160, and 200 kDa, respectively. It has been shown that all three subunits play roles in the growth and the stability of axons, with NfL forming the backbone of Nf polymers, whereas NfM and NfH extend their tails in a side‐arm shape, providing structural stability to the polymers. 2 These tails typically show relevant levels of phosphorylation, and it is usually in this form that NfM and NfH (labeled pNfH) are detected in the CSF and blood.
Even in healthy subjects, a certain amount of Nf exits the neuronal system to the surrounding fluids and is transported to the CSF and blood, with an increase of average Nf levels with age. 3 , 4 Numerous studies have reported that peripheral NfL and pNfH levels increase in the presence of neurological diseases, 5 , 6 , 7 , 8 such as multiple sclerosis, 9 spinal muscular atrophy, 10 and amyotrophic lateral sclerosis 11 , 12 as well as Alzheimer’s disease 13 , 14 and Parkinson’s disease. 15 Therefore, they are considered potential biomarkers of disease onset and severity. NfM is less studied, but increasing concentrations in the CSF and blood have been associated with brain injury. 16 Nfs are also considered promising biomarkers of treatment efficacy in specific neurodegenerative diseases, as in the case of recently developed antisense oligonucleotides. 10 , 17 Analytical assays and platforms have been developed and improved to detect Nf in the CSF and blood, reaching the very high sensitivity necessary to discriminate between healthy and disease conditions. 18 Nevertheless, standardization of analytical platforms is not complete, and relevant differences can be observed among some concentration ranges reported in the literature. 10
Given the important role of Nfs in the maintenance and support of the cytoskeleton, modeling efforts have been devoted to study Nf transport along axons as well as Nf pathological segregation from other cytoskeletal components or abnormal accumulation. 19 , 20 However, to date there are no computational models describing the leakage of Nfs from the nervous system to peripheral compartments. The objective of this work was to fill this gap by developing, calibrating, and analyzing a mathematical model describing Nf trafficking in healthy conditions.
METHODS
Model development and calibration
The diagram reported in Figure 1 shows the compartments and the reactions that were considered to model Nf trafficking in healthy subjects. Nfs (labeled NfL,M,H to indicate that the trafficking scheme is common for the subunits) are synthetized and degraded in the neurons of the CNS (r1, synthesis reaction; r2, degradation reaction) and of the PNS (r3 and r4). Reactions r5 and r6 describe the physiological leakage of Nf from neurons into the interstitial fluid (ISF) and endoneurial fluid, respectively. These two reactions are meant to represent in a phenomenological but effective way the combined behavior of all the complex mechanisms underlying the release of Nfs from neurons. Nfs are transported by convective motion to the cranial and spinal CSF (CSF cranial, CSF spinal; r7, r8, r9 and r10) and finally to the blood (r11 and r12), from where they are cleared (r13). The reactions from r7 to r12 work in the model as passive transports for Nfs without any distinction among the different Nf subunits (see the references in Table S1 for more details about the derivation of the corresponding rates). The blood–brain barrier between ISF and blood and the blood–nerve barrier between endoneurial fluid and blood are assumed to prevent any direct exchange of Nfs between these compartments.
FIGURE 1.
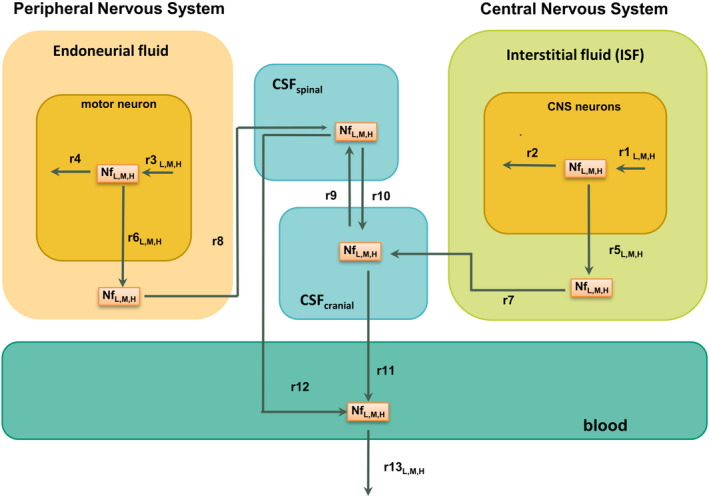
Model diagram representing the structure of the Nf trafficking model. Nf subunits move from neurons to the CSF and to the blood, where they are detected. Reaction rates for which a different estimate has been identified for the three Nf subunits have an additional label “L,M,H”; all of the other rates share the same estimate for neurofilament light chain, neurofilament medium chain, and phosphorylated neurofilament heavy chain and are unlabeled. CNS, central nervous system; CSF, cerebrospinal fluid; Nf, neurofilament
The mathematical implementation of the model consists of three independent replicas, one for each Nf subunit, of the same seven ordinary differential equations (ODEs) (see Figure S1). Most of the volumes were identified directly from the literature. Only the volumes of PNS neurons and endoneurial fluid needed an extrapolation from different literature sources, as reported in the Supplementary Material. The degradation rate of NfL in neurons was set following the indication of Yuan et al. 21 and was obtained by a weighted average of 90% stable and longer living (90 days half‐life) filaments and 10% slowly transported and shorter living (22 days half‐life) filaments. Given the absence of direct measurements, we assumed the same degradation rate for the other subunits, even if the real rates could be different, given their chemical differences. Synthesis rates are not known, and they were computed from the Nf concentration in neurons to keep the initial concentration at equilibrium in the absence of leakage toward the peripheral compartments. To overcome a lack of information on the PNS parameters, we imposed symmetry between the initial conditions and the rates pertaining to the CNS and to the PNS (from r1 to r8) by assuming that the two parts of the model differ only in the volumes. In the same way, we also imposed that the Nf leakage reactions r5 and r6 are equal. The rates of these reactions were multiplied by an age‐dependent function f(age) modeled to reproduce the population data of the average spinal CSF NfL concentration in the range of 20–90 years 3 and validated on the blood data from Khalil et al. 4 (see the Results and Figure 2b). Lacking any additional information, we also used the same age‐dependent function for NfM and pNfH.
