Abstract
Machine learning is able to leverage large amounts of data to infer complex patterns that are otherwise beyond the capabilities of rule-based systems and human experts. Its application to laboratory medicine is particularly exciting, as laboratory testing provides much of the foundation for clinical decision making. In this article, we provide a brief introduction to machine learning for the medical professional in addition to a comprehensive literature review outlining the current state of machine learning as it has been applied to routine laboratory medicine. Although still in its early stages, machine learning has been used to automate laboratory tasks, optimize utilization, and provide personalized reference ranges and test interpretation. The published literature leads us to believe that machine learning will be an area of increasing importance for the laboratory practitioner. We envision the laboratory of the future will utilize these methods to make significant improvements in efficiency and diagnostic precision.
Keywords: Artificial Intelligence, Clinical Pathology, Biochemistry, Precision Medicine, Clinical Decision Support
1. Introduction
The application of machine learning in medicine has garnered enormous attention over the past decade [1–3]. Novel computational methods provide a way to learn from past examples in order to infer complex patterns beyond the capabilities of rule-based algorithms. Along with this attention comes expectations and promises that advances in computation will transform the way that medicine is practiced. In fact, there are already several examples of machine learning methods that have been approved for use by the US Food and Drug Administration (FDA), most recognizably in the field of radiology, cardiology, and pathology [4,5].
The use of machine learning in laboratory medicine has also gained traction and is an increasingly important area of which practitioners should stay abreast [6–8]. The numerical and structured format of data in laboratory medicine lends itself well to computational methods such as machine learning. Such advances harbor promise for the future of medicine, where laboratory testing provides much of the basis for clinical decision making.
In this review we provide a practical introduction to machine learning for the laboratory medicine specialist and a survey of ongoing work using machine learning in routine laboratory testing and laboratory information systems. While there has been extensive work in the use of machine learning in the greater field of clinical pathology, this review will focus on its application in routine laboratory testing including clinical chemistries and common laboratory tests such as blood counts and urinalysis [9,10]. Similarly excluded are machine learning algorithms that rely on laboratory data to make clinical predictions [11–14]. Although this is another growing interest in the medical application of machine learning, we believe such algorithms pertain more to the clinical specialty related to the model’s use case rather than the practice of laboratory medicine. The use of machine learning in these related fields is briefly covered in section 4.3 of the text, to serve as a reference for readers who may be interested in further exploring these areas.
2. A Brief Primer on Machine Learning
2.1. Overview
In contrast to traditional programs that are defined by precoded rules, machine learning refers to computer algorithms that learn from prior examples. The objective for most supervised machine learning models is to take input data and output a predicted result. The algorithm that performs this prediction is trained on large datasets of prior observations. These observations (often referred to as samples) usually consist of features (or predictors), which are the input variables, and a label, which is the dependent variable (the outcome of interest that we wish to predict in the future).
In order to train a model, large amounts of structured data are required. Processing this data involves cleaning and organizing data tables, imputing missing values, and reshaping or combining observations so that they can be summarized and fed into a model. Furthermore, these prior observations must be labeled so that the computer can learn from them (Figure 1). The majority of the work of developing a machine learning model is typically spent in the data preparation phase. Important decisions must be made about which data to include as model inputs and how they should be processed. Furthermore, the creation of accurate labels also requires careful consideration and time. In many cases, labels are created through manual enumeration by an expert; for example, a physician who reviews prior cases and assigns diagnoses.
Figure 1.
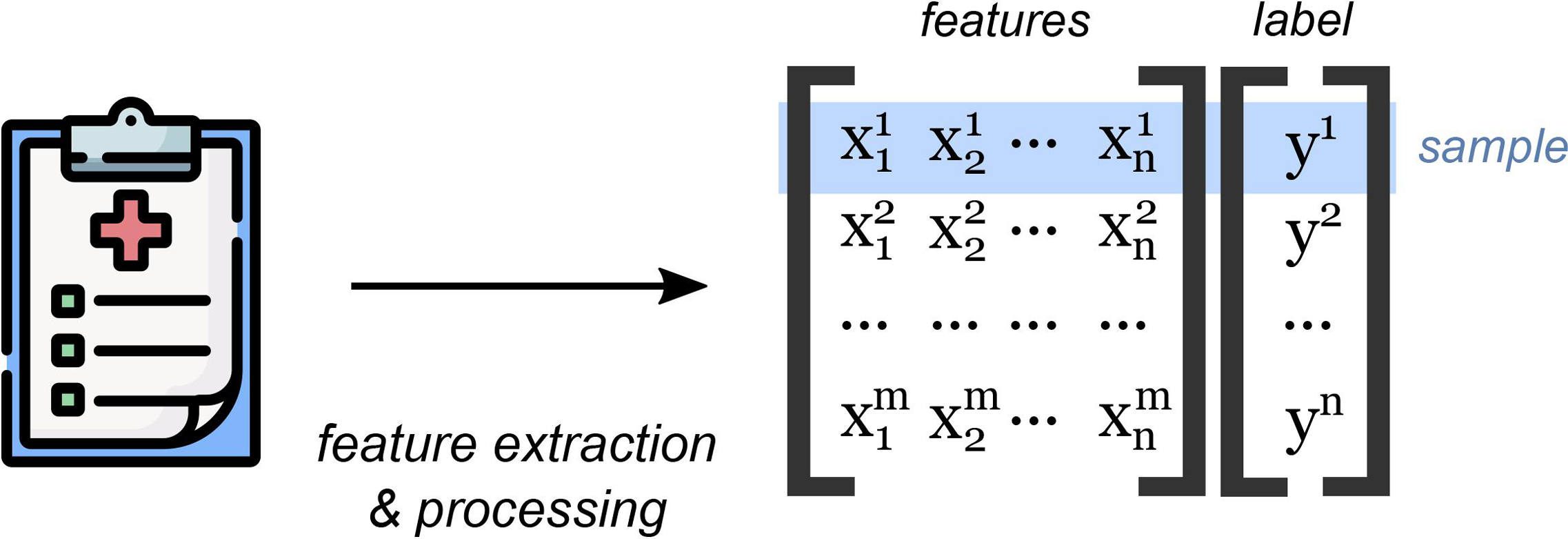
Machine learning models are trained using prior observations (samples). Features from prior observations are extracted and processed into a data matrix. In supervised machine learning, each observation is labeled with an outcome.
