Abstract
Germinal centers (GCs) are the site of immunoglobulin somatic hypermutation and affinity maturation, processes essential to an effective antibody response. The formation of GCs has been studied in detail, but less is known about what leads to their regression and eventual termination, factors that ultimately limit the extent to which antibodies mature within a single reaction. We show that contraction of immunization-induced GCs is immediately preceded by an acute surge in GC-resident Foxp3+ T cells, attributed at least partly to upregulation of the transcription factor Foxp3 by T follicular helper (Tfh) cells. Ectopic expression of Foxp3 in Tfh cells is sufficient to decrease GC size, implicating the natural upregulation of Foxp3 by Tfh cells as a potential regulator of GC lifetimes.
Effective, high-affinity antibodies arise via a Darwinian process of somatic hypermutation of immunoglobulin (Ig) genes and affinity-dependent selection of mutant B cells that takes place in germinal centers (GCs) (1). Prolonged residency of B cells in GCs, either in a single reaction or over multiple rounds of re-entry, can lead to the extraordinary levels of somatic hypermutation (SHM) and affinity maturation typical of broadly neutralizing antibodies (bNAbs) to HIV (2). GC reactions range in duration from 1 to 2 weeks when triggered by haptenated proteins in prime-boost (3) to several months in response to certain infections or to challenge with particulate antigens (4–8). In spite of this wide variability and of the critical importance of GC durability to antibody maturation, our understanding of the factors that determine the timing of GC contraction remains limited (1).
A key determinant of the GC life-course are the CD4+ T cells present within that structure. T follicular helper (Tfh) CD4+ T cells, characterized by expression of chemokine receptor CXCR5, inhibitory receptor PD-1, and transcription factor Bcl6, control the progression and output of the GC reaction by selectively driving proliferation of B cells with affinity-enhancing mutations (9–11). The positive effect of Tfh cells is counterbalanced by GC-resident T cells that express Foxp3, the master transcription factor of the regulatory T cell (Treg) lineage (12). The best characterized of these is a population referred to as T follicular regulatory (Tfr) cells (13–15), thought to regulate various aspects of the GC reaction such as B cell specificity and affinity, isotype switching, and emergence of self-reactivity (16–18). Given this central role of CD4+ T cells in sustaining the GC reaction, we sought to determine whether the dynamics of Foxp3 expression by GC T cells could play a role in GC contraction and termination.
Results
A surge in Foxp3+ GC-resident T cells precedes GC contraction
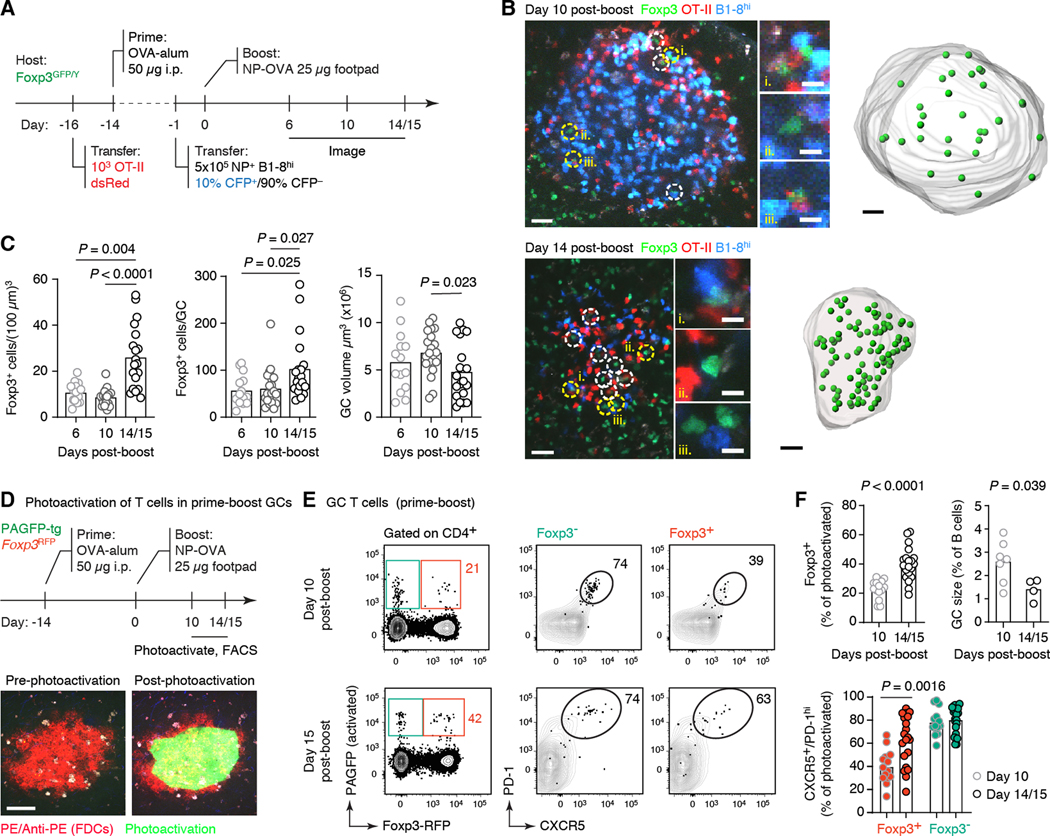
To follow the dynamics of Foxp3 expression by GC-resident T cells throughout the course of the GC reaction, we generated GCs in Foxp3GFP reporter mice (19) using the model antigen 4-hydroxy-3-nitrophenyl acetyl-ovalbumin (NP-OVA). To achieve tighter kinetics of GC dissolution, we employed a well-characterized adoptive transfer–prime–boost strategy (9, 20) (Fig. 1A). We quantified the number and density of recipient-derived GFP+ cells within popliteal lymph node (pLN) GCs, delineated based on the presence of adoptively transferred fluorescent B and T cells. Both parameters remained stable between days 6 and 10 post-boost (corresponding approximately onset and midpoint of the boost-induced GC reaction) (Fig. 1, B and C), averaging 56 and 62 cells per GC at a density of 9 and 11 cells per (100 μm)3, respectively. On day 14/15 post-boost, when GCs began to dissipate under these conditions, the GFP+ population increased in both density and number, to an average of 103 cells per GC, equivalent to 26 cells per (100 μm)3 (Fig. 1, B and C). This coincided with a decrease in the average volume of GCs determined by imaging, confirming that this time point corresponds to the contraction phase of the GC reaction (Fig. 1, B and C).
Fig. 1. Kinetics of Foxp3-expressing T cells throughout the course of the GC reaction.
(A) Experimental setup. (B) Multiphoton image of single optical slices of GCs at 10 and 14 days post-boost, followed by computational rendering of the entire GC volume. Foxp3+ cells are marked with dashed circles (images; yellow circles indicate cells magnified in the insets to the right) or green spheres (renderings). Scale bars: 30 μm (large panels) and 10 μm (insets). (C) Quantification of data as in (B). Each symbol corresponds to one GC. Two-to-three GCs were counted per mouse in at least three independent experiments per timepoint with two mice per group. (D) Experimental setup. GCs were identified by labeling follicular dendritic cells (FDCs) with anti-phycoerythrin (PE)–PE immune complexes prior to imaging and photoactivation (bottom). Scale bars: 100 μm. (E) Frequency and phenotype of photoactivated RFP+ and RFP– CD4+ T cells in single GCs. Each symbol represents one GC. Data are pooled from at least four independent experiments with the mean value indicated. Data for GC size in (F) are from two independent experiments, each symbol representing one mouse. All P-values are for paired Student’s t test, performed only for the comparisons indicated.
To validate these findings using an orthogonal approach, we performed in situ photoactivation (9, 20) of mice carrying a carrying a Foxp3RFP reporter (21) (Fig. 1D) to identify T cells based on their localization to the GC niche by flow cytometry. The proportion of Foxp3+ cells among photoactivated (PA+) GC-localized CD4+ T cells increased from 22% at day 10 to 39% at days 14–15 post-boost as measured using this system (Fig. 1, E and F). This increase coincided with a decrease in the proportion of B cells with GC phenotype by flow cytometry (Fig. 1F), mirroring the decrease in GC volume observed by imaging. Although only a minority of Foxp3+ CD4+ T cells inside early GCs expressed the very high levels of CXCR5 and PD-1 typical of the Tfh cell phenotype (20), this fraction increased towards the contraction phase of the GC (Fig. 1E). We observed similar trends in a primary immunization model, where the proportion of GC-localized T cells expressing Foxp3 increased from 24% at day 10 to 58% at day 20 post-immunization, the time point at which GCs recede in this setting, also while acquiring a more Tfh-like phenotype (fig. S1, A and B). The increase in Foxp3+ cells was concomitant with a decrease in the Foxp3– Tfh cell population, such that, while the total B-to-Tfh cell ratio remained fairly steady over time, the ratio of Foxp3– Tfh cells per B cell decreased by approximately 50% from early to late GCs (from 0.16 to 0.085 Tfh cells per GC B cell), indicative of lower T cell help availability (fig. S1C). Thus, contraction of the GC reaction is associated with a surge in GC-localized Foxp3-expressing T cells accompanied by loss of Foxp3– Tfh cells, such that the former account for a large fraction of the total T cells present in end-stage GCs.
