Vasquez-Limeta et al. use human cells engineered for fast degradation of centrosomal protein CPAP. Using superresolution microscopy, they show that CPAP insufficiency leads to centrioles with incomplete microtubule triplets that convert to centrosomes and duplicate, but fragment owing to loss of cohesion between microtubule blades.
Abstract
Centrioles are structures that assemble centrosomes. CPAP is critical for centrosome assembly, and its mutations are found in patients with diseases such as primary microcephaly. CPAP’s centrosomal localization, its dynamics, and the consequences of its insufficiency in human cells are poorly understood. Here we use human cells genetically engineered for fast degradation of CPAP, in combination with superresolution microscopy, to address these uncertainties. We show that three independent centrosomal CPAP populations are dynamically regulated during the cell cycle. We confirm that CPAP is critical for assembly of human centrioles, but not for recruitment of pericentriolar material on already assembled centrioles. Further, we reveal that CPAP insufficiency leads to centrioles with incomplete microtubule triplets that can convert to centrosomes, duplicate, and form mitotic spindle poles, but fragment owing to loss of cohesion between microtubule blades. These findings further our basic understanding of the role of CPAP in centrosome biogenesis and help understand how CPAP aberrations can lead to human diseases.
Introduction
Centrosomes, small membraneless organelles, facilitate the formation of mitotic spindle poles and faithful segregation of genetic material during cell division. They are composed of a microtubule (MT)-based structure called a centriole and surrounding proteinaceous pericentriolar material (PCM; LeGuennec et al., 2021; Vasquez-Limeta and Loncarek, 2021; Vorobjev and Chentsov Yu, 1982; Woodruff et al., 2014). In addition to organizing centrosomes, centrioles convert into basal bodies and build primary and motile cilia, organelles with sensory and motile functions, respectively (Mitchison and Valente, 2017; Wang and Dynlacht, 2018). Defects in centriole number or structure cause mitotic errors and perturbations in tissue architecture and cilia formation and function (Godinho et al., 2009; Goundiam and Basto, 2021; LoMastro and Holland, 2019; Nigg et al., 2014, 2017). Centriole and centrosome abnormalities can also promote cellular transformation (Basto et al., 2008; Levine et al., 2017) and are a hallmark of most human tumors. Mutations in centrosome components underlie hereditary diseases such as microcephaly and ciliopathies (Marthiens and Basto, 2020; Marthiens et al., 2013).
The number and the assembly of centrioles are strictly controlled (Gonczy and Hatzopoulos, 2019; Nigg et al., 2014; Sullenberger et al., 2020). In cycling somatic cells, there are two mother centrioles, which duplicate at the beginning of S phase by forming a new centriole (procentriole). Procentrioles form orthogonal to and in the vicinity of a mother centriole, where they remain during interphase and mitosis. During cell division, each sister cell inherits one mother centriole–procentriole pair, which ultimately separates and forms individual centrosomes. To maintain centrosome homeostasis, new centrioles must be properly assembled, which is the result of a gradual buildup of their MT wall and recruitment of dozens of proteins inside and outside the MT wall (Sullenberger et al., 2020).
Centrosomal protein 4.1–associated protein (CPAP, in vertebrates also called centromere protein J [CENPJ]; SAS-4 in Drosophila and Caenorhabditis elegans; Hung et al., 2000) is a conserved protein found in all species that have centrioles (Carvalho-Santos et al., 2010; Hodges et al., 2010). Homozygous mutations in the human CPAP gene are associated with autosomal recessive primary microcephaly, a debilitating disease characterized by reduced neuron numbers and small brain size (Bond et al., 2005; Gul et al., 2006; Kitagawa et al., 2011; Lin et al., 2020). CPAP/SAS-4 incorporates into procentrioles from the early stage of their formation (Basto et al., 2006; Kleylein-Sohn et al., 2007; Leidel and Gönczy, 2003). The CPAP C-terminus interacts with centriole assembly initiation factors STIL (Ana2 in Drosophila; Cottee et al., 2013; Moyer and Holland, 2019; Tang et al., 2011; Vulprecht et al., 2012), Sas-6 (Lin et al., 2013a), and Cep135 (Carvalho-Santos et al., 2012; Comartin et al., 2013; Lin et al., 2013a; Ohta et al., 2002), and with centriole elongation factors Cep120 (Comartin et al., 2013; Lin et al., 2013b) and Centrobin (Gudi et al., 2015). CPAP/SAS-4 removal causes centriole duplication failure (Basto et al., 2006; Cho et al., 2006; Gogendeau et al., 2011; Kirkham et al., 2003; Leidel and Gönczy, 2003).
Toward its N terminus, CPAP contains an MT-binding domain and a PN2-3 tubulin-binding domain (Cormier et al., 2009; Hsu et al., 2008; Hung et al., 2004; Zheng et al., 2016), which is thought to regulate the rate of tubulin incorporation to centriole MTs (Sharma et al., 2016; Zheng et al., 2016). Consistent with this, CPAP overexpression results in over-elongated centriolar MTs (Kohlmaier et al., 2009; Schmidt et al., 2009; Tang et al., 2009), and expression of CPAP mutants impeding CPAP MT-related functions results in the formation of aberrant centrioles (Kitagawa et al., 2011; Zheng et al., 2016). Conversely, partial depletion of CPAP in S phase–arrested human cells reduces centriole length (Comartin et al., 2013). Similarly, partial depletion of SAS-4 resulted in shorter centrioles with reduced PCM levels in C. elegans (Kirkham et al., 2003) and in basal bodies with incomplete MTs in Paramecium (Gogendeau et al., 2011). In addition, CPAP/SAS-4 is suggested to act as a scaffold for the assembly of cytoplasmic PCM complexes and for their tethering to centrosomes (Gopalakrishnan et al., 2011; Zheng et al., 2014). Phosphorylation of CPAP by Aurora A (Chou et al., 2016) and Plk1 (Ramani et al., 2018) was reported to regulate centrosomal PCM recruitment during mitosis. Disruption of centrosome integrity and mitotic spindle morphology has been detected after prolonged CPAP depletion by siRNA (Cho et al., 2006; Lee et al., 2014; Lee and Rhee, 2015). However, in Drosophila embryos, PCM recruitment did not require the preassembly of multiprotein SAS-4/PCM cytosolic complexes (Conduit et al., 2015), and depletion of CPAP in the presence of intact mother centrioles did not lead to immediate mitotic problems in human U2OS cells (Kitagawa et al., 2011). Because a detailed ultrastructural analysis of human centrioles depleted for CPAP is lacking, the exact role of CPAP in the organization of interphase and mitotic centrosomes remains controversial. Moreover, a detailed centrosomal localization map of CPAP and its dynamics during the cell cycle has yet to be understood. In this work, we answer these important outstanding questions using superresolution microscopy approaches and human cells genetically engineered for fast and controllable degradation of CPAP.
Results
CPAP’s localization to three independent centrosomal populations is cell cycle regulated
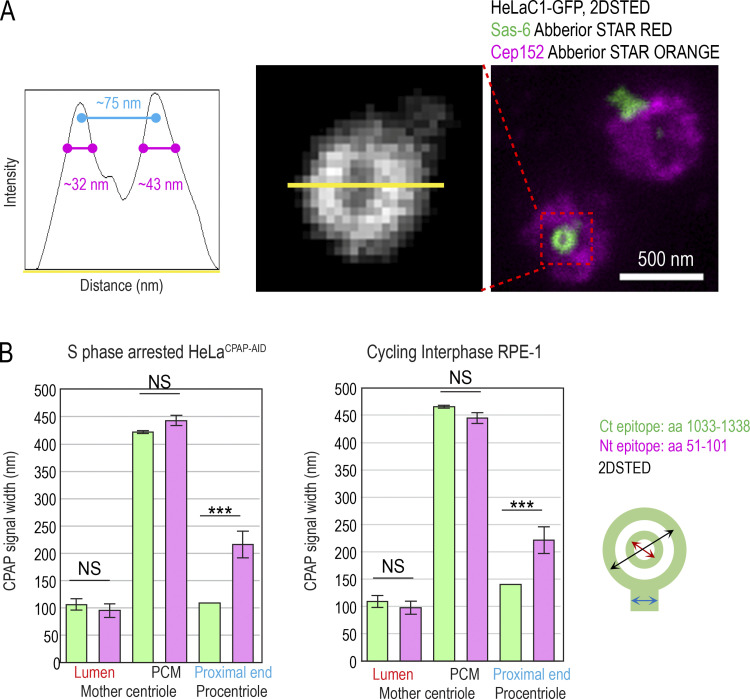
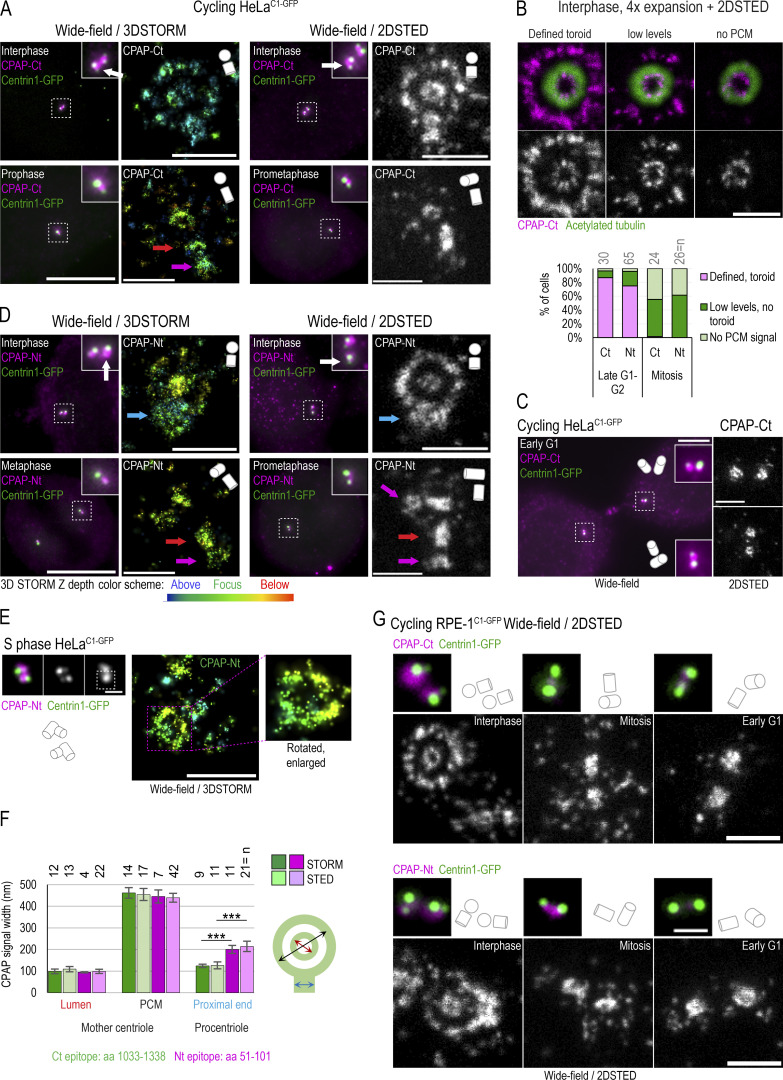
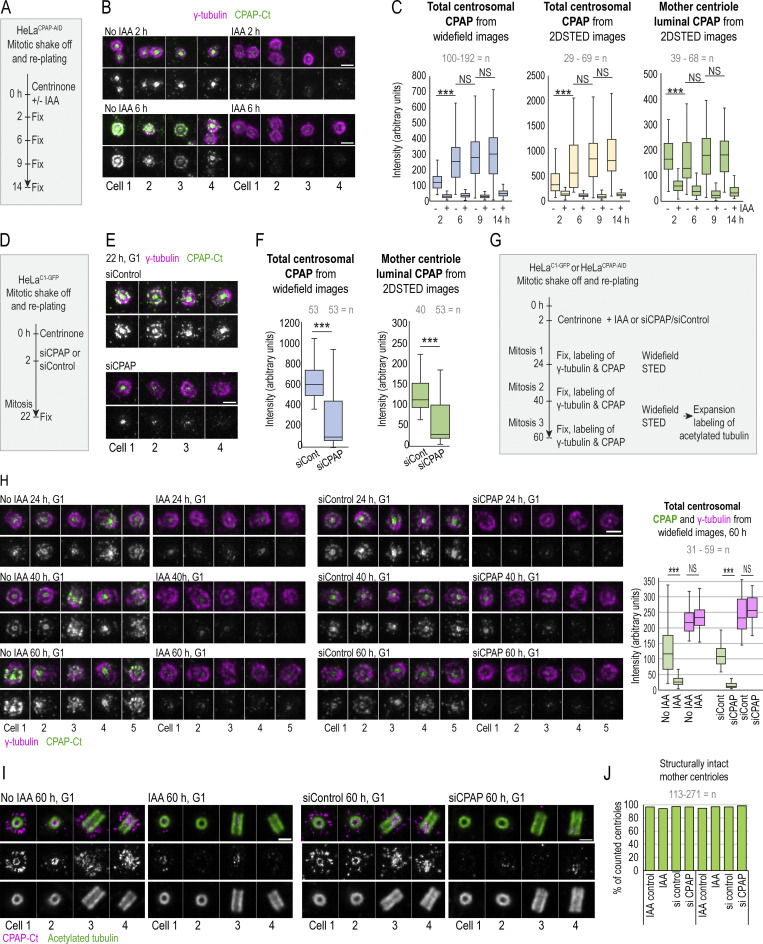
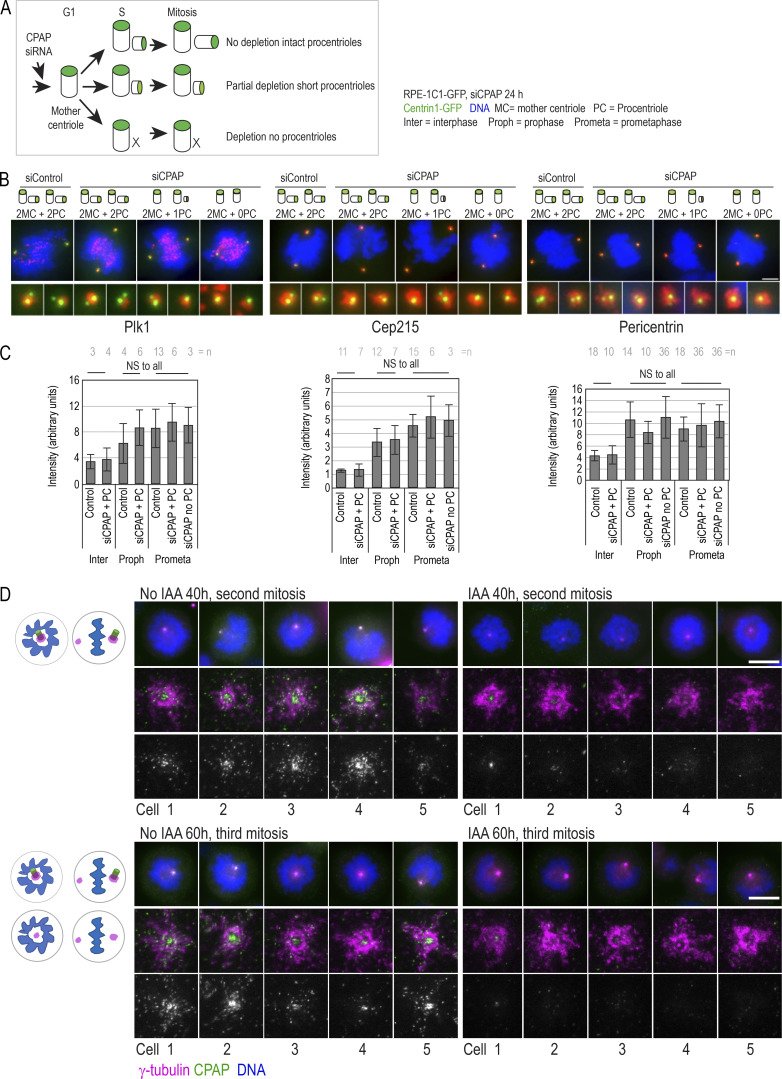
To dissect the centrosomal distribution of CPAP in nanoscale resolution, we used HeLa and RPE-1 cell lines expressing a centriolar marker, Centrin1-GFP (C1-GFP). Cells were first immunolabeled using antibodies directed toward the CPAP C-terminus (Ct) and imaged using two modalities of superresolution microscopy: 3D stochastic optical reconstruction microscopy (3DSTORM) and 2D stimulated emission depletion (2DSTED). The typical resolution of 3DSTORM, in our hands, is ∼25 nm (Bowler et al., 2019), and of 2DSTED, ∼30–40 nm (Fig. S1 A). First, we analyzed the distribution of CPAP in interphase S/G2 cells. The Ct antibody distinguished a CPAP toroid ∼460 nm in diameter around mother centrioles and a smaller toroid ∼100 nm in the lumina of both mother centrioles and procentrioles (Fig. 1, A, B, and F). This CPAP pattern is consistent with previous CPAP immuno-EM and structured illumination microscopy analyses (Kleylein-Sohn et al., 2007; Sonnen et al., 2012). Although ∼80% of interphase mother centrioles formed the PCM toroid, the abundance of the signal within toroids was variable between cells and between centrosomes of the same cell. In ∼20% of centrosomes, the PCM CPAP toroid was not defined or was undetectable (Fig. 1 B). In some mother centrioles, luminal CPAP was organized to nine discrete lobules underlying and slightly overlapping with the centriole’s inner wall, although the luminal signal could also appear irregular and of various intensities (Fig. 1 B). In PCM, CPAP lacked ninefold organization. Before and during mitosis, the PCM CPAP toroid in most centrosomes showed reduced and more scattered signal, or it was entirely absent (Fig. 1, A and B), while the luminal CPAP was preserved in all centrioles. In early G1 (Fig. 1 C), CPAP was entirely absent from the PCM, while the luminal CPAP localized with similar intensity within the proximal ends of both centrioles, suggesting that the PCM and luminal populations are independent.
