Abstract
For more than a hundred years, the fruit fly, Drosophila melanogaster, has served as a powerful model organism for biological and biomedical research due to its many genetic and physiological similarities to humans and availability of sophisticated technologies used to manipulate its genome and genes. The Drosophila research community quickly adopted CRISPR technologies and, in the 8 years since the first CRISPR publications in flies, has explored and innovated methods for mutagenesis, precise genome engineering, and beyond. Moreover, the short lifespan and ease of genetics have made Drosophila an ideal testing ground for in vivo applications and refinements of the rapidly evolving set of CRISPR-Cas tools. Here, we review innovations in delivery of CRISPR reagents, increased efficiency of cutting and HDR, and alternatives to standard Cas9-based approaches. While the focus is primarily on in vivo systems, we also describe the role of Drosophila cultured cells as both an indispensable first step in the process of assessing new CRISPR technologies and a platform for genome-wide CRISPR pooled screens.
Keywords: Drosophila, CRISPR, genome engineering
CRISPR transformed Drosophila as a model organism
For more than a hundred years, the fruit fly, Drosophila melanogaster, has served as a powerful model organism for biological and biomedical research. Drosophila has endured in part due to its many genetic and physiological similarities to humans but perhaps above all due to the availability of sophisticated technologies used to manipulate its genome and genes. From the first description of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Cas (see Glossary) as a programmable system for generating targeted double strand breaks (DSBs) in DNA [1], Drosophila geneticists have recognized the incredible potential of this technique. Targeted DSBs were not new: programmable zinc-finger nucleases and transcription activator-like effector nucleases (TALENs) had been previously described [2,3]. However, the CRISPR-Cas system is simpler and more practical for use as a lab tool, and opened up nearly limitless avenues to manipulate the fly genome. The Drosophila research community quickly adopted CRISPR technologies and, in the 8 years since the first CRISPR publications in flies, has explored and innovated methods for mutagenesis, precise genome engineering, and beyond.
Initially identified as a bacterial defense system against viruses, the basic CRISPR-Cas system is composed of a CRISPR-RNA (crRNA) containing a unique spacer sequence and a CRISPR-associated nuclease (Cas)[4]. Hybridization between the crRNA spacer and the complementary target (protospacer) leads to activation of the Cas nuclease, which creates a DSB in the target DNA. A short 2–5 bp sequence located close to the protospacer sequence called the PAM (Protospacer Adjacent Motif) is essential for efficient targeting. Those are the basics but as we will review below, there are many nuances depending on the type of Cas protein and the application.
Cutting the Genome
Researchers have explored many different CRISPR/Cas systems for genome targeting, but the most widely used is the type II CRISPR-Cas9 system from Streptococcus pyogenes (SpCas9) [4]. In the presence of the crRNA and a trans-activating crRNA (tracrRNA), SpCas9 cleaves any DNA containing a 20-nucleotide (nt) target sequence adjacent to the PAM that is complementary to crRNA sequence. A chimeric single guide RNA (sgRNA), which combines the crRNA and tracrRNA into a single RNA transcript, simplifies the system [1]. By changing the target-specific spacer sequence within the sgRNA, this system can be programmed to target any DNA sequence of interest in the genome and generate a DSB. The break is then repaired either by error prone non-homologous end joining (NHEJ) resulting in small insertion-deletions (indels), or by homology directed repair (HDR) which can be used to generate precise genomic modifications if a homologous repair template is provided [5–7]. The simple NGG PAM sequence requirement of SpCas9 made the enzyme widely applicable to many different experiments. However, researchers are still actively exploring other CRISPR systems to identify Cas9-like effector proteins that may have differences in their sizes, PAM requirements, and substrate preferences. The development of genome cutting by CRISPR-Cas in Drosophila has focused on two areas: (1) Improving Cas9 cutting efficiency in the germline and soma; and (2) exploring alternatives to the typical Cas9-induced DSBs.
Improving cutting with Cas9
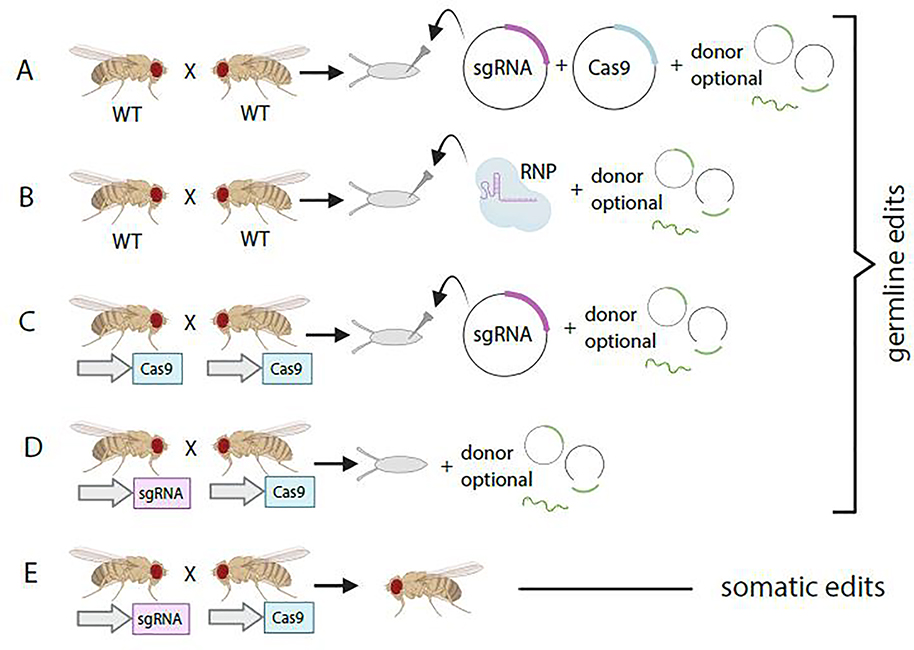
Making cuts in the germline requires delivery of Cas9 and sgRNA (Figure 1). Early attempts at delivery of reagents involved co-injection of sgRNA directly or encoded on a plasmid, plus plasmid derived Cas9 into the germline [8,9]. It was soon discovered that injection of sgRNA into embryos with transgenic Cas9 expressed specifically in the germline produced higher rates of cutting [10,11], and this remains the most common delivery approach for targeting the germline. Recently, microinjection of sgRNA and Cas9 in the form of in vitro assembled ribonucleoproteins (RNPs) was shown to be a highly efficient method for inducing DSBs in flies [12]. Unlike transgenic Cas9, injection of RNPs can be done in a variety of genetic backgrounds, and in other Drosophila species. Several groups have also produced large collections of transgenic sgRNA lines targeting gene coding sequence [13–15]. Mutant animals can be produced by simply crossing sgRNA-expressing flies to germline-specific Cas9 flies. The fully transgenic methods work at least as well as injection and can be implemented by any standard fly genetics lab, including those without access to specialized microinjection equipment.
Figure 1. In vivo methods for introducing CRISPR reagents into Drosophila.
A. Plasmids encoding sgRNA and Cas9 are injected into Drosophila embryos to induce DSB in the germline. Donors can be added as circular plamsids, in vivo linearized dsDNA or ssDNA. B. Recombinant Cas enzymes and chemically synthesized gRNA are combined into a Ribonucleoprotein (RNP) for injection. C. Plasmid encoding sgRNA is injected into embryos from flies expressing Cas9 from a germline promoter. D. Transgenic sgRNA-expressing flies are crossed to transgenic germline-expressing Cas9 flies to produce germline edits. E. Transgenic sgRNA-expressing flies are crossed to transgenic tissue-specific Cas9 flies to produce somatic tissue edits.
Once mature, flies are outcrossed and screened for successful editing events by sequencing of the locus. While reasonably reliable, this step remains the most labor-intensive part of the process of identifying indels, particularly for sgRNAs with low editing efficiency. Thus, strategies have been developed to enrich for desired editing events by co-selecting for an independent edit that produces an easily detectable phenotype in a common marker gene [16,17]. This CRISPR co-selection is based on the fact that the edits produced by two sgRNAs tend to co-occur at the target loci at a rate greater than expected by chance. An interesting twist on this method involves co-selection for rescue of a dominant female sterile allele, ovoD [18]. Here, edits in the gene of interest are enriched, as only germ cells that have successfully edited the ovoD mutation will produce viable eggs [18].