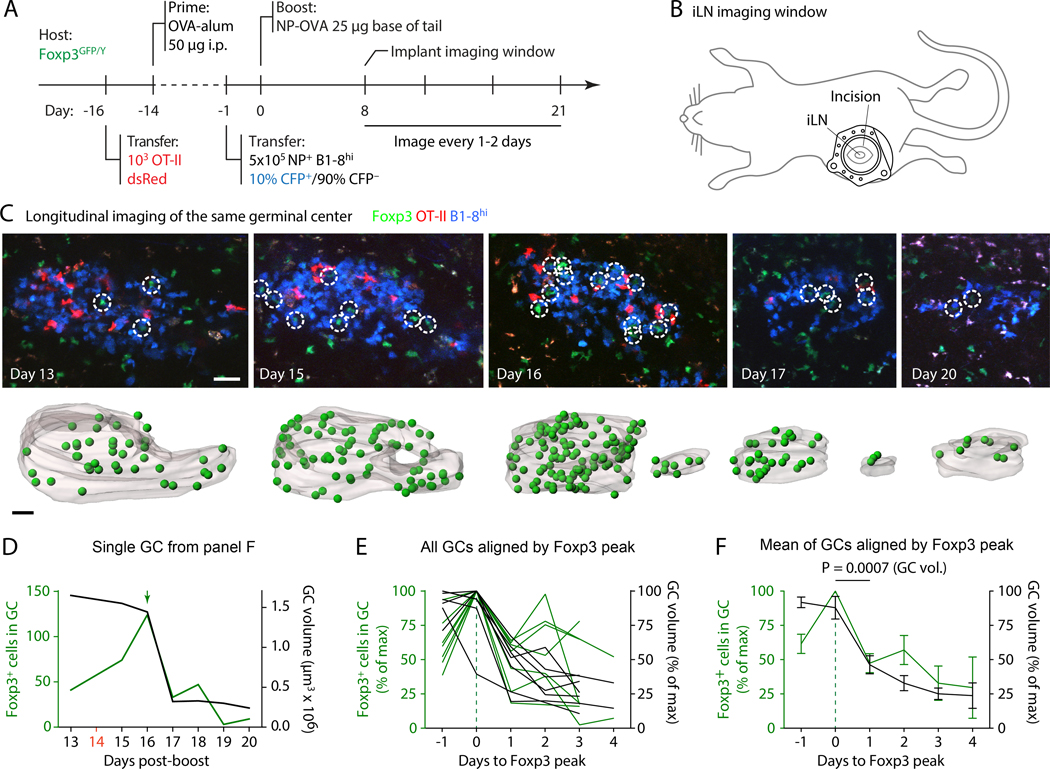
Immunization-induced GCs form with well-defined kinetics, whereas GC resolution is less stereotyped, and individual GCs shut down at different times within the same immune reaction (22). This lack of synchronicity prevents a precise definition of the temporal relationship between the increase in Foxp3+ T cells and end-stage GC contraction by cross-sectional analysis. To time the surge in Foxp3 T cells more precisely with respect to GC contraction, we imaged the same GCs longitudinally by implanting mice with an inguinal (i)LN imaging window (22). This enabled us to quantify changes in Foxp3+ T cell numbers in individual GCs as they approached their dissolution phase (Fig. 2, A and B; fig. S2A; and movie S1). Longitudinal imaging showed that the surge in Foxp3+ T cells most often takes place immediately prior to the onset of GC contraction (Fig. 2, C to F, and fig. S2, A and B). Aligning GCs in time by the peak of the number of Foxp3+ T cells revealed a doubling in the number of Foxp3+ T cells within the 24-hour period preceding the peak, which was directly followed by an almost 50% decrease in GC volume over the next 24 hours (Fig. 2, E and F). Thus, the surge in GC-localized Foxp3+ T cells immediately precedes GC contraction, consistent with these cells playing a direct role in this process.
Fig. 2. Longitudinal imaging of the evolution of Foxp3+ T cell numbers in the same GC.
(A) Experimental setup. (B) Cartoon depicting approximate placement of inguinal imaging window. (C) Images are collapsed 10-μm, 3-slice z-stacks, renderings are entire GCs. Foxp3+ cells are indicated by circles (images) or green spheres (renderings). Scale bars: 30 μm. (D) Quantification of the single GC shown in (A). Peak number of GFP+ cells is indicated by a green arrow, timepoints not imaged are shown in red on the x-axis. (E,F) Combined data for seven mice from six independent experiments in which the GFP peak could be identified, aligned by the peak of GFP+ T cells. Values given as percent of the maximum value in the dataset. Aggregate data in (F) are mean±SEM. Individual graphs for all GCs are shown in fig. S2B. P-value is for paired Student’s t test, performed only for the comparison indicated.
Foxp3+ T cells engage in prolonged interactions with B cells in end-stage GCs
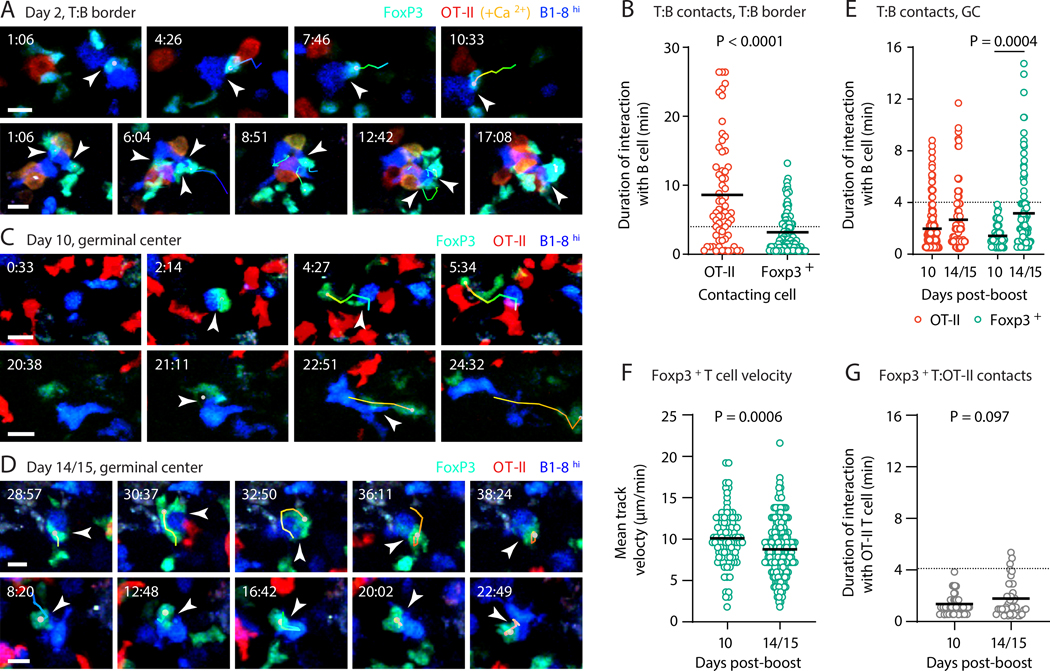
Given the evidence that direct contacts between Treg cells and GC B cells may contribute to the regulation of humoral responses (23), we investigated the interactions between B cells and Foxp3+ T cells at different times during GC evolution by intravital multiphoton microscopy. To establish that our system is capable of discerning Treg–B cell interactions, we first imaged cells at day 2 post-immunization, when effector T and B cell contacts taking place at the T cell zone–B cell follicle (T:B) border are long-lasting (24). We transferred CFP+B1–8hi B cells along with RFP+ OT-II T cells also expressing the fluorescent Ca2+ reporter GCaMP3 into Foxp3GFP hosts, and imaged pLNs at 2 days after footpad immunization with NP-OVA in alum. In this setting, Foxp3+ T cells and B1–8hi B cells engaged in clearly identifiable interactions, which, although generally briefer than those seen between B cells and helper T cells, could occasionally last for 10 min or longer (Fig. 3, A and B, and Movie 1). These interactions fell into two distinct modalities: either multiple Foxp3+ T cells “swarmed” over specific B cells, or B1–8hi B cells dragged a single Foxp3+ T cell behind them, as previously described for interactions with helper T cells (Fig. 3A and Movie 1) (24, 25). Thus, prolonged direct interactions between B cells and Foxp3+ T cells do take place and are detectable using our intravital imaging set-up.
Fig. 3: Dynamics of the interaction between B cells and Foxp3+ T cells.
(A) CFP+B1-8hi B cells and dsRed+GCaMP3+ OT-II T cells were adoptively transferred into Foxp3GFP hosts, which were then immunized in the footpad with 8 μg of NP-OVA in alum. Intravital imaging on pLNs was performed 48 hours later. Time series show interactions (indicated by arrowheads) between Foxp3+ T cells (green) and B1-8hi B cells (blue). OT-II T cells are in red (turning orange/yellow when fluxing calcium). Tracks of interacting Foxp3+ cells are shown. (B) Quantification of interactions between B1-8hi B cells and OT-II or Foxp3+ T cells at different time points. Each symbol represents one interaction. Only interactions lasting two frames or longer were included. A dotted line is placed at 4 min for reference. (C and D) Experimental setup as in Fig. 1A. Intravital imaging on pLN was performed on days 10 or 14/15 post-boost. Time series as described in (A). (E) Quantification of interactions in (C and D), as described in (B). (F) Mean track velocity for Foxp3+ T cells at early and late time points. Each symbol represents one track. (G) Quantification of interactions between host Foxp3+ T cells and transferred OT-II T cells at different time points. Details as in (B). Scale bars: 10 μm. Three movies from three independent experiments were analyzed for each dataset. (B), (E), (F) and (G) show pooled data, with each dot representing a cell and a bar representing the mean. Time stamps are in reference to the start of the movie (see Movies 1, 2, and 3). P-values are for Student’s t test.
Imaging of early (day 10) prime-boost GCs generated as in Fig. 1A revealed interactions between GC B cells and Foxp3+ T cells that were invariably short-lived, never exceeding 4 min of contact (Fig. 3, C to E, and Movie 2). On the other hand, we observed long-lived cognate interactions between GC B cells and OT-II Tfh cells at this time point. Both findings are largely consistent with previous reports (23, 26–28). Notably, a small number of GC-resident Foxp3+ T cells were stationary for extended periods at this time point. Crossing Foxp3GFP mice to the CD11c-YFP reporter strain (29) showed that many of these stationary cells were interacting with CD11c+ partners, most likely tingible body macrophages (TBMs) (Movie S2). In contrast to day 10, Foxp3+ T cell–B cell interactions at day 14/15 post-boost were much longer-lived (20% of observed contacts exceeded 4 min in duration), resembling the level of interaction between GC B cells and Tfh cells (Fig. 3, D and E, and Movie 3). Qualitatively, Foxp3+ T cells also engaged in cellular “entanglement” (26) morphologies characteristic of cognate Tfh–B cell interactions (Fig. 3D, Movie 3). Increased interaction with B cells was associated with a slight decrease in the mean velocity of Foxp3+ GC T cells at the late time point (Fig. 3F). However, this decrease in speed alone was not responsible for the increased interaction with B cells, since contacts between Foxp3+ T cells and transferred OT-II T cells did not increase over the same period (Fig. 3G). Thus, Foxp3+ T cells in early GCs engage only in limited interactions with GC B cells, but these interactions become more pronounced in end-stage GCs, where they resemble those of Tfh–B cell interactions in both duration and morphology.