Figure S1.
Resolution of 2DSTED and quantification of CPAP signals. (A) Mother centriole–specific PCM protein Cep152 and procentriole-specific protein Sas-6 were immunolabeled and imaged with 2DSTED. Sas-6 forms a small toroid within proximal procentriole lumina, which can be resolved using 2DSTED. Intensity plot through the Sas-6 toroid is shown. The length of the Sas-6 signal at half width at half maxima indicates resolution of 2DSTED of ≥32–43 nm (magenta). (B) Quantification of CPAP signal dimensions from superresolution images in S phase–arrested HeLaCPAP-AID and cycling RPE-1C1-GFP, using criteria illustrated by the cartoon. n = centrosome number. Scale bar in A. 0.5 µm. ***, P ≤ 0.001.
Figure 1.
Centrosomal localization of CPAP and its cell cycle dynamics. Cycling cells were immunolabeled with antibodies recognizing Ct or Nt of endogenous CPAP, and the centrosomal distribution of CPAP signals was analyzed using 3DSTORM and 2DSTED. Low-magnification images of corresponding cells and centrosomes, in which brighter Centrin1-GFP signals correspond to mother centrioles and dimmer signals to procentrioles, are shown next to superresolved images. Centriole schemes depicting approximate centriole orientation are included to facilitate image interpretation. (A) Analysis of CPAP distribution in interphase and mitotic cells using anti CPAP-Ct antibody. (B) Image: Illustration of three typical PCM CPAP configurations. Centrioles were expanded fourfold, immunolabeled for CPAP and centriole MT marker acetylated tubulin, and imaged by 2DSTED (only CPAP is superresolved). Graph: Quantification of centrosomes based on three observed PCM CPAP configurations. n = centrosomes analyzed by 2DSTED, no expansion. (C) A pair of early G1 cells with centrioles containing luminal CPAP. (D) Analysis of CPAP distribution in interphase and mitotic cells using CPAP-Nt antibody. Magenta and red arrows in A and D point to CPAP signal on centriole’s distal and proximal ends, respectively. Blue arrows point to procentriole-associated Nt CPAP signal. (E) 3DSTORM analysis of procentriole-associated CPAP Nt signal, to illustrate its toroidal organization. (F) Quantification of CPAP signal dimensions from superresolution images using criteria illustrated by the cartoon. Histogram shows the average width of CPAP signal ± SD, n = centriole number. (G) 2DSTED analysis of CPAP signal in RPE-1C1-GFP cells using CPAP-Ct and -Nt antibody. Scale bars: 5 µm (wide-field), 0.5 µm for 3DSTORM and 2DSTED, 0.2 µm for rotated 3DSTORM (E), and 1 µm for cropped centrosomes in E. ***, P ≤ 0.001. Source data are available for this figure: SourceData F2.
The lack of the PCM CPAP in mitosis could occur because of obstruction of the antibody epitope in mitosis. To verify the localization pattern obtained with the Ct antibody, we repeated the analysis using an antibody directed toward CPAP’s N-terminus (Nt). The Nt antibody revealed a similar CPAP pattern. In interphase, the Nt CPAP signal was detected in toroids ∼450 nm in diameter around mother centrioles and ∼100 nm in centrioles’ lumina (Fig. 1, D and F). With similar frequency as observed with the Ct signals, the PCM CPAP population was reduced or undetectable in mitosis (Fig. 1 B, graph). Interestingly, unlike the Ct CPAP signal, procentriole-associated Nt signal in S phase showed a toroid of wider diameter (∼200–214 nm; Fig. 1, D, E, and F, blue arrows in D). On more advanced mitotic procentrioles, which have completed structuring of proximal triplet MTs (Vorobjev and Chentsov Yu, 1982), the diameter of the proximal Nt signal adopted a more central luminal-like localization (Fig. 1 D, red arrows) with a diameter that was indistinguishable from that of the luminal Nt signal of mature centrioles and the luminal Ct signals. In addition, a distally localized population of CPAP on procentrioles and some mother centrioles could be detected in mitosis (Fig. 1 D, pink arrows). Similar CPAP pattern was detected in cycling RPE-1 cells and in S phase–arrested HeLa cells (Fig. 1, F and G; and Fig. S1 B).
The absence of the PCM CPAP population from mitotic centrosomes, and its large variability in interphase, is not compatible with the proposed critical role of CPAP in recruitment of PCM components by mother centrioles. Thus we hypothesized that in human cells, CPAP may not be universally needed for PCM organization. We tested this hypothesis by exploring the consequence of acute and long-term CPAP removal on the organization and function of interphase and mitotic centrosomes and on centriole stability and assembly.
Removal of CPAP by auxin-induced degradation (AID)
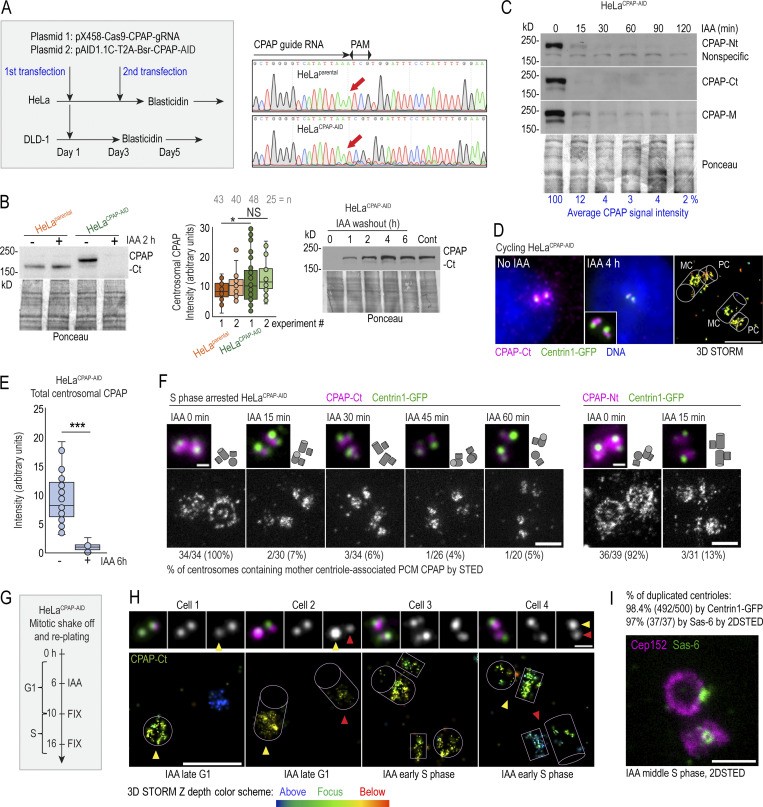
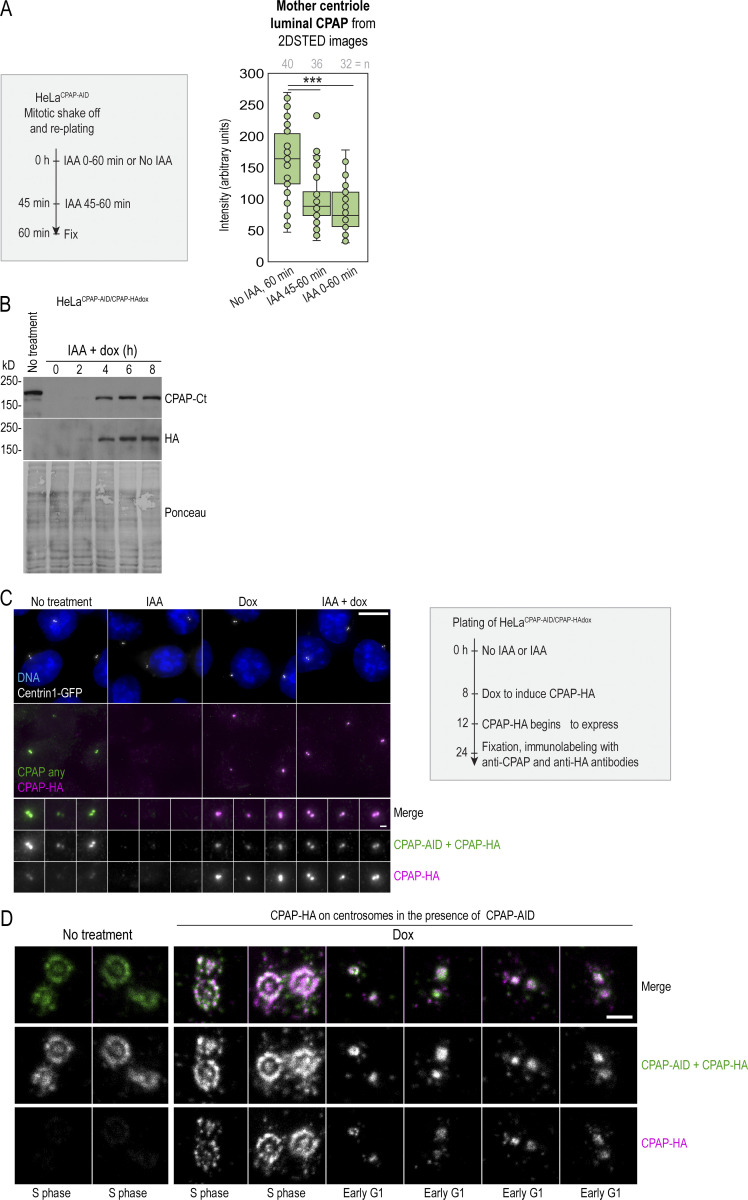
Depletion of proteins using siRNA or shRNA usually results in gradual and uncontrollable depletion, and it would not be suitable for precise and rapid manipulations of CPAP levels in specific phases of the cell cycle. To overcome this limitation, we generated HeLaCPAP-AID and DLD-1CPAP-AID cell lines in which CPAP is rapidly degraded using an auxin-based degradation system in which addition of indole-3-acetic acid (IAA) to the culture medium induces CPAP degradation by the proteasome (Nishimura and Fukagawa, 2017; Nishimura et al., 2009). In these cells, all endogenous CPAP alleles were inactivated by a 1-nucleotide deletion using CRISPR/Cas9 (Figs. 2 A and S3 A), whereas CPAP cDNA, tagged with an AID (CPAP-AID), and blasticidin selection markers were randomly integrated into the genome. Blasticidin-resistant monoclonal colonies were screened for the levels and degradation of CPAP-AID, and the clones used in the study were sequenced to ensure the lack of endogenous CPAP (Fig. 2, A–E; and Fig. S3 A). Immunoblotting showed that CPAP-AID was degraded within 30 min after addition of IAA (Figs. 2 C and S3 A) and restored within 2 h after IAA washout (Fig. 2 B, right). Quantification of CPAP immunoblot signals revealed that only ∼3% of the original CPAP-AID remained in IAA-treated cells (Fig. 2 C). In agreement, immunofluorescence showed a reduction of centrosome-associated CPAP signal in IAA-treated cells (Fig. 2, D and E; and Fig. S3 A). Microscopy revealed that in IAA-treated cells, the PCM CPAP population was entirely absent, and residual CPAP signal localized exclusively in centrioles’ lumina (Fig. 2 D). The loss of CPAP from PCM was rapid, within 15 min of IAA addition (Fig. 2 F). Procentrioles’ luminal CPAP remained detectable in IAA-treated cells, whereas mother centrioles’ luminal CPAP was often reduced or even undetectable after IAA treatment (Fig. 2, F and H (cell 4); Fig. 3, A–C; and Fig. S2 A). Similarly, depletion of CPAP by siRNA resulted in the loss of CPAP from PCM and mother centriole lumina (Fig. 3, D–F).
Figure 2.
Fast degradation of CPAP fused to an AID. (A) Left: The transfection and selection strategy (described in Materials and methods) used to generate HeLa and DLD-1 cell lines expressing CPAP fused to an AID (HeLaCPAP-AID and DLD-1CPAP-AID). Right: Sequencing result confirming the inactivation of CPAP alleles in HeLaCPAP-AID cells by a point mutation (red arrows). (B) Left and middle: Immunoblot and quantification of centrosomal CPAP in parental and HeLaCPAP-AID cells with and without IAA treatment. n = centrosome number. Right: Re-accumulation of CPAP-AID after IAA washout by immunoblot. (C) The efficiency and dynamics of CPAP-AID degradation after IAA addition. Antibodies recognizing Ct, Nt, and the middle (M) region of CPAP were used for detection. The residual levels of CPAP (shown in blue) represent an average signal intensity from the above blots. (D) Centrosomal CPAP levels and localization after 4 h of IAA treatment. Centrosome-associated CPAP is reduced, and residual CPAP signal is localized to centriole’s lumina and is absent from PCM. (E) Quantification of centrosomal CPAP from wide-field images before and after 6 h of IAA treatment. (F) 2DSTED analysis of centrosome-associated CPAP within first 60 min of IAA treatment. The wide-field images of corresponding superresolved centrioles are shown in insets. Centriole schemes depicting approximate centriole orientation are included. (G) Experimental strategy used in H and I. (H) 3DSTORM analysis of mother centrioles (MC) in late G1 (left) and in early S (right) in IAA-treated cells. CPAP signals analyzed by 3DSTORM are indicated by yellow and red arrows. (I) Centrosomes were labeled for mother centriole–specific protein Cep152 and procentriole-specific protein Sas-6. The percentage of duplicated mother centrioles was determined by scoring the number of MC-PC Centrin1-GFP pairs, and additionally by the presence of Sas-6 signal in association with Cep152 ring from 2DSTED recordings. Scale bars: 5 µm (wide-field); 0.5 µm (2DSTED and 3DSTORM); 1 µm (cropped centrosomes in F and H). *, P ≤ 0.05; ***, P ≤ 0.001.
Figure S3.
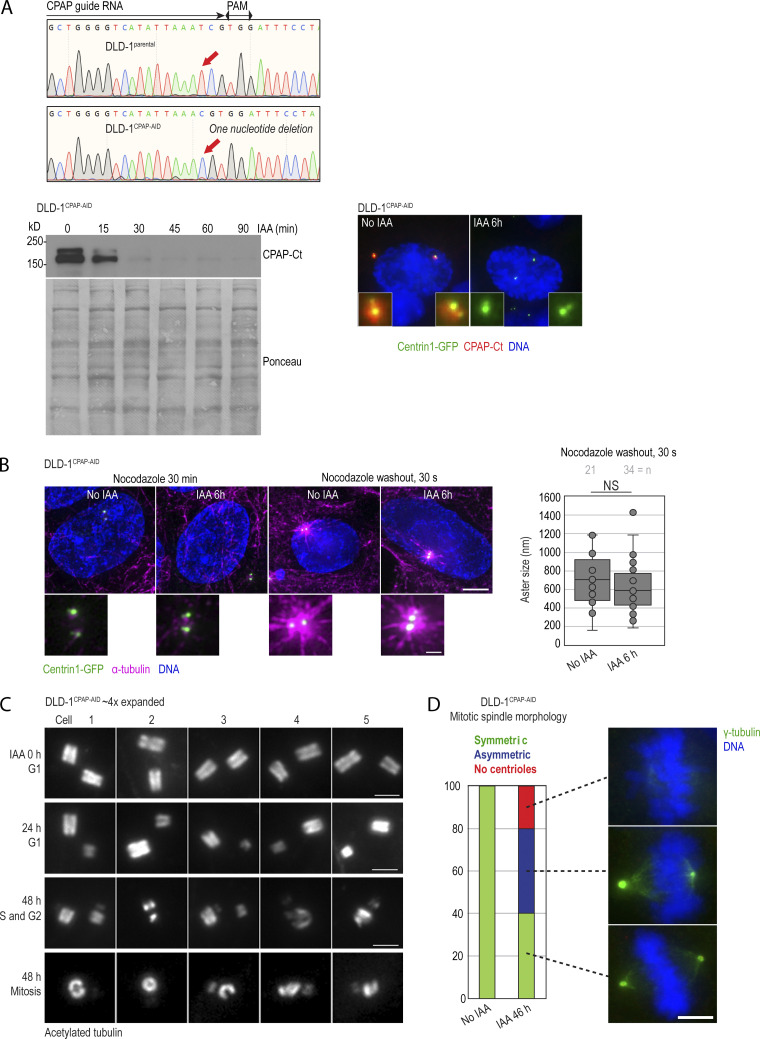
Characterization of DLD-1CPAP-AID cells and the effect of CPAP degradation on centriole structure in this cell line. (A) Sequencing result confirming the inactivation of endogenous CPAP alleles in DLD-1CPAP-AID cells by a point mutation (red arrows). Immunoblot represents the efficiency and dynamics of CPAP-AID degradation in DLD-1CPAP-AID cells after IAA addition. Microscopy image shows decreased centrosomal CPAP levels after 6 h of IAA treatment. Centrosome-associated CPAP-AID signal is significantly reduced in IAA-treated cells. (B) MT nucleation recovery after nocodazole washout. IAA treatment for 6 h does not change the size of MT asters after nocodazole washout. (C) Examples of expanded centrioles from control and IAA-treated DLD-1CPAP-AID cells. Cycling cells were treated with IAA for 24 and 48 h, expanded approximately fourfold, immunolabeled using acetylated tubulin to label centriole MT walls, and imaged using a conventional wide-field microscope. Control G1 cells contain two intact centrioles of similar sizes. After 24 h of IAA treatment, most G1 cells contain one full-length mother centriole and one short and/or narrow daughter centriole. The lower two panels illustrate cells containing short and/or broken duplicated mother centrioles in cells treated with IAA for 48 h. (D) Quantification of mitotic spindle morphology in DLD-1CPAP-AID cells treated with IAA for 46 h and immunolabeled for γ-tubulin. Graph shows percentage of cells. Examples of symmetric, asymmetric, and acentrosomal mitoses are shown. Scale bars: 5 µm (B); 0.5 µm (inset in B); 2 µm (C); 5 µm (D). Source data are available for this figure: SourceData FS3.