FIGURE 2.
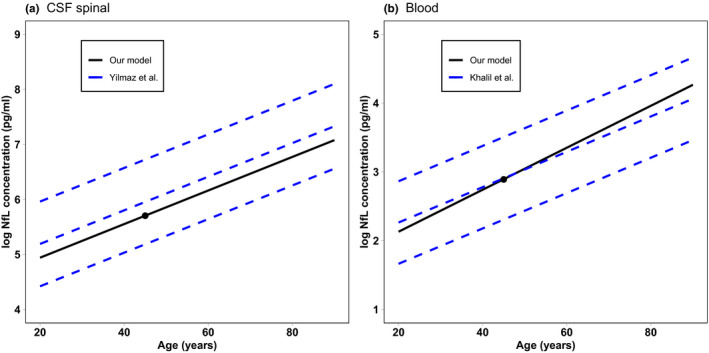
NfL concentration as a function of age computed from the model compared with the two standard deviation intervals from population data in the CSF 3 (a) and the 90% interval in serum 4 (b). The black dot represents the model‐calibrated values at 45 years. CSF, cerebrospinal fluid; NfL, neurofilament light chain
We used values provided by Ferrer‐Alcon et al. 22 , reporting Nf concentration of all subunits measured in the frontal cortex, to derive the model initial state. All model parameter values are reported in Table S1, Table S2 and Table S3, where an additional subscript “(L,M,H)” is used, when necessary, to distinguish among the values relative to the different Nf subunits. The only unknown rates are those of reactions r5 and r6 (assumed equal) and of reaction r13 (see Table 1). These rates were calibrated by optimizing the rates until the model Nf levels at steady state in the CSF and blood matched the experimental levels observed in healthy population as described in the literature. Concentrations in other compartments were initially set to zero and then determined from the same steady‐state condition.
TABLE 1.
Estimated model parameters (the full list of parameters is provided in Table S1)
| Rate | Estimate (1/day) | Estimated 95% CI |
|---|---|---|
| r5L, r6L | 7.3 × 10−6 | (0.47–12) × 10−6 |
| r5M, r6M | 0.60 × 10−6 | (0.3–1.5) × 10−6 |
| r5H, r6H | 0.58 × 10−6 | (0.3–1.4) × 10−6 |
| r13L | 10.1 | 0.5–27.7 |
| r13M | 0.018 | 0.002–0.04 |
| r13H | 7.4 | 1.1–20.1 |
Note: All of the estimates are computed considering the data reported in Byrne et al. 23 Martinez‐Morillo et al. 16 and De Schaepdryver et al. 24 for neurofilament light chain, neurofilament medium chain, and phosphorylated neurofilament heavy chain, respectively. The estimate values are derived from the median neurofilament concentrations, whereas the CIs were obtained by repeated calibrations of neurofilament levels randomly selected from the measured ranges.
Abbreviation: CI, confidence interval.
The model calibration was performed by fitting the following steady‐state data: for NfL, we considered the values 300 pg/ml in the CSF and 9.9 pg/ml in blood 23 ; for NfM, the values of 230 pg/ml in the CSF and 4170 pg/ml in blood 16 ; for pNfH, the values of 225 pg/ml in the CSF 10.5 pg/ml in blood. 24 We selected these values following two criteria: we considered work that measured Nf levels both in the CSF and in blood and in which the median age of the enrolled subjects was close to 45 years. This is the age at which we calibrated the unknown rates of our model. We also considered the concentration ranges reported in these reference articles to perform multiple calibrations by fitting Nf levels selected randomly in their ranges. In this way, we estimated 95% confidence intervals of the optimized parameter values based on population data.
