Abstract
DNA N6-methyladenine (6mA) is the most prevalent DNA modification in prokaryotes. However, its presence and significance in eukaryotes remain elusive. Recently, with methodology advances in detection and sequencing of 6mA in eukaryotes, 6mA is back in the spotlight. While multiple studies reported that 6mA is an important epigenetic mark in eukaryotes and plays a regulatory role in DNA transcription, transposon activation, stress response and other bioprocesses, there are some discrepancies in the current literature. Here we review the recent advances in 6mA research in eukaryotes, especially in mammals. In particular, DNA 6mA’s abundance/distributions, potential role in regulating gene expression, identified regulators, and pathological roles in human diseases, especially in cancer, are described. The limitations faced by the field along with future perspectives in 6mA research are also discussed.
Keywords: Adenine methylation, DNA 6mA, m6dA, ALKBH1, METTL4
DNA 6mA research is reviving in eukaryotes
DNA methylation is a fundamental DNA epigenetic process. For instance, 5-methylcytosine (5mC) is the most well-characterized DNA methylation in eukaryotes that has been shown to regulate various processes in development and disease [1]. Similarly, DNA N6-adenine methylation (6mA) functions as the most prevalent DNA modification in prokaryotes, playing a critical role in regulating a diverse array of biological processes [2]. However, the presence of 6mA and its potential role as another DNA epigenetic marker in eukaryotes have not been recognized until recent years.
Over the past few years, the development of highly sensitive detection methods, such as mass spectrometry, allowed for 6mA identification in multiple types of multicellular eukaryotes, including mammals [3]. Whole genome-wide 6mA sites profiling methods propelled research of 6mA in eukaryotes by allowing for mapping 6mA sites to specific genes. Although the characteristics and functions of 6mA still require more inquest, recent findings have contributed substantial evidence in supporting 6mA’s vital roles in eukaryotic biology. In this review, we discuss the latest discoveries of 6mA in the presence, distribution and regulation in mammals, as well as 6mA’s roles in human diseases.
DNA 6mA exists in eukaryotes
DNA 6mA was first reported as a novel abundant base by Dunn and Smith in a 1957 study, in which they found that the level of 6mA can reach up to 1.75% and 2.5% of total adenine in Escherichia coli and Aerobacter aerogenes, respectively [4]. With accumulating evidence, 6mA then got recognized as the most prevalent DNA modification in prokaryotes [5], akin to 5mC in eukaryotes. Extensive studies reported the functions of 6mA in prokaryotes, including protecting host DNA from restriction enzyme digestion during bacterial host defense [6] and participating in DNA repair, replication, transcription, nucleoid segregation, and gene expression regulation in bacteria [2,7].
The presence of 6mA in eukaryotes was initially reported in unicellular protists, including the ciliates, Tetrahymena pyriformis in 1973 [8] and Paramecium aurelia in 1974 [9], where 6mA is also the primary DNA modification, despite the abundance (~0.4-0.8% of total adenine) subsisting several times lesser than what it is in prokaryotes [10]. Afterwards, although there were sporadic reports indicating the existence of 6mA in higher eukaryotes, such as mosquitos [11], plants [12] and even mammals [13], 6mA did not attract sizeable attention due to the meagre level of abundance and unrevealed genome-wide location. In 2015, multiple approaches demonstrated detectable levels and genome-wide localization of 6mA in Chlamydomonas [14] , Drosophila [15] and C. elegans [16], implying the functional importance of 6mA in eukaryotes [17,18]. Then, the search for the presence of 6mA in vertebrates and the study of its functional significance took off. With the prevalence of advanced mass spectrometry (MS) in detecting DNA modifications, 6mA has been reported to be present in a plethora of multicellular organisms, including Drosophila [15,19,20], Xenopus laevis [21], zebrafish [22], rice [13,23,24], a wide range of fungi [25], and mammals, including mice [13,21,26-28], pigs [22] and humans [13,21,29,30]. While it is very clear and conclusive that 6mA exists in some lower eukaryotic organisms, including protists, the debating for its presence and abundance is generally focusing on mammals [31-33], which will be discussed below. In short, most studies demonstrated that 6mA exists in eukaryotes but with a wide range of abundance and tissue/biological process specificity, especially in mammals.
The abundance and distribution of DNA 6mA in mammals and its potential role in regulating gene expression
Abundance and dynamics
Unlike its high abundance in prokaryotes, 6mA modifications in eukaryotes, especially in mammalian cells, are relatively rare and initially believed to be controversial. With the development and prevalence of ultra-high-performance liquid chromatography-triple quadrupole-tandem mass spectrometry (UHPLC-QQQ-MS/MS), which is highly sensitive in the detection of 6mA and capable of distinguishing DNA 6mA from RNA N6-adenine methylation (m6A) or DNA m1A, more researchers identified 6mA in mammalian cells. Here, we summarize the findings related to the identification of 6mA and its quantification in mammalian cells, based on the publications that applied mass spectrometry, and found growing consensus rather than controversy.
6mA is present at low levels in various mouse tissues
6mA abundance in mouse genome was first reported in mouse TT2 ES cells and several adult mouse tissues by Wu et al. [28]. In TT2 ES cells, H2A.X deposition regions were first enriched by chromatin immunoprecipitation (ChIP) and then subjected to single molecular real-time sequencing (SMRT). This SMRT-ChIP approach identified over one thousand 6mA sites in the H2A.X deposition regions with a high level of confidence. Their highly sensitive MS method later confirmed 6mA abundance in the whole genome and H2A.X deposition regions to be 6-7 Parts Per Million (p.p.m.) and 25-30 p.p.m., respectively. In mouse brain, kidney and thymus, they detected similar levels of 6mA modification with 1.9-3.8 p.p.m [28]. Nevertheless, Schiffers et al. argued that they could not detect 6mA in mouse TT2 ES cells genome using UHPLC-QQQ-MS, even after adding 13CD3-methionine, a reagent that produces signals for 13CD3-m6dA if there were any 6mA in the genome, to ES cell culture [33]. The inability to detect 6mA, however, might be due to the reported rare abundance of 6mA in mouse embryonic stem cells (mESCs) genome and/or the inefficient incorporation of 13CD3-methionine to the 6mA site. Indeed, the presence and quantification of 6mA in mESC genome were later substantiated by four independent studies, which showed similar ranges of 6mA abundance (1.0-8.6 p.p.m) (Table 1) [26,32,34,35], O’Brown et al reported the potential artifacts that could cause inaccurate 6mA quantification during DNA sample preparation which includes enzyme-derived bacterial DNA contamination, operation-introduced bacterial DNA contamination and incorporation of nucleosides from bacterial in food or microbiota [32]. Through UHPLC-MS/MS designed to minimize and subtract artifacts, they detected 6mA levels in ten common mouse tissue samples from gnotobiotic mice to exclude the potential bacterial contamination and found similar 6mA levels (~1.0-5.0 p.p.m) [32] to those reported earlier [25]. Several later studies reported 6mA presence and abundance in other mouse tissues, including mouse brain (3.0-25.5 p.p.m) [27], cortical neurons (15.0-50.0 p.p.m) [36], and primary fibroblasts (~2.0 p.p.m) [37].
Table 1.