Figure 3.
Centriole-associated CPAP is dynamic, and its loss does not destabilize already-assembled mother centrioles. (A) Experimental strategy used to explore the dynamics of mother centriole luminal CPAP in IAA-treated cells in B and C. Cells were treated with IAA to degrade CPAP and with centrinone to prevent centriole duplication. CPAP and γ-tubulin were immunolabeled, and centrosomes were imaged by wide-field and 2DSTED. (B) Centrosomes in IAA-treated samples have reduced or undetectable CPAP and normal γ-tubulin levels. (C) Quantification of CPAP signals. (D) Experimental strategy used to explore the dynamics of mother centriole luminal CPAP in CPAP siRNA–treated cells in E and F. (E) Centrosomes in CPAP siRNA–treated samples have reduced or undetectable CPAP and normal γ-tubulin levels. (F) Quantification of CPAP signals. (G) Experimental strategy used to explore long-term stability of mother centrioles after CPAP removal by IAA or siRNA used in H–J, Fig. 5 D, and Fig. S4 D. (H) Centrosomes in IAA or siRNA-treated samples have reduced or undetectable CPAP and normal γ-tubulin levels. (I) Expansion/2DSTED analysis of G1 centrioles after 60 h of IAA or siRNA treatment. Acetylated tubulin labels centriole MT walls. Acetylated tubulin is not superresolved. (J) Quantification of mother centriole phenotypes from expanded samples after 40 and 60 h of IAA and siRNA treatment. Scale bars: 0.5 µm (B, E, and H); 1 µm (I). n = centrosome number. *, P ≤ 0.05; ***.
Figure S2.
Centriole-associated CPAP can be degraded from centriole lumen and is dynamically associated with centrioles. (A) Loss of the luminal CPAP from G1 centrioles in HeLaCPAP-AID cells after 15 min and 1 h of IAA treatment. Scheme depicts experimental strategy. Mitotic cells were shaken off and replated, and some cells were immediately treated with IAA to degrade CPAP. The second sample of cells was left untreated, and the third sample was treated for the last 15 min before fixation. All samples were fixed 60 min after shake-off, immunolabeled for CPAP and γ-tubulin, and imaged using 2DSTED. The intensity of luminal CPAP was determined from 2DSTED images. γ-tubulin was used as a marker for centriole orientation and centriole identification during imaging. The analysis shows that centriole-associated CPAP (which in early G1 consists of only luminal CPAP, as the PCM CPAP population is absent from early G1 centrioles; Fig. 1 C) is already reduced in IAA-treated cells after 15 min of IAA treatment. (B–D) Characterization of HeLaCPAP-AID/CPAP-HAdox cell line that expresses CPAP-AID constitutively and CPAP-HA inducibly, after doxycycline (dox) addition. (B) Immunoblot showing CPAP-AID levels and expression dynamics of CPAP-HA after dox addition. Newly synthesized CPAP-HA is expressed in cells 4 h after dox addition. (C) Cells were treated as depicted by the scheme. Wide-field images of centrosomal total CPAP (detected by anti CPAP-Ct antibody) and newly synthesized CPAP-HA (detected by anti HA antibody) are shown. (D) 2D STED images of centrosomes showing incorporation of CPAP-HA to centrosomes of all ages. Note that the faint CPAP-HA signal visible in untreated samples in S2D is due to the cross talk of STARORANGE and STARRED dies used to detect total CPAP and HA by STED. Scale bars: 10 µm (C); 1 µm (inset in C); 0.5 µm (D). ***, P ≤ 0.001. Source data are available for this figure: SourceData FS2.
Centriole-bound SAS-4 has previously been shown to be stably incorporated to centrioles of C. elegans (Balestra et al., 2015; Kirkham et al., 2003). However, our analysis indicated that, in human cells, both CPAP populations are dynamic and exchange with the cytoplasmic pool. To further investigate this finding, we modified the HeLaCPAP-AID cell line so that it also inducibly expresses CPAP-HA after addition of doxycycline (Fig. S2 B). We induced CPAP-HA in cells pretreated (or not) with IAA and fixed them after ∼12 h of CPAP-HA expression. Cells were then coimmunolabeled using anti-CPAP Ct and anti-HA antibody to detect total CPAP and CPAP-HA, respectively (Fig. S2 C). Newly synthesized CPAP-HA was associated with centrioles of all ages, and it was regularly localized in both lumina and PCM of mother centrioles even without previous degradation of CPAP-AID (Fig. S2 D), confirming that mother centrioles’ CPAP populations are dynamic and exchange with the cytosolic population.
To test whether mother centrioles in IAA-treated cells can still initiate duplication, we collected mitotic cells by shake-off, replated them, and treated them with IAA 6 h later, while still in G1 (Fig. 2 G). We fixed cells after 10 h (when control cells normally begin entering S phase and centrioles start to duplicate) and after 16 h (when most control centrioles are duplicated) and analyzed CPAP by immunofluorescence. In late G1, mother centrioles harbored low levels of luminal CPAP (Fig. 2 H, cells 1 and 2), traces of which were barely detectable or undetectable in ∼30% of cells by STORM. Yet mother centrioles deprived of CPAP initiated duplication, as evidenced by the second adjacent CPAP signal colocalizing with the emerging C1-GFP signal near mother centrioles (Fig. 2 H, cells 3 and 4). 16 h after shake-off, 98% of mother centrioles initiated duplication (Fig. 2 I). This result was further verified by the presence of a cartwheel protein Sas-6 within the ring of PCM protein Cep152 in 97% of mother centrioles (Fig. 2 I). We concluded that the three CPAP populations (i.e., mother centriole’s PCM, mother centriole’s luminal, and procentriole’s luminal population) localize independently on the centrosome, and that removal of CPAP from mother centrioles is not consequential for initiation of their duplication. In addition, these experiments showed that the PCM and mother centriole luminal CPAP populations are present in a dynamic equilibrium with the cytosolic CPAP population, although the luminal CPAP binds with higher affinity. This conclusion is consistent with previous work in C. elegans (Dammermann et al., 2008). Remarkably, procentriole initiation could still be initiated with only ∼3% of residual CPAP. So, we reasoned that HeLaCPAP-AID and DLD-1CPAP-AID cells were an ideal model to deplete CPAP rapidly from cytosolic and centrosomal populations and to study the effects of acute and prolonged CPAP deficiency on centriole assembly and centrosome organization and function.
Assembled mother centrioles are stable after CPAP removal
To understand how the loss of CPAP affects already assembled mother centrioles long-term, HeLaCPAP-AID and parental HeLaC1-GFP cells were synchronized by mitotic shake-off and treated with IAA or CPAP-siRNA, respectively, 2 h later. Centrinone, an inhibitor of centriole-initiating kinase Plk4 (Wong et al., 2015), was added to the culture medium to inhibit formation of new centrioles (Fig. 3 G). Cells were fixed at the time of the first, second, and third mitotic/postmitotic wave (24, 40, and 60 h after shake-off), and CPAP and γ-tubulin were labeled by immunostaining and imaged. To analyze structural centriolar features, some samples were expanded and labeled for acetylated tubulin. Microscopy revealed that after three consecutive cell cycles, including three mitoses, mother centrioles with significantly reduced or undetectable CPAP normally recruit γ-tubulin (Fig. 3 H) and remain structurally intact (Fig. 3, I and J).
Acute degradation of CPAP does not perturb PCM organization and MT nucleation in interphase
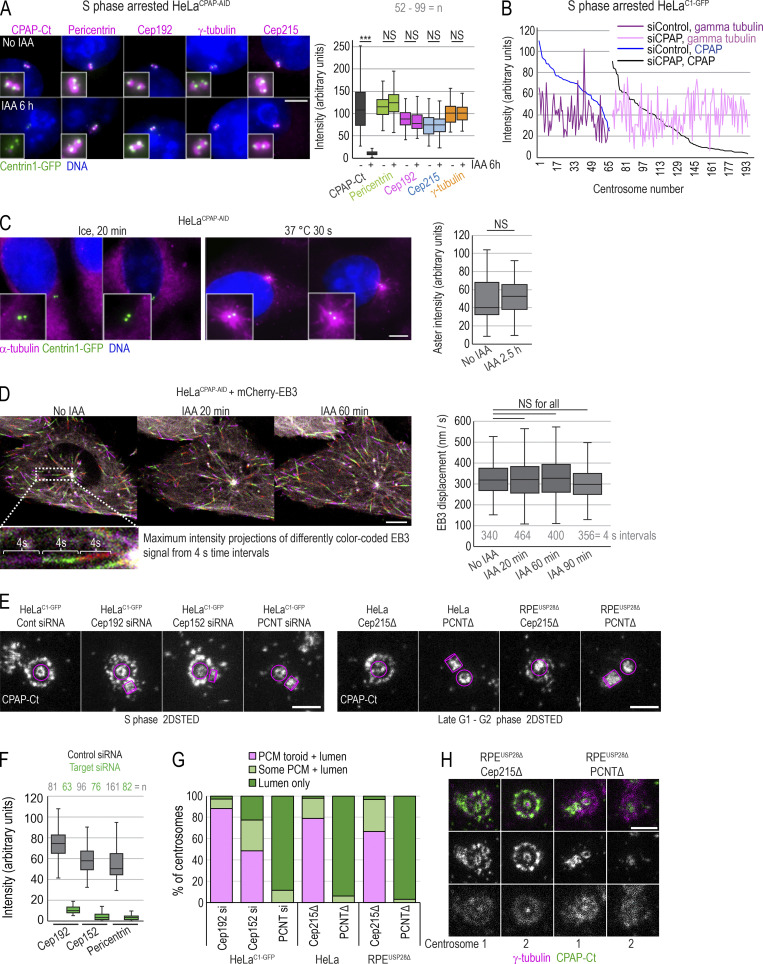
CPAP has been implicated in PCM recruitment and organization in both Drosophila and human cells (Chou et al., 2016; Gopalakrishnan et al., 2011; Ramani et al., 2018; Zheng et al., 2014), by forming cytoplasmic complexes with PCM and promoting their delivery to centrosomes. Now, HeLaCPAP-AID and DLD-1CPAP-AID cells allowed us to directly test this claim. For acute removal of CPAP, we treated S phase cells with IAA for 6 h and analyzed centrosomal levels of four major PCM components: pericentrin, Cep192, Cep215, and γ-tubulin (Dictenberg et al., 1998; Fong et al., 2008; Gomez-Ferreria et al., 2007; Woodruff et al., 2014). We found that IAA-treated cells retained control levels of PCM components on their centrosomes (Fig. 4 A). Similarly, we found no correlation between the levels of centriole-bound CPAP and γ-tubulin in CPAP siRNA-treated cells (Fig. 4 B).
Figure 4.
Acute removal of CPAP does not perturb centrosome PCM recruitment or MT nucleation in interphase. (A) S phase–arrested cells were treated or not with IAA for 6 h, immunolabeled for indicated PCM proteins, and imaged. Plot: Quantification of PCM proteins is shown in box-and-whiskers plot. n = cell number. (B) Cells were treated with CPAP siRNA, arrested in S phase for 26 h, immunolabeled for γ-tubulin and CPAP, and imaged. The levels of CPAP and γ-tubulin were determined and plotted for each centrosome. No correlation between centrosome-associated CPAP and γ-tubulin was observed after plotting. (C) MT nucleation recovery after cold treatment. IAA treatment for 2.5 h does not change the intensity of MT asters, labeled by α-tubulin, after recovery. n = number of asters. (D) Cells expressing mCherry-EB3 were imaged every second over 1 min to record the position of growing MT tips. Maximum-intensity projections of four 1-s frames were generated and color coded (as shown in inset). Displacement of mCherry-EB3 in 4-s intervals was measured for MTs showing linear growth. Box-and-whiskers plot represents displacement of mCherry-EB3 signals within the same cell before and after 20, 60, or 90 min of IAA treatment. n = number of measured 4-s intervals. (E) CPAP was detected by immunofluorescence in HeLaC1-GFP cells depleted for Cep192, Cep152, or pericentrin (PCNT) and in RPE1-USP28Δ cells knocked out for pericentrin or Cep215. CPAP signal was imaged by 2DSTED. Removal of pericentrin results in the loss of the PCM CPAP population. Magenta lines delineate centriole’s position and orientation. (F) Levels of indicated protein on centrosomes of cells transfected with control and targeting siRNA. (G) Plot: Characterization of CPAP phenotypes observed from 2DSTED recordings. (H) Interphase centrosomes in Cep215 and pericentrin knockouts associate with γ-tubulin. n = centrosome number. Scale bars: 5 µm (A, C, and D); 0.5 µm (E and H). ***, P ≤ 0.001.
Consistently, centrosome-mediated MT nucleation, that is mediated by PCM, appeared unchanged after IAA addition, as determined by a MT regrowth assay after MT removal by cold or nocodazole treatment (Figs. 4 C and S3 B). To further test whether CPAP removal affects MT dynamics, we expressed fluorescently tagged EB3, a protein that dynamically binds the growing plus ends of MTs (Alieva et al., 2013), and calculated displacement of EB3 before and after CPAP depletion from time-lapse recordings. The analysis showed similar dynamics of EB3 before and after CPAP depletion (Fig. 4 D).
Based on proximity labeling and coimmunoprecipitation assays, three PCM proteins, Cep192, Cep152, and pericentrin, interact with CPAP (Chou et al., 2016; Dzhindzhev et al., 2010; Firat-Karalar et al., 2014; Zheng et al., 2014). To understand whether these PCM proteins mediate recruitment of CPAP to PCM, we removed each of these proteins and analyzed CPAP localization. HeLaC1-GFP cells were transfected with the corresponding siRNA. Mitotic cells were collected 24 h later, and hydroxyurea was added to synchronize cells at the G1/S border. 24 h later (∼48 h of siRNA transfection), cells were fixed, immunolabeled for CPAP, and imaged by 2DSTED. Parallel coverslips were labeled for each depleted protein to determine the level of depletion. Imaging revealed that depletion of pericentrin abrogated the PCM CPAP population, whereas depletion of Cep192 did not have an effect (Fig. 4, E–I). Depletion of Cep152 reduced overall CPAP levels on centrosomes by ∼30% and partially abrogated its presence within the PCM (Fig. 4, E and G), consistent with the previous observation that removal of Cep152 decreases centrosomal CPAP signal (Cizmecioglu et al., 2010; Dzhindzhev et al., 2010; Sonnen et al., 2013). Additionally, we analyzed CPAP distribution in previously characterized (Watanabe et al., 2020) pericentrin and Cep215-knockout HeLa and RPEUSP28Δ cells. In pericentrin knockouts, CPAP was entirely absent from PCM, whereas Cep215 knockout did not affect its localization (Fig. 4 E). Centrioles in both knockouts were still associated with γ-tubulin (Fig. 4 I), Cep152, and Cep192 (not depicted). Thus, while pericentrin, and to some extent Cep152, mediate CPAP localization to PCM, the localization of PCM components studied here is not mediated by CPAP in the cell lines we explored.
Intact mother centrioles organize functional mitotic spindle poles in the absence of CPAP
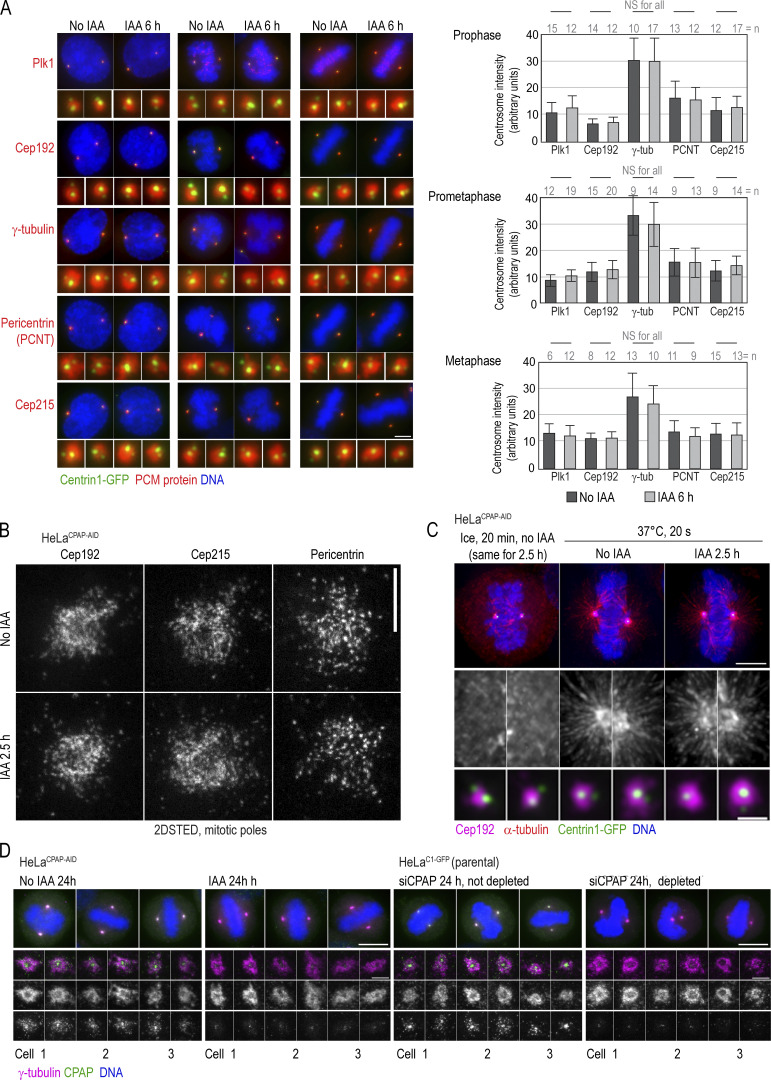
To understand whether CPAP is required for mitotic centrosome maturation and the assembly of mitotic spindle poles, we acutely removed CPAP by IAA treatment for 2.5–6 h and analyzed PCM components on prophase, prometaphase, and metaphase centrosomes. Quantifications revealed that centrosomes lacking CPAP accumulated PLK1, Cep192, γ-tubulin, pericentrin, and Cep215 at levels comparable to controls (Fig. 5 A). High-resolution imaging revealed no discernable differences in the pattern of Cep192, Cep215, and pericentrin, three structural components of the mitotic PCM lattice, after CPAP removal (Fig. 5 B). Consistently, in MT regrowth assays, both control cells and IAA-treated mitotic cells regrew MT asters of comparable sizes (Fig. 5 C).
