Abstract
Background
In September 2018, Burkholderia cepacia complex (BCC) infections in 3 patients associated with exposure to a mouthwash solution (MWS) were reported to the Robert Koch Institute (RKI). As the product was still on the market and the scale of the outbreak was unclear, a nation-wide investigation was initiated.
Methods
We aimed to investigate BCC infections/colonizations associated with MWS. Hospitals, laboratories, and public health services were informed that BCC isolates should be sent to the RKI. These isolates were typed by pulsed-field gel electrophoresis (PFGE) and whole-genome sequencing (WGS) including development of an ad hoc core genome MLST (cgMLST) scheme.
Results
In total, 36 patients from 6 hospitals met the case definition, the last patient in November 2018. Twenty-nine isolates from 26 of these patients were available for typing. WGS analysis revealed 2 distinct cgMLST clusters. Cluster 1 (Burkholderia arboris) contained isolates from patients and MWS obtained from 4 hospitals and isolates provided by the manufacturer. Patient and MWS isolates from another hospital were assigned to cluster 2 (B. cepacia).
Conclusions
The combined clinical, epidemiological, and microbiological investigation, including whole-genome analysis, allowed for uncovering a supraregional BCC outbreak in health care settings. Strains of B. arboris and B. cepacia were identified as contaminating species of MWS bottles and subsequent colonization and putative infection of patients in several hospitals. Despite a recall of the product by the manufacturer in August 2018, the outbreak lasted until December 2018. Reporting of contaminated medical products and recalls should be optimized to protect patients.
Keywords: Burkholderia arboris, outbreak, clonal transfer, mouthwash solution, medical device
Burkholderia is a diverse genus of gram-negative and obligate aerobic bacteria that are ubiquitously found in the environment and have beneficial biotechnological properties, but at the same time represent well-known pathogens of plants, animals, and humans [1–4]. Members of the Burkholderia cepacia complex (BCC) have frequently been reported to cause health care–associated infections in critically ill patients and colonizations and infections in patients suffering from cystic fibrosis (CF) [5–8].
In August 2018, BCC infections in 3 patients that were associated with exposure to a nonalcoholic mouthwash solution (MWS) were detected in a German hospital [9]. This nonalcoholic MWS was a product to decontaminate the mouth and throat and to support wound healing. It was produced and used as a cosmetic product in the general population as well as a medical product especially in intensive care unit (ICU) patients. In Germany, the use of medical MWS is recommended for prevention of ventilator-associated pneumonia [10]. The contract manufacturer of the MWS had discovered BCC in reserve samples and in the production line and had informed purchasers about a voluntary recall of the product on August 9, 2018. Notably, products of the contaminated batch were exported into several EU countries. Although a recall had been initiated by the manufacturer, it seemed likely that patients were still exposed to the contaminated product. Thus, we implemented a nation-wide surveillance of BCC infections/colonization associated with MWS exposure in September 2018 [11]. Here we report on the outbreak investigation across multiple hospitals in Germany and the implications for future prevention of outbreaks caused by contaminated medical products.
METHODS
Data Collection
Since 2011, Germany has been conducting a National Surveillance of Healthcare Associated Outbreaks (https://doi.org/10.1371/journal.pone.0098100). It is mandatory to report 2 or more health care–associated infections with a suspected epidemiological link to the health service; thus all BCC infections with an association to MWS were reportable. In addition, we established a surveillance program for BCC isolates and asked all stakeholders to send BCC isolates to the RKI or the National Reference Center for Multidrug-resistant Gram-negative Bacteria (NRC) for typing. The contract manufacturer investigated the production line in Germany for the presence of BCC.
Case Definition
A confirmed case was defined as a person carrying 1 of the BCC outbreak clones as confirmed by molecular typing AND having been exposed to MWS. A probable case was defined as a person in whom BCC was detected after exposure to MWS, but with no isolate available for typing.
Communication
Public health authorities in Germany were informed in the weekly epidemiological teleconference (EpiLAG from September 18, 2018), and hospitals were informed by short communications and announcements [11, 12]. International public health authorities were informed by means of the Epidemic Intelligence Information System (EPIS; https://www.ecdc.europa.eu/en/publications-data/epidemic-intelligence-information-system-epis) and the Early Warning and Response System of the European Union (EWRS; https://www.ecdc.europa.eu/en/publications-data/early-warning-and-response-system-european-union-ewrs).