Making cuts in somatic cells is possible by tissue-specific expression of Cas9 with a specific promoter or by GAL4/UAS control (Box 1). Within the tissue, CRISPR/Cas9 is targeted to the beginning of target gene ORFs to generate frameshift mutations that strongly reduce or eliminate gene function. In mammalian cells, roughly 80% of indels induced at a given site disrupt the gene coding frame [19]. However, some indels will only cause loss of or changes in a small number of amino acids, resulting in a weak loss-of-function allele. Furthermore, the altered genome will no longer match the sgRNA sequence and thus will be resistant to additional cleavage. For this reason, it is generally better to use multiple guides targeting the same gene as this can result in deletions between the guide sequences or frameshifts at both target sites, changes that are more likely to produce strong loss-of-function mutations in most of the cells of the tissue.
Text Box 1: Use of CRISPR-Cas technologies with binary system control.
For the past three decades, the Gal4-UAS system has been the predominant technique in Drosophila to produce tissue-specific genetic manipulations [97]. The method allows control of gene expression in any cell type and at any time of development. This technique has been further enhanced by introducing a number of modifications, for example, the temperature-sensitive GAL80 (GAL80ts), which inhibits GAL4 activation at permissive temperatures, allows the expression of the UAS gene construct at restrictive temperatures, thus enabling stage and cell-specific expression of the gene of interest [98].
CRISPR approaches can be combined with the GAL4/UAS system for spatial and temporal control of gene perturbation. For example, a GAL4 line can be used to drive expression of UAS-Cas9 in a specific cell population. When crossed to an appropriate sgRNA fly stock, this results in mutations in the target gene only in GAL4-expressing cells. GAL4 + Cas9 lines with and without GAL80ts have been produced for most major tissues of the fly [15] and are available to the community. Likewise, for CRISPR-activation studies in vivo, GAL4+ dCas9-activator + sgRNA-gene are combined in a fly such that the gene of interest is overexpressed in the GAL4 domain. As other Cas proteins have been introduced into the fly, GAL4-UAS remains the method of choice for in vivo control of expression.
Several methods for multiplexing guides have been developed to enable more efficient single gene cutting or simultaneous cutting of multiple genes. In Drosophila, up to two sgRNAs can be expressed from a single plasmid or transgenic line using multiple U6 RNA pol III promoters [20,21]. Up to six sgRNAs can be effectively processed from a single transcript when sgRNAs are linked by self-cleaving ribozymes or tRNA precursors [22–24]. These methods enable use of RNA pol II promoters, allowing for tighter control of CRISPR mutagenesis by regulating sgRNA synthesis in time and space, a feature not possible with ubiquitously expressed RNA pol III-based promoters. Indeed, in somatic CRISPR mutagenesis experiments, slight leakiness of tissue specific Cas9 expression combined with ubiquitous sgRNA expression can lead to cutting outside the desired tissue [23]. Alternative tRNA sequences and mutations in the Cas9-binding portion of the sgRNA have increased the efficiency of these multi-guide constructs even more [25].
Cas9 alternatives
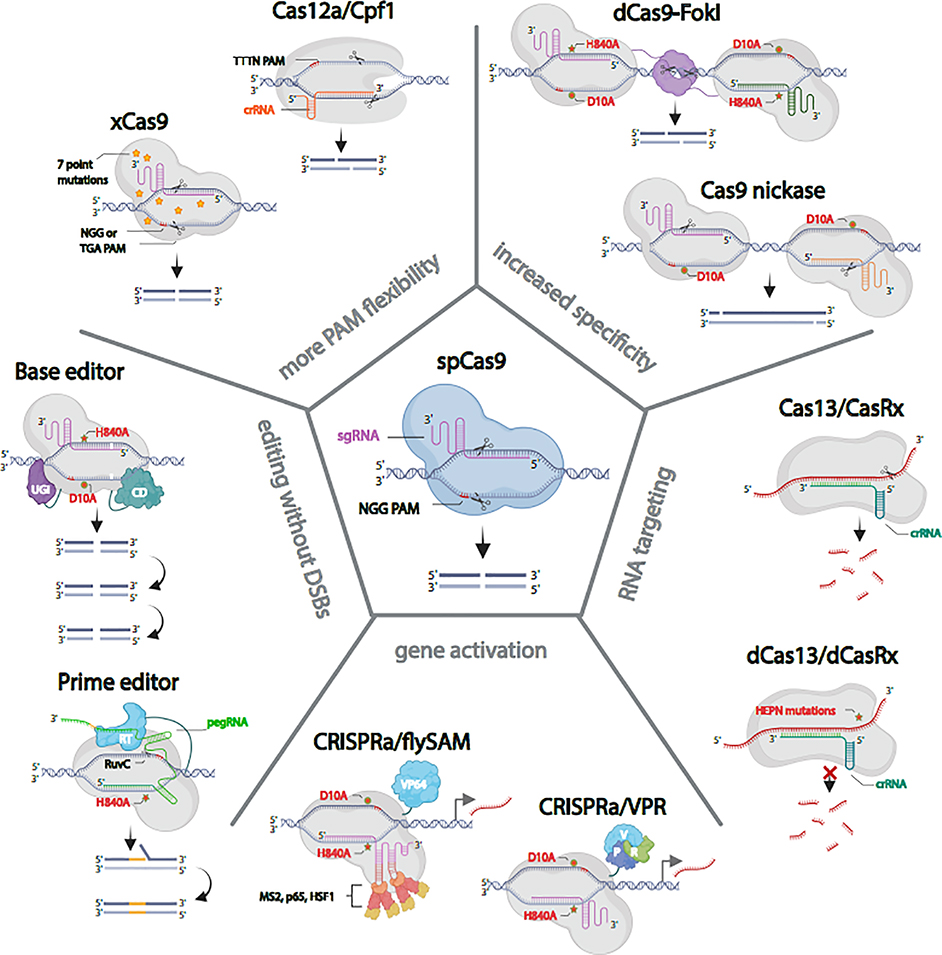
Although the standard SpCas9 is incredibly powerful for cutting the genome, it does have some deficiencies; for example, the NGG PAM requirement does not always enable targeting of the desired locus, generation of DSBs in general can stress the cell [26,27], and off-target DSBs can generate mutations in the wrong locus [28]. This last issue has driven technology innovation in the mammalian CRISPR field, as therapeutic use requires minimal off-target disruption. In flies, the ease of outcrossing to remove deleterious second site mutations means that off-target mutations are not as great of a concern. Nonetheless, several labs have tested alternatives to Cas9 (Figure 2).
Figure 2. In vivo CRISPR in Drosophila with Cas9 and its alternatives.
spCas9, the most commonly used Cas protein in Drosophila produces reliable double strand breaks (DSBs). xCas9 and Cas12a/Cpf1 provide greater PAM flexibility. Paired Cas9 nickase and dCas9-FokI fusions minimize the possibility of off-target DSBs. A cytosine base editor (C>T, G>A) and prime editor allow for precise genome editing without making DSBs. dCas9-fused to VPR or synergistic activation mediator (SAM) produce robust gene activation in vivo. Cas13/CasRx binds and degrades target RNA. The catalytically inactive dCas13/dCasRx binds to RNA but does not cut it.
I. Cas9 Nickase
Cas9 has a RuvC nuclease domain that targets the DNA strand noncomplementary to the sgRNA and a HNH nuclease domain that targets the complementary strand [1]. Either of these nuclease domains can be mutated independently to create DNA ‘nickases’ capable of introducing a single strand cut with the same specificity as an unmodified CRISPR/Cas9 nuclease [29]. DSBs can be introduced using paired nickases for cooperative genome engineering [30]. Because both nicking Cas9 enzymes must effectively nick their target sites to generate a DSB, using paired nickases is less likely to generate off-target DSBs. In Drosophila, the RuvC mutant Cas9 nickase (Cas9D10A) efficiently generates indel mutations in vivo with a pair of sgRNAs [20,31]. Cas9 nickase has not been widely adopted in flies as a means for generating indels, but, as we will discuss below, nickases have been used in Drosophila for alternative editing approaches.
II. dCas9-FokI
To increase specificity, new Cas9 versions that only cleave when dimerized have been developed. For example, a nuclease dead Cas9 (dCas9) fused to the endonuclease FokI maintains sgRNA-directed specificity but relies on obligate FokI dimerization for cleavage [32–34]. Dimerization of FokI depends on recruitment of two dCas9 molecules guided by two distinct sgRNAs that are 15–25 bp apart. In vivo experiments in Drosophila showed that dCas9-FokI was as efficient as SpCas9 in generating loss of function alleles [35].
III. xCas9
A recent study reported that an evolved SpCas9 variant, xCas9(3.7), which prefers various NG-PAM sequences, has broad PAM compatibility, greater DNA specificity, and lower off-target activity than SpCas9 in mammalian cells [36]. In Drosophila, xCas9 showed less activity than SpCas9 on an NGG PAM site but was able to also cleave one non-NGG PAM site with comparable efficiency [37]. However, for other non-NGG PAM sites shown to work in mammalian cells, xCas9 showed no activity. Additional research will be needed to identify the most efficient PAM sites for xCas9 in Drosophila, increasing the targeting range for the applications of genome editing in insects.