To put this all in context, consider a prior study that attempted to predict serum ferritin levels based on other iron panel components [15]. In this example, the ferritin value was the label. The other laboratory components were predictors. In cases where necessary laboratory results were missing, the value was imputed using a statistical formula.
The retrospective data that is used to develop the machine learning model is broken up into a training set of data and a testing set of data. The algorithm learns from the training set, and then its performance is evaluated based on how well it runs on the testing data. This is similar to how a student might study for a test based on published practice questions, but a set of new questions is reserved for the actual evaluation—critical in preventing the computer from simply memorizing the “practice questions” in the training data observations.
2.2. Subcategories within Machine Learning
Machine learning can largely be broken down into two subcategories: supervised machine and unsupervised machine learning. Supervised machine learning, which is the most common application of machine learning in medicine and what is described above, is when a computer infers patterns from prior labeled data—data where the target label is known. These labels provide feedback to the computer program as to what the correct answer is so that the model can improve its predictions.
There are a broad suite of supervised machine learning models suitable for a variety of tasks in medicine, including linear and logistic regression, support vector machines, and tree-based models such as random forest and XGBoost. Tree-based algorithms were commonly encountered in this literature review and in general have achieved good performance in medical applications [16]. Such models use a decision tree that consists of a complex series of decision points. The decision points are inferred during the model development based on training data used to develop the model (Figure 2).
Figure 2.
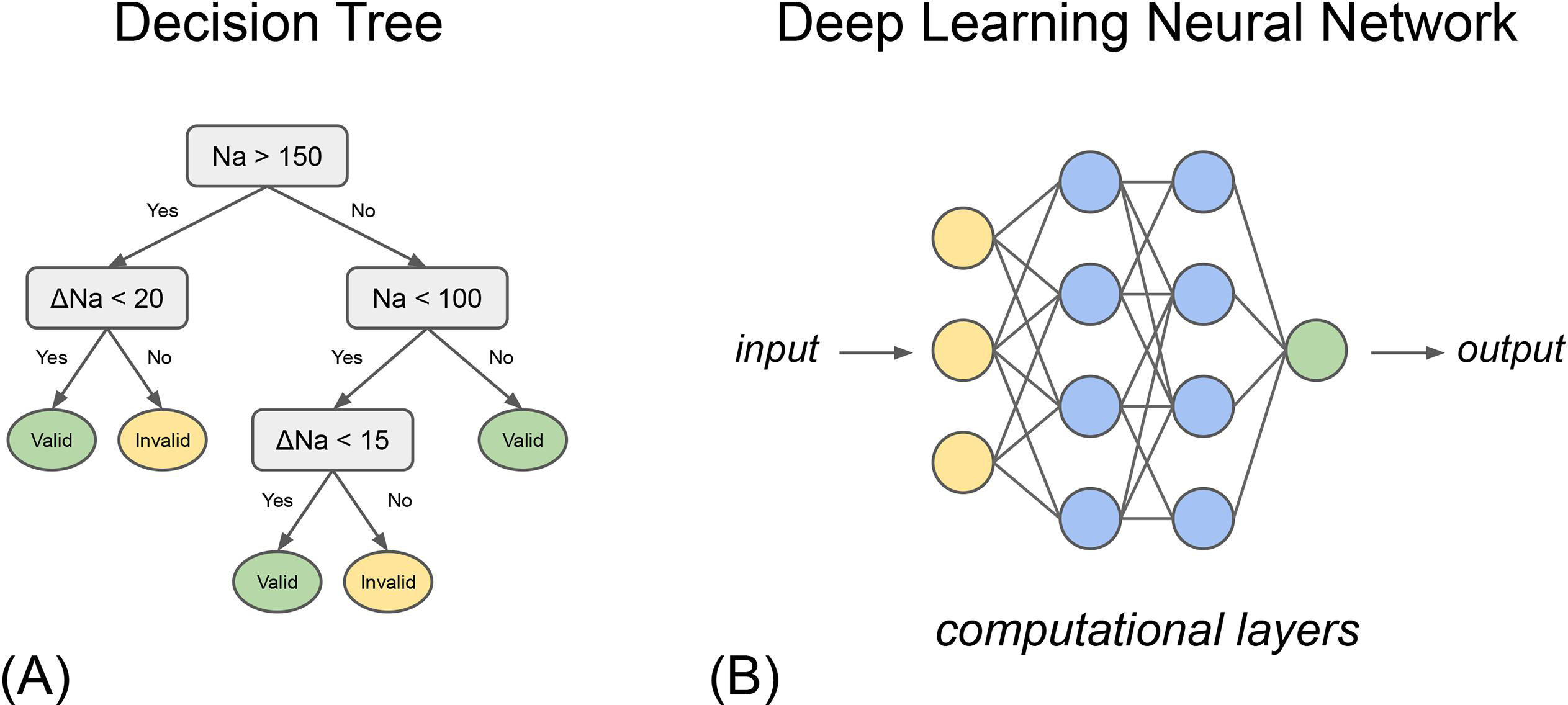
Graphical representation of types of machine learning models: (A) a simple decision tree and (B) deep learning neural network.
In contrast, unsupervised machine learning is when models are provided with an unlabeled dataset. The model is left to describe relationships in the data according to patterns or trends that it observes. Unsupervised machine learning can be used to discover previously unknown patterns [17,18]. Examples of unsupervised machine learning models are k-means clustering, k-nearest neighbors, and principal component analysis.
Finally, it is important to place machine learning in context alongside artificial intelligence and deep learning. Artificial intelligence (AI) refers to the broader field of using computers to perform human-like tasks such as problem-solving. Machine learning is a subset of AI. Meanwhile, deep learning is a subset of machine learning inspired by neuronal networks of the brain. In deep learning algorithms, each layer of the neural network builds upon the last to extract increasingly complex insights from the input data (Figure 2). The emergent property of these methods allows for the performance of complex tasks, such as image recognition and language interpretation, explaining its rising popularity in healthcare [19,20].
2.3. Evaluation Metrics
In scenarios where a model is predicting a binary output (typically referred to as binary classification), evaluation metrics are similar to what is used to evaluate diagnostic tests in medicine: sensitivity, specificity, positive-predictive value, and so on. The area under the receiver operator curve (AUROC), another commonly used evaluation metric, illustrates how a model balances its false positive rate (1 - specificity) with its true positive rate (sensitivity). The closer the AUROC is to 1, the higher the performance of the model. Table 1 outlines some of the most common metrics used by studies in this review.
Table 1.