Abundance of 6mA in mammals by mass spectrum
| Year | Species | Tissues | Abundance (p.p.m) | Dynamic | Reference |
|---|---|---|---|---|---|
| 2015 | Human | HEK293T | 1.7 | [13] | |
| Jurkat-T | 2.3 | ||||
| Rat | Heart, liver, spleen, brain,, kidney, subcutis | 0.5-3.5 | |||
| 2016 | Pig | Embryos | 500-1700 | [22] | |
| 2016 | Mouse | TT2 ES cells | 6.0-7.0 | [28] | |
| 2017 | Mouse | TT2 ES cells | N.D. | [33] | |
| Liver, brain | Very low | ||||
| Human | Hela | N.D. | |||
| 2017 | Mouse | Brain | 3.0-25.0 | Increased in PFC after stress | [27] |
| 2018 | Human | HEK293T | wcDNA: <1; mtDNA: ~15 | [38] | |
| 2018 | Human | Blood | 560 | [30] | |
| Gastric | T:100-300; NT: 500-600 | Decreased in tumor | |||
| Liver | T: 50-200; NT: 500-600 | Decreased in tumor | |||
| 2018 | Human | Astrocytes | 1.0-5.0 | [29] | |
| GSCs | 900-1200 | Increased in tumor | |||
| 2019 | Mouse | ESCs | 8.6 | [35] | |
| Spleen | Very low | ||||
| 2019 | Mouse | Cortical neurons | 15-50 | Increased after activity-induction | [36] |
| 2019 | Mouse | Liver, sm. intestine, colon, cecum, heart, muscle, brain, testis, spleen, kidney | ~1-5 | [32] | |
| ESCs | 1 | ||||
| Human | HEK293T, Hela, LCL, CHM1 | ~1-3 | |||
| 2020 | Human | HepG2 | wcDNA: 0.3; mtDNA: 400 | [37] | |
| 143B | wcDNA: N.D; mtDNA: ~180 | ||||
| MDA-MB-231 | wcDNA: ~2; mtDNA: ~130 | ||||
| Mouse | Spleen, Testis | wcDNA: N.D; mtDNA: ~18 | |||
| Primary fibroblast | wcDNA: ~2; mtDNA: ~18 | ||||
| 2020 | Mouse | iCdx2 ES cells | wcDNA: ~8; MAR-DNA: ~80 | ||
| TSC like cells | wcDNA: ~40; MAR-DNA: ~110 | Increased during TSC development | [26] | ||
| 2020 | Human | HEK293T | 1.3 | [34] | |
| Hela | 1 | ||||
| ESCs | 0.6 |
Note: p.p.m, parts per million (6mA sites per million dAs); PFC, prefrontal cortex; wcDNA, whole cell DNA samples; mtDNA, mitochondria DNA samples; from whole cells; GSCs, glioblastoma stem cells; ESCs, Embryonic stem cells; TSC, trophoblast stem cell.
6mA abundance varies in human cell lines and tissues
Detection of 6mA in the human genome by MS was first performed in HEK293T and Jurkat-T cells [13]. To remove all possibility of bacterial DNA contamination, human genomic DNA samples underwent DpnI digestion followed by size-exclusion ultrafiltration. The samples were first digested by Dpn I restriction enzyme, which cut bacterial genomic DNA into fragments less than 2500 bp but left human genomic DNA in much larger fragments due to their vast difference in 6mA abundance. Afterwards, ultrafiltration removed bacterial genomic DNA fragments. The 6mA levels were detected at 2.3 p.p.m and 1.7 p.p.m in Jurkat-T and 293T genome, respectively [13], the latter of which was further confirmed by several studies without Dpn I treatment [32,34,38]. Besides HEK293T cells, other human cell lines that possess 6mA modifications at comparable levels include Hela (~1-3 p.p.m) [32,34], lymphoblastoid cell line (hLCL) (~1 p.p.m) [32], telomerase immortalized cell line (CHM1) (3 p.p.m) [32], HepG2 (0.3 p.p.m) [37], MDA-MB-231 (~2 p.p.m) [37], and human astrocytes cell lines (~5 p.p.m) [29]. However, Xiao et al. detected much higher abundance of 6mA in human liver cancer cell lines including BEL-7402 (560 p.p.m), MHCC-LM3 (550 p.p.m) and SK-hep1 (570 p.p.m), as well as in human primary liver tumor tissues (50-300 p.p.m) and the gastric/liver adjacent non-tumoral tissues (500-600 p.p.m) [30]. Xie et al. also reported a high 6mA abundance in glioblastoma stem cell (GSC) (1,000 p.p.m) [29]. The vast difference in 6mA abundance may be due to the disease/tissue specificity, which suggests the importance to identify biological conditions where 6mA levels are appreciably varied.
6mA abundance shows dynamic changes in normal development and disease progression, with a particular high abundance in mitochondrial DNA (mtDNA)
Although 6mA has been typically reported to be scarce in the mammalian genome under normal conditions, several studies indicated dynamic changes in 6mA levels during development and disease. During in vitro fertilization and early embryonic development of pig embryos, the 6mA/A ratio first increased from 0.09% (in oocyte) to ~0.17% (from the four-cell to the morula stage) and then decreased to 0.05% at the blastocyst stage [22]. In response to chronic environmental exposure-induced stress, the prefrontal cortex (PFC), the region of the brain involved with stress response in mice, displayed a significant increase of 6mA abundance from an average of 6.6 p.p.m to 25.5 p.p.m [24]. Correspondingly, in an in vitro model of neuronal activation, 6mA level rose from approximately 16 p.p.m to 50 p.p.m within mouse cortical neurons in response to neural activation [36]. During human gastric and liver tissue tumorigenesis, 6mA abundance decreased from 500-600 p.p.m in adjacent non-tumoral tissues to 50-300 p.p.m in tumor samples [30]. In contrast, 6mA levels in GSCs are over 100-folds higher than it is in human normal astrocytes (~1000 p.p.m vs ~5 p.p.m) [29].
Whereas 6mA is sparse in the HEK293T genome (less than 1 p.p.m) as aforementioned, its abundance in HEK293T mtDNA was detected at a much higher level (~15 p.p.m) [38]. Indeed, Hao et al. reported the specific enrichment of 6mA in mammalian mitochondrial DNA through a systematic study [37]. By comparing 6mA levels in gDNA with its levels in mtDNA from several human cell lines, including HepG2, 143B, and MDA-MB-231, and multiple primary mouse tissues (e.g., spleen, testis, and primary fibroblast), Hao et al. observed much higher 6mA abundances in mtDNA than in genomic DNA from all the tested samples [37]. Remarkably, 6mA level is at least 1,300-fold higher in mtDNA than in gDNA in HepG2 cells [37]. Given that the mtDNA only accounted for 40% in their “pure” mtDNA [37], which is already the highest purity reported so far though, real 6mA abundance in mtDNA could be even higher. Similarly, Xiao et al. also reported a higher 6mA density (0.184%) in the mitochondria genome than in autosomal chromosomes (0.050%–0.064%) [30]. Thus, it would be important to study the function of 6mA in mtDNA, given the much higher abundance of 6mA in mtDNA than that in genomic DNA.