Figure 5.
Acute removal of CPAP does not perturb centrosome PCM recruitment or organization of PCM components during mitosis. (A) Cells were treated with IAA for 6 h and immunolabeled for indicated PCM proteins. Imaging and quantification of the intensities of PCM proteins show comparable centrosomal levels in control and IAA-treated cells. Histograms show the average intensity ± SD, n = centrosome number. (B) 2DSTED imaging shows that the patterns of localization of three major components of the mitotic PCM lattice, Cep192, Cep215, and pericentrin, are not changed after 2.5 h of IAA treatment. (C) MT nucleation recovery after cold treatment. IAA treatment for 2.5 h does not change the recovery of MT nucleation or centrosomal levels of Cep192. (D) Structurally intact mother centrioles organize control-looking mitotic spindles. Cells were treated with IAA or depleted by siRNA, and centrinone was added to prevent centriole duplication, as described in Fig. 3 G. Cells were fixed at 24 h when they were undergoing mitosis, immunolabeled for γ-tubulin and CPAP, and imaged by wide-field microscopy; spindle poles were imaged by STED. Scale bars: 5 µm (A); 0.5 µm (B); 5 µm (C); 1 µm (enlarged centrosomes in C); 10 µm (wide-field in D); 1 µm (STED in D).
To eliminate the possibility that the 3% of residual CPAP in IAA-treated cells assisted with PCM recruitment, we depleted CPAP using siRNA in RPE-1 cells (Fig. S4, A–C). In our hands, siRNA transfection, in some cells, leads to an even greater drop in CPAP, preventing centriole duplication in ∼10% of cells within the first cell cycle (∼24 h after transfection). At that time point, mitotic cells contain intact mother centrioles formed before siRNA transfection (Fig. S4 A). Analysis of mitotic spindle poles containing unduplicated mother centrioles revealed that they recruit control levels of PLK1, Cep215, and pericentrin (Fig. S4, B and C).
Figure S4.
The effect of CPAP removal on recruitment of PCM in mitosis by structurally intact mother centrioles. (A–C) Characterization of PCM recruitment by intact mother centrioles in RPE-1 cells after CPAP depletion. RPE-1 cells were collected by shake-off, replated, and transfected with CPAP siRNA or control non-targeting siRNA ∼2 h later. Cells were fixed 24 h after shake-off, when they were undergoing mitosis. Cells were immunolabeled for PLK1, Cep215, and pericentrin, and analyzed by microscopy. Please note that all cells at the time of fixation harbor structurally intact mother centrioles formed before CPAP depletion. (A) Cartoon depicting the effect of various levels of CPAP depletion by siRNA on centriole duplication. (B) Examples of mitotic cells immunolabeled for indicated PCM proteins after CPAP depletion. Mother centriole duplication status is indicated above the image. Cells with unduplicated mother centrioles were considered depleted for CPAP. The images show that both control and CPAP-depleted mother centrioles contain similar levels of PCM components. (C) Quantification of PCM signals on centrosomes formed by control or CPAP-depleted mother centrioles. (D) Characterization of γ-tubulin recruitment by intact mother centrioles in HeLaCPAP-AID cells after CPAP degradation. HeLaCPAP-AID were shaken off and 2 h later treated with IAA to degrade CPAP and with centrinone to prevent duplication of mother centrioles (see Fig. 3 G). Cells were fixed 40 or 60 h after shake-off, when they were undergoing their second and third mitosis, respectively. Cells were immunolabeled for γ-tubulin and CPAP and imaged by wide-field. Corresponding centriole-containing spindle poles were additionally imaged using 2DSTED. Images show examples of mother centriole-containing spindle poles. In IAA-treated cells, mother centriole-associated CPAP is reduced or undetectable. However, γ-tubulin is accumulated to mitotic spindle poles at levels comparable to the controls. Cartoons depict mitotic figures present in the population at indicated times. n = centrosome number. Scale bars: 5 µm (B); 10 µm (D); 1 µm (enlarged centrioles in D).
Finally, using the same treatment scheme shown in Fig. 3 G, we analyzed the long-term ability of unduplicated mother centrioles to assemble functional mitotic spindle poles. Cells were treated with IAA or CPAP siRNA, and centriole duplication was inhibited by centrinone. Cells were fixed 24, 40, and 60 h later and labeled for γ-tubulin and CPAP, and mitotic cells were analyzed. At 24 h, mitotic cells contained symmetrical bipolar mitotic spindles, and mother centrioles formed functional spindle poles with levels of γ-tubulin comparable to control cells (Fig. 5 D). The second and third mitoses were initially monopolar, as cells contained one mother centriole (at 40 h) or one or no mother centrioles (at 60 h; Fig. S4 D). However, mother centrioles were still able to assemble control-looking spindle poles (Fig. S4 D), and mitotic spindles eventually bipolarized (not depicted).
We concluded that intact mother centrioles of cultured human cells can assemble functional mitotic spindle poles in the absence of cytosolic or centriole-bound CPAP.
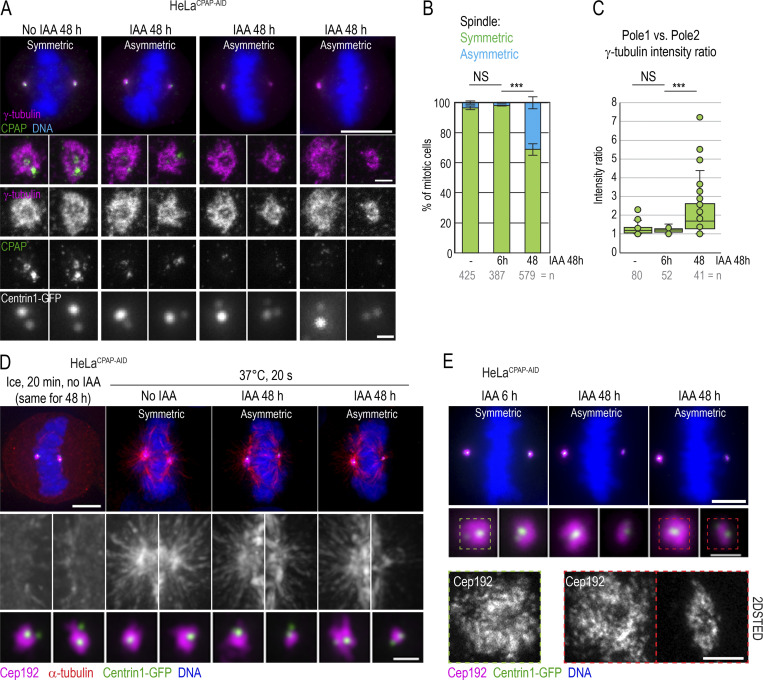
Abnormal mitotic spindles occur in cells that assemble centrioles under low CPAP levels
Because intact mother centrioles repeatedly organized control-looking spindle poles after CPAP removal, we reasoned that aberrant mitotic figures reported after prolonged CPAP siRNA treatment in human cells originate from structural aberrations acquired during centriole assembly after partial CPAP depletion, as previously documented in other species (Gogendeau et al., 2011; Kirkham et al., 2003; Pelletier et al., 2006). So, we analyzed mitotic spindle poles in cells that were allowed to duplicate centrioles during 48-h treatment with IAA. As controls, we used untreated cells or cells treated with IAA for only 6 h. We found that control mitotic cells contained symmetric bipolar mitotic spindles, with control levels of γ-tubulin (Fig. 6, A–C) and Cep192 (Fig. 6, D and E) on each spindle pole. However, ∼30% of mitotic HeLaCPAP-AID cells treated with IAA for 48 h exhibited asymmetric spindles, containing mostly one control-looking spindle pole and the second pole with lower levels of γ-tubulin (Fig. 6, A–C) and Cep192 (Fig. 6, D and E). The “weaker” spindle poles harbored two C1-GFP signals, indicating duplicated mother centrioles, although C1-GFP signals were often dimmer in comparison to the poles with control PCM levels (Fig. 6, A, D, and E). The weaker spindle poles also nucleated smaller α-tubulin asters after MT regrowth assay (Fig. 6 D). High-resolution imaging revealed no discernable differences in the pattern of Cep192 on the dominant centrosomes of asymmetric spindles (Fig. 6 E). The occurrence of the control-looking and abnormal centrosome/spindle pole in the same cell further signified that the observed centrosome defects were inherent to the centrosomes and not driven by the perturbation of stoichiometry between CPAP and other PCM proteins. We moved on to characterize the ultrastructure of centrioles formed under low CPAP levels.
Figure 6.
Asymmetric mitotic spindle poles with reduced PCM in cells after prolonged CPAP degradation. Asynchronous cells were treated with IAA for the indicated time, fixed, and immunolabeled for γ-tubulin + CPAP or α-tubulin + Cep192. (A) Examples of asymmetric spindle poles with decreased recruitment of γ-tubulin after 48 h of IAA treatment. Cells were imaged by wide-field microscopy, and the spindle poles were additionally imaged by 2DSTED. (B) Quantification of mitotic cells harboring mitotic spindle asymmetry. n = cell number. (C) Calculated γ-tubulin intensity ratios between two poles of the same mitotic spindle reveals poles of similar intensity in control cells but of asymmetric intensity in cells treated with IAA for 48 h. n = cell number. (D) MT nucleation recovery after cold treatment. In mitotic cells treated with IAA for 48 h, spindle poles associated with lower amounts of PCM protein Cep192 recover smaller MT asters after cold treatment. (E) The pattern of Cep192 localization on mitotic spindle poles in cells treated with IAA for 6 or 48 h, as analyzed by 2DSTED. CPAP degradation does not affect the pattern of Cep192 (compare to Fig. 5 B). Scale bars: 10 µm (A); 0.5 µm (centrosomes in A); 5 µm (D); 1 µm (centrosomes in D); 5 µm (E); 0.5 µm (centrosomes in E). ***, P ≤ 0.001.
CPAP deficiency results in short and broken centrioles that convert to centrosomes and duplicate
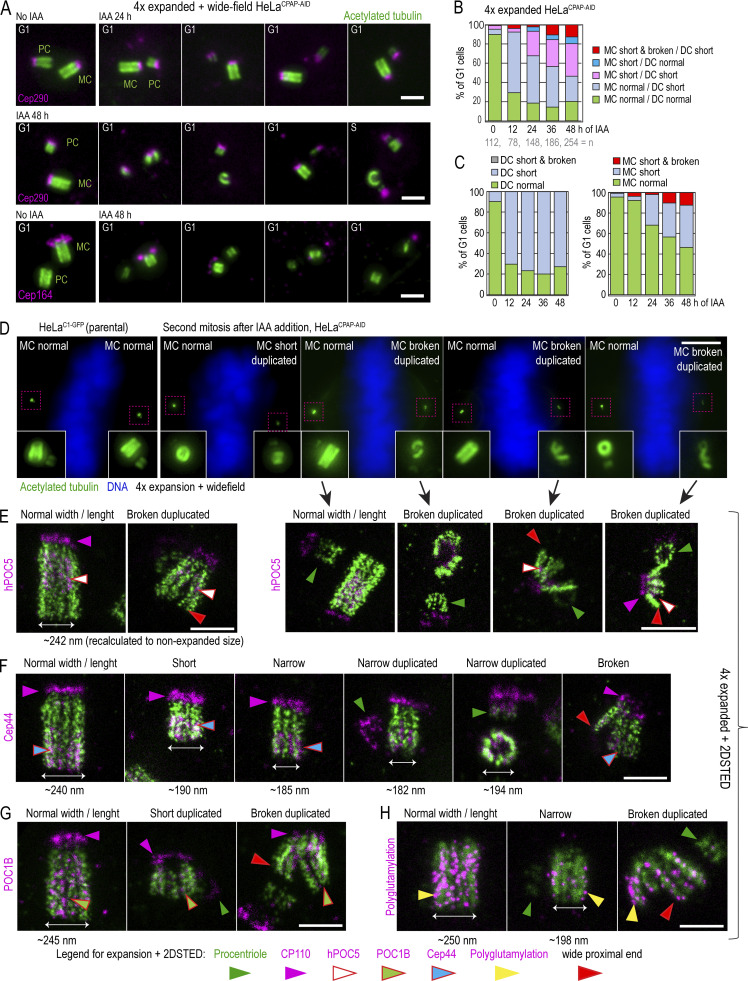
To understand the effect of prolonged CPAP deficiency on centriole structure, we analyzed the structure of centrioles assembled in IAA-treated cells by expansion microscopy. Cells were collected after increasing time spent in IAA (12–48 h; Fig. 7), expanded, and immunolabeled using anti-acetylated tubulin antibody to mark centriole MT walls. As needed, centrioles were colabeled with centriole distal cap protein Cep290 (Tsang et al., 2008) or distal appendage protein Cep164 (Graser et al., 2007).
Figure 7.
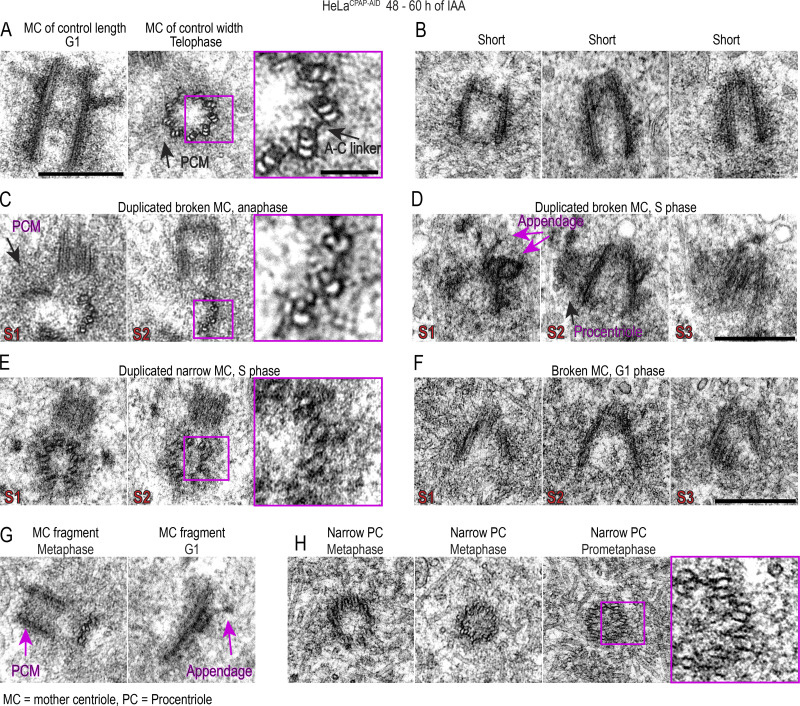
CPAP removal leads to the assembly of structurally defective centrioles that destabilize on proximal ends. Cells were synchronized by mitotic shake-off (t = 0 h) and treated with IAA 6 h after shake-off. Cells were fixed when noted, expanded approximately fourfold, immunolabeled using acetylated tubulin to label centriole MT walls (green) and various centrosomal proteins (magenta), and imaged. (A) Examples of centriole configurations in control and IAA-treated cells. (B and C) Characterization of the phenotypes of MC-DC pairs in G1 cells at indicated times. Phenotypes of MCs and DCs are additionally shown individually in C. (D) Wide-field images of mitotic cells after 48 h of IAA treatment containing one normal-looking and one aberrant mother centriole. 2DSTED high-resolution images of four centrioles indicated by arrows are shown below. (E–H) Examples of expanded centrioles after 36–48 h of IAA treatment, labeled with acetylated tubulin and colabeled with centriole cap protein CP110, centriole distal protein hPOC5 (E), centriole proximal protein Cep44 (F), centriole luminal protein POC1B (G), and polyglutamylated tubulin (H). The panels illustrate mother centrioles that lost cohesion on proximal ends (red arrows) and that have shorter and narrower acetylated tubulin signal than controls. M, mitosis. Scale bars: 2 µm (A); 20 µm (wide-field in D); 2 µm (2DSTED in D); 1 µm (E–H).