The model is implemented as a collection of R scripts, and it is provided in Supplementary File 2. The R libraries deSolve and optim were used for ODE integration and optimization, respectively.
Sensitivity analysis and parameter perturbation analysis
Starting from the rates obtained by our model calibration, we performed a global sensitivity analysis to identify the model parameters that mostly affect Nf dynamics at steady state. For a general review of sensitivity analysis methods, we refer to the work by Simoni et al. and Zi. 25 , 26 In our case, a global sensitivity analysis of the ith model parameter, with respect to the jth model variable, has been computed as the maximum value obtained by running a set of 100,000 local sensitivity analyses starting from different points of the model parameter space:
where k = 1, 2, … , 100,000. Parameters were sampled in the range 1/10 to 10 times the nominal value using the Latin hypercube sampling method to obtain a uniform distribution of sampled points. 27 Each local sensitivity analysis is defined as:
where xj is the model predicted steady‐state value of the jth model variable computed considering the ith parameter estimate pi or its variations pi + ϵ⋅pi and pi − ϵ⋅pi , respectively (ϵ = 0.001).
In addition to a sensitivity analysis of model steady states, it was also useful to perform a time‐dependent parameter perturbation analysis. To this end, for each rate of the model, we started a model simulation from the steady state calibrated on Nf data, and at time zero, and then scaled the rate by a factor of 1/20, 1/10, 1/5, 5, 10, or 20. Next, we simulated the model until the system reached a new steady state and analyzed the computed time series.
Finally, for each variable, we computed the percent variation of the steady state if each rate is scaled to a given percentage of the nominal value.
RESULTS
Calibration of Nf levels under healthy conditions at steady state
The model was successfully calibrated using previously published data as described in the Methods section. Calibrated rates of the reactions describing the physiological exit of Nf from neurons into the ISF and endoneurial fluid (r5 and r6 in Figure 1) are reported in Table 1. We observe that the optimized values of NfM and pNfH are similar, whereas the value for NfL is 10 times higher. Table 1 also reports the optimized values of the blood clearance rate r13. NfL and pNfH have similar values, whereas NfM shows a much smaller value, reflecting its higher availability, as shown in the experimental data used for calibration. In the literature, there is experimental evidence supporting that phosphorylation protects Nfs from degradation, 28 but this was not evident from our estimated pNfH clearance rate.
Figure 2 compares the age dependency of NfL concentration predicted by our model to the regression curves from population data. As reported in the Methods section, the age function multiplying rates r5 and r6 were modeled to reproduce CSF data from Yilmaz et al. 3 This allowed us to obtain a good agreement of the slope of the curve with the employed experimental data (Figure 2a), even if the calibration at 45 years of age was derived from another source. 23 As an independent validation, we compared the predicted growth of NfL in blood with the age‐dependent growth from the whole data set of Khalil et al. 4 As shown in Figure 2b, the modeled NfL curve is compatible with the 90% interval computed from the population data.
Sensitivity analysis and parameter perturbation analysis
The computed sensitivity analysis shows that the responses of the different Nf subunits seem to be very similar (Table S4). This is because the three Nf subunits share the same model structure and the model rates that are different do not lead to relevant differences of the sensitivity values. For conciseness purposes, in Table 2 we report the results for NfL and draw conclusions also valid for the other subunits. The concentration of all compartments is particularly sensitive to the parameters determining Nf concentration in neurons, such as synthesis and degradation rates, and the volume of CNS and PNS. At the same time, all compartments, except those of neurons, are also significantly affected by the rates of Nf leakage. This reflects the fact that the Nf concentration in the periphery is fundamentally driven by Nf flux coming from the CNS and PNS, defined as the Nf concentration in neurons times the leakage rate. We observe that there is an asymmetric effect from the CNS and the PNS, where the Nf concentration in the CSF and blood are more sensitive to the former. Considering that the two parts are symmetric with regard to the initial concentrations and rates, we conclude that it is an effect of the much larger volume of the CNS.
TABLE 2.