Reference Laboratory Methods
Bacterial strain typing of all BCC isolates was performed by SpeI-macrorestriction followed by PFGE and interpreted according to the criteria of Tenover et al. [13]. Whole-genome sequencing (WGS) analyses were carried out following library preparation, as recommended by the manufacturer, using the NexteraXT library prep kit (Illumina) and a read-out of either 2 × 250 bp or 2 × 300 bp on a HiSeq or MiSeq instrument, respectively. Raw reads were subjected to quality assessment by QCumber-2 (https://gitlab.com/RKIBioinformaticsPipelines/QCumber), and genome sequences were reconstructed by applying an in-house-developed pipeline utilizing the SPAdes assembler [14]. Sequence types were extracted using pubmlst.org. An ad hoc core genome MLST (cgMLST) was developed using the software SeqSphere+ with reference strain B. cepacia ATCC25416 to define target core genome genes. These genes were further filtered by 10 completed B. cepacia genomes available for download from NCBI via refDownloader (https://gitlab.com/s.fuchs/refDownloader) as of April 10, 2018, resulting in 1.856 defined target genes for evaluation of strain relatedness by cgMLST. To this end, minimum spanning trees were generated in SeqSphere+ using the parameter of “pairwise ignore missing values” for tree calculation. Isolates from the outbreak published by Becker et al. [9] were included for comparative reasons. Species identification was carried out with Geneious following a ClustalW alignment of the recA genes of all isolates with a database containing recA sequences for 39 type strains. A Neighbor-Joining tree was calculated in Geneious using the Tamura-Nei genetic distance model and 1.000 iterations for branch validation.
Raw read data were submitted to the Sequence Read Archive and are available under accession number PRJNA525973.
Ethical Statement
A formal ethical review was not required for the outbreak investigation in accordance with Article 25, Section 1, of the German Protection against Infection Act of 2001.
Patient Consent
Data were gathered in a public health emergency response to the outbreak. The data were collected in the hospitals, and only selected anonymized data were forwarded to RKI. This study does not include factors necessitating patient consent.
RESULTS
Epidemiological Investigation
We identified 36 cases from 6 different hospitals (A–F) in 4 federal German states.
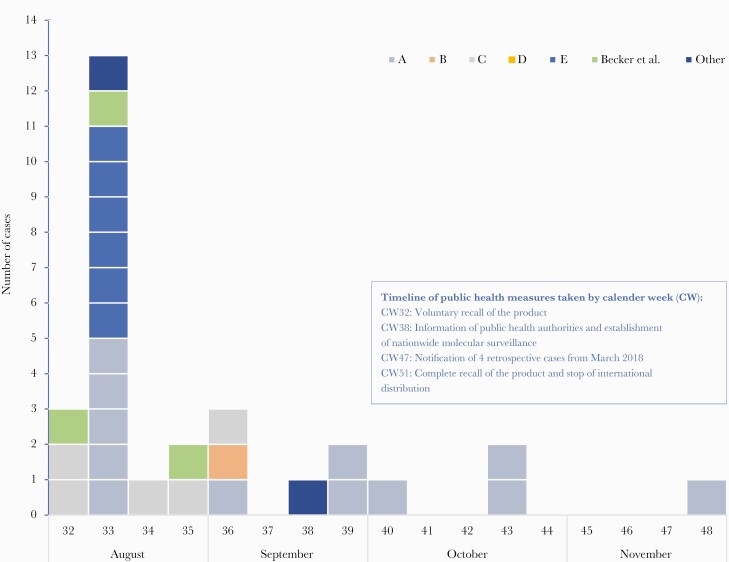
The epidemic curve (data available for 33 cases: 26 confirmed cases and 7 probable cases) illustrates the course of the outbreak (Figure 1). The peak of the outbreak was in calendar week 33, the last case was notified in November 2018. Four retrospectively reported cases were identified in March 2018, 5 months before the recalled batch of MWS was produced.
Figure 1.
Epidemiologic curve for 29 cases from hospitals A–E and cases notified from other sources (August–November 2018, calendar weeks 32–48; n = 29). Four additional cases from hospital F from March 2018 (calendar weeks 11 and 12) were notified retrospectively (data not shown).
Sociodemographic and clinical characteristics of 34 patients for whom data were available are summarized in Table 1. The age of confirmed and suspected cases ranged from 33 to 85 years, with a median age of 73 years. Sixty-five percent of the cases were male. Most clinical cultures were derived from respiratory samples. Seven patients were infected with BCC, and 29 cases were colonized. The 5 cases who died throughout hospital stay were confirmed cases, but were all hospitalized due to an underlying disease and may also have died of other reasons.
Table 1.
Characteristics of Cases/Patients With BCC Colonization/Infections Associated With Contaminated Mouthwash Solution (n = 34a)
| Patient Characteristics | Confirmed (n = 27) | Probable (n = 7) | Total (n = 34a) |
|---|---|---|---|
| Age | |||
| Median (range), y | 74 (44–85) | 70 (33–84) | 73 (33–85) |
| Sex | |||
| Male | 19 (70) | 3 (43) | 22 (65) |
| Female | 8 (30) | 4 (57) | 12 (35) |
| Status | |||
| Colonization | 22 (81) | 5 (71) | 27 (79) |
| Infection | 5 (19) | 2 (29) | 7 (21) |
| ICU | n = 12 | n = 7 | n = 19 |
| Yes | 12 (100) | 2 (30) | 14 (74) |
| No | 0 (0) | 5 (70) | 5 (26) |
| Deceased | 5 (19) | 0 | 5 (15) |
| Site of cultures | n = 20 | n = 7 | n = 27 |
| Anal smear | 2 | 0 | 2 |
| Bronchial lavage | 5 | 1 | 6 |
| Nasal/throat swab | 8 | 1 | 9 |
| Tracheal secretion | 2 | 3 | 5 |
| Other sites | 3 | 2 | 5 |
Abbreviations: BCC, Burkholderia cepacia complex; ICU, intensive care unit.