IV. Cas12a/Cpf1
Unlike Cas9 and its variants, which require a tracrRNA and RNase III for maturation of its guide RNA, Cas12a (also known as Cpf1) can process its own CRISPR array [38]. Therefore, multiple genes can be controlled with a single CRISPR array plus a Cas12a nuclease. Cas12a also requires a T-rich PAM sequence and can target different genomic regions than SpCas9. Port et al. showed that Cas12a from Lachnospiraceae bacterium (LbCas12a) efficiently edits Drosophila genes in vivo [39]. Interestingly, LbCas12a activity is high at 29°C, but low at 18°C, enabling temperature-based control of cutting. The ease of constructing the compact Cas12a crRNA arrays—Port et al. simply ordered 8X arrays from a commercial vendor—is particularly exciting for the prospect of multi-gene loss-of-function analysis. Of all the SpCas9 alternatives so far discussed here, the Cas12a system may be the most suitable for widespread adoption by the Drosophila research community, as it offers new capabilities while maintaining cutting at an efficiency equal to or above that of SpCas9.
Precise Genome Editing
Above, we discussed the use of RNA-guided endonucleases such as Cas9 to generate DNA DSBs followed by repair by the error-prone NHEJ pathway. Alternatively, if a researcher provides a separate DNA template containing homology arms – sequences homologous to the regions flanking the DSB – the template can be incorporated into the locus by homology-directed repair (HDR). Precise genome editing by HDR can introduce gene sequences for protein tags, delete genes, make point mutations, gene reporters, etc. Innovation in precise genome editing in Drosophila has focused on two areas: (1) testing different forms of donor DNA as the HDR template, and (2) exploring alternatives to the typical Cas9-induced HDR.
Types of Donor DNA for repair templates
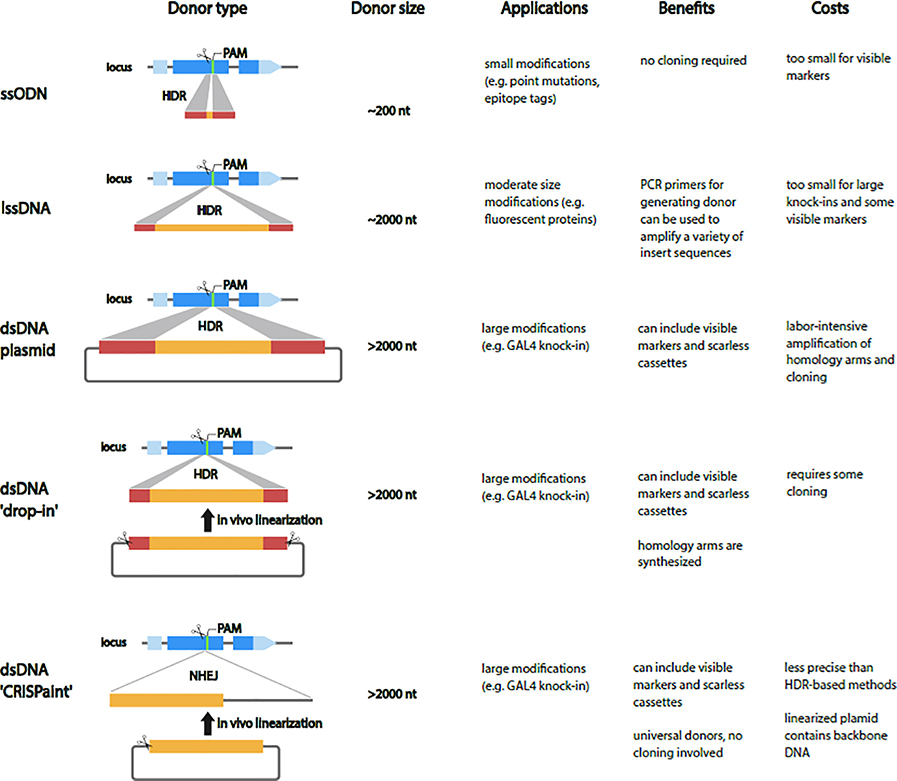
Precise genome editing by HDR requires delivery of Cas9, sgRNA, and donor DNA. Methods for delivery of these reagent into the germline are the same as we have reviewed above for NHEJ (i.e., injection of plasmids or RNPs, or use of transgenic Cas9 and sgRNAs). Typically, donor DNA is co-injected with sgRNA into embryos from a transgenic fly line engineered to express Cas9 in the germline, although this necessitates microinjection into one of a limited set of such lines. Much of the focus of technology innovation has been on testing the efficiency of this approach using different types of donor DNA as the HDR template (Figure 3).
Figure 3. Types of Donor DNA for repair templates.
For small modifications short single-strand oligodeoxynucleotide (ssODN) repair templates are cheap and fast but are limited to a few hundred nucleotides. In the PCR-based long ssDNA (lssDNA) synthesis method, short homology arms are incorporated into PCR primers which amplify from a plasmid template that contains the insert sequence. The PCR product is treated with an exonuclease, which degrades one of the strands leaving behind a lssDNA homology donor. For large modifications, the most-used donors are double-strand DNA (dsDNA) donors supplied as circular plasmids. The main drawback of standard dsDNA donors is that they are labor intensive to generate and require long homology arms (0.5–1.5 kb) for efficient editing. The ‘drop-in’ strategy in which a plasmid containing a full-length dsDNA cassette with short homology arms is linearized in vivo combines the speed and cost effectiveness of ssODNs with the ability to include large repair templates. An alternative to the HDR knock-ins in Drosophila is the ‘CRISPaint’ method, which involves linearization of a universal circular donor plasmid, and integration into the target site by the NHEJ repair pathway.
I. single-stranded DNA (ssDNA) donors.
For small modifications (e.g. point mutations, epitope tags), short single-strand oligodeoxynucleotide (ssODN) repair templates offer a cheap and fast method [9]. Synthesized ssODN donors are limited to a few hundred bases, so the edit must be small with no visible markers included in the repair template. This is the current method-of-choice for changes such as introduction of a variant sequence or other single-nucleotide changes.
II. long ssDNA (lssDNA) donors.
Although it is possible to synthesize lssDNAs to insert longer sequences and lssDNA fragments of up to ~2000 bps can be commercially produced, this approach is costly. Kanca et al. (2019) established a cheaper, PCR-based lssDNA synthesis method in which 100 nt gene-specific homology arms are included in PCR primers which amplify the insert sequence from a plasmid template [40]. The resulting PCR product is treated with an exonuclease, which degrades one of the strands leaving behind a lssDNA homology donor. As the donor is generated directly by PCR, the same primers can potentially be used to amplify a variety of insert sequences. lssDNA donors up to ~2000 bases were shown to be efficient at generating knock-in alleles in Drosophila (REF)
III. Double-stranded DNA (dsDNA) donors.
For large modifications, dsDNA supplied as circular plasmids are the most common donors. The main drawback of dsDNA donors is that they are labor intensive to generate and require long homology arms (0.5–1.5 kb) for efficient editing. A solution to this problem is the recently described ‘drop-in’ strategy in which a plasmid containing a full-length dsDNA cassette with short homology arms is linearized in vivo [40,41]. This approach combines the speed and cost effectiveness of ssODNs with the ability to include large repair templates, including marker genes, and is thus particularly well-suited to large scale production.
Homology-independent dsDNA knock-in
An alternative to the typical Cas9-induced HDR knock-ins is the homology-independent knock-in technology known as ‘CRISPaint’ [42]. This approach involves simultaneous production of DSBs in a target genomic site and in an exogenous circular donor plasmid, leading to integration of the linearized donor into the target site by the NHEJ repair pathway (Figure 3). Recently, CRISPaint was adapted for use in the Drosophila germ line to generate knock-in fly stocks [43]. The main potential advantage of the homology-independent approach is that it is easier, faster, and cheaper, when the goal is to knock in a common insert sequence such as a fluorescent protein ORF or GAL4. Many universal donor plasmids compatible with Drosophila transgenesis are already available from public repositories (Addgene, DGRC). Bosch et al. generated 13 CRISPaint donor plasmids that contain a selectable transgenesis marker (e.g. 3xP3-RFP), many of which contain other useful inserts (e.g. Gal4, LexA, QF2)[43]. A major disadvantage of homology-independent insertion is that the molecular nature of the insertion is less predictable than HDR, so the technique is best suited for when precise insertions are not absolutely required. We note that universal donors generated in Bosch et al. can also be used with other arthropods, as the 3xP3-RFP marker gene is known to drive expression in other arthropod species [44].