The genomic distribution and potential gene regulation role of 6mA in mammalian genome
Revealing the location and distribution of 6mA sites in the genome is essential to uncovering potential functions of 6mA. Several next-generation sequencing (NGS)-based strategies have been developed to identify 6mA in prokaryote and eukaryote genomes [17,39]. Based on methods of enriching/distinguishing 6mA modification, these “NGS”-based techniques can be divided into three categories: including (1) 6mA antibody-based, like 6mA-DIP-seq; (2) restriction enzyme digestion-based, such as 6mA-RE-seq and Dpn1-seq; and (3) direct recognition of 6mA modification, such as third-generation sequencing method SMRT sequencing and the recently developed nitrite sequencing, which employs sodium nitrite and acetic acid to diazotize and deaminate unmethylated adenine, while methylated adenine is left intact [40]. These methods have been used to detect genome-wide 6mA distribution and motifs in prokaryotes [17] and lower eukaryotes [14-16].
While several independent studies have characterized 6mA distribution in mouse and human genome since 2015 [21,26-30,35,36,38], Nestor and colleagues pointed out limitations in those published 6mA-“DIP-seq” datasets [31,41]. They argued that a large portion of 6mA signals were caused by specific binding of mouse IgG antibody (IgG control) to unmodified repetitive DNA elements in the genome [41]. Although it remains debatable to compare 6mA “DIP-seq” data with IgG “DIP-seq” data from different studies and tissues, their conclusion from 5mC/5hmC “DIP-seq” data re-analysis highlights the importance of including IgG controls in DIP-based assays, as well as applying non-antibody-based techniques for validation when studying low-abundance DNA modifications. Another criticism is that the potential detection limits of those 6mA sequencing methods were not carefully assessed, especially in DNA samples with low 6mA levels [28,42]. With the above concerns in mind, we re-evaluated the latest reports about the distribution of 6mA in mammalian genome (summarized in Table 2).
Table 2.
Distribution of 6mA in mammals by genome-wide profiling (Key table)
| Year | Species | Tissues | Sequencing methods |
Distribution | Gene regulation |
References |
|---|---|---|---|---|---|---|
| 2016 | Mouse | Kidney | 6mA-DIP-seq | intergenic, introns | [21] | |
| 2016 | Mouse | ESCs | SMRT-ChIP | intergenic regions of H2A.X deposition region |
Repress transcription | [28] |
| 6mA-DIP-seq | intergenic region, LINE elements, chroX | |||||
| 2017 | Mouse | Brain PFC | 6mA-DIP-seq | intergenic regions (65%), introns (33%) | Repress expression | [27] |
| 2018 | Human | HEK293T | 6mACE-seq | ~16% non-transposable protein coding genes | [38] | |
| ~60% intergenic including LINE1, SINE and Alu evenly distributed across mitochondria genome asymmetrically methylated on negative heavy band | Destabilize dsDNA | |||||
| 2018 | Human | Blood | SMRT-seq | broad distribution across autosomal, half the density in chrX and chrY. intergenic regions (70.61%), introns (26.19%), 6mA density enriched in the exon-coding regions | Activate transcription | [30] |
| 6mA-DIP-seq | broad distribution across autosomal, half the density in chrX and chrY intergenic regions (60.7%), introns (36.29%) 6mA density enriched in the exon-coding regions | |||||
| 2018 | Human | GSCs | 6mA-DIP-seq | intergenic (70.6%), colocalizes with heterochromatin marks and 5mC | Repress expression | [29] |
| 2019 | Mouse | ESCs | MeDIP-seq | enrichend in intergenic regions | Repress expression by proteolysis | [35] |
| 2019 | Mouse | ILPFC | Dpn1-seq | most peaks in repetitive elements. Learning extinction-induced 6mA increased in TSS and start codon regions | Positive correlation with gene expression | [36] |
| 2020 | Mouse | ESCs, TSC like cells | 6mA-DIP-seq | intergenic regions (75.3%), introns (23%) enriched at SIDD regions at boundaries of euchromatin | Repress expression | [26] |
Note: 6mA-DIP-seq, 6mA DNA immunoprecipitation sequencing; SMRT-ChIP-seq, single molecule chromatin immunoprecipitation sequencing; 6mACE-seq, 6mA-Crosslinking-Exonuclease-sequencing; MeDIP-seq, methylated DNA immunoprecipitation sequencing; ILPFC, infralimbic prefrontal cortex.
The genomic distribution of 6mA and potential regulatory role in gene expression in mouse genome
Koziol et al. were the first to identify 6mA modifications at the genomic level in mouse kidney tissues using 6mA-“DIP-seq” [21]. After overlapping 6mA sites from three biological replicates, a total of 2,726 6mA sites (Ab/input) were identified and they mainly localized in the intergenic and intronic regions. Interestingly, the transcription start site (TSS) plots showed a sharp decrease in 6mA level just after the TSSs of genes. Gene vicinity analysis demonstrated 6mA mark depletion in exonic regions. Of note, mouse kidney’s low 6mA abundance (Table 1) may compromise the resolution of 6mA distribution decoded by 6mA-“DIP-seq”. Although an IgG control was employed to confirm their findings in X. laevis tissues, it was not applied in the 6mA-“DIP-seq” of mouse kidney tissue samples [21].
In three other independent studies of 6mA distribution in mESCs, 6mA enrichment in intergenic regions has been consistently reported. In the first study, Wu et al. performed SMRT-ChIP-seq to examine 6mA distribution in H2A.X deposition regions and found elevated 6mA levels at the intergenic regions over gene-rich regions [28]. After evaluating detection limits and lineage response of their 6mA-DIP approach (20-120 p.p.m), they repeated 6mA-“DIP-seq” in Alkbh1 knockout cells (30-35 p.p.m). They found that 6mA sites were still enriched in the intergenic regions in the whole genome but preferentially in full-length L1 elements. DNA 6mA marks are negatively correlated with young full-length “L1 elements” and neighboring protein-coding gene expression [28]. In the second study, Kweon et al. profiled the genomic distribution of 6mA in mESCs using MeDIP-seq and similarly observed 6mA enrichment in the intergenic regions [35]. They also noted decreased 6mA density in TSS and a negative correlation between 6mA density and gene expression. It’s worth noting that neither the limits of detection for MeDIP-seq in mESCs (8.6 p.p.m), nor the utilization of IgG controls were mentioned [35]. In the third study, Li et al. observed the highest 6mA level in trophoblast stem cell (TSC)-like cells during TSC development of mESCs [26]. Through 6mA-“DIP-seq” and ssDNA-seq, they showed that 6mA peaks were mainly located in the intergenic regions, consistent with previous findings [28,35], and that 6mA peaks also significantly overlapped with ssDNA-seq peaks and correlated with repressed gene expression [26]. Moreover, Li et al. addressed the limitations of 6mA-“DIP-seq” [31,41] with a series of assays/analysis [26]. They first treated DNA samples with extensive RNase digestion, followed by 6mA-DIP-qPCR, proving that this 6mA-DIP protocol efficiently enriched only 6mA-containing DNA samples and not any significant signal from m6A-RNA-DNA hybrids compared to background (unmodified DNA controls) [26]. 95.55% of the reads generated from the 6mA-“DIP-seq” could be uniquely mapped to mouse genome with no microbe contamination. Finally, in addition to the input control, IgG and whole-genome amplification (WGA) controls were also applied to identify highly confident 6mA peaks, which substantially overlapped with each other (DIP vs. Input, DIP vs. IgG, and DIP vs. “WGA”) [26].
