SUMMARY
Fallopian tube (FT) homeostasis requires dynamic regulation of heterogeneous cell populations and is disrupted in infertility and ovarian cancer. Here we applied single-cell RNA-seq to profile 59,738 FT cells from 4 healthy pre-menopausal subjects. The resulting cell atlas contains 12 major cell types representing epithelial, stromal and immune compartments. Re-clustering of epithelial cells identified 4 ciliated and 6 non-ciliated secretory epithelial subtypes, two of which represent potential progenitor pools: one leading to mature secretory cells, while the other contributing to either ciliated cells or one of the stromal cell types. To understand how FT cell numbers and states change in a disease state, we analyzed 17,798 cells from two hydrosalpinx samples and observed shifts in epithelial and stromal populations, and cell type-specific changes in extracellular matrix and TGF-β signaling, underscoring fibrosis pathophysiology. This resource is expected to facilitate future studies to understand fallopian tube homeostasis in normal development and disease.
Graphical Abstract
eTOC:
Fallopian tube homeostasis requires dynamic regulation of heterogeneous cell populations, which are disrupted in disease. Ulrich et. al apply single-cell RNA sequencing to untangle cellular heterogeneity within healthy and disease samples, identifying major cell types, epithelial subtypes, cell population shifts, and cell type-specific transcriptomic changes in disease states.
INTRODUCTION
In mammals, the fallopian tube (FT) is a highly specialized organ that is essential for the transport of gametes and embryos during unassisted fertilization. Anatomically, the tube can be partitioned into three segments with distinct functions: fimbriae, the finger-like structures responsible for oocyte capture after ovulation; ampulla, the site of oocyte fertilization by the sperm and the first 3 days of embryo development; and isthmus, which serves as the reservoir for sperm prior to fertilization, and through which the embryo is transported to the uterus for implantation (Barton et al., 2021; Crow et al., 1994).
Each segment of the fallopian tube contains both epithelial and stromal layers. The epithelial layer consists of four main types of cells: ciliated, secretory, peg and basal cells (Paik et al., 2015; Peters, 1986). The ciliated cells (FOXJ1+/CAPS+), the most abundant and well defined, control gamete and embryo movement during fertilization and implantation (Paik et al., 2015; Barton et al., 2021). The secretory cells (PAX8+/KRT7+) produce and secrete factors required for fertilization and early embryo development. The peg and basal cells are less abundant and their biological functions are poorly understood. The peg cells are often defined by EPCAM+/CD44+/ITGA6+ (Paik et al., 2015; Peters, 1986). In vitro, the peg cells are capable of expanding and generating tube-like organoids consisting of both ciliated and secretory cells, suggesting that peg cells may have stem cell or progenitor properties (Paik et al., 2015). Basal cells are defined histologically by their round morphology with a cytoplasmic ring, and by expression of epithelial (EPCAM) and T-cell or resident T-cell markers, indicative of potential immune function (Peters, 1986; Hu et al., 2020). Compared to the epithelial layer, the stromal layer contains a larger number of distinct cell types, including lymphocytes, macrophages, dendritic cells, mast cells, smooth muscle cells, fibroblasts, blood and lymphatic endothelial cells, and possibly Cajal-like cells (Ardighieri et al., 2015; Popescu et al., 2007).
Although epithelial and stromal cell populations are present in all anatomic segments of the fallopian tube, their relative proportions may differ depending on hormonal states, menopause status, diseases such as ovarian cancer, and surgical disruption such as tubal ligation (Crow et al., 1994; Donnez, 1985; Ely and Mireille, 2013; Gaitskell et al., 2016; Paik et al., 2015; Tiourin et al., n.d.). For example, in fertile women, ciliated cells are most abundant in the fimbriae, and reach their highest numbers around the time of ovulation; but their numbers decrease during long-term progestin therapy (Donnez, 1985). Conversely, ciliated cells are decreased in number in the fimbriae of post-menopausal women, but are restored to pre-menopausal levels with estrogen therapy (Crow et al., 1994). Similarly, after tubal ligation, the numbers of peg cells - presumed progenitors of the fallopian epithelium - decrease in the fimbriae. This decrease in peg cells was associated with a decreased risk of developing ovarian cancer in both pre- and post-menopausal women (Ely and Mireille, 2013; Gaitskell et al., 2016; Paik et al., 2015; Tiourin et al., n.d.), which led to the hypothesis that peg cells may be the cells of origin for high-grade serous ovarian cancer (Dinh et al., 2021; Hu et al., 2020).
Additionally, changes in tubal epithelium composition can also lead to a form of female infertility known as tubal factor infertility, which in some women can be associated with hydrosalpinx - an enlarged and fluid-filled fallopian tube (Ajonuma et al., 2005; Sapmaz et al., 2019; Yohannes et al., 2019). For these women, tubal factor infertility treatment involves the surgical removal of hydrosalpinx, followed by in vitro fertilization and embryo transfer (Johnson et al., 2010; Sapmaz et al., 2019; Strandell, 2000). We currently have a limited understanding of the etiology of hydrosalpinx, and we lack alternative therapies (other than surgical removal) that could retain tubal function and allow unassisted fertility. Hence, gaining a greater understanding of the cellular heterogeneity of the healthy fallopian tube and the key changes in diseases can help us develop better therapeutic strategies for treating reproductive organ pathologies.
In this study, we applied single-cell RNA-sequencing (scRNA-seq) to analyze 59,738 unselected cells from 10 specimen of human fallopian tubes from 4 healthy subjects. This unbiased characterization of >59K single cells allowed us to define 12 major cell types in healthy FT. Through iterative re-clustering of the two epithelial cell types, we identified 4 ciliated epithelial (CE) and 6 non-ciliated secretory epithelial (NCSE) subtypes. We validated their presence in native tissues using immunofluorescence of marker proteins in FFPE sections and/or by flow cytometry analysis for select populations. Importantly, we identified two NCSE subtypes as likely progenitor cells that could play a key role in fallopian tube homeostasis. To better understand how cell populations change in hydrosalpinx, we sequenced 17,798 cells in surgically removed FT samples from two patients with this condition. In hydrosalpinx FT, several cell populations, including immune cell and stem cell/progenitor populations, differed significantly from the healthy state. In addition, extracellular matrix (ECM) genes were altered, and TGF-β signaling pathways were upregulated, indicative of tissue fibrosis. Overall, the catalog of cell types, subtypes, and specific markers represent a new cell atlas of healthy human fallopian tube and identify potential progenitor cells responsible for the highly regulated maintenance of this dynamic organ. The comparison with samples from a diseased FT provides the first glimpse of the pathophysiology of hydrosalpinx, for both altered cell number and cell type-specific dysregulation of ECM and TGF-β genes. These insights may help guide the development of new therapies as alternatives to surgical intervention.
RESULTS
Systematic analysis of fallopian tube from 4 healthy individuals revealed 12 major cell types
To ensure that our tissue dissociation protocol was sufficiently robust to include the full range of FT cell heterogeneity, we dissected a benign surgical sample (FT1) into three segments, fimbria, isthmus and ampulla. We collected live cells from both the epithelial and the stromal compartments and performed scRNA-seq analysis (Figure 1A). Through unsupervised clustering of 3,722 cells from isthmus, 4,944 cells from ampulla, and 1,861 cells from fimbria we identified an initial set of 11 molecular clusters from each segment, which were highly concordant in their distributions across the 3 segments (Supplemental Figure 1A) and in their expression profiles (Supplemental Figure 1B). By analyzing cluster-specific RNA markers (Supplemental Table 1) we annotated the 11 clusters and found that they represent all known major cell types in FT, thus validating that our dissociation protocol captured the expected cellular diversity of the organ. Next, we extended the same analysis to 3 tubal segments from a second benign surgical sample (FT3), and from two cadaveric samples with normal anatomy (FT2 and FT4) (see their clinical features in Supplemental Table 2A). Systematic comparisons of the clusters found independently in FT1, FT2, FT3 and FT4 confirmed high individual-to-individual correlations of matched cell types (Supplemental Figure 1C) and tissue composition (Supplemental Figure 1D) which was preserved when individual segments were compared among the 4 samples analyzed by segment (Supplemental Figures 1E), indicating that the heterogeneous cell populations have been reproducibly observed in three independent experiments, which included both surgical (FT1, FT3) and cadaveric (FT2, FT4) samples.
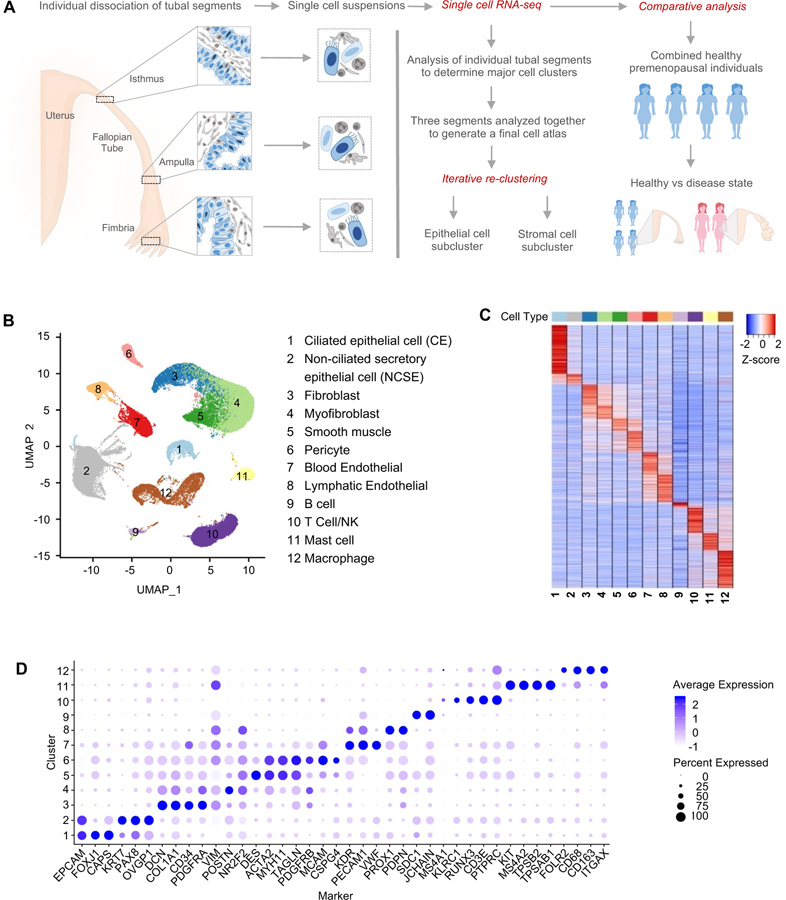
Figure 1. Major cell types and markers identified from single-cell RNA-Seq analysis of healthy human fallopian tubes.
A. Overview of the study, including the fallopian tube sections from which the samples were taken, and the data collection and analysis processes.
B. Identification of 12 major cell types from global clustering of cells from healthy subjects, visualized in UMAP space.
C. Marker gene expression pattern in the 12 major cell types, with values for each gene averaged within each cell type (i.e., the centroid), then standardized over the 12 centroids.
D. Average expression level and prevalence of selected markers used to annotate the 12 major fallopian tube cell types. Details are in Table S3.
To exploit the full power of the entire data we pooled the total set of 59,738 pass-QC cells from the 4 subjects, at an average of ~2.6K detected genes per cell. Through batch-correction and re-clustering (see Computational Methods) of 4 healthy donor samples we defined a stable set of 12 clusters (Figure 1B), annotated them by marker genes (Figure 1C–D), with their average expression levels provided in Supplemental Table 3. The individual samples contributed comparably to each cell type (Supplemental Figure 1D, E). Most of the major cell types are distinctly clustered. However, to examine the potential impact of artificially formed doublets we ran DoubletFinder (Methods), and found that most of the 12 clusters had <1% called doublet rates (Supplemental Figure 1F), with the only exception being “Cluster-13”, which we had identified and removed in early data processing steps. Mainly by relying on known marker genes (Figure 1D) we identified two major epithelial cell types (EPCAM), consisting of ciliated cells (FOXJ1, CAPS) and non-ciliated cells (PAX8, KRT7, OVGP1). Within the stromal compartment we assigned 6 cell types: fibroblast (DCN, COL1A1, PDGFRA), myofibroblast (POSTN, PDGFRB, ACTA2), smooth muscle (DES, MYH11, ACTA2), pericyte (MCAM, CSPG4), blood endothelial (VWF, KDR, PECAM1), and lymphatic endothelial (PDPN, SCD1). Lastly, we identified 4 immune cell types: B cell (JCHAIN, SDC1), T/NK cell (RUNX3, CD3E, KLRC1), mast cell (KIT, MS4A2, TPSB2), and macrophage (CD68, ITGAX, CD163). The only cell type previously reported in the fallopian tube (Popescu et al., 2007) that we failed to identify is the Cajal-like cell. This may be due to their long cytoplasmic processes and small cell body, causing these interstitial cells to be damaged or lost during dissociation. Despite this, our collection of ~59K high-quality single cells portrays the functional and molecular diversity of the premenopausal human fallopian tube and covers all known major cell types of the organ. In the following, we describe focused clustering of the ciliated cells, and separately the non-ciliated epithelial cells, to uncover finer subtypes and their potential lineage relationships.
Identification and characterization of four ciliated epithelial subtypes.