Untreated G1 cells largely contained two centrioles of normal length (Fig. 7, A–C). After 12 h of IAA treatment, the number of G1 cells harboring a control-looking mother centriole and one short and/or narrow daughter centriole gradually increased (Fig. 7, A [upper panel] and B). The number of G1 cells harboring two short centrioles further increased by 48 h. In addition, at 48 h, ∼30% of cells (n = 300) contained short centrioles with some type of structural abnormality, including uneven and reduced acetylated tubulin signal, a gap in acetylated tubulin signal, a mother centriole with reduced levels of appendage proteins (prevalent on centrioles shorter than ∼250 nm; Fig. 7 A, lower panel), or acetylated tubulin fragments associated with Cep290 and Cep164 (Fig. 7 A, middle and bottom panels). Surprisingly, even drastically short (∼230-nm), narrow, and otherwise defective centrioles duplicated in S phase.
Consistent with aberrant images of interphase centrioles after 48 h, in ∼30% of mitotic cells, one spindle pole contained a structurally aberrant or broken mother centriole, and another pole contained a normal or shorter but otherwise intact mother centriole (illustrated in Fig. 7 D, wide-field). Both normally structured and aberrant centrioles were associated with procentrioles. More rarely, mitotic poles contained only a centriole fragment associated with a procentriole. Similar trends and centrosome phenotypes were observed when IAA was added to DLD-1CPAP-AID cells (Fig. S3, C and D).
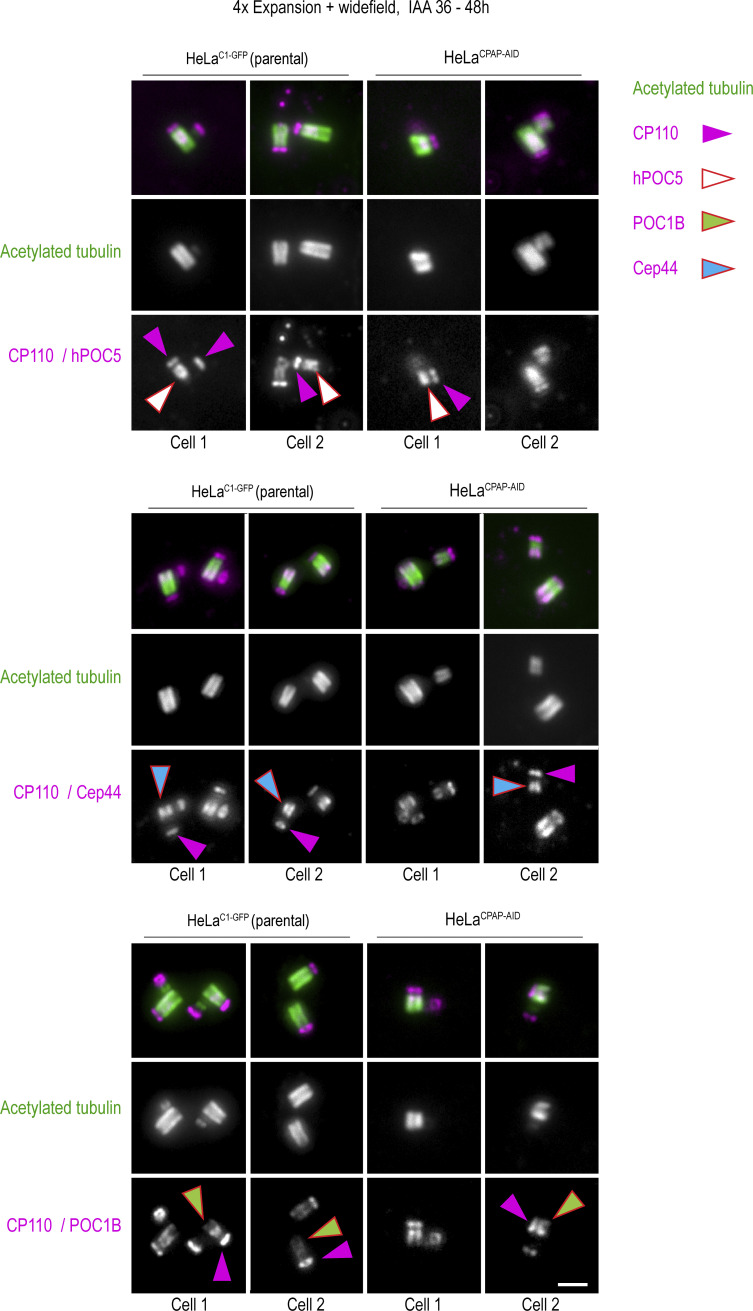
Centrioles assembled in CPAP-depleted cells lose cohesion between MT blades on proximal ends
To better understand the nature of centriolar defects caused by CPAP insufficiency, we labeled additional centriolar proteins on expanded centrioles and imaged them using 2DSTED to further increase resolution. Specifically, we labeled CP110, which localizes distally above centriole MTs; Cep44, which localizes proximally (Atorino et al., 2020); hPOC5, which is enriched toward distal ends (Azimzadeh et al., 2009; Steib et al., 2020); and POC1B (Pearson et al., 2009; Venoux et al., 2013), which, in our hands, spans almost the entire centriole length except the very distal end (Fig. S5). hPOC5 and POC1B are putative inner centriole scaffold elements (Le Guennec et al., 2020), and Cep44 contributes to procentriole structuring and maturation (Atorino et al., 2020).
Figure S5.
Localization of centrosomal proteins on expanded HeLaC1-GFP and HeLaCPAP-AID centrioles after 36–48 h of IAA treatment. Cells were treated with IAA for 36–48 h, fixed, expanded, and immunolabeled for indicated centriolar proteins (magenta) and acetylated tubulin (green). Both control centrioles and short centrioles contain indicated centrosomal proteins. Arrows point to individual proteins, to facilitate interpretation. Proximal-distal polarity is preserved on short and narrow centrioles in IAA-treated cells. On short centrioles, proximal and distal portions are both shorter than normal. Scale bar: 2 µm (corresponding to ∼0.25 µm in unexpanded sample). This figure is associated with Fig. 6 of the main text.
As in parental cells, in IAA-treated HeLaCPAP-AID cells, all proteins were present on structurally intact, narrow, short, and broken centrioles (Fig. 7, D–H; and Fig. S5). Interestingly, of 35 broken centrioles analyzed by 2DSTED after 36–48 h of IAA treatment, all but 2 showed widened proximal ends (Fig. 7, D–H, red arrows). In such centrioles, Cep44, hPOC5, and POC1B were still associated with detached MT blades. In addition, in IAA-treated cells, centrioles, including short, narrow, and broken ones, could acquire/maintain polyglutamylation (Fig. 7 H), which is normally acquired by centrioles at the time of their first mitosis (Sullenberger et al., 2020).
To understand at which stage centrioles of IAA-treated cells structurally destabilize, we scored centrioles in their first G1 and S phases after their first mitosis (24–36 h of IAA treatment), and then in their second mitosis (∼48 h of IAA treatment). In their first G1 phase, the MTs of short and narrow centrioles appeared parallel (n = 51). At 36 h, ∼16% (n = 87) of short S phase centrioles exhibited widened proximal ends, which was consistent with detachment of MT blades. The number of such centrioles slightly increased in the following mitosis to 23% (n = 122). In mitotic cells, defects appeared more severe, indicating a progressive loss of cohesion between centriole MT blades.
Intriguingly, some centrioles went on to duplicate even though they retained a narrow diameter of acetylated tubulin (Fig. 7 F). It is important to mention that normally, the width of acetylated tubulin signal increases on younger centrioles from early to late G1, so both S phase mother centrioles usually have similar widths (Sahabandu et al., 2019), which is ∼240 nm at the proximal end (Fig. 7, E–G).
Human centrioles with incomplete MT triplets can undergo centriole-to-centrosome conversion
Duplicated, narrow centrioles in IAA-treated cells could either lack acetylation on their outer MTs or have incomplete MT triplets. Thus we additionally analyzed centrioles from cells treated with IAA for 48–60 h by EM. As expected, we detected numerous centrioles of smaller than average length (Fig. 8). In addition, we detected centrioles in late G1 and S phase cells with smaller than normal diameters, which was determined by measuring the distance between centrioles’ outmost MTs. In addition, two of four cross-sectioned duplicated mother centrioles showed MT doublets instead of triplets in anaphase and S phase (Fig. 8, C and E). Finally, although in human procentrioles MT triplets are completed by mitosis (Vorobjev and Chentsov Yu, 1982), we found multiple examples of mitotic and early G1 procentrioles containing MT doublets instead of triplets (Fig. 8 H). The inner lumen of narrow centrioles was not changed with respect to control centrioles. In addition, EM revealed broken centrioles, with loss of cohesion between centriole blades and widening of the proximal ends (Fig. 8, D and E), mirroring defects detected by expansion microscopy. Some broken centrioles and centriole fragments were found in association with short but otherwise intact procentrioles, and isolated MT blades could carry distal appendage-like structures (Fig. 8 G), indicating that structurally unstable centrioles matured, as suggested in expansion experiments.
Figure 8.
Prolonged CPAP removal results in centrioles with incomplete MT triplets that can convert to centrosomes and duplicate. CPAP was degraded by addition of IAA to culture medium for 48–60 h. Cells were fixed and analyzed by EM. The figure illustrates various centriolar phenotypes detected during analysis. (A) Examples of normally structured centrioles. (B) Short and narrow centrioles from late G1 cells. (C) Broken mother centriole built of MT doublets instead of MT triplets in anaphase. Mother centriole is associated with a procentriole. (D) Short mother centriole with unparallel MT blades that lost cohesion on proximal end. Mother centriole is associated with a procentriole, and appendage-like structures are visible on its distal end. (E) Duplicated narrow mother centriole in S phase built of MT doublets. (F) A short centriole with unparallel MT blades and widened proximal end. (G) Centriole fragments. Left: Centriole fragment built of MT doublets associated with a normal-looking procentriole. Right: Centriole fragment associated with a distal appendage. (H) Narrower-than-normal mitotic procentrioles with incomplete MT triplets. Scale bars: 0.4 µm; 0.1 µm (insets marked in magenta). S1–S3: the number of a consecutive 80-nm-thick serial section.
EM data clarified that human centrioles could persist through mitosis and stabilize in the absence of a complete set of MT triplets, even if they were entirely built of MT doublets. Considering that these centrioles can also duplicate and acquire polyglutamylation, we concluded that they undergo conversion to centrosomes.
In summary, we show that in cells deficient for CPAP, centrioles are shorter, and they frequently fail to incorporate the outer tubules to their MT blades. Because the number of centrioles with detached MT blades increases from their first G1 until second mitosis, we reason that such converted centrioles gradually destabilize, as depicted in Fig. 9.
Figure 9.
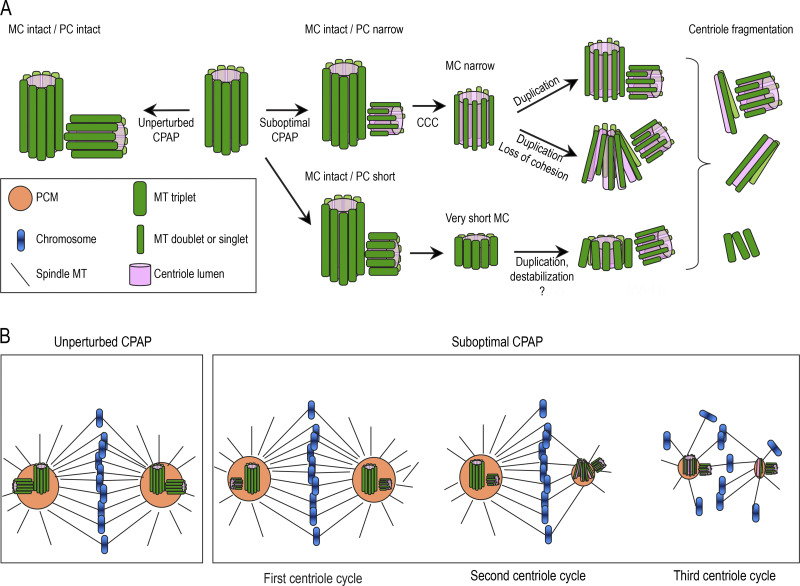
Schematic summary of the consequences of suboptimal CPAP levels in human cells. The scheme illustrates centrioles and mitotic spindles in human cells with CPAP levels reduced to ∼3% of their original levels using AID system (Fig. 2). (A and B) Unperturbed levels of CPAP result in the assembly of intact centrioles during centriole duplication (A). Suboptimal CPAP levels allow centriole duplication but result in the assembly of shorter and sometimes narrower procentrioles (PC). Instead of nine MT triplets, narrower procentrioles contain some or all MT doublets, and possibly singlets. Although built of incomplete MT triplets, procentrioles can undergo centriole-to-centrosome conversion (CCC), and in the subsequent cell cycle, they can acquire PCM and duplicate. However, structurally aberrant centrioles are unstable, lose cohesion between MT blades, destabilize, and fragment. MC, mother centriole. Consequences of acute and prolonged CPAP levels on mitotic spindles (B). In cells with unperturbed CPAP levels, mitotic spindles are largely symmetric. Reduction of CPAP within one centriole cycle affects the assembly of nascent centrioles but does not affect the accumulation of PCM on mitotic spindle poles by previously assembled and structurally intact MCs. Reduction of CPAP levels for longer than one centriole cycle results in asymmetric mitotic spindle poles with reduced levels of PCM, which are organized by structurally defective centrioles or centriole fragments.
Discussion
Herein, we analyze centrosomal localization of CPAP and its cell cycle dynamics in human cultured cells at ∼30-nm resolution. We found that CPAP localizes to three independent centrosomal populations that are differently regulated during the cell cycle. Within the mother centriole lumen, a population of CPAP organizes as a toroid ∼100 nm in diameter, the lateral localization of which roughly corresponds to the position of the cartwheel during early stages of centriole structuring, when CPAP is first accumulated at the centriole. The role of this population is not clear to us at this point, as its reduction due to CPAP degradation or CPAP siRNA transfection does not affect mother centrioles’ stability or ability to duplicate or organize PCM.
On mother centrioles, CPAP additionally localizes to the PCM (in agreement with Kleylein-Sohn et al. [2007]; Sonnen et al. [2012]; Fig. 1), where it can form a toroid ∼460 nm in diameter, which is similar regardless of whether it is detected using the antibody against Nt or Ct, suggesting that CPAP is compactly organized. The levels of luminal and PCM CPAP vary considerably, and the toroidal organization is not always detectable, especially in mitosis. This is consistent with a previous live-cell analysis in fly embryos showing that Sas-4 levels drop as embryos enter mitosis (Novak et al., 2014) and with the observation that, in mitosis, CPAP is more focused than γ-tubulin (Kitagawa et al., 2011).
CPAP/SAS-4 is, reportedly, required for tethering/recruitment of PCM to centrosomes and for organization of mitotic spindle poles in both Drosophila and human cells (Gopalakrishnan et al., 2011; Ramani et al., 2018; Zheng et al., 2016). However, in our hands, removal of ≥97% of CPAP and its loss from centrioles was inconsequential for the assembly of interphase and mitotic PCM by structurally intact mother centrioles. In contrast, the analysis of mitotic spindle poles after several consecutive centriole cycles with decreased CPAP levels recapitulated mitotic defects previously described after CPAP depletion by siRNA. But it also revealed that the aberrant spindle poles, characterized by low PCM levels, consistently harbored structurally aberrant centrioles, whereas control-looking spindle poles harbored intact centrioles (formed before CPAP was reduced). The absence of mitotic spindle defects after acute CPAP removal, and the co-occurrence of the control- and aberrant-looking mitotic spindle poles within the same cytoplasm, do not support the idea that CPAP mediates delivery or tethering of analyzed PCM components to the centrosome of cultured human cells. Our conclusion aligns with several previous observations. In U2OS cells, accumulation of PCM to centrosomes is normal after shorter CPAP depletion by siRNA (Kitagawa et al., 2011). Drosophila cells lacking CPAP/SAS-4 can recruit PCM during mitosis (Riparbelli and Callaini, 2011; Stevens et al., 2007), and in Drosophila syncytial embryos, PCM proteins do not recruit to the mitotic centrosomes as a part of the SAS-4 complex (Conduit et al., 2015). Finally, in C. elegans embryo, it was centriole duplication, but not PCM recruitment, that failed in the presence of a mature centriole after SAS-4 removal (Kirkham et al., 2003). More work will be needed to unravel the role of CPAP on mature centrioles of somatic human cells.
CPAP also localizes in the lumina of procentrioles (Fig. 1 A; Kleylein-Sohn et al., 2007). Using Nt- and Ct-targeting CPAP antibodies, we further dissected this CPAP population and showed that during S phase, the Ct of CPAP localizes within the procentriole lumen, while its Nt extends further outward, past the centriole MTs. However, this radial CPAP distribution on procentriole proximal ends appears temporary, since after S phase, both Nt and Ct signals localize within the procentriole’s proximal lumen. Endogenous CPAP was previously found to be present primarily within the proximal centriole ends (Kohlmaier et al., 2009; Schmidt et al., 2009; Tang et al., 2009), suggesting that it might be critical only during the early stages of centriole elongation (Sharma et al., 2016). However, per our analysis, in prophase and mitosis, CPAP localizes to the distal procentriole ends, where it could participate in elongation of the procentriole distal part, which, in human cells, is accelerated during these cell cycle phases (Kong et al., 2020). Indeed, the proximal and distal end can both be shorter in IAA-treated cells, judging by Cep44 and hPOC5 as markers for the proximal and distal centriole compartment, respectively (Figs. 7 and S5).