Common evaluation metrics used to describe machine learning model performance.
| Evaluation Metric | Definition |
|---|---|
| Accuracy | Proportion of correct predictions over total number of predictions |
| Positive Predictive Value | Probability that a predicted positive is truly positive |
| Negative Predictive Value | Probability that a predicted negative is truly negative |
| True Positive Rate (Sensitivity) | Probability that a truly positive result is predicted to be positive |
| True Negative Rate (Specificity) | Probability that a truly negative result is predicted to be negative |
| False Positive Rate = 1-Specificity | Probability that a truly negative result is falsely predicted to be positive |
3. Current State of Machine Learning in Routine Laboratory Medicine
3.1. Search Methodology
In order to identify relevant articles, a comprehensive PubMed query was designed. The query, shown below, utilizes text word and Medical Subject Headings (MeSH) matching to capture articles at the intersection of machine learning and general clinical laboratory techniques and clinical laboratory information systems. The addition of MeSH inclusion criteria captures articles that may not explicitly mention “machine learning” or “artificial intelligence” in the title or abstract but are indexed into PubMed as pertaining to these topics. The exclusion criteria of the query exclude MeSH children of “Clinical Laboratory Techniques” that do not pertain to common laboratory tests such as serum chemistries, urinalysis, and routine hematologic tests.
( (“Artificial intelligence”[MeSH Major Topic] OR “Artificial intelligence”[Title/Abstract] OR “machine learning”[MeSH Major Topic] OR “machine learning”[Title/Abstract] OR “deep learning”[Title/Abstract])
AND
(“clinical lab*”[tw] OR “clinical chemistr*”[tw] OR “Laboratory medicine”[tw] OR “Clinical Laboratory Techniques”[majr] OR “Clinical Laboratory Information Systems”[majr]) NOT (“covid-19 testing”[mesh] OR “genetic testing”[mesh] OR “histological techniques”[mesh] OR “immunologic tests”[mesh] or “metabolic clearance rate”[mesh] or “microbiological techniques”[mesh] or “molecular diagnostic techniques”[mesh] or “neonatal screening”[mesh] or “occult blood”[mesh] or “parasite load”[mesh] or “pregnancy tests”[mesh] or “radioligand assay”[mesh] or “semen analysis”[mesh] or “sex determination analysis”[mesh] or “specimen handling”[mesh]) )
The query was executed with a date range filter from 1 October 2011 to 30 September 2021. Only English-language articles were included. This search returned 583 articles within the 10-year search period. As evidenced by the number of articles returned by our literature search query over the past decade, this topic has received increasing attention over recent years (Figure 3). Through manual title and abstract review, 544 of the original 583 articles were excluded. Of those excluded articles, 108 did not primarily pertain to the field of laboratory medicine or clinical pathology. Other major excluded themes included laboratory imaging such as microscopy and cytology (166 articles), clinical prediction algorithms (90 articles), molecular medicine (47 articles), and microbiology (21 articles). The remaining 39 articles underwent full manuscript review, after which a total of 18 were included (Figure 4).
Figure 3.
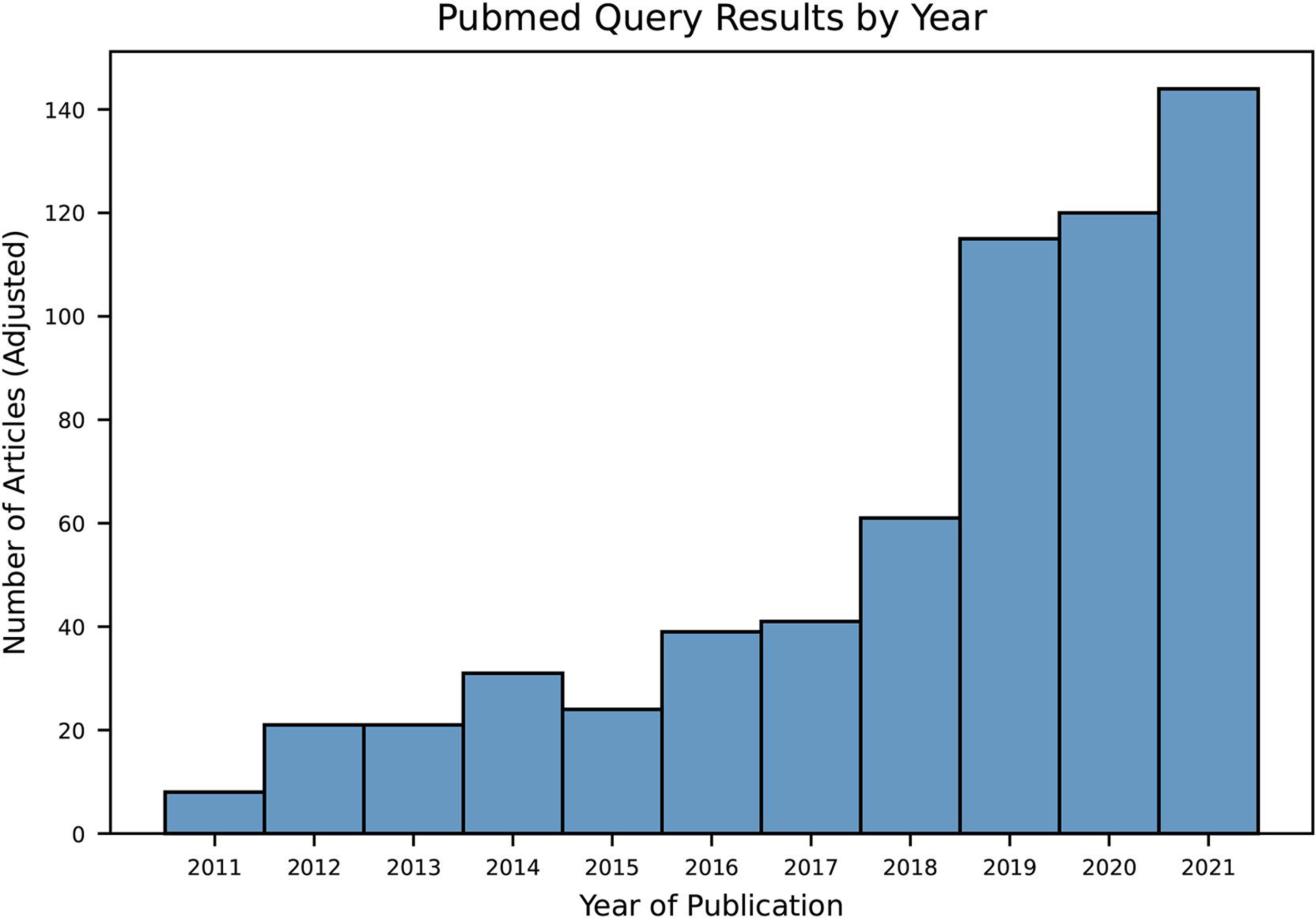
Bar plot showing pubmed query results by year, adjusted by number of months included in the queried year (i.e. only October to December of 2011 and January to September of 2021 are included in the search query date range).