Sociodemographic and clinical characteristics were missing or incomplete for 2 of the 36 notified cases.
Clinical data such as underlying chronic diseases or admission diagnosis were available only for few patients (data not shown). In 5 cases, an atypical pneumonia (in 1 case complicated by sepsis) was reported; of these, 3 patients had an underlying immunosuppressive condition and 2 died of an underlying disease. One patient was reported to have malignant disease, and 1 more patient underwent heart surgery.
Investigation of Isolates From Multiple Hospitals and from the Manufacturer
From September until December 2018, the RKI and NRC received 39 BCC isolates including 3 BCC type strains from the German Consiliary Laboratory on Cystic Fibrosis Bacteriology as internal references for subsequent molecular investigations (Table 2). Hospital A sent isolates from 10 patients and 1 from MWS. Three BCC isolates from patients in hospital D and 1 patient from hospital B (located in the same city as hospital A) were not exposed to a mouthwash solution of the manufacturer and thus were not counted as cases. However, they were included as outgroup isolates in all further analyses.
Table 2.
Characteristics of 39 BCC Isolates From Germany (March–November 2018)
| Isolate No. | BCC Species | Hospital | Sample | Date of Isolation/Notification | PFGE Typeg | cgMLST Cluster | Case Definition |
|---|---|---|---|---|---|---|---|
| 725-18 | B. cepacia | A | Patient 1c | 17.08.2018 | B2 | 2 | Confirmed |
| 726-18 | B. cepacia | A | Patient 2 | 16.08.2018 | B2 | 2 | Confirmed |
| 728-18 | B. cepacia | A | Patient 3d | 17.08.2018 | B2 | 2 | Confirmed |
| 729-18 | B. cepacia | A | Patient 4d | 17.08.2018 | B2 | 2 | Confirmed |
| 730-18 | B. cepacia | A | Patient 5 | 16.08.2018 | B2 | 2 | Confirmed |
| 760-18 | B. cepacia | A | Patient 6 | 07.09.2018 | B2 | 2 | Confirmed |
| 782-18 | B. cepacia | A | Patient 7 | 25.09.2018 | B2 | 2 | Confirmed |
| 841-18 | B. cepacia | A | Patient 8 | 25.10.2018 | B2 | 2 | Confirmed |
| 842-18 | B. cepacia | A | Patient 9d | 26.10.2018 | B2 | 2 | Confirmed |
| 879-18 | B. cepacia | A | Patient 10 | 30.11.2018 | B2 | 2 | Confirmed |
| 727-18 | B. cepacia | A | MWS | 17.08.2018 | B2 | 2 | Not applicable |
| 761-18 | B. cepacia | B | Patient 1 | 07.09.2018 | B4 | 4 (singleton ST922)h | Confirmed |
| 789-18 | B. arboris | C | Patient 1 | Unknown/missing | B1 | 1 | Confirmed |
| 790-18 | B. arboris | C | Patient 2 | Unknown/missing | B1 | n.a. | Confirmed |
| 791-18 | B. arboris | C | Patient 3 | Unknown/missing | B1 | 1 | Confirmed |
| 797-18 | B. cenocepacia IIIB | D | Patient 1 | Unknown/missing | B3 | 3 | No case |
| 798-18 | B. cenocepacia IIIB | D | Patient 2 | Unknown/missing | B3 | 3 | No case |
| 799-18 | B. cenocepacia IIIB | D | Patient 3 | Unknown/missing | B3 | 3 | No case |
| 811-18 | B. arboris | E | MWS | 14.08.2018 | B1 | 1 | Not applicable |
| 812-18 | B. arboris | E | MWS | 22.08.2018 | B1b | 1 | Not applicable |
| 813-18 | B. arboris | E | Patient 1 | 11.08.2018 | B1 | 1 | Confirmed |
| 814-18 | B. arboris | E | Patient 1 | 13.08.2018 | B1 | 1 | Duplicate |
| 815-18 | B. arboris | E | Patient 1 | 15.08.2018 | B1 | 1 | Duplicate |
| 816-18 | B. arboris | E | Patient 2 | 15.08.2018 | B1 | 1 | Confirmed |
| 817-18 | B. arboris | E | Patient 2 | 23.08.2018 | B1 | 1 | Duplicate |
| 818-18 | B. arboris | E | Patient 3 | 15.08.2018 | B1 | 1 | Confirmed |
| 819-18 | B. arboris | E | Patient 4 | 15.08.2018 | B1 | 1 | Confirmed |
| 820-18 | B. arboris | E | Patient 5 | 15.08.2018 | B1 | 1 | Confirmed |
| 821-18 | B. arboris | E | Patient 6 | 15.08.2018 | B1 | 1 | Confirmed |
| 822-18 | B. arboris | E | Patient 7 | 15.08.2018 | B1 | 1 | Confirmed |
| 26-19 | B. arboris | F | Patient 1 | 15.03.2018 | B1 | 1 | Confirmed |
| 27-19 | B. arboris | F | Patient 2 | 02.04.2018 | B1 | 1 | Confirmed |
| 775-18 | B. arboris | -a | Environmente | Unknown/missing | B1 | 1 | Not applicable |