Summary of the sensitivity analysis for NfL
| NfL CNS neurons | NfL PNS | NfL ISF | NfL endoneurial fluid | NfL CSF cranial | NfL CSF spinal | NfL blood |
|---|---|---|---|---|---|---|
| r1 | r3 | Age | Age | Age | Age | Age |
| r2 | r4 | r7 | r8 | r1 | r1 | Blood vol. |
| Age | Age | r1 | r3 | r2 | r2 | r13 |
| r5 | r6 | CNS neurons vol. | Motor neurons vol. | r3 | r3 | r1 |
| r7 | r5 | r2 | r4 | r4 | r4 | r2 |
| r9 | r7 | r5 | r6 | CNS neurons vol. | r12 | r3 |
| r10 | r9 | CSF cranial vol. | Endoneurial fluid vol. | r5 | Motor neurons vol. | r4 |
| r11 | r10 | ISF vol. | CSF spinal vol. | r11 | r6 | CNS neurons vol. |
| r12 | r11 | Endoneurial fluid vol. | ISF vol. | r9 | CNS neurons vol. | r5 |
| r6 | r12 | r9 | r12 | r10 | r5 | r6 |
| r8 | r8 | r12 | r10 | Motor neurons vol. | r11 | Motor neurons vol. |
| r13 | r13 | r8 | r9 | r6 | r9 | Endoneurial fluid vol. |
| CNS neurons vol. | CNS neurons vol. | r10 | r5 | r12 | r10 | CSF cranial vol. |
| ISF vol. | ISF vol. | Motor neurons vol. | CSF cranial vol. | Endoneurial fluid vol. | Endoneurial fluid vol. | r7 |
| Motor neurons vol. | Motor neurons vol. | r11 | CNS neurons vol. | CSF cranial vol. | CSF cranial vol. | ISF vol. |
| Endoneurial fluid vol. | Endoneurial fluid vol. | r6 | r7 | ISF vol. | ISF vol. | r10 |
| CSF cranial vol. | CSF cranial vol. | CSF spinal vol. | r11 | r7 | r7 | r9 |
| CSF spinal vol. | CSF spinal vol. | r13 | r13 | r8 | r8 | r12 |
| Blood vol. | Blood vol. | Blood vol. | Blood vol. | CSF spinal vol. | CSF spinal vol. | r11 |
| r3 | r1 | r3 | r1 | r13 | r13 | r8 |
| r4 | r2 | r4 | r2 | Blood vol. | Blood vol. | CSF spinal vol. |
Note: Similar results can be computed for the other two neurofilament subunits. For each compartment, the parameters are sorted from the most relevant to the least relevant. The color map reflects the computed sensitivity values. White cells correspond to very low or negligible sensitivity values.
Abbreviations: CNS, central nervous system; CSF, cerebrospinal fluid; ISF, interstitial fluid; NfL, neurofilament light chain; PNS, peripheral nervous system; vol., volume.
Focusing on the compartments relevant to the diagnostic use of Nfs, we see that Nf CSF spinal levels are sensitive to other rates, namely, those taking Nfs to blood from spinal and cranial CSF, and those governing the exchange between the cranial and spinal fluid. On the other hand, we see that CSF volume has a negligible impact on the CSF Nf steady‐state concentration. The same happens for reactions from the ISF and from the endoneurial fluid. In the next section, we analyze how such rates lead only to transient effects. Regarding the blood compartment, Nf concentration is particularly sensitive to blood volume and clearance rate. The former is important considering the individual variability of blood volume, which reflects into the individual variability of Nf levels. It is interesting to observe that steady‐state blood levels seem to be fundamentally insensitive to a change in the rate governing the flow from CSF (r11) and those regulating the flow between the cranial and spinal parts of the CSF (r9 and r10).
A particular consideration is reserved to the sensitivity of the model to age. Through the phenomenological leakage function, age demonstrated to be one of the most relevant parameters for all compartments except for neurons of the CNS PNS, where very high Nf concentrations mitigate the effect of leakage. Local sensitivity analyses at different age values suggest that age is a parameter whose sensitivity increases with its own value and becomes the most sensitive parameter from the age of 33, as shown in Table S5 and Figure S2.
Transient dynamics from parameter perturbation
The results of the sensitivity analysis pertain to the system at steady state. Interestingly, some compartment variables seem to be unaffected by the rate variation of reactions from or to the corresponding compartment. This fact suggests that the effect of changing these rates must be only transient without affecting the final steady state. To confirm this hypothesis, we computed a series of time‐course simulations, starting from the calibrated steady state and scaling each parameter as described in the Methods section. Figures 3 and 4 report NfL concentration in the CSF and blood as a function of time for each parameter variation. We decided to focus on these compartments because they are the compartments from which Nf samples are collected. The response of NfM and pNfH is very similar and is not reported. Both CSF and blood present a transient response to reactions r7 and r8 from the ISF and endoneurial fluid. The same happens for blood concentrations for all rates from and between the CSF compartments. This explains why the corresponding sensitivity values were negligible.
FIGURE 3.