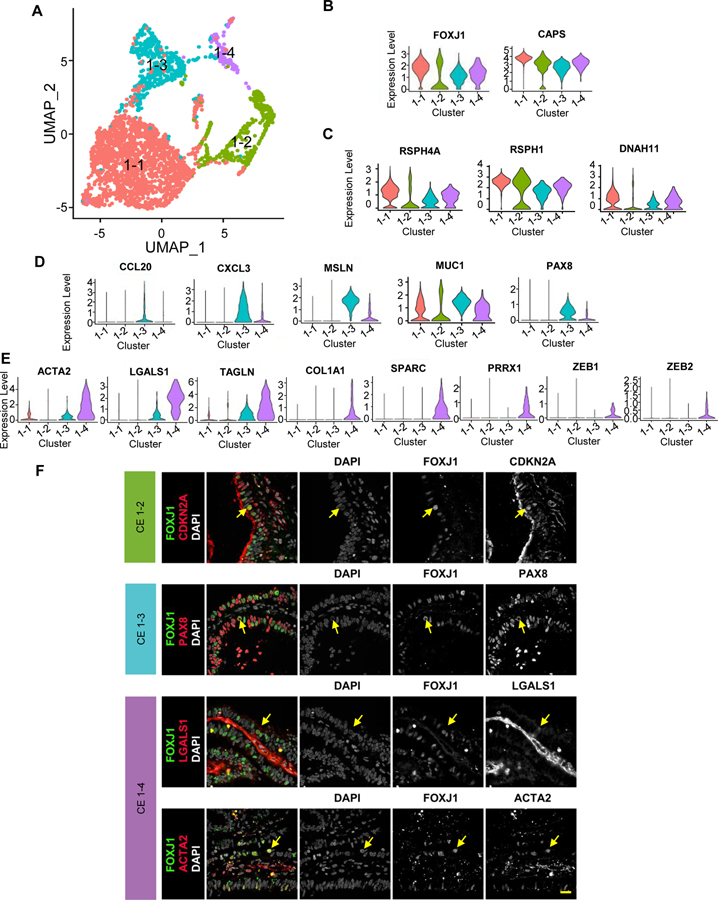
By re-clustering the 2,798 ciliated epithelial cells identified in the global analysis (Figure 1B) we identified four molecularly defined subtypes (Figure 2A), which were positive for FOXJ1 and CAPS (Figure 2B) and were contributed comparably by the four donor samples (Supplemental Figure 2A). We designated them as CE1-1 through CE1-4. Based on subtype-specific RNA markers, CE1-1 cells are likely mature, multi-ciliated cells, as they express genes encoding proteins associated with motile cilia and flagella (CFAP157, CFAP73, CFAP52, and CFAP126), dyneins (DYNLL1, DNAI1, DNAI2, DNAH11, KIF21A), radial spike proteins (RSPH1, 4a, and 9), intraflagellar transport proteins (IFT57), and calcium sensing and binding proteins (CAPS, CALM1) (examples in Figure 2C; full list in Supplemental Table 4). CE1-2 cells express a relatively high level of ribosomal protein-related genes (RPL12, RPL21, RPL31, PRS18 and RPL26) and those involved in apoptosis (Supplemental Table 4). Despite the abundance of ribosomal proteins, these cells have a notably lower number of detected transcripts than other CE subtypes (Supplemental Figure 2B), prompting us to ask whether these cells are damaged or otherwise of lower quality. The relative abundance of mitochondria-encoded transcripts, which tends to be higher in damaged cells, was the lowest in CE1-2 (Supplemental Figure 2C). Furthermore, we estimated the contribution of ambient RNA to each cell by defining an in-silico transcriptome profile for damaged cells – the “soup”– using the thousands of cells that fell below the minimal number of detected genes, and then calculating the “distance” of every cell to the centroids of (1) the 4 CE subtypes and (2) the soup. CE1-2 cells are distinct from “soup” (Supplemental Figure 2D), thus confirming that CE1-2 cells are naturally existing and undamaged. Given their low RNA content and gene markers for apoptosis and p53 pathway (Supplemental Table 4), we speculate that CE1-2 cells are either senescent or executing programmed cell death. The RNA levels of senescence markers such as CDKN2A and CDKN2B did not vary across the four ciliated clusters (not shown); however, they may still be regulated at the protein level. Indeed, our immunofluorescence data suggest that a small number of CDKN2A+/FOXJ1+ cells are scattered in the human fallopian tube epithelium with the majority of FOXJ1+ cells not expressing CDKN2A (Figure 2F), which is consistent with images from the human protein atlas database suggesting that a rare number of CDKN2A cells are scattered in the human fallopian tube epithelium (Supplemental Figure 2E)
Figure 2: Subtypes of ciliated cells.
A. Focused re-clustering 2,798 ciliated cells of healthy fallopian tubes identifies 4 subtypes, 1–1 to 1–4, shown in UMAP space.
B-E. Expression levels of selected markers used to identify the 4 ciliated cell subtypes, with general markers for ciliated cells shown in B; markers for 1–1 and 1–2 in C; those for 1–3 in D; and those for 1–4 in E.
F. Immunofluorescence (IF) staining of the fallopian tube epithelium using antibodies against FOXJ1 and unique markers for ciliated cell subtypes: CDKN2A for 1–2, PAX8 for 1–3, LGALS1 and ACTA2 for 1–4. Arrows indicate double positive cells.
CE1-3 cells express genes involved in antigen processing (CD74), presentation (HLA-DMA, HLA-DRB1), and chemokine/cytokines (CCL20, CXCL1-3) (Supplemental Table 4; examples in Figure 2D). CE1-4 cells express extracellular matrix genes COL1A1, TAGLN, LGALS (Figure 2E), and markers for epithelial-mesenchymal transition (EMT), which were also observed in NCSE2-2 (see below). We validated the presence of additional CE subtypes in vivo with immunofluorescence data (Figure 2F). Indeed, our immunofluorescence data suggests that CE1-3 captures a transitional population co-expressing markers of both ciliated and secretory cells FOXJ1/PAX8, while the FOXJ1/LGALS1 and FOXJ1/ACTA2 cells likely represent CE1-4. Taken together, sub-clustering of ciliated cells reveals multiple ciliated subtypes in the fallopian tube epithelium.
Focused analysis of non-ciliated epithelial cells identified six subtypes.
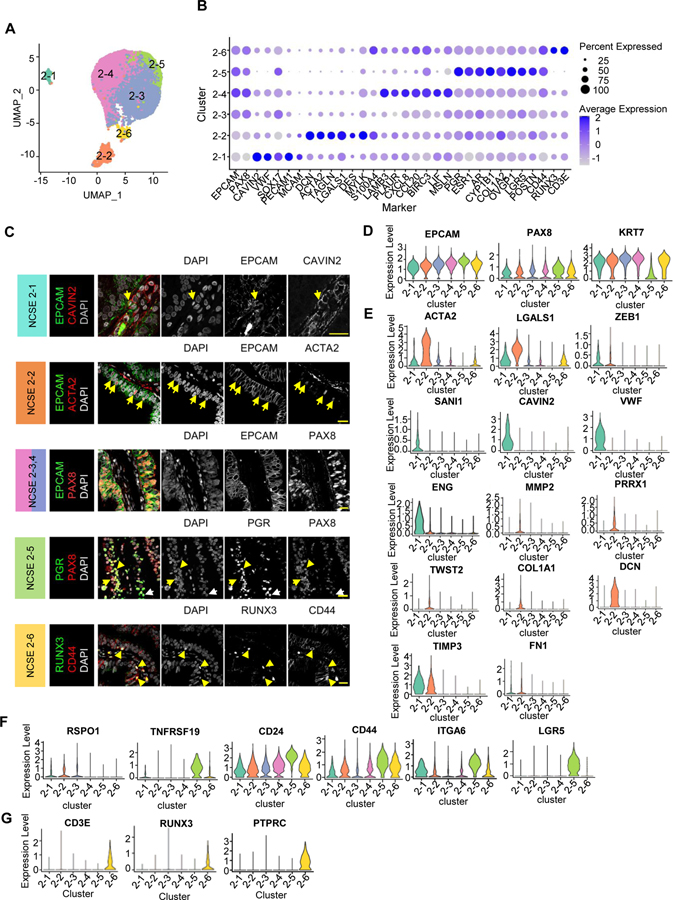
Re-clustering of the ~14,000 non-ciliated epithelial cells from the 4 healthy fallopian tube samples identified 6 molecular subtypes (Figure 3A). They were identified in all subjects (Supplemental Figure 3A), and are named NCSE2-1 through NCSE2-6. As before (Supplemental Figure S1F) we verified that three of the subtypes had 0 doublet rate according to DoubletFinder, while the other three had ~20% apparent doublet rates (Supplemental Figure 3D). This is partly because transitional cells are more prone to be computationally called doublets, yet we didn’t observe the expected number of called doublets involving distant and abundant cell types. All six subtypes express classic secretory cell markers EPCAM, PAX8, and KRT7 (Figure 3B, D). Using differentially expressed markers (Supplemental Table 5; Figure 3B, D) we deduced their potential functional roles.
Figure 3: Subtypes of non-ciliated secretory epithelial cells.
A. Focused re-clustering 14,285 non-ciliated secretory epithelial cells of healthy fallopian tubes identifies 6 subclusters, 2–1 to 2–6, shown in UMAP space.
B. Average expression levels and prevalence of major markers used to annotate the 6 non-ciliated secretory epithelial cell subtypes.
C. IF staining using antibodies against unique markers for non-ciliated secretory epithelial cell subtypes. Arrows indicate double positive cells
D. Expression levels of common markers for non-ciliated epithelial cells.
E-G. Expression levels of selected markers used to identify the 6 subtypes. E. Markers for NCSE 2–1 and 2–2. F. Makers for NCSE 2–3 and NCSE 2–5. G. Markers for NCSE 2–6.
NCSE2-1 cells account for 1.6–2% of the NCSEs (Supplemental Figure 3B), and express EMT markers ACTA2, ZEB1, SNAI, LGALS1, SPARC, and endothelial cell markers CAVIN2, PECAM1, VWF, ENG (Figure 3B, E). To examine whether the epithelial, EMT, and endothelial cell markers are co-expressed in a single cell, or in different cells, we evaluated pairwise co-expression patterns between EPCAM, ACTA2, and CAVIN2, over the individual NCSE cells (Supplemental Figure 3C) and confirm that EPCAM-ACTA2 and EPCAM-CAVIN2 are co-expressed in NCSE2-1 cells. Consistent with the rarity of NCSE2-1 in scRNA-seq data, EPCAM and CAVIN2 proteins are colocalized in a small fraction of cells in vivo (Figure 3C, first row). These EPCAM/CAVIN2 cells do not appear to be intercalated within the epithelium, rather they abut the epithelium and are close to blood vessels, suggesting that these cells may be an epithelial cell population in the process of transitioning to endothelial cells. To further validate NCSE2-1, we performed fluorescence activated cell sorting analysis of two normal fallopian tube samples using EPCAM and Endoglin (ENG, also known as CD105), a cell surface marker enriched in NCSE 2–1 (Figure 3E), since a CAVIN2 flow cytometry grade antibody is not available. Consistent with our scRNA-seq data, within the EPCAM+CD45− population we identify an EPCAM+/CD105+ double positive population (Supplemental Figure 3H, Gate II) that is absent in the flow minus one control (Supplemental Figure 3I, middle panel), validating the presence of NCSE 2–1 population using orthogonal approaches and markers.
Like NCSE2-1, NCSE2-2 cells express EMT markers ACTA2, COL1A1, MMP2, PRRX1, TWIST2, DES, DCN, FN1, ZEB1, TIMP3, LGALS1, SPARC (Figure 3C, E; Supplemental Table 5), but lack any terminal differentiation markers of epithelial cells, consistent with a progenitor-like cell. NCSE2-2 cells account for 5–8% of NCSE cells (Supplemental Figure 3B). To validate their presence in vivo, we confirmed the presence of EPCAM+/ACTA2+/RUNX3−/CD105− population by fluorescence activated cell sorting (Supplemental Figure 3H,I) and EPCAM/ACTA2 double positive cells by immunofluorescence in tissue cross-sections (Figure 3C, second row). The identification of a population with EMT marker expression in the fallopian tube led us to examine its distribution in a larger collection of 11 benign surgical samples varying by age and hormonal status (Note: premenopausal women covering broad phases of the menstrual cycle; see subject demographic and clinical information in Supplemental Table 2B). By immunofluorescence-based counting of EPCAM/ACTA2 double positive cells in pre- and post-menopausal samples, we found that these cells were present in all 3 anatomic segments, although their frequency varied across subjects representing different phases of the cycle and varied by anatomic segments (Supplemental Figure 3E). Despite the variability, samples from post-menopausal women appear to have fewer EPCAM/ACTA2 cells, except for one woman known to be using vaginal estrogen therapy (Supplemental Figure 3E; boxed). To assess if these cells might be directly regulated by estrogen or progesterone, we immuno-stained the tissue sections for ACTA2 and estrogen receptor (ESR) or progesterone receptor (PGR) and found very few ACTA2 cells co-expressing either ESR or PGR (Supplemental Figure 3F), consistent with the relatively low ESR and PGR RNA expression levels in NCSE2-2 (Supplemental Figure 3G). While these results suggest that the EPCAM/ACTA2 cell population is not directly regulated by estrogen or progesterone, it remains possible that the frequency of these cells is indirectly modulated by estrogen or other hormones.
NCSE2-3 to 2–5 are the most abundant subtypes, collectively comprising ~80% of NCSE (Supplemental Figure 3B). In addition to expressing PAX8, KRT7 and OVGP1, NCSE2-3 cells can be distinguished by expression of SOX17 and WNT-signaling regulator RSPO1 (Figure 3B, F). NCSE2-4 cells express genes related to ovarian cancer (LAMB3, MSLN), and LIF (Figure 3B), an interleukin 6 class cytokine essential for normal embryo implantation (Hassan et al., 2006; Ribeiro et al., 2018). Notably, NCSE2-5 cells express stem/progenitor markers such as LGR5, CD24, and TNFRSF19 (Figure 3F) (Blanpain et al., 2007; La Manno et al., 2018; Lamouille et al., 2014; Mabbott et al., 2013; Ng et al., 2014), as well as CD44 and ITGA6 (Figure 3F), markers previously used to define the peg cell population. This population may be hormonally regulated, due to its high expression levels of hormone receptors ESR, PGR, and AR (Supplemental Figure 3G), and CYP1B1 (Figure 3B), a P450 enzyme known to metabolize 17β-estradiol to a 2- and 4-OH estradiol.
Finally, NCSE2-6 cells express lymphoid lineage markers RUNX3, CD3E, PTPRC (Figure 3B, G), consistent with a basal or resident memory T-cells identity. In vivo, we validate the presence of EPCAM+/RUNX3+/CD45− cells by fluorescence activated cell sorting (Supplemental Figure 3H, Gate I; I) and by immunofluorescence (Figure 3C). In the epithelium, the EPCAM+/RUNX3+ cells appear round, and basally located with a cytoplasmic halo (Figure 3C), likely corresponding to the previously described basal cells (Peters, 1986)). In contrast to NCSE2-2, the percentage of NCSE2-6 cells in the 11-subject cohort appears to be relatively constant across premenopausal individuals regardless of menstrual cycle phase, but the number of cells were more variable and lower in post-menopausal women than premenopausal women (Supplemental Figure 3E).
Taken together, our analysis of ~16,500 NCSE and CE cells provided a fine-grained catalog of cellular diversity of the fallopian tube epithelium. The large number of high-quality cells allowed us to delineate 10 subtypes that would have been impossible to separate using previous methodology.
Subtle transcriptomic differences are observed across anatomic segments.
Although cell type composition is largely similar across the fallopian tube segments, we explored if there are between-segment transcriptomic differences within each cell type. We performed differential expression analysis for each defined cell type, and generally found very few genes with significant difference between segments (Supplemental Table 6). Nonetheless, the differentially expressed genes that did emerge reflect subtle differences between segments. For ciliated cells, the genes that show a higher expression in isthmus include those encoding the progesterone receptor (PGR), blood clotting component Fibrinogen Alpha Chain (FGA), apoptosis regulator Immediate Early Response 3 (IER3), receptor tyrosine kinase regulator Leucine-rich alpha-2-glycoprotein 1 (LRG1), and metabolic regulator thioredoxin-interacting protein (TXNIP) (examples shown in Supplemental Figure 4A). The higher expression of PGR in isthmus is notable, as progesterone is known to be required for the highly coordinated movements of cilia in the isthmus, which is responsible for embryo transfer to the uterus for implantation after fertilization occurs in the ampulla (Barton et al., 2021). For both CE and NCSE populations, the expression of stemness markers ALDH1A1 and ALDH1A2 tends to be higher in fimbriae. In NCSEs, some ovarian cancer-associated markers and genes regulating cellular redox homeostasis (e.g., BIRC3, FGA, IER3) are elevated in fimbriae (Supplemental Figure 4B; Supplemental Table 6), supporting the hypothesis that the cell of origin of ovarian cancer resides in the fimbriae, and highlighting the increased oxidative stress that occurs in the distal tube in response to ovulation (Sowamber et al., 2020). Further, the non-ciliated cells express higher levels of inflammatory cytokines CXCL1 (IL-1), CXCL8 (IL-8), and the sperm chemoattractant CCL20 in the fimbriae (Supplemental Figure 4A), consistent with previous reports (Duan et al., 2020; Palter et al., 2001). Taken together, although transcriptomic differences are subtle, the genes identified point to functional differences across segment compartments, serving as a baseline for future studies exploring the genes dynamically regulated to achieve normal reproduction or in response to disease.