Reduction of the cytosolic pool of CPAP to 3–5% of its original level allowed procentriole initiation. But procentrioles formed in these cells were not only short, as previously documented after CPAP siRNA treatments, but were also narrow. EM revealed that narrow centrioles failed to incorporate the outer C tubules to their MT blades. Human centrioles lacking intact MT triplets also occur after depletion of Cep44, POC1B, and Cep295 (Atorino et al., 2020; Chang et al., 2016). The long-term fate of these centrioles is unknown, but after first mitosis, such centrioles reportedly were not able to recruit PCM components and incorporate glutamylation detectable by GT335 antibodies (Atorino et al., 2020). Structurally unstable centrioles were also documented in δ- and ε-tubulin knockout RPE-1 cells (Wang et al., 2017). In these knockouts, multiple centrioles initiated in S phase de novo but failed to form MT triplets, destabilized in mitosis, and became undetectable by G1. These observations prompted the intriguing idea that human centrioles cannot convert to centrosomes without structurally intact MT triplets (Atorino et al., 2020; Wang et al., 2017).
Here we report that human centrioles lacking a complete set of MT triplets can convert to centrosomes, duplicate, and accumulate PCM. Therefore, we propose that, in human cells, the assembly of an intact set of MT triplets is not a prerequisite for centriole-to-centrosome conversion (Wang et al., 2011). In agreement, stabilized and duplicated basal bodies with all or occasional MT doublets or singlets instead of triplets have been previously documented in Paramecium (Garreau de Loubresse et al., 2001; Gogendeau et al., 2011) and Chlamydomonas (O’Toole et al., 2003) and in Drosophila spermatocytes (Zheng et al., 2016), although in these cells basal bodies are, as in human cells, normally built of MT triplets.
Although converted, structurally aberrant centrioles formed under low CPAP levels appear unstable and break. Interestingly, almost all broken centrioles showed a uniform pattern of breakdown, characterized by loss of cohesion between MT blades on the proximal end, while still maintaining cohesion on their distal lends (Figs. 7 and 8). We speculate that the partial or complete lack of C tubules could prevent the adjacent centriole MT blades from establishing the A–C linker (Fig. 8 A; Klena et al., 2020; LeGuennec et al., 2021; Nazarov et al., 2020; Vorobjev and Chentsov Yu, 1982) and stabilizing the proximal end of the MT barrel. Ultimately, passage through the cell cycle, and specifically through dynamic mitosis, in the absence of the A–C linker could lead to detachment of MT blades. Although centrioles as short as ∼230 nm can propagate in culture and even duplicate, there is a possibility that some of the overly short centrioles disintegrate due to their small size, and not due to lack of triplet MTs, which does not allow for the incorporation of sufficient amounts of proximal and distal elements needed for long-term stability.
In perspective, disruption of multiple centriolar proteins can result in ultrastructural defects and ultimately lead to centriole loss and mitotic spindle defects. Nevertheless, our data and published ultrastructural data show that depletion of each protein carries a specific structural and biochemical signature, which can help in understanding their role within the centriole and ultimately in human diseases. Our detailed analysis of centriolar defects caused by perturbed CPAP stoichiometry can now be used as a stepping stone for future, in-depth characterization of centriolar defects caused by specific CPAP mutations identified in association with human diseases.
Materials and methods
Cell culture and drug treatments
Retinal pigment epithelial RPE-1C1-GFP (Uetake et al., 2007), and HeLaC1-GFP (Piel et al., 2000) constitutively express Centrin1 fused with GFP (C1-GFP). RPE1-USP28Δ/pericentrinΔ, RPE1-USP28Δ/Cep215Δ, HeLa/215Δ, and HeLa/pericentrinΔ cells, a kind gift from Dr. Arshad Desai lab (University of California San Diego), were previously established and characterized (Watanabe et al., 2020). DLD-1C1-GFP were generated by infecting adenocarcinoma DLD-1 cells (ATCC) with lentivirus carrying Centrin1-GFP, using the same strategy as in Uetake et al. (2007). All cells were grown in DMEM (10569010; Thermo Fisher Scientific) supplemented with 10% FBS and 1% penicillin/1% streptomycin, at 37°C, in a humified environment with 5% CO2. For various microscopy analyses, cells were plated on round 25-mm, 1.5-mm-thick, high-tolerance coverslips (64-0735; Warner Instruments). To prevent centriole duplication, 0.1 μM centrinone (CEN; 5687; Tocris) was added to the culture medium.
Depletion of proteins using siRNA
To deplete CPAP, mitotic cells were shaken off and replated onto round coverslips. 2 h later, cells were transfected with CPAP siRNA (5′-AGAAUUAGCUCGAAUAGAA-3′; Dharmacon) using Oligofectamine (12252011; Invitrogen) following the manufacturer’s instructions. Cells were fixed as indicated in figure legends and in the text and immunolabeled. To deplete Cep192, pericentrin and Cep152 for Fig. 4, E–G, mitotic cells were shaken off and replated to 75-cm2 cell culture flasks; 2 h later, they were transfected with Cep192 siRNA (5′-GCUAGUAUGUCUGAUACUUGG-3′; Dharmacon), Cep152 siRNA (sc-90225; Santa Cruz), or pericentrin siRNA (sc-45456; Santa Cruz). 20–24 h after transfection, mitotic cells were collected and replated to 25-mm round glass coverslips. 2 mM hydroxyurea (H8627; Sigma-Aldrich) was added 2 h later to synchronize cells in S phase and minimize cell cycle–related variability in the levels of centrosomal PCM proteins. After 12 h of S phase arrest, cells were fixed and immunolabeled for CPAP or depleted proteins. Nontargeting siRNA (5′-UGGUUUACAUGUCGACUAA-3′; Dharmacon) was used in all siRNA experiments as a control.
Generation of HeLaCPAP-AID and DLD-1CPAP-AID cell lines
To generate monoclonal cell lines expressing auxin-dependent degradable CPAP, two plasmids were generated. The first is a CRISPR plasmid (pX458-Cas9-CPAP-gRNA) that contains a nuclease gene Cas9 and a specific sgRNA sequence (5′-GCTGGGGTCATATTAAATCGTGG-3′) targeting exon 2 of the endogenous CPAP gene. The plasmid was generated based on the CRISPR plasmid pX458 (Addgene plasmid 48138; Ran et al., 2013). The second plasmid (pAID1.1C-T2A-Bsr-CPAP-AID), generated based on pAID1.1C-T2A-Bsr (a kind gift from Dr. Tatsuo Fukagawa’s Lab; Nishimura and Fukagawa, 2017), contained TIR1 (transport inhibitor response 1) gene, blasticidin S resistance gene, and AID-tagged CRISPR-resistant CPAP cDNA (with several sense mutations at the sgRNA site). To make CPAP-AID resistant to CPAP gRNA, a part of the CPAP cDNA sequence was mutated as follows: 5′-ATGACCAATCCTTCTCGGGCTGGGGTCATATTAAATCGTGG-3′ into 5′-ATGACAAACCCCTCCAGAGCCGGCGTGATCCTGAATCGTGG-3′. CPAP cDNA also contained silent mutations in sequence 5′-AGAATTAGCTCGAATAGAAGA to AGAGTTAGCTAGGATCGAAGA-3′, which rendered it resistant to siRNA (Kohlmaier et al., 2009). Plasmids were cotransfected at a molar ratio of 4:1 (CPAP-AID:CRISPR) using Lipofectamine 2000 (11668027; Thermo Fisher Scientific) following the manufacturer’s instructions. To facilitate inactivation of all copies of CPAP genes, HeLaC1-GFP cells were transfected twice (Fig. 2 A). 2 d after transfection, cells were replated in medium containing Blasticidin (1 and 5 µg/ml for HeLa and DLD-1, respectively). Blasticidin-resistant clones were isolated and individually screened by Western blot and immunofluorescence, comparing CPAP-AID levels before and after addition of 500 µM of IAA (I5148; Sigma-Aldrich) for 2 h. To confirm inactivation of endogenous CPAP loci by a frame-shift mutation, a DNA fragment covering the CPAP gRNA targeting site was amplified from positive HeLaCPAP-AID clones and sequenced. To degrade CPAP-AID in established cell lines, 500 µM of IAA was added to the culture medium for the time indicated in the figure panels.
Generation of HeLaCPAP-AID/CPAP-HAdox cell line
To generate a polyclonal cell line stably expressing inducible CPAP fused with HA tag, we used PiggyBac Transposon/Transposase genetic modification system. A cargo plasmid pCE0934_Dox-PB-CPAP-WT-2X-HA (modified from Wang et al. [2014]) contained an siRNA resistant CPAP cDNA (with silent mutations in sequence 5′-AGAATTAGCTCGAATAGAAGA3′ to 5′-AGAGTTAGCTAGGATCGAAGA3′; Kohlmaier et al., 2009), tagged with two HA sequences on its C terminus and was flanked with inverted terminal repeats. The cargo plasmid and SPB plasmid (PB210PA-1; System Biosciences) expressing transposase were cotransfected to HeLaCPAP-AID cell line at a 4:1 ratio using FuGENE-HD (E2311; Promega), following manufacturer’s instructions. 2 d later, 2 µg/ml of puromycin was added to the culture medium for 10 d until puromycin-resistant colonies emerged. CPAP-HA was induced by 1 µg/ml doxycycline, and the presence of CPAP-HA was confirmed using anti-HA antibody (sc-7392; Santa Cruz) and CPAP-Ct antibody.
MT regrowth after cold and nocodazole treatment
HeLaCPAP-AID or DLD1CPAP-AID cells were plated on coverslips 1 d before the experiment. IAA was added to some plates 2.5, 6, or 48 h before MT depolymerization. The coverslips with cells were incubated for 20 min on ice to depolymerize MTs. The medium was rapidly replaced with prewarmed (37°C) medium, and the coverslips were transferred to 37°C for 20 s and fixed with ice-cold methanol for 8 min at −20°C before immunostaining. DLD-1 cells were treated with 4 µM nocodazole for 30 min to depolymerize MTs. To allow MT regrowth, cells were incubated at 37°C for 30 s with PHEM buffer (100 mM Pipes, 10 mM Hepes, 5 mM EGTA, and 2 mM MgCl2, pH 7.0) + 0.05% Triton X-100, incubated in 1.5% glutaraldehyde for 5 min, rinsed for 10 min in 1× PBS, and incubated for 20 min in NaBH4. Cells were rinsed 3× for 10 min in 1× PBS, followed by immunolabeling. Images of MT asters were recorded in wide-field mode collecting 0.2-µm z sections and deconvoluted before being assembled in image panels. The total intensities (for HeLa) or the area occupied by the α-tubulin asters surrounding centrosomes (for DLD-1) were measured from un-deconvoluted projected z sections. Two different quantification methods yielded similar results.
EB3 comet displacement assay
Cells were plated on coverslips and transfected with a plasmid encoding mCherry-EB3 (a kind gift from Dr. Irina Kaverina, Vanderbilt University, Nashville, TN) using GenJet (SignaGen Laboratories) following the manufacturer’s instructions. ∼24 h after transfection, cells were mounted in Attofluor Cell chambers (A7816; Thermo Fisher Scientific) and imaged using an Eclipse Ti inverted microscope (Nikon) equipped with a Yokogawa spinning disc (Yokogawa Electric Corp.), 405-, 488-, 561-, and 640-nm laser launch (MCL-400; Agilent Technology), and a back-illuminated EMCCD camera (DU888; Andor), 100×, NA 1.42, Plan Apo objective, with a 2× relay lens placed before the spinning disc. A time-lapse over 1 min was recorded imaging one z plane each 1 s, using 200-ms exposure time. 4-s time projections were generated in Fiji (National Institutes of Health). The length of the comet over a 4-s period was determined if the same comet appeared over more than one 4-s timeframes. The displacement of the EB3 comets in 1 s was calculated and plotted.
Sample preparation for expansion microscopy
Samples for expansion microscopy were prepared using the protocol established previously (Kong and Loncarek, 2021; Sahabandu et al., 2019). Briefly, cells growing on glass coverslips or conventionally immunolabeled cells on coverslips (see Immunofluorescence) were fixed or postfixed, respectively, with 4% formaldehyde (15686; Electron Microscopy Sciences) in 1× PBS at RT for 1 h. After fixation, the coverslips were incubated at 40°C for 16 h in a solution containing 30% acrylamide (A4058; Sigma-Aldrich) and 4% formaldehyde in 1× PBS. Cells were washed three times in 1× PBS (10 min each at RT). Coverslips were placed on a parafilm-covered Petri dish floating in an ice-cold water bath. Precooled gelling mixture (20% acrylamide, 7% sodium acrylate [408220; Sigma-Aldrich], 0.04% bis-acrylamide [A9926; Sigma-Aldrich], 0.5% ammonium persulfate [248614; Sigma-Aldrich], and 0.5% tetramethylethylenediamine [411019; Sigma-Aldrich]) was pipetted onto the coverslips and incubated on ice for 20 min and then 1–2 h at RT. After gel polymerization, a 4-mm biopsy puncher (33-34-P/25; Integra Miltex) was used to excise multiple punches from the gel. Excised punches were placed in a 50-ml tube and dry-preheated at >90°C for 10 min. SDS solution (200 mM SDS, 200 mM NaCl, and 50 mM Tris, pH 9.0), preheated to >90°C, was added to the punches, which were boiled in SDS solution for 1 h at >90°C, with swirling every 10 min. After boiling, SDS solution containing punches was cooled to RT, and SDS was removed by exchanging 1× PBS every 20 min for the first 2–3 h, followed by an overnight wash in 1× PBS at 4°C. Punches were immunolabeled as described in Immunofluorescence below.
Immunofluorescence
Cells were fixed in 1.5% formaldehyde for 4 min, postfixed in 100% methanol at −20°C for 1–4 min, washed in 1× PBS, blocked in IF buffer (1% BSA [A9647; Sigma-Aldrich] and 0.05% Tween-20 [P9416; Sigma-Aldrich], in 1× PBS) for 15 min, and incubated with primary antibody diluted in IF buffer at 37°C for 1–2 h. After washing, cells were incubated with secondary antibody, diluted in IF buffer, and incubated at 37°C for 1 h. To visualize DNA, cells were stained with a 1:3,000 dilution of Hoechst 33342 (H3570; Thermo Fisher Scientific) in 1× PBS.
Immunolabeling of excised punches for expansion microscopy was conducted as follows: punches were blocked in IF buffer for 1–2 h at RT and incubated with primary antibody diluted in IF buffer for 48 h at 4°C. Punches were washed in 1× PBS for 1–2 h and incubated with secondary antibody and DAPI (1:10,000 final dilution; D1306; Thermo Fisher Scientific) in IF buffer for 24 h at 4°C. After immunostaining, the samples were expanded with deionized H2O for 2 h at RT with deionized H2O exchanged every 10 min, and additionally overnight at 4°C. Before imaging, expanded punches were mounted in Rose chambers, Attofluor cell chambers, or glass-bottom Microwell dishes (P35G-1.5-14-C; MatTek). A complete list of primary and secondary antibodies and their dilutions are provided in Tables S1 and S2.
Wide-field microscopy
Wide-field images were acquired using an inverted Eclipse Ti or Ti2 inverted microscope (Nikon), equipped with an ORCA-Flash4.0 V3 (Hamamatsu), Intensilight C-HGFIE illuminator, and Lumencor SPECTRA light engine (Lumencor), using a 100×, NA 1.42 and 1.45 Plan Apo objective, and with a 1.5× magnifying tube lens. 200-nm-thick z sections spanning the entire cell or the entire centrosome, as needed, were acquired. Fiji and NIS Elements software were used to make maximal-intensity projections and assemble image panels. AutoQuant X3 software (MediaCybernetics) was used for deconvolution.
2DSTED microscopy
Coverslips with immunolabeled cells were mounted in a homemade mounting medium (90% glycerol, 100 mM Tris, pH 8.0, and 0.1 mg/ml p-phenylenediamine [695106; Sigma-Aldrich]). 2DSTED imaging was performed using STEDYCON (Abberior Instruments) assembled on an Eclipse Ti2 inverted microscope (Nikon) with a 100×, NA 1.45 Plan Apo objective. Avalanche photo detectors (650–700, 575–625, and 505–545 nm; DAPI detection) were used to detect the signals. Browser-based control software (Abberior Instruments) was used to generate 2DSTED images. Imaging conditions were as follows: excitation/detector wavelength, 561/616 nm and 640/660 nm for STAR ORANGE and STAR RED (Table S2), respectively; gate delay and width, 1 and 6 ns. Wide-field images of the centrosomes were acquired before 2DSTED.
3DSTORM
Cells plated on coverslips were fixed and labeled with primary antibodies. CF647-conjugated FAB2 antibodies (Biotium) were used at 1:800 dilution to label primary antibodies. Before STORM imaging, samples were layered with 100 nm tetra-spectral fluorescent spheres (T7279; Thermo Fisher Scientific), which served as fiducial markers. Coverslips were mounted in Attofluor Cell chambers (Thermo Fisher Scientific) and immersed in imaging buffer (25 mM β-mercaptoethylamine [30070; Sigma-Aldrich], 0.5 mg/ml glucose oxidase [G2133; Sigma-Aldrich], 67 μg/ml catalase [C40; Sigma-Aldrich], and 10% dextrose [D9434; Sigma-Aldrich], in 100 mM Tris, pH 8.0). 3DSTORM imaging was performed on a N-STORM4.0 assembled on an Eclipse Ti2 inverted microscope (Nikon); a superresolution high-power Apo TIRF 100×, NA 1.49 oil objective; 405-, 561-, 488-, and 647-nm excitation laser launch; and a back-illuminated EMCCD camera (DU897; Andor). The 647-nm laser line (150 mW out of the fiber and ∼90 mW before the objective lens) was used to promote fluorophore blinking. The 405-nm laser was used to reactivate fluorophores throughout the imaging process. The 561-nm laser was used to record the signals of fiducial markers. 15,000–30,000 time points were acquired at a 50-Hz frame rate each 16–20 ms. NIS Elements (Nikon) was used to analyze and present the data. Before STORM imaging, centrosomes were recorded in wide-field mode. 3DSTORM data is presented as a projection of the entire 3D volume. A rainbow z-color-coding scheme, typically spanning ∼650 nm of an imaging z range, was used for signal presentation. The signals closer to the coverslip were presented in red, and those further from the coverslip in blue. When needed, 3DSTORM images were overlaid with the mask indicating centriole orientation. 3D images were rotated in the volume viewer of the original Nikon software.