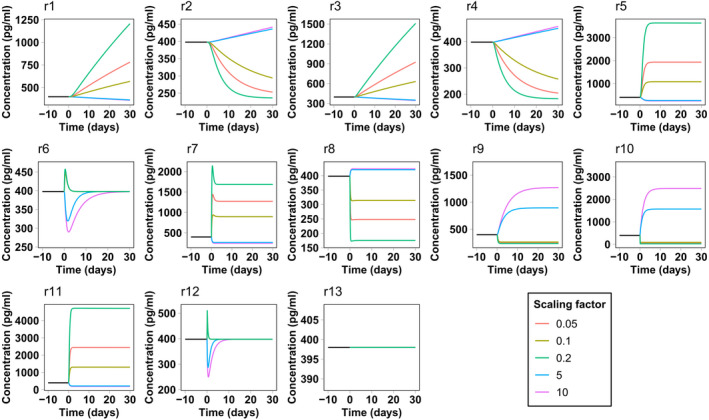
Simulated neurofilament light chain concentration in the spinal cerebrospinal fluid in the presence of a perturbation in the model reaction rates. The simulation starts from the calibrated steady state. At time zero the rate of the corresponding reaction is scaled by different scaling factors
FIGURE 4.
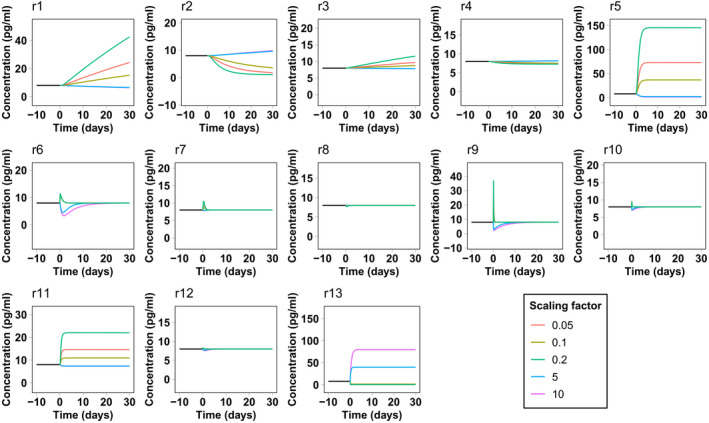
Simulated neurofilament light chain concentration in blood in the presence of a perturbation in the model reaction rates. The simulation starts from the calibrated steady state. At time zero the rate of the corresponding reaction is scaled by different scaling factors
This type of analysis also gives an estimate of the velocity of the Nf level response to the change of rates. In most cases, the system takes less than 30 days to reach a new steady state or to return to the previous steady state, even for the maximum (or minimum) scaling factors considered here. In some cases, such as for leakage variations from neurons, the response is even faster, taking only a few days. In the case of Nf increase attributed to underlying pathology or a traumatic injury, represented by an increase in reaction r5 and r6, the model predicts that Nf levels in the CSF and blood should very rapidly reflect neuronal damage. This is in line with the surveillance of NfL levels after sports‐related concussions 29 or in contact sports such as boxing, 30 where an increase of NfL in blood is already observed from several hours to a few days after the occurrence of the traumatic events.
The percent variation of the steady state in the CSF and blood as a response to the percentage variation of each rate is reported in detail in Table S6 and Table S7. The results suggest classifying the model variables into three classes according to their response to the variation of each rate. The classification is reported in Table 3 and could be a valuable tool to estimate the impact of the rate uncertainties on Nf concentrations and prioritize future experiments.
TABLE 3.
Classification of the model variables according to their percent change as a response to a 50% variation of the rates
| Rate | Strong effect | Weak effect | Negligible effect |
|---|---|---|---|
| r1 | CNS neurons, ISF, CSF cranial, blood | CSF spinal | PNS neurons, endoneurial fluid |
| r2 | CNS neurons, ISF, CSF cranial, CSF spinal, blood | PNS neurons, endoneurial fluid | |
| r3 | PNS neurons, endoneurial fluid, CSF spinal | CSF cranial, blood | CNS neurons, ISF |
| r4 | PNS neurons, endoneurial fluid, CSF spinal | CSF cranial, blood | CNS neurons, ISF |
| r5 | ISF, CSF cranial, blood | CSF spinal, CNS neuron | PNS neurons, endoneurial fluid |
| r6 | Endoneurial fluid, CSF spinal | PNS neurons, CSF cranial, blood | CNS neurons, ISF |
| r7 | ISF | CNS neurons, PNS neurons, endoneurial fluid, CSF cranial, CSF spinal, blood | |
| r8 | Endoneurial fluid | CNS neurons, PNS neurons, ISF, CSF cranial, CSF spinal, blood | |
| r9 | CSF cranial, CSF spinal | CNS neurons, PNS neurons, ISF, endoneurial fluid, Blood | |
| r10 | CSF cranial, CSF spinal | CNS neurons, PNS neurons, ISF, endoneurial fluid, blood | |
| r11 | CSF cranial | CSF spinal | CNS neurons, PNS neurons, ISF, endoneurial fluid, blood |
| r12 | CSF spinal | CSF cranial | CNS neurons, PNS neurons, ISF, endoneurial fluid, blood |
| r13 | Blood | CNS neurons, PNS neurons, ISF, endoneurial fluid, CSF cranial, CSF spinal |
Note: Strong effect represents an absolute variation of the concentration larger than 25%, weak effect an absolute concentration variation between 0 and 25%, whereas the negligible effect category includes the rates for which the concentration change is zero.