Two studies in mouse brain tissues, however, showed different 6mA distribution patterns and the opposite effects of 6mA on gene regulation, albeit also sharing some similarity between them. In one study, Yao et al. observed a significant 6mA level increase in the PFC of stressed mice. Their genome-wide analysis (6mA-“DIP-seq”) showed that 65% of the regions that gained 6mA upon stress were associated with intergenic regions, while 33% in introns [27]. A non-antibody-based method (Dpn1 digestion followed by qPCR) further validated the 6mA elevated regions identified by the 6mA-“DIP-seq”. Similar to previous reports [26,28,35], they also found that 6mA level is negatively correlated with LINE transposon and expression of a group of neuronal genes [27]. In the second study, Li et al. found that 6mA accumulated in adult mouse brains in response to extinction learning; Dpn1-seq approach was then used to identify extinction learning-induced genome-wide increase of 6mA at single-base resolution [36]. Remarkably, the total 6mA sites identified by Dpn1-seq represents 0.16% of total adenines in the mouse genome of the specific neuronal populations [36], which is almost an order of magnitude larger than a previous report [27]. They explained that the big discrepancy is probably due to the specific neuronal populations investigated and that Dpn1-seq is more sensitive than 6mA “DIP-seq” in identifying 6mA sites [36]. Nevertheless, due to the motif (i.e., GATC) limitation of DpnI, Dpn1-seq may not capture all the 6mA sites in the genome. In addition, they didn’t provide QQQ-MS evidence to confirm the potentially very high 6mA abundance in the genomes of investigated neuronal populations. For 6mA sites common to both extinction-trained (EXT) and retention control (RC) groups, although ~50% located within repetitive regions, extinction learning-induced 6mA accumulated within the promoter, 5’ untranslated region (UTR) and coding regions (CDS) [36], but not within LINE1 elements as was reported previously [27]. The activity-induced 6mA accumulation at proximal GATC in Bdnf P4 promoter is believed to be strongly correlated with increased Bdnf exon IV mRNA expression (a direct regulator of extinction-related learning and memory) [36]. The differential 6mA distribution and effects on gene regulation between these two studies can be possibly attributed to tissue specificity and different genome-wide 6mA identification methods applied. Additional investigation is thus warranted for further clarification.
The genomic distribution of 6mA and potential regulatory role in gene expression in human genome
Thus far, only three publications reported genome-wide 6mA distribution in humans. Due to the difference in 6mA identification methods and tissues tested, each demonstrated discrete findings. Through the analysis of PacBio sequencing datasets generated from human blood, Xiao et al. reported that 6mA sites were broadly distributed across autosomal chromosomes (0.050%-0.064%) but decreased to half the density in sex chromosomes (0.023%-0.024%) [30]. Additionally, they observed a much higher 6mA density (0.184%) in mitochondrial genomes than that in autosomal chromosomes. Although most of the 6mA sites were in intergenic (70.6%) and intronic (26.19%) regions, 6mA sites were significantly enriched (relative to the genomic background) only in exon-coding regions. The above findings were confirmed by additional 6mA-“DIP-seq” [30]. Moreover, Xiao et al. demonstrated a positive correlation between 6mA abundance in exons (identified by 6mA-“DIP-seq”) and RNA expression level of associated genes [30]. However, after aligning Xiao et al.’s data with splice-aware aligner, Nestor et al. argued that up to 20% of the reads in their 6mA-“DIP-seq” data might originate from RNA [31]. In the second study, Koh et al. utilized 6mA-Crosslinking-Exonuclease-sequencing (6mACE-seq) to map genome-wide 6mA profiles in 293T cells [38]. They found that ~60% of 6mA sites were associated with various repetitive sequences and ~14.3% located in intronic regions. Relative to the genomic background, 6mA sites were enriched in intergenic regions and promoters (1 kb region upstream of TSS). Strikingly, they observed that 6mA site density is over 8000 times higher in the mitochondrial genome than in the nuclear genome, showing a strong preference for the negative heavy strand over the positive light strand [38]. Notably, 6mA levels in 293T cells were shown to be rare (~1 p.p.m) in this study as well as in other reports [13,32,34]; since 6mACE-seq is still a 6mA antibody-based method, whether 6mA level in 293T cells is within the limit of detection of 6mACE-seq remains to be justified. In the last study, Xie et al. performed 6mA-“DIP-seq” in GSCs (6mA: ~900 p.p.m) and identified 7,282-17,263 6mA peaks per GSC model and 2,119 common peaks overlapping in three GSC models [29]. 6mA peaks were located across autosomal chromosomes and, most commonly, in intergenic regions (70.6%). In addition, they found that over 80% of 6mA peaks intersected with regions containing heterochromatic histone modifications (H3K9me3 and H3K27me3), suggesting that 6mA co-localizes with heterochromatic histone modifications and acts as a repressive mark [29].
6mA regulators in mammals
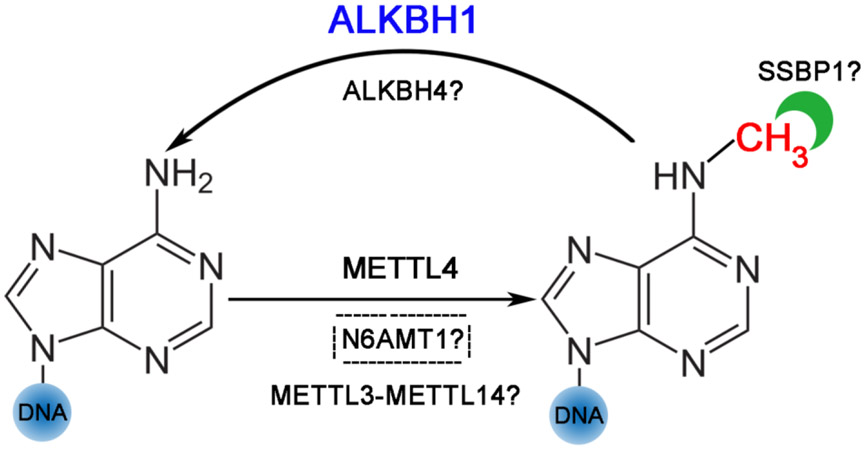
DNA 6mA studies in eukaryotes are still at early stage. Identification of enzymes that are responsible for adding (“Writers”) and removing (“Eraser”) methylation is still under investigation. Here is the summary of the reports as to 6mA regulators so far (Table 3; Figure 1).
Table 3.
Experimental evidence for 6mA regulators
| Types | Regulators |
In vitro methylation/ demethylation assay |
In vivo regulation on 6mA level |
References |
|---|---|---|---|---|
| (Y: support, N: against, N/A: no evidence) | ||||
| Writer | N6AMT1 | Y | Y, human liver cancer cells | [30] |
| N | N, human GSCs | [29] | ||
| N/A | Y, mouse primary cortical neurons | [36] | ||
| N | N/A | [45] | ||
| METTL4 | N/A | Y, 293T and mESCs | [35] | |
| Y | Y, HepG2 mtDNA | [37] | ||
| METTL3-ME | Y | N/A | [52] | |
| TTL14 heterocomplex | Y | N/A | [53] | |
| Eraser | ALKBH1 | Y | Y, mESCs | [28] |
| N/A | N, mESCs and MEFs | [63] | ||
| Y | Y, 293T mtDNA | [38] | ||
| Y | Y, human liver cancer cells | [30] | ||
| Y | Y, human GSCs | [29] | ||
| Y | N/A | [57] | ||
| Y | N/A | [56] | ||
| N/A | Y, mouse primary VSMCs | [64] | ||
| N/A | Y, mouse TSC-like cells | [26] | ||
| ALKBH4 | Y | N/A | [35] | |
| Reader | SSBP1 | N/A | N/A | [38] |
Note: MEFs, Mouse Embryonic Fibroblasts; VSMCs, Vascular smooth muscle cells.