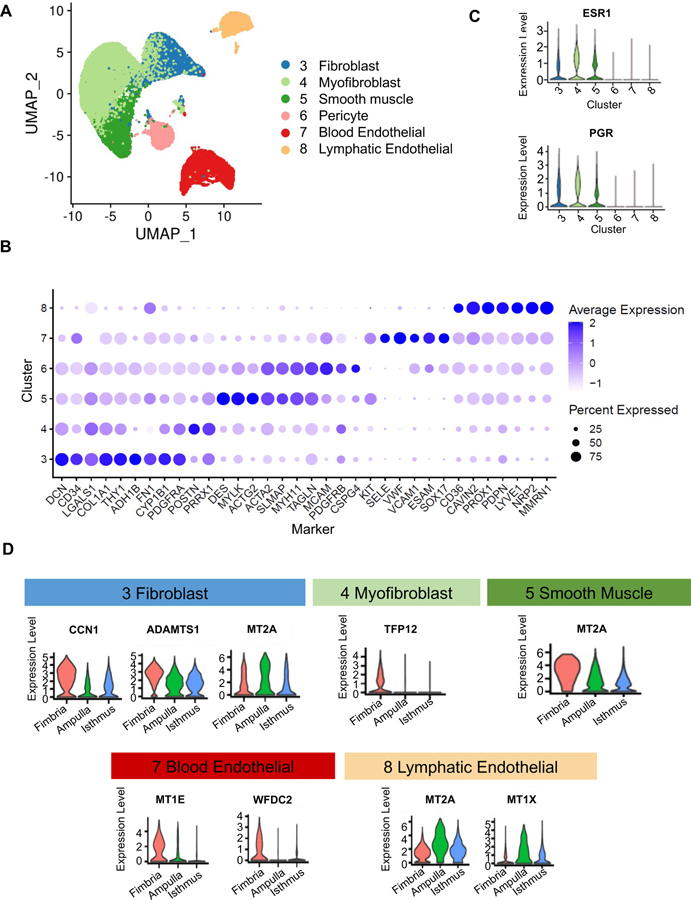
Identification of six stromal subtypes in human fallopian tube.
The global clustering (Figure 1B) involved ~26,400 stromal cells and identified six stromal cell types (re-projected in Figure 4A), which are evenly contributed by the three subjects (Supplemental Figure 5A). Cluster-specific genes (Figure 4B) allowed annotation of fibroblast (DCN, PDGFRA), myofibroblast (PRRX1, POSTN), and smooth muscle cell (ACTA2, MYH11); and we validated their presence in vivo using immunofluorescence or immunohistochemistry of representative markers (Supplemental Figure 5B). These three cell types appear to be the most similar to each other based on the location of the clusters in UMAP space (Figure 4A) and have the highest expression of hormone receptors among the stromal cells (Figure 4C), consistent with their function being influenced by hormonal status. The next three cell types are pericyte (MCAM, PDGFRB, CSPG4), blood endothelial (VWF, SELE), and lymphatic endothelial (PPROX1, LYVE1, CAVIN2) cells (Figure 4B), which we identified in vivo using MCAM, VWF, and CAVIN2 immunocytochemistry (Supplemental Figure 5B).
Figure 4: Stromal cell classification.
A. Visualization of 28,691 stromal cells in a UMAP projection of only the stromal cells, colored by the 6 stromal clusters from global clustering shown in Fig.1B
B. Average expression levels and prevalence of major markers used to annotate the stromal cell types.
C. Expression levels of genes encoding hormone receptors, ESR1 and PGR, in the 6 stromal cell types.
D. Expression levels of select markers differentially expressed across the three fallopian tube segments for different stromal clusters.
Like the epithelial cells, the stromal cell types display subtle differences in gene expression across segments (Figure 4D, Supplemental Table 6). For fibroblast cells, CCN1, a matrix-associated angiogenesis and cell proliferation regulator, is higher in the fimbriae, as expected given their dense vascularity. Additional genes that show segmental differences in specific cell types include serine protease inhibitor TFPI2 (myofibroblasts), metalloproteinase ADAMTS1 (fibroblast), stress proteins metallothionein 2A (MT2A) (smooth muscle), metallothionein 1X MT1X (lymphatic endothelial), and pro-angiogenic and immunosuppressive STAT3 pathway regulator WFDC2 (HE4) (expressed in pericytes and blood endothelial). Many of these genes are involved in ECM deposition or remodeling, suggesting possible differences in ECM composition across the tube. Taken together, the gene expression variation of the stromal cell populations across segments reveals a deeper complexity to gene regulation than previously appreciated.
Identification of multiple progenitor populations that possibly contribute to epithelial and stromal cell homeostasis.
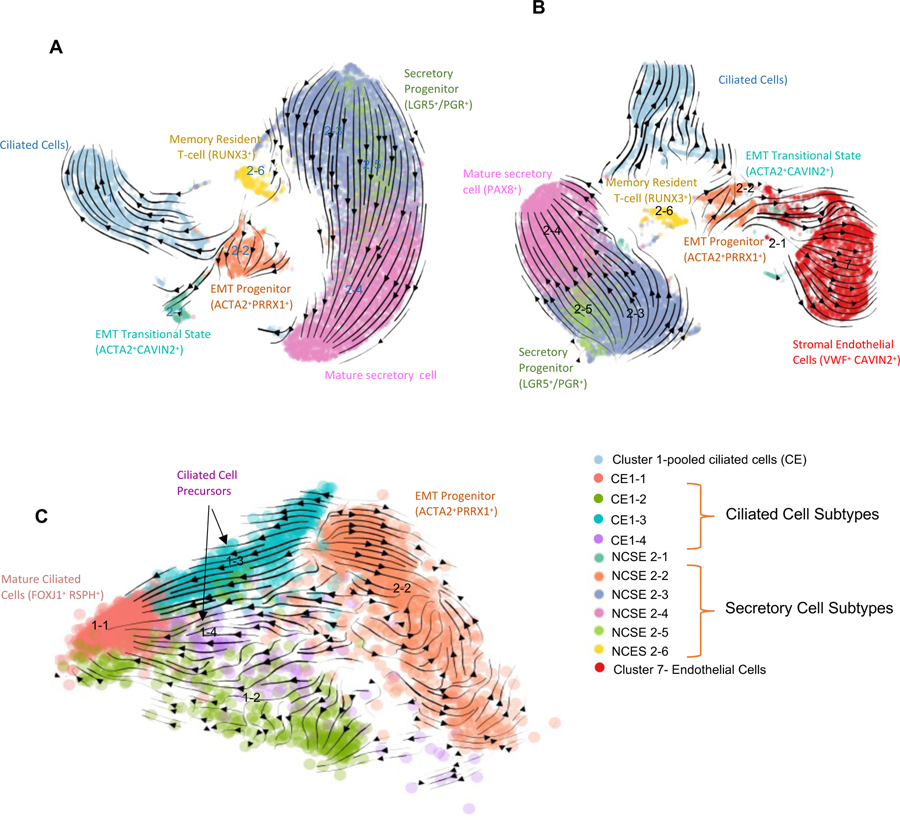
Marker genes of NCSE2-5 and NCSE2-2 suggest that these populations may be progenitor cell populations in the fallopian tube epithelium. Next, we asked whether these populations are (1) part of a single, continuous developmental trajectory, (2) can contribute to different epithelial cell subtypes, or (3) can contribute to some of the stromal cell populations. As shown above, cluster analysis (Figure 3A) revealed a discontinuous distribution, such that NCSE2-2 is not directly connected to other NCSE subtypes, and NCSE2-5 is linked to 2–3, 2–4, but not with other populations. To overcome the limitations of cluster analysis, we carried out RNA “velocity” analysis, which took advantage of the unspliced transcripts in scRNA-seq data to compare with the cell’s spliced transcripts (La Manno et al., 2018). The ratio of unspliced to spliced transcripts for each gene represent the difference between nascent and mature RNA and is used as a proxy measurement for the cell’s “velocity” in its differentiation trajectory, as depicted by the vectors in UMAP plots. First, when we jointly analyzed the six NCSE subtypes and the CE cells (Figure 5A), the velocity vectors for NCSE2-5 cells point unidirectionally towards NCSE2-4, a mature secretory cell population, while making no contribution to other epithelial cell types, suggesting that the LGR5+ NCSE2-5 cells are likely precursors for mature secretory cells. Running Velocity analysis for the 3 FT samples separately (Methods) did not alter the observed patterns (Supplemental Figure 4C). In contrast, NCSE2-2 cells, those with quasi-EMT features, contribute to NCSE2-1 cells, an intermediate between epithelial cells and endothelial cells (Figure 5B). This intermediate population subsequently projects to the blood endothelial cell population (cluster 7) in the stroma – raising the possibility that an epithelial progenitor undergoes complete EMT to give rise to a stromal cell type (Figure 5B).
Figure 5. Velocity analysis that uncovers two potential progenitor populations.
A. Velocity plot for CE and 6 subtypes of NCSE in UMAP view. Similar results were found when running Velocity analysis for the 3 FT samples separately while using common UMAP projections (Figure S4C. see Methods).
B. Velocity plot for CE, 6 subtypes of NCSE, and Blood endothelial cells (global cluster 7) in UMAP view.
C. Velocity plot for 4 subtypes of CE and NCSE 2–2 in PCA view.
See also Figure S4.
In addition to contributing to the stroma, NCSE2-2 cells also feed into the ciliated cells; thus, they are potentially responsible for the replenishment of ciliated cells. We repeated the velocity analysis by combining the NCSE2-2 cells with the ciliated cells, while distinguishing the 4 CE subtypes (Figure 5C). NCSE2-2 cells project to CE1-4, which in turn project to CE1-1, suggesting that CE1-4 are descendants of NCSE2-2, while at the same time represent precursors of CE1-1, the more mature ciliated cells. Marker genes of CE1-4 also supports its role as an intermediate population: they retain residual expression of EMT markers and lower levels of terminally differentiated ciliated cell markers such as FOXJ1, CAPS, RSPH genes, DNAH11 (Figure 2B–F). Curiously, we also observe strong flow from CE1-3 to CE1-4, suggesting a parallel route to CE1-4 cells, although the source of CE1-3 cells remains unclear.
In sum, our trajectory analysis suggests that the fallopian tube has multiple progenitor populations that maintain multiple epithelial and stromal cell subpopulations in this dynamically regulated organ. Future spatial mapping and lineage tracing studies are needed to fully validate these putative developmental relationships.
The hydrosalpinx tube had altered cell type proportions and cell states.
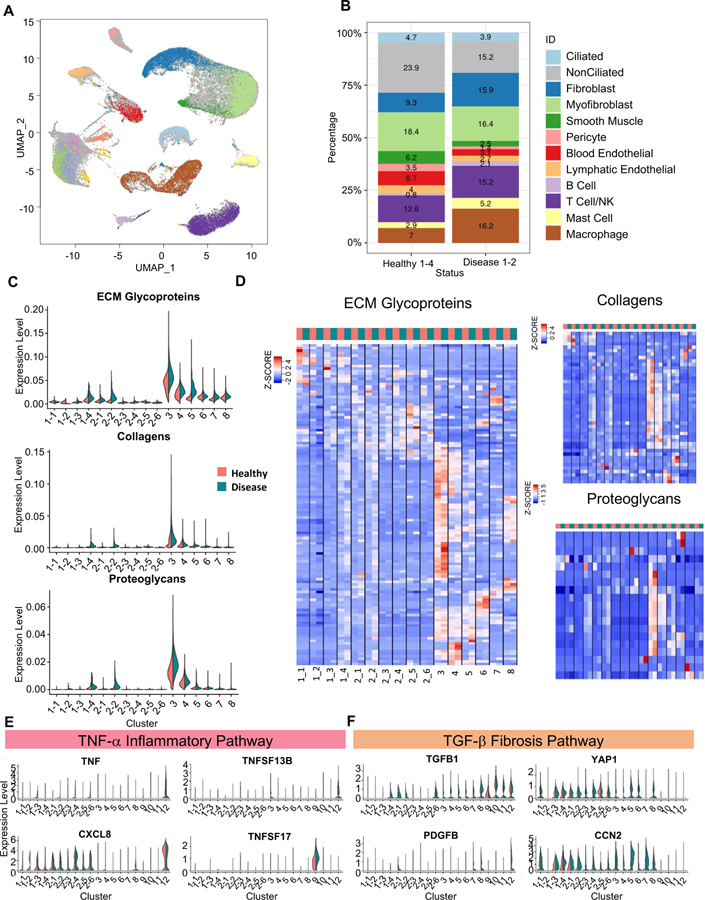
In ultrastructure studies, the hydrosalpinx tube appears as a denuded epithelium with loss of cilia and microvilli, containing disordered edematous stroma with atrophic and distorted muscle fibers, and engorged blood vessels (Ajonuma et al., 2005). To investigate the cellular changes accompanying the abnormal structure and function of hydrosalpinx we performed scRNA-seq on surgically removed hydrosalpinx tubes from two individuals (FT5, FT6) with tubal factor infertility and without evidence of endometriosis. Supervised assignment of the ~18K cells from the diseased samples reveals the same 12 clusters found in the healthy tubes, with no additional cell types (Figure 6A). Comparison of the relative cell number across the 12 cell types found that the hydrosalpinx tubes had fewer ciliated and non-ciliated epithelial cells than the healthy tubes (Figure 6B). Among the stroma and immune cell types, fibroblast and macrophage populations are expanded, while the smooth muscle, pericyte, and endothelial cell populations are reduced (Figure 6B), indicating drastic tissue remodeling in the disease state.
Figure 6: Comparison of epithelial and stromal populations between healthy and disease samples.
A. Visualization of cells from two diseased fallopian tube samples (FT5 and FT6) in a global UMAP projection, colored by supervised assignment of the FT5 and FT6 cells into clusters found for the healthy samples. Cells from healthy samples are colored as grey background.
B. Composition of the 12 cell types, compared between the 4 healthy samples and the 2 disease samples.
C. Expression level distribution of three ECM-related gene sets, for the 4 CE subtypes. 6 NCSE subtypes, and 6 stromal cell types, compared between healthy (red) and disease (blue) samples.
D. Heatmaps of expression levels of three sets of genes, for ECM glycoproteins, collagens, and proteoglycans, respectively, compared across cell types and between healthy (red) and disease (blue) samples. Average expression of each gene is calculated for each cell type (i.e., the centroid), then standardized over the centroids shown.
E, F. Expression levels of genes in TNFα pathway (E) and TGFβ pathway (F), compared across cell types and between healthy (red) and disease (blue) samples.
See also Figure S6, Table S2, Table S7, Table S8 and Table S9.