Abbreviations: CNS, central nervous system; CSF, cerebrospinal fluid; ISF, interstitial fluid; PNS, peripheral nervous system.
DISCUSSION
The interest of the scientific community in Nfs as a potential biomarker for several neurodegenerative diseases has been increasing in past years. 2 This is motivated by the assumption that Nf levels could be an indicator of neuronal health, related to the effect of aging, pathological conditions, or brain injuries. Numerous clinical studies have investigated the association between CSF/blood Nf levels and specific diseases 9 , 12 or treatment effects. 10 , 17 Although the role of Nfs in the formation, stability, conductivity, transport, and degeneration of the cytoskeleton is widely investigated, also with computational and modeling techniques, 19 , 20 to our knowledge inadequate attention has been paid to the Nf transport mechanism to peripheral tissues. The aim of this work was to model this important process under physiological conditions by also including the effect of aging. The model describes the transport of Nf subunits between different compartments, whereas the leakage reactions represent the complex, mostly unknown mechanisms of Nf release in a phenomenological way. 7
Most of the parameter estimates were derived from the literature, whereas parameters of leakage reaction and of the rate of blood clearance were estimated by fitting experimental data. The computed sensitivity analysis showed similar results for the different Nf subunits because they share the same model structure and similar model rates. The reaction rates that have been highlighted as particularly relevant for the compartments used for experiments (CSF and blood) are synthesis and degradation rates of Nf in neurons, leakage reactions, and clearance in blood. Although additional information on them would be very useful, the values used in our model can already consistently reproduce average Nf levels in the CSF and blood.
An experimental Nf increase with age has been reproduced by a data‐regression formula included in the leakage reaction. The formula was successfully validated on Nf blood levels reported by Khalil et al. 4 The hypothesis reported in Khalil et al. 4 that the slowing of CSF turnover with age could increase serum Nf levels, is disfavored by our model, where variations in the corresponding rates induce only transient effects. Therefore, our model suggests that Nf levels in blood are mainly ruled by neuronal leakage and the blood clearance rate.
The calibrated clearance rate of NfL and pNfH in blood is faster than the degradation rate in neurons. This is expected because in the latter case, Nfs are protected as they are a part of the cytoskeletal structure and are generally thought to be transported to synapses for degradation by proteolysis. 31 , 32 On the other hand, we could expect a smaller clearance rate for pNfH with respect to NfL because of its higher level of phosphorylation protecting it from degradation, 28 but the calibration provided similar values. In general, the calibration of the clearance rate is highly dependent on the accuracy of the data in plasma or serum. It was previously noted that the standardization of the assays, especially for pNfH, is still an open question. 33 The values considered for this work were chosen because Nf levels were measured consistently both in the CSF and blood. Different analyses that measured much higher pNfH levels (e.g., Table 1 in Darras et al. 10 ) would lead to a different calibration, resulting in a slower blood clearance. The clearance value obtained for NfM, reflecting the very high measured concentration in blood, looks out of scale with respect to the other data. Because we could find only one study measuring it, 16 it remains a matter that deserves further investigation.
The model suffers from some limitations because of the requirement of defining several a priori assumptions and would highly benefit from additional data, especially concentration values and rates in the PNS. Initial Nf concentrations in neurons and the estimated leakage rate could suffer from uncertainties attributed to the limited amount of available data. Nevertheless, the total flux of r5 and r6, defined as the product of the calibrated rate and of the Nf concentration in neurons, is tightly constrained in our model by CSF and blood data. If we consider the fluxes of the three subunits at steady state, we get similar values. This is a reasonable result, considering the similar stoichiometry of the three subunits in the process of filament polymerization in the cytoskeleton. 1 Additional details regarding Nfs in neurons, including the measurements of the synthesis and degradation rates of the different Nf subunits and the leakage rates to the ISF, could be helpful and provide in the future a firmer link with the physiological and pathological mechanisms underlying Nf release, which are currently accounted only from a phenomenological perspective. Moreover, the model could be easily extended to include a non‐null permeability of blood–brain and blood–nerve barriers. These barriers are filters made of endothelial cells preventing harmful substances in the circulatory system from entering the ISF and endoneurial fluid. 34 A non‐null permeability of the barriers to Nfs could be associated either to an immature barrier, as in the first months after birth, or to brain injuries and secondary effects of neurodegeneration. 35 , 36 None of these conditions pertain to the healthy model developed in this work, and hence were not considered. However, possible permeability of the barriers in different conditions, such as some neurodegenerative disease or brain injuries, could be easily included with additional reactions among model compartments.