Figure 4.
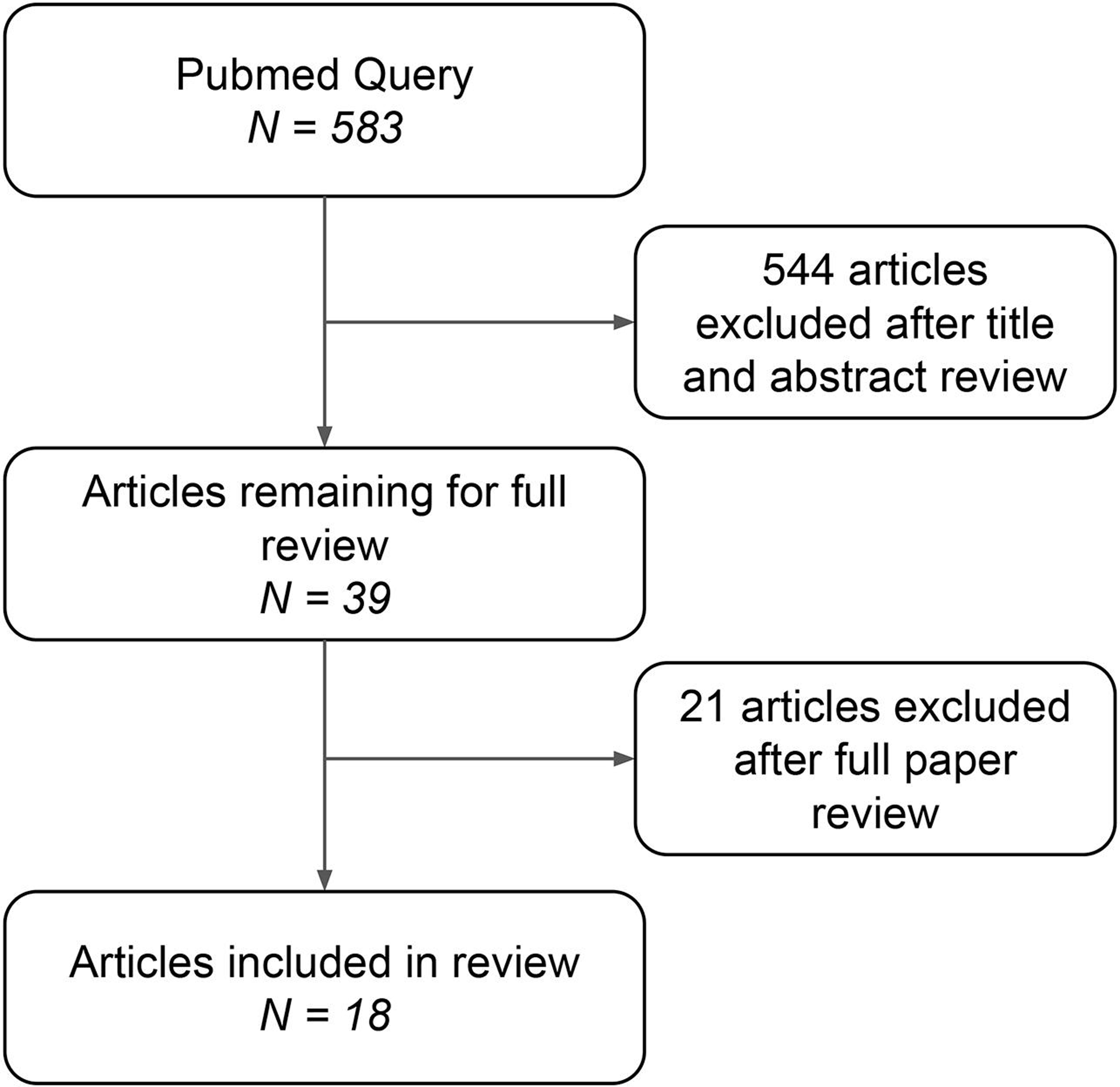
Diagram showing manuscript inclusion and exclusion criteria for review.
3.2. Literature Review
Our review of the current state of machine learning in laboratory medicine reveals several exciting applications including predicting laboratory test values, improving laboratory utilization, automating laboratory processes, promoting precision laboratory test interpretation, and finally improving laboratory medicine information systems. In these studies, tree-based learning algorithms and neural networks often achieved the best performance. Table 2 summarizes the characteristics and themes from this literature review. Articles are described below, by application.
Table 2.
Summary of characteristics of machine learning algorithms included for review.
| Author and Year | Objective and Machine Learning Task | Best Model | Major Themes |
|---|---|---|---|
| Azarkhish (2012) | Predict iron deficiency anemia and serum iron levels from CBC indices | Neural Network | Prediction |
| Cao (2012) | Triage manual review for urinalysis samples | Tree-based | Automation |
| Yang (2013) | Predict normal reference ranges of ESR for various laboratories based on geographic and other clinical features | Neural Network | Interpretation |
| Lidbury (2015) | Predict liver function test results from other tests in the panel, highlighting redundancy in the liver function panel | Tree-based | Prediction, Utilization |
| Demirci (2016) | Classify whether critical lab result is valid or invalid using other lab values and clinical information | Neural Network | Automation, Interpretation, Validation |
| Luo (2016) | Predict ferritin from other tests in iron panel | Tree-based | Prediction, Utilization |
| Poole (2016) | Create personalized reference ranges that take into account patients’ diagnoses | Unsupervised learning | Interpretation |
| Parr (2018) | Automate mapping of Veterans Affair laboratory data to LOINC codes | Tree-based | Information systems, Automation |
| Wilkes (2018) | Classify urine steroid profiles as normal or abnormal, and further interpret into specific disease processes | Tree-based | Interpretation, Automation |
| Fillmore (2019) | Automate mapping of Veterans Affair laboratory data to LOINC codes | Tree-based | Information systems, Automation |
| Lee (2019) | Predict LDL-C levels from a limited lipid panel more accurately than current gold standard equations | Neural Network | Interpretation, Prediction |
| Xu (2019) | Identify redundant laboratory tests and predict their results as normal or abnormal | Tree-based | Prediction, Utilization |
| Islam (2020) | Use prior ordering patterns to create an algorithm that can recommend best practice tests for specific diagnoses | Neural Network | Utilization |
| Peng (2020) | Interpret newborn screening assays based on gestational age and other clinical information to reduce false positives | Tree-based | Interpretation, Utilization |
| Wang (2020) | Automatically verify if lab test result is valid or invalid | Tree-based | Validation, Automation |
| Dunn (2021) | Predict laboratory test results from wearable data | Tree-based | Prediction |
| Fang (2021) | Classify blood specimen as clotted or not clotted based on coagulation indices | Neural Network | Quality control |
| Farrell (2021) | Automatically identify mislabelled laboratory samples | Neural Network | Quality control, Automation |
3.2.1. Laboratory Test Value Prediction and Laboratory Utilization
One of the prevailing themes in how machine learning has been applied to laboratory medicine is for the prediction of laboratory results based on other clinically available data. Authors of these studies propose that such models can be used to power clinical decision support tools for ordering providers and optimize laboratory testing utilization.