Next, we performed differential expression analysis between the healthy and disease samples within each epithelial and stromal cell type. Many of the genes upregulated in the disease state were involved in cell growth and proliferation (see Supplemental Table 7); and pathway analyses highlighted ERK1/ERK2 and the WNT signaling pathways (Supplemental Table 8). Consistent with the gene and pathway results, the percentage of MKI67+ cells were higher in the diseased than in the healthy tissue (Supplemental Figure 6A), reflecting a compensatory mechanism of higher proliferation in multiple epithelial subtypes after the loss of CE and NCSE cells (Figure 6B). Among the differentially expressed genes, many markers previously associated with adult somatic and cancer stem cell populations (ALDH1 and 2, THY1, LGR5, PDGFRA, PDGFRB), and EMT markers (ZEB1, LIF, POSTN, LGALS, MMP2, ACTA2) are elevated in the disease state (Supplemental Figure 6B). In particular, the expression of LGR5, which marks NCSE2-5 – an epithelial cell progenitor, is lower in the diseased sample, whereas levels of THY1 and PDGFRA, markers of mesenchymal stemness, and ALDH, are higher in the diseased state (Figure 3B, Supplemental Figure 6B). Importantly, these markers are generally higher in the NCSE2-2 progenitor population, suggesting that the balance between the two progenitor populations, NCSE2-5 and NCSE 2–2, is altered in the hydrosalpinx tubes. Hydrosalpinx by itself has not been recognized as an ovarian cancer risk factor; but our data revealed changes in EMT and progenitor signature, thus possibly consistent with an elevated risk of cell transformation and tumorigenesis. This is supported by an increased expression of ovarian cancer markers in the fallopian tube (Supplemental Figure 6C).
In addition to the differences in progenitor markers, the differentially expressed genes revealed changes in the complement pathway, cellular redox, and WNT signaling pathways (Supplemental Table 8), consistent with previous analyses of hydrosalpinx. Specifically, the development of tubal disease and ultimately hydrosalpinx are associated with an inflammatory response to urogenital pathogens via the TNF-α pathway (Morales et al., 2006; Rasmussen et al., 1997; Xu et al., 2019). Proteomics analysis of tubal fluid uncovered abnormalities in the complement and redox pathways (Yohannes et al., 2019). We did not observe in the disease state an increase in TNF, TNFSF13B, or its receptor TNFRSF17 (Figure 6E), reported to be elevated in cases of salpingitis, or in active infections of the tube (Xu et al., 2019). However, the expression of CXCL8, an inflammatory cytokine involved in the immune response for urogenital infections, is increased in several cell types in the disease state (Figure 6E).
Unexpectedly, the hydrosalpinx sample showed differences in genes associated with cell-to-cell adhesion and ECM-related processes. Drawing from earlier work that quantified molecular changes in chronic kidney disease and fibrosis (Kuppe et al., 2021), we calculated the average expression for each of three sets of genes associated with ECM glycoproteins, collagen, and proteoglycan production, respectively, for each epithelial and stromal subtypes and compared between healthy and disease samples (Figure 6C). As expected, the overall expression of the three ECM related gene sets is higher in the stromal cell types than in the epithelial cell subtypes. However, while CE1-4, NSCE2-1 and NCSE2-2 express ECM glycoproteins, collagen, and proteoglycan at low levels, their levels are higher in the disease state (Figure 6C). By displaying gene expression heatmaps for the cell type centroids and individual genes, we observed finer patterns of heterogeneity involving cell type-specific changes, and specifically in subsets of genes in the three gene sets (Figure 6D; Supplemental Table 9).
Changes in ECM genes in hydrosalpinx raised the possibility that it represents a fibrotic disease. Fibrosis is a known consequence of tissue damage and has been observed in multiple organs. But its underlying molecular and cellular processes in the fallopian tube are unknown (Ajonuma et al., 2005). The TGF-β pathway has been proposed as a main driver of fibrosis in multiple disease processes in many tissues (Biernacka et al., 2011). Consistent with the fibrosis model, the hydrosalpinx sample show increased expression of TGF-β pathway effector genes TGFB1, PDGFB, CCN2 (CTGF), and YAP1 in the epithelial and stromal cells (Figure 6F). These findings suggest that hydrosalpinx may share a similar mechanism with pathogenic fibrosis, and could potentially benefit from therapies developed for other tissues (Pinto et al., 2017). Unlike previous studies, our scRNA-seq data reveal cell type-specific changes in hydrosalpinx, serving as a resource for identifying more specific biomarkers for earlier intervention and development of targeted therapies.
DISCUSSION
A refined cell atlas of the pre-menopausal fallopian tube
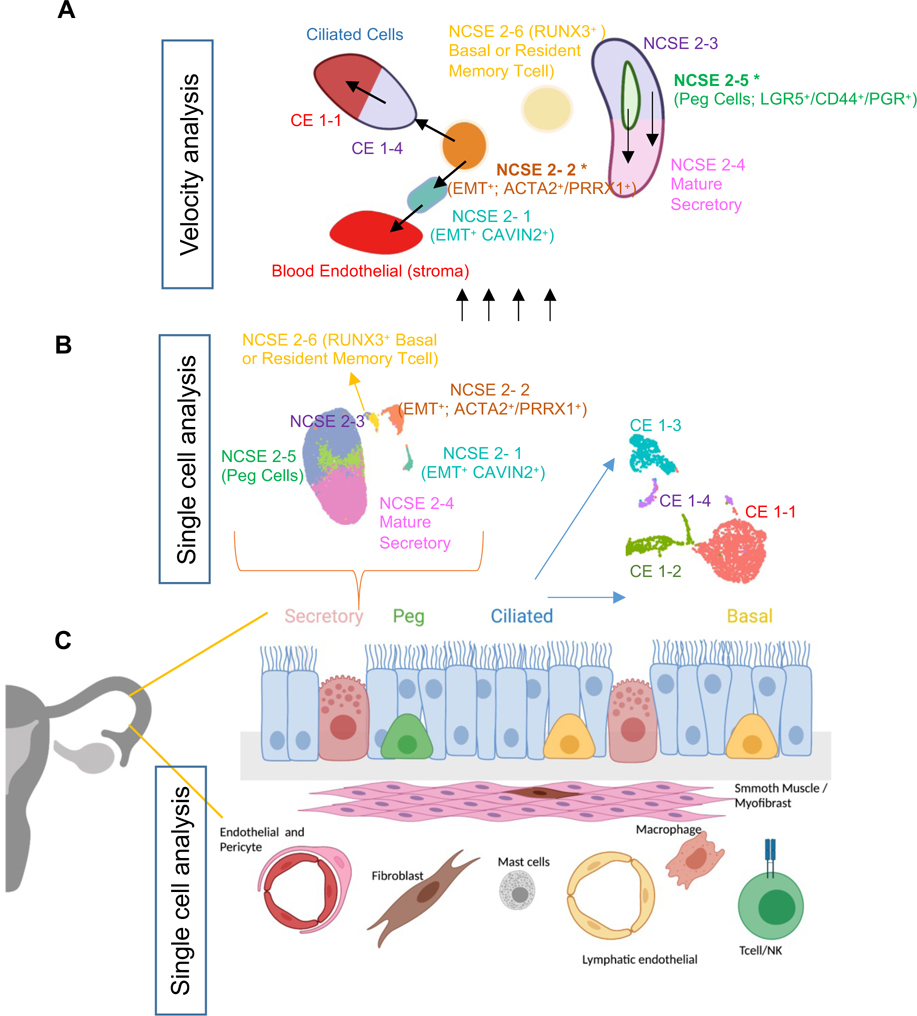
We used single-cell RNA-seq to establish a new catalog of cell types, subtypes, and mRNA markers for the healthy human fallopian tube, and identified 12 major cell types (Figure 7C). While earlier studies have documented four epithelial cell subtypes: ciliated, secretory, peg, and basal cells, the large cell number and relatively high sequencing depth of our data allowed us to identify 10 subtypes of epithelial cells (Figure 7B), thus providing a more refined view of the functional heterogeneity of the pre-menopausal fallopian tube. In particular, the gene expression profiles of these subtypes give a glimpse of multiple progenitor cell populations and their associated transitional or mature cell populations. First, we identified two secretory cell progenitors: (1) an epithelial progenitor (LGR5/PGR) population, NCSE2-5, predicted to give rise to mature secretory cells (i.e., NCSE2-4), and (2) a bipotential progenitor population, NCSE2-2, expressing several EMT markers, predicted to generate mature ciliated epithelial cells CE1-4 on one hand, and the blood endothelial cells in the stromal compartment on the other (Figure 7A).
Figure 7: Model for epithelial differentiation trajectory.
A. Illustration of the differentiation trajectories of NCSE cells originated from two secretory cell progenitors (NCSE2-2 (ACTA2/PRRX1) and NCSE2-5 (LGR5/PGR)) revealed by velocity analysis.
B. scRNA-seq of thousands of fallopian tube cells expands cellular taxonomy of the epithelial cell subtypes from four to ten.
C. Schematic of 12 major cell types for healthy human fallopian tubes established by scRNA-seq analysis.
See also Figure S7.
The identification of an EMT expressing progenitor cell population in a simple epithelium is unexpected, since these tissues are believed to be maintained by epithelial stem cells (e.g. LGR5+ cells), which are reliant on active WNT signaling (Clevers, 2013). Therefore, in a simple epithelium, the activation of an EMT program is typically only observed in response to wound healing, or during cancer invasion and metastasis in tissue (Nieto et al., 2016). Recently, a study by Hu et. al., analyzed ~1,800 epithelial cells sorted from pre- and post-menopausal fallopian tube samples from ovarian cancer or endometrium cancer patients, and also identified a small number of cells (n~20) bearing an EMT signature (Hu et al., 2020), suggesting that this population can be present in the healthy epithelium, and may be a population vulnerable for cancer initiation. Therefore, the presence of an EMT progenitor pool in the healthy epithelium, reported by us and Hu et. al., raises a question as to how this population is regulated to prevent oncogenic transformation. Curiously, in our dataset we find that the EMT progenitor population (NSCE2-2) expresses PRRX1 (Figure 3F), a homeobox factor gene and strong EMT inducer. The loss of PRRX1 leads to tumor initiation and metastasis (Ocaña et al., 2012). Thus, it is possible that in the fallopian tube, PRRX1 serves as a gatekeeper in NSCE2-2 cells, preventing these EMT type cells from adopting a pro-oncogenic phenotype, which could lead to tumor initiation, or even subsequent metastasis. Future studies are needed to validate the functional roles of the two progenitor pools in the healthy and disease state by combining genetic reporters with lineage tracing, preferably at the single-cell level.
A recent study by Dinh et. al. profiled human fallopian tube samples using 10X Chromium (Dinh et al., 2021). The authors analyzed >50,000 cells, albeit with a shallower sequencing depth than in our dataset (an average of ~850 detected genes per cell, compared to 2,600 in our study), and a wider heterogeneity among samples, which contributed highly varied combinations of major cell types. Similar to our study, Dinh et al. reported several subtypes of ciliated and non-ciliated epithelial cell subtypes that correlated with our epithelial cell subtypes (Supplemental Figure 7A). Furthermore, a comparison with a smaller Smart-Seq2 dataset from Hu et. al. also showed that CE cells are most similar to the ciliated cells in Hu et al., while our NCSE cells correspond to the non-ciliated subtypes in that study (Supplemental Figure 7B). However, Dinh et. al. identified two transitional epithelial populations using pseudotime analysis, one of which was predicted as a RUNX3+ progenitor population. However, our data do not support such a model for several reasons. First, in our data, RUNX3 is one of the most distinctive markers for the T/NK cells and is expressed at much lower levels in the other 11 cell types, including the ciliated and non-ciliated epithelial cells (Figure 1D). Second, of the four CE subtypes and 6 NCSE subtypes in our data, RUNX3 is expressed highest in NCSE2-6 (Figure 3G), but NCSE2-6 is not one of the predicted progenitor cells. In our velocity analysis, the RUNX3-expressing epithelial cells, NCSE2-6, did not contribute to any other NCSE or CE populations. Genes highly expressed in this population are more consistent with resident memory T-cells (Ardighieri et al., 2015; Hu et al., 2020). In fact, the Dinh et. al transitional population had the highest expression of markers of immune cell populations (T and NK cells, Mast cells, and macrophage: Supplemental Figure 7C). In short, our data do not support the existence of a RUNX3-marked progenitor population.
Next, we sought to understand whether hormonal states could influence the progenitor populations we identified. Since high-quality LGR5+ antibodies are not available, we could not monitor the protein expression and cell number for NCSE2-5 across the samples covering the cycle. However, we were able to count the number of putative NCSE2-2 cells by an antibody for ACTA2 and compare its frequency across the menstrual cycle for pre-menopausal women, and between pre- and post-menopausal women. The numbers of ACTA2+ cells were higher in pre-menopausal women than in post-menopausal women (Supplemental Figure 3E), except for one woman on postmenopausal hormone therapy, suggesting that the hormonal milieu may regulate the size of this population. However, the number of ACTA2+ cells that co-express PGR or ESR were very small, suggesting there may be some aspects of the hormonal state that indirectly drives the expansion of this population rather than a direct hormone effect. Future studies should include a larger number of benign samples at varying points in the menstrual cycle to examine this question more carefully.
Potential role of TGF-β mediated tissue fibrosis in hydrosalpinx
Our analysis of the hydrosalpinx sample revealed changes in both tissue composition (i.e., cell number, Figure 6B, S6A, S6D) and regulatory states (Figure 6C–F, S6B–C). Importantly we identified significant changes in progenitor cell markers between the healthy and disease state. For example, multiple EMT markers, such as THY1, PRRX1, and LGALS1, are elevated in the disease state; and we consistently find a higher number of NCSE2-2 cells in hydrosalpinx (Supplemental Figure 6B, D). In contrast to EMT markers, LGR5 – an epithelial progenitor marker- levels are significantly reduced in NCSE2-5 cells (Figure S6B). However, this is not accompanied by a decrease in NCSE2-5 cell number (Supplemental Figure 6D), but rather a rewiring of the transcriptional regulatory network in the disease state.
In addition to changes in progenitor cells, the epithelial and stromal populations express higher levels ECM-related genes (Figure 6C), which led us to explore whether the hydrosalpinx disease state may have fibrosis-related tissue damage. Fibrosis has been described for the lung epithelium in idiopathic pulmonary fibrosis (IPF) and many organs in response to tissue damage, whereby epithelial injury activates inflammatory cells, which in turn activate fibroblasts and induce EMT in epithelial cells, leading to the uncontrolled deposition of ECM with abnormal remodeling, and ultimately organ failure (Pinto et al., 2017). In animal models of fibrosis the initial insult is often a brief inflammatory response mediated through the TNF-α pathway, before ultimately progressing to a longer term TGF-β mediated fibrosis (Meng et al., 2016; Morales et al., 2006; Palter et al., 2001; Xu et al., 2019). We have also evaluated a representative sample of the normal and abnormal fallopian tube with H&E staining to evaluate for evidence of inflammatory changes. This revealed the absence of inflammatory changes in the “healthy” samples (FT3, P9) with only one rare plasma cell noted in one sample and clear evidence of widespread inflammatory changes in the hydrosalpinx samples with multiple, frequently observed plasma cells (Supplemental Figure 6E). In our study we did not observe significant differences in TNF-α related genes, but did detect transcriptomic changes in TGFB1, PDGFB, CCN2, and YAP1 in the hydrosalpinx (Figure 6E, F), suggesting that we may have captured a more advanced disease state. To fully test this model would require multiple hydrosalpinx samples at varying time points of disease progression. Nevertheless, several clinical trials are underway for medications targeting individual steps in this fibrosis pathway (Pinto et al., 2017). It is possible that some of these medications could be leveraged as a new pharmacologic therapy for hydrosalpinx and/or tubal factor infertility.