EM
Cells grown on glass coverslips were fixed in 2.5% glutaraldehyde (G5882; Sigma-Aldrich) and 0.25% formaldehyde in 1× PBS (pH 7.4) for 1 h at RT. Centriole position within the target cells was recorded, and the position of the cells was marked on the coverslip. Cells were prestained with 1% osmium tetroxide (19100; Electron Microscopy Sciences) and 1% uranyl acetate (22400; Electron Microscopy Sciences), dehydrated in ethanol, and embedded in EMbed-812 resin (13940; Electron Microscopy Sciences). 80-nm-thick serial sections were sectioned, transferred to the formvar-coated copper grids (2330P-XA; SPI Supplies), and stained with uranyl acetate and lead citrate. Samples were imaged using an FEI Tecnai 12 Spirit transmission electron microscope operating at 80 kV. Image analysis was performed in Photoshop and Fiji. Centriole length was determined from one of the sections of centrioles sectioned longitudinally or near-longitudinally. Centriole width was determined from centrioles in various favorable orientations.
Image acquisition, analysis, processing, and data presentation
Within one experiment, images were acquired under identical imaging settings. The levels of fluorescent signals were sometimes differentially adjusted on image panels to improve the visibility of the dimmer signals, such as Centrin1-GFP signals. Such adjustments were not performed if quantitative comparison between centrosomal proteins was the goal of illustration. Maximum-intensity projections of all acquired z slices are presented for wide-field images. For some expansion microscopy recordings, only the central section through the centriole is presented.
Fluorescence intensity measurement
Florescence intensity measurements in wide-field recordings of centrosome-associated proteins were done using Fiji. 200-nm-thick z sections spanning all centrosomes were recorded. Maximum-intensity projections of all z slices were generated. The integrated density of centrosome-associated signals was measured within a defined area of a constant size encircling the centrosome. The background intensity of an equivalent area adjacent to each centrosome was recorded and subtracted from the signal intensity. The intensities of luminal mother centriole CPAP signals were determined from 2DSTED images. Centrioles were colabeled for CPAP and imaged in two colors. γ-tubulin signal was coimaged and used as a marker for centriole orientation. When the PCM CPAP signal was present on centrosomes, only centrioles oriented vertically or almost vertically to the coverslip were used for quantifications, to eliminate the contamination of the luminal with the PCM signals. Within one experiment, the same labeling and imaging conditions were kept across samples, and imaging conditions were chosen to avoid saturation of the signal. The integrated density of luminal CPAP signals was measured in Fiji within a circular area of 200-nm diameter (pixel size = 10 nm) encircling the luminal CPAP signal. The intensity of an equivalent circular background area adjacent to the centrosome was recorded and subtracted from the measured luminal signal.
Statistical analysis
Statistical difference between two sets of data was determined in Excel using an unpaired, two-tailed Student’s t test with equal or nonequal variance. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. Box-and-whisker plots show the minimum and maximum, median line, upper and lower quartiles, and all or none data points. Histograms show average values ± SD. Sample sizes (the number of analyzed cells, centrosomes, or centrioles) are indicated on the plots or in the figure legends.
Number of independent experiments performed in this study
Fig. 1: STORM, experiment done >3× with similar results; data shown from multiple experiments; STED, experiment done 2× with similar results; data shown from both experiments. Expansion experiment done 2× with similar result. Fig. 2 B: Experiment done 1× for immunoblot. CPAP centrosome intensity quantifications from two experiments. Fig. 2 C: Experiment done 1×. Fig. 2 D: Experiment done 1×. Fig. 2 E: Experiment done >3× with similar results. Quantifications are from one experiment; data align with quantifications in Figs. 3 and 4. Fig. 2 F: Experiment done 1×, data align with immunoblot data. Fig. 2 H: Experiment done 1×, data in agreement with the rest of the data. Fig. 2 I: Experiment done 1× with Cep152/Sas-6 staining and >3× with Centrin-GFP/CPAP staining. Quantifications from one experiment. Fig. 3, A–C: Experiment done 2×, quantifications from one experiment. Fig. 3, D–F: Experiment done 2×, quantifications from one experiment. Fig. 3, H–J: 24- and 40-h repeated 2×, 60-h done 1×. STED images from one experiment. Fig. 4 A: Experiment done 3× with similar results; data shown from one experiment. Fig. 4 B: Experiment done 2× with similar results; data shown from one experiment. Fig. 4 C: Experiment done 3× with similar results; data shown from one experiment. Fig. 4 D: Transfection done 3×. Correlative time-lapse data collected from two cells with similar results (one shown). Recordings were additionally repeated for five control and five IAA-treated cells (2.5–6 h of IAA treatment) and showed similar results. Fig. 4, E–I: siRNA experiment done 1×, HeLa knockout cells analyzed 2× with similar results, RPE-1 knockout cells 1×. Quantifications of CPAP phenotypes from all experiments. Microscopy done 2×; data shown from one experiment. Fig. 5, A and B: Experiment done 3× with similar results; data shown from one experiment. Fig. 5 C: Experiment done 3× with similar results; data shown from one experiment. Fig. 5 D: Experiment done 2× with similar results; data shown from one experiment. Fig. 6 A: Experiment done 2× with similar results; data shown from one experiment. Fig. 6 B: Experiment done 3× with similar results; data shown from three experiments. Fig. 6 C: Experiment done 3× with similar results; data shown from one experiment. Fig. 6 D: Experiment done 2× with similar results; data shown from one experiment. Fig. 6 E: Experiment done 2× with similar results; data shown from one experiment. Fig. 7 A: Expansion repeated 2× with similar results; data shown from one experiment. Data in agreement with the rest of expansion data. From one large, expanded sample (one coverslip with cells = one sample), multiple smaller samples were excised and individually immunolabeled and imaged. Fig. 7 B: Expansion and assessment of centriole morphology after IAA treatment repeated >3× with slight variations in timing. Quantifications in B and C from 1 experiment. Fig. 7, D–H: Expansion done 1×, data in agreement with expansion data in Fig. 7 A. From one large, expanded sample (one coverslip with cells = one sample), multiple smaller samples were excised and individually immunolabeled and imaged. Fig. 8: Experiment done 2× with similar results. Data shown from both experiments, 66 cells analyzed in total. Fig. S1 A: 2DSTED of SAS-6 done >5× with similar resolution. Fig. S1 B: Experiment done 2× with similar results, cells imaged from both experiments. Fig. S2 A: 15-min pulse time point repeated 4× with various intervals after shake-off time, 60-min IAA pulse once. Quantifications shown in A from one experiment. Fig. S2 B: Experiment repeated 1×. Immunoblot of HeLaCPAP-AID/CPAP-HAdox cells under different conditions done two more times (not depicted). Fig. S2 C: Experiment done 2×; images from one experiment. Additional experiment in hydroxyurea-arrested cells done 1×. Fig. S2 E: STED imaging done for two experiments. Images shown in Fig. S2 D from one experiment. Fig. S3 B: Experiment done 2×. Images and quantifications from one experiment. Fig. S3, C and D: Experiment done 2× with similar results; images from one experiment. Fig. S4, B and C: Experiment done 2× with slightly different conditions, with similar results; data from one experiment. Fig. S4 D: 40-h timepoint repeated 2×, 60-h timepoint done 1×. STED images from one experiment. Fig. S5: From the same dataset as in Fig. 7, E–G. Expansion done 1×, data in agreement with Fig. 7 A. From one large, expanded sample (one coverslip with cells = one sample), multiple smaller samples were excised and individually immunolabeled and imaged.
Online supplemental material
Fig. S1 contains resolution of 2DSTED and measurements of CPAP signals. Fig. S2 contains centriole-associated CPAP degradation from centriole lumen and its dynamic association with centrioles. Fig. S3 contains characterization of DLD-1CPAP-AID cells and the effect of CPAP degradation on centriole structures in this cell line. Fig. S4 contains the effect of CPAP removal on recruitment of PCM in mitosis by structurally intact mother centrioles. Fig. S5 contains localization of centrosomal proteins on expanded HeLaC1-GFP and HeLaCPAP-AID centrioles after ∼48 h of IAA treatment. Tables S1 and S2 list primary and secondary antibodies and their dilutions, respectively.
Supplementary Material
is a list of primary antibodies and their dilutions.
is a list of secondary antibodies and their dilutions.
is the source file for Fig. 2.
is the source file for Fig. S2.
is the source file for Fig. S3.
Acknowledgments
We thank Dr. Kevin F. O’Connell for critical reading of the manuscript. We are grateful to Dr. Arshad Desai for pericentrin and Cep215-knockout RPE-1 and HeLa cells, Dr. Tatsuo Fukagawa for sharing pAID1.1C-T2A-Bsr plasmid, and Dr. Irina Kaverina for mCherry-EB3 plasmid.
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
The authors declare no competing financial interests.
Author contributions: A. Vasquez-Limeta and K. Lukasik conducted most experiments and analyzed the data. D. Kong generated CPAP-AID cell lines and conducted EM and expansion microscopy. N. Sahabandu conducted some expansion microscopy. D. Luvsanjav conducted EM. R. Chari prepared CRISPR-Cas9 and PiggyBac plasmids. J. Loncarek supervised the project and conducted some microscopy and image analysis. J. Loncarek, A. Vasquez-Limeta, K. Lukasik, C. Sullenberger, and D. Kong wrote the manuscript. All authors discussed the manuscript.
References
- Alieva, I.B., Zemskov E.A., Smurova K.M., Kaverina I.N., and Verin A.D.. 2013. The leading role of microtubules in endothelial barrier dysfunction: Disassembly of peripheral microtubules leaves behind the cytoskeletal reorganization. J. Cell. Biochem. 114:2258–2272. 10.1002/jcb.24575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atorino, E.S., Hata S., Funaya C., Neuner A., and Schiebel E.. 2020. CEP44 ensures the formation of bona fide centriole wall, a requirement for the centriole-to-centrosome conversion. Nat. Commun. 11:903. 10.1038/s41467-020-14767-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh, J., Hergert P., Delouvée A., Euteneuer U., Formstecher E., Khodjakov A., and Bornens M.. 2009. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J. Cell Biol. 185:101–114. 10.1083/jcb.200808082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestra, F.R., von Tobel L., and Gönczy P.. 2015. Paternally contributed centrioles exhibit exceptional persistence in C. elegans embryos. Cell Res. 25:642–644. 10.1038/cr.2015.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto, R., Brunk K., Vinadogrova T., Peel N., Franz A., Khodjakov A., and Raff J.W.. 2008. Centrosome amplification can initiate tumorigenesis in flies. Cell. 133:1032–1042. 10.1016/j.cell.2008.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto, R., Lau J., Vinogradova T., Gardiol A., Woods C.G., Khodjakov A., and Raff J.W.. 2006. Flies without centrioles. Cell. 125:1375–1386. 10.1016/j.cell.2006.05.025 [DOI] [PubMed] [Google Scholar]
- Bond, J., Roberts E., Springell K., Lizarraga S.B., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O., et al. 2005. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 37:353–355. 10.1038/ng1539 [DOI] [PubMed] [Google Scholar]
- Bowler, M., Kong D., Sun S., Nanjundappa R., Evans L., Farmer V., Holland A., Mahjoub M.R., Sui H., and Loncarek J.. 2019. High-resolution characterization of centriole distal appendage morphology and dynamics by correlative STORM and electron microscopy. Nat. Commun. 10:993. 10.1038/s41467-018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos, Z., Machado P., Alvarez-Martins I., Gouveia S.M., Jana S.C., Duarte P., Amado T., Branco P., Freitas M.C., Silva S.T., et al. 2012. BLD10/CEP135 is a microtubule-associated protein that controls the formation of the flagellum central microtubule pair. Dev. Cell. 23:412–424. 10.1016/j.devcel.2012.06.001 [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos, Z., Machado P., Branco P., Tavares-Cadete F., Rodrigues-Martins A., Pereira-Leal J.B., and Bettencourt-Dias M.. 2010. Stepwise evolution of the centriole-assembly pathway. J. Cell Sci. 123:1414–1426. 10.1242/jcs.064931 [DOI] [PubMed] [Google Scholar]
- Chang, C.-W., Hsu W.-B., Tsai J.-J., Tang C.-J.C., and Tang T.K.. 2016. CEP295 interacts with microtubules and is required for centriole elongation. J. Cell Sci. 129:2501–2513. 10.1242/jcs.186338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, J.H., Chang C.J., Chen C.Y., and Tang T.K.. 2006. Depletion of CPAP by RNAi disrupts centrosome integrity and induces multipolar spindles. Biochem. Biophys. Res. Commun. 339:742–747. 10.1016/j.bbrc.2005.11.074 [DOI] [PubMed] [Google Scholar]
- Chou, E.J., Hung L.Y., Tang C.J., Hsu W.B., Wu H.Y., Liao P.C., and Tang T.K.. 2016. Phosphorylation of CPAP by Aurora-A maintains spindle pole integrity during mitosis. Cell Rep. 14:2975–2987. 10.1016/j.celrep.2016.02.085 [DOI] [PubMed] [Google Scholar]
- Cizmecioglu, O., Arnold M., Bahtz R., Settele F., Ehret L., Haselmann-Weiss U., Antony C., and Hoffmann I.. 2010. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol. 191:731–739. 10.1083/jcb.201007107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comartin, D., Gupta G.D., Fussner E., Coyaud É., L. Pelletier, Archinti M., Cheung S.W.T., Pinchev D., Lawo S., Raught B., et al. 2013. CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr. Biol. 23:1360–1366. 10.1016/j.cub.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Conduit, P.T., Wainman A., Novak Z.A., Weil T.T., and Raff J.W.. 2015. Re-examining the role of Drosophila Sas-4 in centrosome assembly using two-colour-3D-SIM FRAP. eLife. 4:e08483. 10.7554/elife.08483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier, A., Clément M.J., Knossow M., Lachkar S., Savarin P., Toma F., Sobel A., Gigant B., and Curmi P.A.. 2009. The PN2-3 domain of centrosomal P4.1-associated protein implements a novel mechanism for tubulin sequestration. J. Biol. Chem. 284:6909–6917. 10.1074/jbc.m808249200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottee, M.A., Muschalik N., Wong Y.L., Johnson C.M., Johnson S., Andreeva A., Oegema K., Lea S.M., Raff J.W., and van Breugel M.. 2013. Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly. eLife. 2:e01071. 10.7554/eLife.01071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann, A., Maddox P.S., Desai A., and Oegema K.. 2008. SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the gamma-tubulin-mediated addition of centriolar microtubules. J. Cell Biol. 180:771–785. 10.1083/jcb.200709102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictenberg, J.B., Zimmerman W., Sparks C.A., Young A., Vidair C., Zheng Y., Carrington W., Fay F.S., and Doxsey S.J.. 1998. Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J. Cell Biol. 141:163–174. 10.1083/jcb.141.1.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev, N.S., Yu Q.D., Weiskopf K., Tzolovsky G., Cunha-Ferreira I., Riparbelli M., Rodrigues-Martins A., Bettencourt-Dias M., Callaini G., and Glover D.M.. 2010. Asterless is a scaffold for the onset of centriole assembly. Nature. 467:714–718. 10.1038/nature09445 [DOI] [PubMed] [Google Scholar]
- Firat-Karalar, E.N., Rauniyar N., Yates J.R. 3rd, and Stearns T.. 2014. Proximity interactions among centrosome components identify regulators of centriole duplication. Curr. Biol. 24:664–670. 10.1016/j.cub.2014.01.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, K.W., Choi Y.K., Rattner J.B., and Qi R.Z.. 2008. CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol. Biol. Cell. 19:115–125. 10.1091/mbc.e07-04-0371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garreau de Loubresse, N., Ruiz F., Beisson J., and Klotz C.. 2001. Role of delta-tubulin and the C-tubule in assembly of Paramecium basal bodies. BMC Cell Biol. 2:4. 10.1186/1471-2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho, S.A., Kwon M., and Pellman D.. 2009. Centrosomes and cancer: How cancer cells divide with too many centrosomes. Cancer Metastasis Rev. 28:85–98. 10.1007/s10555-008s1059163-6 [DOI] [PubMed] [Google Scholar]
- Gogendeau, D., Hurbain I., Raposo G., Cohen J., Koll F., and Basto R.. 2011. Sas-4 proteins are required during basal body duplication in Paramecium. Mol. Biol. Cell. 22:1035–1044. 10.1091/mbc.E10-11-0901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Ferreria, M.A., Rath U., Buster D.W., Chanda S.K., Caldwell J.S., Rines D.R., and Sharp D.J.. 2007. Human Cep192 is required for mitotic centrosome and spindle assembly. Curr. Biol. 17:1960–1966. 10.1016/j.cub.2007.10.019 [DOI] [PubMed] [Google Scholar]