Figure 1. A summary of 6mA regulators in mammals.
Blue and bigger front means strong evidence. Question mark means lacking evidence from either in vitro (cell-free) methylation/demethylation assays or 6mA DNA binding assays, or in vivo (cellular) experiments. Dotted box means contradiction exists.
“Writers”
N6AMT1 was reported as a methyltransferase for nuclear DNA 6mA methylation from an earlier study [30]. However, two separate studies failed to repeat the in vitro methylation assays which are critical to confirm DNA 6mA as a substrate of N6AMT1 [29,43]. Considering it has also been reported to be responsible for protein methylation [43-46], N6AMT1 may affect 6mA levels in DNA indirectly [36].
METTL4 belongs to a subclade of MT-A70 adenine methyltransferases [47], which includes two reported DNA 6mA methyltransferases, C. elegans’ DAMT-1 [16] and bacterial M.Munl [48]. Mouse Mettl4 is capable of adding 6mA deposition in 293T genomic DNA. Mettl4 deletion caused loss of 6mA signals in genomic DNA but no change of m6A level in mRNA in mESCs [35]. Recently, human METTL4 protein was reported to have 6mA methyltransferase activity on mtDNA [37]. In the in vitro DNA methylation assays, METTL4 significantly increased 6mA signals in both dsDNA and ssDNA of HepG2 mtDNA, with a preference for ssDNA substrates. Depletion of METTL4 in HepG2 cells reduced 6mA level in mtDNA but not m6A level in mtRNA species [37]. As studies typically report low levels of 6mA in the human cell line genome, more evidence supporting METTL4 as a 6mA writer in the mitochondria genome would be intriguing.
METTL3 and METTL14 are two additional members in the MT-A70 family and are well-established as RNA m6A writers [49-51]. Interestingly, through a series of in vitro methylation assays, Woodcock et al. demonstrated that human METTL3-METTL14 heterocomplex methylates single-strand DNA and unpaired regions (with reduced activity) in the context of double-strand DNA with similar recognition sequences (GGACT) as it is on mRNA [52]. Later, the same group reported that the METTL3-METTL14 heterocomplex has methyltransferase activity on dsDNA containing cyclopyrimidine dimers, which are the major UV radiation-induced photoproducts, by in vitro methylation assays [53]. It would be interesting to test its DNA 6mA methyltransferase activity in the context of DNA damage in vivo.
“Erasers”
ALKBH1 belongs to a 2-oxoglutarate (2OG)-dependent dioxygenase family/group, which consists of several well-known DNA/RNA demethylases, such as TET proteins [54] (DNA demethylase of 5mC), FTO, and ALKBH5 [55] (RNA demethylases of m6A) proteins. Recent studies consistently support ALKBH1 as a DNA 6mA demethylase in mammals. In multiple independent in vitro demethylation assays, ALKBH1 was proven to exhibit 6mA demethylation activity [28-30,38,56,57] with preference for bubbled or bulged DNA [56]. In several types of mammalian cells, ALKBH1 depletion increased 6mA levels in DNA [28-30,38]. Similar evidence was demonstrated Alkbh1 knockout mouse samples [26,28].
As a family member of ALKBH1, ALKBH4 was only recently reported to have 6mA demethylation activity on DNA in an in vitro demethylation assay [35]. Factoring that it also demonstrates demethylation activity on protein substrates [58], further investigations are required to clarify the full scope of ALKBH4’s demethylation activity.
6mA in human diseases
In the past 5 years, advancement of 6mA detection techniques and new evidence supporting 6mA as an important DNA modification during mammalian development opened new doors for researchers. Subsequently, several studies explored 6mA dysregulation and the role of 6mA regulators in human diseases, including cancer.
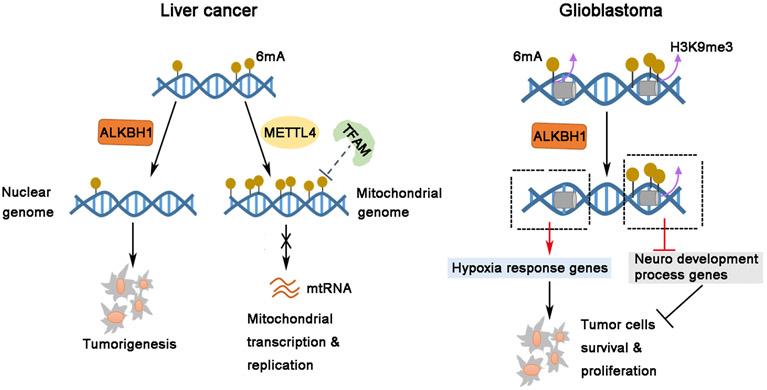
As mass spectrometry and genome-wide 6mA sequencing are essential to generate reliable 6mA data, here we mainly discuss the cancer-related publications with mass spectrometry and 6mA sequencing evidence. Figure 2 depicts the role of 6mA regulators in human cancers. In gastric and liver cancers, ALKBH1 expression increases while 6mA level in genomic DNA decreases in primary tumor samples, relative to those in non-tumor adjacent tissues; ALKBH1 was then shown to play a tumor-promoting role, which is dependent on its 6mA demethylase activity [30]. Conversely, N6AMT1, which was reported as a 6mA methyltransferase in this paper, was shown to suppress tumor progression [30]. However, since later studies suggest that N6AMT1 is not a genuine 6mA methyltransferase [29,43], its role in gastric and liver cancers might not be related to 6mA regulation. In glioblastoma, genomic DNA 6mA level increases in GSCs and primary glioblastoma samples and is correlated with H3K9me3 heterochromatin histone modification levels. ALKBH1 protein level was also modestly elevated in GSCs compared to matched differentiated non-stem tumor cells. Targeting ALKBH1 inhibited glioblastoma carcinogenesis. Mechanistically, ALKBH1 transcriptionally regulates hypoxia response pathway expression during glioblastoma carcinogenesis via reducing 6mA levels located close to genes in the hypoxia response pathway [29]. In HepG2 cells (a liver cancer cell line), METTL4 also functions as an oncogene by regulating 6mA levels in the mitochondria genome, thereby affecting mitochondrial replication and transcription [37]. In addition to the aforementioned systematic studies, DNA 6mA has also been reported as a biological maker in triple negative breast cancer (TNBC) (lower than normal tissues) and correlates with Olaparib resistance [59]. On the contrary, DNA 6mA level was demonstrated to be higher in human esophageal squamous cell carcinoma relative to the matched adjacent normal tissue samples [60]. While the opposite 6mA patterns in primary patient samples could be due to tissue specificity, the DNA 6mA antibody chosen (not verified by other studies before) [59] and lack of RNA removal during DNA sample preparation [60] would also affect DNA 6mA quantification by dot-blot.
Figure 2. Oncogenic role of 6mA regulators in human cancers.