In summary, by sequencing >59K cells from three tubal segments in four healthy subjects we produced a detailed cell atlas of the human fallopian tube, allowing comparisons across anatomic segments of the tube. The identification of 10 subtypes of epithelial cells advanced our knowledge of the maintenance and dynamic regulation of the FT epithelium and established a baseline to study changes of cell number and cell state across many types of biological variation: among tubal segments, during the hormone cycle, from pre-menopausal to peri- and post-menopausal, and for diseases such as tubal factor infertility and ovarian cancer. Here we analyzed hydrosalpinx as one example of such comparisons; and the same approach can be extended to other variations of interest. Furthermore, by using computational trajectory analysis we were able to extract information regarding the direction of cell differentiation, and this allowed us to define multiple putative progenitor cells, which will likely be a focus of future studies of tissue homeostatic as well as its dysregulation in diseases.
LIMITATIONS
Given the rise in COVID cases and the halt of elective surgeries at Michigan Medicine, procuring additional surgical specimens has been challenging. As a result, the CE subtypes identified by scRNAseq data were only validated by immunofluorescence from existing formalin fixed paraffin embedded tissue samples. We unfortunately did not have the opportunity to validate CE subtypes by flow cytometry, which was done for NCSE subtypes. Another limitation of the current work is that our Velocity analysis is based 3’ end single-cell RNA-sequencing technologies; the inferred differentiation trajectories can be refined in the future by using full-length single-cell RNAseq. Nevertheless, the patterns observed are reproducible across the individual patient samples analyzed. Future expansions of the reference cell atlas will allow a comparative analysis of cell types, states, and regulation dynamics across the menstrual cycle, and between pre- and postmenopausal subjects.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for reagents should be directed to and will be fulfilled by the lead contact S. Sue Hammoud (hammou@med.umich.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Single-cell RNA-seq data have been deposited at GEO and HCA and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. The data visualization is available in the interactive explorer cellxgene. Microscopy data reported in this paper will be shared by the lead contact upon request.
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti CD326-Brillianat Violet 510 (EPCAM) | Biolegend | Cat#324235; PRID: AB_2632934 |
| Mouse monoclonal anti CD45- PE/Cy7 | Biolegend | Cat#368531; PRID: AB_2715891 |
| Mouse monoclonal anti alpha-Smooth Muscle Actin-APC | Novus Biologicals | Cat#NBP2-47698APC; PRID: N/A |
| Mouse monoclonal anti ENG (CD105)-PE | Abcam | Cat#ab53321; PRID: AB_868769 |
| Mouse monoclonal anti RUNX3-Alexa 488 | R&D Systems | Cat#IC3765G-100UG; PRID: N/A |
| Mouse monoclonal anti mouse IgG2b, κ isotype control antibody-Brilliant Violet 510 | Biolegend | Cat#400345; PRID: N/A |
| Mouse monoclonal anti mouse IgG2a, κ isotype control antibody-Alexa 488 | Biolegend | Cat#400233; PRID: N/A |
| Rat monoclonal anti mouse IgG2a antibody-APC | Biolegend | Cat#407109; PRID: AB_2561754 |
| Mouse monoclonal anti mouse IgG1, κ isotype control antibody-PE/Cy7 | Biolegend | Cat#400126; PRID: AB_326448 |
| Mouse monoclonal anti mouse IgG2a, κ isotype control (FC) antibody-PE | Biolegend | Cat#400213; PRID: AB_2800438 |
| Mouse monoclonal anti FOXJ1 | Invitrogen | Cat#14-9965-82; PRID: AB_1548835 |
| Mouse monoclonal anti FOXJ1 | Sigma | Cat#AMAB91254; PRID: AB_2665866 |
| Rabbit polyclonal anti PAX8 | Proteintech | Cat#10336-1-AP; PRID: AB_2236705 |
| Rabbit monoclonal anti PAX8 | Abcam | Cat#ab239363; PRID: N/A |
| Rabbit monoclonal anti EPCAM-Alexa 488 | Abcam | Cat#ab237395; PRID: N/A |
| Rabbit polyclonal anti CAPS | Sigma | Cat#HPA043520; PRID: AB_10964138 |
| Mouse monoclonal anti alpha-Smooth Muscle Actin (ACTA2)-594 | Abcam | Cat#ab202368; PRID: N/A |
| Rabbit polyclonal anti CAVIN 2 (SDPR) | Proteintech | Cat#12339-1-AP; PRID: AB_2183305 |
| Rat monoclonal anti CD44 | Invitrogen | Cat#14-0441-82; PRID: AB_467246 |
| Mouse monoclonal anti Estrogen receptor 1 (ESR1) | Invitrogen | Cat#MA5-13191; PRID: AB_10986080 |
| Rabbit polyclonal anti Progesterone receptor (PGR) | Sigma | Cat#HPA008428; PRID: AB_1079680 |
| Rabbit polyclonal anti RUNX3 | Sigma | Cat#HPA059006; PRID: AB_2683877 |
| Rabbit monoclonal anti CD34 | Abcam | Cat#ab81289; PRID: AB_1640331 |
| Rabbit polyclonal anti LGALS1 | Sigma | Cat#HPA000646; PRID: AB_1078935 |
| Rabbit monoclonal anti CDKN2A | Abcam | Cat#ab108349; PRID: AB_10858268 |
| Biological samples | ||
| Adult Human 1 (FT1, Age 52 yrs) Fallopian Tube Tissue For single cell RNAseq and statining |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 2 (FT2, Age 30 yrs) Fallopian Tube Tissue For single cell RNAseq |
International Institute for the Advancement of Medicine | N/A |
| Adult Human 3 (FT3, Age 46 yrs) Fallopian Tube Tissue For single cell RNAseq, staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 4 (FT4, Age 31 yrs) Fallopian Tube Tissue For single cell RNAseq |
International Institute for the Advancement of Medicine | N/A |
| Adult Human 5 (FT5, Age 34 yrs) Fallopian Tube Tissue For single cell RNAseq, staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 6 (FT6, Age 32 yrs) Fallopian Tube Tissue For single cell RNAseq |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 7 (Age 71 yrs) Fallopian Tube Tissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 8 (Age 52 yrs) Fallopian Tube Tissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 9 (Age 72 yrs) Fallopian Tube Tissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 10 (Age 68 yrs) Fallopian TubeTissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 11 (Age 58 yrs) Fallopian Tube Tissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 12 (Age 47 yrs) Fallopian Tube Tissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 13 (Age 36 yrs) Fallopian Tube Tissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 14 (Age 49 yrs) Fallopian Tube Tissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 15 (Age 36 yrs) Fallopian Tube Tissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 16 (Age 41 yrs) Fallopian Tube Tissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 17 (Age 38 yrs) Fallopian Tube Tissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Proteinase | Sigma | Cat#P5147-1G |
| Deoxyribonuclease I | Sigma | Cat#4716728001 |
| Collagenase D | Sigma/Roche | Cat#11088866001 |
| Hyalurondase | Sigma | Cat#H3884-1G |
| HBSS | Invitrogen | Cat#14025092 |
| DAPI | Sigma | Cat#D9542 |
| FBS | Sigma | Cat#F4135 |
| Vectashield Antifade Mounting Medium | Vector Laboratories | Cat#H-1000-10 |
| Tween-20 | Sigma | Cat# P1379 |
| Triton X-100 | Sigma | Cat# T8787 |
| BSA | Sigma | Cat# A7030 |
| Paraformaldehyde | MilliporeSigma | Cat# 818715 |
| Bloxall Endogenous Peroxidase and Alkaline Phosphatase Blocking Solution | Vector Laboratories | Cat# SP-6000 |
| Sucrose | Sigma | Cat# S9378 |
| Citric Acid | Sigma | Cat#791725 |
| SignalBoostImmunoreaction Enhancer Kit | MilliporeSigma | Cat#407207 |
| EasySep RBC Depletion Reagent | Stemcell Technologies | Cat#18170 |
| Bacto™ FA Buffer | BD Biosciences | Cat#223142 |
| Deposited data | ||
| Raw data files for scRNA-seq | This paper or NCBI Sequence Read Archive or Human Cell Atlas | SRA: SRP323939; HCA: https://data.humancellatlas.org/explore/projects/21ea8ddb-525f-4f1f-a820-31f0360399a2 |
| Gene expression data for scRNA-seq | This paper or NCBI Gene Expression Omnibus or Human Cell Atlas | GEO: GSE178101; HCA: https://data.humancellatlas.org/explore/projects/21ea8ddb-525f-4f1f-a820-31f0360399a2 |
| Data visualization for scRNA-seq | This paper or cellxgene | https://cellxgene.cziscience.com/collections/fc77d2ae-247d-44d7-aa24-3f4859254c2c |
| Deposited code | Zenodo or Github | DOI:10.5281/zenodo.5883840; https://github.com/qianqianshao/FallopianTube/ |
| Software and algorithms | ||
| Cell Ranger (v4.0.0) | https://github.com/10XGenomics/cellranger | |
| SoupX (v1.4.5) | Young and Behjati, 2020 | https://github.com/constantAmateur/SoupX |
| DoubletFinder (v2.0.3) | McGinnis et al., 2019 | https://github.com/chris-mcginnis-ucsf/DoubletFinder |
| Seurat (v3.2.3) | Satija et al., 2019 | https://github.com/satijalab/seurat |
| SeuratWrappers (v0.1.0) | https://github.com/satijalab/seurat-wrappers | |
| velocyto.py (v0.17.17) | Manno et at., 2018 | https://github.com/velocyto-team/velocyto.py |
| scVelo (v0.2.3) | Bergen et al., 2020 | https://github.com/theislab/scvelo |
| LRpath | Kim et al., 2012 | http://lrpath.ncibi.org/ |
All original code has been deposited at Zenodo and GitHUB. all codes are publicly available as of the date of publication. DOIs and links are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Healthy human specimens collection process
Fallopian tube samples were obtained from four female subjects of premenopausal ages (30–52). Detailed information about the subjects is in Supplemental Table 2A. Two subjects were undergoing surgery for benign indications. Prior to surgery, the subjects were enrolled in the University of Michigan Reproductive Subject Registry and Sample Repository (Schon et al., 2021). The collection of fresh tissue was approved by the University of Michigan Institutional Review Board (HUM00167998). In accordance with the standard procedures, the donors were prepped for organ recovery while maintained on a ventilator. During the procedure the sternum was opened to enable access to the thoracic and abdominal cavities, which were packed with ice. The ventilator was turned off prior to aortic cross-clamp and an arterial line was opened to start the flow of one of three preservation media (Belzer UW®Cold Storage Solution (Bridge of Life, SC, USA); Custodiol® HTK (Histidine-Tryptophan-Ketoglutarate) Solution (Essential Pharmaceuticals, NC, USA); or SPS-1 Static Preservation Solution (Organ Recovery Systems, IL, US)). This serves to flush the organs and maintain their viability during recovery.
The two cadaveric samples were collected as whole organs from medically healthy subjects procured by the International Institute for the Advancement of Medicine (IIAM) through standard procedures. Based on the information provided by IIAM these donors had 46XX karyotypes, negative for infection and pregnancy screens, and were without known disease in the reproductive system such as endometriosis, significant uterine surgery, systemic disease, or previous obstetric complications. Fallopian tubes were surgically removed from deceased donors and packed on ice using one of the tissue preservation media. They were shipped and delivered to the lab within 24 hours and immediately processed for cell dissociation.
Hydrosalpinx specimen collection process
Two fallopian tube samples were obtained from premenopausal subjects ages 32 and 34 with tubal factor infertility and known hydrosalpinx. Detailed information about the subjects is in Supplemental Table 2A. The abnormal fallopian tubes were removed prior to planned fertility treatment. Prior to surgery, the subjects were enrolled in the University of Michigan Reproductive Subject Registry and Sample Repository (Schon et al., 2021). The collection of fresh tissue was approved by the University of Michigan Institutional Review Board (HUM00167998).
Ethical approval process for cadaveric samples
The IIAM procures tissue and organs for non-clinical research from Organ Procurement Organizations (OPOs), which comply with state Uniform Anatomical Gift Acts, are certified and regulated by the Centers for Medicare and Medicaid Services. These OPOs are members of the Organ Procurement and Transplantation Network and the United Network for Organ Sharing (UNOS), and operate under a set of standards established by the Association of Organ Procurement Organizations and UNOS. Written informed assent from the next-of-kin of the deceased donor was obtained. A biomaterial transfer agreement is in place between IIAM and the authors that restricts the use of the tissue for pre-clinical research. The use of deceased donor tissue in this research is categorized as ‘not regulated’, per 45 CFR 46.102 and the ‘Common Rule’, as it does not involve human subjects and complies with the University of Michigan’s IRB requirements as such.
METHOD DETAILS
Tissue processing and preparation for single-cell RNA sequencing
Surgical samples were collected in Hank’s Balanced Salt Solution (HBSS) in the operating room. The samples were transported at room temperature to the lab and processed immediately for dissociation. The cadaveric samples were processed within 24–36 hours of removal due to logistical constraints, including travel and availability of core laboratory services (see Supplemental Table 2). If a delay in processing for dissociation was needed, the sample was maintained at 4°C in tissue preservation media (UW or HTK). For most samples, the fallopian tube was dissected into three anatomic segments (isthmus, ampulla, fimbria), and bivalved prior to dissociation and processing. For the ampulla and isthmus samples, the tissue was initially incubated in 1.8 mg/ml pronase (Sigma P5147, diluted in Opti-mem, Gibco 31985-062; 10 ml solution and 100–200 mg of tissue per tube) in the 37°C shaker for 5–10min. Samples were filtered using a 100 micron strainer, and tissue sections were transferred to a digestion buffer containing 1 mg/ml collagenase D (Roche 11088866001), 1.5 mg/ml hyaluronidase (Sigma H3884), and 2U/ml DNase I in HBSS, and incubated for 30 minutes before quenched with 10% fetal bovine serum (FBS). The undigested tissue was used for a second round of digest and quenching. Dissociated cells were washed in 10% FBS/DMEM, centrifuged at 400g for 4 minutes, and resuspended in 100 μl 10% FBS/DMEM. For fimbriae samples, tissue processing did not include the pronase step. Cell suspensions collected from both digestion steps were combined, and treated with EasySep RBC depletion reagent (Stemcell Technologies #18170) according to the manufacturer’s instruction. After centrifugation at 400g for 4 minutes, cells were washed with 0.04% BSA/PBS 2–3 times and filtered through a 40 μm filter, centrifuged at 400g for 4 minutes, and resuspended in 1 ml 10% FBS/DMEM. Live cells were collected by flow cytometry, and submitted to the University of Michigan Advanced Genomics Core for processing on the 10X Genomics Chromium platform. Sequencing libraries were sequenced by Illumina NovaSeq to create 151-nt reads for the transcript.