| 776-18 | B. arboris | -a | MWSf | Unknown/missing | B1 | 1 | Not applicable |
| 795-18 | B. arboris | -a | MWSf | Unknown/missing | B1 | 1 | Not applicable |
| 796-18 | B. arboris | -a | MWSf | Unknown/missing | B1 | 1 | Not applicable |
| 808-18 | B. latens | NCL-CFb | Type strain DSM 23436 | - | B6 | 5 (singleton) | Not applicable |
| 809-18 | B. arboris | NCL-CFb | Type strain DSM 23435 | - | B5 | 6 (singleton) | Not applicable |
| 810-18 | B. cenocepacia IIIA | NCL-CFb | Type strain LMG 16656 | - | B7 | 7 (singleton) | Not applicable |
Patients from hospitals A, C, E, and F had contact with MWS from 2 different batches; patients from hospital D and hospital B (located in the same city as hospital A) had no contact with any MWS and were used as outgroup cases; the index patient of hospital A is not listed because no isolate was available for WGS analysis; isolates from 2 of the 4 patients in hospital F were available for WGS analysis. “BCC species” indicates species of the Burkholderia cepacia complex identified by recA gene sequence analyses (Figure 3).
Abbreviations: BCC, Burkholderia cepacia complex; cgMLST, ad hoc core genome MLST; MWS, mouthwash solution; NCL-CF, National Consiliary Laboratory on Cystic Fibrosis Bacteriology; PFGE, pulsed-field gel electrophoresis; WGS, whole-genome sequencing.
Isolates provided by the MWS manufacturer.
Three BCC reference isolates (type strains) provided by the National Consiliary Laboratory on Cystic Fibrosis Bacteriology, Germany, were included in the present analyses.
Patients with BCC infection.
Patients were colonized with BCC isolates and died due to underlying diseases.
Swab of MWS filling system from the manufacturer.
Sample from an MWS-containing bottle directly from the manufacturer (unopened batch).
PFGE analyses were performed repeatedly in 3 German institutions (Robert Koch Institute, Wernigerode; National Reference Center for Multidrug-resistant Gram-negative Bacteria, Bochum; Bavarian Health and Food Safety Authority, Erlangen).
Multilocus sequence type was determined using a published scheme (pubmlst.org), but yielded positive results for indicated isolates only; “n.a.” indicates no analysis possible due to contamination with Moraxella spp.
We additionally received 4 isolates from the manufacturer of the MWS, which were collected from the filling plant and from an MWS bottle of the affected batch. Hospital F provided 2–4 patient isolates that were exposed to a different batch of MWS from the same provider. Hospital F had previously sent the 4 isolates to the Bavarian Health and Food Safety Authority (LGL) for PFGE typing. Because identical PFGE patterns were observed, the LGL only provided 2 isolates to the RKI for confirmatory PFGE and WGS analysis.
Many isolates showed reduced growth on Müller-Hinton agar supplemented with sheep blood, but the usage of tryptic soy agar and BD Cepacia Medium (Becton Dickinson) accelerated growth and yielded sufficient colony material after 24–48 hours of incubation. PFGE analyses (illustrative example in Supplementary Figure 1) assigned 11 isolates (9 from patients, 2 from MWS) from hospital E, 3 clinical isolates from hospital C, and 2 clinical isolates from hospital F to the major outbreak group with PFGE type B1 (Table 2).
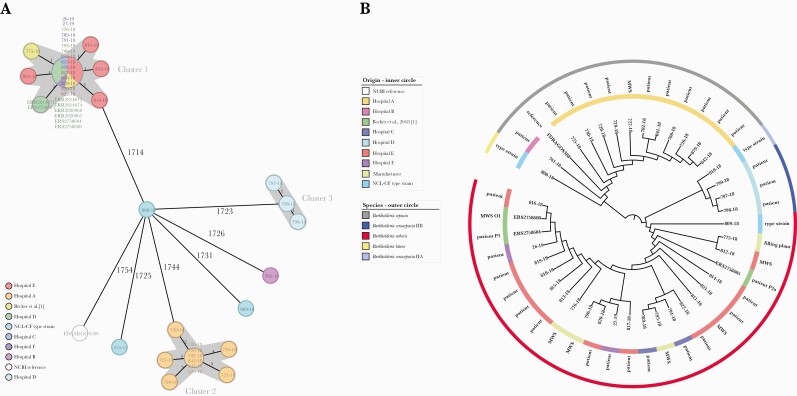
Isolates from the filling plant and from previously unopened MWS bottles of the manufacturer also clustered with PFGE type B1 isolates. A subsequent cgMLST analysis confirmed these results and, moreover, revealed that PFGE type B1 isolates were identical or closely related to the 3 isolates (ERR14669-674, ERR2820960, and ERR2820961) investigated by Becker et al. in August 2018 (Figure 2) [9]. Most importantly, the 2 isolates from patients admitted to hospital F were identical to isolates of cgMLST cluster 1 (samples 26-19 and 27-19) (Figure 2); these patients were exposed to a different batch of MWS from the same provider ~5 months before the initial outbreak was reported.