Left panel delineates cellular processes in liver cancer: ALKBH1 promotes tumorigenesis by mediating 6mA modifications; METTL4 regulated 6mA in mtDNA affects mitochondrial transcription and replication. Right panel depicts cellular phenomena in human glioblastoma: ALKBH1 specifically regulates the 6mA modification near hypoxia response genes, but not 6mA enriched region (neuro development genes). ALKBH1 mediated 6mA modification is critical for cell survival and proliferation.
Very recently, two groups reported that 6mA and ALKBH1 play important roles in non-neoplastic diseases. One group showed that in patients with chronic kidney disease (CDK), 6mA levels displayed negative correlations with aortic arch calcification, while ALKBH1 expression was positively correlated with aortic arch calcification; ALKBH1 is demonstrated to regulate vascular calcification by promoting BMP2 transcription via demethylating DNA 6mA modification and thus facilitating the binding of Oct4 to BMP2 promoter and activating BMP2 transcription [61]. The other group reported that DNA 6mA level in leukocytes decreased in patients with hypertension; ALKBH1 is negatively correlated with 6mA level and regulates angiotensin II-induced vascular smooth muscle phenotypes in hypertension mouse and rat models by directly affecting hypoxia inducible factor 1 α (HIF1a) expression [62]. Of note, in the aforementioned two studies, 6mA level was quantified by ELISA assay or IHC, neither of which could distinguish DNA 6mA from RNA m6A due to the specificity limitation of the antibody. As RNase treatment was not described in the DNA sample preparation or IHC methods, RNA m6A contamination could not be ruled out in the DNA 6mA quantification results. More rigorous methods, such as mass spectrometry or extensive RNA digestion of the samples, are essential to confirm the clinical relevance of 6mA level in diseases.
Concluding Remarks:
With 6mA emerging as a critical DNA epigenetic mark in prokaryotes and eukaryotes [3], the features of 6mA in mammals and its roles in human diseases have attracted much attention in the past five years. Current research revealed 6mA abundance in mammalian nuclear and mitochondria genome, dynamics of 6mA levels in development and disease, location and distribution patterns of 6mA sites, effects of 6mA marks on gene expression, and the significance of 6mA in human diseases, especially in cancer. In this review, we focused on the progress of 6mA studies in mammals and human diseases. The 6mA levels in mammals are summarized (Table 1). The distribution of 6mA marks and their correlations with gene regulation are also shown (Table 2). The experimental evidence for regulators of 6mA in mammals are presented (Table 3, Figure 1). The study of 6mA in mammals is still in its infancy; some discrepancies due to differences in technology and biological systems are hence somewhat expected, as is the case with other emerging fields. Further research is required to conclude the features of 6mA in mammals and expand our knowledge of the biological functions of 6mA in human diseases, especially its roles in tumorigenesis.
Some controversies regarding 6mA abundance in mammalian tissues or cell lines have been reported. Giving the low abundance of 6mA in mammal cells and the inability of antibodies to distinguish DNA 6mA from RNA m6A, highly sensitive mass spectrum for detecting 6mA levels in new tissues is strongly recommended. Furthermore, antibody-based quantification of 6mA levels requires complete RNA removal from DNA samples by extensive RNase treatment to obtain accurate abundance or patterns of 6mA if mass spectrum is not available.
Currently, researchers commonly utilize “DIP-seq”, SMRT-seq or DpnI-seq for genome-wide identification and characterization of 6mA sites to understand 6mA-mediated gene regulation in different cellular contexts. However, these are not without limitations. Anti-6mA antibody-based sequencing cannot achieve base-pair resolution and the sensitivity and specificity needs to be verified especially in samples with rare 6mA abundance. To generate reliable data with “DIP-seq”, it is important to work in samples with DNA 6mA level above reported limits (~20 p.p.m) [28]; otherwise, it is necessary to test the lineage response range of DIP before utilizing it in samples with lower 6mA level. Likewise, SMRT-seq aptly maps 6mA at base-pair resolution in bacteria but falls short in eukaryotes, especially in those with large genome size and low 6mA abundance, and tends to overestimate the amount of 6mA sites due to interference from other changes/modifications such as DNA damages, 5mC and its derivatives in the process of demethylation [32]. More rigorous bioinformatic approaches are required to guarantee the sensitivity and specificity of SMRT-seq when utilized in mammalian genome. In term of DpnI-seq, it is very sequence specific and may not capture all the 6mA sites due to the motif of DpnI.
The dynamics and roles of 6mA in development and disease conveys the importance in identifying the involved machineries, particularly writers (methyltransferases), erasers (demethylases), and readers (6mA binding proteins). While recent evidence consistently supports ALKBH1 as a DNA 6mA demethylase, additional studies to verify other potential regulators, such as METTL3-METTL14, METTL4 and ALKBH4, is warranted. N6AMT1 was reported as a methyltransferase, but subsequent independent studies did not support it. Although no well-recognized readers of 6mA have been identified thus far, there have been efforts of SILAC (stable isotope labelling by amino acids)-based pulldown and SSBP1 was identified as a potential 6mA reader candidate [38]. The identification of all the 6mA writers, erasers and readers is essential for us to understand how 6mA is regulated by the writers and erasers and how 6mA readers recognize 6mA modification and manipulate the fate of 6mA-modified genomic loci in different cellular contexts. Lastly, determining the roles of 6mA and its regulators in various biological processes, including cancer, through functional studies and whole-genome profiling of 6mA sites could lead to expansion of our knowledge about this new DNA epigenetic mark in normal development and diseases (e.g., cancers) and, eventually, new therapies in disease treatment. The 6mA signatures from patients could serve as biomarkers for early diagnosis, drug response prediction, and prognosis of diseases. The dysregulated 6mA regulators could be attractive therapeutic targets and novel therapeutics targeting such regulators could hold great potential in treating various types of diseases, including cancers.
OUTSTANDING QUESTIONS BOX:
What’s the abundance of DNA 6mA in eukaryotes especially in mammals? Is it dynamic during normal development and disease process?
What’s the genomic distribution pattern of DNA 6mA? Does it play a regulatory role in gene expression similar to 5mC?
What’s the abundance and role of 6mA in mammalian mitochondrial genome?
Can much more effective and sensitive base-resolution 6mA-seq methods that need much less start material be developed?
Are there any crosstalks of DNA 6mA with other DNA modifications, or with RNA modifications or histone modifications in eukaryotes, especially in mammals?
What are the DNA 6mA regulators (writers, erasers and readers) in mammals?
Do DNA 6mA and its regulators play important roles in human diseases, including cancers?
Can DNA 6mA signatures in genomic DNA or mitochondria DNA or in cell-free DNA (cfDNA) serve as biomarkers for early diagnosis, drug response prediction, and prognosis of diseases?
Whether and how DNA 6mA regulators are dysregulated in human diseases (e.g., cancers)? Are their dysregulation is associated with disease progression and clinical outcomes? Are they druggable targets for disease treatment?
Highlights:
DNA N(6)-methyldeoxyadenosine (6mA) is an abundant and functionally important modification in prokaryotes. Recently, emerging evidence support its presence in eukaryotes, including mammals.
In mammals, 6mA level is consistently low in normal mouse tissues, but varies in human cell lines or tissues. In addition, 6mA abundance is dynamic during normal development and disease progression.