FFPE Immunofluorescence
Paraffin sections from the three segments of 11 pre- and post-menopausal human fallopian tubes were obtained from the University of Michigan Reproductive Subject Registry and Sample Repository (See Supplemental Table 2B for patient information – Note patient samples include sequenced and non-sequenced individuals). Five-micron FFPE tissue sections were deparaffinized in Histoclear 3x for 5 minutes, followed by serial EtOH washes, from 100% to 30%, 3 minutes each, and by deionized water 2x for 3 min. Tissue sections were permeabilized by incubation in 0.1% Triton in PBS for 15 minutes. For all antibodies, antigen retrieval was performed by boiling in 10mM sodium citrate, pH 6.0 for 30 minutes. Sections were blocked in 1xPBS supplemented with 3% BSA and 500mM glycine for two hours at room temperature. Endogenous peroxidases and alkaline phosphatases were blocked by a ten-minute incubation in BloxAll solution (VectorLabs, Cat. No: SP-6000). The primary antibodies used are FOXJ1 (Invitrogen 14-9965-82, 1:500; Sigma AMAB91254, 1:100), PAX8 (Proteintech 10336-1-AP, 1:200; Abcam, ab239363, 1:100), CDKN2A (Abcam, Ab108349, 1;200), LAGALS1 (Sigma, HPA000646, 1: 500), EPCAM-488 (Abcam, Ab237395, 1:200), CAPS (Sigma HPA043520, 1:250), CD44 (Invitrogen 14-0441-82, 1:500), smooth muscle actin (ACTA2-594, Abcam ab202368, 1:200), CAVIN 2 (SDPR, Proteintech 12339-1-AP, 1:500), estrogen receptor (ESR, Invitrogen MA5-13191, 1:200), progesterone receptor (PGR, Sigma HPA008428, 1:50), RUNX3 (Sigma HPA059006, 1:100), LGALS1 (Sigma HPA000646, 1: 500), CDKN2A (Abcam, ab108349, 1:300) and CD34 (Abcam Ab81289, 1:200). Alexa-488-, Alexa- 555-, and Alexa-647-conjugated secondary antibodies (LifeTechnologies/Molecular Probes) were all used at 1:1000. DAPI was used as a nuclear counterstain. The images were taken with a Nikon A1R-HD25 confocal microscope and processed with ImageJ. Staining was generally done in a combination of indicated on each panel to distinguish the different cell types.
Flow Cytometry Analysis
Fallopian tube cells were dissociated as described above and resuspended in Bacto™ FA Buffer (BD Biosciences 223142). The single-cell suspensions were initially stained with antibodies against extracellular molecules: anti-CD326 (EPCAM)-Brilliant Violet 510 (1:400; Biolegend, Cat#324235), anti-ENG (CD105)-PE (1:400; Abcam, Cat#ab53321), and anti-CD45- PE/Cy7 (1:400; Biolegend, Cat#368531). The cells were than fixed and permeabilized and stained with intracellular markers such as anti- alpha-Smooth Muscle Actin-APC (1:400; Novus Biologicals, Cat# NBP2-47698APC) and anti-RUNX3-Alexa 488 (1:200; R&D Systems, Cat# IC3765G-100UG) for 30 minutes before sorting. Isotype control antibodies are used as negative controls for gating. These include Brilliant Violet 510™ Mouse IgG2b, κ Isotype Ctrl Antibody (1:400, Biolegend, Cat#400345), Alexa Fluor® 488 Mouse IgG2a, κ Isotype Ctrl Antibody (1:200, Biolegend, Cat#400233), APC anti-mouse IgG2a Antibody (1:400, Biolegend, Cat#407109), PE/Cyanine7 Mouse IgG1, κ Isotype Ctrl Antibody (1:400, Biolegend, Cat#400126), and PE Mouse IgG2a, κ Isotype Ctrl (FC) Antibody (1:400, Biolegend, Cat#400213).
Cell counting for select subtypes
Cell type quantification for nonciliated secretory epithelial cell subtypes (NCSE 2–2 (ACTA2+), NCSE 2–6 (RUNX3+) were performed on tissue sections that were stained using two different combination of markers: EPCAM+/ACTA2+/FOXJ1−andEPCAM+/RUNX3+/CD44+, respectively. After immunofluorescence staining, the whole tissue cross-section was imaged using a wide-field image setting. One thousand cells were randomly selected from four quadrants of the tissue, and the number of CAPS, ACTA2, or RUNX3 positive cells were counted among the EPCAM+ cells, and percentages shown in Supplemental Figure 3H were derived from the total of 1000 cells.
QUANTIFICATION AND STATISTICAL ANALYSIS
Single-cell RNA-seq data processing
Each paired-end 10x scRNA-seq reads contain a cell barcode and a UMI in Read 1, and the transcript-specific read in Read 2 with a read length of 151 nt. The transcript reads were aligned to human reference genome GRCh38 and the barcodes and UMIs were processed by Cell Ranger v4.0.0, which generated single-cell gene expression count matrices for each sample. For healthy donor fallopian tubes, we produced 10 scRNA-seq datasets involving 4 healthy human subjects (FT1-FT4), with 3 anatomic segments from Fimbria, Ampulla, and Isthmus for each of the first 3 donors, and a whole fallopian tube dataset from FT4. For diseased fallopian tubes, we produced 3 datasets, representing Fimbria/Ampulla combined and Isthmus from a hydrosalpinx patient (FT5), and the whole fallopian tube from another patient (FT6).
For scRNA-seq read count data, cells were selected with 1) the cell size factor and integrity filter: cells with >500 detected genes and with <10% mitochondria transcripts were kept (N=60,035 for 4 healthy subjects, and 17,905 for 2 diseased subjects); 2) the doublets filter: cells in a cluster that corresponded to doublets from 2 distant clusters in the global clustering (N=297 for 4 healthy subjects, and 107 for 2 diseased subjects) were removed. In all there were 59,738 cells for the 4 healthy human fallopian tubes with an average of 2,603 detected genes and 10,530 UMIs per cell; and 17,798 cells for the 2 diseased fallopian tubes with an average of 2,780 detected genes and 11,683 UMIs per cell. For each cell, raw transcript counts were normalized by (1) dividing by the total number of UMIs per cell and (2) multiplying by 10,000 to obtain a transcripts-per-10K measure, and then log-transformed by E=ln(transcripts-per-10K+1). For some downstream analyses, the normalized gene expression matrix was standardized for genes by centering and scaling for each gene using (E-mean(E))/sd(E).
QC for the impact of ambient RNA
An early step of data processing was to select cells with >500 detected genes, as those with fewer UMIs are more likely to be droplets containing only ambient RNAs, aka “soup”. To evaluate the potential contribution of ambient RNAs to the selected cells, we first defined the transcriptional profile of “soup” by pooling 3,000 cell barcodes with <100 UMIs, and then calculated the correlation of each selected cell and each of the 3000 soup barcodes to 1) the centroid of each identified cell type and 2) the soup centroid, using either 2K HVG for the identified cell types, or the markers of the cell types that were also highly expressed in soup. This analysis confirmed that the selected cells had low correlation with the soup centroid. We also estimated the soup contamination rate using SoupX v1.4.5 for the selected cells in FT1, and found very low average rates: 0.3%, 0.6% and 0.3%, for cells from Fimbria, Ampulla, and Isthmus, respectively. Furthermore, since the ciliated cell subtype (1–2) had much lower UMI counts (Figure S2B) we ran a similar analysis (Figure S2D), and concluded that CE1-2 cells are not contaminated by ambient RNA.
Evaluation of between-segment and between-subject reproducibility
For the 3 healthy fallopian tubes samples (FT1-3), each with 3 segments, data for each tubal segment contained 1,861–8,332 pass-QC cells. For each segment, we standardized the gene expression matrix, selected 2K highly-variable genes (HVG) to performed PCA, selected the top 6 – 8 PCs based on elbow point of scree plot, and did t-SNE, UMAP and Louvain-Jaccard clustering using Seurat v3.2.3 (Satija et al., 2015), resulting in 11 clusters for each segment. For each subject, the 11 clusters were similar among the 3 segments in the PCA, t-SNE and UMAP projections (Figure S1A), indicating reproducible cell types found across the 3 segments. The pairwise rank correlation coefficients among the 11 cluster centroids also demonstrated consistent cell types across the 3 segments (Figure S1B). Next, to compare among the 4 healthy subjects, we merged the cells from the 3 segments for each of the 3 healthy subjects, re-standardized the gene expression matrix, re-selected 2K HVG, performed PCA, selected top 10 – 12 PCs, and did t-SNE, UMAP and Louvain-Jaccard clustering, resulting in 15–16 clusters for each subject. The rank correlation coefficients (Figure S1C) and the UMAP projections (Figure S1D) confirmed that the same cell types were reproducibly observed in the 4 subjects.
Global analyses of major cell types for healthy fallopian tubes
We combined the gene expression data for 53,376 cells from the 3 healthy fallopian tube samples (FT1-3) by correcting for batch effect across subjects using IntegrateData() function in Seurat3, specifying Subject as the batch factor. We standardized the integrated gene expression matrix, performed PCA using 2K HVG, and chose top 9 PCs based on elbow point of scree plot. We performed tSNE and UMAP projections using top 9 PCs and did Louvain-Jaccard clustering with the top PCs. We initially detected 17 clusters. Since our first goal was to identify major cell types, and then the second step was to identify subtypes using focused clustering, we merged some of the initial clusters based on statistical evaluation and biological marker interpretations. To do so, we first identified markers for each cluster by comparing the expression level in one cluster against that in all other clusters using binomial test and selected those with 1) at least a 20% difference in detection rate; 2) a minimum of 2-fold difference in average expression levels. Based on the markers, we annotated cluster 1 as the ciliated epithelial cells, the clusters 2–6 as the non-ciliated epithelial cells, cluster 7 as the fibroblasts, cluster 8 as myofibroblasts, cluster 9 as smooth muscle cells, cluster 10 as pericytes, cluster 11 as blood endothelial cells, cluster 12 as lymphatic endothelial cells, cluster 13 as doublets of myofibroblast and immune cells, which were removed from further analysis, cluster 14 as B cells, cluster 15 as T cell/NK cells, cluster 16 as mast cells, and cluster 17 as macrophages. After removing the doublet cluster-13 and combining 2–6 we arrived at the 12 major cell types (with UMPA projection shown in Figure 1B). Cells in the combined 2–6 group were subsequently re-clustered, as described below in “Focused subset analysis...”. To display the expression levels of markers in the cell type centroids (Figure 1C), we centered the expression levels of each marker across the 12 centroids and scaled by its standard deviation. To evaluate the differences among the 3 tubal segments, we identified segment-specific markers for each cell type with at least 1.6-fold change in average expression levels between any one segment and the other two segments.
FT4 was analyzed last in our study, when the first three samples (FT1-FT3) had been analyzed extensively. It also had the fewest number of cells among the four samples (n=10447, 25387, 17365, and 6560 cells respectively). To evaluate cell number distribution across cell types, we preformed supervised assignment of the cells from the 4th fallopian tube sample (FT4) to one of 12 global clusters defined by FT1-FT3, based on the highest rank correlation of each cell across the 12 cluster centroids using 3,083 HVG. The counts of assigned cells are in Figure S1E. UMAP projection in Figure 1B were created for all 4 samples. Clustering results from the first 3 samples were changed very little when reanalyzed by adding the 4th sample.
QC to assess the potential presence of doublets
Unlike some scRNA-seq datasets in literature, the level of doublets in our data is extremely low, based on the following three lines of evidence. First, the global clustering pattern in Figure S1A indicates that scRNA-seq data for the fallopian tube revealed distinct, well separated clusters, with no evidence of a large fraction of “in-between” cells that could be doublets. Second, we calculated the rank correlation of each of the >35,000 single cells in FT1-FT3 with each of the 12 centroids shown in Figure 1C, and asked: how often do we see an individual cell that could have been assigned to two centroids? The results confirmed that an overwhelming majority of the cells (<99%) had the highest correlation to the cluster they have been assigned to, with little evidence that some of them could have been plausibly assigned to a second cluster. While two similar cell types may both show a higher level of correlation with a given single cell, this is nearly complete absence of such apparent doublets involving distant cell types. The only exception was a cluster of 198 cells (out of >53K), previously assigned to the old cluster 13, that express marker genes for the old clusters 8 and 15. Third, we adopted a published doublet-detection tool, DoubletFinder1, and confirmed that most of old-13 cells were doublets, and most doublets were in this cluster (Supplemental Figure 1F). We also simulated doublet cells and confirmed that they were detected at the same rate as cluster-13. For the six NCSE subtypes, 2_1 and 2_2 had ~20% doublet rate according to DoubletFinder, and this may seem to suggest that they could be doublets between #2 and one of the stromal cell types (3 to 8). But this scenario is unlikely, because we don’t see the expected number of doublets formed, or at comparable degree of similarity, with cells in clusters 9–12 (the immune cells). The more likely explanation is that these are transitional cells.
Focused subset analysis for ciliated cells and non-ciliated epithelial cells
For the 2,510 ciliated cells and the 14,002 non-ciliated epithelial cells we performed focused reclustering using the same approach and obtained 4 subclusters (Figure 2A) and 6 subclusters (Figure 3A), respectively. We selected markers for each subcluster by comparing one against all other subclusters at a threshold of at least 10% detection rate difference and 1.6-fold difference in average expression levels. For non-ciliated epithelial subclusters with few markers, we also selected markers for each subcluster by comparing it against its neighboring clusters with the same threshold. To test for significant co-expression of pairs of known markers in the 6 non-ciliated epithelial subclusters (Supplemental Figure 3C), we calculated rank correlation between the expression levels of the marker pair in cells expressing both markers. For the six types of stromal cells (clusters 3–8), we re-projected the cells using focused analysis (Figure 4A) but kept the cell type assignment from the global analysis (shown in Figure 1B).
Using RNA Velocity analysis to infer differentiation relationships
For the 53,178 cells in the 3 healthy fallopian tube samples (FT1-3) we obtained the spliced transcript and unspliced transcript counts matrices by velocyto.py v0.17.17, using the GRCh38 genome annotation and repeat mask annotation files. For the cell types analyzed in Figure 5A, 5B, and 5C, respectively, we first created a Seurat object using the spliced and unspliced gene expression matrix, processed the subset Seurat object following SeuratWrappers v0.1.0, and replaced the cluster identity, PCA, UMAP and t-SNE coordinates from the previous focused subset analysis. We then estimated RNA velocity using scVelo v0.2.3 by modeling transcriptional dynamics of splicing kinetics. The velocity estimates were visualized in pre-computed PCA, t-SNE, UMAP coordinates and colored by annotated cell types.
Note: The Velocity results, showing the “flux” between cell groups as curved vector lines, came from a series of upstream analysis steps, chiefly the selection of “highly variable genes” (HVGs), the projection method (UMAP, TSNE, or PCA), and the velocity summaries, which aggregate each cell’s predicted “flow” into coherent lines. The Velocity analysis in Figure 5A–C is based on selecting highly variable genes and running UMAP projection using the three samples jointly. In per-sample Velocity analysis (Supplemental Figure 4C) using the three-sample joint projection, the direction of the velocity lines is consistent across samples, especially with regards to the interpretation for 2–2, and the order among the 4 NC subtypes (1–1 to 1–4). Projecting the 3 samples separately before Velocity analysis would not inform the consistency of Velocity results, as the projected clusters are not align-able across samples.