- Gonczy, P., and Hatzopoulos G.N.. 2019. Centriole assembly at a glance. J. Cell Sci. 132:jcs228833. 10.1242/jcs.228833 [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan, J., Mennella V., Blachon S., Zhai B., Smith A.H., Megraw T.L., Nicastro D., Gygi S.P., Agard D.A., and Avidor-Reiss T.. 2011. Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nat. Commun. 2:359. 10.1038/ncomms1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goundiam, O., and Basto R.. 2021. Centrosomes in disease: How the same music can sound so different? Curr. Opin. Struct. Biol. 66:74–82. 10.1016/j.sbi.2020.09.011 [DOI] [PubMed] [Google Scholar]
- Graser, S., Stierhof Y.-D., Lavoie S.B., Gassner O.S., Lamla S., Le Clech M., and Nigg E.A.. 2007. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179:321–330. 10.1083/jcb.200707181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudi, R., Haycraft C.J., Bell P.D., Li Z., and Vasu C.. 2015. Centrobin-mediated regulation of the centrosomal protein 4.1-associated protein (CPAP) level limits centriole length during elongation stage. J. Biol. Chem. 290:6890–6902. 10.1074/jbc.m114.603423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gul, A., Hassan M.J., Hussain S., Raza S.I., Chishti M.S., and Ahmad W.. 2006. A novel deletion mutation in CENPJ gene in a Pakistani family with autosomal recessive primary microcephaly. J. Hum. Genet. 51:760–764. 10.1007/s10038-006-0017-1 [DOI] [PubMed] [Google Scholar]
- Hodges, M.E., Scheumann N., Wickstead B., Langdale J.A., and Gull K.. 2010. Reconstructing the evolutionary history of the centriole from protein components. J. Cell Sci. 123:1407–1413. 10.1242/jcs.064873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, W.B., Hung L.Y., Tang C.J., Su C.L., Chang Y., and Tang T.K.. 2008. Functional characterization of the microtubule-binding and -destabilizing domains of CPAP and d-SAS-4. Exp. Cell Res. 314:2591–2602. 10.1016/j.yexcr.2008.05.012 [DOI] [PubMed] [Google Scholar]
- Hung, L.Y., Chen H.L., Chang C.W., Li B.R., and Tang T.K.. 2004. Identification of a novel microtubule-destabilizing motif in CPAP that binds to tubulin heterodimers and inhibits microtubule assembly. Mol. Biol. Cell. 15:2697–2706. 10.1091/mbc.e04-02-0121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, L.Y., Tang C.J., and Tang T.K.. 2000. Protein 4.1 R-135 interacts with a novel centrosomal protein (CPAP) which is associated with the gamma-tubulin complex. Mol. Cell Biol. 20:7813–7825. 10.1128/mcb.20.20.7813-7825.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham, M., Müller-Reichert T., Oegema K., Grill S., and Hyman A.A.. 2003. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell. 112:575–587. 10.1016/s0092-8674(03)00117-x [DOI] [PubMed] [Google Scholar]
- Kitagawa, D., Kohlmaier G., Keller D., Strnad P., Balestra F.R., Fluckiger I., and Gonczy P.. 2011. Spindle positioning in human cells relies on proper centriole formation and on the microcephaly proteins CPAP and STIL. J. Cell Sci. 124:3884–3893. 10.1242/jcs.089888 [DOI] [PubMed] [Google Scholar]
- Klena, N., Le Guennec M., Tassin A.-M., van den Hoek H., Erdmann P.S., Schaffer M., Geimer S., Aeschlimann G., Kovacik L., Sadian Y., et al. 2020. Architecture of the centriole cartwheel-containing region revealed by cryo-electron tomography. EMBO J. 39:e106246. 10.15252/embj.2020106246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn, J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y.D., and Nigg E.A.. 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 13:190–202. 10.1016/j.devcel.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Kohlmaier, G., Loncarek J., Meng X., McEwen B.F., Mogensen M.M., Spektor A., Dynlacht B.D., Khodjakov A., and Gonczy P.. 2009. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr. Biol. 19:1012–1018. 10.1016/j.cub.2009.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, D., and Loncarek J.. 2021. Analyzing centrioles and cilia by expansion microscopy. Methods Mol. Biol. 2329:249–263. 10.1007/978-10716-1538-6_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, D., Sahabandu N., Sullenberger C., Vasquez-Limeta A., Luvsanjav D., Lukasik K., and Loncarek J.. 2020. Prolonged mitosis results in structurally aberrant and over-elongated centrioles. J. Cell Biol. 219. 10.1083/jcb.201910019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guennec, M., Klena N., Gambarotto D., Laporte M.H., Tassin A.M., van den Hoek H., Erdmann P.S., Schaffer M., Kovacik L., Borgers S., et al. 2020. A helical inner scaffold provides a structural basis for centriole cohesion. Sci. Adv. 6:eaaz4137. 10.1126/sciadv.aaz4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M., Chang J., Chang S., Lee K.S., and Rhee K.. 2014. Asymmetric spindle pole formation in CPAP-depleted mitotic cells. Biochem. Biophys. Res. Commun. 444:644–650. 10.1016/j.bbrc.2014.01.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M., and Rhee K.. 2015. Determination of mother centriole maturation in CPAP-depleted cells using the ninein antibody. Endocrinol. Metabol. 30:53–57. 10.3803/EnM.2015.30.1.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeGuennec, M., Klena N., Aeschlimann G., Hamel V., and Guichard P.. 2021. Overview of the centriole architecture. Curr. Opin. Struct. Biol. 66:58–65. 10.1016/j.sbi.2020.09.015 [DOI] [PubMed] [Google Scholar]
- Leidel, S., and Gönczy P.. 2003. SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell. 4:431–439. 10.1016/s1534-5807(03)00062-5 [DOI] [PubMed] [Google Scholar]
- Levine, M.S., Bakker B., Boeckx B., Moyett J., Lu J., Vitre B., Spierings D.C., Lansdorp P.M., Cleveland D.W., Lambrechts D., et al. 2017. Centrosome amplification is sufficient to promote spontaneous tumorigenesis in mammals. Dev. Cell. 40:313–322.e5. 10.1016/j.devcel.2016.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y.C., Chang C.W., Hsu W.B., Tang C.J., Lin Y.N., Chou E.J., Wu C.T., and Tang T.K.. 2013a. Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. EMBO J. 32:1141–1154. 10.1038/emboj.2013.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y.N., Lee Y.S., Li S.K., and Tang T.K.. 2020. Loss of CPAP in developing mouse brain and its functional implication for human primary microcephaly. J. Cell Sci. 133:jcs243592. 10.1242/jcs.243592 [DOI] [PubMed] [Google Scholar]
- Lin, Y.N., Wu C.T., Lin Y.C., Hsu W.B., Tang C.J., Chang C.W., and Tang T.K.. 2013b. CEP120 interacts with CPAP and positively regulates centriole elongation. J. Cell Biol. 202:211–219. 10.1083/jcb.201212060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoMastro, G.M., and Holland A.J.. 2019. The emerging link between centrosome aberrations and metastasis. Dev. Cell. 49:325–331. 10.1016/j.devcel.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthiens, V., and Basto R.. 2020. Centrosomes: The good and the bad for brain development. Biol. Cell. 112:153–172. 10.1111/boc.201900090 [DOI] [PubMed] [Google Scholar]
- Marthiens, V., Rujano M.A., Pennetier C., Tessier S., Paul-Gilloteaux P., and Basto R.. 2013. Centrosome amplification causes microcephaly. Nat. Cell Biol. 15:731–740. 10.1038/ncb2746 [DOI] [PubMed] [Google Scholar]
- Mitchison, H.M., and Valente E.M.. 2017. Motile and non-motile cilia in human pathology: From function to phenotypes. J. Pathol. 241:294–309. 10.1002/path.4843 [DOI] [PubMed] [Google Scholar]
- Moyer, T.C., and Holland A.J.. 2019. PLK4 promotes centriole duplication by phosphorylating STIL to link the procentriole cartwheel to the microtubule wall. eLife. 8:e46054. 10.7554/elife.46054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarov, S., Bezler A., Hatzopoulos G.N., Nemčíková Villímová V., Demurtas D., Le Guennec M., Guichard P., and Gönczy P.. 2020. Novel features of centriole polarity and cartwheel stacking revealed by cryo-tomography. EMBO J. 39:e106249. 10.15252/embj.2020106249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg, E.A., Cajanek L., and Arquint C.. 2014. The centrosome duplication cycle in health and disease. FEBS Lett. 588:2366–2372. 10.1016/j.febslet.2014.06.030 [DOI] [PubMed] [Google Scholar]
- Nigg, E.A., Schnerch D., and Ganier O.. 2017. Impact of centrosome aberrations on chromosome segregation and tissue architecture in cancer. Cold Spring Harbor Symp. Quant. Biol. 82:137–144. 10.1101/sqb.2017.82.034421 [DOI] [PubMed] [Google Scholar]
- Nishimura, K., and Fukagawa T.. 2017. An efficient method to generate conditional knockout cell lines for essential genes by combination of auxin-inducible degron tag and CRISPR/Cas9. Chromosome Res. 25:253–260. 10.1007/s10577-017-9559-7 [DOI] [PubMed] [Google Scholar]
- Nishimura, K., Fukagawa T., Takisawa H., Kakimoto T., and Kanemaki M.. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods. 6:917–922. 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- Novak, Z.A., Conduit P.T., Wainman A., and Raff J.W.. 2014. Asterless licenses daughter centrioles to duplicate for the first time in Drosophila embryos. Curr. Biol. 24:1276–1282. 10.1016/j.cub.2014.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole, E.T., Giddings T.H., McIntosh J.R., and Dutcher S.K.. 2003. Three-dimensional organization of basal bodies from wild-type and delta-tubulin deletion strains of Chlamydomonas reinhardtii. Mol. Biol. Cell. 14:2999–3012. 10.1091/mbc.e02-11-0755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, T., Essner R., Ryu J.H., Palazzo R.E., Uetake Y., and Kuriyama R.. 2002. Characterization of Cep135, a novel coiled-coil centrosomal protein involved in microtubule organization in mammalian cells. J. Cell Biol. 156:87–99. 10.1083/jcb.200108088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, C.G., Osborn D.P.S., Giddings T.H. Jr., Beales P.L., and Winey M.. 2009. Basal body stability and ciliogenesis requires the conserved component Poc1. J. Cell Biol. 187:905–920. 10.1083/jcb.200908019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier, L., O’Toole E., Schwager A., Hyman A.A., and Müller-Reichert T.. 2006. Centriole assembly in Caenorhabditis elegans. Nature. 444:619–623. 10.1038/nature05318 [DOI] [PubMed] [Google Scholar]
- Piel, M., Meyer P., Khodjakov A., Rieder C.L., and Bornens M.. 2000. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 149:317–330. 10.1083/jcb.149.2.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani, A., Mariappan A., Gottardo M., Mandad S., Urlaub H., Avidor-Reiss T., Riparbelli M., Callaini G., Debec A., Feederle R., and Gopalakrishnan J.. 2018. Plk1/Polo phosphorylates sas-4 at the onset of mitosis for an efficient recruitment of pericentriolar material to centrosomes. Cell Rep. 25:3618–3630.e6. 10.1016/j.celrep.2018.11.102 [DOI] [PubMed] [Google Scholar]
- Ran, F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riparbelli, M.G., and Callaini G.. 2011. Male gametogenesis without centrioles. Dev. Biol. 349:427–439. 10.1016/j.ydbio.2010.10.021 [DOI] [PubMed] [Google Scholar]
- Sahabandu, N., Kong D., Magidson V., Nanjundappa R., Sullenberger C., Mahjoub M.R., and Loncarek J.. 2019. Expansion microscopy for the analysis of centrioles and cilia. J. Microsc. 276:145–159. 10.1111/jmi.12841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, T.I., Kleylein-Sohn J., Westendorf J., Le Clech M., Lavoie S.B., Stierhof Y.D., and Nigg E.A.. 2009. Control of centriole length by CPAP and CP110. Curr. Biol. 19:1005–1011. 10.1016/j.cub.2009.05.016 [DOI] [PubMed] [Google Scholar]
- Sharma, A., Aher A., Dynes N.J., Frey D., Katrukha E.A., Jaussi R., Grigoriev I., Croisier M., Kammerer R.A., Akhmanova A., et al. 2016. Centriolar CPAP/SAS-4 imparts slow processive microtubule growth. Dev. Cell. 37:362–376. 10.1016/j.devcel.2016.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen, K.F., Gabryjonczyk A.M., Anselm E., Stierhof Y.D., and Nigg E.A.. 2013. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J. Cell Sci. 126:3223–3233. 10.1242/jcs.129502 [DOI] [PubMed] [Google Scholar]
- Sonnen, K.F., Schermelleh L., Leonhardt H., and Nigg E.A.. 2012. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open. 1:965–976. 10.1242/bio.20122337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steib, E., Laporte M.H., Gambarotto D., Olieric N., Zheng C., Borgers S., Olieric V., Le Guennec M., Koll F., Tassin A.M., et al. 2020. WDR90 is a centriolar microtubule wall protein important for centriole architecture integrity. eLife. 9:e57205. 10.7554/eLife.57205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, N.R., Raposo A.A., Basto R., St Johnston D., and Raff J.W.. 2007. From stem cell to embryo without centrioles. Curr. Biol. 17:1498–1503. 10.1016/j.cub.2007.07.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullenberger, C., Vasquez-Limeta A., Kong D., and Loncarek J.. 2020. With age comes maturity: Biochemical and structural transformation of a human centriole in the making. Cells. 9. 10.3390/cells9061429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, C.J.C., Fu R.H., Wu K.S., Hsu W.B., and Tang T.K.. 2009. CPAP is a cell-cycle regulated protein that controls centriole length. Nat. Cell Biol. 11:825–831. 10.1038/ncb1889 [DOI] [PubMed] [Google Scholar]
- Tang, C.J.C., Lin S.Y., Hsu W.B., Lin Y.N., Wu C.T., Lin Y.C., Chang C.W., Wu K.S., and Tang T.K.. 2011. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J. 30:4790–4804. 10.1038/emboj.2011.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang, W.Y., Bossard C., Khanna H., Peränen J., Swaroop A., Malhotra V., and Dynlacht B.D.. 2008. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev. Cell. 15:187–197. 10.1016/j.devcel.2008.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake, Y., Loncarek J., Nordberg J.J., English C.N., La Terra S., Khodjakov A., and Sluder G.. 2007. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J. Cell Biol. 176:173–182. 10.1083/jcb.200607073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez-Limeta, A., and Loncarek J.. 2021. Human centrosome organization and function in interphase and mitosis. Semin. Cell Dev. Biol. 117:30–41. 10.1016/j.semcdb.2021.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venoux, M., Tait X., Hames R.S., Straatman K.R., Woodland H.R., and Fry A.M.. 2013. Poc1A and Poc1B act together in human cells to ensure centriole integrity. J. Cell Sci. 126:163–175. 10.1242/jcs.111203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev, I.A., and Chentsov Yu S.. 1982. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 93:938–949. 10.1083/jcb.93.3.938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulprecht, J., David A., Tibelius A., Castiel A., Konotop G., Liu F., Bestvater F., Raab M.S., Zentgraf H., Izraeli S., and Kramer A.. 2012. STIL is required for centriole duplication in human cells. J. Cell Sci. 125:1353–1362. 10.1242/jcs.104109 [DOI] [PubMed] [Google Scholar]
- Wang, G., McCain M.L., Yang L., He A., Pasqualini F.S., Agarwal A., Yuan H., Jiang D., Zhang D., Zangi L., et al. 2014. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 20:616–623. 10.1038/nm.3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.T., Kong D., Hoerner C.R., Loncarek J., and Stearns T.. 2017. Centriole triplet microtubules are required for stable centriole formation and inheritance in human cells. eLife. 6:e29061. 10.7554/eLife.29061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., and Dynlacht B.D.. 2018. The regulation of cilium assembly and disassembly in development and disease. Development. 145:dev151407. 10.1242/dev.151407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W.J., Soni R.K., Uryu K., and Tsou M.F.B.. 2011. The conversion of centrioles to centrosomes: Essential coupling of duplication with segregation. J. Cell Biol. 193:727–739. 10.1083/jcb.201101109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, S., Meitinger F., Shiau A.K., Oegema K., and Desai A.. 2020. Centriole-independent mitotic spindle assembly relies on the PCNT-CDK5RAP2 pericentriolar matrix. J. Cell Biol. 219:e202006010. 10.1083/jcb.202006010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, Y.L., Anzola J.V., Davis R.L., Yoon M., Motamedi A., Kroll A., Seo C.P., Hsia J.E., Kim S.K., Mitchell J.W., et al. 2015. Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 348:1155–1160. 10.1126/science.aaa5111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff, J.B., Wueseke O., and Hyman A.A.. 2014. Pericentriolar material structure and dynamics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130459. 10.1098/rstb.2013.0459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X., Gooi L.M., Wason A., Gabriel E., Mehrjardi N.Z., Yang Q., Zhang X., Debec A., Basiri M.L., Avidor-Reiss T., et al. 2014. Conserved TCP domain of Sas-4/CPAP is essential for pericentriolar material tethering during centrosome biogenesis. Proc. Natl. Acad. Sci. USA. 111:E354–E363. 10.1073/pnas.1317535111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X., Ramani A., Soni K., Gottardo M., Zheng S., Ming Gooi L., Li W., Feng S., Mariappan A., Wason A., et al. 2016. Molecular basis for CPAP-tubulin interaction in controlling centriolar and ciliary length. Nat. Commun. 7:11874. 10.1038/ncomms11874 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is a list of primary antibodies and their dilutions.
is a list of secondary antibodies and their dilutions.
is the source file for Fig. 2.
is the source file for Fig. S2.
is the source file for Fig. S3.