The healthy calibration provides the basis for modeling Nf behavior in neurodegenerative disease onset, progression, and treatment. We are actively working on model extensions to describe Nf dynamics in several neurodegenerative diseases with increased peripheral Nf levels. 10 , 17 Preliminary results illustrating an extension to simulate the Nf dynamics during the progress and the treatment of amyotrophic lateral sclerosis were recently presented 37 and will constitute the content of a future article.
CONFLICT OF INTEREST
M.M., X.T., S.E., E.M., T.F., A.M., D.G., and I.N. are employees/shareholders of Biogen at the time of this work. A.P., P.B., S.P., E.D., and L.M. were contracted by Biogen while this research was conducted.
AUTHOR CONTRIBUTIONS
A.P., S.P., P.B., and L.M. wrote the manuscript. L.M., E.D., I.N., and E.M. designed the research. A.P., S.P., P.B., L.M., and M.M. performed the research. S.E., A.M., T.F., D.G., and X.T. analyzed the data.
Supporting information
Supplementary Material
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Jeannette Stankowski, Dr. Christine Nelson, and Beatrix Bevernage for providing useful suggestions and editing the article.
Paris A, Bora P, Parolo S, et al. An age‐dependent mathematical model of neurofilament trafficking in healthy conditions. CPT Pharmacometrics Syst Pharmacol. 2022;11:447‐457. doi: 10.1002/psp4.12770
Funding information
This work was funded by Biogen
Alessio Paris is first author.
REFERENCES
- 1. Perrot R, Berges R, Bocquet A, Eyer J. Review of the multiple aspects of neurofilament functions, and their possible contribution to neurodegeneration. Mol Neurobiol. 2008;38:27–65. [DOI] [PubMed] [Google Scholar]
- 2. Bomont P. The dazzling rise of neurofilaments: physiological functions and roles as biomarkers. Curr Opin Cell Biol. 2021;68:181–191. [DOI] [PubMed] [Google Scholar]
- 3. Yilmaz A, Blennow K, Hagberg L, et al. Neurofilament light chain protein as a marker of neuronal injury: review of its use in HIV‐1 infection and reference values for HIV‐negative controls. Exp Rev Mol Diagn. 2017;17(8):761–770. [DOI] [PubMed] [Google Scholar]
- 4. Khalil M, Pirpamer L, Hofer E, et al. Serum neurofilament light levels in normal aging and their association with morpohologic brain changes. Nat Commun. 2020;11:812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gaiottino J, Norgren N, Dobson R et al. Increased neurofilament light chain blood levels in neurodegenerative neurological diseases. PLoS One. 2013;8(9):e75091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Håkansson I, Tisell A, Cassel P, et al. Neurofilament levels, disease activity and brain volume during follow‐up in multiple sclerosis. J Neuroinflammation. 2018;15:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khalil MT, Teunissen CE, Otto M, et al. Neurofilaments as biomarkers in neurological disorders. Nat Rev Neurol. 2018;14(10):577–589. [DOI] [PubMed] [Google Scholar]
- 8. Yuan A, Rao MV, Veeranna V, Nixon RA. Neurofilaments and neurofilament proteins in health and disease. Cold Spring Harb Perspect Biol. 2017;9(4):a018309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barro C, Benkert P, Disanto G, et al. Serum neurofilament as a predictor of disease worsening and brain and spinal cord atrophy in multiple sclerosis. Brain. 2018;141:2382–2391. [DOI] [PubMed] [Google Scholar]
- 10. Darras BT, Crawford TO, Finkel RS, et al. Neurofilament as a potential biomarker for spinal muscular atrophy. Ann ClinTransl Neurol. 2019;6(5):932–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Poesen K, De Schaepdryver M, Stubendorff B, et al. Neurofilament markers for ALS correlate with extent of upper and lower motor neuron disease. Neurology 2017;88(6):2302–2309. [DOI] [PubMed] [Google Scholar]
- 12. Poesen K, Van Damme P. Diagnostic and prognostic performance of neurofilaments in ALS. Front Neurol. 2019;9:1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zetterberg H, Skillbäck T, Mattsson N, et al. Association of cerebrospinal fluid neurofilament light concentration with Alzheimer disease progression. JAMA Neurology. 2016;73(1):60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mattsson N, Andreasson U, Zetterberg H, Blennow K. Association of plasma neurofilament light with neurodegeneration in patients with Alzheimer disease. JAMA Neurology. 2017;74(5):557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin YS, Lee WJ, Wang SJ, Fuh JL. Levels of plasma neurofilament light chain and cognitive function in patients with Alzheimer or Parkinson disease. Sci Rep. 2018;8(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Martinez‐Morillo E, Childs C, Prieto Garcia B, et al. Neurofilament medium polypeptide (NFM) protein concentration is increased in CSF and serum samples from patients with brain injury. Clin Chem Lab Med. 2015;53(10):1575–1584. [DOI] [PubMed] [Google Scholar]