Efficiency of the different knock-in approaches
In general, the use of HDR-directed repair to generate precise edits is significantly less efficient than NHEJ-based mutagenesis [45]. There has been no systematic comparison of all or most of the different HDR approaches, so there is no consensus on which approach is most efficient. If we simply look at the percentage of knock-in experiments that give at least one correctly edited fly, we find reported rates of 60–80% for dsDNA plasmids, drop-in dsDNAs, and homology-independent dsDNA knock-ins [40,43,46]. However, within a successful experiment, the percentage of injected embryos that give rise to the correct edit can vary significantly, with reports for dsDNA plasmids at 5–22% [46], 46–88% [47], and 7–42% [48]. Homology-independent insertion performs similarly, with 5–21% of injected embryos producing the correct edit [43]. Finally, for all the approaches, nearly all the modifications were found at the intended target site.
Detection of successful events
The lssDNA and dsDNA approaches allow for inclusion of markers for visible screening, such as 3xP3-DsRed, which enables rapid screening for DsRed expression in the eye. The size limit of ssODNs precludes incorporation of markers in the inserted sequence, such that molecular analysis by PCR and sequencing or phenotypic screening indicative of the engineered change is required.
HDR can sometimes result in integration of donor plasmid backbone sequences due to crossover repair [49]. The O’Connor-Giles group reported a donor plasmid that contains a mini-white transgene in the plasmid backbone that is useful to detect imprecise integration events (flyCRISPR.molbio.wisc.edu). Furthermore, Nyberg et al. developed a similar approach, placing an RNAi hairpin that causes loss of the fly eye (GMR-eyashRNA) on the donor plasmid backbone [12]. Two advantages of the GMR-eyashRNA transgene over mini-white are the smaller size of the reagent that results in a visible phenotype (875 bp for the shRNA vs 2 kb for mini-white) and that the GMR-eyashRNA transgene can be used in a white+ genetic background and in non-D. melanogaster strains.
Scarless editing
Engineering a precise edit without leaving any other modifications, referred to as “scarless” editing, is challenging. Scarless editing can be performed using HDR and donor DNA that only contains the edit and, in most cases, a mutated sgRNA target site that does not otherwise disrupt gene function. If a marker gene is included on the donor insert the process takes two steps, first isolating a knock-in line by screening flies for the transgenesis marker, and then removing the marker to produce the final edit. Some groups developed marker genes that can be removed by Cre/lox recombination [46,50], which leaves behind a loxP site “scar” that is acceptable for some applications. Two truly scarless, two-step methods were subsequently developed, each with its own tradeoffs. The methods reported by Lamb and Li-Kroeger [51,52] involve integration of a marker gene into the target locus, followed by a second HDR targeting event that is used to insert the final desired sequence. This requires production of two sets of donor plasmids and sgRNAs and two rounds of microinjections into embryos. The O’Connor-Giles group i() developed a two-step scarless editing method based on precise excision of the 3xP3-RFP marker contained in a PiggyBac transposon. Marker removal is performed by simply crossing the flies to a strain carrying the PiggyBac transposase. Importantly, intentional inclusion of mutations in the target site are required to prevent cutting of the donor unless the knock-in element itself disrupts the site. As such, for some applications this method is not truly scarless. Researchers should carefully consider the tradeoffs of each two-step method given their experimental goals. We also note that new technologies such as prime editing and base editing have the potential to enable scarless editing without a need for donor plasmids (see below).
Alternative editing tools
There has been intense scrutiny of the possibility that by generating DSBs Cas9 causes unintended changes to the genome. For example, many groups have found that Cas9 + sgRNA in mammalian cells can generate indels at off-target locations [53]. In addition, a variety of undesired on-target effects can occur, such as genome rearrangements or integration of plasmid DNA into a cut site. The genome editing field is exploring alternative editing tools that do not cause DSBs and have new functionalities. Two prominent tools are base editing and prime editing, both of which have been tested in Drosophila.
Base editing
Base editing is a CRISPR/Cas9-based method in which a catalytically dead Cas9 (dCas9) or Cas9 nickase (nCas9) is fused with enzymes that generate specific single base pair changes. A cytosine base editor (C>T, G>A) was recently tested in Drosophila [54] by ubiquitous expression throughout development (Act5c-BE2). When crossed with an sgRNA-expressing transgenic line, the progeny underwent cytosine editing at levels high enough to detect by Sanger sequencing of target regions amplified from genomic DNA. Of 30 sgRNAs tested, 15 resulted in editing in the target region, all edits were the expected C>T change, most edits occurred in the expected editing window of ~11 bases, and the editing efficiency in some cases was extremely high (near 100%). Therefore, cytosine base editing appears to be functional in Drosophila somatic cells. The potential applications of this method in Drosophila are not yet clear, particularly considering that which cytosine is edited is unpredictable, and given the fact that editing was not tested in the germ line. Nevertheless, the demonstration that there are no obvious barriers to cytosine base editing in Drosophila opens the doors to testing of other base editors, which include adenine base editors (A>T, T>C) [55], dual cytosine/adenine base editors [56,57], cytosine to guanine base editors (C>G, G>C) [58], and glycosylase base editors [59].
Prime editing
Prime editing is another CRISPR/Cas9-based method used to engineer precise nucleotide changes without DSBs [60]. Like base editing, this system uses a modified Cas9 enzyme. For prime editing, Cas9 nickase is fused with an engineered reverse transcriptase (RT) domain, together referred to as prime editor 2 (PE2). Prime editing also uses a modified guide RNA, called a prime editing guide RNA (pegRNA), which contains the intended edit and short regions of flanking homology sequence. The pegRNA directs PE2 to the target location, where it causes a single strand nick and anneals a portion of the pegRNA to the exposed genome. The RT domain then transcribes the edit from the pegRNA into the genome.
Prime editing is functional in Drosophila in both somatic cells and the germ line [61]. In somatic cells, the efficiency of making 4 bp changes in the ebony, white, or forked genes varied from 10–40%. In germ cells, a 4 bp change in ebony was successfully made and transmitted to 36% of progeny. As expected, based on mammalian studies, co-expressing a nicking sgRNA with the pegRNA (referred to as the PE3 system) successfully increased the editing efficiency; however, this also resulted in isolation of flies with indels at the target site. Furthermore, transgenic crosses were significantly more efficient than microinjection-based editing. Improvements in prime editing systems, such as the recently described engineered pegRNAs [62] promise to further increase efficiency.
One novel application of prime editing tested in Drosophila is induction of precise edits in somatic cells via crossing transgenic animals with a pegRNA to a line expressing the modified Cas protein [61]. By restricting expression of the PE2 enzyme to specific cell types, editing will likewise be restricted to that cell type. A limitation of the approach is that unlike HDR, prime editing has only been used to edit or insert small regions (up to ~100 bp), although this has not yet been determined experimentally in flies.
Beyond DNA cuts and edits
CRISPR-Cas systems can do more than target the genome to produce indels and edits. Some Cas proteins can target RNA and researchers have explored a variety of applications for Cas proteins and Cas fusion proteins to target and modify both RNA and DNA.
Cutting and modifying RNA
Uniquely among the Class 2 Cas proteins, Cas13 can cut RNA rather than DNA. Like Cas9, Cas13 forms a complex with a crRNA, identifies its target by the protospacer present in the crRNA, and then cleaves its substrate. To date, there are currently four subtypes identified in the Cas13 family, including Cas13a (aka C2c2), Cas13b, Cas13c, and Cas13d (aka CasRx). Two groups recently evaluated Cas13 in Drosophila and identified Cas13d/CasRX as efficiently able to degrade RNA in vivo [63,64]. Hunyh et al further generated a Drosophila-optimized variant, CasFX, that generated RNA knockdown levels comparable to RNAi. Both groups looked at off-target effects, with Buchman et al. but not Hunyh et al. detecting non-specific RNA degradation. Further research will be needed to establish the degree of specificity of Cas13 in Drosophila. Another CRISPR effector, Cas7-11, was recently identified as a programmable RNAse in mammalian cells [65]. Unlike, Cas13, Cas7-11 showed no evidence of off-target activity or toxicity, making it an attractive alternative system.
Cas13 may be useful in a broad range of applications other than RNA cleavage. Hunyh et al. introduced quadruple mutations in the catalytic HEPN domains of CasFx (R239A, H244A, R858A, and H863A) to abolish nuclease activity but not RNA-binding activity. This catalytically inactive dCasFX could be used to detect protein interactions on RNAs. By fusing dCasFX with the RNA-modifying domain of Adenosine Deaminase Acting on RNA 2 (ADAR2) they were able to perform programmable adenosine-to-inosine editing on target transcripts with an overall low off-target rate. In theory, Cas13 can be modified for many approaches to study RNA, including splicing, transcript stabilization, or RNA localization.