Late-GC Foxp3+ cells arise through upregulation of Foxp3 by Tfh cells
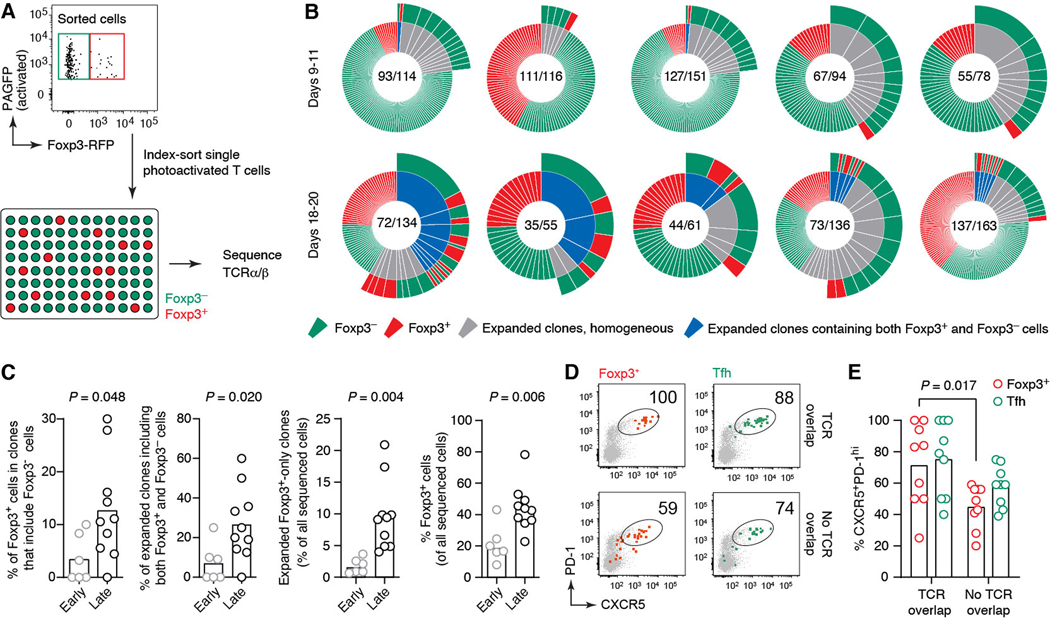
The more Tfh-like surface phenotype and dynamic behavior of late Foxp3+ GC T cells raises the possibility that at least a subset of these may have a distinct ontogeny from the canonical Tfr cells described at earlier time points, which arise primarily via acquisition of a Tfh-like program by thymic-derived (t)Treg cells (13–15). The idea of multiple ontogenies is supported by a prior report of differentiation of Tfr cells from conventional naïve T cells via a peripherally induced (p)Treg cell intermediate (30). To determine the lineage of late-GC Foxp3+ T cells, we sequenced the TCR rearrangements of Foxp3+ and Foxp3– T cells obtained from the same GCs by photoactivation (Fig. 4A). TCR diversity within single GCs was remarkably high, with an average of 75.5 distinct TCRs per 100 cells sequenced, corresponding to a D50 (percent of clones accounting for 50% of sequenced cells) of 0.34 (Fig. 4B). Despite this diversity, our sequencing data revealed a shift over time in the clonal relatedness of Tfh and Foxp3+ cells within the same GC: although Foxp3+ T cells whose TCRs overlapped with those of Foxp3– Tfh cells were rare at early time points (representing 3.4% of Foxp3+ or 0.8% of all T cells), they became much more frequent in late GC samples (12.6% of Foxp3+ or 4.7% of all T cells) (Fig. 4, B and C, and fig. S3A). The fraction of Foxp3+ T cells displaying detectable clonal expansion also increased over time, from 0.9% of all cells at the GC peak to 9.3% in end-stage GCs (Fig. 3, B and C). Index-sorting information showed that Foxp3+ GC T cells whose TCRs overlapped with Foxp3– Tfh expressed Tfh-like levels of CXCR5 and PD-1. This was not the case for the Foxp3+ GC T cells from the same GCs that were not clonally related to Tfh (Fig. 4. D and E). Overlap was also observable, albeit to a lesser extent, between TCR sequences obtained from RFP+ and RFP– CXCR5+PD-1hi T cells sorted from whole LN (rather than single GCs) of immunized Foxp3RFP mice (fig. S3B). Intranuclear staining of sorted CXCR5+PD-1hiRFP+ cells showed that >95% of these cells expressed Foxp3 protein, whereas none of the similarly sorted RFP– Tfh cells did so, confirming the accuracy of the fluorescent reporter (fig. S4). Thus, the late GC Foxp3+ cell surge appears to arise at least partially via the upregulation of Foxp3 expression by Tfh cells.
Fig. 4: A subset of late-GC Foxp3+ T cells arises via upregulation of Foxp3 by Tfh cells.
(A) Experimental setup. RFP+ and RFP– T cells from single photoactivated GCs obtained from Foxp3RFP+ PAGFP-tg mice at days 9–11 (early) or 18–20 (late) after primary immunization with NP-OVA were index-sorted for TCRα and β sequencing. (B) Clonal distribution of T cells within single GCs. Each pie chart represents one GC. Numbers are (number of clones/number of cells sequenced). Expanded clones (defined as those found more than once within the same GC) are colored in gray when including only one cell type or blue when including both cell types, with the cell type composition of the clone indicated in the outer circle. Additional pie charts can be found in fig. S3; full TCR sequences are available as Supplementary Data S1. (C) Quantification of data in (B), comparing early and late time points. Each symbol represents one GC. Includes additional late GCs not shown in (A). (D) Expression of Tfh markers among RFP+ and RFP– cells stratified by TCR overlap, obtained from index-sorting data. “Overlapping” cells are defined as those belonging to clones that contain both RFP+ and RFP– cells. (E) Quantification of data in (D). Each symbol represents one GC. Data are for 6 GCs from 5 independent experiments (days 9–11) and 10 GCs from 9 independent experiments (days 18–20). Bar indicates the median. P-values are for Student’s t test.
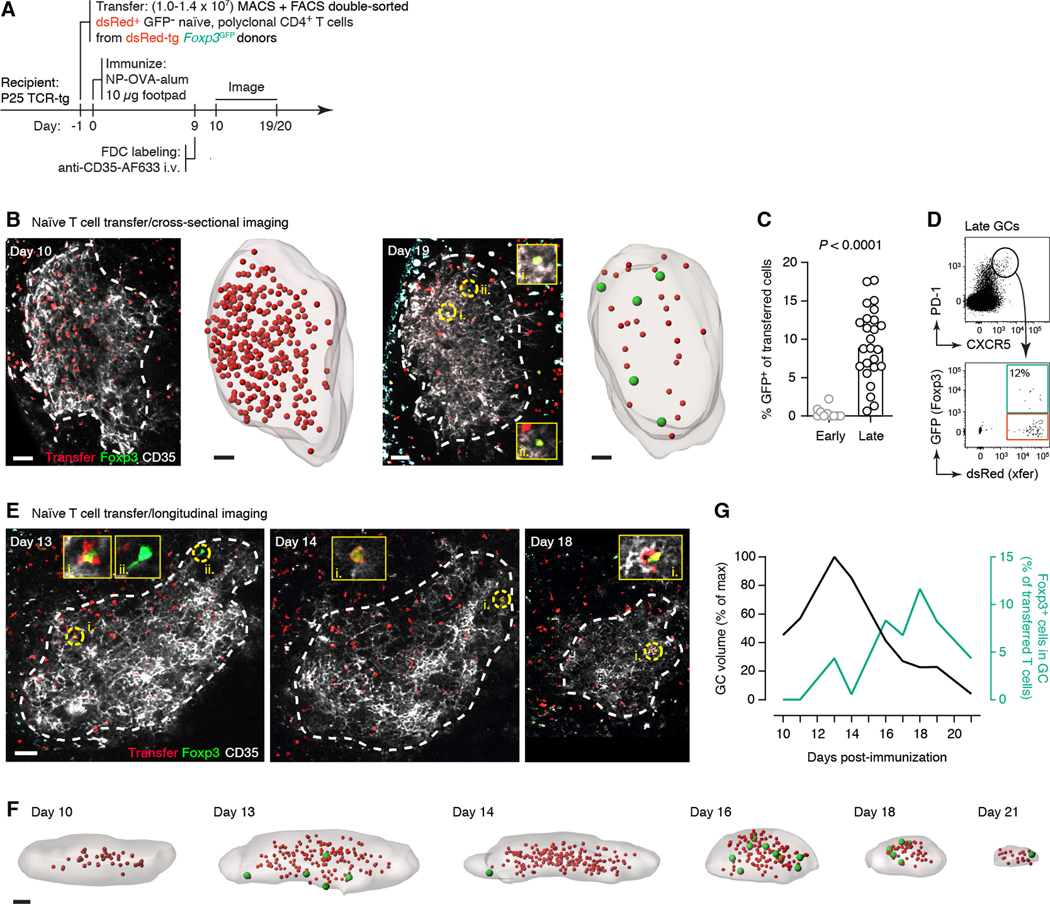
We next sought to determine whether we could achieve conversion of Foxp3– Tfh cells to a Foxp3+ state experimentally. Sorted Foxp3– Tfh cells readily became Foxp3+ when cultured in vitro in the presence of TGF-β (fig. S5), indicating that there is no impediment, epigenetic or otherwise, to acquisition of Foxp3 expression by this population. In vivo, polyclonal naïve CD4+dsRed+Foxp3– T cells from Foxp3GFP, dsRed-transgenic donors adoptively transferred into non-fluorescent recipients with irrelevant TCR specificity (Fig. 5A) were also able to upregulate Foxp3 expression in late GCs. In close agreement with our TCR sequencing results, GFP expression among adoptively-transferred GC-resident T cells increased from barely detectable (0.4% GFP+) in peak GCs to substantial (9.4% GFP+) at the end-stage time point (Fig. 5, B to D). To obtain better temporal resolution, we performed the same experiment longitudinally using an iLN imaging window. Corroborating our previous data, Foxp3 expression began to rise prior to the onset of GC collapse, peaking at approximately 10% of transferred cells at day 18 post-immunization (Fig. 5E–G). Thus, upregulation of Foxp3 by Tfh cells contributes to the surge in Foxp3+ cells that takes place in end-stage GCs.
Fig. 5. Upregulation of Foxp3 in late GCs by adoptively transferred naïve T cells.
(A) Experimental setup. (B) Sorted RFP+ GFP(Foxp3)– CD4+ T cells were transferred into P25 TCR-tg recipients, which were immunized with NP-OVA in alum and analyzed by multiphoton microscopy at days 10 (left) or 19–20 (right) post-immunization, as in fig. S3D. Images show single optical slices of GCs. GC cross-section (dotted white line) was defined based on in vivo FDC (CD35) labeling. RFP+GFP+ cells are indicated as yellow circles and magnified in the inset panels. Rendering shows full GC volumes with RFP+ cells shown as smaller red spheres and RFP+GFP+ cells shown as larger green spheres. (C) Quantification of data as in (F). (D) Representative flow cytometry plot from a late GC generated as in (A). (E and F) Longitudinal imaging of transferred RFP+ GFP(Foxp3)– T cells. Details as in (A). iLN window was mounted on day 8 post-immunization. Data is for a single experiment. All scale bars are 50 μm. (G) Quantification of data in (E and F). P-values are for Student’s t test. In (C), each dot represents one GC from 3 mice in 3 independent experiments (early) and 5 mice in 3 independent experiments (late). Bar indicates the median. Data in (E and F) are for a single experiment.