Figure 2.
Determination of genetic relatedness by means of cgMLST analysis of BCC isolates from Germany March 2018–September 2018. Minimum spanning tree (A) and neighbor joining tree (B) of BCC isolates from hospitals A–F, BCC isolates from mouthwash solution and filling plant from the manufacturer, and 3 BCC isolates from the previously reported outbreak in Southern Germany [9]. Type strains for BCC species, B. arboris, B. latens, and B. cenocepacia IIIA, were included as outgroups. aCluster 3 contains isolates from patients without epidemiological linkage to the MWS of the manufacturer. Abbreviations: BCC, Burkholderia cepacia complex; cgMLST, ad hoc core genome MLST; MWS, mouthwash solution.
PFGE type B2 was represented by 11 isolates from hospital A (10 isolates from patients and 1 from MWS). According to the cgMLST data, these isolates were assigned to cluster 2, which was found to be clearly distinct from cgMLST cluster 1 (PFGE type B1 isolates) (Figure 2). Three reference strains (DSM 23436 B. latens, DSM 23435 B. arboris, and LMG 16656 B. cenocepacia IIIA), which were carried along as internal controls, 3 patient isolates from hospital D, and 1 patient isolate from hospital B are not related to either cluster 1 or cluster 2 isolates (Figure 2).
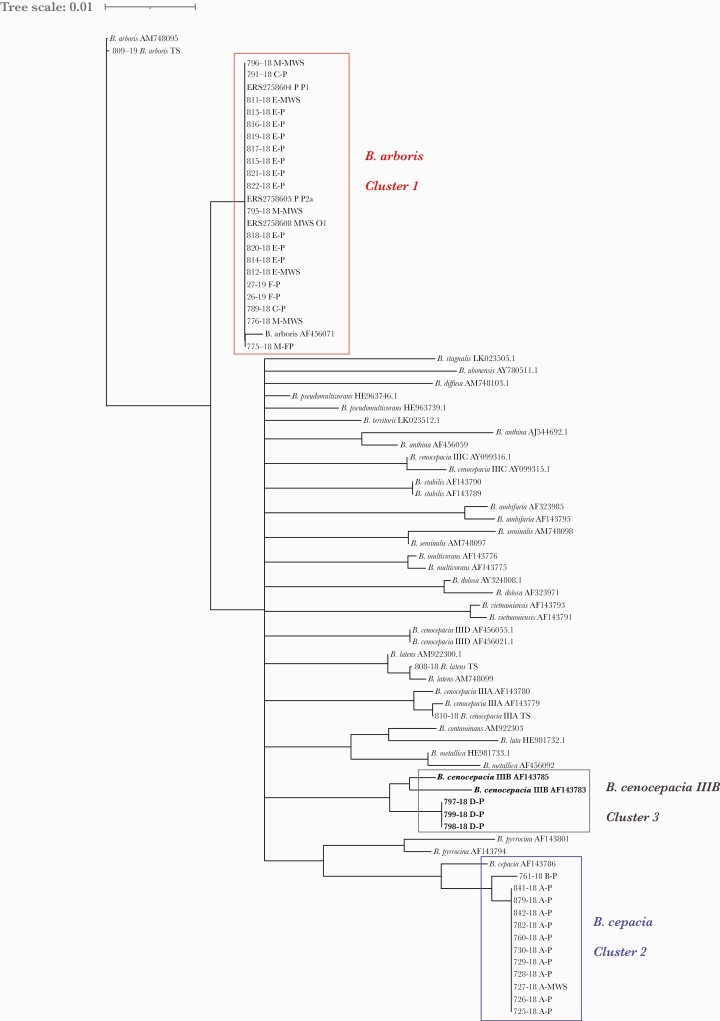
To determine the species within BCC that were responsible for the different outbreak clusters, recA open reading frames were extracted from the reconstructed genomes and compared with 39 recA reference sequences. Cluster 1, including isolates from hospitals C, E, and F, isolates from the MWS manufacturer, and data from Becker et al. [9] were identified as B. arboris. Cluster 2 isolates from hospital A and the single, unrelated isolate from hospital B belonged to B. cepacia, while recA analysis of outgroup isolates from hospital D identified B. cenocepacia IIIB (Figure 3). Interestingly, isolates from MWS of the same batch (isolated in hospitals A and E) were assigned to 2 different species, B. arboris (cgMLST cluster 1) and B. cepacia (cgMLST cluster 2). Furthermore, the B. arboris strain was putatively present in 2 different batches from the MWS manufacturer, although in hospital F B. arboris was isolated from patients with contact with MWS only, but not directly from MWS. As a consequence, all batches produced since March 2018 and available in Germany and in adjacent European countries were recalled from the market, and further distribution was stopped by December 2018.