- 17. Miller T, Cudkowicz M, Shaw PJ, et al. Phase 1–2 trial of antisense oligonucleotide tofersen for SOD1 ALS. N Engl J Med. 2020;383:109–119. [DOI] [PubMed] [Google Scholar]
- 18. Kuhle J, Barro C, Andreasson U, et al. Comparison of three analytical platforms for quantification of the neurofilament light chain in blood samples: ELISA, electrochemiluminescence immunoassay and Simoa. Clin Chem Lab Med. 2016;54(10):1655–1661. [DOI] [PubMed] [Google Scholar]
- 19. Li Y, Jung P, Brown A. Axonal transport of neurofilaments: a single population of intermittently moving polymers. J Neurosci. 2012;32(2):746–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xue C, Shtylla B, Brown A. A stochastic multiscale model that explains the segregation of axonal microtubules and neurofilaments in neurological diseases. PloS Comput Biol. 2015;11(8):e1004406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yuan A, Sasaki T, Rao MV, et al. Neurofilaments form a highly stable stationary cytoskeleton after reaching a critical level in axons. J Neurosci. 2009;29(9):11316–11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ferrer‐Alcon M, Garcia‐Sevilla JA, Jaquet PE, et al. Regulation of nonphosphorylated and phosphorylated forms of neurofilament proteins in the prefrontal cortex of human opioid addicts. J Neurosci Res. 2000;61(3):338–349. [DOI] [PubMed] [Google Scholar]
- 23. Byrne LM, Rodrigues FB, Blennow K, et al. Neurofilament light protein in blood as a potential biomarker of neurodegeneration in Huntington's disease: a retrospective cohort. Lancet Neurol. 2017;16(8):601–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. De Schaepdryver M, Jeromin A, Gille B et al. Comparison of elevated phosphorylated neurofilament heavy chains in serum and cerebrospinal fluid of patients with amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2018;89(4):367–373. [DOI] [PubMed] [Google Scholar]
- 25. Simoni G, Vo HT, Corrado P, Marchetti L. A comparison of deterministic and stochastic approaches for sensitivity analysis in computational systems biology. Brief Bioinform. 2020;21(2):527–540. [DOI] [PubMed] [Google Scholar]
- 26. Zi Z. Sensitivity analysis approaches applied to systems biology models. IET Syst Biol. 2011;5(6):336–346. [DOI] [PubMed] [Google Scholar]
- 27. McKay M, Beckman R, Conover WJ. A comparison of three methods for selecting values of input variables in the analysis of output from a computer code. Technometrics. 2000;42(1):55–61. [Google Scholar]
- 28. Goldsteinab ME, Sternbergerab NH, Sternbergerab LA. Phosphorylation protects neurofilaments against proteolysis. J Neuroimmunol. 1987;14(2):149–160. [DOI] [PubMed] [Google Scholar]
- 29. Shahim P, Tegner Y, Marklund N, Blennow K, Zetterberg H. Neurofilament light and tau as blood biomarkers for sports‐related concussion. Neurology. 2018;90:e1780–e1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shahim P, Zetterberg H, Tegner Y, Blennow K. Serum neurofilament light as a biomarker for mild traumatic brain injury in contact sports. Neurology. 2017;88(19):1788–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fasani F, Bocquet A, Robert P, Peterson A, Eyer J. The amount of neurofilaments aggregated in the cell body is controlled by their increased sensitivity to tripsin‐like proteases. J Cell Sci. 2004;117(6):861–869. [DOI] [PubMed] [Google Scholar]
- 32. Yuan A, Sershen H, Veeranna B, et al. Neurofilament subunits are integral components of synapses and modulate neurotransmission and behavior in vivo. Mol Psychiatry. 2015;20:986–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wilke C, Pujol‐Calderon F, Barro C, et al. Correlations between serum and CSF pNfH levels in ALS, FDT and controls: a comparison of three analytical approaches. Clin Chem Lab Med. 2019;57(10):1556–1564. [DOI] [PubMed] [Google Scholar]
- 34. Sano Y, Kanda T. Blood‐brain barrier and blood‐nerve barrier. In: Kusunoki S, ed. Neuroimmunological Diseases. Springer; 2016:55–69. [Google Scholar]
- 35. Abbott N, Patabendige AA, Dolman DE, Yusof SR, Begley DJ. Structure and function of the blood‐brain barrier. Neurobiol Dis. 2010;37(1):13–25. [DOI] [PubMed] [Google Scholar]
- 36. Kanda T. Biology of the blood‐nerve barrier and its alteration in immune mediated neuropathies. J Neurol Neurosurg Psychiatry. 2013;84(2):208–212. [DOI] [PubMed] [Google Scholar]
- 37. Paris A, Bora P, Parolo S, et al. A QSP model of neurofilament trafficking in SOD1 amyotrophic lateral sclerosis. ACoP11, ISSN:2688‐3953, 2020, Vol 2.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Supplementary Material