Late-GC Foxp3+ T cells display an intermediate phenotype between Tfh and Tfr cells
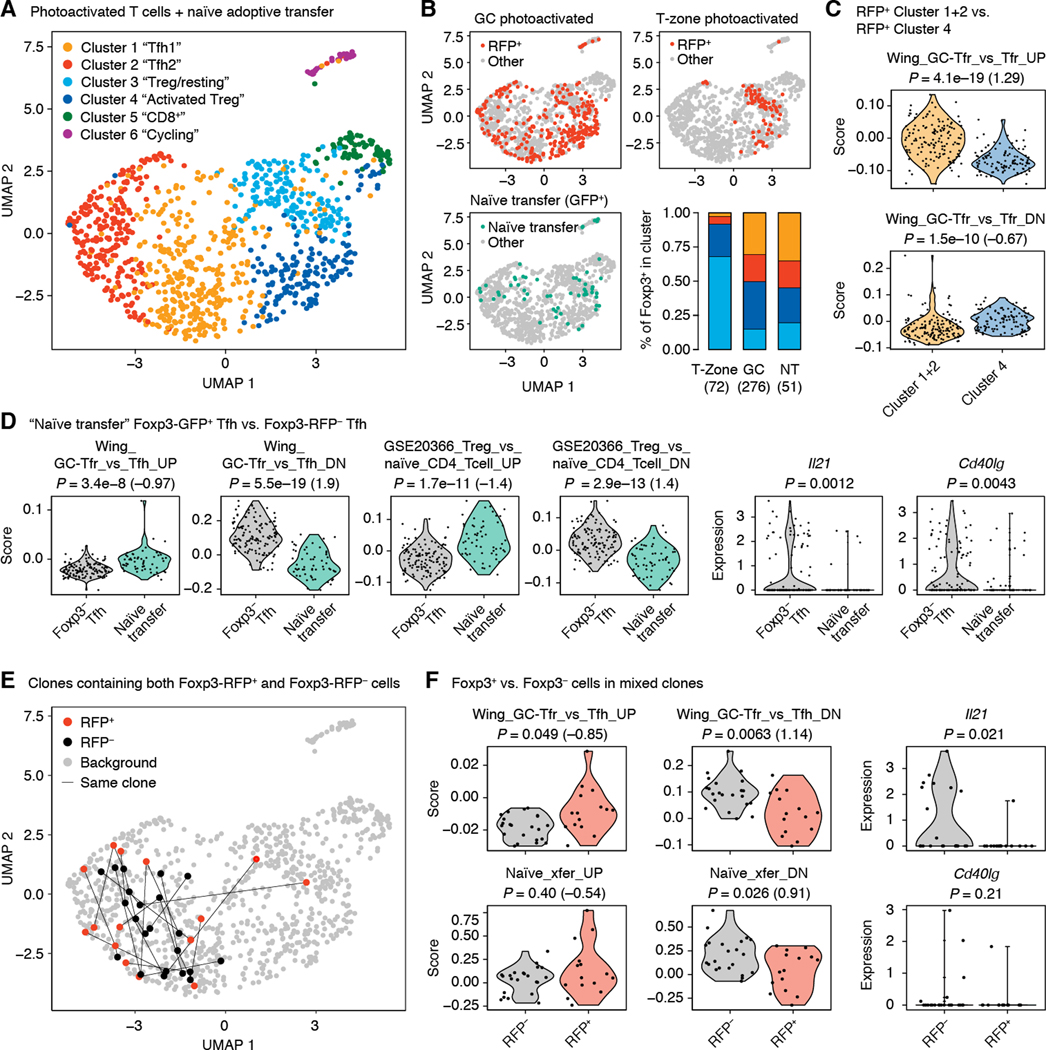
To determine what effect Foxp3 expression has on Tfh cells, we carried out whole-transcriptome single-cell RNA-sequencing (scRNA-seq) on T cells sorted from individual photoactivated GCs at days 10 or 20 post-immunization as described earlier (fig. S1). For reference, we also sorted a sample of Foxp3+ Tfh-phenotype T cells converted from adoptively transferred naïve precursors (Fig. 5) and one plate of Foxp3+ T cells photoactivated in the T-zone (table S1). The 968 cells that passed the quality threshold fell into six major clusters (Fig. 6A, and fig. S6, A to D). RFP+ T cells were distributed across Clusters 1 and 2 (Tfh1 and Tfh2, respectively), 3 (Treg/resting), and 4 (Activated Treg), indicative of heterogeneity among this population (Fig. 4B). As expected, T-zone Treg cells were found almost exclusively in Clusters 3 and 4. By contrast, RFP+ cells from photoactivated GCs were also often found in the two Tfh Clusters, as were naïve transfer-derived CXCR5+PD-1hiFoxp3+ cells (Fig. 6B). This was confirmed when only cells with detectable Foxp3 mRNA were analyzed (fig. S6E). Thus, two major populations of Foxp3+ T cells are present in GCs, those that are Tfh-like (Clusters 1 and 2) and those that resemble T-zone Treg cells (Clusters 3 and 4). The relative absence of prototypical Treg transcript Il2ra expression (encoding for CD25) (fig. S6C) in Foxp3+ T cells from Clusters 1 and 2 suggested these cells may resemble a previously described GC-resident CD25–Foxp3+ population (“GC-Tfr”) with a hybrid Tfh/Treg phenotype (31). Indeed, transcriptional signature analysis showed that Foxp3+ cells within Tfh Clusters 1/2 resemble GC-Tfr cells, whereas Cluster 4 Foxp3+ cells resemble the canonical CD25+ Tfr population (Fig. 6C). Thus, although Cluster 1/2 Foxp3+ T cells are Tfh-like, expression of Foxp3 shifts these cells towards a transcriptional state similar to that of CD25– GC-Tfr cells.
Fig. 6. scRNA-seq of GC-resident T cells.
Total T cells (Foxp3RFP+ and Foxp3RFP– combined) from single photoactivated GCs were sorted either as B220–/CD4+/PA+ or as B220–/TCRβ+/PA+ (which also includes CD8+ T cells) in different samples at days 10 or 20 after primary immunization with NP-OVA. For comparison, we also sorted Treg cells photoactivated in the T-zone of an unimmunized mouse, sorted as B220–CD4+GFP+RFP+ and Foxp3+CXCR5+PD-1hi Tfh cells derived from transferred naïve precursors as in Fig. 5. (A) UMAP plot showing clustering of T cells according to whole transcriptome analysis. (B) Distribution of different subsets of Foxp3-reporter+ cells in UMAP space. Cells in color are those that are Foxp3-reporter+ in the sample indicated in the graph title. All other analyzed cells are shown in gray. Bar graph shows distribution of T-zone and GC RFP+ cells among clusters 1–3. (C) Expression of selected gene signatures by Foxp3RFP+ and Foxp3RFP– cells within Tfh Clusters 1 and 2. (D) Expression of selected genes or gene signatures by Foxp3+ Tfh cells derived from transferred naïve precursors compared to photoactivated Foxp3– Tfh cells. Only cells from day 20 post-immunization are included in the Foxp3– Tfh group. “GSE20366” signatures are from the Broad Institute’s MSigDB database. (E) Distribution of clonal expansions (defined as TCRs detected more than once in the same GC) containing both RFP+ and RFP– cells (“mixed clones”). Cells carrying identical TCRs are linked by lines. (F) Expression of selected genes or gene signatures by Foxp3-RFP+ and Foxp3-RFP – cells within mixed clones. (A-F) Each symbol represents one cell. P-values are for the Wilcoxon signed-rank test. Numbers in parentheses are Cohen’s d for effect size. The number of GCs included and number of independent experiments are indicated in Table S1.
To determine whether this hybrid transcriptional state is also observable in Foxp3+ cells known to be derived from Foxp3– Tfh precursors, we took two approaches. We first compared gene expression between Foxp3– Tfh cells in photoactivated late GCs and spiked in Foxp3+ Tfh cells derived from transferred naïve T cells. This comparison showed evidence of acquisition by naïve-transferred Foxp3+ cells of both the CD25– GC-Tfr program (31) and of other Treg-associated signatures (32) (Fig. 6D), as well as loss of expression of T cell help-associated genes such as Il21 (33) and Cd40lg (34). As a second, more strict approach, we defined Foxp3+ T cells of Tfh origin by determining the TCR sequences of day 20 GC T cells using scRNA-seq data confirmed by long-read PCR based sequencing. Eleven clonal expansions contained both Foxp3+ and Foxp3– cells. With few exceptions, Foxp3+ and Foxp3– cells within these “mixed” clones remained predominantly within Tfh Clusters 1 and 2 and were not found within Cluster 4, likely to include most of the canonical Tfr cell population (Fig. 6E). However, despite the small number of cells in this analysis, Foxp3+ cells showed statistically detectable modulation of GC-Tfh signature genes and of a set of 30 genes downregulated by Foxp3+ Tfh derived from transferred naïve precursors (Fig. 6F and fig. S7). This included detectable downregulation of Il21, while Cd40lg mRNA was not well captured in this sample (Fig. 6F). Thus, although Foxp3 expression is insufficient to fully convert late Tfh cells into the Treg or Tfr phenotype, it is associated with the induction of Treg-associated transcriptional changes, suggesting that Foxp3 may play a functional role in these cells.