One of the first examples of this type of work is the study by Azarkhish et al. [21] in which a neural network model predicted iron deficiency anemia and serum iron levels based on features from a routine complete blood count. The model achieved an impressive AUROC of 98% for the binary classification of iron-deficiency anemia. It predicted the actual serum iron level with less accuracy, achieving a root-mean squared error of 0.136 mcg/dL and R2 of 0.93. It is important to note, however, this study was limited by the relatively small number of participants, with 149 subjects in the training group and 54 subjects in the testing group.
This work continued with Luo et al. [15], who conceived a clinical decision support tool capable of predicting laboratory test results from related laboratory results and other clinical information. As a proof of concept, they demonstrated a machine learning algorithm that was capable of predicting whether serum ferritin level was abnormal with considerable accuracy—achieving an AUROC of 97% using a random forest imputation method to fill in required missing laboratory features that were then fed into a logistic regression model. Meanwhile, Lidbury et al. [22] also studied the redundancy of laboratory test panels, with a focus on liver function tests. They were able to predict whether ɣ-glutamyl transferase (GGT) was normal or abnormal using other components of the liver function panel, achieving an accuracy of 90% with a tree-based machine learning model. They concluded that GGT offered little additional value beyond the other components of a typical liver function panel.
Along similar lines of test result prediction and lab utilization, Xu et al. [23] studied a machine learning model to predict laboratory test results as normal or abnormal in order to identify low-yield, repetitive laboratory tests. Their group performed a multi-site study of nearly 200,000 inpatient laboratory testing orders to identify the most repetitive laboratory tests, and then attempted to predict each one. They were able to achieve an AUROC of >90% for 20 common laboratory tests including sodium, hemoglobin, and lactate dehydrogenase. They proposed a sensitive decision threshold pertaining to a negative predictive value of 95% to power a clinical decision support tool aimed at reducing low-yield, repetitive testing.
In the same realm of clinical decision support and laboratory utilization, Islam et al. [24] developed a deep learning machine learning model capable of recommending what laboratory tests a provider should order. Rather than predicting specific test results, their model predicted what tests should be ordered in the first place. Using features such as clinical diagnoses, medications, prior laboratory tests, and demographic information, their neural network model was able to achieve moderate performance with AUROCmacro of 0.76 and AUROCmicro of 0.87. One important limitation of this study, however, is that the algorithm learned from prior testing patterns, but no expert determination was made about whether these prior ordering behaviors were optimal in the first place. Thus a model like this is prone to learning undesirable practices from historic testing patterns.
Lee et al. [25] from South Korea proposed a neural network deep learning model to predict low density lipoprotein cholesterol (LDL-C) from high density lipoprotein cholesterol (HDL-C), total cholesterol, and triglycerides model compared to a ground truth of fractionated LDL-C measurement. They showed that their model achieved better performance than the historical Friedewald equation [26] and Martin’s “novel method” [27], with a root mean squared error of 8.1 mg/dL versus 10.8 mg/dL and 8.3 mg/dL respectively.
Finally, Dunn et al. [28] completed an experimental study using a machine learning regression model to predict common laboratory tests using data from wearable devices such as accelerometers and electrodermal probes sensors. Unfortunately, this futuristic take on laboratory test prediction was unable to achieve meaningful performance. For example, their model using wearable data was able to explain only 21% of the variability in hematocrit level, which was the laboratory test for which the model performed best.
3.2.2. Validation and Quality Assurance/Quality Control
Our review of the literature also revealed many examples of using machine learning for test result validation and quality control in routine laboratory medicine. For example, Demirci et al. [29] developed a neural network machine learning algorithm capable of classifying whether a critical lab result was valid or invalid. They studied several common biochemical assays such as electrolytes and liver function tests and used prior test results, lab indices such as hemolytic index, and demographic information in order to predict whether a critical result was valid. Model results were compared to expert opinion of a group of biochemists. The model was able to correctly classify critical values as valid with a sensitivity of 91% at a specificity of 100%, meaning the model could drastically reduce the number of critical results requiring manual validation while keeping the rate of incorrectly validated tests to a minimum.
Similarly, Wang et al. [30] developed an ensemble of tree-based algorithms to automate the test result verification process. Their model was able to automatically verify laboratory results with a sensitivity of 99.9% and specificity of 98%. On their retrospective data, this would have led to an 80% reduction in laboratory reports requiring manual verification compared to their current rule-based verification system.
Meanwhile, Cao et al. [31] used a tree-based machine learning model to reduce the volume of samples flagged for manual review. Their model, which used features from a 10-point dipstick and urine cytometry measurements, called for a manual review rate of 32%, which corresponded to a sensitivity of 92% and specificity of 81.5% compared against expert-label ground truth manual urine microscopy results.
As for quality assurance/quality control, Farrell et al. [32] showed that a machine learning algorithm for identifying mislabeled lab samples was able to outperform manual verification. Their best performing algorithm was a neural network that achieved an AUROC of 98%. A limitation of this study, however, is that they do not compare their performance against rule-based delta checks, which are the current gold standard. Meanwhile, a neural network machine learning algorithm by Fang et al. [33] was able to classify if a blood specimen was clotted or not with moderate accuracy (AUROC 91%). Their algorithm used coagulation testing results from the sample and compared model outputs to a ground truth of manual inspection for clotting by laboratory technicians.