Regulating gene expression
Catalytically inactive Cas proteins can be used to carry other proteins and enzymes to a desired DNA or RNA target. CRISPR activation (CRISPRa), in which catalytically dead Cas9 (dCas9) recruits transcriptional activation machinery to a DNA sequence upstream of the transcriptional start site (TSS) of a target gene, is a highly scalable method for gene activation [66]. Importantly, the CRISPRa system has several advantages as compared with overexpression of an open reading frame (ORF) using the GAL4-UAS system, as CRISPRa allows: (1) overexpression of all of the relevant splice isoforms for a given cell type; (2) preservation of the 3′UTR, which can contain regulatory information such as microRNA binding sites; and (3) overexpression of difficult-to-clone ORFs, such as those that are long, contain repeat sequences, or are otherwise intractable to cloning. With CRISPRa, target specificity is conferred by 20-bp protospacer sequences in the sgRNA, such that production of transgenic fly reagents for CRISPRa at genome-wide scale is feasible [15]. Two systems for CRISPRa in vivo have been developed in flies. With one, gene activation is triggered by co-expression of the sgRNA and dCas9 fused to VP64-p65-Rta (VPR) [67,68], and with the other, via co-expression of a modified sgRNA and the Synergistic Activation Mediator (SAM) system [69]. Both systems use GAL4/UAS to control tissue specificity of the dCas9-activators such that the gene of interest is only overexpressed in the GAL4 domain. To date, over 2500 transgenic sgRNA lines compatible with CRISPRa have been produced [15].
Several groups have shown that dCas9 can repress transcription when it is targeted downstream of the TSS by hindering the elongation activity of RNA polymerase II [70,71]. In mammalian cells, this CRISPR interference (CRISPRi) method is more effective when dCas9 is fused to the KRAB repressor domain and sgRNAs can also be targeted slightly upstream of the TSS [72]. However, while dCas9 has been shown in a small number of cases to transcriptionally repress target genes in Drosophila [35,73], whether the system is broadly applicable in vivo remains to be seen.
Imaging and lineage tracing
CRISPR has been adapted in vivo in Drosophila as a tool for cell labeling and lineage tracing. The CaSSA technology, for Cas9 and Single Strand Annealing, [74] uses CRISPR-based single strand annealing repair of non-functional fluorescent proteins to label only cells that express Cas9 and a specific sgRNA. This technique was subsequently extended to create a tool called CLADES, which similarly uses SSA repair of a Cas9-induced DSBs to reconstitute an active sgRNA [75]. The method can be used to create a sequential cascade of reporters that is inherited by the progeny of the target cell, recording serial biological events. Recently, cell lineage reconstruction based on CRISPR/Cas9 editing of genomic target sequences was tested in Drosophila [76], however the authors conclude that much optimization is still required to achieve accurate lineage tracing. The CRISPR/Cas9 system has also been applied to generate mosaic tissues in Drosophila. The recently described Mosaic analysis by gRNA-induced crossing-over (MAGIC) technique induces DSBs and crossover between homologous pairs of chromatids, resulting in genetically distinct marked clones in both the Drosophila soma and germline [77].
CRISPR technologies in Drosophila cell lines
Drosophila cells are widely used in functional genomic, transcriptomic, and proteomic analyses, and provide an important complement to in vivo studies. Significant progress has been made towards adapting CRISPR technologies to generate knock out or knock in cell lines, regulate gene expression, and perform genome-scale forward genetic screens. Selected examples of the use of CRISPR in Drosophila cells include constructing knockout cell lines for subsequent use in RNAi-based synthetic lethality screens [78,79]; constructing a reporter cell line as the basis of microscopy-based RNAi screens [80]; visualizing sub-cellular organelles and compartments with fluorescent protein tags [40]; engineering the insect protein glycosylation pathway [81]; and use of a genome-wide knockout screen to identify a novel ecdysone transporter [82].
NHEJ-mediated knockout cell lines
Perhaps the most straightforward application of CRISPR in Drosophila cells is the generation of knockout cell lines by co-transfection of Cas9 with an sgRNA or by introducing an sgRNA into a cell line that expresses Cas9. Notably, most Drosophila cell lines are polyploid, such that following NHEJ-mediated CRISPR editing, the edited cells will be a mixed population that can include null mutations, edited but functionally wild-type cells, and unedited cells [83]. As a result, following introduction of the CRISPR reagents, cells are typically single-cell cloned and edits verified by PCR, qPCR, or next-generation sequencing (NGS) of targeted regions [78,79,81,84,85].
HDR-mediated knockout cell lines
Knockout cells can also be generated using an HDR-based approach, for example using a knock-in construct that deletes or otherwise disrupts the gene. Because NHEJ predominates even in the presence of a donor template, the resulting cells often contain multiple types of edits, including one or more allele generated via HDR-mediated knock-in and other(s) generated via NHEJ-induced editing. This can result in functionally wild-type cells, since NHEJ-induced mutations may result in one or more allele with an in-frame indel. In such experiements, the rate of HDR can be boosted by simultaneous knockdown of DNA ligase 4 (lig4) the DNA polymerase θ ortholog, mus308, both of which are important for end-joining based repair [86]. An alternative HDR-based method was described in which a donor cassette with both a selectable marker and multiple sgRNAs is introduced into Cas9-expressing cells to facilitate enrichment for edited cells. With this method, the editing rate is higher, making single cell cloning unnecessary in some cases [87].
Knock-in cell lines
Similar to in vivo knock-in, several methods are available for insertion of tags or other sequences into the genome of Drosophila cultured cells. N- or C-tags have been added via HDR using dsDNA donors [85,86] or using lssDNA donors that provide a fluorescent protein open reading frame as an artificial exon [40,88]. Homology-independent dsDNA knock-in can generate C-terminal but not N-terminal tags [43,88]. Extending beyond knock-in of protein tags, Kunzelmann et al. reported insertion of a metallothionine promoter upstream of an endogenous gene to make it copper-inducible [86], while Mariyappa et al. inserted attP docking sites into embryonic and larval CNS derived cell lines for site-specific recombination [89].
Regulating gene expression in cell lines
A few groups have explored the use of CRISPRa, CRISPR interference (CRISPRi), or Cas13 in Drosophila cell lines. CRISPRa initially used the dCas9-VPR system [90]. More recently, Sajwan and Mannervik (2019) compared dCas9-VPR, dCas9-SAM, and dCas9-CBD, in which dCas9 is fused to the histone acetyl transferase, CBD, and found that SAM and CBD resulted in higher levels of activation, whereas there was significant variation from locus to locus [91]. Gene knockdown using CRISPRi [73] or Cas13 have also been demonstrated, although a precise comparison to RNAi has yet to be reported. Cas13 was shown to be broadly effective for RNA knock down in Drosophila cells [63,92]. Interestingly, Huynh et al. (2020) report knockdown of mitochondrial mRNAs using Cas13, to our knowledge the first demonstration of any CRISPR technology to alter mitochondrial genes [63].