Foxp3 upregulation by Tfh cells promotes contraction of late GCs
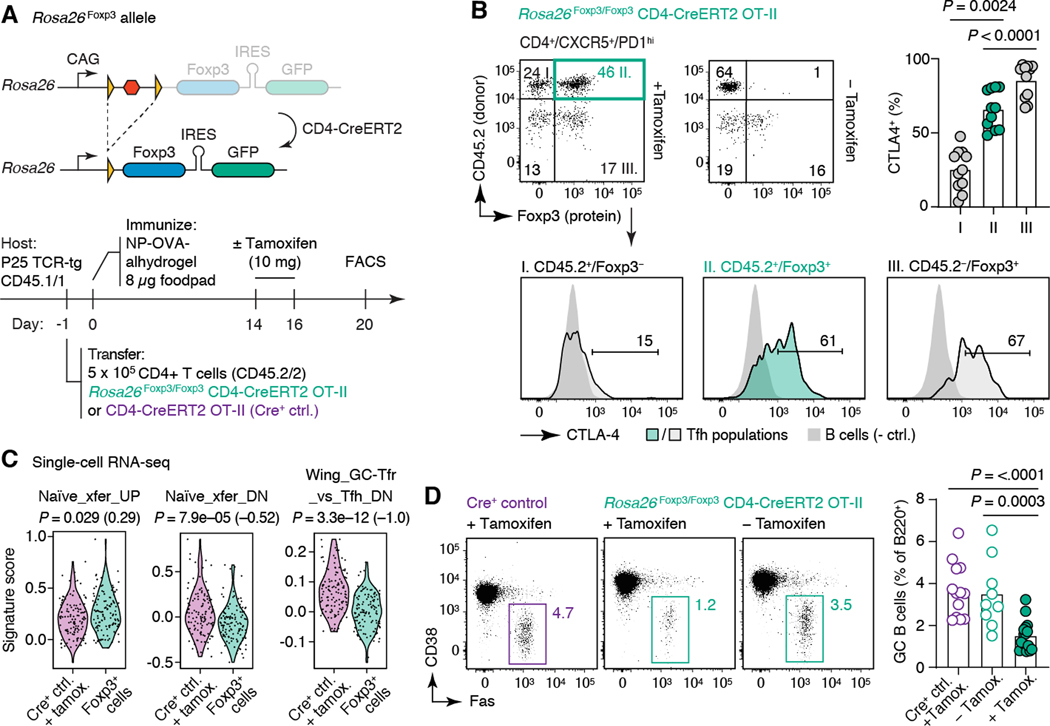
To determine whether acquisition of Foxp3 by late-stage Tfh cells can promote GC shutdown, we generated mice carrying an inducible Rosa26Foxp3 allele, where near-physiological expression of Foxp3 protein, followed by a GFP reporter, is conditional upon removal of a loxP-flanked stop cassette by cre-mediated recombination (Fig. 7A and fig. S8). To acutely induce Foxp3 expression by Tfh cells, we adoptively transferred CD4+ T cells from Rosa26Foxp3 CD4-CreERT2 OT-II mice into allelically marked CD45.1 P25 TCR-transgenic hosts, which we then immunized in the footpad with NP-OVA in alhydrogel (Fig. 7A). This protocol generates larger, longer-lived GCs, in which roughly 70% of Tfh cells derive from the donor mouse. Notably, OT-II Tfh cells are refractory to spontaneously upregulating Foxp3 expression even at later timepoints (fig. S9), in line with previous studies of Tfr differentiation using this TCR (13, 35). Tamoxifen administration led to detectable expression of Foxp3 protein in approximately 40% of transferred T cells (corresponding to approximately 30% of all Tfh cells), at levels that matched those of T cells naturally expressing Foxp3 in the Tfh gate (Fig. 7B). Rosa26Foxp3-expressing cells upregulated Treg markers cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) and, to a lesser extent, glucocorticoid-induced tumor necrosis factor receptor-related protein (GITR), but not CD25 (Fig. 7B and fig. S10). scRNA-seq of OT-II Tfh cells forced to express Foxp3 compared to control OT-II CD4-CreERT2+ T cells lacking the Rosa26Foxp3 allele recapitulated the changes observed during physiological upregulation of Foxp3+ within mixed Foxp3+–Foxp3– Tfh clones (Fig. 6, E and F). Although expression of Foxp3 alone was again not sufficient to segregate Tfh cells into different nearest-neighbor clusters (fig. S10C), Foxp3+ cells showed clear changes in the expression of both “naïve transfer” (fig. S7) and “GC-Tfh” (31) signatures, including limited but detectable loss of Il21 and Cd40lg mRNAs (Fig. 7C and fig. S10D). Thus, forced expression of Foxp3 induces Tfh cells to modulate expression of a Treg-associated transcriptional program similar to that acquired under physiological conditions. Most importantly, ectopic expression of Foxp3 in peak GC Tfh cells led to a roughly 60% reduction in GC size compared to control mice that did not receive tamoxifen or tamoxifen-treated mice receiving Cre+ control T cells (Fig. 7C). Thus, Foxp3 expression by Tfh cells is sufficient to both promote a Treg-like phenotype in Tfh cells and to accelerate the contraction of the GC reaction.
Fig. 7. Ectopic expression of Foxp3 in peak-GC Tfh cells promotes GC contraction.
(A) Top, design of the Rosa26Foxp3 allele for inducible expression of Foxp3. Further details in fig. S8. Bottom, experimental setup. (B) Induction of Foxp3 protein and CTLA-4 in Tfh cells. CTLA-4 MFI is quantified in the top-right panel; each symbol represents one mouse from three independent experiments. P-values are for one-way ANOVA with Dunnett’s post-test (comparing the experimental group to all other groups). (C) Expression by scRNA-seq of selected genes or gene signatures by Foxp3+ OT-II Tfh cells (GFP+CXCR5+PD-1hi) from Rosa26Foxp3 mice compared to Foxp3– OT-II Tfh cells from Cre+ control mice. Each symbol represents one cell, pooled from three experimental and three control mice sorted from a single experiment. The Sakaguchi_GCTfr_vs_Tfh_UP signature was not included because only three genes from this signature were detected the scRNA-seq dataset. P-values are for Wilcoxon signed-rank test, numbers in parentheses are Cohen’s d for effect size. (D) GC contraction upon forced expression of Foxp3 in Tfh cells. Quantified in bottom-right panel; each symbol represents one mouse from 3–4 independent experiments. Bar indicates the mean. P-values as in (B).
Discussion
Our data support a model in which the contraction and eventual shutdown of late-stage GCs is driven at least in part by a surge in Foxp3+ GC T cells. This surge is driven at least in part by acquisition of Foxp3+ by Tfh cells, which join pre-existing tTreg-derived Tfr and GC-Tfr populations to dramatically increase Foxp3+ T cell density in late GCs.
Two major considerations underlie our conclusion that late-GC Foxp3+ T cells originate partly from Tfh precursors. First, intravital imaging showed that the quality of interactions between Foxp3+ T cells and GC B cells changes markedly with time, from an early preponderance of the brief interactions typical of Tfr cells (23) to the appearance of the long-lived entanglements typical of Tfh cells (26) at the time of the Foxp3 surge. This suggests either that Tfr cells undergo a dramatic change in dynamic behavior at late stages, or, more likely, that they are joined in their ranks by a second population of Foxp3+ T cells with Tfh-like behavior. Second, and most important, end-stage Foxp3+ T cells share TCR sequences with Foxp3– Tfh cells, indicating a common precursor. Our finding that adoptively-transferred Foxp3– naïve T cells upregulate Foxp3 in late but not early GCs suggests that, in this particular case, the direction of change is from Foxp3– to Foxp3+, rather than in the opposite direction as observed in early GCs using a fate-mapping model (36). This timing also suggests that late-GC Foxp3+ Tfh cells are unlikely to be derived from peripherally-induced (p)Treg cells, since these would be expected to be present in earlier-stage GCs as well (30).
Critically, expression of Foxp3 is not sufficient to trigger Tfh cells to adopt a full-fledged Tfr or Treg phenotype. Instead, Tfh cells resemble more closely a previously described CD25– “GC-Tfr” population (31), which has an intermediate phenotype between Tfh and canonical Tfr cells. Given that GC-Tfr cells are present also in early GCs and are thought to derive from Tfr precursors (31), it is likely that late-GC Foxp3+ Tfh cells are not identical to GC-Tfr cells, but rather assume some of their transcriptional characteristics and possibly regulatory properties.
It is unclear what triggers the upregulation of Foxp3 by Tfh cells in end-stage GCs. TGF-β was capable of inducing Foxp3 expression in Tfh cells cultured in vitro (fig. S5) and has been shown to be active within the GC environment in vivo (37), suggesting that this cytokine at least has the potential to play a role in Foxp3 upregulation by Tfh cells. Another potential factor is the availability of antigen for TCR stimulation. Although antigen is known to be maintained on follicular dendritic cells for long periods (38), its progressive consumption by GC B cells may eventually lead to suboptimal TCR engagement, which has been shown to favor Treg generation from naïve T cells (39). A role for TCR signals in controlling Foxp3 upregulation is supported by the finding that this does not take place in adoptively-transferred OT-II TCR-transgenic T cells, although it is readily observable when polyclonal naïve T cells are transferred.
Two possible explanations for how Foxp3 expression by Tfh cells could favor GC contraction are passive loss of B cell helper capacity by the Foxp3+ Tfh cells themselves, or an active suppressive effect of these cells on Foxp3– Tfh cells. In our gain-of-function model, forcing physiological levels of expression of Foxp3 in Tfh cells led to upregulation of CTLA-4, a hallmark of the Treg phenotype that has been suggested to contribute to Tfr-mediated suppression in trans (40). However, such upregulation was not noted in late-GC Tfh cells naturally expressing Foxp3 (not shown), possibly because Ctla4 expression by Tfh cells is already high at this time point. On the other hand, loss of Tfh help in late GCs is suggested quantitatively by the increased GC B cell to Foxp3– Tfh cell ratio seen in late GCs (fig. S1B). Foxp3+ Tfh cells also lose expression of the two key effectors of T cell help to B cells, Il21 and Cd40lg (34, 41–43), both under physiological conditions and upon ectopic expression. As a true loss-of-function model where Foxp3 can be ablated specifically in Tfh but not Tfr cells is currently unavailable, the mechanism of action, as well as the relative contribution to GC contraction of Tfh cell Foxp3 versus other factors, remain to be determined.
The termination of the GC reaction is a relatively understudied phenomenon that is likely to play an important role in controlling the extent of somatic hypermutation and affinity maturation achievable by a B cell clone (1). The finding that the state of Tfh cells can play a role in GC shutdown raises the possibility that this is an active process rather than simply a result of the progressive consumption of antigen by GC B cells. Manipulating this process by interfering with normal Tfh kinetics may thus provide an avenue towards extending GC lifetime, potentially contributing to the induction of highly mutated antibodies by vaccination.
Materials and Methods
Mice
Wild-type C57BL6/J mice, transgenic mice with ubiquitous expression of CFP (B6.129(ICR)-Tg(CAG-ECFP)CK6Nagy/J) (44) and dsRed (B6.Cg-Tg(CAG-DsRed*MST)1Nagy/J) (45), P25 TCR-transgenic mice (C57BL/6-Tg(H2-Kb-Tcra,-Tcrb)P25Ktk/J) (46), and Foxp3IRES-RFP mice (C57BL/6-Foxp3tm1Flv/J) (21) where purchased from Jackson Laboratories. PAGFP-transgenic (Tg(UBC-PA-GFP)1Mnz/J) (9), B1–8hi (CBy.129P2(B6)-Ightm1Mnz/J) (47), and Y-chromosome OT-II TCR transgenic (Tg(TcraTcrb)426–6Cbn) (48) mice were bred and maintained in our laboratory. Mice ubiquitously expressing GCaMP3 were generated by crossing the Rosa26Lox-Stop-Lox-GCaMP3 strain purchased from Jackson Laboratories (Gt(ROSA)26Sortm1(CAG-GCaMP3)Dbe) (49) to a germline cre deleter allele, which was subsequently bred out. Foxp3IRES-GFP (Foxp3tm1Kuch) (50) mice were a kind gift from V. Kuchroo (Brigham and Women’s Hospital). This strain was crossed to the autosomal OT-II TCR (B6.Cg-Tg(TcraTcrb)425Cbn/J) and CD4-CreERT2 (B6(129X1)-Tg(Cd4-cre/ERT2)11Gnri/J) (48) alleles, both obtained from Jackson Laboratories. “Cre-control” donors used for Rosa26Foxp3 experiments were CD4-CreERT2 crossed to the Y-chromosome OT-II TCR transgenic (Tg(TcraTcrb)426–6Cbn) mice. There are reportedly no differences in T cell responses between the autosomal and Y-chromosome OT-II strains (48). All strains were either generated on the C57BL/6 background or backcrossed a sufficient number of times to allow adoptive transfer between mice. Mice were maintained under SPF conditions in the Rockefeller University’s Comparative Biosciences Center or in the MSKCC animal facility (Rosa26Foxp3 strains). All procedures were approved by the Rockefeller University or MSKCC animal research ethics committees.