3.2.3. Test Result Interpretation and Personalized Reference Ranges
Additionally, there are several recent studies aimed at using machine learning in laboratory medicine for test interpretation and personalized reference ranges—efforts towards achieving precision medicine. With regards to test interpretation, Wilkes et al. [34] developed a tree-based machine learning model capable of classifying a urine steroid profile as either normal or potentially abnormal (compared to manually expert-labeled interpretations) with an AUROC of 96%. They then sought to interpret urinary steroid profiles into specific pathophysiologic conditions such as “adrenal suppression” or “congenital adrenal hyperplasia.” Their model was able to achieve modest performance at this more complicated multiclass classification problem with an accuracy of 87%.
Peng et al. [35] were able to achieve significant test performance improvements with a tree-based (random forest) machine learning model capable of reducing false positives from newborn screening, a common issue with the highly sensitive assay. Their model, which used 39 metabolic analytes and clinical variables such as weight and gestational age was able to reduce false positives by 98% for ornithine transcarbamylase deficiency and 89% for glutaric acidemia type 1, without sacrificing any test sensitivity. They published their tool online for providers to use freely.
Finally, one of the most promising applications of machine learning in medicine is the general development of “personalized” medical diagnosis and interpretation. To this effect, Poole et al. [36] demonstrated that a series of statistical learning methods can be used to create more personalized reference ranges by analyzing test result distributions against clinical features such as diagnosis codes. In an earlier study from China, Yang et al. [37] also demonstrated a neural network model capable of predicting reference ranges for erythrocyte sedimentation rate (ESR) testing, which is known to vary based on geographic factors such as altitude. Their algorithm uses a number of environmental variables and is able to predict ESR reference ranges for laboratories across China, differing only up to 3% from established reference ranges (which vary from 4 to 21 mm/hr).
3.2.4. Laboratory Information Systems
Machine learning has also been applied to laboratory information systems to improve operations and enable clinical research. For example, two studies in this review proposed machine learning methods to map laboratory data to standard LOINC (Observation Identifiers Names and Codes) codes to enable interoperability and clinical research. LOINC codes are the current industry standard ontology for representing measurement data in healthcare, including laboratory testing healthcare [38].
In one such article, Fillmore et. al [39] studied a group of models for mapping 7 common laboratory concepts to the United States Department of Veterans Affairs (US VA) medical records system, where LOINC mappings are imperfect. The best performance was achieved by a tree-based (random forest) model with an accuracy of 98%, presenting a significant improvement over what was an otherwise tedious task of manually reviewing hundreds of possible conceptual links.
Similarly, Parr et al. [40] developed a machine learning model to assign missing LOINC codes and improve the accuracy of existing codes in the US VA medical records data warehouse. Their tree-based machine learning algorithm was able to correctly identify the LOINC code with a rate of 85% in unlabeled laboratory tests and correctly identify the LOINC code in 96% of randomly selected previously labeled laboratory tests. In cases where the algorithm differed from the currently assigned LOINC code, manual review revealed that the machine learning algorithm was correct 83% of the time, compared to the 72% accuracy rate of the incumbent label.
4. Discussion
4.1. Reflections and Future Direction
Machine learning is able to leverage large amounts of data to infer complex relationships and patterns that may otherwise be beyond the capabilities of a rule-based system or human expert. Furthermore, while static rule-based algorithms are based on previously established knowledge, machine learning can identify new patterns and applications, and continuously use new data to improve its performance.
Along those lines, one of the most promising aspects of artificial intelligence in laboratory medicine has been its success in automation. The reviewed work demonstrates significant advancements in using machine learning algorithms to improve upon current rule-based methods for identifying samples for manual verification or validation. Such algorithms have already achieved excellent performance and we anticipate will soon be commonplace in the modern laboratory.
Another exciting application of machine learning is its ability to leverage large amounts of prior medical data to create more personalized interpretation of test results. Although still in its early stages, we see the foundations for this in the work by Poole et al. who propose a relatively simple method of using diagnosis codes to achieve slightly more personalized reference ranges [36]. We envision future work into more comprehensive algorithms that consider the entire clinical context of the patient to provide personalized laboratory test reference ranges to enable precision diagnostics. The paradigm will shift from “What is a normal hemoglobin?” to “What is a normal hemoglobin for you.”
Finally, while it is the focus of many articles considered in this review, considerable work must still be done until machine learning prediction of laboratory test results can be utilized to make changes in clinical practice. While studies have demonstrated a high level of redundancy in lab panels and ordering practices, attempts at predicting laboratory test results still fail to achieve consistently high performance across a variety of tests. Despite the lack of a generalizable solution in this space, there are opportunities for smaller gains to be made by optimizing testing utilization in specific situations.
4.2. Challenges to the Field
There is great excitement regarding the future of machine learning in laboratory medicine, however, there are significant challenges that must be addressed as well. From a technical perspective, one of the biggest limitations faced by machine learning algorithms in medicine is data quality. Laboratory information systems are plagued with incorrectly labeled or missing data, limiting the maximum performance that any algorithm can achieve. Another technical and financial challenge includes the cost of the computational infrastructure, along with the cost of personnel with the right computational expertise to develop, deploy, maintain, and update the machine learning algorithms and the software tools needed to run them.
From a clinical point of view, as a relatively young field, machine learning in laboratory medicine requires standardization and regulation. Currently, there are no guidelines regarding the best practices for the clinical validation of machine learning algorithms. Even in pathology fields where clinical machine learning tools are developing rapidly, such as digital pathology, there are no well-established guidelines for laboratory-developed applications or for the verification of vendor-developed software [41]. In fact, just recently, the College of American Pathologists (CAP) assembled a committee to start addressing this gap, including the creation of laboratory standards for AI applications [42].
Similarly, regulatory entities, such as the FDA, have not completely determined what their role will be in the regulation of laboratory-developed machine learning applications. In 2021, the FDA published an action plan to update the proposed regulatory framework for artificial intelligence/machine learning-based software as a medical device [43]. This is an important step to regulate the market of such software in medicine. However, it is still unclear what the position of federal and state regulators will be in regard to laboratory-developed machine learning tools. Finally, more studies assessing the actual implementation of these applications in clinical laboratories and demonstrating their safety and reliability will be necessary to have laboratory medicine professionals fully embrace this technology.
4.3. Machine Learning in Related Fields
This review focuses on the use of machine learning in routine laboratory testing. However, there has been much attention given to the application of machine learning to the broader field of pathology. Several related applications are briefly discussed here to serve as a reference for laboratory medicine practitioners who may wish to explore these topics further.