Genome-wide CRISPR screening
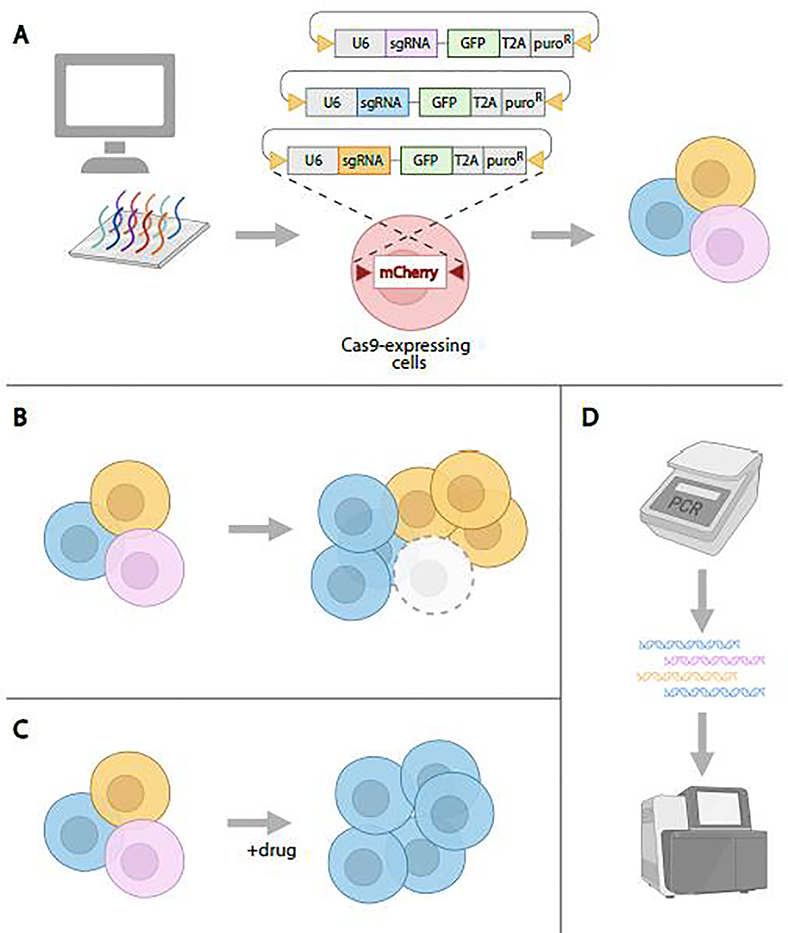
CRISPR knockout screens can be conducted in a pooled format in which a large sgRNA library (>10,000 sgRNAs) is delivered into a population of cells so that each cell gets ~1 sgRNA [80,93]. The resulting pool of KO cells is then outgrown and subjected to a selection that separates cells with a phenotype of interest from the rest of the cell population. The distribution of sgRNAs in starting and outgrown cell populations, or selected and non-selected populations, is subsequently revealed by PCR amplification of the sgRNAs inserted into the cells, followed by next-generation sequencing to determine the identity and relative proportions of sgRNAs in control and phenotypically selected cell populations. Key to performing CRIPSR screens in Drosophila cells was the development of an approach that results in integration of sgRNAs into the genome (Figure 4). Viswanatha et al. accomplished this using site-specific recombination and showed that pooled CRISPR knockout screening in fly cells can be used to identify cell essential genes more reliably than genome-wide RNAi and can be used to identify genes that when knocked out, confer resistance to a drug or other treatment that perturbs cell growth or viability [82,94]. Although thus far only demonstrated for knockout, the approach is in theory extensible to screening using CRISPRa, CRISPRi, or other CRISPR-based methods. One thing that is particularly exciting about CRISPR pooled screening in Drosophila cells is that the method is more accessible and of a lower cost as compared with arrayed-format RNAi screens.
Figure 4. CRISPR pooled-format screening in Drosophila cultured cells.
A. To generate a pool of cells with knockout mutations, sgRNAs are designed and synthesized, then cloned into recombination mediated cassette-exchange (RMCE)-compatible expression vectors. Next, the sgRNA library plasmids are introduced into Cas9-expressing cells via RMCE, resulting in a population of cells in which sgRNAs are integrated into the genome, expressed, and generate knockouts via NHEJ. B. Outgrowth, followed by identification of sgRNAs that ‘drop out’ in the outgrown population as compared with the starting population, can be used to identify essential genes. C. Treatment of the cell population with a drug or other cytotoxin can be used to select for cells in which knockout confers resistance. Genes are identified by comparing sgRNAs in the treated population to sgRNAs in an untreated control. D. Following the pooled cell assay, genomic DNA is extracted and PCR is used to amplify sgRNA sequences. Next-generation sequencing and data analysis is then used to uncover the identity and proportion of sgRNAs in control and experimental knockout cell populations, followed by gene-level analyses.
Concluding Remarks
Due to the ease of generating transgenic animals and its rapid life cycle, Drosophila has been an important testing ground for new CRISPR technologies. Drosophila is well suited going forward as an in vivo system in which to assess the multitude of Cas variants and fusion proteins that continue to be identified and engineered. Drosophila cultured cells have been an indispensable first step in the process of assessing new CRISPR technologies. Moreover, the new availability of CRISPR pooled screening in fly cells has expanded the ease, precision, and types of genome-wide screens that are now feasible to do. Altogether, CRISPR technologies have further expanded the Drosophila genetic toolkit, improving the efficiency and precision with which we can engineer the genome and manipulate genes or RNA (Table 1). Among the resources that make this possible are bioinformatic tools for Drosophila sgRNA design [46,78,95] new in vivo CRISPR resources for targeted knock-out or knockdown of gene expression [13–15] and resources for manipulating the activity and expression of genes with tight spatial and temporal control, including expression of wild-type or variant alleles [96]. We anticipate further expansion of these and other resources for precise and/or large-scale Drosophila genome editing. In particular, the development of more efficient in vivo techniques for HDR, as well as tools for base editing and prime editing will provide a powerful supplement to currently available resources for modeling conserved human disease variants.
Table 1:
Drosophila focused CRISPR resources
| Resource | Type | Comments | Refs | Webpage |
|---|---|---|---|---|
| DRSC Find CRISPR Tool | guide RNA design | Fly sgRNA designs with genome view | [78] | https://www.flyrnai.org/crispr3/web/ |
| SNP-CRISPR | guide RNA design | Accepts variant annotations as the input | [95] | https://www.flyrnai.org/tools/snp_crispr/web/ |
| CRISPR Optimal Target Finder | guide RNA design | Includes multiple Drosophila and insect species | [46] | http://targetfinder.flycrispr.neuro.brown.edu |
| TRiP-CRISPR | transgenic CRISPR fly stocks and plasmids | Search for and nominate CRISPR-a and CRISPR-KO fly stocks | [15] | https://www.flyrnai.org/tools/grna_tracker/web/ |
| CRISPR Fly Design | transgenic CRISPR fly stocks and plasmids | CRISPR stocks and protocols for genome engineering | https://www.crisprflydesign.org/ | |
| FlyCRISPR | transgenic CRISPR fly stocks and plasmids | CRISPR stocks and protocols for genome engineering | https://flycrispr.org | |
| FlyCas9 | transgenic CRISPR fly stocks and plasmids | CRISPR stocks and protocols for genome engineering | https://shigen.nig.ac.jp/fly/nigfly/cas9/ | |
| DRSC/TRiP | cell based and transgenic CRISPR reagents | Includes online tools, protocols, and large scale resources | https://fgr.hms.harvard.edu/fly-cellcrispr-cas | |
| DGRC | cells lines and plasmids | Collects and distributes CRISPR plasmids and cell lines | https://dgrc.bio.indiana.edu/Home | |
| BDSC | transgenic CRISPR fly stocks | Collects and distributes transgenic CRISPR fly stocks | https://bdsc.indiana.edu | |
| VDRC | transgenic CRISPR fly stocks | Collects and distributes transgenic CRISPR fly stocks | https://stockcenter.vdrc.at/control/main |
Outstanding questions.
SpCas9 remains by far the most widely used Cas protein in the Drosophila model system. Other Cas proteins, particularly Cas12a, offer new capabilities while maintaining cutting and efficiency equal to or above that of SpCas9. Will Cas12a prove effective and reliable enough to become a widespread alternative to Cas9 for Drosophila researchers?
Cytosine base editing is functional in Drosophila somatic cells, but which cytosine is edited is unpredictable. Is this system functional in the germline, and will other base editor systems be accurate enough for precision genome engineering in flies?
Similarly, prime editing is functional in both somatic and germline cells in Drosophila but is only moderately efficient and can cause indels. Will new innovations in Prime editors and pegRNAs increase the accuracy and efficiency enough to make this method an attractive alternative to traditional HDR?
The RNA-targeting CRISPR-Cas13 has been reported to have off-target effects and cell toxicity. To what degree can new RNA-targeting CRISPR systems like Cas7-11 overcome this problem?
CRISPR pooled screens in Drosophila cells have thus far only been demonstrated for knockout. Can the approach be extened to screening using CRISPRa, CRISPRi, or other CRISPR-based methods?
Highlights.
The fruit fly Drosophila melanogaster has long served as both an important biological and biomedical research model and an in vivo platform for testing of molecular genetic technologies.
In recent years, CRISPR-Cas9 approaches have been added to the Drosophila tool box and there are now robust methods for in vivo engineering of Drosophila via CRISPR knockout and knock-in approaches, as well as for CRISPR knockout, knock-in, and genome-wide pooled screening in Drosophila cells.
The adoption, development, and optimization of new CRISPR technologies are further improving the efficiency of CRIPSR engineering in Drosophila and expanding the repertoire of molecular genetic perturbations that can be done in this exemplary genetic model system.
Acknowledgements.
This work was supported by NIH NIGMS P41 GM132087 and Damon Runyon (J.B). NP is an investigator of HHMI.