Generation of the Rosa26LoxP-Stop-LoxP-Foxp3 (Rosa26Foxp3) allele
The targeting vector was generated by modifying a previously reported targeting vector expressing Cre-inducible STAT5b-CA (51) by substituting the Stat5b-CA sequence with the Foxp3 coding sequence. The targeting vector was linearized and electroporated into albino C57BL/6 ES cells. After neomycin selection, Southern blot screen and karyotyping, correctly targeted clones were injected into WT C57BL/6 blastocysts. The resulting chimeric mice were bred to albino C57BL/6 mice. Founders identified based on the coat color and genotyping were then bred to WT C57BL/6 mice.
Adoptive cell transfers
Spleens were homogenized by filtering through a 70-μm cell strainer and red blood cells were lysed with ACK buffer (Thermo Scientific). Resting T and B cells were purified by negative magnetic-activated cell sorting (MACS) using the mouse CD4+ T cell Isolation Kit and anti-CD43 Microbeads (Miltenyi Biotec), respectively, according to the manufacturer’s protocol. For all imaging experiments except intravital imaging of T–B border interactions, NP-binding B cells were quantified by flow cytometry using NP-PE (ThermoFisher), and total B cells containing the specified number of NP-binding B cells were transferred. When imaging T–B border interactions, we enriched for Igλ+ B cells by incubating splenocytes with anti-Igκ PE (Clone 187.1, BD Biosciences) at 0.7 μg/ml for 30 min prior to MACS using anti-CD43 and anti-PE Microbeads combined (Miltenyi Biotec). For transfers of resting CD4+ Foxp3– cells, negative MACS isolation of CD4+ T cells was followed by FACS of unstained GFP– dsRed+ cells prior to adoptive transfer. For characterization of the Rosa26Foxp3 mouse, CD4+ T cells were enriched using the Dynabeads Untouched Mouse CD4 Cells Kit (Life Technologies 11415D). A total of 1.5×107 enriched CD45.2+ CD4 T cells from Foxp3RFPRosa26 Foxp3 mice positive or negative for the CD4-CreERT2 transgene were transferred retro-orbitally into CD45.1+ recipients.
Immunizations and treatments
For the induction of primary GCs, mice were immunized subcutaneously in the hind footpad with 10 μg of NP-OVA (Biosearch Technologies) precipitated in alum (Imject® Alum, Thermo Scientific) at a 2:1 antigen (PBS):alum ratio, for a final volume of 25 μl. To generate longer-lived primary GCs for Rosa26Foxp3 experiments, mice were immunized with 60 μg NP-OVA in alhydrogel (InvivoGen) at a 1:1 antigen (PBS):hydrogel ratio. For Foxp3 induction in Tfh ex vivo, mice were immunized with 10 μg or 20 μg of NP-OVA in alhydrogel in the footpad and base of tail respectively. For the induction of prime-boost GCs, mice were first primed i.p. with 50 μg of OVA (Sigma) precipitated in alum at a 2:1 antigen (PBS):alum ratio for a final volume of 100 μl. Two-to-three weeks later, mice were boosted with 25 μg of a 1 μg/ml solution of NP-OVA (Biosearch Technologies) in PBS the hind footpad. For CD80 blocking experiments, mice were injected i.v. with 350 μg anti-CD80 (16–10A1, Bio-X-cell) or with the same amount of the recommended isotype control (InVivoMAb polyclonal Armenian hamster IgG, Bio-X-cell) on days 16, 17, and 19 post-immunization. FDCs were labeled by i.v. injection of 10 μg of non-blocking monoclonal antibody to CD35 (clone 8C12, either a generous gift from M. Carroll, Harvard Medical School or produced in our laboratory from a hybridoma kindly provided by J. Cyster, UCSF) conjugated to Alexa Fluor 633 or Alexa Fluor 594 (Thermo Fisher), 24 hours prior to imaging (52). For photoactivation experiments, FDCs were labeled 4 days prior to imaging by intraperitoneal injection of 20 μg of rabbit polyclonal anti-B-PE antibody (Rockland) followed by footpad injection of 10 μg of B-PE (Molecular Probes/ThermoFisher), as described previously (28). Recombination of the Rosa26Foxp3 floxed allele by CD4-CreERT2 was induced by gavage with 10 mg of tamoxifen (Sigma) dissolved in 100 μl of corn oil (Sigma) on days 14 and 15 post-immunization (Fig. 7) or by 4 mg of tamoxifen on days 1 and 2 post-transfer (Fig. S8).
Sample processing for flow cytometry and cell sorting
LNs or fragments were placed into microcentrifuge tubes containing 100 μl of PBS supplemented with 0.5% BSA and 1 mM EDTA (PBE) and macerated using disposable micropestles (Axygen). One hundred microliters of 2X antibody stain containing Fc-block plus fluorescent antibodies (see table S2) was added to the cell suspension, which was incubated on ice for 40 min. Cells were filtered and washed before analysis on BD FACS LSR II or sorting on BD ARIA II (BD Biosciences). Intracellular stains were performed according to the manufacturer’s protocols using the Foxp3/Transcription Factor Staining Buffer Set (eBioscience). Data were analyzed with Flowjo v. 10.0.7r2 (Tristar) or FCS express v. 7 (DeNovo Software).
Multiphoton imaging and photoactivation
Imaging was performed on an Olympus FV1000 upright microscope with a 25X 1.05NA Plan water-immersion objective, a Mai-Tai DeepSee Ti-Sapphire laser (Spectraphysics), and four photomultiplier tubes. Fluorescence emission from CFP, GFP, and YFP was collected in two channels, using a pair of CFP (480/40 nm) and YFP (525/50 nm) filters separated by a 505 nm dichroic mirror, with GFP appearing as positive in both channels. A third filter was used for RFP, PE, or Alexa Fluor 495 (605/70 nm) and Alexa Fluor 633 (665/40 nm). The excitation wavelength was 910 nm for all fluorochromes except Alexa Fluor 633, which was imaged at 810 nm.
Single-timepoint intravital imaging was performed as described (9). 4D datasets were acquired as nine 40 μm-deep z-slices (at 5-μm increments) with 1.5× zoom and 512 × 512 x–y resolution. Anesthesia was induced by inhalation of 4% isoflurane and maintained on 1.25% isoflurane. For pLN exposure, hind legs were shaved using a razor blade and mice were restrained on a stage warmer set to 37°C (BioTherm Micro S37; Biogenics). pLN were exposed by an incision behind the knee joint and held in position using a metallic strap. Mice were placed under the microscope objective, connected to an objective heater set to 40°C.
Longitudinal iLN window imaging was performed as previously described (22, 53). Mice were approximately 8 weeks of age. Windows were mounted at 8 days post-boost (or post-immunization for naïve transfer experiments) on mice prepared using standard surgical procedures and placed in supine position. The surgical area bounded by the femoral region and the hypochondriac region and the ventral and dorsal midlines was shaved and washed with ethanol and betadine. The inguinal lymph node was exposed by an incision in the regio inguinalis after locating the node from the ventral side by shining a bright light through the skin from the dorsal side and/or using the nipple of the fourth mammary fat pad to estimate the approximate position. The iLN window was mounted as described (22), with subsequent intravital imaging starting at 48 hours post-surgery under isoflurane anesthesia as above on a specially designed stage with a fixture for window positioning.
Photoactivation was performed as described (9, 20, 54). pLN from PAGFP mice were first imaged at 950 nm, at which no photoactivation is observed, to visualize PE/anti-PE immune complexes indicating FDCs. A 3D region of interest internal to the boundaries of the FDC network was photoactivated at 820 nm. Either one or two GCs were photoactivated in each LN. In either case the LNs were cut into fragments containing a single GC using a disposable razor blade (Astra) under a Fluorescent Stereomicroscope (Leica M165 FC). For scRNA-seq analysis, total T cells (RFP+ and RFP– combined) were sorted either as B220–CD4+PA+ or as B220–TCRβ+PA+ (which includes GC-localized CD8+ T cells) in different samples.
Image analysis
ImageJ v. 1.52a (NIH) and Imaris v. 9.1.2 and 9.5.1 (Bitplane) were used for Image analysis. Cells were counted manually using the Spots and OrthoSlicer functions in Imaris. GC size was estimated as a volume calculated from the manual surface renderings based the boundaries set by GC B cells and T cells or by GC B cells and FDC stain depending on the experimental setup. To quantify T cell–B cell contacts, T cells were first manually tracked as surfaces (T–B border) or spots (GC). B cells were rendered automatically by first creating a new CFP-only channel using the colocalization tool. B cells were then rendered using this new channel as either surfaces (T–B border) or spheres (GC). To estimate contacts between B cells and T cells, we used an Imaris XTension that applies a Euclidian Distance Transformation (DT) function to the surfaces (Imaris 9.1.2). The DT function was visualized as a new channel based on B cell surfaces/spheres. Each pixel intensity value in this channel is a measure of the closest distance in μm away from the B cell surface/sphere. As T cells were already rendered as Imaris objects, their distance to the nearest B cell at any given timepoint is represented by DT channel intensity. We set the threshold for a B cell interaction as <11 μm from a B cell center to the same T cell center for a duration >2 frames (for spheres) and as <2 μm from surface of a B cell to the surface of the same T cell for a duration of >2 frames (for surfaces). For track visualization in all movies we used dragon tails displaying the last 300 s of track. All movies were acquired at 30 frames/s and are presented at 7 frames/s (210× real-time). Adobe Photoshop CC was used for final movie editing.