The digitization of histopathology slides have allowed for the widespread utilization of computer vision and other artificial intelligence methods for image interpretation. Many studies in this area focus on histopathology in cancer [44–46]. Along these lines, the FDA recently approved the first artificial intelligence product in pathology that can identify areas of interest in prostate biopsy slides [47].
Similarly, digital image acquisition in microscopy has spawned additional work in this field. Identification of cellular events such as mitosis or apoptosis can be used to flag areas of dysregulated growth or quantify response to chemotherapy [48,49]. Applications of artificial intelligence in specialized cytometry have also allowed for more precise detection of neoplastic cell lines and cellular markers of interest [50,51].
Finally, related to laboratory medicine is the use of machine learning for point-of-care testing. In this field, there has been considerable emphasis on the use of predictive algorithms in continuous glucose monitoring for patients with diabetes [52].
5. Conclusion
Machine learning promises exciting advancements in medicine, but its application in laboratory medicine is still nascent. As a young field, there is additional need for standardization of how these algorithms are developed and presented. Regardless, several machine learning models have achieved excellent performance in automating test result validation and triaging samples for manual review. There is also exciting, ongoing work in using machine learning for optimizing laboratory utilization, predicting laboratory test results, and providing personalized laboratory test interpretation.
Acknowledgements:
The authors wish to thank Connie Wong, medical education librarian, for her help in forming our literature search query.
Funding:
Jonathan H Chen was supported in part by the NIH/National Library of Medicine Award R56LM013365, the Stanford Artificial Intelligence in Medicine and Imaging and Human-Centered Artificial Intelligence (AMIA-HAI) Partnership Grant, Stanford Aging and Ethnogeriatrics Research Center (under NIH/National Institute on Aging grant P30AG059307), the Stanford Clinical Excellence Research Center (CERC), and the Stanford Departments of Medicine and Pathology.
Footnotes
Conflicts of Interest: Jonathan H Chen is the co-founder of Reaction Explorer LLC, which develops and licenses organic chemistry software. He has received consulting fees from Sutton Pierce and Younker Hyde MacFarlane PLLC. Naveed Rabbani has received consulting fees from Atropos LLC.
Ethics Statement: This study does not involve the use of human subjects.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Darcy AM, Louie AK, Roberts LW. Machine Learning and the Profession of Medicine. JAMA 2016;315:551–2. [DOI] [PubMed] [Google Scholar]
- [2].Obermeyer Z, Emanuel EJ. Predicting the Future - Big Data, Machine Learning, and Clinical Medicine. N Engl J Med 2016;375:1216–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Beam AL, Kohane IS. Big Data and Machine Learning in Health Care. JAMA 2018;319:1317–8. [DOI] [PubMed] [Google Scholar]
- [4].Benjamens S, Dhunnoo P, Meskó B. The state of artificial intelligence-based FDA-approved medical devices and algorithms: an online database. NPJ Digit Med 2020;3:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cui M, Zhang DY. Artificial intelligence and computational pathology. Lab Invest 2021;101:412–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lippi G, Bassi A, Bovo C. The future of laboratory medicine in the era of precision medicine. J Lab Precis Med 2016;1:1–5. [Google Scholar]
- [7].Cabitza F, Banfi G. Machine learning in laboratory medicine: waiting for the flood? Clin Chem Lab Med 2018;56:516–24. [DOI] [PubMed] [Google Scholar]
- [8].Paranjape K, Schinkel M, Hammer RD, Schouten B, Nannan Panday RS, Elbers PWG, et al. The Value of Artificial Intelligence in Laboratory Medicine. Am J Clin Pathol 2021;155:823–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Pillay TS. Artificial intelligence in pathology and laboratory medicine. J Clin Pathol 2021;74:407–8. [DOI] [PubMed] [Google Scholar]
- [10].Niazi MKK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence. Lancet Oncol 2019;20:e253–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Goldstein BA, Navar AM, Pencina MJ, Ioannidis JPA. Opportunities and challenges in developing risk prediction models with electronic health records data: a systematic review. J Am Med Inform Assoc 2017;24:198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Brnabic A, Hess LM. Systematic literature review of machine learning methods used in the analysis of real-world data for patient-provider decision making. BMC Med Inform Decis Mak 2021;21:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tomašev N, Glorot X, Rae JW, Zielinski M, Askham H, Saraiva A, et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature 2019;572:116–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fleuren LM, Klausch TLT, Zwager CL, Schoonmade LJ, Guo T, Roggeveen LF, et al. Machine learning for the prediction of sepsis: a systematic review and meta-analysis of diagnostic test accuracy. Intensive Care Med 2020;46:383–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Luo Y, Szolovits P, Dighe AS, Baron JM. Using Machine Learning to Predict Laboratory Test Results. Am J Clin Pathol 2016;145:778–88. [DOI] [PubMed] [Google Scholar]
- [16].Uddin S, Khan A, Hossain ME, Moni MA. Comparing different supervised machine learning algorithms for disease prediction. BMC Med Inform Decis Mak 2019;19:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang Y, Zhao Y, Therneau TM, Atkinson EJ, Tafti AP, Zhang N, et al. Unsupervised machine learning for the discovery of latent disease clusters and patient subgroups using electronic health records. J Biomed Inform 2020;102:103364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Roohi A, Faust K, Djuric U, Diamandis P. Unsupervised Machine Learning in Pathology: The Next Frontier. Surg Pathol Clin 2020;13:349–58. [DOI] [PubMed] [Google Scholar]
- [19].Yu K-H, Beam AL, Kohane IS. Artificial intelligence in healthcare. Nat Biomed Eng 2018;2:719–31. [DOI] [PubMed] [Google Scholar]
- [20].Shrestha A, Mahmood A. Review of Deep Learning Algorithms and Architectures. IEEE Access 2019;7:53040–65. [Google Scholar]
- [21].Azarkhish I, Raoufy MR, Gharibzadeh S. Artificial intelligence models for predicting iron deficiency anemia and iron serum level based on accessible laboratory data. J Med Syst 2011;36:2057–61. [DOI] [PubMed] [Google Scholar]