Figure 3.
Speciation of BCC clinical and MWS isolates by means of recA comparison. Thirty-nine reference recA sequences were provided by the NCL-CF and aligned to the recA sequences of this study, which were extracted from whole-genome data. Legend for sequences of BCC isolates (this study): isolate number (and species), first letter hospitals A–F, manufacturer (M), second letter patient (P) or mouthwash solution (MWS) isolate. Reference for “ERS” isolates [9]: MWS or P isolate and isolate name. Legend for BCC reference strains: species and strain identification number. Abbreviations: BCC, Burkholderia cepacia complex; cgMLST, ad hoc core genome MLST; M, manufacturer; MWS, mouthwash solution; NCL-CF, National Consiliary Laboratory on Cystic Fibrosis Bacteriology; P, patient; TS, type strain.
DISCUSSION
Contamination of diverse medical products with BCC species has been described multiple times in recent decades. Medical devices such as central venous catheters and washing gloves and contamination of medical liquids such as MWS, eye drops, and/or cosmetic products such as moisturizing body milk have already been identified as sources for severe and putatively fatal health care–associated BCC infections [15–20]. BCC species isolates are environmentally widely distributed. Their capability to grow under laboratory conditions and thus to be diagnosed adequately is limited. As such, BCC-associated outbreaks often have delayed recognition and become protracted. They constitute a public health problem in health care settings in many countries, affecting quality of treatment and quality of life of susceptible patients, in particular patients with cystic fibrosis and chronic granulomatous disease [3, 21].
The outbreaks in German hospitals described herein correspond to the existing evidence on similar outbreaks in high-, low-, and middle-income countries. A first systematic review of BCC outbreak reports published between 1971 and 2019 reported that BCC was associated with 111 nosocomial outbreaks worldwide (20 in Europe), with 2390 affected patients and 240 fatalities (28 directly attributed to BCC) [22]. Most outbreaks occurred either exclusively in ICUs or with the involvement of the ICU, which is in line with our results. In 53.2% of reports (82 outbreaks with known causes), outbreaks were caused by medical preparations, with contaminated chlorhexidine and alcohol-free mouthwash solution being responsible for 6.5% and 4.8% of cases, respectively. About one-third of identified sources of BCC in outbreaks were due to intrinsically contaminated products. A comprehensive review conducted by Tavares et al. additionally provided a microbiological and clinical overview of the complex features of BCC, its capacity to survive and proliferate in water-based environments, and the ability of BCC bacteria to survive in the presence of antimicrobials and disinfectants, such as the biocides used in pharmaceutical products’ formulations [21].
BCC outbreaks often extend beyond 1 region or federal state of a country (due to the manufacturer supplying to different customers), making a multistate outbreak investigation necessary [23, 24]. The most recent report from the United States revealed contaminated saline flush syringes being associated with 162 BCC bloodstream infections across 59 nursing facilities in 5 states and occurring during September 2016–January 2017. It led to a nationwide recall of the product [23]. More than one-third of the cases occurred before the official notification to health authorities, similar to the outbreak described in the present report.
However, not every species of the BCC is recovered at the same frequency from a given source [1], and thus species identification is of great importance. Due to the close genetic relationship, speciation within the BCC is a challenging task especially when relying on phenotypical characterization only [25]. PCR- and sequencing-based examination of the 16S rRNA cannot be applied to distinguish all members of the BCC at species level [26, 27]. At the moment, the most reliable taxonomic method for discrimination is a comparison of the recA sequences [28, 29], which was also applied in the present study. As WGS data were available, we additionally bypassed potential dropouts due to failure of recA amplification of certain Burkholderia spp. [27]. Thereby, our analyses revealed B. cepacia and B. arboris as the contaminating species of MWS bottles from hospitals A and E, respectively, which caused the subsequent colonization and putative infection of patients. Samples of patients from 3 hospitals throughout Germany contained the B. arboris strain that was also identified directly from the retained samples of the manufacturer. However, the B. cepacia strain that caused the outbreak in hospital A was not found among the retained samples of MWS that we analyzed, but might have contaminated batches that were not tested in the present study. B. cenocepacia IIIB was identified as another potential pathogen in 3 patients without any proven association with the MWS. Previous studies reported the isolation of the above-mentioned BCC species from CF patients and/or the (hospital) environment [1, 29, 30].