The 6mA genomic distribution and its roles in gene regulation are tissue/biological process dependent. Common features, distinct findings and research limitations are discussed.
Several proteins have been reported to function as 6mA writers or erasers in mammals, whereas some needs further confirmation.
Dysregulated 6mA levels and its regulators contribute to the development of human diseases including cancers. Targeting 6mA and its regulators holds potential in cancer treatment.
Acknowledgments
This work was supported in part by the U.S. National Institutes of Health (NIH) grants CA243386 (J.C.), R01 CA214965 (J.C.), R01 CA236399 (J.C.), R01 CA211614 (J.C.) and R01 DK124116 (J.C), and the Simms/Mann Family Foundation. J.C. is a Leukemia & Lymphoma Society (LLS) Scholar.
Glossary
- 6mA-Crosslinking-Exonuclease-sequencing (6mACE-seq)
Method for identifying 6mA sites at whole genome level. Anti-6mA antibodies are first crosslinked onto 6mA-containing dsDNA fragments, which are thus protected from subsequent 5’-to-3’ exonuclease treatment. Exonuclease degradation of all genomic DNA other than regions proximal to the protected 6mA is achieved.
- DNA immunoprecipitation sequencing (DIP-seq)
Technique that uses antibody binding and next-generation sequencing to measure genome-wide enrichment of a specific DNA modification.
- L1 elements (LINE1)
Class I transposable elements in the DNA of some organisms. Active L1 elements can interrupt the genome through insertions, deletions, rearrangements, and copy number variations (CNV). LINE1 are tightly regulated in the germline by DNA methylation, histone modifications, and piRNA.
- Next-generation sequencing (NGS)
A technique that performs sequencing of millions of small fragments of DNA in parallel. Bioinformatics analyses are used to piece together these fragments by mapping the individual reads to the reference genome.
- RNA-DNA hybrids
A non-canonical nucleic acid structure that forms when nascent transcripts anneal to the DNA template strand or any homologous DNA region. It could lead to DNA damage and genome instability with improper turnover and/or unscheduled formation.
- Whole-genome amplification (WGA)
A method for robust amplification of an entire genome. As the WGA doesn’t contain any DNA modifications, it could be used as a negative control for studies of genome-wide distribution of a specific DNA modification like DIP-seq.
Footnotes
Competing interests
J.C. is a Scientific Advisor for Race Oncology.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eleftheriou M and Ruzov A (2021) Modified Forms of Cytosine in Eukaryotes: DNA (De)methylation and Beyond. Methods Mol Biol 2198, 3–13. 10.1007/978-1-0716-0876-0_1 [DOI] [PubMed] [Google Scholar]
- 2.Wion D and Casadesus J (2006) N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat Rev Microbiol 4, 183–192. 10.1038/nrmicro1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu KJ (2020) The epigenetic roles of DNA N(6)-Methyladenine (6mA) modification in eukaryotes. Cancer Lett 494, 40–46. 10.1016/j.canlet.2020.08.025 [DOI] [PubMed] [Google Scholar]
- 4.Dunn DB and Smith JD (1958) The occurrence of 6-methylaminopurine in deoxyribonucleic acids. Biochem J 68, 627–636. 10.1042/bj0680627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vanyushin BF et al. (1968) 5-methylcytosine and 6-methylamino-purine in bacterial DNA. Nature 218, 1066–1067. 10.1038/2181066a0 [DOI] [PubMed] [Google Scholar]
- 6.Naito T et al. (1995) Selfish behavior of restriction-modification systems. Science 267, 897–899. 10.1126/science.7846533 [DOI] [PubMed] [Google Scholar]
- 7.Vasu K and Nagaraja V (2013) Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol Mol Biol Rev 77, 53–72. 10.1128/MMBR.00044-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorovsky MA et al. (1973) ( 6 N)methyl adenine in the nuclear DNA of a eucaryote, Tetrahymena pyriformis. J Cell Biol 56, 697–701. 10.1083/jcb.56.3.697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cummings DJ et al. (1974) Methylated bases in DNA from Paramecium aurelia. Biochim Biophys Acta 374, 1–11. 10.1016/0005-2787(74)90194-4 [DOI] [PubMed] [Google Scholar]
- 10.Ratel D et al. (2006) N6-methyladenine: the other methylated base of DNA. Bioessays 28, 309–315. 10.1002/bies.20342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams RL et al. (1979) Methylation of mosquito DNA. Biochim Biophys Acta 563, 72–81. 10.1016/0005-2787(79)90008-x [DOI] [PubMed] [Google Scholar]
- 12.Ashapkin VV et al. (2002) The gene for domains rearranged methyltransferase (DRM2) in Arabidopsis thaliana plants is methylated at both cytosine and adenine residues. FEBS Lett 532, 367–372. 10.1016/s0014-5793(02)03711-0 [DOI] [PubMed] [Google Scholar]
- 13.Huang W et al. (2015) Determination of DNA adenine methylation in genomes of mammals and plants by liquid chromatography/mass spectrometry. RSC Advances 5, 64046–64054. 10.1039/C5RA05307B [DOI] [Google Scholar]
- 14.Fu Y et al. (2015) N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell 161, 879–892. 10.1016/j.cell.2015.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang G et al. (2015) N6-methyladenine DNA modification in Drosophila. Cell 161, 893–906. 10.1016/j.cell.2015.04.018 [DOI] [PubMed] [Google Scholar]
- 16.Greer EL et al. (2015) DNA Methylation on N6-Adenine in C. elegans. Cell 161, 868–878. 10.1016/j.cell.2015.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo GZ et al. (2015) DNA N(6)-methyladenine: a new epigenetic mark in eukaryotes? Nat Rev Mol Cell Biol 16, 705–710. 10.1038/nrm4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo GZ and He C (2017) DNA N(6)-methyladenine in metazoans: functional epigenetic mark or bystander? Nat Struct Mol Biol 24, 503–506. 10.1038/nsmb.3412 [DOI] [PubMed] [Google Scholar]
- 19.Shah K et al. (2019) Adenine Methylation in Drosophila Is Associated with the Tissue-Specific Expression of Developmental and Regulatory Genes. G3 (Bethesda) 9, 1893–1900. 10.1534/g3.119.400023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao B et al. (2018) Active N(6)-Methyladenine Demethylation by DMAD Regulates Gene Expression by Coordinating with Polycomb Protein in Neurons. Mol Cell 71, 848–857 e846. 10.1016/j.molcel.2018.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koziol MJ et al. (2016) Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat Struct Mol Biol 23, 24–30. 10.1038/nsmb.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J et al. (2016) Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig. Nat Commun 7, 13052. 10.1038/ncomms13052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang K et al. (2021) The dynamics of N(6)-methyladenine RNA modification in interactions between rice and plant viruses. Genome Biol 22, 189. 10.1186/s13059-021-02410-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou C et al. (2018) Identification and analysis of adenine N(6)-methylation sites in the rice genome. Nat Plants 4, 554–563. 10.1038/s41477-018-0214-x [DOI] [PubMed] [Google Scholar]
- 25.Seidl MF (2017) Adenine N6-methylation in diverse fungi. Nat Genet 49, 823–824. 10.1038/ng.3873 [DOI] [PubMed] [Google Scholar]