Comparison between diseased and healthy fallopian tube samples
To use the healthy fallopian tube samples as the reference, we preformed supervised assignment of the cells from the diseased samples to one of global clusters of healthy fallopian tubes, based on the highest rank correlation using 3,083 HVG selected for the 12 reference cluster centroids. This led to the identification of 559 ciliated cells and 2,507 non-ciliated epithelial cells in FT5, and 143 ciliated cells and 207 non-ciliated epithelial cells in FT6, respectively. We then performed supervised assignment of these epithelial cells to either the 4 ciliated cell subtypes using 4,057 HVG for CE or the 6 non-ciliated epithelial cell subtypes using 3,742 HVG for NCE, respectively. For visualization purpose, we integrated all 6 fallopian tube samples together, selected HVG and standardized for genes, and then performed UMAP projection using top 11 PCs (Figure 6A). To identify differentially-expressed genes between healthy and diseased samples for each cell type, we applied the threshold of at least 1.5-fold difference in average expression levels and 10% difference in detection rate. We visualized the differentially-expressed markers between healthy and diseased samples in split violin plots. After obtaining the fold change and p-value for each gene between healthy and diseased sample, we performed gene set enrichment analysis for Gene Ontology using LRpath on 3/22/2021, and removed GO terms containing >2,000 genes or <1,000 genes. To compare the extracellular matrix (ECM) expression pattern between healthy and diseased samples in the ciliated cell subtypes, epithelial cell subtypes, and stromal cell types, we obtained three lists of ECM genes from (Naba et al., 2016). – ECM glycoproteins (127 genes), collagens (45 genes), and proteoglycans (20 genes), and calculated 3 ECM scores (Figure 6C) for each cell as the total transcript counts for the set of ECM genes divided by total transcript counts of all genes for that cell. We also displayed the expression levels for the genes in the 3 ECM gene sets, compared between healthy and diseased samples, across the 10 epithelial subtypes (4 ciliated and 6 non-ciliated epithelial) and 6 stromal cell types (Figure 6D).
Supplementary Material
A. Markers of 11 clusters for Fimbria of FT1. Markers were obtained by comparing each cluster against all other clusters using binomial likelihood test. In each selection, only genes that satisfied two conditions were considered: 1) with a minimum of 20% difference in detection rate in the two groups; 2) with a minimum 2-fold effect size. Each row is a marker for each cluster ranked by p-value; from left to right, the columns include p-value, log-scale fold change, fraction of cells expressing the marker in this cluster, fraction of cells expressing the marker in all other clusters, adjusted p-value, cluster, and gene name.
B. Markers of 11 clusters for Ampulla of FT1. Marker detection method, cutting thresholds and formats are the same as in Table S1A.
C. Markers of 11 clusters for Isthmus of FT1. Marker detection method, cutting thresholds and formats are the same as in Table S1A.
A. Clinical features of 4 healthy (FT1-4) and 2 diseased (FT5-6) fallopian tube samples.
B. Subject demographic and clinical information of 11 benign surgical fallopian tube samples.
A. Markers for each of 12 global cell types of healthy fallopian tubes. Marker detection method and cutting thresholds are the same as in Table S1A. Each row is a marker for each cluster ranked by p-value; from left to right, the columns include cluster, gene name, p-value, log-scale fold change, fraction of cells expressing the marker in this cluster, fraction of cells expressing the marker in all other clusters, and adjusted p-value,.
B. Average expression of markers in each global cell type of healthy fallopian tubes standardized across all 12 cell types. For each detected gene in healthy fallopian tubes, we calculated its centroids in each of the 12 major cell types by averaging the normalized expression of the gene within each cell type and then plus 1 and log-transformed. Each row is a detected gene; each column is one of the 12 major cell types for healthy fallopian tubes; each cell is log (average (normalized expression) +1) for a gene within a cell type.
A. Markers for 4 ciliated subtypes of healthy fallopian tubes. Marker detection method and formats are the same as in Table S3A. In each selection, only genes that satisfied two conditions were considered: 1) with a minimum of 10% detection rate; 2) with a minimum 1.6-fold effect size.
B. GO for ciliated subtype 1–1 of healthy fallopian tubes. Gene ontology terms enriched in each gene cluster.
C. GO for ciliated subtype 1–2 of healthy fallopian tubes.
D. GO for ciliated subtype 1–3 of healthy fallopian tubes.
E. GO for ciliated subtype 1–4 of healthy fallopian tubes.
Marker detection method, cutting thresholds and formats are the same as in Table S4A.
A-L. Differential expressed genes across 3 anatomic segments in global cell type 1 – 12 of healthy fallopian tubes, respectively. Marker detection method and formats are the same as in Table S1A. In each selection, only genes with a minimum 1.6-fold effect size were considered.
A-Q. Differential expressed markers between diseased and healthy fallopian tubes in global cell type 1, non-ciliated epithelial subtypes 1–6, and global cell types 3 – 12, respectively. Marker detection method and formats are the same as in Table S3A. In each selection, only genes that satisfied two conditions were considered: 1) with a minimum of 10% detection rate; 2) with a minimum 1.5-fold effect size.
A-Q. Enrichment between diseased and healthy fallopian tubes in global cell type 1, non-ciliated epithelial subtypes 1–6, and global cell types 3 – 12, respectively, using LRpath.
A. Expression of ECM glycoproteins in healthy and disease states across the 10 epithelial subtypes and 6 stromal clusters.
B. Expression of collagens in healthy and disease states across the 10 epithelial subtypes and 6 stromal clusters.
C. Expression of proteoglycans in healthy and disease states across the 10 epithelial subtypes and 6 stromal clusters.
INCLUSION AND DIVERSITY.
We worked to ensure ethnic or other types of diversity in the recruitment of human subjects. We worked to ensure that the study questionnaires were prepared in an inclusive way. We worked to ensure diversity in experimental samples through the selection of the cell lines. We worked to ensure diversity in experimental samples through the selection of the genomic datasets. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Highlights:
Analysis of ~60K cells of healthy fallopian tubes identifies 12 major cell types.
Defines 4 ciliated and 6 nonciliated subtypes, including two putative progenitors.
Disease samples reveal cell population shifts and changes in gene regulation.
ACKNOWLEDGMENTS
We thank members of the Hammoud, and Li Labs for scientific discussions and manuscript comments. This research was supported by National Institute of Health (NIH) 1DP2HD091949-01 (S.S.H.), training grants T32-HD079342 (A.J.), T32-GM70449 (D.F.H.), fellowship F31-HD106626 (A.J.), Open Philanthropy grant 2019-199327 (5384) (S.S.H.) and Chan Zuckerberg Foundation Grant CZF2019-002428 (A.S., E.E.M., J.Z.L. and S.S.H.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ajonuma LC, Ng EHY, Chan LN, Chow PH, Kung LS, Cheung ANY, Ho LS, Briton-Jones C, Lok IH, Haines CJ, Chan HC, 2005. Ultrastructural characterization of whole hydrosalpinx from infertile Chinese women. Cell Biol. Int. 8. [DOI] [PubMed] [Google Scholar]
- Ardighieri L, Lonardi S, Moratto D, Facchetti F, Shih I-M, Vermi W, Kurman RJ, 2015. Characterization of the Immune Cell Repertoire in the Normal Fallopian Tube 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton BE, Herrera GG, Anamthathmakula P, Rock JK, Willie AM, Harris EA, Takemaru K-I, Winuthayanon W, 2021. Roles of steroid hormones in oviductal function 25. [PMC free article] [PubMed] [Google Scholar]
- Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ, 2020. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414. 10.1038/s41587-020-0591-3 [DOI] [PubMed] [Google Scholar]
- Biernacka A, Dobaczewski M, Frangogiannis NG, 2011. TGF-β signaling in fibrosis. Growth Factors Chur Switz. 29, 196–202. 10.3109/08977194.2011.595714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Horsley V, Fuchs E, 2007. Epithelial Stem Cells: Turning over New Leaves. Cell 128, 445–458. 10.1016/j.cell.2007.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, 2013. The Intestinal Crypt, A Prototype Stem Cell Compartment. Cell 154, 274–284. [DOI] [PubMed] [Google Scholar]
- Crow J, Amso NN, Lewin J, Shaw, Robert W, 1994. Morphology and ultrastructure of fallopian tube epithelium at different stages of the menstrual cycle and menopause. Hum. Reprod. 9, 2224–33. 10.1093/oxfordjournals.humrep.a138428 [DOI] [PubMed] [Google Scholar]
- Dinh HQ, Lin Xianzhi, Abbasi Forough, Nameki Robbin, Haro Marcela, Olingy Claire E., Chang Heidi, Hernandez Lourdes, Gayther Simon A., Wright Kelly N., Aspuria Paul-Joseph, Karlan Beth Y., Corona Rosario I., Li Andrew, Rimel BJ, Siedhoff, Matthew T, Fabiola Medeiros, Kate Lawrenson, 2021. Single-cell transcriptomics identifies gene expression networks driving differentiation and tumorigenesis in the human fallopian tube. Cell Rep. 35, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnez J, 1985. Cyclic changes in ciliation, cell height, and mitotic activity in human tubal epithelium during reproductive life. Fertil. Steril. 43, 6. [DOI] [PubMed] [Google Scholar]
- Duan Y-G, Wehry UP, Buhren BA, Schrumpf H, Oláh P, Bünemann E, Hirchenhain J, van AL, Novak N, Cai Z-M, Krüssel JS, 2020. CCL20-CCR6 axis directs sperm–oocyte interaction and its dysregulation correlates/associates with male infertility 103, 13. [DOI] [PubMed] [Google Scholar]
- Ely LK, Mireille T, 2013. The Role of Opportunistic Bilateral Salpingectomy vs Tubal Occlusion or Ligation for Ovarian Cancer Prophylaxis 371–378. [DOI] [PubMed] [Google Scholar]
- Gaitskell K, Coffey Kate, Green Jane, Pirie Kirstin, Reeves, Gillian K, Ahmed, Ahmed A, Isobel Barnes, Valerie Beral, 2016. Tubal ligation and incidence of 26 site-specific cancers in the Million Women Study. Br. J. CANCER 114, 1033–1037. 10.1038/bjc.2016.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan R, Remaley AT, Sampson ML, Zhang J, Cox DD, Pingpank J, Alexander R, Willingham M, Pastan I, Onda M, 2006. Detection and Quantitation of Serum Mesothelin, a Tumor Marker for Patients with Mesothelioma and Ovarian Cancer. Clin. Cancer Res. 12, 447–453. [DOI] [PubMed] [Google Scholar]
- Hu Z, Artibani M, Alsaadi A, Wietek N, Morotti M, Shi T, Zhong Z, Santana Gonzalez L, El-Sahhar S, KaramiNejadRanjbar M, Mallett G, Feng Y, Masuda K, Zheng Y, Chong K, Damato S, Dhar S, Campo L, Garruto Campanile R, Soleymani majd H, Rai V, Maldonado-Perez D, Jones S, Cerundolo V, Sauka-Spengler T, Yau C, Ahmed AA, 2020. The Repertoire of Serous Ovarian Cancer Non-genetic Heterogeneity Revealed by Single-Cell Sequencing of Normal Fallopian Tube Epithelial Cells. Cancer Cell 37, 226–242.e7. 10.1016/j.ccell.2020.01.003 [DOI] [PubMed] [Google Scholar]
- Johnson N, van Voorst S, Sowter MC, Strandell A, Mol BWJ, 2010. Surgical treatment for tubal disease in women due to undergo in vitro fertilisation. Cochrane Database Syst. Rev 2010, CD002125. 10.1002/14651858.CD002125.pub3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Karnovsky A, Mahavisno V, Weymouth T, Pande M, Dolinoy DC, Rozek LS, Sartor MA, 2012. LRpath analysis reveals common pathways dysregulated via DNA methylation across cancer types. BMC Genomics 13, 526. 10.1186/1471-2164-13-526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppe C, Ibrahim MM, Kranz J, Zhang X, Ziegler S, Perales-Patón J, Jansen J, Reimer KC, Smith JR, Dobie R, Wilson-Kanamori JR, Halder M, Xu Y, Kabgani N, Kaesler N, Klaus M, Gernhold L, Puelles VG, Huber TB, Boor P, Menzel S, Hoogenboezem RM, Bindels EMJ, Steffens J, Floege J, Schneider RK, Saez-Rodriguez J, Henderson NC, Kramann R, 2021. Decoding myofibroblast origins in human kidney fibrosis. Nature 589, 281–286. 10.1038/s41586-020-2941-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, Fan J, Borm LE, Liu Z, van Bruggen D, Guo J, He X, Barker R, Sundström E, Castelo-Branco G, Cramer P, Adameyko I, Linnarsson S, Kharchenko PV, 2018. RNA velocity of single cells. Nature 560, 494–498. 10.1038/s41586-018-0414-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S, Jian Xu, Rik Derynck, 2014. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabbott NA, Donaldson DS, Ohno H, Williams IR, 2013. Microfold (M) cells: important immunosurveillance posts in the intestinal epithelium 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis CS, Murrow LM, Gartner ZJ, 2019. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst 8, 329–337.e4. 10.1016/j.cels.2019.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Nikolic-Paterson DJ, Lan HY, 2016. TGF-β: the master regulator of fibrosis. Nat. Rev. Nephrol. 12, 325–338. 10.1038/nrneph.2016.48 [DOI] [PubMed] [Google Scholar]
- Morales P, Reyes P, Vargas M, Rios M, Imarai M, Cardenas H, Croxatto H, Orihuela P, Vargas R, Fuhrer J, Heckels JE, Christodoulides M, Velasquez L, 2006. Infection of Human Fallopian Tube Epithelial Cells with Neisseria gonorrhoeae Protects Cells from Tumor Necrosis Factor Alpha-Induced Apoptosis. Infect. Immun. 74, 3643–3650. 10.1128/IAI.00012-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naba A, Clauser KR, Ding H, Whittaker CA, Carr SA, Hynes RO, 2016. The Extracellular Matrix: Tools and Insights for the “Omics” Era. Matrix Biol. J. Int. Soc. Matrix Biol 49, 10–24. 10.1016/j.matbio.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng A, Tan S, Singh G, Rizk P, Swathi Y, Tan TZ, Huang RY-J, Leushacke M, Barker N, 2014. Lgr5 marks stem/progenitor cells in ovary and tubal epithelia 16, 23. [DOI] [PubMed] [Google Scholar]
- Nieto MA, Huang RY-J, Jackson RA, Thiery JP, 2016. EMT: 2016. Cell 166, 21–45. 10.1016/j.cell.2016.06.028 [DOI] [PubMed] [Google Scholar]
- Ocaña OH, Corcoles Rebeca, Fabra Angels, Moreno-Bueno Gema, Herve Acloque, Vega Sonia, Barrallo-Gimeno Alejandro, Cano Amparo, Nieto, Angela M, 2012. Metastatic Colonization Requires the Repression of the Epithelial-Mesenchymal Transition Inducer Prrx1. Cancer Cell 22, 709–724. [DOI] [PubMed] [Google Scholar]
- Paik DY, Janzen DM, Schafenacker AM, Velasco VS, Cheng D, Huang J, Witte ON, Memarzadeh S, 2015. Stem-like epithelial cells are concentrated in the distal end of the fallopian tube: a site for injury and serous cancer initiation 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palter SF, Mulayim N, Senturk L, Arici A, 2001. Interleukin-8 in the Human Fallopian Tube 86, 8. [DOI] [PubMed] [Google Scholar]
- Peters W, 1986. Nature of “basal” and “reserve” cells in oviductal and cervical epithelium in man. J. Clin. Pathol. 39, 306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto S, Hoek M, Huang Y, Costet P, Ma L, Imbriglio JE, 2017. Drug Discovery in Tissue Fibrosis, in: Comprehensive Medicinal Chemistry III. Elsevier, pp. 694–713. 10.1016/B978-0-12-409547-2.12432-1 [DOI] [Google Scholar]
- Popescu LM, Ciontea SM, Cretoiu D, 2007. Interstitial Cajal-Like Cells in Human Uterus and Fallopian Tube. Ann. N. Y. Acad. Sci. 1101, 139–165. 10.1196/annals.1389.022 [DOI] [PubMed] [Google Scholar]
- Rasmussen SJ, Eckmann L, Quayle AJ, Shen L, Zhang YX, Anderson DJ, Fierer J, Stephens RS, Kagnoff MF, 1997. Secretion of proinflammatory cytokines by epithelial cells in response to Chlamydia infection suggests a central role for epithelial cells in chlamydial pathogenesis. J. Clin. Invest. 99, 77–87. 10.1172/JCI119136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JR, Gaudet HM, Khan M, Schorl C, James NE, Oliver MT, DiSilvestro PA, Moore RG, Yano N, 2018. Human Epididymis Protein 4 Promotes Events Associated with Metastatic Ovarian Cancer via Regulation of the Extracelluar Matrix. Front. Oncol. 7, 332. 10.3389/fonc.2017.00332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapmaz T, Gündoğdu LS, Çetin MT, Ürünsak F, Polat S, 2019. The ultrastructural effects of surgical treatment of hydrosalpinx on the human endometrium: a light. Ultrastruct. Pathol. 43, 99–109. 10.1080/01913123.2019.1600087 [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A, 2015. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon SB, Raja N, Xu M, Cameron H, Yang K, Reynolds J, Fenner D, Marsh EE, 2021. Establishing a reproductive biorepository for basic and translational research: experience developing the reproductive subjects registry and sample repository. J. Assist. Reprod. Genet. 10.1007/s10815-021-02165-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowamber R, Nelson O, Dodds L, DeCastro V, Paudel I, Milea A, Considine M, Cope L, Pinto A, Schlumbrecht M, Slomovitz B, Shaw PA, George SHL, 2020. Integrative Transcriptome Analyses of the Human Fallopian Tube: Fimbria and Ampulla—Site of Origin of Serous Carcinoma of the Ovary. Cancers 12, 1090. 10.3390/cancers12051090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strandell A, 2000. The infuence of hydrosalpinx on IVF and embryo transfer: a review. Hum. Reprod. Update 6, 387–395. [DOI] [PubMed] [Google Scholar]
- Tiourin E, Velasco VS, Rosales MA, Sullivan PS, Janzen DM, Memarzadeh S, n.d. Tubal Ligation Induces Quiescence in the Epithelia of the Fallopian Tube Fimbria. Reprod. Sci 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA-K, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist P-H, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F, 2015. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Xu J, Luo X, Qu S, Yang G, Shen N, 2019. B cell activation factor (BAFF) induces inflammation in the human fallopian tube leading to tubal pregnancy. BMC Pregnancy Childbirth 19, 169. 10.1186/s12884-019-2324-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohannes E, Kazanjian Avedis A., Lindsay Morgan E., Fujii Dennis T., Ieronimakis Nicholas, Chow Gregory E., Beesley Ronald D., Heitmann Ryan J., Burney Richard O., 2019. The human tubal lavage proteome reveals biological processes that may govern the pathology of hydrosalpinx. Sci. Rep. 9. 10.1038/s41598-019-44962-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Behjati S, 2020. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. GigaScience 9, giaa151. 10.1093/gigascience/giaa151 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Markers of 11 clusters for Fimbria of FT1. Markers were obtained by comparing each cluster against all other clusters using binomial likelihood test. In each selection, only genes that satisfied two conditions were considered: 1) with a minimum of 20% difference in detection rate in the two groups; 2) with a minimum 2-fold effect size. Each row is a marker for each cluster ranked by p-value; from left to right, the columns include p-value, log-scale fold change, fraction of cells expressing the marker in this cluster, fraction of cells expressing the marker in all other clusters, adjusted p-value, cluster, and gene name.