Glossary Box
- Cas
Abbreviation for “CRISPR associated”, describes the proteins that act in concert with CRISPR arrays to target foreign nucleic acid for destruction
- Cas9
The first Cas protein adapted for use in genome editing. The CRISPR/Cas9 complex requires both crRNA and tracrRNA to form an active complex, in which Cas9 cuts both strands of the target DNA
- CRISPR
Acronym for “Clustered Regularly Interspaced Short Palindromic Repeats.” Part of the prokaryotic immune system, these repeating DNA sequences alternate with “spacer” sequences from past invading viruses. They are used to destroy viral DNA during subsequent infections
- crRNA
Abbreviation for “CRISPR RNA.” A small RNA molecule encoded by the CRISPR locus. It directs the Cas protein to cut a specific DNA sequence. In some CRISPR/Cas systems (e.g. Cas9), the crRNA complexes with the tracrRNA
- HDR
Acronym for “Homology-Directed Repair.” A type of repair of a break in DNA which relies on a DNA donor “template” with homology to the region surrounding the break
- NHEJ
Acronym for “Non-Homologous End Joining.” A type of repair of a break in DNA by connecting free DNA ends. This pathway is more error-prone than HDR, often causing small insertions and/or deletions near the DNA break
- PAM
Acronym for “Protospacer Adjacent Motif.” A short sequence downstream of the target sequence that is essential for binding of the Cas protein to the DNA and cleavage of the target
- sgRNA
Abbreviation for “single guide RNA”. A combination of the crRNA and tracrRNA into a single molecule for use in CRISPR-Cas genome editing
- tracrRNA
Abbreviation for “trans-activating CRISPR RNA.” A small looping RNA sequence which, in the CRISPR/Cas9 system pairs with the crRNA to form a functional guide RNA
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Resources:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jinek M et al. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibikova M et al. (2002) Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161, 1169–1175. 10.1093/genetics/161.3.1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J et al. (2012) Efficient and specific modifications of the Drosophila genome by means of an easy TALEN strategy. J Genet Genomics 39, 209–215. 10.1016/j.jgg.2012.04.003 [DOI] [PubMed] [Google Scholar]
- 4.Charpentier E and Doudna JA (2013) Biotechnology: Rewriting a genome. Nature 495, 50–51. 10.1038/495050a [DOI] [PubMed] [Google Scholar]
- 5.Paques F and Haber JE (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63, 349–404. 10.1128/MMBR.63.2.349-404.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sung P and Klein H (2006) Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol 7, 739–750. 10.1038/nrm2008 [DOI] [PubMed] [Google Scholar]
- 7.Pannunzio NR et al. (2018) Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J Biol Chem 293, 10512–10523. 10.1074/jbc.TM117.000374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassett A and Liu JL (2014) CRISPR/Cas9 mediated genome engineering in Drosophila. Methods 69, 128–136. 10.1016/j.ymeth.2014.02.019 [DOI] [PubMed] [Google Scholar]
- 9.Gratz SJ et al. (2013) Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194, 1029–1035. 10.1534/genetics.113.152710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kondo S and Ueda R (2013) Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195, 715–721. 10.1534/genetics.113.156737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren X et al. (2013) Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci U S A 110, 19012–19017. 10.1073/pnas.1318481110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nyberg KG et al. (2020) A pipeline for precise and efficient genome editing by sgRNA-Cas9 RNPs in Drosophila. Fly (Austin) 14, 34–48. 10.1080/19336934.2020.1832416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Port F et al. (2020) A large-scale resource for tissue-specific CRISPR mutagenesis in Drosophila. Elife 9. 10.7554/eLife.53865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meltzer H et al. (2019) Tissue-specific (ts)CRISPR as an efficient strategy for in vivo screening in Drosophila. Nat Commun 10, 2113. 10.1038/s41467-019-10140-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zirin J et al. (2020) Large-Scale Transgenic Drosophila Resource Collections for Loss- and Gain-of-Function Studies. Genetics 214, 755–767. 10.1534/genetics.119.302964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ge DT et al. (2016) Rapid Screening for CRISPR-Directed Editing of the Drosophila Genome Using white Coconversion. G3 (Bethesda) 6, 3197–3206. 10.1534/g3.116.032557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kane NS et al. (2017) Efficient Screening of CRISPR/Cas9-Induced Events in Drosophila Using a Co-CRISPR Strategy. G3 (Bethesda) 7, 87–93. 10.1534/g3.116.036723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ewen-Campen B and Perrimon N (2018) ovo(D) Co-selection: A Method for Enriching CRISPR/Cas9-Edited Alleles in Drosophila. G3 (Bethesda) 8, 2749–2756. 10.1534/g3.118.200498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakrabarti AM et al. (2019) Target-Specific Precision of CRISPR-Mediated Genome Editing. Mol Cell 73, 699–713 e696. 10.1016/j.molcel.2018.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Port F et al. (2014) Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci U S A 111, E2967–2976. 10.1073/pnas.1405500111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xue Z et al. (2014) CRISPR/Cas9 mediates efficient conditional mutagenesis in Drosophila. G3 (Bethesda) 4, 2167–2173. 10.1534/g3.114.014159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao Y and Zhao Y (2014) Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J Integr Plant Biol 56, 343–349. 10.1111/jipb.12152 [DOI] [PubMed] [Google Scholar]
- 23.Port F and Bullock SL (2016) Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat Methods 13, 852–854. 10.1038/nmeth.3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie K et al. (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci U S A 112, 3570–3575. 10.1073/pnas.1420294112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koreman GT et al. (2021) Upgraded CRISPR/Cas9 tools for tissue-specific mutagenesis in Drosophila. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2014255118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haapaniemi E et al. (2018) CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med 24, 927–930. 10.1038/s41591-018-0049-z [DOI] [PubMed] [Google Scholar]
- 27.Ihry RJ et al. (2018) p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med 24, 939–946. 10.1038/s41591-018-0050-6 [DOI] [PubMed] [Google Scholar]
- 28.Alkan F et al. (2018) CRISPR-Cas9 off-targeting assessment with nucleic acid duplex energy parameters. Genome Biol 19, 177. 10.1186/s13059-018-1534-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gasiunas G et al. (2012) Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A 109, E2579–2586. 10.1073/pnas.1208507109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mali P et al. (2013) RNA-guided human genome engineering via Cas9. Science 339, 823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren X et al. (2014) Performance of the Cas9 nickase system in Drosophila melanogaster. G3 (Bethesda) 4, 1955–1962. 10.1534/g3.114.013821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wah DA et al. (1997) Structure of the multimodular endonuclease FokI bound to DNA. Nature 388, 97–100. 10.1038/40446 [DOI] [PubMed] [Google Scholar]
- 33.Guilinger JP et al. (2014) Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol 32, 577–582. 10.1038/nbt.2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai SQ et al. (2014) Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol 32, 569–576. 10.1038/nbt.2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huynh N et al. (2018) A Drosophila CRISPR/Cas9 Toolkit for Conditionally Manipulating Gene Expression in the Prothoracic Gland as a Test Case for Polytene Tissues. G3 (Bethesda) 8, 3593–3605. 10.1534/g3.118.200539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu JH et al. (2018) Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63. 10.1038/nature26155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ni XY et al. (2020) Targeted gene disruption by CRISPR/xCas9 system in Drosophila melanogaster. Arch Insect Biochem Physiol 104, e21662. 10.1002/arch.21662 [DOI] [PubMed] [Google Scholar]
- 38.Zetsche B et al. (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771. 10.1016/j.cell.2015.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Port F et al. (2020) Multiplexed conditional genome editing with Cas12a in Drosophila. Proc Natl Acad Sci U S A 117, 22890–22899. 10.1073/pnas.2004655117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanca O et al. (2019) An efficient CRISPR-based strategy to insert small and large fragments of DNA using short homology arms. Elife 8. 10.7554/eLife.51539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cristea S et al. (2013) In vivo cleavage of transgene donors promotes nuclease-mediated targeted integration. Biotechnol Bioeng 110, 871–880. 10.1002/bit.24733 [DOI] [PubMed] [Google Scholar]
- 42.Schmid-Burgk JL et al. (2016) CRISPaint allows modular base-specific gene tagging using a ligase-4-dependent mechanism. Nat Commun 7, 12338. 10.1038/ncomms12338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bosch JA et al. (2020) Gene Knock-Ins in Drosophila Using Homology-Independent Insertion of Universal Donor Plasmids. Genetics 214, 75–89. 10.1534/genetics.119.302819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berghammer AJ et al. (1999) A universal marker for transgenic insects. Nature 402, 370–371. 10.1038/46463 [DOI] [PubMed] [Google Scholar]