Based on evidence from four systematic reviews, seven meta-analysis studies and several randomized clinical trials, the German Commission for Hospital Hygiene and Infection Prevention recommends the use of medical MWS to prevent ventilator-associated pneumonia [10]. The safety of medical products is of high value. Benefits and harms of medical products must be weighed against each other. A recall of involved medical products is one of the most common reported interventions in outbreaks caused by BCC. Notably, in about one-fourth of the reviewed BCC outbreaks, no source of the outbreak could be detected, making it difficult to introduce targeted infection prevention and control measures and other interventions [22]. Investigations of the patients’ records and the environment were conducted only in 59.5% of the reviewed outbreaks. Overall, the quality of outbreak reporting in the literature varied largely [21, 22]. Standardization of outbreak reporting in the future would significantly contribute to better understanding of dynamics of such infections and appropriate IPC measures.
BCC species show extraordinary ability to thrive under versatile environmental conditions and, most importantly, to withstand/persevere in disinfecting agents. Therefore, these medical products should be considered as a potential source when BCC species are detected in patients without typical risk factors. Given difficulties in timely detection and identification of BCC bacteria, prevention of BCC contamination on-site by pharmaceutical manufacturers and further development of detection methods of species in case of suspected contamination are urgently needed [9, 15, 21, 22]. In Germany, reporting of such incidents does not necessarily include the involvement of epidemic intelligence services. In the investigation presented here, time was lost because the national public health institute was notified about the first reported cases about 4 weeks after the manufacturer became aware of the contamination and had initiated a product recall. The present outbreaks revealed further weaknesses in communication and transfer of information, with the result that the voluntary recall of the product by the manufacturer had not reached all product users (since then, the medical product safety act was amended to ensure that a responsible person in facilities is named to receive such notifications: https://www.gesetze-im-internet.de/mpbetreibv/__6.html). In addition, our investigation revealed that the outbreak had started before production of the contaminated batch, most likely in spring 2018 (Table 2), and thus more batches of MWS were likely contaminated. It takes time and effort to gather sufficient evidence for a product recall. But during this time patients may be exposed to contaminated products. May more preemptive measures be needed in the field of medical product safety? Follow-up of the distribution is also cumbersome, as for instance products are bought by resellers. As a consequence, we could not establish a comprehensive list of all final consumers.
In conclusion, we present a comprehensive nationwide outbreak investigation that included in-depth microbiological analysis in addition to extensive epidemiological investigation, source identification, and public health action. Epidemiological and clinical case data hypothesized a common source of BCC contamination and a common cause of the infections in all cases. Genome-based analyses allowed us to differentiate 2 separate outbreak scenarios, which were not identifiable by standard microbial diagnostics. Additionally, our investigation emphasizes the importance of national and international efforts to follow up on the distribution of all possible contaminated products, which we found to be a bottleneck for immediate and timely action. Our results raise awareness of possible contaminants of medical products used for highly vulnerable patients. Thus, medical product recalls should be optimized to inform public health services and consumers in a timely manner, and cooperation and communication between all involved institutions should be further strengthened.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Acknowledgments
We thank Sibylle Müller-Bertling and Kirstin Ganske for excellent technical assistance. We further thank Regina Selb and Giuseppe Valenza, who were involved in outbreak analysis and bacterial strain typing. We thank Hans Peter Blank and Muna Abu Sin for their support in the epidemiological outbreak investigation. We thank the manufacturer for cooperation and for making isolates available for further analyses.
Financial support. This work was supported by funds from the German Federal Ministry of Health to the National Consiliary Laboratory.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Chiarini L, Bevivino A, Dalmastri C, Tabacchioni S, Visca P.. Burkholderia cepacia complex species: health hazards and biotechnological potential. Trends Microbiol 2006; 14:277–86. [DOI] [PubMed] [Google Scholar]
- 2. Engledow AS, Medrano EG, Mahenthiralingam E, LiPuma JJ, Gonzalez CF.. Involvement of a plasmid-encoded type IV secretion system in the plant tissue watersoaking phenotype of Burkholderia cenocepacia. J Bacteriol 2004; 186:6015–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coenye T, Vandamme P.. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol 2003; 5:719–29. [DOI] [PubMed] [Google Scholar]
- 4. Berriatua E, Ziluaga I, Miguel-Virto C, et al. Outbreak of subclinical mastitis in a flock of dairy sheep associated with Burkholderia cepacia complex infection. J Clin Microbiol 2001; 39:990–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wiener-Well Y, Segonds C, Mazuz B, Kopuit P, Assous MV.. Successful outbreak investigation of Burkholderia cepacia complex bacteremia in intensive care patients. Am J Infect Control 2014; 42:580–1. [DOI] [PubMed] [Google Scholar]