T cell culture
Foxp3-RFP– Tfh and non-Tfh CD4+ T cells were sorted from Foxp3IRES-RFP reporter mice immunized with NP-OVA in alhydrogel, on day 10 post-immunization. T cells were added to 96-well round bottom well plates with antigen-presenting cells at a 1:2 ratio, with 5000 cells/well in 200 μl of RPMI (Corning 10–040-CV) supplemented with 10% FBS (Gemini 100–106), 1 mM sodium pyruvate (Gibco 1136–070), 100 mM non-essential amino acids (Gibco 11140–050), 5 mM HEPES (Gibco 25–060-Cl), 55 μM β-mercaptoethanol (Gibco 21985023), and 100 U/ml penicillin/streptomycin (Corning 30–002-Cl). CD11c+ cells were purified according to the manufacturer’s protocol (Miltenyi Biotec 130–125-853). Igλ+ B cells from B1–8hi mice were purified as described in Adoptive cell transfers. In various combinations, we added the following: 1:1 (beads:cells) ratio of CD3/CD28 T-activator beads (Gibco 11456D), 2 ng/μl TGF-β (R&D systems 243-B3–002), 10 ng/μl IL-2 (Biolegend 575406), 10 nM retinoic acid (Sigma-Aldrich R2625), 50 μg/ml NP-OVA (Biosearch Technologies N-5051).
Retroviral transduction of Rosa26Foxp3 T cells with Cre recombinase
293T cells were transfected with MigR1-iCre-IRES-Thy1.1 (transfer) and pCL-Eco (packaging) plasmids at 1:1 ratio using FuGENE® HD (Promega E2311). Virus-containing supernatant was collected 48 hours after transfection and filtered through a 0.45-μm syringe filter. Sorted CD62LhiCD44loCD25– naïve CD4 T cells were activated with plate-bound anti-CD3/anti-CD28 antibody and transduced on day 2 post-activation with the virus-containing supernatant, supplemented with 4 mg/ml of polybrene (Sigma-Aldrich H9268) by centrifugation at 800g for 90 min.
Single-cell TCR sequencing and analysis
Single T cells were index-sorted into 96-well PCR plates containing 5 μl of TCL buffer (Qiagen) with 1% β-mercaptoethanol. Nucleic acids were extracted using SPRI beads as described (55). RNA was reverse-transcribed using RT maxima reverse transcriptase (Thermo Scientific) and oligo(dT) as a primer. Initial amplification was based on previously described primers and conditions (56). Subsequently, a nested PCR was performed to incorporate a common adaptor sequence. Finally, a third PCR adding plate, row, and column identifiers together with paired-end primers enabling Illumina sequencing was performed. All primers were essentially as previously detailed (57), see Data S2. Amplicons were pooled by plate and purified using SPRI beads (0.7X volume ratio). Pooled amplicon libraries were sequenced with a 500-cycle Reagent Nano v2 kit on the Illumina Miseq platform. Additional GCs were sequenced using Sanger sequencing, as previously described (56).
All TCRβ chains were sequenced, with the additional amplification of TCRα chains in expanded clones to ascertain clonality. Paired-end sequences were demultiplexed using PandaSeq (58) and processed with the FASTX toolkit. The resulting reads were assigned to wells according to barcodes. Highest-count sequences for every single cell were analyzed. TCRα and β sequences were aligned to the IMGT database (59) (http://www.imgt.org). In the resulting annotation, sequences with common Vα/Jα and Vβ/Dβ/Jβ and identical CDR3 sequences were assigned to the same clone.
Single-cell RNA sequencing and analysis
Single cells were sorted and processed as previously described (60). Raw FASTQ sequence files generated from Smartseq2 libraries were aligned to the mouse genome (v. mm10) with the annotated transcriptome (v. gencode M22) using STAR (v. 2.6) (61). Subsequently, genome-mapped BAM files were processed using RSEM (v. 1.3.1) (62) for gene quantification. The matrix of gene counts was used as input for analysis using the R package Seurat (v. 3.1.4.) (63). To control unwanted sources of experimental variation, we eliminated any experiment-specific variables by regressing out the batch effect factors as described (63). Additionally, cells containing more than 10% of sequence reads aligned to mitochondrial genes were excluded before normalization. We used the ‘JackStraw’ method to determine the number of significant principal components present in the dataset, and 11 were chosen for downstream procedures. Finally, single cells were clustered using the Shared Nearest Neighbor (SNN) Graph method, and gene expression was evaluated using the Seurat workflow. To calculate gene signature scores among single cells, we used the AddModuleScore function from the Seurat workflow using various gene sets as input. We used gene sets from GSE20366 (TREG_VS_NAIVE_CD4_TCELL: UP and DN), Wing et al., 2017 (Top 300 genes from: CD25M_TFR_VS_TFH: UP and DN; CD25M_TFR_VS_TFH: UP and DN), our study (Top 30 genes from: Naive_xfer_VS_ExpandedRFP-: UP and DN). All cells used in the scRNA-seq analysis were from male mice, except for the “naïve transfer” experiment, in which the use of only males is impractical given a large number of donors needed. The only noticeable effect of this was that the female-specific RNA Xist was differentially expressed between “naïve transfer” and other cell types. This gene was removed from downstream comparisons.
For scRNA-seq analysis, we performed full TCR reconstruction in silico using the TRACER protocol (64). Briefly, FASTQ files were trimmed for adapter removal and aligned to the mouse TCRα and β sequences obtained from IMGT (59). Aligned reads were then assembled into full TCR transcripts with Trinity (v2.9.1) and the VDJ features annotated using IgBlast (65). We defined T cell populations as part of the same clonal lineage when displaying identical V(D)J gene annotation and identical CDR3 sequence at the nucleotide level. In silico TCR reconstruction was cross-validated by de novo TCRα and β amplification and sequencing as detailed above.
Statistical analysis
Except when otherwise noted, statistical analysis were performed in GraphPad Prism version 8.3.1 for Windows (GraphPad Software, San Diego, California USA). Differences between two individual groups were compared using a two-tailed Student’s t test. In the case of three groups, one-way ANOVA followed by Dunnett’s multiple comparisons test was performed. The statistical test and details about group number and replicates are indicated in the figure legends. Statistically different gene markers or gene signatures between single-cell populations were detected using the Wilcoxon rank test and considered as significant when showing an adjusted P-value of less than 0.05. Cohen’s d, a measure of effect size, was calculated for changes in expression of gene signatures, as d= [(mean of first group)–(mean of second group)]/(S.D. of whole sample).
Supplementary Material
Collapsed 4D datasets showing pLN intravital imaging of endogenous Foxp3-GFP+ T cells (green) along with adoptively-transferred CFP+ NP-specific (B1–8hi) B cells (blue) and RFP+ OT-II T cells (red; yellow when fluxing calcium) at the T:B border, two days after footpad immunization with NP-OVA in alum. The first (overview) movie is shown twice, once as raw data and the second time with interactions between B1–8hi B cells and Foxp3+ T cells exceeding 5 min in duration indicated by tracks. This is followed by magnified examples of “swarming” and “dragging”-type interactions between these two cell types. Videos displayed at 210X real time.
Collapsed 4D datasets of pLN intravital imaging showing endogenous Foxp3-GFP+ T cells (green) and adoptively transferred CFP+ NP-specific (B1–8hi) B cells (blue/cyan) and RFP+ OT-II T cells (red) at within GCs, 10 days after footpad boosting with NP-OVA (experimental setup as in Fig. 1A). The first (overview) movie is shown twice, once as raw data and the second time with short interactions between B1–8hi B cells and Foxp3+ T cells indicated by tracks. This is followed by magnified examples of the observable interactions between these two cell types. Videos displayed at 210X real time.
Collapsed 4D datasets of pLN intravital imaging showing endogenous Foxp3-GFP+ T cells (green) and adoptively transferred CFP+ NP-specific (B1–8hi) B cells (blue/cyan) and RFP+ OT-II T cells (red) at within GCs, 15 days after footpad boosting with NP-OVA (experimental setup as in Fig. 1A). The first (overview) movie is shown twice, once as raw data and the second time with interactions between B1–8hi B cells and Foxp3+ T cells exceeding 5 min in duration indicated by tracks. This is followed by magnified examples of long-lived “entanglement”-type interactions between these two cell types. Videos displayed at 210X real time.
ACKNOWLEDGEMENTS
We thank D. Mucida (Rockefeller University) and C. Vinuesa (Australian National University) for critical reading of our manuscript; M. Carroll and D. Firl (Harvard University Medical School) for help with implantation and imaging of iLN lymph node windows; K. Gordon and K. Chhosphel for cell sorting; P. Strogies, D. Gross, and J. Petrillo at Rockefeller University’s Precision Instrumentation Technologies Facility and V. Sherman at Rockefeller University’s High Energy Physics Instrument Shop for adapting and producing parts for longitudinal inguinal lymph node imaging; the Rockefeller University Genomics Center for RNA sequencing and Comparative Biosciences Center for animal housing; and Rockefeller University employees for their continuous assistance.
Funding: This work was supported by NIH grants R21AI138020, R01AI119006, and R01AI139117, Human Frontier of Science Program Research Grant RPG003/2015, and a March of Dimes Basil O’Connor Starting Scholar Award to G.D.V; NIH grants DP2GM119419 and RM1HG006193 to A.K.S.; and NCI Cancer Center Support Grant P30 CA008748 and NIH grant R37AI034206 to A.Y.R. Work in the Victora laboratory is further supported by an NIH Director’s Pioneer Award (DP1AI144248) and by the Robertson Foundation. J.T.J. was supported by Fripro mobility grant # 239757, jointly funded by the Research Council of Norway and the Co-funding of Regional, National, and International Programmes (COFUND) – Marie Curie Actions under the EU Seventh Framework Programme (FP7) and South-Eastern Norway Regional Health Authority grant #2012026. W.H. was supported by an Irvington-Cancer Research Institute Postdoctoral Fellowship. A.Y.R. is an HHMI Investigator. A.K.S. was supported by a Sloan Fellowship in Chemistry. A.K.S and G.D.V. are Pew-Stewart Scholars. G.D.V. is a Burroughs-Wellcome Investigator in the Pathogenesis of Infectious Disease, a Searle Scholar, and a MacArthur Fellow.
Footnotes
Competing interests: A.K.S. reports compensation for consulting and/or SAB membership from Merck, Honeycomb Biotechnologies, Cellarity, Repertoire Immune Medicines, Third Rock Ventures, Ochre Bio, and Dahlia Biosciences unrelated to this work. A.Y.R. is a co-founder of Sonoma Biotherapeutics and Vedanta Biosciences. He is an SAB member and holds stock options in Sonoma Biotherapeutics, RAPT Therapeutics and Vedanta Biosciences, and holds IP for therapeutic antibody depleting Treg cells licensed to Takeda.