- [22].Lidbury BA, Richardson AM, Badrick T. Assessment of machine-learning techniques on large pathology data sets to address assay redundancy in routine liver function test profiles. Diagnosis (Berl) 2015;2:41–51. [DOI] [PubMed] [Google Scholar]
- [23].Xu S, Hom J, Balasubramanian S, Schroeder LF, Najafi N, Roy S, et al. Prevalence and Predictability of Low-Yield Inpatient Laboratory Diagnostic Tests. JAMA Netw Open 2019;2:e1910967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Islam MM, Yang H-C, Poly TN, Li Y-CJ. Development of an Artificial Intelligence-Based Automated Recommendation System for Clinical Laboratory Tests: Retrospective Analysis of the National Health Insurance Database. JMIR Med Inform 2020;8:e24163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lee T, Kim J, Uh Y, Lee H. Deep neural network for estimating low density lipoprotein cholesterol. Clin Chim Acta 2018;489:35–40. [DOI] [PubMed] [Google Scholar]
- [26].Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 1972;18:499–502. [PubMed] [Google Scholar]
- [27].Martin SS, Blaha MJ, Elshazly MB, Toth PP, Kwiterovich PO, Blumenthal RS, et al. Comparison of a novel method vs the Friedewald equation for estimating low-density lipoprotein cholesterol levels from the standard lipid profile. JAMA 2013;310:2061–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Dunn J, Kidzinski L, Runge R, Witt D, Hicks JL, Schüssler-Fiorenza Rose SM, et al. Wearable sensors enable personalized predictions of clinical laboratory measurements. Nat Med 2021;27:1105–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Demirci F, Akan P, Kume T, Sisman AR, Erbayraktar Z, Sevinc S. Artificial Neural Network Approach in Laboratory Test Reporting: Learning Algorithms. Am J Clin Pathol 2016;146:227–37. [DOI] [PubMed] [Google Scholar]
- [30].Wang H, Wang H, Zhang J, Li X, Sun C, Zhang Y. Using machine learning to develop an autoverification system in a clinical biochemistry laboratory. Clin Chem Lab Med 2020;59:883–91. [DOI] [PubMed] [Google Scholar]
- [31].Cao Y, Cheng M, Hu C. UrineCART, a machine learning method for establishment of review rules based on UF-1000i flow cytometry and dipstick or reflectance photometer. Clin Chem Lab Med 2012;50:2155–61. [DOI] [PubMed] [Google Scholar]
- [32].Farrell C-J. Identifying mislabelled samples: Machine learning models exceed human performance. Ann Clin Biochem 2021:45632211032991. [DOI] [PubMed] [Google Scholar]
- [33].Fang K, Dong Z, Chen X, Zhu J, Zhang B, You J, et al. Using machine learning to identify clotted specimens in coagulation testing. Clin Chem Lab Med 2021;59:1289–97. [DOI] [PubMed] [Google Scholar]
- [34].Wilkes EH, Rumsby G, Woodward GM. Using Machine Learning to Aid the Interpretation of Urine Steroid Profiles. Clin Chem 2018;64:1586–95. [DOI] [PubMed] [Google Scholar]
- [35].Peng G, Tang Y, Cowan TM, Enns GM, Zhao H, Scharfe C. Reducing False-Positive Results in Newborn Screening Using Machine Learning. Screening 2020;6. 10.3390/ijns6010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Poole S, Schroeder LF, Shah N. An unsupervised learning method to identify reference intervals from a clinical database. J Biomed Inform 2015;59:276–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yang Q, Mwenda KM, Ge M. Incorporating geographical factors with artificial neural networks to predict reference values of erythrocyte sedimentation rate. Int J Health Geogr 2013;12:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Huff SM, Rocha RA, McDonald CJ, De Moor GJ, Fiers T, Bidgood WD Jr, et al. Development of the Logical Observation Identifier Names and Codes (LOINC) vocabulary. J Am Med Inform Assoc 1998;5:276–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Fillmore N, Do N, Brophy M, Zimolzak A. Interactive Machine Learning for Laboratory Data Integration. Stud Health Technol Inform 2019;264:133–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Parr SK, Shotwell MS, Jeffery AD, Lasko TA, Matheny ME. Automated mapping of laboratory tests to LOINC codes using noisy labels in a national electronic health record system database. J Am Med Inform Assoc 2018;25:1292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Baxi V, Edwards R, Montalto M, Saha S. Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod Pathol 2022;35:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].College of American Pathologists. Artificial Intelligence (AI) Committee. College of American Pathologists; 2021. https://www.cap.org/member-resources/councils-committees/artificial-intelligence-ai-committee/ (accessed January 24, 2022). [Google Scholar]
- [43].US Food and Drug Administration. Artificial intelligence/machine learning (ai/ml)-based software as a medical device (SAMD) action plan 2021. https://www.fda.gov/media/145022/download (accessed January 24, 2022).
- [44].Korbar B, Olofson AM, Miraflor AP, Nicka CM, Suriawinata MA, Torresani L, et al. Deep Learning for Classification of Colorectal Polyps on Whole-slide Images. J Pathol Inform 2017;8:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Nagpal K, Foote D, Liu Y, Chen P-HC, Wulczyn E, Tan F, et al. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. NPJ Digit Med 2019;2:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Couture HD, Williams LA, Geradts J, Nyante SJ, Butler EN, Marron JS, et al. Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype. NPJ Breast Cancer 2018;4:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Office of the Commissioner. FDA Authorizes Software that Can Help Identify Prostate Cancer 2021. https://www.fda.gov/news-events/press-announcements/fda-authorizes-software-can-help-identify-prostate-cancer (accessed October 27, 2021).
- [48].Grys BT, Lo DS, Sahin N, Kraus OZ, Morris Q, Boone C, et al. Machine learning and computer vision approaches for phenotypic profiling. J Cell Biol 2016;216:65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Falk T, Mai D, Bensch R, Çiçek Ö, Abdulkadir A, Marrakchi Y, et al. U-Net: deep learning for cell counting, detection, and morphometry. Nat Methods 2018;16:67–70. [DOI] [PubMed] [Google Scholar]
- [50].Wang S, Zhou Y, Qin X, Nair S, Huang X, Liu Y. Label-free detection of rare circulating tumor cells by image analysis and machine learning. Sci Rep 2020;10:12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Syed-Abdul S, Firdani R-P, Chung H-J, Uddin M, Hur M, Park JH, et al. Artificial Intelligence based Models for Screening of Hematologic Malignancies using Cell Population Data. Sci Rep 2020;10:4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Perkins BA, Sherr JL, Mathieu C. Type 1 diabetes glycemic management: Insulin therapy, glucose monitoring, and automation. Science 2021;373:522–7. [DOI] [PubMed] [Google Scholar]