- 45.Frit P et al. (2014) Alternative end-joining pathway(s): bricolage at DNA breaks. DNA Repair (Amst) 17, 81–97. 10.1016/j.dnarep.2014.02.007 [DOI] [PubMed] [Google Scholar]
- 46.Gratz SJ et al. (2014) Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971. 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Port F et al. (2015) Systematic Evaluation of Drosophila CRISPR Tools Reveals Safe and Robust Alternatives to Autonomous Gene Drives in Basic Research. G3 (Bethesda) 5, 1493–1502. 10.1534/g3.115.019083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gratz SJ et al. (2015) CRISPR-Cas9 Genome Editing in Drosophila. Curr Protoc Mol Biol 111, 31 32 31–31 32 20. 10.1002/0471142727.mb3102s111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bier E et al. (2018) Advances in Engineering the Fly Genome with the CRISPR-Cas System. Genetics 208, 1–18. 10.1534/genetics.117.1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen HM et al. (2020) Enhanced Golic+: highly effective CRISPR gene targeting and transgene HACKing in Drosophila. Development 147. 10.1242/dev.181974 [DOI] [PubMed] [Google Scholar]
- 51.Lamb AM et al. (2017) Tools and strategies for scarless allele replacement in Drosophila using CRISPR/Cas9. Fly (Austin) 11, 53–64. 10.1080/19336934.2016.1220463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li-Kroeger D et al. (2018) An expanded toolkit for gene tagging based on MiMIC and scarless CRISPR tagging in Drosophila. Elife 7. 10.7554/eLife.38709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fu Y et al. (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31, 822–826. 10.1038/nbt.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marr E and Potter CJ (2021) Base editing using CRISPR/Cas9 in Drosophila. bioRxiv, 2021.2003.2024.436868. 10.1101/2021.03.24.436868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gaudelli NM et al. (2017) Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471. 10.1038/nature24644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grunewald J et al. (2020) A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing. Nat Biotechnol 38, 861–864. 10.1038/s41587-020-0535-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakata RC et al. (2020) Base editors for simultaneous introduction of C-to-T and A-to-G mutations. Nat Biotechnol 38, 865–869. 10.1038/s41587-020-0509-0 [DOI] [PubMed] [Google Scholar]
- 58.Kurt IC et al. (2021) CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat Biotechnol 39, 41–46. 10.1038/s41587-020-0609-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao D et al. (2021) Glycosylase base editors enable C-to-A and C-to-G base changes. Nat Biotechnol 39, 35–40. 10.1038/s41587-020-0592-2 [DOI] [PubMed] [Google Scholar]
- 60.Anzalone AV et al. (2019) Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bosch JA et al. (2021) Precise genome engineering in Drosophila using prime editing. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2021996118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nelson JW et al. (2021) Engineered pegRNAs improve prime editing efficiency. Nat Biotechnol. 10.1038/s41587-021-01039-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huynh N et al. (2020) A versatile toolkit for CRISPR-Cas13-based RNA manipulation in Drosophila. Genome Biol 21, 279. 10.1186/s13059-020-02193-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buchman AB et al. (2020) Programmable RNA Targeting Using CasRx in Flies. CRISPR J 3, 164–176. 10.1089/crispr.2020.0018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ozcan A et al. (2021) Programmable RNA targeting with the single-protein CRISPR effector Cas7-11. Nature 597, 720–725. 10.1038/s41586-021-03886-5 [DOI] [PubMed] [Google Scholar]
- 66.Konermann S et al. (2015) Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588. 10.1038/nature14136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chavez A et al. (2016) Comparison of Cas9 activators in multiple species. Nat Methods 13, 563–567. 10.1038/nmeth.3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ewen-Campen B et al. (2017) Optimized strategy for in vivo Cas9-activation in Drosophila. Proc Natl Acad Sci U S A 114, 9409–9414. 10.1073/pnas.1707635114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jia Y et al. (2018) Next-generation CRISPR/Cas9 transcriptional activation in Drosophila using flySAM. Proc Natl Acad Sci U S A 115, 4719–4724. 10.1073/pnas.1800677115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gilbert LA et al. (2013) CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451. 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qi LS et al. (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gilbert LA et al. (2014) Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159, 647–661. 10.1016/j.cell.2014.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ghosh S et al. (2016) Effective knockdown of Drosophila long non-coding RNAs by CRISPR interference. Nucleic Acids Res 44, e84. 10.1093/nar/gkw063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Garcia-Marques J et al. (2019) Unlimited Genetic Switches for Cell-Type-Specific Manipulation. Neuron 104, 227–238 e227. 10.1016/j.neuron.2019.07.005 [DOI] [PubMed] [Google Scholar]
- 75.Garcia-Marques J et al. (2020) A programmable sequence of reporters for lineage analysis. Nat Neurosci 23, 1618–1628. 10.1038/s41593-020-0676-9 [DOI] [PubMed] [Google Scholar]
- 76.Salvador-Martinez I et al. (2019) Is it possible to reconstruct an accurate cell lineage using CRISPR recorders? Elife 8. 10.7554/eLife.40292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Allen SE et al. (2021) Versatile CRISPR/Cas9-mediated mosaic analysis by gRNA-induced crossing-over for unmodified genomes. PLoS Biol 19, e3001061. 10.1371/journal.pbio.3001061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Housden BE et al. (2015) Identification of potential drug targets for tuberous sclerosis complex by synthetic screens combining CRISPR-based knockouts with RNAi. Sci Signal 8, rs9. 10.1126/scisignal.aab3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Housden BE et al. (2017) Synthetic Lethality Screens Using RNAi in Combination with CRISPR-based Knockout in Drosophila Cells. Bio Protoc 7. 10.21769/BioProtoc.2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang T et al. (2014) Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84. 10.1126/science.1246981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mabashi-Asazuma H et al. (2015) Modifying an Insect Cell N-Glycan Processing Pathway Using CRISPR-Cas Technology. ACS Chem Biol 10, 2199–2208. 10.1021/acschembio.5b00340 [DOI] [PubMed] [Google Scholar]
- 82.Okamoto N et al. (2018) A Membrane Transporter Is Required for Steroid Hormone Uptake in Drosophila. Dev Cell 47, 294–305 e297. 10.1016/j.devcel.2018.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bassett AR et al. (2014) Mutagenesis and homologous recombination in Drosophila cell lines using CRISPR/Cas9. Biol Open 3, 42–49. 10.1242/bio.20137120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Franz A et al. (2017) Generation of genome-modified Drosophila cell lines using SwAP. Fly (Austin) 11, 303–311. 10.1080/19336934.2017.1372068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ishizu H et al. (2017) Use of the CRISPR-Cas9 system for genome editing in cultured Drosophila ovarian somatic cells. Methods 126, 186–192. 10.1016/j.ymeth.2017.05.021 [DOI] [PubMed] [Google Scholar]
- 86.Kunzelmann S et al. (2016) A Comprehensive Toolbox for Genome Editing in Cultured Drosophila melanogaster Cells. G3 (Bethesda) 6, 1777–1785. 10.1534/g3.116.028241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xia B et al. (2020) CRISPR-based engineering of gene knockout cells by homology-directed insertion in polyploid Drosophila S2R+ cells. Nat Protoc 15, 3478–3498. 10.1038/s41596-020-0383-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bosch JA et al. (2020) Use of the CRISPR-Cas9 System in Drosophila Cultured Cells to Introduce Fluorescent Tags into Endogenous Genes. Curr Protoc Mol Biol 130, e112. 10.1002/cpmb.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mariyappa D et al. (2021) Generation of Drosophila attP containing cell lines using CRISPR-Cas9. G3 (Bethesda). 10.1093/g3journal/jkab161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chavez A et al. (2015) Highly efficient Cas9-mediated transcriptional programming. Nat Methods 12, 326–328. 10.1038/nmeth.3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sajwan S and Mannervik M (2019) Gene activation by dCas9-CBP and the SAM system differ in target preference. Sci Rep 9, 18104. 10.1038/s41598-019-54179-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Viswanatha R et al. (2020) CRISPR-Cas13 mediated Knock Down in Drosophila cultured cells. bioRxiv, 2020.2011.2001.364166. 10.1101/2020.11.01.364166 [DOI] [Google Scholar]
- 93.Shalem O et al. (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Viswanatha R et al. (2018) Pooled genome-wide CRISPR screening for basal and context-specific fitness gene essentiality in Drosophila cells. Elife 7. 10.7554/eLife.36333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen CL et al. (2020) SNP-CRISPR: A Web Tool for SNP-Specific Genome Editing. G3 (Bethesda) 10, 489–494. 10.1534/g3.119.400904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee PT et al. (2018) A gene-specific T2A-GAL4 library for Drosophila. Elife 7. 10.7554/eLife.35574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brand AH and Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 [DOI] [PubMed] [Google Scholar]
- 98.Lee T and Luo L (1999) Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451–461. 10.1016/s0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]