Data and materials availability: All data are available in the main text or supplementary materials. scRNA-seq data is available from Gene Expression Omnibus, under accession number GSE173227.
REFERENCES
- 1.Mesin L, Ersching J, Victora GD, Germinal Center B Cell Dynamics. Immunity 45, 471–482 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corti D, Lanzavecchia A, Broadly neutralizing antiviral antibodies. Annu Rev Immunol 31, 705–742 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Liu YJ, Zhang J, Lane PJ, Chan EY, MacLennan IC, Sites of specific B cell activation in primary and secondary responses to T cell-dependent and T cell-independent antigens. Eur J Immunol 21, 2951–2962 (1991). [DOI] [PubMed] [Google Scholar]
- 4.Bachmann MF, Odermatt B, Hengartner H, Zinkernagel RM, Induction of long-lived germinal centers associated with persisting antigen after viral infection. The Journal of experimental medicine 183, 2259–2269 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adachi Y. et al. , Distinct germinal center selection at local sites shapes memory B cell response to viral escape. The Journal of experimental medicine 212, 1709–1723 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dogan I. et al. , Multiple layers of B cell memory with different effector functions. Nat Immunol 10, 1292–1299 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Krishnamurty AT et al. , Somatically Hypermutated Plasmodium-Specific IgM(+) Memory B Cells Are Rapid, Plastic, Early Responders upon Malaria Rechallenge. Immunity 45, 402–414 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasturi SP et al. , Programming the magnitude and persistence of antibody responses with innate immunity. Nature 470, 543–547 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Victora GD et al. , Germinal Center Dynamics Revealed by Multiphoton Microscopy with a Photoactivatable Fluorescent Reporter. Cell 143, 592–605 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crotty S, Follicular helper CD4 T cells (TFH). Annu Rev Immunol 29, 621–663 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Vinuesa CG, Linterman MA, Yu D, MacLennan IC, Follicular Helper T Cells. Annu Rev Immunol 34, 335–368 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Josefowicz SZ, Lu LF, Rudensky AY, Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol 30, 531–564 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linterman MA et al. , Foxp3(+) follicular regulatory T cells control the germinal center response. Nat Med 17, 975–982 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wollenberg I. et al. , Regulation of the germinal center reaction by Foxp3+ follicular regulatory T cells. Journal of immunology 187, 4553–4560 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Chung Y. et al. , Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med 17, 983–988 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sage PT, Sharpe AH, T follicular regulatory cells in the regulation of B cell responses. Trends in immunology 36, 410–418 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fonseca VR, Ribeiro F, Graca L, T follicular regulatory (Tfr) cells: Dissecting the complexity of Tfr-cell compartments. Immunol Rev 288, 112–127 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez-Figueroa P. et al. , Follicular regulatory T cells produce neuritin to regulate B cells. Cell 184, 1775–1789 e1719 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Korn T. et al. , Myelin-specific regulatory T cells accumulate in the CNS but fail to control autoimmune inflammation. Nat Med 13, 423–431 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shulman Z. et al. , T follicular helper cell dynamics in germinal centers. Science 341, 673–677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan YY, Flavell RA, Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proceedings of the National Academy of Sciences of the United States of America 102, 5126–5131 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Firl DJ, Degn SE, Padera T, Carroll MC, Capturing change in clonal composition amongst single mouse germinal centers. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benet ZL et al. , CCL3 Promotes Germinal Center B Cells Sampling by Follicular Regulatory T Cells in Murine Lymph Nodes. Front Immunol 9, 2044 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okada T. et al. , Antigen-engaged B cells undergo chemotaxis toward the T zone and form motile conjugates with helper T cells. PLoS Biol 3, e150 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwickert TA et al. , A dynamic T cell-limited checkpoint regulates affinity-dependent B cell entry into the germinal center. The Journal of experimental medicine 208, 1243–1252 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu D. et al. , T-B-cell entanglement and ICOSL-driven feed-forward regulation of germinal centre reaction. Nature 517, 214–218 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Shulman Z. et al. , Dynamic signaling by T follicular helper cells during germinal center B cell selection. Science 345, 1058–1062 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allen CD, Okada T, Tang HL, Cyster JG, Imaging of germinal center selection events during affinity maturation. Science 315, 528–531 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Lindquist RL et al. , Visualizing dendritic cell networks in vivo. Nat Immunol 5, 1243–1250 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Aloulou M. et al. , Follicular regulatory T cells can be specific for the immunizing antigen and derive from naive T cells. Nat Commun 7, 10579 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wing JB et al. , A distinct subpopulation of CD25(−) T-follicular regulatory cells localizes in the germinal centers. Proc Natl Acad Sci U S A 114, E6400–E6409 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feuerer M. et al. , Genomic definition of multiple ex vivo regulatory T cell subphenotypes. Proc Natl Acad Sci U S A 107, 5919–5924 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chtanova T. et al. , T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J Immunol 173, 68–78 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Liu YJ et al. , Mechanism of antigen-driven selection in germinal centres. Nature 342, 929–931 (1989). [DOI] [PubMed] [Google Scholar]
- 35.Maceiras AR et al. , T follicular helper and T follicular regulatory cells have different TCR specificity. Nat Commun 8, 15067 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hou S. et al. , FoxP3 and Ezh2 regulate Tfr cell suppressive function and transcriptional program. J Exp Med 216, 605–620 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albright AR et al. , TGFbeta signaling in germinal center B cells promotes the transition from light zone to dark zone. J Exp Med 216, 2531–2545 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandel TE, Phipps RP, Abbot A, Tew JG, The follicular dendritic cell: long term antigen retention during immunity. Immunol Rev 53, 29–59 (1980). [DOI] [PubMed] [Google Scholar]
- 39.Kretschmer K. et al. , Inducing and expanding regulatory T cell populations by foreign antigen. Nat Immunol 6, 1219–1227 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Wing JB, Ise W, Kurosaki T, Sakaguchi S, Regulatory T cells control antigen-specific expansion of Tfh cell number and humoral immune responses via the coreceptor CTLA-4. Immunity 41, 1013–1025 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Zotos D. et al. , IL-21 regulates germinal center B cell differentiation and proliferation through a B cell-intrinsic mechanism. J Exp Med 207, 365–378 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Linterman MA et al. , IL-21 acts directly on B cells to regulate Bcl-6 expression and germinal center responses. J Exp Med 207, 353–363 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han S. et al. , Cellular interaction in germinal centers. Roles of CD40 ligand and B7–2 in established germinal centers. J Immunol 155, 556–567 (1995). [PubMed] [Google Scholar]
- 44.Hadjantonakis AK, Macmaster S, Nagy A, Embryonic stem cells and mice expressing different GFP variants for multiple non-invasive reporter usage within a single animal. BMC Biotechnol 2, 11 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vintersten K. et al. , Mouse in red: red fluorescent protein expression in mouse ES cells, embryos, and adult animals. Genesis 40, 241–246 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Wolf AJ et al. , Initiation of the adaptive immune response to Mycobacterium tuberculosis depends on antigen production in the local lymph node, not the lungs. J Exp Med 205, 105–115 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shih TA, Roederer M, Nussenzweig MC, Role of antigen receptor affinity in T cell-independent antibody responses in vivo. Nat Immunol 3, 399–406 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Barnden MJ, Allison J, Heath WR, Carbone FR, Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunology and cell biology 76, 34–40 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Zariwala HA et al. , A Cre-dependent GCaMP3 reporter mouse for neuronal imaging in vivo. J Neurosci 32, 3131–3141 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bettelli E. et al. , Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Chinen T. et al. , An essential role for the IL-2 receptor in Treg cell function. Nat Immunol 17, 1322–1333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roozendaal R. et al. , Conduits mediate transport of low-molecular-weight antigen to lymph node follicles. Immunity 30, 264–276 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meijer EFJ et al. , Murine chronic lymph node window for longitudinal intravital lymph node imaging. Nature protocols 12, 1513–1520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jacobsen JT, Victora GD, Microanatomical Labeling of Germinal Center Structures for Flow Cytometry Using Photoactivation. Methods Mol Biol 1623, 51–58 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Tas JM et al. , Visualizing antibody affinity maturation in germinal centers. Science 351, 1048–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dash P. et al. , Paired analysis of TCRalpha and TCRbeta chains at the single-cell level in mice. The Journal of clinical investigation 121, 288–295 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han A, Glanville J, Hansmann L, Davis MM, Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol 32, 684–692 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.A. P. Masella, A. K. Bartram, J. M. Truszkowski, D. G. Brown, J. D. Neufeld, PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics 13, 31 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lefranc MP et al. , IMGT, the international ImMunoGeneTics information system. Nucleic acids research 37, D1006–1012 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trombetta JJ et al. , Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing. Curr Protoc Mol Biol 107, 4 22 21–24 22 17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobin A. et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li B, Dewey CN, RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stuart T. et al. , Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stubbington MJT et al. , T cell fate and clonality inference from single-cell transcriptomes. Nat Methods 13, 329–332 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haas BJ et al. , De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature protocols 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Collapsed 4D datasets showing pLN intravital imaging of endogenous Foxp3-GFP+ T cells (green) along with adoptively-transferred CFP+ NP-specific (B1–8hi) B cells (blue) and RFP+ OT-II T cells (red; yellow when fluxing calcium) at the T:B border, two days after footpad immunization with NP-OVA in alum. The first (overview) movie is shown twice, once as raw data and the second time with interactions between B1–8hi B cells and Foxp3+ T cells exceeding 5 min in duration indicated by tracks. This is followed by magnified examples of “swarming” and “dragging”-type interactions between these two cell types. Videos displayed at 210X real time.
Collapsed 4D datasets of pLN intravital imaging showing endogenous Foxp3-GFP+ T cells (green) and adoptively transferred CFP+ NP-specific (B1–8hi) B cells (blue/cyan) and RFP+ OT-II T cells (red) at within GCs, 10 days after footpad boosting with NP-OVA (experimental setup as in Fig. 1A). The first (overview) movie is shown twice, once as raw data and the second time with short interactions between B1–8hi B cells and Foxp3+ T cells indicated by tracks. This is followed by magnified examples of the observable interactions between these two cell types. Videos displayed at 210X real time.
Collapsed 4D datasets of pLN intravital imaging showing endogenous Foxp3-GFP+ T cells (green) and adoptively transferred CFP+ NP-specific (B1–8hi) B cells (blue/cyan) and RFP+ OT-II T cells (red) at within GCs, 15 days after footpad boosting with NP-OVA (experimental setup as in Fig. 1A). The first (overview) movie is shown twice, once as raw data and the second time with interactions between B1–8hi B cells and Foxp3+ T cells exceeding 5 min in duration indicated by tracks. This is followed by magnified examples of long-lived “entanglement”-type interactions between these two cell types. Videos displayed at 210X real time.