B. Markers of 11 clusters for Ampulla of FT1. Marker detection method, cutting thresholds and formats are the same as in Table S1A.
C. Markers of 11 clusters for Isthmus of FT1. Marker detection method, cutting thresholds and formats are the same as in Table S1A.
A. Clinical features of 4 healthy (FT1-4) and 2 diseased (FT5-6) fallopian tube samples.
B. Subject demographic and clinical information of 11 benign surgical fallopian tube samples.
A. Markers for each of 12 global cell types of healthy fallopian tubes. Marker detection method and cutting thresholds are the same as in Table S1A. Each row is a marker for each cluster ranked by p-value; from left to right, the columns include cluster, gene name, p-value, log-scale fold change, fraction of cells expressing the marker in this cluster, fraction of cells expressing the marker in all other clusters, and adjusted p-value,.
B. Average expression of markers in each global cell type of healthy fallopian tubes standardized across all 12 cell types. For each detected gene in healthy fallopian tubes, we calculated its centroids in each of the 12 major cell types by averaging the normalized expression of the gene within each cell type and then plus 1 and log-transformed. Each row is a detected gene; each column is one of the 12 major cell types for healthy fallopian tubes; each cell is log (average (normalized expression) +1) for a gene within a cell type.
A. Markers for 4 ciliated subtypes of healthy fallopian tubes. Marker detection method and formats are the same as in Table S3A. In each selection, only genes that satisfied two conditions were considered: 1) with a minimum of 10% detection rate; 2) with a minimum 1.6-fold effect size.
B. GO for ciliated subtype 1–1 of healthy fallopian tubes. Gene ontology terms enriched in each gene cluster.
C. GO for ciliated subtype 1–2 of healthy fallopian tubes.
D. GO for ciliated subtype 1–3 of healthy fallopian tubes.
E. GO for ciliated subtype 1–4 of healthy fallopian tubes.
Marker detection method, cutting thresholds and formats are the same as in Table S4A.
A-L. Differential expressed genes across 3 anatomic segments in global cell type 1 – 12 of healthy fallopian tubes, respectively. Marker detection method and formats are the same as in Table S1A. In each selection, only genes with a minimum 1.6-fold effect size were considered.
A-Q. Differential expressed markers between diseased and healthy fallopian tubes in global cell type 1, non-ciliated epithelial subtypes 1–6, and global cell types 3 – 12, respectively. Marker detection method and formats are the same as in Table S3A. In each selection, only genes that satisfied two conditions were considered: 1) with a minimum of 10% detection rate; 2) with a minimum 1.5-fold effect size.
A-Q. Enrichment between diseased and healthy fallopian tubes in global cell type 1, non-ciliated epithelial subtypes 1–6, and global cell types 3 – 12, respectively, using LRpath.
A. Expression of ECM glycoproteins in healthy and disease states across the 10 epithelial subtypes and 6 stromal clusters.
B. Expression of collagens in healthy and disease states across the 10 epithelial subtypes and 6 stromal clusters.
C. Expression of proteoglycans in healthy and disease states across the 10 epithelial subtypes and 6 stromal clusters.
Data Availability Statement
Single-cell RNA-seq data have been deposited at GEO and HCA and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. The data visualization is available in the interactive explorer cellxgene. Microscopy data reported in this paper will be shared by the lead contact upon request.
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti CD326-Brillianat Violet 510 (EPCAM) | Biolegend | Cat#324235; PRID: AB_2632934 |
| Mouse monoclonal anti CD45- PE/Cy7 | Biolegend | Cat#368531; PRID: AB_2715891 |
| Mouse monoclonal anti alpha-Smooth Muscle Actin-APC | Novus Biologicals | Cat#NBP2-47698APC; PRID: N/A |
| Mouse monoclonal anti ENG (CD105)-PE | Abcam | Cat#ab53321; PRID: AB_868769 |
| Mouse monoclonal anti RUNX3-Alexa 488 | R&D Systems | Cat#IC3765G-100UG; PRID: N/A |
| Mouse monoclonal anti mouse IgG2b, κ isotype control antibody-Brilliant Violet 510 | Biolegend | Cat#400345; PRID: N/A |
| Mouse monoclonal anti mouse IgG2a, κ isotype control antibody-Alexa 488 | Biolegend | Cat#400233; PRID: N/A |
| Rat monoclonal anti mouse IgG2a antibody-APC | Biolegend | Cat#407109; PRID: AB_2561754 |
| Mouse monoclonal anti mouse IgG1, κ isotype control antibody-PE/Cy7 | Biolegend | Cat#400126; PRID: AB_326448 |
| Mouse monoclonal anti mouse IgG2a, κ isotype control (FC) antibody-PE | Biolegend | Cat#400213; PRID: AB_2800438 |
| Mouse monoclonal anti FOXJ1 | Invitrogen | Cat#14-9965-82; PRID: AB_1548835 |
| Mouse monoclonal anti FOXJ1 | Sigma | Cat#AMAB91254; PRID: AB_2665866 |
| Rabbit polyclonal anti PAX8 | Proteintech | Cat#10336-1-AP; PRID: AB_2236705 |
| Rabbit monoclonal anti PAX8 | Abcam | Cat#ab239363; PRID: N/A |
| Rabbit monoclonal anti EPCAM-Alexa 488 | Abcam | Cat#ab237395; PRID: N/A |
| Rabbit polyclonal anti CAPS | Sigma | Cat#HPA043520; PRID: AB_10964138 |
| Mouse monoclonal anti alpha-Smooth Muscle Actin (ACTA2)-594 | Abcam | Cat#ab202368; PRID: N/A |
| Rabbit polyclonal anti CAVIN 2 (SDPR) | Proteintech | Cat#12339-1-AP; PRID: AB_2183305 |
| Rat monoclonal anti CD44 | Invitrogen | Cat#14-0441-82; PRID: AB_467246 |
| Mouse monoclonal anti Estrogen receptor 1 (ESR1) | Invitrogen | Cat#MA5-13191; PRID: AB_10986080 |
| Rabbit polyclonal anti Progesterone receptor (PGR) | Sigma | Cat#HPA008428; PRID: AB_1079680 |
| Rabbit polyclonal anti RUNX3 | Sigma | Cat#HPA059006; PRID: AB_2683877 |
| Rabbit monoclonal anti CD34 | Abcam | Cat#ab81289; PRID: AB_1640331 |
| Rabbit polyclonal anti LGALS1 | Sigma | Cat#HPA000646; PRID: AB_1078935 |
| Rabbit monoclonal anti CDKN2A | Abcam | Cat#ab108349; PRID: AB_10858268 |
| Biological samples | ||
| Adult Human 1 (FT1, Age 52 yrs) Fallopian Tube Tissue For single cell RNAseq and statining |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 2 (FT2, Age 30 yrs) Fallopian Tube Tissue For single cell RNAseq |
International Institute for the Advancement of Medicine | N/A |
| Adult Human 3 (FT3, Age 46 yrs) Fallopian Tube Tissue For single cell RNAseq, staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 4 (FT4, Age 31 yrs) Fallopian Tube Tissue For single cell RNAseq |
International Institute for the Advancement of Medicine | N/A |
| Adult Human 5 (FT5, Age 34 yrs) Fallopian Tube Tissue For single cell RNAseq, staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 6 (FT6, Age 32 yrs) Fallopian Tube Tissue For single cell RNAseq |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 7 (Age 71 yrs) Fallopian Tube Tissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 8 (Age 52 yrs) Fallopian Tube Tissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 9 (Age 72 yrs) Fallopian Tube Tissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 10 (Age 68 yrs) Fallopian TubeTissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 11 (Age 58 yrs) Fallopian Tube Tissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 12 (Age 47 yrs) Fallopian Tube Tissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 13 (Age 36 yrs) Fallopian Tube Tissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 14 (Age 49 yrs) Fallopian Tube Tissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 15 (Age 36 yrs) Fallopian Tube Tissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 16 (Age 41 yrs) Fallopian Tube Tissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Adult Human 17 (Age 38 yrs) Fallopian Tube Tissue For staining and quantification |
University of Michigan Michigan Medicine RSRSR | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Proteinase | Sigma | Cat#P5147-1G |
| Deoxyribonuclease I | Sigma | Cat#4716728001 |
| Collagenase D | Sigma/Roche | Cat#11088866001 |
| Hyalurondase | Sigma | Cat#H3884-1G |
| HBSS | Invitrogen | Cat#14025092 |
| DAPI | Sigma | Cat#D9542 |
| FBS | Sigma | Cat#F4135 |
| Vectashield Antifade Mounting Medium | Vector Laboratories | Cat#H-1000-10 |
| Tween-20 | Sigma | Cat# P1379 |
| Triton X-100 | Sigma | Cat# T8787 |
| BSA | Sigma | Cat# A7030 |
| Paraformaldehyde | MilliporeSigma | Cat# 818715 |
| Bloxall Endogenous Peroxidase and Alkaline Phosphatase Blocking Solution | Vector Laboratories | Cat# SP-6000 |
| Sucrose | Sigma | Cat# S9378 |
| Citric Acid | Sigma | Cat#791725 |
| SignalBoostImmunoreaction Enhancer Kit | MilliporeSigma | Cat#407207 |
| EasySep RBC Depletion Reagent | Stemcell Technologies | Cat#18170 |
| Bacto™ FA Buffer | BD Biosciences | Cat#223142 |
| Deposited data | ||
| Raw data files for scRNA-seq | This paper or NCBI Sequence Read Archive or Human Cell Atlas | SRA: SRP323939; HCA: https://data.humancellatlas.org/explore/projects/21ea8ddb-525f-4f1f-a820-31f0360399a2 |
| Gene expression data for scRNA-seq | This paper or NCBI Gene Expression Omnibus or Human Cell Atlas | GEO: GSE178101; HCA: https://data.humancellatlas.org/explore/projects/21ea8ddb-525f-4f1f-a820-31f0360399a2 |
| Data visualization for scRNA-seq | This paper or cellxgene | https://cellxgene.cziscience.com/collections/fc77d2ae-247d-44d7-aa24-3f4859254c2c |
| Deposited code | Zenodo or Github | DOI:10.5281/zenodo.5883840; https://github.com/qianqianshao/FallopianTube/ |
| Software and algorithms | ||
| Cell Ranger (v4.0.0) | https://github.com/10XGenomics/cellranger | |
| SoupX (v1.4.5) | Young and Behjati, 2020 | https://github.com/constantAmateur/SoupX |
| DoubletFinder (v2.0.3) | McGinnis et al., 2019 | https://github.com/chris-mcginnis-ucsf/DoubletFinder |
| Seurat (v3.2.3) | Satija et al., 2019 | https://github.com/satijalab/seurat |
| SeuratWrappers (v0.1.0) | https://github.com/satijalab/seurat-wrappers | |
| velocyto.py (v0.17.17) | Manno et at., 2018 | https://github.com/velocyto-team/velocyto.py |
| scVelo (v0.2.3) | Bergen et al., 2020 | https://github.com/theislab/scvelo |
| LRpath | Kim et al., 2012 | http://lrpath.ncibi.org/ |
All original code has been deposited at Zenodo and GitHUB. all codes are publicly available as of the date of publication. DOIs and links are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.