- 26.Li Z et al. (2020) N(6)-methyladenine in DNA antagonizes SATB1 in early development. Nature 583, 625–630. 10.1038/s41586-020-2500-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao B et al. (2017) DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat Commun 8, 1122. 10.1038/s41467-017-01195-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu TP et al. (2016) DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature 532, 329–333. 10.1038/nature17640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie Q et al. (2018) N(6)-methyladenine DNA Modification in Glioblastoma. Cell 175, 1228–1243 e1220. 10.1016/j.cell.2018.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao CL et al. (2018) N(6)-Methyladenine DNA Modification in the Human Genome. Mol Cell 71, 306–318 e307. 10.1016/j.molcel.2018.06.015 [DOI] [PubMed] [Google Scholar]
- 31.Douvlataniotis K et al. (2020) No evidence for DNA N (6)-methyladenine in mammals. Sci Adv 6, eaay3335. 10.1126/sciadv.aay3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Brown ZK et al. (2019) Sources of artifact in measurements of 6mA and 4mC abundance in eukaryotic genomic DNA. BMC Genomics 20,445. 10.1186/s12864-019-5754-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schiffers S et al. (2017) Quantitative LC-MS Provides No Evidence for m(6) dA or m(4) dC in the Genome of Mouse Embryonic Stem Cells and Tissues. Angew Chem Int Ed Engl 56, 11268–11271. 10.1002/anie.201700424 [DOI] [PubMed] [Google Scholar]
- 34.Musheev MU et al. (2020) The origin of genomic N(6)-methyl-deoxyadenosine in mammalian cells. Nat Chem Biol. 10.1038/s41589-020-0504-2 [DOI] [PubMed] [Google Scholar]
- 35.Kweon SM et al. (2019) An Adversarial DNA N(6)-Methyladenine-Sensor Network Preserves Polycomb Silencing. Mol Cell 74, 1138–1147 e1136. 10.1016/j.molcel.2019.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X et al. (2019) The DNA modification N6-methyl-2'-deoxyadenosine (m6dA) drives activity-induced gene expression and is required for fear extinction. Nature neuroscience 22, 534–544. 10.1038/s41593-019-0339-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hao Z et al. (2020) N(6)-Deoxyadenosine Methylation in Mammalian Mitochondrial DNA. Mol Cell. 10.1016/j.molcel.2020.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koh CWQ et al. (2018) Single-nucleotide-resolution sequencing of human N6-methyldeoxyadenosine reveals strand-asymmetric clusters associated with SSBP1 on the mitochondrial genome. Nucleic Acids Res 46, 11659–11670. 10.1093/nar/gky1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heyn H and Esteller M (2015) An Adenine Code for DNA: A Second Life for N6-Methyladenine. Cell 161, 710–713. 10.1016/j.cell.2015.04.021 [DOI] [PubMed] [Google Scholar]
- 40.Mahdavi-Amiri Y et al. (2020) Single-nucleotide resolution of N (6)-adenine methylation sites in DNA and RNA by nitrite sequencing. Chemical science 12, 606–612. 10.1039/d0sc03509b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lentini A et al. (2018) A reassessment of DNA-immunoprecipitation-based genomic profiling. Nat Methods 15, 499–504. 10.1038/s41592-018-0038-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mondo SJ et al. (2017) Widespread adenine N6-methylation of active genes in fungi. Nat Genet 49, 964–968. 10.1038/ng.3859 [DOI] [PubMed] [Google Scholar]
- 43.Woodcock CB et al. (2019) Human HemK2/KMT9/N6AMT1 is an active protein methyltransferase, but does not act on DNA in vitro, in the presence of Trm112. Cell discovery 5, 50. 10.1038/s41421-019-0119-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Metzger E et al. (2019) KMT9 monomethylates histone H4 lysine 12 and controls proliferation of prostate cancer cells. Nat Struct Mol Biol 26, 361–371. 10.1038/s41594-019-0219-9 [DOI] [PubMed] [Google Scholar]
- 45.Li W et al. (2019) Structural insight into human N6amt1-Trm112 complex functioning as a protein methyltransferase. Cell Discov 5, 51. 10.1038/s41421-019-0121-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu P et al. (2010) Deficiency in a glutamine-specific methyltransferase for release factor causes mouse embryonic lethality. Mol Cell Biol 30, 4245–4253. 10.1128/MCB.00218-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bujnicki JM et al. (2002) Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m(6)A methyltransferase. J Mol Evol 55, 431–444. 10.1007/s00239-002-2339-8 [DOI] [PubMed] [Google Scholar]
- 48.Iyer LM et al. (2016) Adenine methylation in eukaryotes: Apprehending the complex evolutionary history and functional potential of an epigenetic modification. Bioessays 38, 27–40. 10.1002/bies.201500104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu J et al. (2014) A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10, 93–95. 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang H et al. (2020) m(6)A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell 37, 270–288. 10.1016/j.ccell.2020.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang H et al. (2020) The Biogenesis and Precise Control of RNA m6A Methylation. Trends in Genetics, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woodcock CB et al. (2019) Human MettL3-MettL14 complex is a sequence-specific DNA adenine methyltransferase active on single-strand and unpaired DNA in vitro. Cell Discov 5, 63. 10.1038/s41421-019-0136-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu D et al. (2021) Human MettL3-MettL14 RNA adenine methyltransferase complex is active on double-stranded DNA containing lesions. Nucleic Acids Res. 10.1093/nar/gkab460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cimmino L et al. (2015) TET1 is a tumor suppressor of hematopoietic malignancy. Nat Immunol 16, 653–662. 10.1038/ni.3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang H et al. (2020) The Biogenesis and Precise Control of RNA m(6)A Methylation. Trends Genet 36, 44–52. 10.1016/j.tig.2019.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang M et al. (2020) Mammalian ALKBH1 serves as an N(6)-mA demethylase of unpairing DNA. Cell Res. 10.1038/s41422-019-0237-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tian LF et al. (2020) Structural basis of nucleic acid recognition and 6mA demethylation by human ALKBH1. Cell Res. 10.1038/s41422-019-0233-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li MM et al. (2013) ALKBH4-dependent demethylation of actin regulates actomyosin dynamics. Nat Commun 4, 1832. 10.1038/ncomms2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sheng X et al. (2020) DNA N6-Methyladenine (6mA) Modification Regulates Drug Resistance in Triple Negative Breast Cancer. Front Oncol 10, 616098. 10.3389/fonc.2020.616098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen L et al. (2020) DNA N6-methyladenine increased in human esophageal squamous cell carcinoma. Discov Med 29, 85–90 [PubMed] [Google Scholar]
- 61.Zhu K and Reiser J (2021) ALKBH1 reduces DNA N6-methyladenine to allow for vascular calcification in chronic kidney disease. J Clin Invest 131. 10.1172/JCI150966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo Y et al. (2020) DNA N(6)-methyladenine modification in hypertension. Aging 12, 6276–6291. 10.18632/aging.103023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu F et al. (2016) ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell 167, 1897. 10.1016/j.cell.2016.11.045 [DOI] [PubMed] [Google Scholar]
- 64.Ouyang L et al. (2021) ALKBH1-demethylated DNA N6-methyladenine modification triggers vascular calcification via osteogenic reprogramming in chronic kidney disease. J Clin Invest 131. 10.1172/JCI146985 [DOI] [PMC free article] [PubMed] [Google Scholar]