- 6. Baul SN, De R, Mandal PK, Roy S, Dolai TK, Chakrabarti P.. Outbreak of Burkholderia cepacia infection: a systematic study in a hematolooncology unit of a tertiary care hospital from eastern India. Mediterr J Hematol Infect Dis 2018; 10:e2018051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rastogi N, Khurana S, Veeraraghavan B, et al. Epidemiological investigation and successful management of a Burkholderia cepacia outbreak in a neurotrauma intensive care unit. Int J Infect Dis 2019; 79:4–11. [DOI] [PubMed] [Google Scholar]
- 8. Mahenthiralingam E, Baldwin A, Vandamme P.. Burkholderia cepacia complex infection in patients with cystic fibrosis. J Med Microbiol 2002; 51: 533–8. [DOI] [PubMed] [Google Scholar]
- 9. Becker SL, Berger FK, Feldner SK, et al. Outbreak of Burkholderia cepacia complex infections associated with contaminated octenidine mouthwash solution, Germany, August to September 2018. Euro Surveill 2018; 23:1800540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. KRINKO. Commission for Hospital Hygiene and Infection Prevention. Bundesgesundheitsblatt 2013; 56:1578–90. doi: 10.1007/s00103-013-1846-7 [DOI] [PubMed] [Google Scholar]
- 11. Robert Koch Institute. Überregionale häufung von Burkholderia-cepacia-complex-nachweisen bei intensivpatienten. Epidemiologisches Bulletin 2018; 38:422. [Google Scholar]
- 12. Aerzteblatt. Mundspüllösung wahrscheinliche ursache für infektionen von intensivpatienten. Aerzteblatt.de. 21 September 2018. [Google Scholar]
- 13. Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 1995; 33:2233–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19:455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sommerstein R, Fuhrer U, Lo Priore E, et al. Burkholderia stabilis outbreak associated with contaminated commercially-available washing gloves, Switzerland, May 2015 to August 2016. Euro Surveill 2017; 22:17-00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lalitha P, Das M, Purva PS, et al. Postoperative endophthalmitis due to Burkholderia cepacia complex from contaminated anaesthetic eye drops. Br J Ophthalmol 2014; 98:1498–502. [DOI] [PubMed] [Google Scholar]
- 17. Alvarez-Lerma F, Maull E, Terradas R, et al. Moisturizing body milk as a reservoir of Burkholderia cepacia: outbreak of nosocomial infection in a multidisciplinary intensive care unit. Crit Care 2008; 12:R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Martin M, Winterfeld I, Kramme E, Ewert I, Sedemund-Adib B, Mattner F.. Outbreak of Burkholderia cepacia complex caused by contaminated alcohol-free mouthwash. Anaesthesist 2012; 61:25–9. [DOI] [PubMed] [Google Scholar]
- 19. Gupta P, Jain V, Hemrajani M, Gupta A, Sharma U.. Outbreak of Burkholderia cepacia catheter-related bloodstream infection in cancer patients with long-term central venous devices at a tertiary cancer centre in India. Indian Anaesth Forum 2018; 19:1–5. [Google Scholar]
- 20. Zurita J, Mejia L, Zapata S, et al. Healthcare-associated respiratory tract infection and colonization in an intensive care unit caused by Burkholderia cepacia isolated in mouthwash. Int J Infect Dis 2014; 29:96–9. [DOI] [PubMed] [Google Scholar]
- 21. Tavares M, Kozak M, Balola A, Sa-Correia I.. Burkholderia cepacia complex bacteria: a feared contamination risk in water-based pharmaceutical products. Clin Microbiol Rev 2020; 33:e00139-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hafliger E, Atkinson A, Marschall J.. Systematic review of healthcare-associated Burkholderia cepacia complex outbreaks: presentation, causes and outbreak control. Infect Prev Pract 2020; 2:100082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Akinboyo IC, Sick-Samuels AC, Singeltary E, et al. Multistate outbreak of an emerging Burkholderia cepacia complex strain associated with contaminated oral liquid docusate sodium. Infect Control Hosp Epidemiol 2018; 39:237–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brooks RB, Mitchell PK, Miller JR, et al. Multistate outbreak of Burkholderia cepacia complex bloodstream infections after exposure to contaminated saline flush syringes: United States, 2016-2017. Clin Infect Dis 2019; 69:445–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Coenye T, Vandamme P, Govan JR, LiPuma JJ.. Taxonomy and identification of the Burkholderia cepacia complex. J Clin Microbiol 2001; 39:3427–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. LiPuma JJ, Dulaney BJ, McMenamin JD, et al. Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J Clin Microbiol 1999; 37:3167–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mahenthiralingam E, Bischof J, Byrne SK, et al. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J Clin Microbiol 2000; 38:3165–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Payne GW, Vandamme P, Morgan SH, et al. Development of a recA gene-based identification approach for the entire Burkholderia genus. Appl Environ Microbiol 2005; 71:3917–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vanlaere E, Lipuma JJ, Baldwin A, et al. Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov. and Burkholderia metallica sp. nov., novel species within the Burkholderia cepacia complex. Int J Syst Evol Microbiol 2008; 58:1580–90. [DOI] [PubMed] [Google Scholar]
- 30. Manno G, Dalmastri C, Tabacchioni S, et al. Epidemiology and clinical course of Burkholderia cepacia complex infections, particularly those caused by different Burkholderia cenocepacia strains, among patients attending an Italian cystic fibrosis center. J Clin Microbiol 2004; 42:1491–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.